Abstract
SARS-CoV-2 has caused the global COVID-19 pandemic. Although passively delivered neutralizing antibodies against SARS-CoV-2 show promise in clinical trials, their mechanism of action in vivo is incompletely understood. Here, we define correlates of protection of neutralizing human monoclonal antibodies (mAbs) in SARS-CoV-2-infected animals. Whereas Fc effector functions are dispensable when representative neutralizing mAbs are administered as prophylaxis, they are required for optimal protection as therapy. When given after infection, intact mAbs reduce SARS-CoV-2 burden and lung disease in mice and hamsters better than loss-of-function Fc variant mAbs. Fc engagement of neutralizing antibodies mitigates inflammation and improves respiratory mechanics, and transcriptional profiling suggests these phenotypes are associated with diminished innate immune signaling and preserved tissue repair. Immune cell depletions establish that neutralizing mAbs require monocytes and CD8+ T cells for optimal clinical and virological benefit. Thus, potently neutralizing mAbs utilize Fc effector functions during therapy to mitigate lung infection and disease.
Keywords: SARS-CoV-2, antibody, effector function, monocytes, CD8+ T cells, therapy, pathogenesis, mouse model, RNA sequencing, lung
Graphical abstract
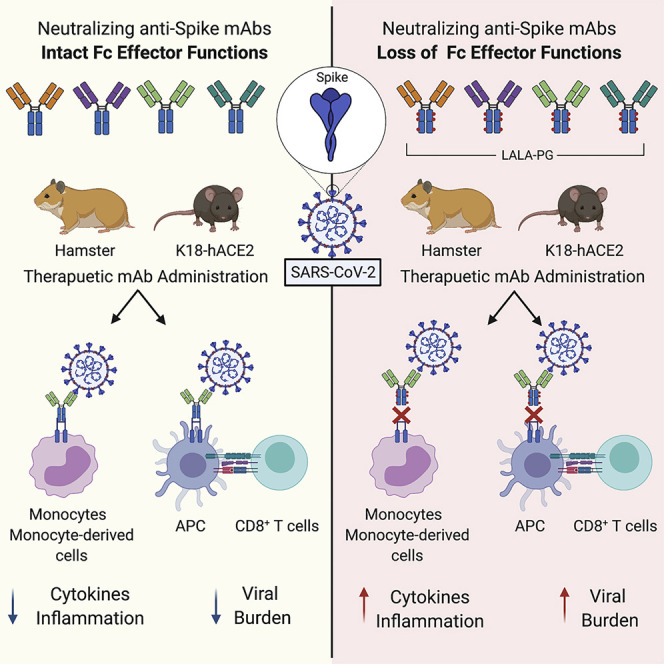
Neutralizing human monoclonal antibodies (mAbs) against SARS-CoV-2 require Fc effector functions for optimal protection during post-exposure therapy, with intact mAbs reducing SARS-CoV-2 burden and lung disease in rodent models better than LALA-PG loss-of-function Fc variant mAbs and requiring monocytes and CD8+ T cells for optimal clinical and virological benefit.
Introduction
Severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) is the recently emerged RNA virus responsible for the coronavirus disease 2019 (COVID-19) pandemic that has led to over 116 million infections and over 2.5 million deaths (Dong et al., 2020). The need for effective countermeasures of COVID-19 is urgent, because only dexamethasone and remdesivir have demonstrated clinical efficacy in specific indications. One approach for both prevention and treatment of COVID-19 has been the development of SARS-CoV-2-neutralizing monoclonal antibodies (mAbs) isolated from the B cells of individuals with recent SARS-CoV-2 infection (Abraham, 2020; Marovich et al., 2020). Multiple neutralizing mAbs directed against the spike (S) glycoprotein of SARS-CoV-2 have been identified that target non-overlapping epitopes and show differences in neutralization potency (Alsoussi et al., 2020; Cao et al., 2020; Hansen et al., 2020; Ju et al., 2020; Liu et al., 2020; Noy-Porat et al., 2020; Pinto et al., 2020; Robbiani et al., 2020; Zost et al., 2020a). The majority of these mAbs bind to the S1 subunit, specifically the receptor binding domain (RBD), and inhibit virus engagement with its cell surface receptor, angiotensin converting enzyme (ACE)2.
Prophylactic and therapeutic efficacy of anti-S mAbs has been demonstrated in vivo in murine, hamster, and non-human primate models of SARS-CoV-2 pathogenesis (Alsoussi et al., 2020; Baum et al., 2020; Fagre et al., 2020; Hansen et al., 2020; Hassan et al., 2020; Kreye et al., 2020; Rogers et al., 2020; Shi et al., 2020; Zost et al., 2020a), with varying degrees of reduction in viral burden and dampened inflammation of the lung. However, the mechanisms of protection in vivo can be due to multiple factors including direct virus neutralization and engagement of complement or Fc gamma receptors (FcγRs) on leukocytes. Fc effector functions of antibodies can promote immune-mediated cellular clearance, enhance antigen presentation and CD8+ T cell responses, and reshape inflammation through engagement of FcγRs on specific cells (Lu et al., 2018). In contrast, under certain circumstances, Fc-FcγR interactions can promote antibody-dependent enhancement of virus infection (ADE) (Halstead, 1994) or pathological immune skewing (Bolles et al., 2011; Ruckwardt et al., 2019), which is at least a theoretical concern of antibody-based therapies and vaccines against SARS-CoV-2 (Diamond and Pierson, 2020). Thus, a more thorough understanding of the contribution of Fc effector functions in the context of antibody-based therapies is needed.
Here, we use the K18-hACE2 transgenic mouse model of SARS-CoV-2 pathogenesis (Golden et al., 2020; Winkler et al., 2020) and a Fc region genetic variant of immunoglobulin G (IgG) (LALA-PG) of potent RBD-binding neutralizing mAbs that cannot engage FcγRs or complement to define the role of Fc effector functions in antibody protection. We find that Fc effector functions are dispensable when neutralizing mAbs are administered as prophylaxis but are required for optimal protection when given as post-exposure therapy. When administered after SARS-CoV-2 infection, intact, but not LALA-PG, mAbs reduce viral burden and lung disease. Fc engagement by antibodies decreases immune cell activation and levels of inflammatory cytokines, and this activity is linked to improved respiratory mechanics and outcome. RNA sequencing analysis of lung homogenates reveals distinct gene signatures associated with Fc engagement including decreased innate immune signaling and extracellular matrix remodeling but maintained expression of genes that mediate tissue repair. Immune cell depletions showed that neutralizing mAbs require monocytes and CD8+ T cells for maximal clinical and virological benefit. We confirmed these findings in hamsters, a second mammalian species, that also required Fc effector functions of a neutralizing mAb to prevent weight loss, control viral infection, and limit inflammation. Overall, these studies establish that Fc effector functions of neutralizing antibodies are necessary for optimal therapeutic outcome after SARS-CoV-2 infection.
Results
Pre-exposure protection against SARS-CoV-2 infection in mice by a neutralizing mAb does not require Fc effector functions
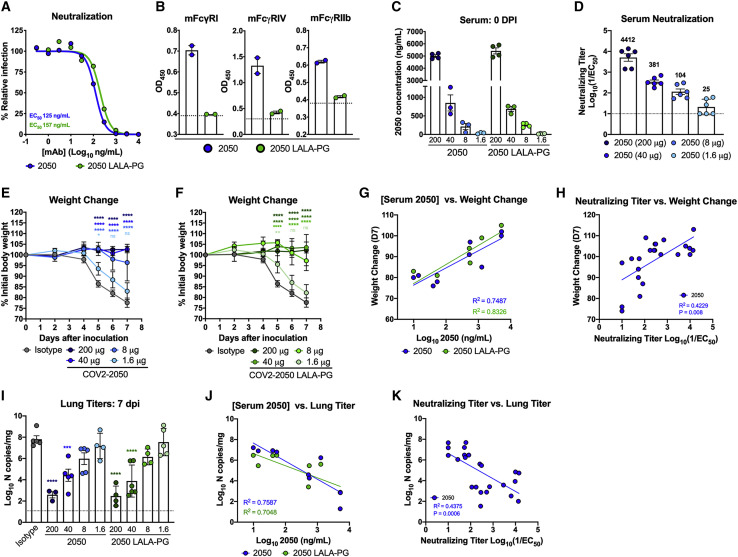
Passive transfer of neutralizing mAbs targeting the S protein confers protection in multiple pre-clinical models of SARS CoV-2 infection (Alsoussi et al., 2020; Baum et al., 2020; Hansen et al., 2020; Hassan et al., 2020; Kreye et al., 2020; Rogers et al., 2020; Zost et al., 2020a). However, in vivo antiviral efficacy can be due to multiple mechanisms including Fab-dependent direct virus neutralization and Fc-dependent engagement that promotes opsonization of virus, clearance of virally infected cells, and modulation of innate and adaptive immune responses (Bournazos et al., 2014b, 2019; DiLillo et al., 2014, 2016; Fox et al., 2019). To evaluate the contribution of Fc effector functions, we introduced loss-of-function LALA-PG (L234A, L235A, and P329G) mutations into the Fc region of the human IgG1 heavy chain of COV2-2050, a neutralizing human mAb that binds the RBD of S and blocks ACE2 binding (Zost et al., 2020a), to abolish antibody Fc interactions with FcγRs and complement proteins (Lo et al., 2017). Intact or LALA-PG variants of COV2-2050 neutralized SARS-CoV-2 equivalently in cell culture (Figure 1 A), and introduction of the LALA-PG mutation prevented binding to mouse FcγRI, FcγRIV, or FcγRIIb (Figure 1B). To test their relative efficacy in vivo, we used the K18-hACE2 transgenic mouse model of SARS-CoV-2 pathogenesis (McCray et al., 2007; Winkler et al., 2020). In prophylaxis studies, K18-hACE2 transgenic mice received decreasing doses of COV2-2050 or COV2-2050 LALA-PG (200 μg, 40 μg, 8 μg, or 1.6 μg) by intraperitoneal injection 16 h prior to intranasal inoculation with SARS-CoV-2 (103 plaque-forming unit [PFU], strain 2019 n-CoV/USA_WA1/2020). An isotype control human mAb (DENV-2D22) was delivered at a single 200 μg dose for comparison. At the time of SARS-CoV-2 challenge, the serum levels of COV2-2050 or COV2-2050 LALA-PG were equivalent (Figure 1C). As expected, lower doses of COV2-2050 resulted in decreasing neutralizing antibody titers in serum at the time of virus inoculation (Figure 1D).
Figure 1.
Neutralizing activity is sufficient for prophylactic efficacy of mAbs against SARS-CoV-2 infection in K18-hACE2 mice
(A) MAbs (COV2-2050 and COV2-2050 LALA-PG) were incubated with 102 focus-forming units (FFU) of SARS-CoV-2 for 1 h followed by addition to Vero E6 cells. Wells containing mAb were compared to wells without mAb to determine relative infection. One experiment with the mean of 2 technical replicates is shown.
(B) Binding of COV2-2050 or COV2-2050 LALA-PG to murine FcγRI, FcγRIV, or FcγRIIb by ELISA (two experiments). The dotted line indicates the limit of detection (LOD), as determined by background binding to a negative control.
(C–K) Eight-week-old female and male K18-hACE2 mice received 200, 40, 8, or 1.6 μg of COV2-2050 or COV2-2050 LALA-PG by intraperitoneal injection 1 day prior to intranasal inoculation with 103 PFU of SARS-CoV-2.
(C) Serum concentrations (ng/mL) of COV2-2050 or COV2-2050 LALA-PG at the time of challenge (0 dpi) (mean ± SEM; n = 3–4, 2 experiments).
(D) Neutralizing titers in serum of indicated groups at 0 dpi as measured by FRNT (mean ± SEM; n = 6, 2 experiments).
(E and F) Weight change following COV2-2050 (E) or COV2-2050 LALA-PG (F) administration (mean ± SEM; n = 4–6, 2 experiments).
(G) Correlation analyses comparing COV2-2050 or COV2-2050 LALA-PG serum concentrations (day 0 [D0]) against weight change (D+7) (n = 4–6, 2 experiments).
(H) Correlation analyses comparing COV2-2050 neutralizing titers in serum (D0) against weight change (D+7) (n = 6–8, 3 experiments).
(I) Viral RNA levels at 7 dpi in the lung (n = 4–6, 2 experiments).
(J) Correlation analyses comparing COV2-2050 or COV2-2050 LALA-PG serum concentrations (D0) against lung viral titer (D+7) (n = 4–6, 2 experiments).
(K) Correlation analyses comparing serum neutralizing titers (D0) against lung viral titer (D7) (n = 6–8, 2 experiments).
(E and F) Two-way ANOVA with Sidak’s post-test: ns not significant, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗∗p < 0.0001; comparison to the isotype control mAb-treated group. (G, H, J, and K) Pearson’s correlations: (G) COV2-2050, p = 0.0026; COV2-2050 LALA-PG, p = 0.0006; (H) COV2-2050, p = 0.0008; (J) COV2-2050, p = 0.0022; COV2-2050 LALA-PG, p = 0.0095; and (K) COV2-2050, p = 0.0006. (I) One-way ANOVA with Turkey’s post-test: ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001, comparison to the isotype control mAb-treated group.
Effects on SARS-CoV-2-induced weight loss were comparable between COV2-2050 or COV2-2050 LALA-PG at all doses with a loss of protection observed only at the 1.6 μg dose (Figures 1E and 1F). Levels of COV2-2050 correlated with clinical protection (Figures 1G and 1H), with a minimum serum neutralizing titer (NT50) of 104 and concentration of 212 ng/mL required to prevent weight loss in this stringent challenge model. Although weight loss was prevented at a dose of 86 μg per animal, a higher 40 μg dose was required to reduce viral burden in the lung at 7 days post-infection (dpi) (Figure 1I). Serum levels of COV2-2050 also correlated inversely with SARS-CoV-2 RNA in the lung (Figures 1J and 1K) with a minimum neutralizing titer of 381 and mAb concentration of 851 ng/mL required to reduce viral infection at 7 dpi. Differences in viral burden were not observed at several different doses of COV2-2050 and COV2-2050 LALA-PG treatment. Importantly, no evidence of antibody-dependent enhancement of clinical disease or viral burden (Lee et al., 2020; Taylor et al., 2015) was detected even when levels were below the protective dose. Thus, the Fc-dependent effector functions of COV2-2050 are dispensable for clinical and virological protection when a potently neutralizing antibody is administered as prophylaxis.
Fc effector functions enhance therapeutic activity of neutralizing mAbs
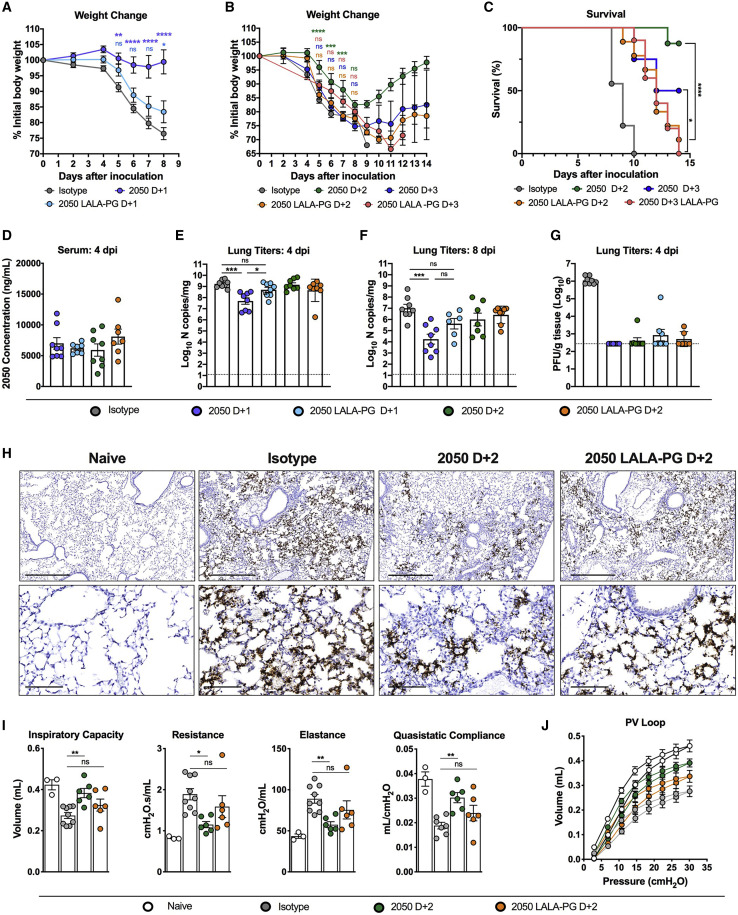
Although we did not detect a requirement for Fc effector functions when COV2-2050 was administered as prophylaxis, we re-evaluated this in the setting of post-exposure administration. We inoculated K18-hACE2 mice with SARS-CoV-2 by the intranasal route and then delivered a single 200 μg dose of COV2-2050 or COV2-2050 LALA-PG mAbs at 1 (D+1), 2 (D+2), or 3 (D+3) dpi by intraperitoneal injection. Whereas passive transfer of the intact COV2-2050 prevented weight loss compared to isotype control mAb-treated animals, this protection was lost in animals treated with the COV2-2050 LALA-PG variant (Figures 2A and 2B). Although treatment beginning at 3 dpi with either COV2-2050 or COV2-2050 LALA-PG did not prevent weight loss, therapy with COV2-2050, but not COV2-2050 LALA-PG, reduced lethality (Figures 2B and 2C). The differences in protection were not due to disparate levels of COV2-2050 and COV2-2050 LALA-PG, as equivalent amounts were detected in serum at 0 and 4 dpi (Figure 2D). Treatment at 1 dpi with COV2-2050, but not COV2-2050 LALA-PG, reduced SARS-CoV-2 viral RNA levels in the lung at 4 and 8 dpi substantially (Figures 2E and 2F). In contrast, D+2 treatment with either intact COV2-2050 or COV2-2050 LALA-PG did not reduce viral RNA levels in the lung when compared to isotype-treated controls. However, COV2-2050 and COV2-2050 LALA-PG mAbs both reduced levels of infectious virus in the lung at 4 dpi (Figure 2G). Given the disparity in viral RNA levels, this effect might be due in part to ex vivo neutralization after lung tissue homogenization, as reported for other respiratory viruses (Subbarao et al., 2004; Wells et al., 1981). Visualization of viral RNA by in situ hybridization showed that although expression of SARS-CoV-2 RNA was spread diffusely throughout the lung in groups treated at D+2 with isotype control mAb or 2050 LALA-PG, viral RNA was more focal in the lungs of mice receiving intact COV2-2050 at D+2 with evidence of cleared regions and a smaller affected proportion (Figure 2H). Additionally, K18-hACE2 mice receiving intact COV2-2050 at D+2, but not COV2-2050 LALA-PG, showed functional improvement in pulmonary mechanics (e.g., inspiratory capacity, resistance, and elastance), and lung compliance and distensibility (Figures 2I and 2J).
Figure 2.
Fc effector functions enhance the therapeutic activity of a neutralizing antibody against SARS-CoV-2 in K18-hACE2 mice
(A–J) Eight-week-old K18-hACE2 mice were inoculated by the intranasal route with 103 PFU of SARS-CoV-2. At 1 (D+1), 2 (D+2), or 3 (D+3) dpi, mice were given 200 μg of COV2-2050 or COV2-2050 LALA-PG by intraperitoneal injection. Naive animals were mock-infected with sterile PBS.
(A and B) Weight change. For (B), statistical analysis was performed only at time points when all mice were alive to avoid survivor bias (mean ± SEM; n = 8–10, 3 experiments).
(C) Survival analysis (n = 8–19, 3 experiments).
(D) Serum concentrations (ng/mL) of COV2-2050 or COV2-2050 LALA-PG at 4 dpi (mean ± SEM; n = 8, 2 experiments).
(E and F) Viral RNA levels at 4 and 8 dpi in the lung (n = 6–10, 3 experiments).
(G) Infectious virus at 4 dpi in the lung (n = 6–8, 2 experiments).
(H) SARS-CoV-2 RNA in situ hybridization of lung sections. Images show low- (top; scale bars, 500 μm) and high-power magnification (bottom; scale bars, 100 μm). Images are representative of n = 4 per group.
(I) Parameters of respiratory mechanics: inspiratory capacity, resistance, elastance, tissue damping, and quasistatic compliance measured at 8 dpi (n = 3–6, 2 experiments).
(J) Pressure volume loops (n = 3–6, 2 experiments).
(A and B) Two-way ANOVA with Sidak’s post-test: ∗p < 0.05, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001; comparison to the isotype control mAb-treated group. (C) Mantel-Cox log-rank test for survival: ∗p < 0.05, ∗∗∗∗p < 0.0001. (E, F, and I) One-way (E and F) or two-way (I and J) ANOVA with Turkey’s post-test: ns not significant, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001, comparison to the isotype control mAb-treated group.
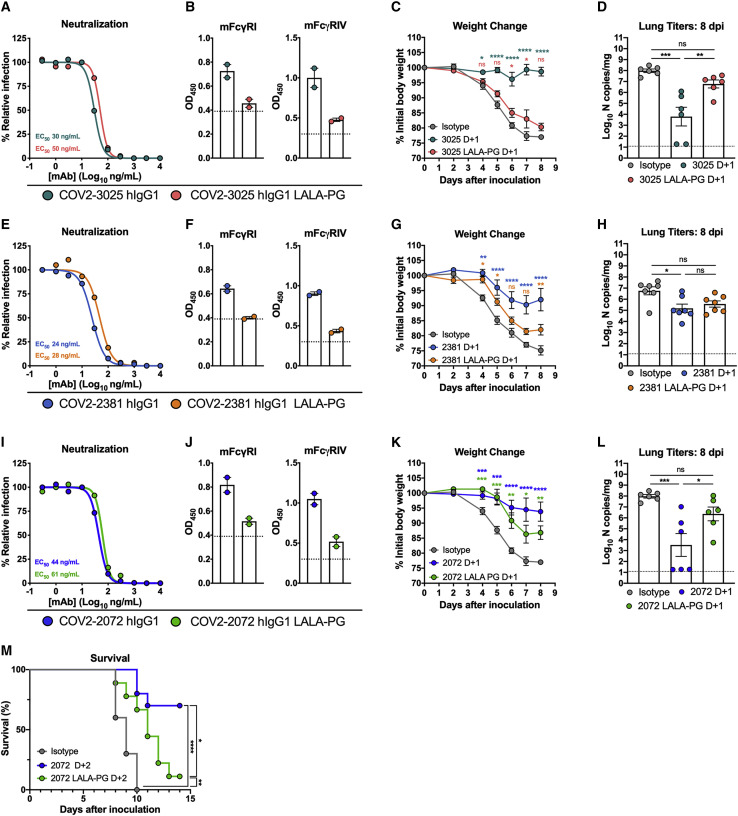
To expand on these results, we tested the therapeutic activity of three additional neutralizing human mAbs that bind the RBD, COV2-3025, COV2-2072, and COV2-2381 (Zost et al., 2020b), and compared them to LALA-PG variants of each mAb. COV2-3025, COV2-2072, and COV2-2381 neutralized SARS-CoV-2 equivalently compared to their respective LALA-PG variants (Figures 3A, 3E, and 3I), and introduction of the LALA-PG mutation abolished binding to murine FcγRI or FcγRIV (Figures 3B, 3F, and 3J). Administration of intact COV2-3025 at D+1 recapitulated the same pattern of protection observed with COV2-2050 treatment: COV2-3025 LALA-PG failed to protect mice from weight loss or reduce viral titers compared to the intact COV2-3025 (Figures 3C and 3D). In comparison, COV2-2381 LALA-PG still showed a modest level of protection against weight loss, and viral burden was reduced in mice receiving either COV2-2381 or COV2-2381 LALA-PG compared to isotype-treated controls (Figures 3G and 3H). Administration of COV2-2072 LALA-PG at D+1 or D+2 partially protected against SARS-CoV-2-induced weight loss (D+1) or lethality (D+2) compared to the isotype-treated controls, but failed to reduce viral titers, unlike the intact COV2-2072 (Figures 3K–3M). The differences in therapeutic activity of COV2-2050 LALA-PG and COV2-3025 LALA-PG compared to COV2-2381 LALA-PG and COV2-2072 LALA-PG could be due to differential neutralizing activity, pharmacokinetics, or bioavailability in vivo. Regardless, these results indicate that intact Fc effector functions of neutralizing antibodies are needed for optimal therapeutic activity to mitigate clinical disease caused by SARS-CoV-2 infection.
Figure 3.
Requirement of Fc effector functions for different anti-RBD mAb in vivo
(A, E, and I) Anti-SARS-CoV-2 mAbs: COV2-3025 and COV2-3025 LALA-PG (A); COV2-2381, COV2-2381 LALA-PG (E); and COV2-2072, and COV2-2072 LALA-PG (I); MAbs were incubated with 102 focus-forming units (FFU) of SARS-CoV-2 before adding to Vero E6 cells. Wells containing mAb were compared to those without mAb to determine relative infection. One experiment with the mean of 2 technical replicates is shown.
(B, F, and J) Binding to recombinant murine FcγRI or FcγRIV of COV2-3025 or COV2-3025 LALA-PG (B); COV2-2381 or COV2-2381 LALA-PG (F); and COV2-2072 or COV2-2072 LALA-PG (J). Data are from 2 experiments. The dotted line indicates the LOD.
(C, D, G, H, K, and L) Eight-week-old K18-hACE2 mice were inoculated by the intranasal route with 103 PFU of SARS-CoV-2. At 1 dpi (D+1), mice were given 200 μg of each respective mAb by intraperitoneal injection. (C, G, and K) Weight change (mean ± SEM; n = 6–8, 2 experiments: two-way ANOVA with Sidak’s post-test: ∗p < 0.05, ∗∗p < 0.01, ∗∗∗∗p < 0.0001; comparison to the isotype control mAb-treated group). (D, H, and L) Viral RNA levels at 8 dpi in the lung (n = 6–8, 2 experiments, one-way ANOVA with Dunnett’s test; ∗∗∗∗p < 0.0001). (K) Eight-week-old male K18-hACE2 mice were inoculated by the intranasal route with 103 PFU of SARS-CoV-2. At 2 (D+2) dpi, mice were given 200 μg of COV2-2072 or COV2-2072 LALA-PG by intraperitoneal injection and survival was monitored (n = 8–10, 2 experiments; Mantel-Cox log-rank test for survival: ∗p < 0.05, ∗∗p < 0.01, ∗∗∗∗p < 0.0001).
See also Figure S1.
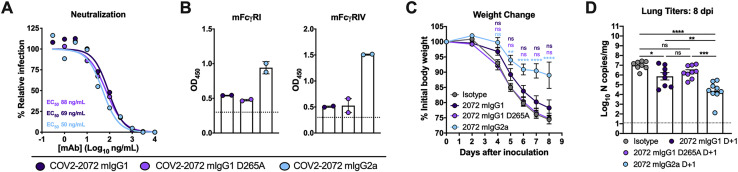
To confirm the requirement of Fc-mediated effector functions in protection in a species-homologous experimental system, we generated chimeric versions of COV2-2072 in which the human IgG1 Fc region was replaced with ones from murine (m)IgG2a, murine (m)IgG1, or mIgG1 containing a loss-of-function D265A mutation that abolishes binding to murine FcγRs (Baudino et al., 2008; Lo et al., 2017). Chimeras of COV2-2072 with murine Fc regions neutralized SARS-CoV-2 equivalently compared to parental COV2-2072 human IgG1 mAb (Figure S1 A). As expected, mIgG1 and mIgG1 D265A bound mFcγRI or mFcγRIV poorly compared to the mIgG2a (Figure S1B). Notably, D+1 administration of COV2-2072 mIgG1 or COV2-2072 mIgG1 D265A failed to protect mice from weight loss. In contrast, COV2-2072 mIgG2a prevented weight loss and reduced viral titers similarly to the human IgG1 form of COV2-2072 (Figures S1C and S1D). These data establish the importance of Fc effector functions to antibody protection when administered in a corresponding system with murine Fc regions and murine FcγRs.
Figure S1.
Protective effects of chimeric COV2-2072 with murine Fc regions, related to Figure 3
(A) Chimeric COV2-2072 (mIgG1, mIgG1 D265A, or mIgG2a) were incubated with 102 FFU of SARS-CoV-2 for 1 h at 37°C before adding to Vero E6 cells for a FRNT. Wells containing mAb were compared to wells without mAb to determine relative infection. One experiment of two, with similar results, is shown. The mean of two technical replicates is shown. (B) Binding of COV2-2072 mIgG1, mIgG1 D265A, and mIgG2a to recombinant mouse FcγRI or FcγRIV as measured by ELISA (two independent experiments). The dotted line indicates the limit of detection, as determined by background binding to a negative control. For C-D, eight-week-old female or male K18-hACE2 transgenic mice were inoculated by the intranasal route with 103 PFU of SARS-CoV-2. At D+1, mice were given a single 200 μg dose of COV2-2072 mIgG1, mIgG1 D265A, and mIgG2a by intraperitoneal injection. (C) Weight change (mean ± SEM; n = 7-8, two experiments: two-way ANOVA with Sidak’s post-test: ns not significant, ∗∗p < 0.01, ∗∗∗∗p < 0.0001; comparison to the isotype control mAb-treated group). (D) Viral RNA levels at 8 dpi in the lung as determined by qRT-PCR (n = 8, two experiments, one-way ANOVA with Dunnett’s test; ns not significant, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001).
Fc effector functions of a neutralizing mAb modulate the immune responses to SARS-CoV-2 infection
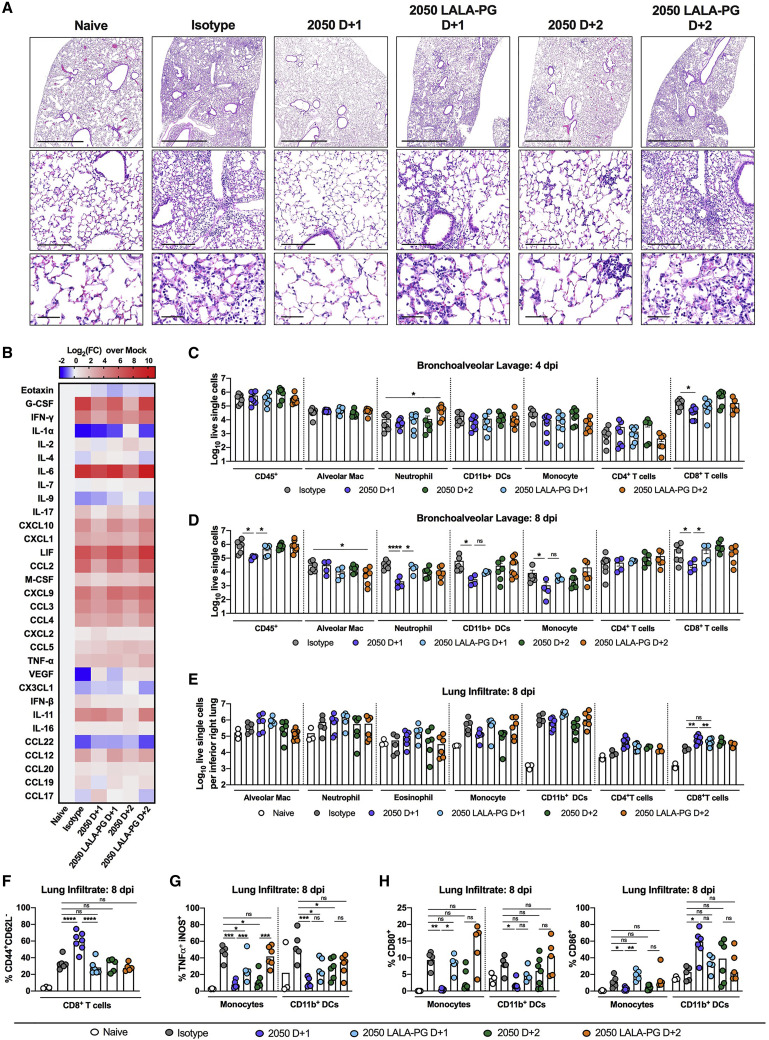
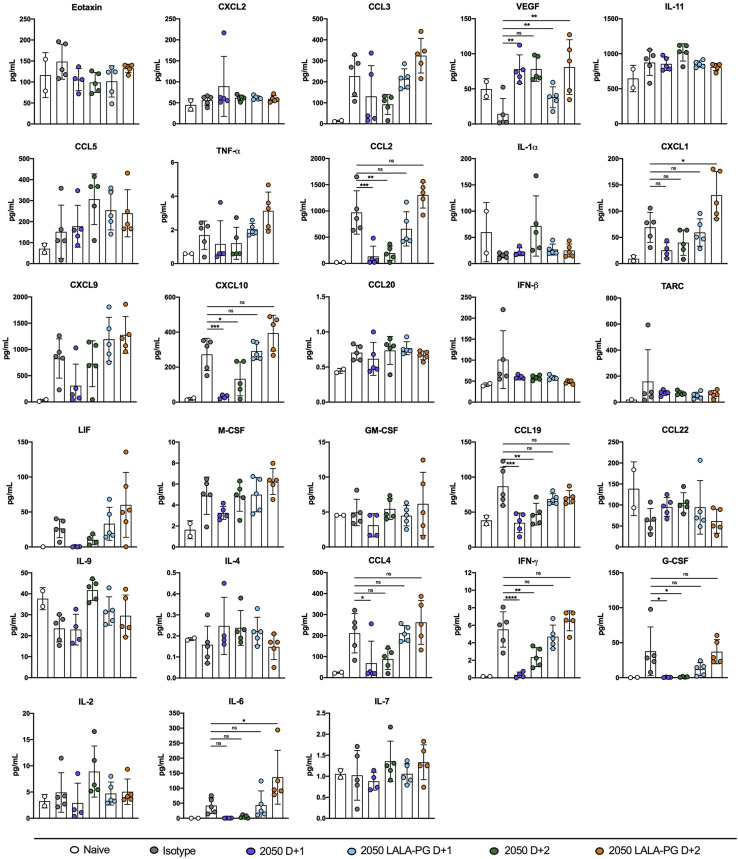
An excessive pro-inflammatory host response to SARS-CoV-2 infection is hypothesized to contribute to pulmonary pathology and severe COVID-19 (Giamarellos-Bourboulis et al., 2020; Tay et al., 2020). Given that COV2-2050-mediated improvement in pulmonary function was not associated with reduced viral RNA levels in the lung when given at D+2, we speculated that the intact mAb might modulate immune cell and inflammatory responses. Analysis of hematoxylin and eosin-stained lung sections from isotype mAb-treated mice at 8 dpi showed perivascular and parenchymal immune cell accumulation with accompanying edema and lung consolidation (Figure 4 A). Animals receiving COV2-2050 at D+1 showed markedly reduced lung inflammation, but this protection was lost in mice receiving COV2-2050 LALA-PG at D+1 or D+2. Although COV2-2050 treatment at D+2 did not completely reduce pathology, immune cell infiltration was more focal with patches of clear airspaces throughout the lung. Measurements of pro-inflammatory cytokine and chemokines in the lung at 8 dpi showed decreased levels of CXCL10, G-CSF, interleukin (IL)-6, interferon (IFN)-γ, CCL2, CCL3, CCL4, CCL19, and CXCL1 following both D+1 and D+2 COV2-2050 treatment, which did not occur in animals receiving COV2-2050 LALA-PG (Figures 4B and S2 ).
Figure 4.
Fc effector functions of a neutralizing antibody modulate the immune responses to SARS-CoV-2 infection
Eight-week-old K18-hACE2 mice were inoculated by the intranasal route with 103 PFU of SARS-CoV-2. At 1 (D+1) or 2 (D+2) dpi, mice were given 200 μg of COV2-2050 or COV2-2050 LALA-PG by intraperitoneal injection. Naive animals were mock-infected with sterile PBS.
(A) Hematoxylin and eosin staining of lung sections at 8 dpi or from a mock-infected animal. Images show low- (top; scale bars, 1 mm), medium-power (middle; scale bars, 200 μm), and high-power (bottom; scale bars, 50 μm). Representative images from n = 5 per group.
(B) Heat-maps of cytokine levels in lung tissue of SARS-CoV-2-infected mice at 8 dpi. Fold-change was calculated compared to mock-infected animals, and log2(fold-change) was plotted in the corresponding heat-map (3 experiments, n = 5 per group except naive (n = 2), statistics reported in Figure S2).
(C and D) Flow cytometric analysis of cells harvested from BAL fluid at (C) 4 and (D) 8 dpi (2 experiments, n = 4–8 per group; Bars, mean number of cells). Gating scheme in Figure S3.
(E–H) Flow cytometric analysis of lung tissues at 8 dpi. Gating scheme in Figure S3. (E) Number of immune cells harvested from the right inferior lobe. Bars, mean number of cells. (F) Proportion of CD44+CD62L− CD8+ T cells. (G and H) Proportion of CD80+, CD86+, or TNFα+iNOS+ monocytes or CD11b+ DCs. Bars, mean percentage of positive cells. (E–H) Two experiments, n = 3–8 per group. (C–H) One-way ANOVA with Dunnett’s test; ∗p < 0.05, ∗∗p < 0.01, ∗∗∗∗p < 0.0001.).
See also Figures S2 and S3.
Figure S2.
Cytokine induction following SARS-CoV-2 infection, related to Figure 4
Cytokine levels as measured by multiplex platform in the lungs of SARS-CoV-2 infected mice at 8 dpi following isotype, COV2-2050, or COV2-2050 LALA-PG treatment at 1 dpi (D+1) or 2 dpi (D+2) (three experiments, n = 5 per group except naive in which n = 2. One-way ANOVA with Dunnett’s test; ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001.) Asterisks indicate statistical significance compared to the isotype-control mAb-treated group.
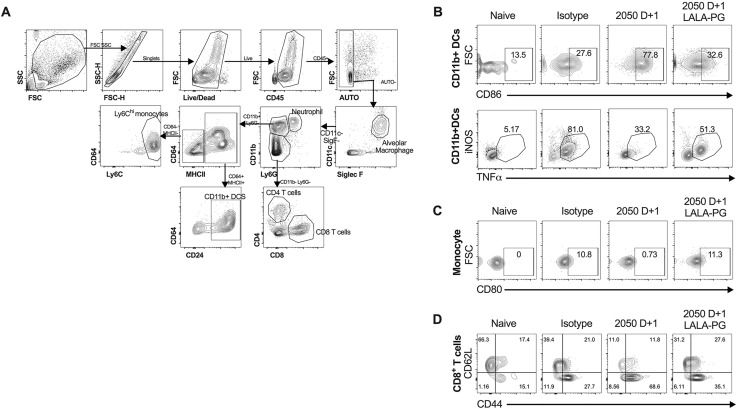
Cellular analysis of bronchoalveolar lavage (BAL) fluid at 4 dpi (Figure S3 ) showed significantly reduced numbers of CD8+ T cells with a trend toward fewer monocytes after administration of intact COV2-2050 at D+1. Differences in BAL fluid cell numbers were not observed at 4 dpi when treatment with COV2-2050 or COV2-2050 LALA-PG was started at D+2 (Figure 4C). At 8 dpi, we observed a significant reduction in the numbers of CD45+ cells, neutrophils, CD11b+DCs, Ly6Chi monocytes, and CD8+ T cells in the BAL fluid of mice treated at D+1 with COV2-2050 compared to COV2-2050 LALA-PG (Figure 4D) but no difference in BAL cell number when comparing COV2-2050 compared to COV2-2050 LALA-PG following D+2 treatment (Figure 4D). In comparison, flow cytometric analysis of the lung tissues at 8 dpi showed increased numbers of CD8+ T cells (Figure 4E) and a higher percentage of activated CD44+CD62L− CD8+ T cells (Figure 4F) associated with intact COV2-2050 D+1 treatment compared to animals treated with isotype control mAb or COV2-2050 LALA-PG at D+1. Additionally, monocytes and monocyte-derived CD11b+ DCs showed decreased expression of tumor necrosis factor alpha (TNF-α) and inducible nitric oxide synthase (iNOS) as well as the activation marker CD80+ following COV2-2050 D+1 treatment (Figures 4G and 4H). These results showing that greater antibody-dependent CD8+ T cell responses and skewing of myeloid cell inflammatory responses in the lung correlate with protection are consistent with studies showing that Fc engagement can enhance antiviral CD8+ T cell responses (Bournazos et al., 2020), and myeloid cell dysfunction may drive severe COVID-19 (Merad and Martin, 2020; Sánchez-Cerrillo et al., 2020).
Figure S3.
Flow cytometric gating strategy for BAL fluid analysis, related to Figure 4
(A) For BAL fluid and lung tissue flow cytometric analysis, cells were gated on live, single, autofluorescent-negative CD45+ cells to identify hematopoietic cells. Alveolar macrophages were identified as SiglecFhi CD11chi cells. Neutrophils were identified as Ly6Ghi CD11bhi cells. CD11b- cells were gated further into CD4+ and CD8+ T cells. CD11bhiLy6G- cells were gated subsequently using CD64, CD24, and MHC-II. MHCIIhi CD24hi were defined as CD11b+ DCs. MHCIIlo Ly6hi cells were defined as monocytes. (B) Representative flow cytometry plots of CD86 expression and TNFα/iNOS expression on CD11b+ DCs isolated from the lung tissues of indicated treatment groups. (C) Representative flow cytometry plots of CD80 expression on monocytes isolated from the lung tissues of indicated treatment groups. (D) Representative flow cytometry plots of CD44 and CD62L expression on CD8+ T cells isolated from the lung tissues of indicated treatment groups.
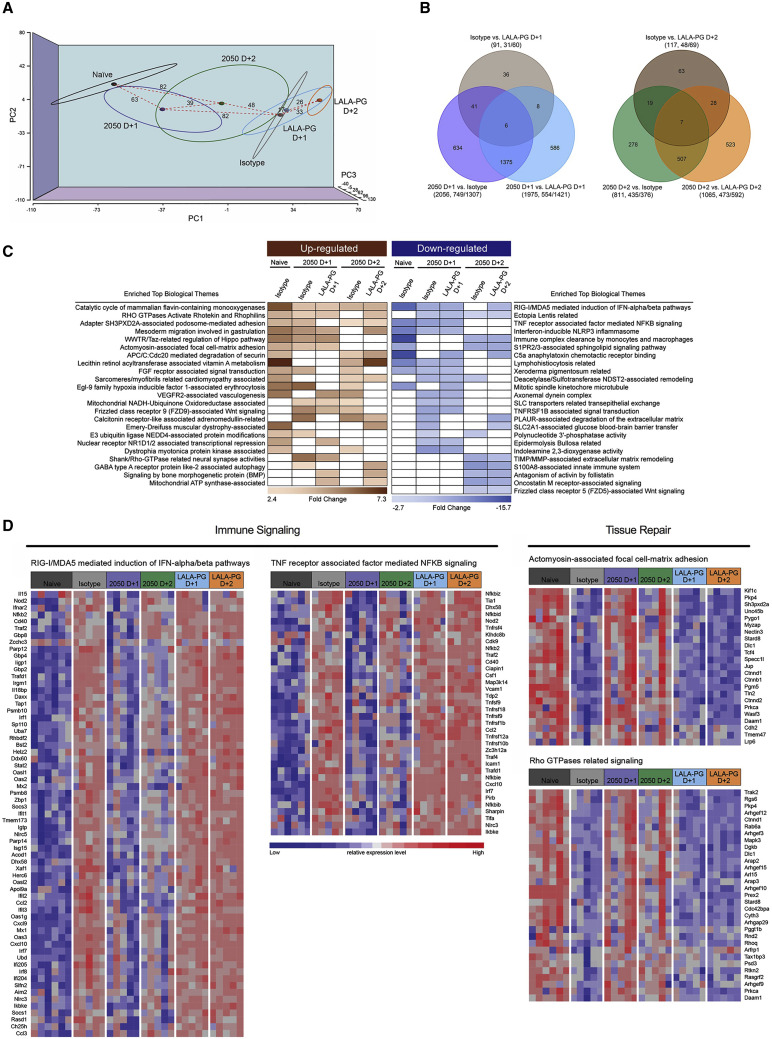
Distinct transcriptional signatures in the lung after treatment with COV2-2050 or COV2-2050 LALA-PG
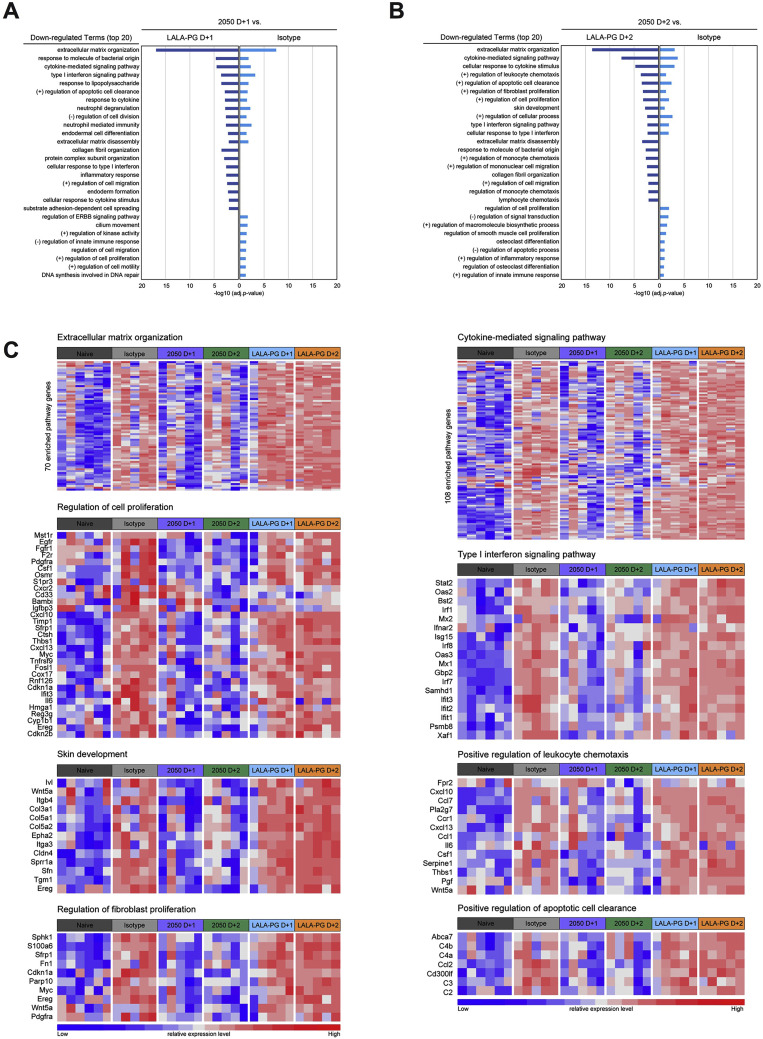
To interrogate further the impact of Fc effector functions on protection, we performed RNA sequencing of lung homogenates at 8 dpi in mice receiving an isotype control mAb, COV2-2050, or COV2-2050 LALA-PG mAbs at 1 (D+1) or 2 (D+2) and compared the results to those from samples from naive mice. Principal component analysis (PCA) revealed distinct transcriptional signatures associated with SARS-CoV-2 infection when compared to mock-infected naive animals. Treatment with intact COV2-2050 demonstrated a clear transcriptional shift toward the naive group, whereas the profiles from COV2-2050 LALA-PG-treated mice were more similar to isotype control mAb-treated animals (Figure 5 A). Indeed, only 91 differentially expressed genes (DEGs) were identified when comparing the isotype control mAb to the COV2-2050 LALA-PG D+1 group, whereas 2,056 and 1,975 DEGs were detected when comparing COV2-2050 D+1 to isotype control and COV-2050 LALA-PG D+1, respectively (Figure 5B). A similar large number of DEGs was observed in the lungs at 8 dpi from mice treated at D+2 with COV2-2050 or COV2-2050 LALA-PG (Figure 5B). Gene ontology analysis of the top downregulated genes comparing COV2-2050 to COV2-2050 LALA-PG showed immune gene clusters including cytokine-mediated signaling (e.g., Il10ra, Il15, Il17ra, Socs1, and Jak3), type I IFN signaling (e.g., Ifnar2, Stat2, Ifit1, Ifit2, Ifit3, and Irf7), and leukocyte chemotaxis (e.g., Ccl2, Cxcl10, and Ccl7) as well as genes involved in cell proliferation (e.g., Egfr, Fgfr1, Fosl1, Myc, and Cdkn2b) and metalloproteinase-mediated extracellular matrix organization (e.g., Adam15, Adam19, Col1a, Mmp14, and Itgam) (Figure S4 ; Table S1).
Figure 5.
Distinct transcriptional signatures in the lung are associated with COV2-2050 with intact Fc effector functions
RNA sequencing analysis from the lung homogenates of naive K18-hACE2 mice or mice infected with SARS-CoV-2 infection at 8 dpi. At 1 (D+1) or 2 (D+2) dpi, mice were given 200 μg of COV2-2050 or COV2-2050 LALA-PG by intraperitoneal injection (n = 5 per group except naive where n = 6).
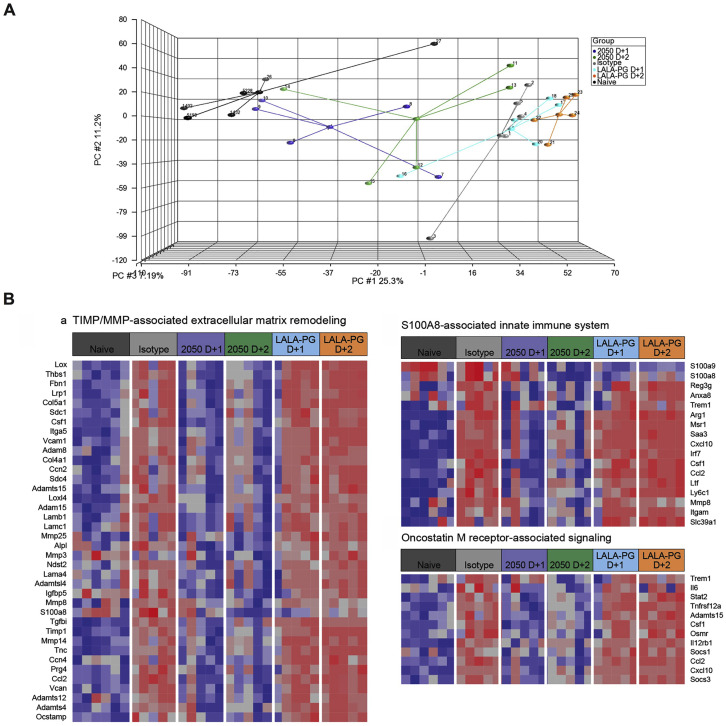
(A) Three-dimensional map from principal component analysis (PCA) of the RNA sequencing data in the study. The PCA has been performed using 12,157 unique genes with count per million reads ≥1 in at least 5 of the study samples (n = 31). Each group is represented by an ellipse and the color-matched solid circle, which is the centroid of each group. The size of the ellipse is the centroid with 1 SD. The dashed red lines with numbers indicate the spatial distance between centroids of the 6 groups, which is calculated by using the three-dimensional coordinates for the centroids.
(B) Venn diagrams of overlapping genes identified in differential expression analysis when comparing isotype control, COV2-2050 D+1, and COV2-2050 LALA-PG D+1 or isotype control, COV2-2050 D+2, and COV2-2050 LALA-PG D+2. Numbers in the parenthesis under each comparison indicate the number of differentially expressed genes (fold-change ≥2 at p < 0.05) followed by the proportion that are up- or downregulated.
(C) The significantly enriched biological themes defined by a “CompBio” pathway analysis tool comparing treatments with isotype control mAb, COV2-2050 (D+1 and D+2), and COV2-2050 LALA-PG (D+1 and D+2). Only those themes enriched in at least two comparisons are displayed. These themes either are upregulated (brown) or downregulated (blue) in the COV2-2050-treated group (at D+1 or D+2) when compared to the isotype control or COV2-2050 LALA-PG-treated groups. A comparison between the naive and isotype mAb-treated animals for each identified theme also was made. The scaled color blocks represent the mean fold-change of enriched genes with an enrichment score of 10 or greater in the comparison.
(D) Heatmaps of selected relevant biological themes (RIG-I/MDA-5 mediated signaling, TNF receptor-associated signaling, actinomyosin cell adhesion, Rho GTPases related signaling) enriched in COV2-2050 D+1 versus isotype control or COV2-2050 LALA-PG D+1. Genes shown are common in the pair of comparisons having an enrichment score of 100 or greater.
See also Figures S4 and S5 and Tables S1 and S2.
Figure S4.
Gene ontology analysis of RNA-seq data, related to Figure 5
(A-C) RNA sequencing analysis from the lung homogenates of naive K18-hACE2 mice or mice inoculated with SARS-CoV-2 at 8 dpi. At 1 (D+1) or 2 (D+2) dpi, mice were given a single 200 μg dose of COV2-2050 or COV2-2050 LALA-PG by intraperitoneal injection. (A-B) Gene Ontology (GO) Enrichment Analysis of biological process terms enriched in downregulated genes from comparisons of isotype control, COV2-2050, and COV2-2050 LALA-PG when given at D+1 (A) or isotype control, COV2-2050, and COV2-2050 LALA-PG when given at D+2 (B). Terms were ranked by the false discovery rate (q-value), and the top 20 are listed after eliminating redundant terms. (C) Heatmaps of significantly downregulated gene sets corresponding with intact COV2-2050 treatment identified through GO analysis. Genes shown in each pathway are the union of the differentially expressed genes (DEGs) from the five comparisons (isotype control, COV2-2050 D+1, COV2-2050 D+2, COV2-2050 LALA-PG D+1, or COV2-2050 LALA-PG D+2 versus mock-infected). Columns represent samples and rows represent genes. Gene expression levels in the heatmaps are z score-normalized values determined from log2cpm values.
We next performed CompBio analysis (v2.0, PercayAI), as we did previously (Adamo et al., 2020; Gehrig et al., 2019), to identify biological themes uniquely enriched in the COV2-2050-treated groups compared to the isotype control and COV2-2050 LALA-PG-treated groups (Figure 5C; Table S2). Pathways unique to the COV2-2050 D+1 treatment group compared to the isotype-treated group included genes involved in actinomyosin-associated cell adhesion (e.g., Kif1c, Ctnnd1, Nectin3, Unc45b, and Prkca) and Rho GTPase signaling (e.g., Rhoq, Mapk3, Prkca, and Cdc42bpa), processes that are typically associated with wound repair programs (Verboon and Parkhurst, 2015) (Figure 5D). Pathways downregulated in the COV2-2050 D+1 group included genes involved in type I IFN and nuclear factor κB (NF-κB)-dependent signaling (e.g., Irf7, Stat2, Nfkb2, Bst2, Isg15, and Ikbk3) (Figure 5D), which may in part be due to the lower levels of viral RNA detected (Figures 2E and 2F). The expression pattern of down- or upregulated gene sets in the COV2-2050 D+1 treated animals was similar to that in naive animals, suggesting that the intact antibody limited the transcriptional changes usually seen during SARS-CoV-2 infection.
Pathways that were downregulated in the D+2 COV2-2050-treated group compared to the isotype control or D+2 COV2-2050 LALA-PG-treated animals included S100A8-associated innate immune signaling (e.g., Reg3g, Saa3, Itgma, Mmp8, and S100a8), oncostatin M receptor associated signaling (e.g., Il6, Osmr, Csf1, and Socs3), and extracellular matrix remodeling (e.g., Adamts15, Col5a1, Vcam1, and Lama4) (Figure S5 ; Table S2). Thus, COV2-2050 treatment at D+2 and the resultant Fc-dependent effector responses may differentially modulate neutrophil activation, gp130 signaling, and tissue damage due to matrix metalloproteinase activation. Unlike what was observed in the analysis of gene sets unique to D+1, these downregulated programs at D+2 were not lower in naive animals. Therefore, intact COV2-2050 administration at D+2 may induce a separate protective transcriptional program that is not simply preventing virus-induced transcriptional changes. Of note, we also identified diminished expression of genes involved in immune complex clearance following COV2-2050 versus COV2-2050 LALA-PG administration at D+2, which may reflect a negative feedback loop (Daëron and Lesourne, 2006). Collectively, our results with intact COV2-2050 suggest that Fc engagement by FcγR and/or complement can induce distinct and protective transcriptional programs in the SARS-CoV-2-infected lung depending on the timing of therapy.
Figure S5.
CompBio analysis comparing COV2-2050 D+2, isotype control, and COV2-2050 LALA-PG D+2, related to Figure 5
(A) Three-dimensional map from principal component analysis (PCA) of the RNA-seq data in the study. The PCA was performed using 12,157 unique genes with counts per million reads ≥ 1 in at least 5 of the study samples (n = 31). Each mouse is represented by an individual point that is color-matched to its corresponding experimental group and connected to the centroid point (with a cross bar and ID number) of the corresponding group. (B) Heatmaps of selected relevant biological themes enriched in COV2-2050 D+2 versus isotype control and COV2- LALA-PG D+2. Genes shown are common in the pair of comparisons with an enrichment score of 100 or greater in either of the paired comparisons.
Monocytes and CD8+ T cells are required for optimal therapeutic activity of neutralizing mAb
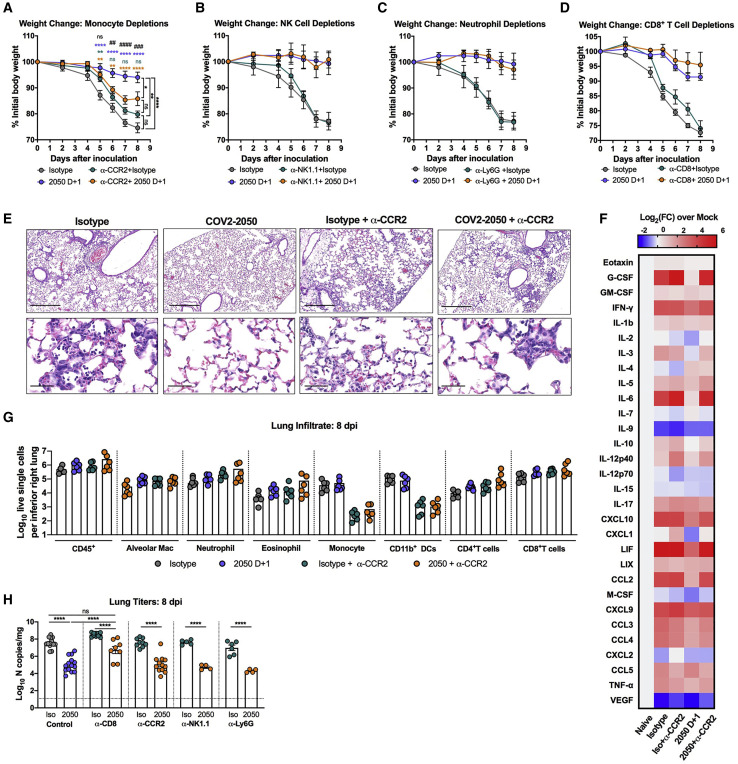
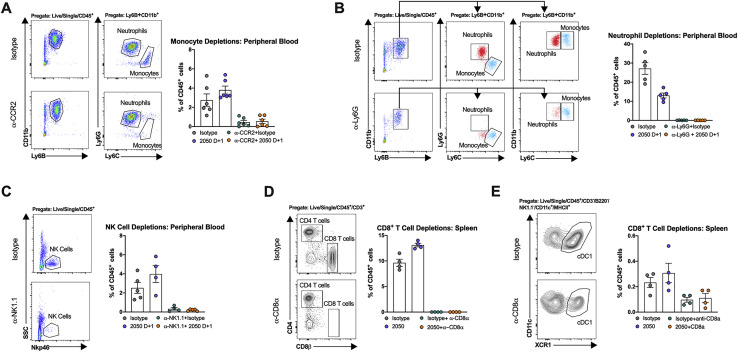
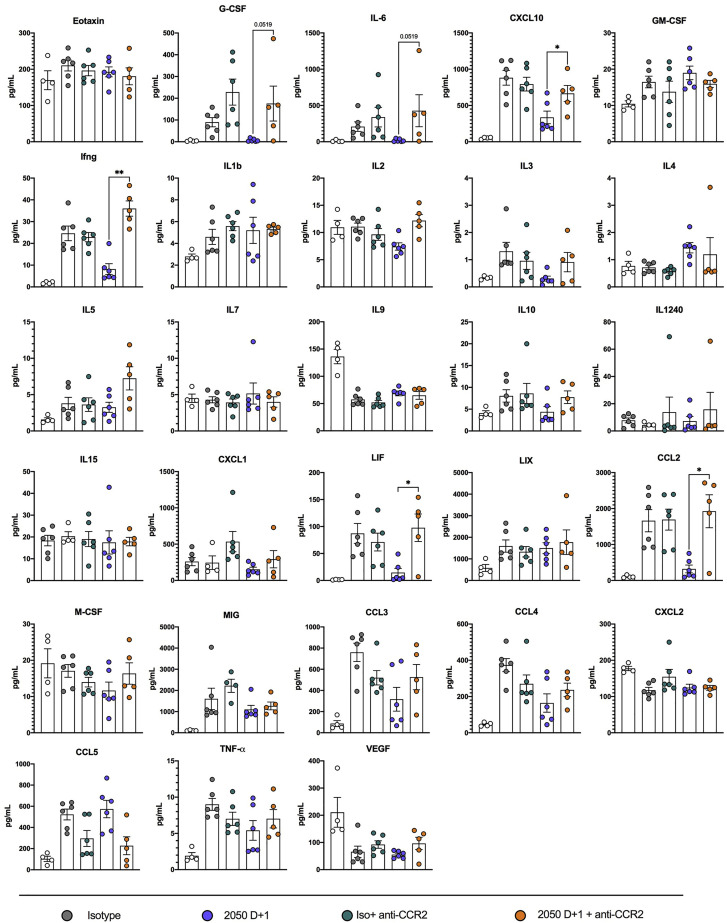
Fc engagement of FcγRs on innate immune cells including macrophages, monocytes, natural killer (NK) cells, and neutrophils can lead to antibody-dependent cellular cytotoxicity (ADCC) or phagocytosis (ADCP), modulation of inflammatory mediators produced by these cells, and enhanced adaptive immunity (Bournazos et al., 2020; Lu et al., 2018). To determine which immune cells contributed to the antibody-mediated protection observed in vivo, we depleted Ly6Chi monocytes (anti-CCR2) (Figures 6 A and S6 ), natural killer (NK) cells (anti-NK1.1) (Figures 6B and S6), mature neutrophils (anti-Ly6G) (Figures 6C and S6) or CD8+ T cells (anti-CD8) (Figures 6D and S6) in combination with COV2-2050 treatment at D+1. Depletion of mature neutrophils, NK cells, or CD8+ T cells had no impact on weight loss in the presence of COV2-2050 or the isotype control mAb (Figures 6B–6D). When monocytes were depleted, COV2-2050 failed to prevent the weight loss phenotype seen in non-depleted, COV2-2050-treated mice (Figure 6A). Monocyte depletion during COV2-2050 therapy also was associated with a loss of improvement in lung pathology (Figure 6E) and higher levels of some cytokines and chemokines (CXCL10, G-CSF, IL-6, IFN-γ, and CCL2) compared to treatment with COV2-2050 without cell depletion (Figures 6F and S7 ). As expected, the numbers of monocytes and CD11b+ DCs in the lung were lower after anti-CCR2 treatment, but other cell subsets were unaffected. Because these myeloid cells show a less inflammatory phenotype after COV2-2050, but not COV2-2050 LALA-PG, treatment (Figures 4G and 4H), FcγR engagement may shape cellular responses to limit immunopathology independently of viral clearance.
Figure 6.
Monocytes and CD8+ T cells are necessary for protection following mAb therapy
(A–H) Eight-week-old K18-hACE2 mice received anti-CCR2 (50 μg/dose) (A), anti-NK1.1 (200 μg/dose) (B), anti-Ly6G (250 μg/dose) (C), or anti-CD8 (500 μg/dose) (D) or corresponding isotype controls at D−1, D+1, D+3, D+5, and D+7 relative to SARS-CoV-2 infection. At D0, animals were inoculated by the intranasal route with 103 PFU of SARS-CoV-2. At 1 (D+1) dpi, mice were administered 200 μg of COV2-2050 by intraperitoneal injection. (A–D) Weight change (mean ± SEM; n = 8–12, 2–3 experiments: two-way ANOVA with Sidak’s post-test: ∗p < 0.001, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001; comparison to the isotype control mAb-treated group).
(E) Hematoxylin and eosin staining of lung sections at 8 dpi. Images show low-power (top; scale bars, 250 μm), or high-power (bottom; scale bars, 50 μm). Representative images from n = 3 per group.
(F) Heat-maps of cytokine levels in lung tissues of SARS-CoV-2-infected mice at 8 dpi. For each cytokine, fold-change was calculated compared to mock-infected animals, and log2 (fold-change) was plotted in the corresponding heat-map (3 experiments, n = 5–6 per group except naive (n = 4).
(G) Flow cytometric analysis of lung tissues at 8 dpi (2 experiments, n = 8 per group; Bars, mean number of cells).
(H) Viral RNA levels at 8 dpi in the lung (n = 4–12, 2–3 experiments: one-way ANOVA with Turkey’s post-test: ns not significant, ∗∗∗∗p < 0.0001, comparison to the isotype control mAb-treated group).
See Figures S6 and S7.
Figure S6.
Confirmation of cellular depletions, related to Figure 6
(A) Representative flow cytometry plots of monocytes and neutrophils from peripheral blood at 8 dpi following intraperitoneal injection of a depleting anti-CCR2 mAb or isotype control mAb and frequency of Ly6Chi monocytes and neutrophils in blood at 8 dpi following anti-CCR2 or isotype control mAb administration in isotype control or COV2-2050-treated mice (two experiments, n = 6 per group). (B) Representative flow cytometry plots of peripheral blood at 8 dpi following intraperitoneal injection of a depleting anti-Ly6G mAb or isotype control mAb and frequency of Ly6Chi monocytes and mature Ly6G+ neutrophils in blood at 8 dpi following anti-Ly6G or isotype control mAb administration in isotype control or COV2-2050-treated mice (two experiments, n = 5-6 per group). In the groups treated with anti-Ly6G, neutrophils were identified as CD11b+ Ly6B+ Ly6Cint cells. Red dots (neutrophils) and monocytes (blue dots) correspond to the same population identified in both plots (neutrophils: CD11b+ Ly6C+Ly6G+ Ly6Cint and monocytes: CD11b+ Ly6C+Ly6G- Ly6Chigh). (C) Representative flow cytometry plots of peripheral blood at 8 dpi following intraperitoneal injection of a depleting anti-NK1.1 mAb or isotype control mAb. Also shown is the frequency of NK cells in blood at 8 dpi following anti-NK1.1 or isotype control mAb administration in isotype control or COV2-2050-treated mice (two experiments, n = 4-5 per group). (D) Representative flow cytometry plots of splenocytes gated on CD4+ and CD8+ T cells at 8 dpi following intraperitoneal injection of a depleting anti-CD8α mAb or isotype control mAb (left). Frequency of CD8+ T cells in the spleen at 8 dpi following anti-CD8α or isotype control mAb administration in isotype mAb control or COV2-2050-treated mice (right) (two experiments, n = 4-5 per group). (E) Representative flow cytometry plots of splenocytes gated to cDC1s at 8 dpi following intraperitoneal injection of a depleting anti-CD8α mAb or isotype control mAb (left). Frequency of cDC1 cells in the spleen at 8 dpi following anti-CD8α or isotype control mAb administration in isotype control mAb or COV2-2050-treated mice (right) (two experiments, n = 4-5 per group).
Figure S7.
Cytokine induction following SARS-CoV-2 infection and monocyte depletion, related to Figure 6
Cytokine levels as measured by multiplex platform in the lungs of mock-infected mice or SARS-CoV-2-infected mice at 8 dpi following treatment with isotype mAb, COV2-2050 D+1, isotype mAb + anti-CCR2, or COV2-2050 D+1 + anti-CCR2 (two experiments, n = 4-6 per group. One-way ANOVA with Dunnett’s test; ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001.) One animal was excluded from cytokine analysis in the 2050 D+1 + anti-CCR2 group due to incomplete monocyte depletion in peripheral blood (monocytes still made up ~3% of CD45+ cells).
Although monocyte depletion in combination with COV2-2050 treatment resulted in increased weight loss, it was not associated with changes in viral RNA levels at 8 dpi (Figure 6H), suggesting that Fc interactions with monocytes do not contribute directly to viral clearance. In contrast, and despite no difference in weight loss, when CD8+ T cells were depleted, COV2-2050 treatment less efficiently reduced viral RNA levels (Figure 6H). These data in conjunction with data showing increased CD8+ T cell activation following COV2-2050 D+1 treatment (Figure 4F) suggest that Fc effector functions can enhance CD8+ T cell responses, possibly through increased antigen presentation (Bournazos et al., 2020), to mediate viral clearance.
Fc effector functions enhance the therapeutic activity of neutralizing mAbs against SARS-CoV-2 in Syrian hamsters
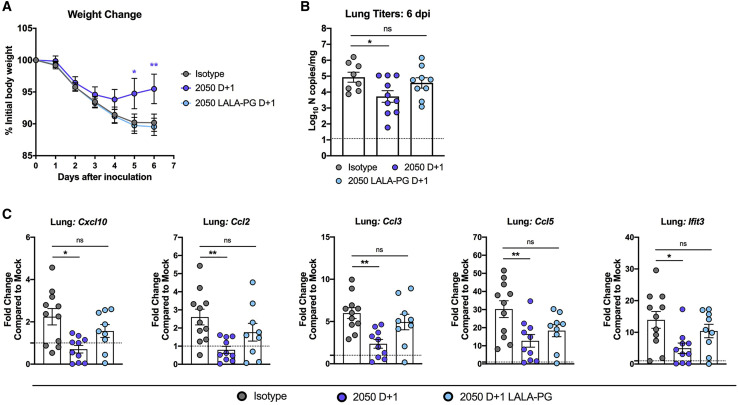
To validate the requirement for Fc effector functions in therapeutic antibody protection, we used a golden Syrian hamster model. This animal model supports viral replication with corresponding weight loss and interstitial pneumonia (Rosenke et al., 2020; Sia et al., 2020) and has been used to evaluate mAb-based therapies and vaccines (Liu et al., 2020; Rogers et al., 2020; Tostanoski et al., 2020) against SARS-CoV-2. We inoculated 7-month-old hamsters with SARS-CoV-2 (5 × 105 PFU, 2019 n-CoV/USA_WA1/2020 strain) by the intranasal route and then delivered a single 1-mg dose of COV2-2050 or COV2-2050 LALA-PG mAbs at 1 dpi (D+1) by intraperitoneal injection. Consistent with observations in mice, passive transfer of intact COV2-2050 prevented weight loss compared to isotype mAb-treated animals at 5 and 6 dpi, and this protection was lost in animals treated with the COV2-2050 LALA-PG variant (Figure 7 A). Furthermore, hamsters treated with intact COV2-2050, but not COV2-2050 LALA-PG, showed reductions in viral RNA levels at 6 dpi (Figure 7B). The improved viral burden with COV2-2050 was associated with lower levels of the inflammatory mediators Cxcl10, Ccl2, Ccl3, Ccl5, and Ifit3 (Figure 7C). Thus, therapeutic efficacy following neutralizing mAb administration also depends on Fc interactions in a second, relevant small animal model.
Figure 7.
Fc effector functions enhance the therapeutic activity of neutralizing antibodies against SARS-CoV-2 in Syrian hamsters
(A–C) Seven-month-old female Syrian hamsters were inoculated by the intranasal route with 5 × 105 PFU of SARS-CoV-2. At 1 dpi (D+1), hamsters were given 1 mg of COV2-2050 or COV2-2050 LALA-PG by intraperitoneal injection.
(A) Weight change (mean ± SEM; n = 8–10, 2 experiments: two-way ANOVA with Sidak’s post-test: ∗p < 0.05, ∗∗p < 0.01; comparison to the isotype control mAb-treated group).
(B) Viral RNA levels at 6 dpi in the lung (n = 8–10, 2 experiments: one-way ANOVA with Turkey’s post-test: ns not significant, ∗p < 0.05, comparison to the isotype control mAb-treated group).
(C) Fold change in gene expression of indicated cytokines and chemokines in lung homogenates. Data are normalized to Rpl18 and compared to naive controls (2 experiments, n = 8–10 per group, one-way ANOVA with Dunnett’s test; ∗p < 0.05; ∗∗p < 0.01). Dotted lines indicate the mean cytokine or chemokine transcript levels in naive hamsters.
Discussion
Neutralizing mAbs against SARS-CoV-2 are a promising option for the treatment of COIVD-19 and have demonstrated efficacy in a variety of pre-clinical models (Abraham, 2020; Marovich et al., 2020). Antibody programs have reported encouraging results in clinical trials (Chen et al., 2021), and the Food and Drug Administration (FDA) granted Emergency Use Authorization to the anti-SARS-CoV-2 S mAbs bamlanivimab and casirivimab/imdevimab for mild-to-moderately ill, high-risk patients. However, the mechanisms of protection in vivo have not been fully investigated and are assumed to be related to the neutralizing capacity of the mAb. In this study, we examined the contributions of Fc effector functions for clinical and virological protection in murine and hamster models of SARS-CoV-2 infection. Whereas intact Fc effector functions were not required when neutralizing mAbs were given as prophylaxis, for several anti-RBD human mAbs tested in mice, a functional Fc region was required for optimal protection as post-exposure therapy. This enhanced therapeutic efficacy of intact compared to their LALA-PG IgG loss-of-function Fc region variants correlated with decreased viral burden, improved pulmonary function, diminished inflammatory responses, and preserved tissue repair processes. In mice, cell depletion studies identified both monocytes and CD8+ T cells as key immune cell type for antibody-dependent clinical and virological protection.
Fc effector function interactions were not required when neutralizing mAbs were administered as prophylaxis, suggesting their mechanism of protection in a pre-exposure setting depends largely on the neutralizing capacity of the antibody to prevent initial viral infection and limit dissemination. In the stringent K18-hACE2 model of SARS-CoV-2 pathogenesis (Golden et al., 2020; Winkler et al., 2020), we defined minimum serum neutralizing titers (NT50) and concentrations of 104 and 212 ng/mL for the prevention of weight loss and 381 and 851 ng/mL for reduction of viral burden in the lung. Establishing serum correlates of protection for mAb- and vaccine-based therapies in pre-clinical models is an important step for translation and analysis of human studies. Levels of neutralizing antibody required for protection against disease in humans might be lower due to the slower kinetics of disease pathogenesis and/or a longer half-life of the antibody. Last, even at sub-protective doses of intact COV2-2050, we did not observe evidence of antibody-dependent enhancement (ADE) of infection or immune enhancement, consistent with other studies (Baum et al., 2020; Cao et al., 2020; Halstead and Katzelnick, 2020; Laczkó et al., 2020; Luo et al., 2018; Qin et al., 2006; Rogers et al., 2020).
In contrast to prophylactic administration, Fc effector functions were required for optimal efficacy when neutralizing mAbs were administered as post-exposure therapy. The treatment window to reduce viral RNA levels in the lung in the stringent K18-hACE2 model was essentially 1 day, suggesting that antibody therapy optimally should be given prior to the peak of viral replication, which is 2 dpi in this model (Winkler et al., 2020). The narrow therapeutic window for reducing viral load we observed could be related to the stoichiometry of antibody binding required for virus neutralization (Pierson and Diamond, 2015), such that when too much viral antigen is present in the lung, neutralization is limited. Against this hypothesis, infectious virus levels were neutralized and equivalently low at 4 dpi in mice treated with intact or LALA-PG versions of antibodies at 1 or 2 dpi, although it is possible that this effect is due to ex vivo neutralization of virus after tissue homogenization (Subbarao et al., 2004; Wells et al., 1981). More likely, the Fc effector functions of neutralizing antibodies serve to clear SARS-CoV-2 infected cells through enhanced antigen presentation and induction of antigen-specific CD8+ T cell responses (Bournazos et al., 2020). Although the precise mechanism awaits delineation, our data suggest that in the post-exposure therapeutic setting, the neutralizing activity of most RBD-specific antibodies may no longer be sufficient for optimal protection, and additional mechanisms mediated by Fc effector functions are required. We acknowledge that some anti-RBD mAbs with superior neutralizing activity in vivo may confer protection in the absence of Fc effector functions, at least at early time points post-infection. However, at later time points, once infection is established and high amounts of viral RNA are produced in the lung, Fc effector functions of even “elite” neutralizing mAbs may be needed for immune modulation and the most beneficial clinical outcome.
With the lower levels of viral RNA in lungs of mice treated at 1 dpi with COV2-2050 but not COV2-2050 LALA-PG, we also found reduced levels of pro-inflammatory cytokines, immune cell infiltration, and expression of genes downstream of type I IFN and NF-κB-dependent signaling pathways. Transcriptional signatures associated with COV2-2050 therapy also included enrichment of gene sets involved in tissue repair processes. This same pattern of expression was observed in naive mice, suggesting that Fc effector functions of antibodies may limit virus-induced perturbations and maintain certain reparative homeostatic transcriptional programs. Remarkably, administration of neutralizing mAb at 2 or 3 dpi still improved survival in an Fc-dependent manner without substantive reduction in viral RNA levels. The gene signatures uniquely associated with intact COV2-2050 mAb administration at 2 dpi differed from those at 1 dpi, as they showed reduced expression of genes involved in neutrophil activation, IL-6 signaling, and metalloproteinase-mediated extracellular matrix remodeling pathways. Altogether, these results suggest that at least some neutralizing mAbs protect in vivo through multiple Fc-dependent mechanisms including both viral clearance and modulation of the immune response to enhance resolution of inflammation.
The importance of Fc effector functions for antibody efficacy in vivo has been illustrated in other viral models (Bournazos et al., 2020; DiLillo et al., 2014, 2016; Fox et al., 2019; Hessell et al., 2007; Li et al., 2017; Liu et al., 2017; Vogt et al., 2011), although the specific cells mediating protection have been characterized in relatively few cases (Fox et al., 2019; He et al., 2017; Laidlaw et al., 2013). Our immune cell depletion studies showed that monocytes, but not neutrophils or NK cells were necessary for mAb-dependent clinical protection and diminished pro-inflammatory cytokine responses following SARS-CoV-2 infection in mice, although monocyte depletion did not affect viral burden. Circulating CCR2+ monocytes can differentiate into myeloid cell subsets including interstitial macrophages and monocyte-derived CD11b+ dendritic cells following migration to the lung (Jakubzick et al., 2017). The phenotype of these myeloid cells in the lung can vary from TNF/iNOS producing DCs (Tip-DCs) (Serbina et al., 2003) to interstitial macrophages involved in tissue repair. FcγR-mediated signaling can impact myeloid cell function, differentiation, and polarization (Clynes et al., 1999; Dhodapkar et al., 2007). Moreover, myeloid cell dysregulation has been linked to severe COVID-19 in humans (Schulte-Schrepping et al., 2020), and blocking of TNF-α and IFNγ reduces lethality following SARS-CoV-2 infection in K18-hACE2 mice (Karki et al., 2021). Administration of intact COV2-2050 but not COV2-2050 LALA-PG, even at D+2, when a difference in viral titers was not detected between these two groups, reduced the proportion of monocytes and monocyte-derived CD11b+DCs producing TNF-α and iNOS without impacting the total number of myeloid cells in the lung. Thus, Fc effector engagement on monocyte-derived cells in the lung may not directly impact viral clearance, but rather improve clinical outcome through reprograming of the immune response.
In comparison, CD8+ T cells were required for Fc-effector function-dependent reductions in viral burden. Phagocytosis of IgG immune complexes by activating FcγRs on antigen-presenting cells, such as dendritic cells, can enhance antigen processing (Amigorena et al., 1992; Bergtold et al., 2005; Hoffmann et al., 2012) and accelerate CD8+ T cell priming and cytolysis of virally infected cells (Lu et al., 2016; Niessl et al., 2020). Our data suggest a compartmentalization of cellular interactions mediated by antibody Fc regions in the context of SARS-CoV-2 infection: myeloid cell interactions regulate inflammation, and enhanced CD8+ T cell activity promotes clearance of virus-infected cells. Engineering of the Fc domain sequences to optimize selected effector functions (Bournazos et al., 2020; Lazar et al., 2006; Saunders, 2019) on specific cell types could augment the therapeutic activity of neutralizing mAbs against SARS-CoV-2.
Limitations of study
Consistent with previous findings (Schäfer et al., 2021), our results highlight the importance of Fc effector functions of antibody in two animal models of SARS-CoV-2 infection. Although LALA-PG mutations abrogate binding to both FcγRs and C1q, it remains unknown which of these effector molecules or particular receptors (e.g., FcγRI, FcγRIII, or FcγRIV) dominantly associate with loss of anti-SARS-CoV-2 mAb therapeutic activity. Confirmatory studies also are needed in non-human primates, a model that more closely recapitulates human mAb dosing kinetics in vivo. Although we used human IgG1 anti-SARS-CoV-2 antibodies in mice and hamsters, we validated our findings in a species-homologous system with chimeric versions of COV2-2072 with murine Fc moieties in mice. FcγR expression patterns on immune cells and human IgG-mouse/hamster FcγR interactions or even mouse IgG-mouse FcγR interactions may not fully recapitulate patterns observed with human IgG, human FcγRs, and human cells (Bournazos et al., 2014a). Although animals expressing human FcγRs exist (Smith et al., 2012), they have not been crossed to the K18-hACE2 transgenic mice. Future studies with human FcγR mice, mouse-adapted strains of SARS-CoV-2 (Leist et al., 2020), and additional human anti-SARS-CoV-2 mAbs may be useful for confirmation of the phenotypes described.
The requirement of Fc effector functions for protection by other anti-SARS-CoV-2 mAbs warrants more study. Indeed, in vitro neutralization potency does not uniformly correlate with in vivo protection (Schäfer et al., 2021), and compromised Fc effector functions correlates with increased mortality in humans (Zohar et al., 2020). Our studies used neutralizing antibodies that bind the RBD on S and block ACE2 receptor engagement (Zost et al., 2020b). Other neutralizing mAbs against the N-terminal domain or some classes of non-neutralizing mAbs also may require Fc effector functions for therapeutic activity. Even among anti-RBD mAbs, we observed variation, as clinical protection with COV2-2381 and COV2-2072 was only partially Fc-dependent when administered as post-exposure therapy at D+1, yet virological protection was lost with COV2-2072 LALA-PG, but not COV2-2381 LALA-PG administration. In conclusion, our study highlights the contributions of Fc effector functions for therapeutic activity of neutralizing mAbs through reductions in viral burden and/or immunopathology. Accordingly, the design of antibody-based combinations against SARS-CoV-2 likely should optimize both neutralization and multiple Fc effector function pathways to enhance the window of treatment and provide the greatest antiviral and clinical protection.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| COV2-2050 | Zost et al., 2020b | N/A |
| COV2-2050 LALA-PG | This paper | N/A |
| COV2-2072 | Zost et al., 2020b | N/A |
| COV2-2072 LALA-PG | This paper | N/A |
| COV2-3025 | Zost et al., 2020b | N/A |
| COV2-3025 LALA-PG | This paper | N/A |
| COV2-2381 | Zost et al., 2020b | N/A |
| COV2-2381 LALA-PG | This paper | N/A |
| COV2-2072 mIgG1 | This paper | N/A |
| COV2-2072 mIgG1 D265A | This paper | N/A |
| COV2-2072 mIgG2a | This paper | N/A |
| DENV-2D22 | Fibriansah et al., 2015 | N/A |
| HRP conjugated goat anti-mouse IgG (H + L) | Jackson ImmunoResearch | 115-035-062; RRID: AB_2338504 |
| CR3022, anti-SARS-CoV-2 mAb | Yuan et al., 2020 | N/A |
| Goat anti-human lambda, mouse ads-UNLB | Southern Biotech | RRID:AB_2795760 |
| Goat anti-human kappa, mouse ads-UNLB | Southern Biotech | RRID:AB_2795728 |
| Goat anti-human IgG Fc, Multispecies ads-HRP | Southern Biotech | RRID:AB_2795580 |
| BUV395 anti-CD45 | BD BioSciences | RRID:AB_2651134 |
| Brilliant Violet 650 anti-mouse Ly-6G | BioLegend | RRID:AB_2565881 |
| Pacific Blue anti-mouse Ly-6C | BioLegend | RRID:AB_1732090 |
| PE/Dazzle 594 anti-mouse/human CD11b | BioLegend | RRID:AB_2563648 |
| Brilliant Violet 711 anti-mouse CD3 Antibody | BioLegend | RRID:AB_2563945 |
| Brilliant Violet 421 anti-mouse CD3 Antibody | BioLegend | RRID:AB_10900227 |
| APC anti-mouse CD8b Antibody | BioLegend | RRID:AB_2562774 |
| PE anti-mouse/rat XCR1 Antibody | BioLegend | RRID:AB_2563843 |
| Brilliant Violet 785 anti-mouse CD4 Antibody | BioLegend | RRID:AB_2565843 |
| BV421 Rat Anti-Mouse CD335 | BD BioSciences | RRID:AB_2737837 |
| FITC anti-mouse NK-1.1 Antibody | BioLegend | RRID:AB_313393 |
| PE-Cy7 Hamster Anti-Mouse CD11c | BD BioSciences | RRID:AB_2033997 |
| Alexa Fluor® 700 anti-mouse I-A/I-E | BioLegend | RRID:AB_493726 |
| eBioscience Fixable Viability Dye eFluor 506 | Thermo Fisher | 65-0866-14 |
| FITC Anti-Neutrophil (Anti-Ly6B) Antibody | Abcam | RRID:AB_881408 |
| TruStain FcX (anti-mouse CD16/32) Antibody | BioLegend | RRID:AB_1574973 |
| Brilliant Violet 711 anti-mouse/human CD45R/B220 Antibody | BioLegend | RRID:AB_2563491 |
| Brilliant Violet 711 anti-mouse/human CD11b Antibody | BioLegend | RRID:AB_2563310 |
| FITC anti-mouse CD24 Antibody | BioLegend | RRID:AB_312839 |
| PerCP/Cyanine5.5 anti-mouse Ly-6G Antibody | BioLegend | RRID:AB_1877271 |
| PE/Cyanine7 anti-mouse CD64 (FcγRI) Antibody | BioLegend | RRID:AB_2563904 |
| PE Rat Anti-Mouse Siglec-F | BD BioSciences | RRID:AB_394341 |
| Brilliant Violet 605 anti-mouse CD80 Antibody | BioLegend | RRID:AB_11126141 |
| PE/Cyanine5 anti-mouse CD86 Antibody | BioLegend | RRID:AB_493602 |
| Brilliant Violet 605 anti-mouse CD62L Antibody | BioLegend | RRID:AB_2563058 |
| APC/Cyanine7 anti-mouse/human CD44 Antibod6 | BioLegend | RRID:AB_830785 |
| PE/Cyanine7 anti-mouse TNF-α Antibody | BioLegend | RRID:AB_2256076 |
| Brilliant Violet 421 anti-mouse CD64 (FcγRI) Antibody | BioLegend | RRID:AB_2562694 |
| APC-Cy7 Hamster Anti-Mouse CD11c | BD BioSciences | RRID:AB_10611727 |
| NOS Monoclonal Antibody (CXNFT), APC, eBioscience | Thermo Fisher | RRID:AB_2573244 |
| InVivoMAb rat IgG2b isotype control, anti-keyhole limpet hemocyanin | BioXCell | RRID:AB_1107780 |
| InVivoMAb anti-mouse Ly6G | BioXCell | RRID:AB_1107721 |
| InVivoMAb rat IgG2a isotype control, anti-trinitrophenol | BioXCell | RRID:AB_1107769 |
| InVivoPlus anti-mouse CD8α | BioXCell | RRID:AB_10950145 |
| InVivoMAb anti-mouse NK1.1 | BioXCell | RRID:AB_1107737 |
| InVivoMAb mouse IgG2a isotype control, unknown specificity | BioXCell | RRID:AB_1107771 |
| anti-CCR2 (clone MC-21) | Matthias Mack | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Recombinant Mouse Fc gamma RIIB/CD32b Protein, CF | R and D Systems | 1460-CD-050 |
| Recombinant Mouse Fc gamma RI/CD64 Protein, CF | R and D Systems | 2074-FC-050 |
| Recombinant Mouse FcgR4/CD16-2 Protein, CF | R and D Systems | 1974-CD-050 |
| Critical commercial assays | ||
| GIBCO ExpiCHO Expression System | ThermoFisher Scientific | A29133 |
| MagMAX-96 Viral RNA Isolation Kit | Thermo Fisher | AM1836 |
| MagMAX mirVana Total RNA Isolation Kit | Thermo Fisher | A27828 |
| TaqMan RNA-to-Ct 1-Step Kit | Thermo Fisher | 4392939 |
| Mouse Cytokine Array / Chemokine Array 44-Plex (MD44) | Eve Technologies | MD44 |
| Mouse Cytokine Array / Chemokine Array 31-Plex (MD31) | Eve Technologies | MD31 |
| Ambion DNase I (RNase-free) | Thermo Fisher | AM2222 |
| High-Capacity cDNA Reverse Transcription Kit with RNase Inhibitor | Thermo Fisher | 4374966 |
| Foxp3/Transcription Factor Staining Buffer Set | Thermo Fisher | 00-5523-00 |
| Endotoxin PTS201F cartridge | Charles River | PTS201F |
| SeqPlex RNA Amplification Kit | Millipore Sigma | EQR-500RXN |
| Virus and bacterial strains | ||
| SARS-CoV-2 (strain 2019 n-CoV/USA_WA1/2020) | CDC/BEI Resources | NR52281 |
| Experimental models: Cell lines | ||
| Vero CCL-81 | ATCC | CCL-81; RRID: CVCL_0059 |
| Vero E6 | ATCC | CRL-1586; RRID:CVCL_0574 |
| Hamster: ExpiCHO-S | ThermoFisher Scientific | Cat#A29127; RRID: CVCL_5J31 |
| Vero Furin | Mukherjee et al., 2016 | N/A |
| Experimental models: Organisms/strains | ||
| Mouse: B6.Cg-Tg(K18-ACE2)2Prlmn/J | Jackson Laboratory | Cat# 034860; RRID:IMSR_JAX:03486 |
| Golden Syrian Hamster | Charles River Laboratory | Strain Code: 049 |
| Oligonucleotides | ||
| SARS-CoV-2 N F: 5′-ATGCTGCAATCGTGCTACAA-3′ | 32838945 | N/A |
| SARS-CoV-2 N R: 5′-GACTGCCGCCTCTGCTC-3′ | 32838945 | N/A |
| SARS-CoV-2 N Probe: 5′-/56-FAM/TCAAGGAAC/ZEN/AACATTGCCAA/3IABkFQ/-3′ | 32838945 | N/A |
| SARS-CoV-2 RNA ISH probe (S gene) | Advanced Cell Diagnostics | Cat# 4848561 |
| Hamster Rpl18 TaqMan F: GTTTATGAGTCGCACTAACCG | This paper | N/A |
| Hamster Rpl18 TaqMan R: TGTTCTCTCGGCCAGGAA | This paper | N/A |
| Hamster Rpl18 TaqMan Probe: YAK-TCTGTCCCTGTCCCGGATGATC-BBQ | This paper | N/A |
| Hamster Cxcl10 TaqMan F: GCCATTCATCCACAGTTGACA | This paper | N/A |
| Hamster Cxcl10 TaqMan R: CATGGTGCTGACAGTGGAGTCT | This paper | N/A |
| Hamster Cxcl10 TaqMan Probe: 6FAM-CGTCCCGAGCCAGCCAACGA-BBQ | This paper | N/A |
| Hamster Ccl2 TaqMan F: CTCACCTGCTGCTACTCATTC | This paper | N/A |
| Hamster Ccl2 TaqMan R: CTCTCTCTTGAGCTTGGTGATG | This paper | N/A |
| Hamster Ccl2 TaqMan Probe: 6FAM- CAGCAGCAAGTGTCCCAAAGAAGC-BBQ | This paper | N/A |
| Hamster Ccl3 TaqMan F: CTCACCTGCTGCTACTCATTC | This paper | N/A |
| Hamster Ccl3 TaqMan R: CTCTCTCTTGAGCTTGGTGATG | This paper | N/A |
| Hamster Ccl3 TaqMan Probe: 6FAM- CAGCAGCAAGTGTCCCAAAGAAGC-BBQ | This paper | N/A |
| Hamster Ccl5 TaqMan F: TGCTTTGACTACCTCTCCTTTAC | This paper | N/A |
| Hamster Ccl5 TaqMan R: GGTTCCTTCGGGTGACAAA | This paper | N/A |
| Hamster Ccl5 TaqMan Probe: 6FAM- TGCCTCGTGTTCACATCAAGGAGT-BBQ | This paper | N/A |
| Hamster Ifit3 TaqMan F: CTGATACCAACTGAGACTCCTG | This paper | N/A |
| Hamster Ifit3 TaqMan R: CTTCTGTCCTTCCTCGGATTAG | This paper | N/A |
| Hamster Ifit3 TaqMan Probe: 6FAM- ACCGTACAGTCCACACCCAACTTT-BBQ | This paper | N/A |
| Software and algorithms | ||
| FlowJo | FlowJo, LLC | v10 |
| GraphPad Prism | GraphPad | v 9.0.0 |
| Biorender | biorender.com | N/A |
| flexiWare | SCIREQ Inc. | v8.1.3 |
| STAR program | Dobin et al., 2013 | v 2.5.1a |
| EdgeR | Robinson et al., 2010 | N/A |
| limma | Ritchie et al., 2015 | N/A |
| RSeQC | Liao et al., 2014 | v2.6.2 |
| Nanozoomer Digital Pathology | Hamamatsu | v2 |
| Recombinant DNA | ||
| Plasmid: rCOV2-2050 in pTwist-mCis_hG1 | Zost et al., 2020b | N/A |
| Plasmid: rCOV2-2050 in pTwist-mCis_hG1 LALA-PG | This study | N/A |
| Plasmid: rCOV2-2381 in pTwist-mCis_hG1 | Zost et al., 2020b | N/A |
| Plasmid: rCOV2-2381 in pTwist-mCis_hG1 LALA-PG | This study | N/A |
| Plasmid: rCOV2-2072 in pTwist-mCis_hG1 | Zost et al., 2020b | N/A |
| Plasmid: rCOV2-2072 in pTwist-mCis_hG1 LALA-PG | This study | N/A |
| Plasmid: rCOV2-3025 in pTwist-mCis_hG1 | Zost et al., 2020b | N/A |
| Plasmid: rCOV2-3025 in pTwist-mCis_hG1 LALA-PG | This study | N/A |
| Plasmid: rCOV2-2072 in pTwist-mCis_mG1 | This study | N/A |
| Plasmid: rCOV2-2072 in pTwist-mCis_mG1 D265A | This study | N/A |
| Plasmid: rCOV2-2072 in pTwist-mCis_mG2a | This study | N/A |
| Plasmid: rCOV2-2072 in pTwist-mCis_mK | This study | N/A |
| Deposited data | ||
| Lung RNA-Seq Data | This paper | GEO: GSE161615 |
| Other | ||
| HiTrap MabSelect SuRe | Cytvia | 11003493 |
| CountBright Absolute Counting Beads | Thermo Fisher | C36950 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to the Lead Contact, Michael S. Diamond (diamond@wusm.wustl.edu).
Materials availability
All requests for resources and reagents should be directed to the Lead Contact author. This includes mice, antibodies, viruses, and proteins. All reagents will be made available on request after completion of a Materials Transfer Agreement.
Data and code availability
All data supporting the findings of this study are available within the paper and are available from the corresponding author upon request. RNA sequencing datasets have been uploaded and are available at GSE161615.
Experimental model and subject details
Cells and viruses
Vero E6 (CRL-1586, American Type Culture Collection (ATCC), Vero CCL81 (ATCC), and Vero-furin cells (Mukherjee et al., 2016) were cultured at 37°C in Dulbecco’s Modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 10 mM HEPES pH 7.3, 1 mM sodium pyruvate, 1 × non-essential amino acids, and 100 U/ml of penicillin–streptomycin. The 2019n-CoV/USA_WA1/2019 isolate of SARS-CoV-2 was obtained from the US Centers for Disease Control (CDC). Infectious stocks were propagated by inoculating Vero CCL81 cells and collecting supernatant upon observation of cytopathic effect; debris was removed by centrifugation and passage through a 0.22 μm filter. Supernatant was aliquoted and stored at −80°C. All work with infectious SARS-CoV-2 was performed in Institutional Biosafety Committee approved BSL3 and A-BSL3 facilities at Washington University School of Medicine using appropriate positive pressure air respirators and protective equipment.
Antibodies
The human antibodies studied in this paper were isolated from blood samples from two individuals in North America with previous laboratory-confirmed symptomatic SARS-CoV-2 infection that was acquired in China. The original clinical studies to obtain specimens after written informed consent were previously described (Zost et al., 2020b) and had been approved by the Institutional Review Board of Vanderbilt University Medical Center, the Institutional Review Board of the University of Washington and the Research Ethics Board of the University of Toronto.
Mouse experiments
Animal studies were carried out in accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The protocols were approved by the Institutional Animal Care and Use Committee at the Washington University School of Medicine (assurance number A3381–01). Virus inoculations were performed under anesthesia that was induced and maintained with ketamine hydrochloride and xylazine, and all efforts were made to minimize animal suffering.
Heterozygous K18-hACE c57BL/6J mice (strain: 2B6.Cg-Tg(K18-ACE2)2Prlmn/J) were obtained from The Jackson Laboratory. Animals were housed in groups and fed standard chow diets. Eight- to nine-week-old mice of both sexes were administered 103 PFU of SARS-CoV-2 by intranasal administration.
Hamster experiments
Seven-month-old female Syrian hamsters were purchased from Charles River Laboratories and housed in microisolator units. All hamsters were allowed free access to food and water and cared for under United States Department of Agriculture (USDA) guidelines for laboratory animals. Hamsters were administered with 5 × 105 PFU of SARS-CoV-2 (2019-nCoV/USA-WA1/2020) by the intranasal route in a final volume of 100 μL. One day later, hamsters were administered by intraperitoneal injection COV2-2050, COV2-2050 LALA-PG, or isotype control (10 mg/kg). All hamsters were monitored for body weight loss until humanely euthanized at 6 dpi. All procedures were approved by the University of Georgia Institutional Animal Care and Use Committee (IACUC number A2020 04-024-Y1-A4). Virus inoculations and antibody transfers were performed under anesthesia that was induced and maintained with 5% isoflurane. All efforts were made to minimize animal suffering.
Method details
mAb production and purification
COV2-2050, COV2-2381, COV2-2072, and COV2-3025 were isolated from B cells from a SARS-CoV-2 convalescent patient and described previously (Zost et al., 2020a, 2020b). mAb 2D22 was described previously (Fibriansah et al., 2015). Sequences of mAbs that had been synthesized (Twist Bioscience) and cloned into an IgG1 monocistronic expression vector (designated as pTwist-mCis_G1) or IgG1 monocistronic expression vector containing L234A, L235A, and P329G mutations in the Fc region of the heavy chain (designated as pTwist-mCis_LALA-PG) were used for mAb secretion in mammalian cell culture. This vector contains an enhanced 2A sequence and GSG linker that allows the simultaneous expression of mAb heavy and light chain genes from a single construct upon transfection (Chng et al., 2015). For the design of chimeric mAbs, the human kappa light chain constant region was replaced with mouse kappa light chain constant region, and the human IgG1 heavy chain constant region was exchanged for the murine IgG1, mouse IgG1 D265A, or mouse IgG2a constant region. Sequences of the light and heavy chains for these chimeric mAbs were synthesized (Twist Bioscience), cloned into separate expression vectors (designated as pTwist-mG1 and pTwist-mK), and were used for expression in mammalian cell culture. mAb proteins were produced after transient transfection using the GIBCO ExpiCHO Expression System (ThermoFisher Scientific) following the manufacturer’s protocol. Culture supernatants were purified using HiTrap MabSelect SuRe columns (Cytiva, formerly GE Healthcare Life Sciences) on an AKTA Pure chromatographer (GE Healthcare Life Sciences). Purified mAbs were buffer-exchanged into PBS, concentrated using Amicon Ultra-4 50-kDa centrifugal filter units (Millipore Sigma) and stored at −80 °C until use. Purified mAbs were tested routinely for endotoxin levels (found to be less than 30 EU per mg IgG). Endotoxin testing was performed using the PTS201F cartridge (Charles River), with a sensitivity range from 10 to 0.1 EU per ml, and an Endosafe Nexgen-MCS instrument (Charles River).
Recombinant FcγR protein ELISA
Recombinant murine FcγRI, FcγRIIb, and FcγRIV protein (R&D Systems) was immobilized on Maxisorp ELISA plates (Thermo Fisher) overnight in sodium bicarbonate buffer, pH 9.3. Plates were rinsed with PBS and 0.05% Tween-20 and blocked with 4% BSA for 1 h at 25°C. Intact or LALA-PG mAbs were diluted in 2% BSA and added to plates for 1 h at 25°C. After rinsing, primary mAbs were detected with horseradish peroxidase conjugated goat-anti human IgG F(ab′)2 (1:5,000 dilution, Jackson ImmunoResearch) for 1 h at 25°C. Plates were washed and developed with 3,3′-5,5′ tetramethylbenzidine substrate (Thermo Fisher), stopped with 2N H2SO4, and read at 450 nM using a TriStar Microplate Reader (Berthold).
Human mAb serum ELISA
Goat anti-human kappa (cross-absorbed against mouse IgG) (Southern Biotech) and goat anti-human lambda (cross-absorbed against mouse IgG) (Southern Biotech) were coated onto 96-well Nunc Maxisorp flat-bottomed plates at 2 μg/mL in coating buffer (0.1 M sodium carbonate, 0.1 M sodium bicarbonate, 0.02% sodium azide, pH 9.6) overnight at 4°C. Coating buffers were removed, and wells were blocked with 2% BSA (blocking buffer) (Thermo Fisher), for 1 h at 37°C. Heat-inactivated serum samples were diluted in blocking buffer in a separate polypropylene plate. The plates then were washed four times with PBS + 0.05% Tween-20 (PBST), followed by addition of 50 μL of respective serum dilutions, and then incubated for 1 h at 4°C. The plates were again washed four times in PBST, followed by addition of 50 μL of 1:2,000 Goat Anti-Human IgG Fc, Multi-Species (Southern Biotech). Plates were incubated at room temperature for 1 h. Plates were washed with four times in PBST, followed by 100 μL of TMB-ELISA substrate (Thermo Fisher) and incubated at room temperature for 3 to 5 min. Color development was monitored, and reactions were stopped with 50 μL of 2N H2SO4. Optical density (450 nm) measurements were determined using a microplate reader (Bio-Rad).
Plaque forming assay
Vero-furin cells (Mukherjee et al., 2016) were seeded at a density of 2.5 × 105 cells per well in flat-bottom 12-well tissue culture plates. The following day, medium was removed and replaced with 200 μL of 10-fold serial dilutions of the material to be titered, diluted in DMEM+2% FBS. One hour later, 1 mL of methylcellulose overlay was added. Plates were incubated for 72 h, then fixed with 4% paraformaldehyde (final concentration) in phosphate-buffered saline for 20 min. Plates were stained with 0.05% (w/v) crystal violet in 20% methanol and washed twice with distilled, deionized water.
Measurement of viral burden
Tissues were weighed and homogenized with zirconia beads in a MagNA Lyser instrument (Roche Life Science) in 1000 μL of DMEM media supplemented with 2% heat-inactivated FBS. Tissue homogenates were clarified by centrifugation at 10,000 rpm for 5 min and stored at −80°C. RNA was extracted using the MagMax mirVana Total RNA isolation kit (Thermo Scientific) on the Kingfisher Flex extraction robot (Thermo Scientific). RNA was reverse transcribed and amplified using the TaqMan RNA-to-CT 1-Step Kit (ThermoFisher). Reverse transcription was carried out at 48°C for 15 min followed by 2 min at 95°C. Amplification was accomplished over 50 cycles as follows: 95°C for 15 s and 60°C for 1 min. Copies of SARS-CoV-2 N gene RNA in samples were determined using a previously published assay (Case et al., 2020). Briefly, a TaqMan assay was designed to target a highly conserved region of the N gene (Forward primer: ATGCTGCAATCGTGCTACAA; Reverse primer: GACTGCCGCCTCTGCTC; Probe: /56-FAM/TCAAGGAAC/ZEN/AACATTGCCAA/3IABkFQ/). This region was included in an RNA standard to allow for copy number determination down to 10 copies per reaction. The reaction mixture contained final concentrations of primers and probe of 500 and 100 nM, respectively.
Cytokine and chemokine mRNA measurements
RNA was isolated from lung homogenates as described above. cDNA was synthesized from DNase-treated RNA using the High-Capacity cDNA Reverse Transcription kit (Thermo Scientific) with the addition of RNase inhibitor following the manufacturer’s protocol. Mouse cytokine and chemokine expression was determined using TaqMan Fast Universal PCR master mix (Thermo Scientific) with commercial primers/probe sets specific for IFN-g (IDT: Mm.PT.58.41769240), IL-6 (Mm.PT.58.10005566), CXCL10 (Mm.PT.58.43575827), CCL2 (Mm.PT.58.42151692), and results were normalized to GAPDH (Mm.PT.39a.1) levels. Fold change was determined using the 2-ΔΔCt method comparing treated mice to naive controls. Hamster cytokine and chemokine expression was determined using TaqMan Fast Universal PCR master mix (Thermo Scientific) with primers/probe sets specific for Rpl18 (For: GTTTATGAGTCGCACTAACCG, Rev: TGTTCTCTCGGCCAGGAA, Probe: YAK-TCTGTCCCTGTCCCGGATGATC-BBQ), Cxcl10 (For: GCCATTCATCCACAGTTGACA, Rev: CATGGTGCTGACAGTGGAGTCT, Probe: 6FAM-CGTCCCGAGCCAGCCAACGA-BBQ), Ccl2 (For: CTCACCTGCTGCTACTCATTC, Rev: CTCTCTCTTGAGCTTGGTGATG, Probe: 6FAM- CAGCAGCAAGTGTCCCAAAGAAGC-BBQ), Ccl3 (For: CTCACCTGCTGCTACTCATTC, Rev: CTCTCTCTTGAGCTTGGTGATG, Probe: 6FAM- CAGCAGCAAGTGTCCCAAAGAAGC-BBQ), Ccl5 (For: TGCTTTGACTACCTCTCCTTTAC, Rev: GGTTCCTTCGGGTGACAAA, Probe: 6FAM- TGCCTCGTGTTCACATCAAGGAGT-BBQ), Ifit3 (For: CTGATACCAACTGAGACTCCTG, Rev: CTTCTGTCCTTCCTCGGATTAG, Probe: 6FAM- ACCGTACAGTCCACACCCAACTTT-BBQ). Fold change was determined using the 2-ΔΔCt method comparing treated hamster to naive controls.
Cytokine and chemokine protein measurements
Lung homogenates were incubated with Triton X-100 (1% final concentration) for 1 h at room temperature to inactivate SARS-CoV-2. Homogenates then were analyzed for cytokines and chemokines by Eve Technologies Corporation (Calgary, AB, Canada) using their Mouse Cytokine Array/Chemokine Array 44-Plex (MD44) platform or Mouse Cytokine Array/Chemokine Array 31-Plex (MD31) platform.
Lung histology
Animals were euthanized before harvest and fixation of tissues. The left lung was first tied off at the left main bronchus and collected for viral RNA analysis. The right lung then was inflated with ∼1.2 mL of 10% neutral buffered formalin using a 3-mL syringe and catheter inserted into the trachea. Tissues were embedded in paraffin, and sections were stained with hematoxylin and eosin. RNA in situ hybridization was performed using the RNAscope 2.5 HD Assay (Brown Kit) according to the manufacturer’s instructions (Advanced Cell Diagnostics). Briefly, sections were deparaffinized and treated with H2O2 and Protease Plus before probe hybridization. Probes specifically targeting hACE2 (catalog number 848151) or SARS-CoV-2 spike sequence (catalog number 848561) were hybridized followed by proprietary signal amplification and detection with 3,3′-diaminobenzidine. Tissues were counterstained with Gill’s hematoxylin. An uninfected mouse was used as a negative control and stained in parallel. Images were captured using the Nanozoomer (Hamamatsu) at the Alafi Neuroimaging Core at Washington University.
Flow cytometry analysis of immune cell infiltrates
For analysis of BAL fluid, mice were sacrificed by ketamine overdose, followed by cannulation of the trachea with a 19-G canula. BAL was performed with three washes of 0.8 mL of sterile PBS. BAL fluid was centrifuged, and single cell suspensions were generated for staining. For analysis of lung tissues, mice were perfused with sterile PBS, and the right inferior lung lobes were digested at 37°C with 630 μg/mL collagenase D (Roche) and 75 U μL of DNase I (Sigma–Aldrich) for 2 h. Single cell suspensions of BAL fluid and lung digest were preincubated with Fc Block antibody (BD PharMingen) in PBS + 2% heat-inactivated FBS for 10 min at room temperature before staining. Cells were incubated with antibodies against the following markers: efluor506 Viability Dye, BUV395 anti- CD45, BV711 anti-CD11b, APC-Cy7 anti-CD11c, Percp-Cy5.5 anti-Ly6G, Pacific Blue anti-Ly6C, FITC anti-CD24, PE anti-Siglec F, PE-Cy7 anti-CD64 or BV421 anti-CD64, AF700 anti-MHCII, BV605 anti-CD80, PeCy5 anti-CD86, BV421 anti-CD3, BV785 anti-CD4, APC anti-CD8α, BV711 anti-B220, APC-Cy7 anti-CD44, BV605 anti-CD62L. All antibodies were used at a dilution of 1:200. Cells were stained for 20 min at 4°C, washed, fixed and permeabilized for intracellular staining with Foxp3/Transcription Factor Staining Buffer Set (eBioscience) according to manufacturer’s instructions. Myeloid cells were incubated overnight at 4°C with APC anti-NOS and PeCy7 anti-TNFα, washed, re-fixed with 4% PFA (EMS) for 20 min and resuspended in permeabilization buffer. Absolute cell counts were determined using Trucount beads (BD). Flow cytometric data were acquired on a cytometer (BD-X20; BD Biosciences) and analyzed using FlowJo software (Tree Star).
Antibody depletion of immune cell subsets
For neutrophil depletion, anti-Ly6G (BioXCell; clone 1A8; 250 μg) or an isotype control (BioXCell; clone 2A3; 250 μg) was administered to mice by intraperitoneal injection one day before infection and at D+1, D+3, and D+6 relative to SARS-CoV-2 inoculation. For monocyte depletion, anti-CCR2 (clone MC-21; 50 μg) (Mack et al., 2001) or an isotype control mAb (BioXCell; clone LTF-2; 50 μg) was administered to mice by intraperitoneal injection one day before infection and at D+1, D+3, D+5, and D+7 relative to SARS-CoV-2 inoculation. For NK cell depletion, anti-NK1.1 (clone PK136; 200 μg) or an isotype control mAb (BioXCell; clone C1.18.4; 200 μg) was administered to mice by intraperitoneal injection one day before infection and at D+1, D+3, D+5, and D+7 relative to SARS-CoV-2 inoculation. For CD8+ T cell depletions, anti-CD8a (clone 2.43; 500 μg) or an isotype control mAb (BioXCell; clone LTF-2; 500 μg) was administered to mice by intraperitoneal injection one day before infection and at D+1, D+3, D+5, and D+7 relative to SARS-CoV-2 inoculation.
For analysis of immune cell depletion, peripheral blood or the spleen was collected on the day of harvest. Erythrocytes were lysed with ACK lysis buffer (GIBCO) and resuspended in RPMI supplemented with 10% FBS. For anti-Ly6G or anti-CCR2 depletion, single-cell suspensions were preincubated with Fc Block antibody (BD PharMingen) in PBS + 2% heat-inactivated FBS for 10 min at room temperature and then stained with antibodies against CD45 BUV395, CD11b PE/Dazzle 594, Ly6C Pacific Blue, Ly6B FITC, Ly6G BV650, and fixable viability dye (eFluor 506). For NK cell depletions, single cell suspensions were blocked for FcγR binding and stained with antibodies against CD45 BUV395, CD19 BV711, CD3 BV711, NK1.1 FITC, and Nkp56 Pacific Blue. For CD8+ T cell depletions, single-cell suspensions were blocked for FcγR binding and stained with antibodies against CD45 BUV395, CD3 421, CD4 BV785, CD8b APC, CXCR1-PE, B220 BV711, MHCII-AF700, and CD11c PE-Cy7.
Respiratory mechanics
Mice were anesthetized with ketamine/xylazine (100 mg/kg and 10 mg/kg, i.p., respectively). The trachea was isolated by dissection of the neck area and cannulated using an 18-gauge blunt metal cannula (typical resistance of 0.18 cmH2O.s/mL), which was secured in place with a nylon suture. The mouse then was connected to the flexiVent computer-controlled piston ventilator (SCIREQ Inc.) via the cannula, which was attached to the FX adaptor Y-tubing. Mechanical ventilation was initiated, and mice were given an additional 100 mg/kg of ketamine and 0.1 mg/mouse of the paralytic pancuronium bromide by intraperitoneal route to prevent breathing efforts against the ventilator and during measurements. Mice were ventilated using default settings for mice, which consisted in a positive end expiratory pressure at 3 cm H2O, a 10 mL/kg tidal volume (Vt), a respiratory rate at 150 breaths per minute (bpm), and a fraction of inspired oxygen (FiO2) of 0.21 (i.e., room air). Respiratory mechanics were assessed using the forced oscillation technique, as previously described (McGovern et al., 2013), using the latest version of the flexiVent operating software (flexiWare v8.1.3). Pressure-volume loops and measurements of inspiratory capacity also were done.
Neutralization assay
Serial dilutions of mAbs were incubated with 102 focus-forming units (FFU) of SARS-CoV-2 for 1 h at 37°C. mAb-virus complexes were added to Vero E6 cell monolayers in 96-well plates and incubated at 37°C for 1 h. Subsequently, cells were overlaid with 1% (w/v) methylcellulose in MEM supplemented with 2% FBS. Plates were harvested 30 h later by removing overlays and fixed with 4% PFA in PBS for 20 min at room temperature. Plates were washed and sequentially incubated with 1 μg/mL of a recombinant IgG based on the sequence of CR3022 (Yuan et al., 2020) anti-S antibody and HRP-conjugated goat anti-human IgG in PBS supplemented with 0.1% saponin and 0.1% bovine serum albumin. SARS-CoV-2-infected cell foci were visualized using TrueBlue peroxidase substrate (KPL) and quantitated on an ImmunoSpot microanalyzer (Cellular Technologies).
RNA sequencing
cDNA libraries were constructed starting with 10 ng of total RNA from lung tissues of each sample that was extracted using the MagMax Mirvana kit per manufacturer’s instructions. cDNA was generated using the Seqplex kit (Sigma-Aldrich, St. Louis, MO) with amplification of 20 cycles. Library construction was performed using 100 ng of cDNA undergoing end repair, A-tailing, ligation of universal TruSeq adapters, and 8 cycles of amplification to incorporate unique dual index sequences. Libraries were sequenced on the NovaSeq 6000 (Illumina, San Diego, CA) targeting 40 million read pairs and extending 150 cycles with paired end reads. RNA-seq reads were aligned to the mouse Ensembl GRCh38.76 primary assembly and SARS-CoV-2 NCBI NC_045512 Wuhan-Hu-1 genome with STAR program (version 2.5.1a) (Dobin et al., 2013). Gene counts were derived from the number of uniquely aligned unambiguous reads by Subread:featureCount (version 1.4.6-p5) (Liao et al., 2014). The ribosomal fraction, known junction saturation, and read distribution over known gene models were quantified with RSeQC (version 2.6.2) (Liao et al., 2014). All gene counts were preprocessed with the R package EdgeR (Robinson et al., 2010) to adjust samples for differences in library size using the trimmed mean of M values (TMM) normalization procedure. Viral and ribosomal genes and genes not expressed in at least five samples (the smallest group size) at a level greater than or equal to 1 count per million reads were excluded, resulting 12,157 unique genes in further analysis. The R package limma (Ritchie et al., 2015) with voomWithQualityWeights function (Liu et al., 2015) was utilized to calculate the weighted likelihoods for all samples, based on the observed mean-variance relationship of every gene and sample. Differentially expressed genes were defined as those with at least 2-fold difference between two individual groups at the Benjamini-Hochberg false-discovery rate (FDR) (https://www.jstor.org/stable/2346101?seq=1) adjusted P value (i.e., q-value < 0.05).
Quantification and statistical analysis
Statistical significance was assigned when P values were < 0.05 using Prism version 8 (GraphPad). Tests, number of animals (n), median values, and statistical comparison groups are indicated in the Figure legends. Analysis of weight change was determined by two-way ANOVA. Changes in functional parameters or immune parameters were compared to isotype-treated animals and were analyzed by one-way ANOVA or one-way ANOVA with Dunnett’s test.
Acknowledgments
We thank the Pulmonary Morphology Core at Washington University School of Medicine for tissue sectioning. We also thank Joseph Reidy and Andrew Trivette at Vanderbilt Vaccine Center for human antibody sequence analysis and verification, and Rachel Nargi for assistance with mAb purification. This study was supported by the NIH (75N93019C00062, 75N93019C00074, R01 AI157155, and U01 AI15181), the Defense Advanced Research Projects Agency (HR001117S0019), Fast Grants (Mercatus Center, George Mason University), the Dolly Parton COVID-19 Research Fund, and the Future Insight Prize (Merck KGaA to J.E.C.). E.S.W. is supported by NIH (F30 AI152327). J.B.C. is supported by a Helen Hay Whitney Foundation postdoctoral fellowship. This work was also funded by the University of Georgia and Washington University. T.M.R. is supported by the Georgia Research Alliance as an Eminent Scholar.
Author contributions
P.G., S.J.Z., R.E.S., and R.H.C. designed and generated human antibodies. E.S.W., A.L.B., and R.E.C. performed mouse experiments. E.S.W. and Z.C. analyzed viral burden and performed immune cell processing. E.S.W. and A.L.B. performed pulmonary mechanics analysis. E.S.W. and J.B.C. performed neutralization analyses. J.Y. and R.H. performed RNA sequencing and analysis. H.J., Y.H., J.D.A., and T.M.R. performed hamster experiments. T.L.D. and A.C.M.B. validated reagents for hamster gene expression. M.M. provided anti-CCR2 antibody. J.E.C., T.M.R., A.C.M.B., and M.S.D. obtained funding and supervised research. E.S.W. and M.S.D. wrote the initial draft, with the other authors providing editorial comments.
Declaration of interests
M.S.D. is a consultant for Inbios, Vir Biotechnology, NGM Biopharmaceuticals, and Carnival Corporation, and on the Scientific Advisory Boards of Moderna and Immunome. The Diamond laboratory has received funding support in sponsored research agreements from Moderna, Vir Biotechnology, and Emergent BioSolutions. J.E.C. has served as a consultant for Eli Lilly and Luna Biologics, is a member of the Scientific Advisory Boards of CompuVax and Meissa Vaccines, and is Founder of IDBiologics. The Crowe laboratory has received sponsored research agreements from AstraZeneca and IDBiologics. Vanderbilt University has applied for patents related to antibodies described in this paper. The Boon laboratory has received funding support in sponsored research agreements from AI Therapeutics, GreenLight Biosciences, AbbVie, and Nano Targeting & Therapy Biopharma. R.H. may receive royalty income based on the CompBio technology developed by R.H. and licensed by Washington University to PercayAI.
Published: February 12, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.cell.2021.02.026.
Supplemental information
Lists of upregulated and downregulated genes comparing Isotype control mAb versus COV2-2050 D+1, COV2-2050 D+1 versus COV2-2050 LALA-PG D+1, Isotype control mAb versus COV2-2050 D+2, and COV2-2050 D+2 versus COV2-2050 LALA-PG D+2 in the top pathways identified through Gene Ontology analysis. Associated q-value and fold-change values are shown.
Lists of upregulated and downregulated genes comparing Isotype control mAb versus COV2-2050 D+1, COV2-2050 D+1 versus COV2-2050 LALA-PG D+1, Isotype control mAb versus COV2-2050 D+2, and COV2-2050 D+2 versus COV2-2050 LALA-PG D+2 in the top pathways identified through CompBio analysis. Associated q-value and fold-change values are shown.
References
- Abraham J. Passive antibody therapy in COVID-19. Nat. Rev. Immunol. 2020;20:401–403. doi: 10.1038/s41577-020-0365-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adamo L., Yu J., Rocha-Resende C., Javaheri A., Head R.D., Mann D.L. Proteomic Signatures of Heart Failure in Relation to Left Ventricular Ejection Fraction. J. Am. Coll. Cardiol. 2020;76:1982–1994. doi: 10.1016/j.jacc.2020.08.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alsoussi W.B., Turner J.S., Case J.B., Zhao H., Schmitz A.J., Zhou J.Q., Chen R.E., Lei T., Rizk A.A., McIntire K.M., et al. A Potently Neutralizing Antibody Protects Mice against SARS-CoV-2 Infection. J. Immunol. 2020;205:915–922. doi: 10.4049/jimmunol.2000583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amigorena S., Salamero J., Davoust J., Fridman W.H., Bonnerot C. Tyrosine-containing motif that transduces cell activation signals also determines internalization and antigen presentation via type III receptors for IgG. Nature. 1992;358:337–341. doi: 10.1038/358337a0. [DOI] [PubMed] [Google Scholar]
- Baudino L., Shinohara Y., Nimmerjahn F., Furukawa J., Nakata M., Martínez-Soria E., Petry F., Ravetch J.V., Nishimura S., Izui S. Crucial role of aspartic acid at position 265 in the CH2 domain for murine IgG2a and IgG2b Fc-associated effector functions. J. Immunol. 2008;181:6664–6669. doi: 10.4049/jimmunol.181.9.6664. [DOI] [PubMed] [Google Scholar]
- Baum A., Ajithdoss D., Copin R., Zhou A., Lanza K., Negron N., Ni M., Wei Y., Mohammadi K., Musser B., et al. REGN-COV2 antibodies prevent and treat SARS-CoV-2 infection in rhesus macaques and hamsters. Science. 2020;370:1110–1115. doi: 10.1126/science.abe2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergtold A., Desai D.D., Gavhane A., Clynes R. Cell surface recycling of internalized antigen permits dendritic cell priming of B cells. Immunity. 2005;23:503–514. doi: 10.1016/j.immuni.2005.09.013. [DOI] [PubMed] [Google Scholar]
- Bolles M., Deming D., Long K., Agnihothram S., Whitmore A., Ferris M., Funkhouser W., Gralinski L., Totura A., Heise M., Baric R.S. A double-inactivated severe acute respiratory syndrome coronavirus vaccine provides incomplete protection in mice and induces increased eosinophilic proinflammatory pulmonary response upon challenge. J. Virol. 2011;85:12201–12215. doi: 10.1128/JVI.06048-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bournazos S., DiLillo D.J., Ravetch J.V. humanized mice to study FcγR function. Curr. Top. Microbiol. Immunol. 2014;382:237–248. doi: 10.1007/978-3-319-07911-0_11. [DOI] [PubMed] [Google Scholar]
- Bournazos S., Klein F., Pietzsch J., Seaman M.S., Nussenzweig M.C., Ravetch J.V. Broadly neutralizing anti-HIV-1 antibodies require Fc effector functions for in vivo activity. Cell. 2014;158:1243–1253. doi: 10.1016/j.cell.2014.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bournazos S., DiLillo D.J., Goff A.J., Glass P.J., Ravetch J.V. Differential requirements for FcγR engagement by protective antibodies against Ebola virus. Proc. Natl. Acad. Sci. USA. 2019;116:20054–20062. doi: 10.1073/pnas.1911842116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bournazos S., Corti D., Virgin H.W., Ravetch J.V. Fc-optimized antibodies elicit CD8 immunity to viral respiratory infection. Nature. 2020;588:485–490. doi: 10.1038/s41586-020-2838-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y., Su B., Guo X., Sun W., Deng Y., Bao L., Zhu Q., Zhang X., Zheng Y., Geng C., et al. Potent neutralizing antibodies against SARS-CoV-2 identified by high-throughput single-cell sequencing of convalescent patients’ B cells. Cell. 2020;182:73–84.e16. doi: 10.1016/j.cell.2020.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Case J.B., Bailey A.L., Kim A.S., Chen R.E., Diamond M.S. Growth, detection, quantification, and inactivation of SARS-CoV-2. Virology. 2020;548:39–48. doi: 10.1016/j.virol.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P., Nirula A., Heller B., Gottlieb R.L., Boscia J., Morris J., Huhn G., Cardona J., Mocherla B., Stosor V., et al. SARS-CoV-2 Neutralizing Antibody LY-CoV555 in Outpatients with Covid-19. N. Engl. J. Med. 2021;384:229–237. doi: 10.1056/NEJMoa2029849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chng J., Wang T., Nian R., Lau A., Hoi K.M., Ho S.C., Gagnon P., Bi X., Yang Y. Cleavage efficient 2A peptides for high level monoclonal antibody expression in CHO cells. MAbs. 2015;7:403–412. doi: 10.1080/19420862.2015.1008351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clynes R., Maizes J.S., Guinamard R., Ono M., Takai T., Ravetch J.V. Modulation of immune complex-induced inflammation in vivo by the coordinate expression of activation and inhibitory Fc receptors. J. Exp. Med. 1999;189:179–185. doi: 10.1084/jem.189.1.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daëron M., Lesourne R. Negative signaling in Fc receptor complexes. Adv. Immunol. 2006;89:39–86. doi: 10.1016/S0065-2776(05)89002-9. [DOI] [PubMed] [Google Scholar]
- Dhodapkar K.M., Banerjee D., Connolly J., Kukreja A., Matayeva E., Veri M.C., Ravetch J.V., Steinman R.M., Dhodapkar M.V. Selective blockade of the inhibitory Fcgamma receptor (FcgammaRIIB) in human dendritic cells and monocytes induces a type I interferon response program. J. Exp. Med. 2007;204:1359–1369. doi: 10.1084/jem.20062545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamond M.S., Pierson T.C. The Challenges of Vaccine Development against a New Virus during a Pandemic. Cell Host Microbe. 2020;27:699–703. doi: 10.1016/j.chom.2020.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiLillo D.J., Tan G.S., Palese P., Ravetch J.V. Broadly neutralizing hemagglutinin stalk-specific antibodies require FcγR interactions for protection against influenza virus in vivo. Nat. Med. 2014;20:143–151. doi: 10.1038/nm.3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiLillo D.J., Palese P., Wilson P.C., Ravetch J.V. Broadly neutralizing anti-influenza antibodies require Fc receptor engagement for in vivo protection. J. Clin. Invest. 2016;126:605–610. doi: 10.1172/JCI84428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong E., Du H., Gardner L. An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect. Dis. 2020;20:533–534. doi: 10.1016/S1473-3099(20)30120-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagre A.C., Manhard J., Adams R., Eckley M., Zhan S., Lewis J., Rocha S.M., Woods C., Kuo K., Liao W., et al. A potent SARS-CoV-2 neutralizing human monoclonal antibody that reduces viral burden and disease severity in Syrian hamsters. bioRxiv. 2020 doi: 10.1101/2020.09.25.313601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fibriansah G., Ibarra K.D., Ng T.S., Smith S.A., Tan J.L., Lim X.N., Ooi J.S., Kostyuchenko V.A., Wang J., de Silva A.M., et al. DENGUE VIRUS. Cryo-EM structure of an antibody that neutralizes dengue virus type 2 by locking E protein dimers. Science. 2015;349:88–91. doi: 10.1126/science.aaa8651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox J.M., Roy V., Gunn B.M., Huang L., Edeling M.A., Mack M., Fremont D.H., Doranz B.J., Johnson S., Alter G., Diamond M.S. Optimal therapeutic activity of monoclonal antibodies against chikungunya virus requires Fc-FcγR interaction on monocytes. Sci. Immunol. 2019;4:eaav5062. doi: 10.1126/sciimmunol.aav5062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehrig J.L., Venkatesh S., Chang H.W., Hibberd M.C., Kung V.L., Cheng J., Chen R.Y., Subramanian S., Cowardin C.A., Meier M.F., et al. Effects of microbiota-directed foods in gnotobiotic animals and undernourished children. Science. 2019;365:eaau4732. doi: 10.1126/science.aau4732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giamarellos-Bourboulis E.J., Netea M.G., Rovina N., Akinosoglou K., Antoniadou A., Antonakos N., Damoraki G., Gkavogianni T., Adami M.E., Katsaounou P., et al. Complex Immune Dysregulation in COVID-19 Patients with Severe Respiratory Failure. Cell Host Microbe. 2020;27:992–1000.e3. doi: 10.1016/j.chom.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golden J.W., Cline C.R., Zeng X., Garrison A.R., Carey B.D., Mucker E.M., White L.E., Shamblin J.D., Brocato R.L., Liu J., et al. Human angiotensin-converting enzyme 2 transgenic mice infected with SARS-CoV-2 develop severe and fatal respiratory disease. JCI Insight. 2020;5:e142032. doi: 10.1172/jci.insight.142032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halstead S.B. Cellular Receptors for Animal Viruses. Cold Spring Harbor Laboratory Press; 1994. Antibody-dependent enhancement of infection: a mechanism for indirect virus entry into cells; pp. 493–516. [Google Scholar]
- Halstead S.B., Katzelnick L. COVID 19 Vaccines: Should we fear ADE? J. Infect. Dis. 2020;222:1946–1950. doi: 10.1093/infdis/jiaa518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen J., Baum A., Pascal K.E., Russo V., Giordano S., Wloga E., Fulton B.O., Yan Y., Koon K., Patel K., et al. Studies in humanized mice and convalescent humans yield a SARS-CoV-2 antibody cocktail. Science. 2020;369:1010–1014. doi: 10.1126/science.abd0827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan A.O., Case J.B., Winkler E.S., Thackray L.B., Kafai N.M., Bailey A.L., McCune B.T., Fox J.M., Chen R.E., Alsoussi W.B., et al. A SARS-CoV-2 Infection Model in Mice Demonstrates Protection by Neutralizing Antibodies. Cell. 2020;182:744–753.e4. doi: 10.1016/j.cell.2020.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He W., Chen C.J., Mullarkey C.E., Hamilton J.R., Wong C.K., Leon P.E., Uccellini M.B., Chromikova V., Henry C., Hoffman K.W., et al. Alveolar macrophages are critical for broadly-reactive antibody-mediated protection against influenza A virus in mice. Nat. Commun. 2017;8:846. doi: 10.1038/s41467-017-00928-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hessell A.J., Hangartner L., Hunter M., Havenith C.E., Beurskens F.J., Bakker J.M., Lanigan C.M., Landucci G., Forthal D.N., Parren P.W., et al. Fc receptor but not complement binding is important in antibody protection against HIV. Nature. 2007;449:101–104. doi: 10.1038/nature06106. [DOI] [PubMed] [Google Scholar]
- Hoffmann E., Kotsias F., Visentin G., Bruhns P., Savina A., Amigorena S. Autonomous phagosomal degradation and antigen presentation in dendritic cells. Proc. Natl. Acad. Sci. USA. 2012;109:14556–14561. doi: 10.1073/pnas.1203912109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakubzick C.V., Randolph G.J., Henson P.M. Monocyte differentiation and antigen-presenting functions. Nat. Rev. Immunol. 2017;17:349–362. doi: 10.1038/nri.2017.28. [DOI] [PubMed] [Google Scholar]
- Ju B., Zhang Q., Ge J., Wang R., Sun J., Ge X., Yu J., Shan S., Zhou B., Song S., et al. Human neutralizing antibodies elicited by SARS-CoV-2 infection. Nature. 2020;584:115–119. doi: 10.1038/s41586-020-2380-z. [DOI] [PubMed] [Google Scholar]
- Karki R., Sharma B.R., Tuladhar S., Williams E.P., Zalduondo L., Samir P., Zheng M., Sundaram B., Banoth B., Malireddi R.K.S., et al. Synergism of TNF-α and IFN-γ Triggers Inflammatory Cell Death, Tissue Damage, and Mortality in SARS-CoV-2 Infection and Cytokine Shock Syndromes. Cell. 2021;184:149–168.e17. doi: 10.1016/j.cell.2020.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreye J., Reincke S.M., Kornau H.C., Sánchez-Sendin E., Corman V.M., Liu H., Yuan M., Wu N.C., Zhu X., Lee C.D., et al. A Therapeutic Non-self-reactive SARS-CoV-2 Antibody Protects from Lung Pathology in a COVID-19 Hamster Model. Cell. 2020;183:1058–1069.e19. doi: 10.1016/j.cell.2020.09.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laczkó D., Hogan M.J., Toulmin S.A., Hicks P., Lederer K., Gaudette B.T., Castaño D., Amanat F., Muramatsu H., Oguin T.H., 3rd, et al. A Single Immunization with Nucleoside-Modified mRNA Vaccines Elicits Strong Cellular and Humoral Immune Responses against SARS-CoV-2 in Mice. Immunity. 2020;53:724–732.e7. doi: 10.1016/j.immuni.2020.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laidlaw B.J., Decman V., Ali M.A., Abt M.C., Wolf A.I., Monticelli L.A., Mozdzanowska K., Angelosanto J.M., Artis D., Erikson J., Wherry E.J. Cooperativity between CD8+ T cells, non-neutralizing antibodies, and alveolar macrophages is important for heterosubtypic influenza virus immunity. PLoS Pathog. 2013;9 doi: 10.1371/journal.ppat.1003207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazar G.A., Dang W., Karki S., Vafa O., Peng J.S., Hyun L., Chan C., Chung H.S., Eivazi A., Yoder S.C., et al. Engineered antibody Fc variants with enhanced effector function. Proc. Natl. Acad. Sci. USA. 2006;103:4005–4010. doi: 10.1073/pnas.0508123103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W.S., Wheatley A.K., Kent S.J., DeKosky B.J. Antibody-dependent enhancement and SARS-CoV-2 vaccines and therapies. Nat. Microbiol. 2020;5:1185–1191. doi: 10.1038/s41564-020-00789-5. [DOI] [PubMed] [Google Scholar]
- Leist S.R., Dinnon K.H., 3rd, Schäfer A., Tse L.V., Okuda K., Hou Y.J., West A., Edwards C.E., Sanders W., Fritch E.J., et al. A Mouse-Adapted SARS-CoV-2 Induces Acute Lung Injury and Mortality in Standard Laboratory Mice. Cell. 2020;183:1070–1085.e12. doi: 10.1016/j.cell.2020.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D., He W., Liu X., Zheng S., Qi Y., Li H., Mao F., Liu J., Sun Y., Pan L., et al. A potent human neutralizing antibody Fc-dependently reduces established HBV infections. eLife. 2017;6:e26738. doi: 10.7554/eLife.26738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y., Smyth G.K., Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- Liu R., Holik A.Z., Su S., Jansz N., Chen K., Leong H.S., Blewitt M.E., Asselin-Labat M.L., Smyth G.K., Ritchie M.E. Why weight? Modelling sample and observational level variability improves power in RNA-seq analyses. Nucleic Acids Res. 2015;43:e97. doi: 10.1093/nar/gkv412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q., Fan C., Li Q., Zhou S., Huang W., Wang L., Sun C., Wang M., Wu X., Ma J., et al. Antibody-dependent-cellular-cytotoxicity-inducing antibodies significantly affect the post-exposure treatment of Ebola virus infection. Sci. Rep. 2017;7:45552. doi: 10.1038/srep45552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Wang P., Nair M.S., Yu J., Rapp M., Wang Q., Luo Y., Chan J.F., Sahi V., Figueroa A., et al. Potent neutralizing antibodies against multiple epitopes on SARS-CoV-2 spike. Nature. 2020;584:450–456. doi: 10.1038/s41586-020-2571-7. [DOI] [PubMed] [Google Scholar]
- Lo M., Kim H.S., Tong R.K., Bainbridge T.W., Vernes J.M., Zhang Y., Lin Y.L., Chung S., Dennis M.S., Zuchero Y.J., et al. Effector-attenuating Substitutions That Maintain Antibody Stability and Reduce Toxicity in Mice. J. Biol. Chem. 2017;292:3900–3908. doi: 10.1074/jbc.M116.767749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C.L., Murakowski D.K., Bournazos S., Schoofs T., Sarkar D., Halper-Stromberg A., Horwitz J.A., Nogueira L., Golijanin J., Gazumyan A., Ravetch J.V., Caskey M., Chakraborty A.K., Nussenzweig M.C. Enhanced clearance of HIV-1-infected cells by broadly neutralizing antibodies against HIV-1 in vivo. Science. 2016;352:1001–1004. doi: 10.1126/science.aaf1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu L.L., Suscovich T.J., Fortune S.M., Alter G. Beyond binding: antibody effector functions in infectious diseases. Nat. Rev. Immunol. 2018;18:46–61. doi: 10.1038/nri.2017.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo F., Liao F.L., Wang H., Tang H.B., Yang Z.Q., Hou W. Evaluation of Antibody-Dependent Enhancement of SARS-CoV Infection in Rhesus Macaques Immunized with an Inactivated SARS-CoV Vaccine. Virol. Sin. 2018;33:201–204. doi: 10.1007/s12250-018-0009-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mack M., Cihak J., Simonis C., Luckow B., Proudfoot A.E., Plachý J., Brühl H., Frink M., Anders H.J., Vielhauer V., et al. Expression and characterization of the chemokine receptors CCR2 and CCR5 in mice. J. Immunol. 2001;166:4697–4704. doi: 10.4049/jimmunol.166.7.4697. [DOI] [PubMed] [Google Scholar]
- Marovich M., Mascola J.R., Cohen M.S. Monoclonal Antibodies for Prevention and Treatment of COVID-19. JAMA. 2020;324:131–132. doi: 10.1001/jama.2020.10245. [DOI] [PubMed] [Google Scholar]
- McCray P.B., Jr., Pewe L., Wohlford-Lenane C., Hickey M., Manzel L., Shi L., Netland J., Jia H.P., Halabi C., Sigmund C.D., et al. Lethal infection of K18-hACE2 mice infected with severe acute respiratory syndrome coronavirus. J. Virol. 2007;81:813–821. doi: 10.1128/JVI.02012-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGovern T.K., Robichaud A., Fereydoonzad L., Schuessler T.F., Martin J.G. Evaluation of respiratory system mechanics in mice using the forced oscillation technique. J. Vis. Exp. 2013;(75):e50172. doi: 10.3791/50172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merad M., Martin J.C. Pathological inflammation in patients with COVID-19: a key role for monocytes and macrophages. Nat. Rev. Immunol. 2020;20:355–362. doi: 10.1038/s41577-020-0331-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee S., Sirohi D., Dowd K.A., Chen Z., Diamond M.S., Kuhn R.J., Pierson T.C. Enhancing dengue virus maturation using a stable furin over-expressing cell line. Virology. 2016;497:33–40. doi: 10.1016/j.virol.2016.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niessl J., Baxter A.E., Mendoza P., Jankovic M., Cohen Y.Z., Butler A.L., Lu C.-L., Dubé M., Shimeliovich I., Gruell H., et al. Combination anti-HIV-1 antibody therapy is associated with increased virus-specific T cell immunity. Nat. Med. 2020;26:222–227. doi: 10.1038/s41591-019-0747-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noy-Porat T., Makdasi E., Alcalay R., Mechaly A., Levy Y., Bercovich-Kinori A., Zauberman A., Tamir H., Yahalom-Ronen Y., Israeli M., et al. A panel of human neutralizing mAbs targeting SARS-CoV-2 spike at multiple epitopes. Nat. Commun. 2020;11:4303. doi: 10.1038/s41467-020-18159-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierson T.C., Diamond M.S. A game of numbers: the stoichiometry of antibody-mediated neutralization of flavivirus infection. Prog. Mol. Biol. Transl. Sci. 2015;129:141–166. doi: 10.1016/bs.pmbts.2014.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto D., Park Y.J., Beltramello M., Walls A.C., Tortorici M.A., Bianchi S., Jaconi S., Culap K., Zatta F., De Marco A., et al. Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature. 2020;583:290–295. doi: 10.1038/s41586-020-2349-y. [DOI] [PubMed] [Google Scholar]
- Qin E., Shi H., Tang L., Wang C., Chang G., Ding Z., Zhao K., Wang J., Chen Z., Yu M., et al. Immunogenicity and protective efficacy in monkeys of purified inactivated Vero-cell SARS vaccine. Vaccine. 2006;24:1028–1034. doi: 10.1016/j.vaccine.2005.06.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., Smyth G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbiani D.F., Gaebler C., Muecksch F., Lorenzi J.C.C., Wang Z., Cho A., Agudelo M., Barnes C.O., Gazumyan A., Finkin S., et al. Convergent antibody responses to SARS-CoV-2 in convalescent individuals. Nature. 2020;584:437–442. doi: 10.1038/s41586-020-2456-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers T.F., Zhao F., Huang D., Beutler N., Burns A., He W.T., Limbo O., Smith C., Song G., Woehl J., et al. Isolation of potent SARS-CoV-2 neutralizing antibodies and protection from disease in a small animal model. Science. 2020;369:956–963. doi: 10.1126/science.abc7520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenke K., Meade-White K., Letko M.C., Clancy C., Hansens F., Liu Y., Okumura A., Tang-Huau T.L., Li R., Saturday G., et al. Defining the Syrian hamster as a highly susceptible preclinical model for SARS-CoV-2 infection. Emerg. Microbes Infect. 2020;9:2673–2684. doi: 10.1080/22221751.2020.1858177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruckwardt T.J., Morabito K.M., Graham B.S. Immunological Lessons from Respiratory Syncytial Virus Vaccine Development. Immunity. 2019;51:429–442. doi: 10.1016/j.immuni.2019.08.007. [DOI] [PubMed] [Google Scholar]
- Sánchez-Cerrillo I., Landete P., Aldave B., Sánchez-Alonso S., Sánchez-Azofra A., Marcos-Jiménez A., Ávalos E., Alcaraz-Serna A., de Los Santos I., Mateu-Albero T., et al. REINMUN-COVID and EDEPIMIC groups COVID-19 severity associates with pulmonary redistribution of CD1c+ DCs and inflammatory transitional and nonclassical monocytes. J. Clin. Invest. 2020;130:6290–6300. doi: 10.1172/JCI140335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders K.O. Conceptual Approaches to Modulating Antibody Effector Functions and Circulation Half-Life. Front Immunol. 2019 doi: 10.3389/fimmu.2019.01296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schäfer A., Muecksch F., Lorenzi J.C.C., Leist S.R., Cipolla M., Bournazos S., Schmidt F., Maison R.M., Gazumyan A., Martinez D.R., et al. Antibody potency, effector function, and combinations in protection and therapy for SARS-CoV-2 infection in vivo. J. Exp. Med. 2021;218:e20201993. doi: 10.1084/jem.20201993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulte-Schrepping J., Reusch N., Paclik D., Baßler K., Schlickeiser S., Zhang B., Krämer B., Krammer T., Brumhard S., Bonaguro L., et al. Deutsche COVID-19 OMICS Initiative (DeCOI) Severe COVID-19 Is Marked by a Dysregulated Myeloid Cell Compartment. Cell. 2020;182:1419–1440.e23. doi: 10.1016/j.cell.2020.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serbina N.V., Salazar-Mather T.P., Biron C.A., Kuziel W.A., Pamer E.G. TNF/iNOS-producing dendritic cells mediate innate immune defense against bacterial infection. Immunity. 2003;19:59–70. doi: 10.1016/s1074-7613(03)00171-7. [DOI] [PubMed] [Google Scholar]
- Shi R., Shan C., Duan X., Chen Z., Liu P., Song J., Song T., Bi X., Han C., Wu L., et al. A human neutralizing antibody targets the receptor-binding site of SARS-CoV-2. Nature. 2020;584:120–124. doi: 10.1038/s41586-020-2381-y. [DOI] [PubMed] [Google Scholar]
- Sia S.F., Yan L.M., Chin A.W.H., Fung K., Choy K.T., Wong A.Y.L., Kaewpreedee P., Perera R.A.P.M., Poon L.L.M., Nicholls J.M., et al. Pathogenesis and transmission of SARS-CoV-2 in golden hamsters. Nature. 2020;583:834–838. doi: 10.1038/s41586-020-2342-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith P., DiLillo D.J., Bournazos S., Li F., Ravetch J.V. Mouse model recapitulating human Fcγ receptor structural and functional diversity. Proc. Natl. Acad. Sci. USA. 2012;109:6181–6186. doi: 10.1073/pnas.1203954109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subbarao K., McAuliffe J., Vogel L., Fahle G., Fischer S., Tatti K., Packard M., Shieh W.J., Zaki S., Murphy B. Prior infection and passive transfer of neutralizing antibody prevent replication of severe acute respiratory syndrome coronavirus in the respiratory tract of mice. J. Virol. 2004;78:3572–3577. doi: 10.1128/JVI.78.7.3572-3577.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tay M.Z., Poh C.M., Rénia L., MacAry P.A., Ng L.F.P. The trinity of COVID-19: immunity, inflammation and intervention. Nat. Rev. Immunol. 2020;20:363–374. doi: 10.1038/s41577-020-0311-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor A., Foo S.S., Bruzzone R., Dinh L.V., King N.J., Mahalingam S. Fc receptors in antibody-dependent enhancement of viral infections. Immunol. Rev. 2015;268:340–364. doi: 10.1111/imr.12367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tostanoski L.H., Wegmann F., Martinot A.J., Loos C., McMahan K., Mercado N.B., Yu J., Chan C.N., Bondoc S., Starke C.E., et al. Ad26 vaccine protects against SARS-CoV-2 severe clinical disease in hamsters. Nat. Med. 2020;26:1694–1700. doi: 10.1038/s41591-020-1070-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verboon J.M., Parkhurst S.M. Rho family GTPases bring a familiar ring to cell wound repair. Small GTPases. 2015;6:1–7. doi: 10.4161/21541248.2014.992262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt M.R., Dowd K.A., Engle M., Tesh R.B., Johnson S., Pierson T.C., Diamond M.S. Poorly neutralizing cross-reactive antibodies against the fusion loop of West Nile virus envelope protein protect in vivo via Fcgamma receptor and complement-dependent effector mechanisms. J. Virol. 2011;85:11567–11580. doi: 10.1128/JVI.05859-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells M.A., Ennis F.A., Albrecht P. Recovery from a viral respiratory infection. II. Passive transfer of immune spleen cells to mice with influenza pneumonia. J. Immunol. 1981;126:1042–1046. [PubMed] [Google Scholar]
- Winkler E.S., Bailey A.L., Kafai N.M., Nair S., McCune B.T., Yu J., Fox J.M., Chen R.E., Earnest J.T., Keeler S.P., et al. SARS-CoV-2 infection of human ACE2-transgenic mice causes severe lung inflammation and impaired function. Nat. Immunol. 2020;21:1327–1335. doi: 10.1038/s41590-020-0778-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan M., Wu N.C., Zhu X., Lee C.D., So R.T.Y., Lv H., Mok C.K.P., Wilson I.A. A highly conserved cryptic epitope in the receptor binding domains of SARS-CoV-2 and SARS-CoV. Science. 2020;368:630–633. doi: 10.1126/science.abb7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zohar T., Loos C., Fischinger S., Atyeo C., Wang C., Slein M.D., Burke J., Yu J., Feldman J., Hauser B.M., et al. Compromised Humoral Functional Evolution Tracks with SARS-CoV-2 Mortality. Cell. 2020;183:1508–1519. doi: 10.1016/j.cell.2020.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zost S.J., Gilchuk P., Case J.B., Binshtein E., Chen R.E., Nkolola J.P., Schäfer A., Reidy J.X., Trivette A., Nargi R.S., et al. Potently neutralizing and protective human antibodies against SARS-CoV-2. Nature. 2020;584:443–449. doi: 10.1038/s41586-020-2548-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zost S.J., Gilchuk P., Chen R.E., Case J.B., Reidy J.X., Trivette A., Nargi R.S., Sutton R.E., Suryadevara N., Chen E.C., et al. Rapid isolation and profiling of a diverse panel of human monoclonal antibodies targeting the SARS-CoV-2 spike protein. Nat. Med. 2020;26:1422–1427. doi: 10.1038/s41591-020-0998-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Lists of upregulated and downregulated genes comparing Isotype control mAb versus COV2-2050 D+1, COV2-2050 D+1 versus COV2-2050 LALA-PG D+1, Isotype control mAb versus COV2-2050 D+2, and COV2-2050 D+2 versus COV2-2050 LALA-PG D+2 in the top pathways identified through Gene Ontology analysis. Associated q-value and fold-change values are shown.
Lists of upregulated and downregulated genes comparing Isotype control mAb versus COV2-2050 D+1, COV2-2050 D+1 versus COV2-2050 LALA-PG D+1, Isotype control mAb versus COV2-2050 D+2, and COV2-2050 D+2 versus COV2-2050 LALA-PG D+2 in the top pathways identified through CompBio analysis. Associated q-value and fold-change values are shown.
Data Availability Statement
All data supporting the findings of this study are available within the paper and are available from the corresponding author upon request. RNA sequencing datasets have been uploaded and are available at GSE161615.