Abstract
Coronavirus disease 2019 (COVID-19) is an infectious disease caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). The ability of our cells to secrete type I interferons (IFN-Is) is essential for the control of virus replication and for effective antiviral immune responses; for this reason, viruses have evolved the means to antagonize IFN-I. Inhibition of IFN-I production is pronounced in SARS-CoV-2 infection, which can impair the adaptive immune response and exacerbate inflammatory disease at late stages of infection. However, therapeutic boosting of IFN-I offers a narrow time window for efficacy and safety. Here, we discuss how limits placed on IFN-I by SARS-CoV-2 shape the immune response and whether this might be countered with therapeutic approaches and vaccine design.
IFN-I and Immunity to SARS-CoV-2 Infection
COVID-19 presents a spectrum of clinical manifestations in humans, ranging from asymptomatic infection to severe pneumonia accompanied by multisystemic failure, especially in aged people [1]. The cause of severe COVID-19 is still controversial, but increasing evidence suggests that defects in responsiveness to IFN-I is of prime importance [2]. In this article, we review current literature on IFN-I in COVID-19 patients and present our own perspective on how defective production of IFN-I can impair the adaptive immune response, yet can also exacerbate inflammatory disease at late stages of infection, and how our increasing knowledge of SARS-CoV-2 presents opportunities for therapeutic intervention and vaccine design.
The respiratory virus SARS-CoV-2 initially infects cells lining the upper respiratory tract. To establish infection, SARS-CoV-2 first binds to angiotensin-converting enzyme (ACE)2 and transmembrane serine protease (TMPRSS)2 on the respiratory epithelium [3]. SARS-CoV-2 is recognized in the cytosol of human epithelial cells by single-stranded (ss)RNA sensing Toll-like receptors (see Glossary), including TLR3 and TLR7 in endosomes, and cytosolic RIG-like receptors (RLRs), which engage with the mitochondrial antivirus signaling (MAVS) protein [4]. These events lead to the activation of interferon regulatory factor (IRF)3, IRF7, and nuclear factor (NF)-κB, inducing rapid production of IFN-I, IFN-III, and proinflammatory cytokines [5].
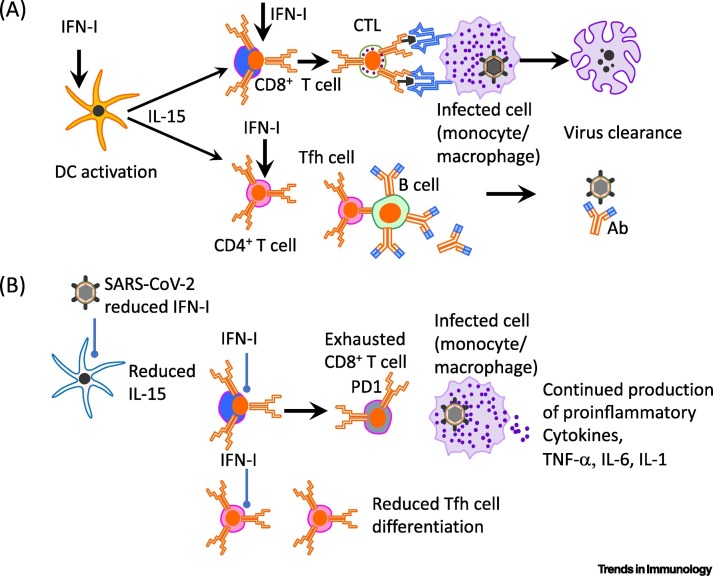
IFN-I α, β and other members of the extended IFN-I family are produced rapidly following virus infection and exhibit key antiviral activity within infected cells, thereby limiting virus proliferation and spread [2,6]. In conjunction with the products of IFN-I-stimulated genes (ISGs), IFN-I potentiates both the innate and adaptive immune responses to clear viral infections [7]. These properties of IFN-I are shared by IFN-III (IFN-λ), although expression of the receptors for these cytokines is different. Thus, whereas receptors for IFN-I (IFNAR) are widely expressed, the receptor for IFN-λ (IFNLR1) is restricted to a few cell types and tissues in mice and humans, namely macrophages, conventional dendritic cells (DCs) and plasmacytoid dendritic cells (pDCs), neutrophils, and respiratory epithelial cells [8., 9., 10.]. Generally speaking, IFN-III can suppress both T helper cell ( Th)2 and Th17 responses and has important antiviral functions in respiratory tissues [11,12]. For IFN-I, these cytokines augment immune responses to viruses in several ways, namely by inducing the expression of MHC II and co-stimulatory molecules, as well as interleukin (IL)-15 synthesis by antigen-presenting cells (APCs) (Figure 1 , Key Figure). The activation of APCs, such as DCs, induces natural killer (NK) and T cell proliferation and differentiation, with enhanced IFN-γ secretion [13., 14., 15., 16.]. In addition to supporting T cell responses via the activation of DCs and IL-15 production, IFN-I acts directly on both CD8+ T cells [14,15] and CD4+ T cells [including both T follicular helper (Tfh) cells and T follicular regulatory cells] [17] for expansion and differentiation (Figure 1). In this manner, IFN-I can positively influence both cellular and humoral immunity (Figure 1).
Figure 1.
Key Figure. Type-I Interferon (IFN-I), Adaptive Immunity, and Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV2) Infection.
Schematic showing (A) the positive effect of IFN-I on activation of antigen-presenting dendritic cells (DCs) that interact with CD8+ T cells for differentiation of CD8+ cytotoxic T lymphocytes [that target and lyse virus-infected cells (shown as infected monocyte/macrophage)]. Similarly, IFN-I acts via DCs or directly on CD4+ T cells to promote the differentiation of T follicular helper (Tfh) cells that interact with B cells in germinal centers within secondary lymphoid organs for the production of affinity-matured antibody (Ab) that binds virus. (B) In the case of SARS-CoV-2 infection in mice and humans, IFN-I production is hindered, leading to poor T cell activation, reduced cytotoxic T lymphocyte (CTL) numbers, and reduced production of high affinity Abs that have undergone somatic hypermutation. This can lead to slower clearance of the virus and infected cells, and continued production of inflammatory mediators from infected (shown, is a monocyte/macrophage) and bystander cells. Abbreviations: IL, interleukin; PD-1, programmed death 1; TNF-α, tumor necrosis factor α.
The timing of IFN-I production is crucial for its influence on the immune response to virus infection. Thus, while early induction of IFN-I is associated with effective virus inhibition, virus clearance, and antiviral immune responses, administration of IFN-I after respiratory virus infection (SARS-CoV-1 or influenza virus) has been established in mice and is associated with increased immunopathology [18,19]; this topic will be discussed in detail. This review examines the ability of SARS-CoV-2 to antagonize IFN-I and how this might affect the course of inflammation following virus infection, with relevant implications on the pathology of COVID-19 disease.
Antagonism of IFNs by SARS-CoV-2
To evade the inhibitory effects of IFN-I, viruses have evolved mechanisms to suppress IFN-I production [7]. This is particularly evident for SARS-CoV-2 as its capacity to elicit IFN-β (and IFN-III) in human cells infected in vitro is weaker or delayed, compared with other viruses and coronaviruses with similar tropism [20,21] (Figure 1). With severe illness, the ability of coronaviruses to evade innate immunity during the first 10 days of infection in humans has been associated with a period of widespread inflammation and steadily increasing viral load [22]. Indeed, a profoundly low production of IFN-β, with consequent minimal induction of ISGs, has been observed in patients with moderate and severe COVID-19 [20]. By contrast, other studies demonstrating increased levels of expression of ISGs following SARS-CoV-2 infection in humans have queried the notion of poor IFN-I induction [23,24]. However, expression of ISGs does not invariably correlate with an abundance of IFN-I and can be influenced by the presence of other cytokines, such as tumor necrosis factor (TNF)-α, which have been reported to correlate with disease severity in COVID-19 patients [25,26]. For instance, the activation of IRF1 by TNF-α in human macrophages in vitro induces picomolar concentrations of IFN-β that increase signal transducer and activator of transcription (STAT)1 expression, thus resulting in high induction of ISGs in these cells [27,28].
In both SARS-CoV-1 and SARS-CoV-2 infections, IFN-I antagonism largely functions through the ability of CoV proteins to interfere with signaling via the RLR/MAVS pathway and modulation of the receptor for IFN-I [29]. SARS-CoV-1 encodes at least 10 proteins that allow the virus to either escape or counteract the induction and antiviral action of IFN [29], and initial observations indicate that SARS-CoV-2 proteins are at least as efficient at inhibiting IFN-I [30]. The 30-kb genome of SARS-CoV-2 has 82% nucleotide identity with SARS-CoV-1 [31]. It includes ORF1a/b, encoding 16 nonstructural proteins (Nsp1–16), structural proteins including spike (S), envelope (E), membrane (M), and nucleocapsid (N), and nine accessory proteins (ORF3a, 3b, 6, 7a, 7b, 8, 9b, 9c, and 10) [32]. SARS-CoV-2 proteins, including N, ORF6, ORF8 and ORF3b, have the capacity to interfere with IFN-I induction, and ORF3a protein decreases the amount of IFNAR expressed on the cell surface [29,33,34]. Thus, confirmation of the most powerful SARS-CoV-2 IFN-I antagonists and design of protein modifications that can prevent IFN-I inhibition might provide a unique opportunity for vaccine design.
In addition to the impaired IFN-I response triggered by SARS-CoV-2, recent studies have demonstrated that further restrictions on IFN-I function can occur through genetic predisposition or the development of autoantibodies that neutralize IFN-I [35]. A recent study revealed that at least 3.5% of patients with life-threatening COVID-19 pneumonia have presented genetic mutations at candidate loci known to be involved in TLR3- and IRF7-dependent induction and amplification of IFN-I [36]. Susceptibility to severe COVID-19 is also associated with the presence of neutralizing autoantibodies against IFN-I (IFN-α and/or IFN-ω), which were found in 10.2% (101/987) of patients with life-threatening COVID-19 and men were more likely than women to carry the rogue antibodies (Abs) [35]. By contrast, anti-IFN-I Abs were not present in 663 patients with asymptomatic or mild COVID-19 and were only found in 0.33% (4/1227) healthy individuals not exposed to SARS-CoV-2 [35]. Whether the 10.2% of patients with severe COVID-19 had preexisting autoantibodies remains an important question. However, none of the patients who produced Abs against IFN-I or had the mutations that limited IFN-I had a history of life-threatening viral illnesses requiring hospitalization, suggesting that the effect of limitation of IFN-I may have been particularly problematic in SARS-CoV-2 infection [35]. However, increased blood concentrations of IFN-α have been observed in some patients with severe COVID-19 relative to healthy controls [37]. Several possibilities may explain this finding, including a delayed onset of IFN-I production or persistence of the cytokine in the bloodstream through binding to IFN-α Abs, as has been shown for IL-2 monoclonal Abs (mAbs) [38].
Since COVID-19 is associated with raised concentrations of IL-1, IL-6, and IL-8, the ability of SARS-CoV-2 to reduce IFN-I production is clearly not matched by impaired production of proinflammatory cytokines [39]. This might be due to the ability of CoV proteins to more selectively inhibit IFN-I signaling than NF-κΒ signaling during SARS-CoV-2 infection [29,33,34]. Skewing of the cytokine milieu away from IFN-I, and towards production of proinflammatory cytokines such as TNF-α and IL-6 might have subsequent effects on immune cells, such as T cells and monocytes, but this possibility remains to be rigorously tested.
IFN-I and T Cells in COVID-19
Due to the importance of neutralizing antibodies (nAbs) in virus clearance, research on COVID-19 has been focused on B cell production of SARS-CoV-2-specific Abs. However, the presence of high titers of nAbs in severe COVID-19 sera, and effective viral clearance in patients lacking nAbs [40], including B cell-deficient patients [41], clearly query the decisive role of nAbs, focusing researchers’ attention on other immune cells, notably T cells [41,42]. In this respect, it is often forgotten that nAbs are of little or no use once viruses enter cells. Once this happens, T cells, especially CD8+ cytotoxic T cells (CTLs) play a major part in the elimination of virus-infected cells.
T lymphopenia is prominent in severe COVID-19 patients, and when considering CD8+ T cells, these can display an exhausted phenotype, although recovery from infection notably leads to rapid restoration of T cell numbers and phenotype in peripheral blood [43,44]. Despite the occurrence of lymphopenia, both CD8+ and CD4+ T cell responses against SARS- CoV-2 are detected in the blood around 1 week after the onset of COVID-19 symptoms with the majority of T cell reactivity in convalescent patients directed towards peptides derived from SARS-CoV-2 M, S, and N proteins [45]. Moreover, the degree of severity of COVID-19 cases has been shown to positively correlate with the breadth and magnitude of memory CD4+ and CD8+ T cell responses to SARS-CoV-2 M, S, N, and ORF proteins, indicating a strong T cell response against SARS-CoV-2 [46]. Of note, specific CD8+ T cell responses have been found to predominate during mild SARS-CoV-2 infection [46., 47., 48.]. Thus, the characterization of T cell responses that are associated with milder disease can further our understanding of protective immunity to SARS-CoV-2.
For CD4+ T cells, SARS-CoV-2-reactive cells have also been observed in a variable proportion of unexposed individuals and this has been suggested to reflect crossreactive responses generated from prior exposure to other coronaviruses [45,47,49]. This finding raises the interesting possibility that these responses might provide crossprotection against COVID-19. Although still unresolved, this issue begs the question of how CD4+ T cells function during COVID-19 pathology. Similar to other viruses, CD4+ T cells are presumed to react against SARS-CoV-2 largely by providing help to both CD8+ T cells for the acquisition of cytolytic functions and B cells for nAb production [50]. In addition, CD4+ T cells synthesize a spectrum of cytokines, some of which, namely IFN-γ and IL-2, are probably of major importance in countering SARS-CoV-2 infection through their actions on T cells and innate cell populations. In this respect, studies on SARS-CoV-1 infection in mice showed that prior immunization with a single peptide recognized exclusively by CD4+ T cells, provided strong protection against subsequent virus infection; this effect suggested the occurrence of not only augmented IFN-γ synthesis, but also the provision of increased CD4+ T cell-mediated help for naïve CD8+ T cells upon infection [51]. In addition, such priming of CD4+ T cells allows their conversion into CD4+ Tfh cells, which then provide help for Ab production by B cells [50] (see later).
Germinal Center Reaction in COVID-19
Similar to other virus infections, the generation of nAbs is thought to be important for inactivating SARS-CoV-2 during infection and preventing its distribution via the bloodstream. Such a general antiviral mechanism is known to be important for protection against re-infection through the generation of long-lived Ab secreting plasma cells and memory B cells that can be quickly reactivated to secrete potent nAbs. B cells undergo a process of somatic hypermutation (SHM) to generate the best virus-binding Abs. Affinity-matured Ab requires the interaction of Tfh cells with B cells in germinal centers (GCs) that form in response to antigen within B cell follicles in secondary lymphoid organs [52]. IFN-I is important for the differentiation of Tfh cells, acting both through direct stimulation of Tfh precursors [17] and also by eliciting cytokine generation from DCs [53]. In addition to Tfh cells, IFN-I is important for the differentiation of T follicular regulatory cells [17] that originate from thymic-derived Foxp3+ precursors and act to suppress the outgrowth of non-antigen-specific B cells in GCs [54].
Despite the limited analyses of Tfh cells in COVID-19, there is some indication that GC output is impaired during infection. Specifically, T cells in peripheral blood that resemble Tfh cells known as circulatory T follicular helper (cTfh) cells (defined as CD3+CD4+CD45RA-CXCR5+) that can provide help to B cells for Ab production have been detected in convalescent COVID-19 patients [55]. However, cTfh cells in the periphery may not be a reliable surrogate for GC-residing Tfh cells, as cTfh cells may instead arise from T helper cells that reside outside of the B cell follicles (extrafollicular) in secondary lymphoid organs [56]. In this regard, studies in critically ill patients with COVID-19 have reported marked expansion of plasmablasts, which is a hallmark of extrafollicular B cell activation [57]. The generation of affinity matured Abs are an effective readout for a successful GC reaction and Abs against SARS-CoV-2 detected in most COVID-19 patients remain close to germline, with low levels of SHM [58,59]. Moreover, for COVID-19 patients, a recent postmortem study showed a reduction of both GC B cells and Tfh cells and an absence of GCs in the thoracic lymph nodes relative to controls [60]; furthermore, there was also a reciprocal increase in Tbet + Th1 cells and aberrant extrafollicular accumulation of TNF-α in these patients, consistent with an extrafollicular immune response [60]. Collectively, these findings suggest conspicuous GC dysfunction in severe COVID-19 patients.
Seroconversion is observed in all COVID-19 patients 2–3 weeks after symptom onset, but early observations indicate that Ab titers can wane significantly as early as 30–50 days after symptom onset [40,61,62]. Thus, the durability of SARS-CoV-2-specific Ab titers exhibits marked heterogeneity, with some patients recovering rapidly from COVID-19, exhibiting longer-lasting Abs and higher frequencies of previously activated CD4+ T cells relative to patients recovering slowly [63]. For SARS-CoV-1 infection, nAb responses in convalescent patients have also declined over time [64]. These findings are not unexpected, however, because similar abbreviated Ab responses also apply to other coronaviruses, including those that cause the common cold [22]. By contrast, at least for SARS-CoV-1, memory T cell responses can persist for up to 6 years [64], suggesting that SARS-CoV-2 vaccines that target T cells as well as B cells may provide durable immunity. However, whether SARS-CoV-2-specific T cells are similarly long-lived, awaits future studies.
Age: IFN -I and COVID-19
As discussed elsewhere, COVID-19 is most often mild or asymptomatic in young people, but severe or lethal in old age, especially in men [65]. It is well documented that immune responses are less effective in old age and are associated with immunosenescence characterized, among several factors, by a loss of T cell clonal diversity and a paucity of naïve T cells with proliferative capacity [66]. In this scenario, a restricted T cell repertoire is likely prone to antigen-mediated exhaustion.
Although the cause of immunosenescence is contentious, there is increasing evidence that ageing per se is associated with IFN-1 dysfunction [67]. Thus, aging is marked by a sharp decline in IFN-I and IFN-III production by both myeloid DCs and pDCs in response to viruses, including influenza virus and West Nile virus (WNV) [68]. This deficit in IFN production is prominent in severe COVID-19 patients and correlates with the earlier-mentioned finding that IFN-I and IFN-III production in response to SARS CoV-2 infection is low and parallels impaired T cell function [20].
At face value, the findings described earlier would seem to imply that severe COVID-19 is primarily a reflection of IFN-I dysfunction potentiated by immunosenescence. Although this might be true, at least to some extent, studies in mice suggest that the role of IFN-I signaling in virus infections is highly complex and, in part, is virus-specific. Specifically, for infection of mice with either influenza virus or Middle East respiratory syndrome coronavirus (MERS-CoV), the generation of an effective adaptive immune response essentially depends on early T cell contact with IFN-I; without such contact, severe disease occurs, but can be prevented by therapy with IFN-I (or IFN-I-inducing agents) given soon after infection, although not at later stages [69]. With SARS-CoV-1 infection, however, the results are similar but much more extreme: although effective when given before infection, IFN-β therapy potentiates disease elicited by SARS-CoV-1 when given as early as 12 h postinfection [19]. Conversely, blocking contact with IFN-1 or using IFN-I-deficient mice as hosts can reduce disease severity [19].
IFN-I therapy delivered after infection might exacerbate inflammation by direct stimulatory effects on both innate immune cell subsets and T cells, but with time, can also act to suppress the adaptive immune response by inducing T cell exhaustion [70,71]. Thus, during chronic infection of mice with lymphocytic choriomeningitis virus (LCMV), clone-13, IFN-I induces the production of suppressive factors, such as IL-10 and programmed cell death 1 ligand 1 (PD-L1) on splenic DCs [71]. Furthermore, blockade of IFNAR with mAb led to the rescue of IFNγ+ CD4+ T cells and promoted LCMV clearance by CD8+ CTLs [71]. These findings suggest that the beneficial effects on the immune response resulting from IFN-1 signaling might change rapidly soon after infection; instead of protection, IFN-I signaling may lead to the onset of immunopathology associated with infiltration of the lungs by immature macrophages synthesizing IL-6 and other proinflammatory cytokines. These findings are most prominent in aged mice compared with young [18,72,73] and could potentially be applicable to SARS-CoV-2 infections [22], although direct data on the consequences of IFN-I signaling in COVID-19 patients awaits future studies.
There is still much to be learned about the effects of aging on the severity of COVID-19. For both myeloid DCs and pDCs, studies in mice have shown that a selective decline in IFN-I and IFN-III production with aging retards responses to influenza virus and WNV relative to young mice [68]. In addition, aging has been associated with poor DC maturation in humans [74] and migration to the lungs following influenza virus infection in mice [75], thereby further impairing T cell responses in aged COVID-19 patients. In mice, impaired DC migration from the lungs to the draining lymph nodes is mediated by the presence of inhibitory macrophages and increased expression of prostaglandin D2, leading to impaired T cell responses [75]. Of note, in SARS-CoV-1 infection in mice, macrophage depletion by clodronate was beneficial in reducing the severity of disease [76]. These data suggest that the prominent immunopathology seen in severe COVID-19 patients might be due, at least in part, to an influx of proinflammatory monocytes and macrophages into the lungs [77], although this remains to be further investigated.
Systemic Inflammation in COVID-19
To prevent the deleterious effects of chronic inflammation, IFN-I during viral infections is important for causing rapid elimination of the virus, thereby curtailing the immune response [2]. For certain viruses, infection of mice that lack expression of IFN-I receptors can lead to heightened pathology associated with elevated production of proinflammatory cytokines [78]. Specifically, IFN-I acts on T effector cells that eliminate virus and regulatory CD4+ T cells that secrete inhibitory cytokines such as IL-10 [79]. In this context, it is tempting to speculate that dysregulation of IFN-I by SARS-CoV-2 may lead to sustained inflammation by constraining T cells.
In systemic inflammatory response syndrome (SIRS), inflammatory cytokines go into overdrive and can leak into the circulation and cause multiorgan dysfunction [80]. This cytokine storm is a feature of many virus infections and sepsis, and is a prospective predictor of morbidity and mortality; yet, the cellular sources of the cytokines involved remain undefined. A cytokine storm resulting from influenza virus infection is thought to have contributed to the high mortality rates associated with the 1918 influenza A virus subtype H1N1 pandemic, and perhaps also with subsequent influenza virus epidemics [81]. In severe SARS cases with rapid fatalities, SARS-CoV-1 was not invariably detected, suggesting that death of the infected individuals was not necessarily the result of viral replication and could potentially reflect exuberant host inflammatory responses [82,83]. Indeed, in this study, a poor outcome following SARS-CoV-1 infection was found to correlate with high concentrations of proinflammatory cytokines and chemokines [82].
It is now well documented that SIRS is also a characteristic feature of COVID-19 [84]. As for SARS, lung pathology in severe COVID-19 patients is associated with excess production of proinflammatory chemokines and cytokines that promote vascular leakage and disseminated intravascular coagulation [85]. SARS-CoV-2 infection of respiratory epithelium leads to cell death by pyroptosis [86,87], which results in the release of proinflammatory cytokines from neighboring cells. This cytokine release, in turn, recruits immune cells, including inflammatory cytokine-producing (e.g., IL-6, TNF-α, and IL-8) macrophages and monocytes to the lungs, to compound the inflammation [88]. This sequence of events is especially pronounced in elderly people, and is accentuated late in disease by the onset of a delayed IFN-I response [19,89]. Furthermore, high IL-6 concentrations inhibit the activation of CD8+ T cells via STAT3 phosphorylation, and impair their function via the induction of inhibitory molecules, including PD-1, PD-L1, and NKG2A/CD94 [90]. Likewise, for CD4+ T cells, increased IL-6 favors IL-4 production and a Th2 phenotype; this allergic-type response occurs in aged mice following SARS-CoV-1 infection, and is associated with Th2 immunopathology in the lung [73]. Consequently, it is tempting to speculate that this cadre might present a potential problem for successful vaccination outcomes of the elderly against COVID-19.
The findings described earlier highlight the hypothesis that the positive effects of IFN-I on the immune response might apply only during the initial stage of the immune response. During this stage, rapid production of IFN-I promotes efficient differentiation of T cells into effector cells and prompts elimination of the virus. However, when IFN-I synthesis is limited and/or delayed, for instance, in elderly people, IFN-I signaling might no longer be protective: instead, signaling may become proinflammatory, with T cell dysfunction, and possibly, prominent immunopathology. The implication, therefore, is that T cells play an important role in regulating the intensity of the proinflammatory response of the innate immune system. In support of this notion, injecting intravenously T cell-depleted nude, or Rag -/- mice, with a dose of MHV-A59 murine coronavirus that was not lethal in T cell-replete wild-type (WT) mice, or with poly:IC (an IFN-I inducer), elicited a prominent cytokine storm (including TNF-α or IL-6) and mortality that could be prevented by injection of purified CD4+ or CD8+ T cells [91]. Of note, the mortality in nude mice did not reflect increased virus titers relative to WT controls, which suggested an immune- rather than virus-induced pathology [91]. Collectively, these findings suggest that in response to viruses, the innate immune system (or cells other than T cells) can elicit an intense cytokine storm that is normally suppressed by the presence of T cells.
Concluding Remarks
Since SARS-CoV-2 strongly inhibits IFN-I induction [29,33,34], yet is highly sensitive to exogenous IFN-I in vitro [92], it has been suggested that boosting IFN-I concentrations might be an effective candidate treatment for COVID-19. However, studies in mice have cautioned that once infection has been established, it may be too late to administer a safe and effective IFN-I therapy [19]. From another angle, because of its local effects in the respiratory system [11], early therapy with IFN-III is currently being investigated. However, as for IFN-1, it is now apparent that IFN-III may have an injurious proinflammatory effect in severe COVID-19 [93]. Hence, therapy with either IFN-I of IFN-III could be potentially dangerous late in disease.
Despite the concerns described earlier, prophylactic use of IFN-1 (or IFN-III) might have potential; a recent uncontrolled exploratory study on hospitalized COVID-19 patients with mild disease found that administering nebulized IFN-α2b reduced the duration of detectable SARS-CoV-2 in the upper respiratory tract, and also reduced the concentrations of both IL-6 and C reactive protein (CRP) in blood [23]. In addition, repeated intranasal administration of IFN-α in volunteers before exposure to common coronaviruses led to reduced viral loads and minimal symptoms [94], and prophylactic intranasal IFN-α treatment prevented subsequent infection with acute viral respiratory viruses [95]. We posit that when used either as a prophylactic or administered early in infection, IFN-I and or IFN-III therapy might show beneficial outcomes, especially for elderly patients that are vulnerable to severe COVID-19. Currently, multiple clinical trials on the prophylactic and therapeutic use of IFNs for COVID-19 are in progress [96].
Regarding other forms of therapy, there is much interest in preventing the prominent rise in proinflammatory cytokines seen in severe disease. Here, multiple clinical trials are in progress to examine the effects of blocking the rise of IL-6, IL-1, or TNFα; however, results from these trials for IL-6 have been disappointing [97]. Hopefully, the results of future trials, notably for TNF-α blockade [26], might be more encouraging.
Needless to say, eliminating the SARS-CoV-2 virus pandemic will hinge on the development of an effective vaccine (see Outstanding Questions) and, as discussed in depth elsewhere, multiple vaccines are in advanced stages of production and distribution [98,99]. The majority of vaccines currently approved for use deliver immunogenic forms of a single (S) protein from SARS-CoV-2 that can generate Abs that block entry of the virus into cells [99]. While these single protein vaccines have shown success at reducing the incidence of COVID-19, the duration of ensuing immunity is not yet known, and SARS-CoV-2 remains in the nasopharyngeal passage of vaccine trial subjects, thus indicating a lack of sterilizing immunity [99]. Furthermore, as SARS-CoV-2 mutates to avoid immunological pressure [100], so does the probability that the efficacy of current vaccines will be reduced. There are a number of possible approaches that could be used to further refine SARS-CoV-2 vaccines to meet these challenges, including targeting the site of infection by oral delivery of a vaccine to promote anti-SARS-CoV-2 IgA at mucosal surfaces. In addition, utilizing a variety of SARS-CoV-2 proteins rather than a single protein to induce immunity may counter emerging SARS-CoV-2 mutations and ensure that both Ab producing B cells and T cells are armed to fight infection. The ability of SARS-CoV-2 proteins to antagonize IFN-I [29,33,34] and thus limit an effective antiviral immune response poses a problem when expanding the repertoire of proteins for vaccination. However, increasing knowledge of SARS-CoV-2 suggests that modification of SARS-CoV-2 proteins to prevent IFN-I antagonism might provide an opportunity to develop a multi-protein vaccine to generate broad immunity against SARS-CoV-2.
The ability of SARS-CoV-2 proteins to antagonize IFN-I [29,33,34] and thus limit an effective antiviral immune response poses a problem for the use of the whole virus as vaccine. Conversely, vaccination with individual SARS-CoV-2 proteins that lack IFN-I regulatory activity, for example, the S-protein, might be expected to lead to enhanced IFN-I production and consequent strong immunogenicity. It is notable that strong induction of IFN-I is a conspicuous feature of mRNA vaccines [101]. Hence, for the currently used mRNA vaccines encoding the S-protein, it is not surprising that these highly-efficient vaccines elicit transient induction of fever, aches etc. – a known hallmark of strong IFN-I induction [102]. Indeed, treatment with a dose of one of these vaccines at the time of initial SARS-CoV-2 diagnosis, before significant clinical symptoms develop, might be a simple and effective method for overcoming the IFN-I deficit seen in older patients, thus boosting the initial antiviral immune response, and potentially preventing the onset of severe disease. Evidently, clinical trials would be needed to test this hypothesis.
Outstanding Questions.
Will vaccines work equally well in young and old COVID-19 infected patients? Phase III clinical trials for current SARS-CoV-2 vaccines will need to be expanded from testing on young to middle aged adults to determine whether they work as well in children and adults aged >65-years.
Will intranasal vaccination prove more effective than injected intramuscular vaccines?
The lingering presence of SARS-CoV-2 in the nasopharyngeal passage following vaccination may be countered by a potent secretory IgA response driven by oral vaccination.
Will novel mRNA and viral vector vaccines prove to be as effective and durable as conventional whole-virus vaccines, and will S-protein vaccines elicit a sufficiently broad B cell and T cell response to induce long-lasting immunity? Increasing the arsenal of SARS-CoV-2 proteins that induce B and T cell immune responses in vaccines may improve long-lasting immunity and counter the threat of emerging SARS-CoV-2 mutations. This approach will require further understanding of T and B cell SARS-CoV-2 protein epitopes and mechanisms underlying IFN-I antagonism by these proteins.
Alt-text: Outstanding Questions
Acknowledgments
Acknowledgments
C.K. is supported by grant APP2004306 from the National Health and Medical Research Council, Australia.
Declaration of Interests
There are no interests to declare.
Glossary
- Affinity-matured antibody
process of selection of surface antibody variants generated from somatic hypermutation; requires B cell interaction with antigen and Tfh cells to produce antibodies with increased binding affinity for antigen.
- Circulatory T follicular helper (cTfh) cells
phenotypically resemble Tfh cells with surface expression of CXCR5 and PD-1, but are found in the blood.
- Exhausted phenotype
observed during virus infection or in cancers due to T cell persistent antigen exposure; characterized by the stepwise and progressive loss of T cell effector functions.
- GC B cells
germinal centers (GC) are specialized sites within B cell follicles of secondary lymphoid organs where B cells proliferate, differentiate, and mutate their antibody genes to make potent antibodies.
- IFN-I-stimulated genes (ISGs)
genes whose expression is stimulated by IFN-I.
- Neutralizing antibodies (nAbs)
can bind to a virus and prevent it from entering a cell.
- Pyroptosis
highly inflammatory form of programmed cell death, commonly seen with cytopathic viruses.
- Somatic hypermutation (SHM)
genetic process of rapid and sequential mutations of surface immunoglobulins (antibodies) by which B cells increase the diversity of their antibody receptors and thus, the chance of improved antibody binding to antigen.
- Systemic inflammatory response syndrome (SIRS)
exaggerated and persistent inflammatory response affecting multiple organs; most often induced by infection.
- T follicular helper (Tfh) cells
specialized subset of CD4+ T cells found within specialized structures within B cell follicles of secondary lymphoid organs, known as GCs, help B cells to produce potent antibodies.
- T follicular regulatory cells
subset of CD4+ T cells found within the GC; express Foxp3 and act to regulate Tfh cells and GC B cells.
- Tbet+Th1 cells
expression of the T-box family molecule, T-bet, is important for CD4+ T helper cell effector functions, including secretion of IFN-γ and cytotoxicity.
- Th2 and Th17 responses
CD4+ T helper cells can be categorized by their secretion of soluble mediators known as cytokines: Th2 cells typically secrete IL-4 and Th17 cells typically secrete IL-17.
- Toll like receptors
evolutionarily conserved receptors expressed in various immune and non-immune cells of the mammalian host that recognize structurally conserved molecules derived from microbes.
References
- 1.Gallo Marin B., et al. Predictors of COVID-19 severity: a literature review. Rev. Med. Virol. 2021;31:1–10. doi: 10.1002/rmv.2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Park A., Iwasaki A. Type I and type III interferons - induction, signaling, evasion, and application to combat COVID-19. Cell Host Microbe. 2020;27:870–878. doi: 10.1016/j.chom.2020.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hoffmann M., et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280. doi: 10.1016/j.cell.2020.02.052. e278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Seth R.B., et al. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell. 2005;122:669–682. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 5.Takeuchi O., Akira S. Pattern recognition receptors and inflammation. Cell. 2010;140:805–820. doi: 10.1016/j.cell.2010.01.022. [DOI] [PubMed] [Google Scholar]
- 6.García-Sastre A., Biron C.A. Type 1 interferons and the virus-host relationship: a lesson in detente. Science. 2006;312:879–882. doi: 10.1126/science.1125676. [DOI] [PubMed] [Google Scholar]
- 7.Hoffmann H.-H., et al. Interferons and viruses: an evolutionary arms race of molecular interactions. Trends Immunol. 2015;36:124–138. doi: 10.1016/j.it.2015.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kotenko S.V., et al. IFN-λs mediate antiviral protection through a distinct class II cytokine receptor complex. Nat. Immunol. 2003;4:69–77. doi: 10.1038/ni875. [DOI] [PubMed] [Google Scholar]
- 9.Zhou Z., et al. Type III interferon (IFN) induces a type I IFN-like response in a restricted subset of cells through signaling pathways involving both the Jak-STAT pathway and the mitogen-activated protein kinases. J. Virol. 2007;81:7749–7758. doi: 10.1128/JVI.02438-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Misumi I., Whitmire J.K. IFN-lambda exerts opposing effects on T cell responses depending on the chronicity of the virus infection. J. Immunol. 2014;192:3596–3606. doi: 10.4049/jimmunol.1301705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johnson T.R., et al. Role for innate IFNs in determining respiratory syncytial virus immunopathology. J. Immunol. 2005;174:7234–7241. doi: 10.4049/jimmunol.174.11.7234. [DOI] [PubMed] [Google Scholar]
- 12.Dai J., et al. IFN-λ1 (IL-29) inhibits GATA3 expression and suppresses Th2 responses in human naive and memory T cells. Blood J. Am. Soc. Hematol. 2009;113:5829–5838. doi: 10.1182/blood-2008-09-179507. [DOI] [PubMed] [Google Scholar]
- 13.Tough D.F., et al. Induction of bystander T cell proliferation by viruses and type I interferon in vivo. Science. 1996;272:1947–1950. doi: 10.1126/science.272.5270.1947. [DOI] [PubMed] [Google Scholar]
- 14.Sun S., et al. Type I interferon-mediated stimulation of T cells by CpG DNA. J. Exp. Med. 1998;188:2335–2342. doi: 10.1084/jem.188.12.2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kolumam G.A., et al. Type I interferons act directly on CD8 T cells to allow clonal expansion and memory formation in response to viral infection. J. Exp. Med. 2005;202:637–650. doi: 10.1084/jem.20050821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Surh C.D., Sprent J. Homeostasis of naive and memory T cells. Immunity. 2008;29:848–862. doi: 10.1016/j.immuni.2008.11.002. [DOI] [PubMed] [Google Scholar]
- 17.Loetsch C., et al. Cytosolic recognition of RNA drives the immune response to heterologous erythrocytes. Cell Rep. 2017;21:1624–1638. doi: 10.1016/j.celrep.2017.10.044. [DOI] [PubMed] [Google Scholar]
- 18.Zhao J., et al. Intranasal treatment with poly(I*C) protects aged mice from lethal respiratory virus infections. J. Virol. 2012;86:11416–11424. doi: 10.1128/JVI.01410-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Channappanavar R., et al. Dysregulated type I interferon and inflammatory monocyte-macrophage responses cause lethal pneumonia in SARS-CoV-infected mice. Cell Host Microbe. 2016;19:181–193. doi: 10.1016/j.chom.2016.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Blanco-Melo D., et al. Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell. 2020;181:1036–1045. doi: 10.1016/j.cell.2020.04.026. e1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chu H., et al. Comparative replication and immune activation profiles of SARS-CoV-2 and SARS-CoV in human lungs: an ex vivo study with implications for the pathogenesis of COVID-19. Clin. Infect. Dis. 2020;71:1400–1409. doi: 10.1093/cid/ciaa410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sariol A., Perlman S. Lessons for COVID-19 immunity from other coronavirus infections. Immunity. 2020;53:248–263. doi: 10.1016/j.immuni.2020.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou Z., et al. Heightened innate immune responses in the respiratory tract of COVID-19 patients. Cell Host Microbe. 2020;27:883–890.e2. doi: 10.1016/j.chom.2020.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wilk A.J., et al. A single-cell atlas of the peripheral immune response in patients with severe COVID-19. Nat. Med. 2020;26:1070–1076. doi: 10.1038/s41591-020-0944-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Del Valle D.M., et al. An inflammatory cytokine signature predicts COVID-19 severity and survival. Nat. Med. 2020;26:1636–1643. doi: 10.1038/s41591-020-1051-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Robinson P.C., et al. Accumulating evidence suggests anti-TNF therapy needs to be given trial priority in COVID-19 treatment. Lancet Rheumatol. 2020;2:e653–e655. doi: 10.1016/S2665-9913(20)30309-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yarilina A., et al. TNF activates an IRF1-dependent autocrine loop leading to sustained expression of chemokines and STAT1-dependent type I interferon–response genes. Nat. Immunol. 2008;9:378–387. doi: 10.1038/ni1576. [DOI] [PubMed] [Google Scholar]
- 28.Venkatesh D., et al. Endothelial TNF receptor 2 induces IRF1 transcription factor-dependent interferon-β autocrine signaling to promote monocyte recruitment. Immunity. 2013;38:1025–1037. doi: 10.1016/j.immuni.2013.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sa Ribero M., et al. Interplay between SARS-CoV-2 and the type I interferon response. PLoS Pathog. 2020;16 doi: 10.1371/journal.ppat.1008737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hadjadj J., et al. Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science. 2020;369:718–724. doi: 10.1126/science.abc6027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou P., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chan J.F., et al. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg. Microbes Infect. 2020;9:221–236. doi: 10.1080/22221751.2020.1719902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Konno Y., et al. SARS-CoV-2 ORF3b is a potent interferon antagonist whose activity is increased by a naturally occurring elongation variant. Cell Rep. 2020;32 doi: 10.1016/j.celrep.2020.108185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li J.Y., et al. The ORF6, ORF8 and nucleocapsid proteins of SARS-CoV-2 inhibit type I interferon signaling pathway. Virus Res. 2020;286 doi: 10.1016/j.virusres.2020.198074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bastard P., et al. Autoantibodies against type I IFNs in patients with life-threatening COVID-19. Science. 2020;370 doi: 10.1126/science.abd4585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang Q., et al. Inborn errors of type I IFN immunity in patients with life-threatening COVID-19. Science. 2020;370 doi: 10.1126/science.abd4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lucas C., et al. Longitudinal analyses reveal immunological misfiring in severe COVID-19. Nature. 2020;584:463–469. doi: 10.1038/s41586-020-2588-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Boyman O., et al. Selective stimulation of T cell subsets with antibody-cytokine immune complexes. Science. 2006;311:1924–1927. doi: 10.1126/science.1122927. [DOI] [PubMed] [Google Scholar]
- 39.Costela-Ruiz V.J., et al. SARS-CoV-2 infection: the role of cytokines in COVID-19 disease. Cytokine Growth Factor Rev. 2020;54:62–75. doi: 10.1016/j.cytogfr.2020.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Robbiani D.F., et al. Convergent antibody responses to SARS-CoV-2 infection in convalescent individuals. Nature. 2020;584:437–442. doi: 10.1038/s41586-020-2456-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Quinti I., et al. A possible role for B cells in COVID-19? Lesson from patients with agammaglobulinemia. J. Allergy Clin. Immunol. 2020;146:211–213.e4. doi: 10.1016/j.jaci.2020.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tarke A., et al. Comprehensive analysis of T cell immunodominance and immunoprevalence of SARS-CoV-2 epitopes in COVID-19 cases. Cell Rep. Med. 2021:100204. doi: 10.1016/j.xcrm.2021.100204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ruan Q., et al. Clinical predictors of mortality due to COVID-19 based on an analysis of data of 150 patients from Wuhan, China. Intensive Care Med. 2020;46:846–848. doi: 10.1007/s00134-020-05991-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sun L., et al. Combination of four clinical indicators predicts the severe/critical symptom of patients infected COVID-19. J. Clin. Virol. 2020;128 doi: 10.1016/j.jcv.2020.104431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Grifoni A., et al. Targets of T cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell. 2020;181:1489–1501.e15. doi: 10.1016/j.cell.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Peng Y., et al. Broad and strong memory CD4(+) and CD8(+) T cells induced by SARS-CoV-2 in UK convalescent individuals following COVID-19. Nat. Immunol. 2020;21:1336–1345. doi: 10.1038/s41590-020-0782-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sekine T., et al. Robust T cell immunity in convalescent individuals with asymptomatic or mild COVID-19. Cell. 2020;183:158–168. doi: 10.1016/j.cell.2020.08.017. e114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liao M., et al. Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19. Nat. Med. 2020;26:842–844. doi: 10.1038/s41591-020-0901-9. [DOI] [PubMed] [Google Scholar]
- 49.Le Bert N., et al. SARS-CoV-2-specific T cell immunity in cases of COVID-19 and SARS, and uninfected controls. Nature. 2020;584:457–462. doi: 10.1038/s41586-020-2550-z. [DOI] [PubMed] [Google Scholar]
- 50.King C. New insights into the differentiation and function of T follicular helper cells. Nat. Rev. Immunol. 2009;9:757–766. doi: 10.1038/nri2644. [DOI] [PubMed] [Google Scholar]
- 51.Zhao J., et al. Airway memory CD4+ T cells mediate protective immunity against emerging respiratory coronaviruses. Immunity. 2016;44:1379–1391. doi: 10.1016/j.immuni.2016.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.King C., Sprent J. Emerging cellular networks for regulation of T follicular helper cells. Trends Immunol. 2012;33:59–65. doi: 10.1016/j.it.2011.11.006. [DOI] [PubMed] [Google Scholar]
- 53.Cucak H., et al. Type I interferon signaling in dendritic cells stimulates the development of lymph-node-resident T follicular helper cells. Immunity. 2009;31:491–501. doi: 10.1016/j.immuni.2009.07.005. [DOI] [PubMed] [Google Scholar]
- 54.Linterman M., et al. Foxp3 (+) follicular regulatory T cells control T follicular helper cells and the germinal centre response. Immunology. 2012;136:4–5. [Google Scholar]
- 55.Juno J.A., et al. Humoral and circulating follicular helper T cell responses in recovered patients with COVID-19. Nat. Med. 2020;26:1428–1434. doi: 10.1038/s41591-020-0995-0. [DOI] [PubMed] [Google Scholar]
- 56.Linterman M.A., Hill D.L. Can follicular helper T cells be targeted to improve vaccine efficacy? F1000Res. 2016;5 doi: 10.12688/f1000research.7388.1. F1000 Faculty Rev-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Woodruff M., et al. Critically ill SARS-CoV-2 patients display lupus-like hallmarks of extrafollicular B cell activation. medRxiv. 2020 doi: 10.1101/2020.04.29.20083717. Published online May 3, 2020. [DOI] [Google Scholar]
- 58.Seydoux E., et al. Analysis of a SARS-CoV-2-infected individual reveals development of potent neutralizing antibodies with limited somatic mutation. Immunity. 2020;53:98–105.e5. doi: 10.1016/j.immuni.2020.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kreer C., et al. Longitudinal isolation of potent near-germline SARS-CoV-2-neutralizing antibodies from COVID-19 patients. Cell. 2020;182:843–854.e12. doi: 10.1016/j.cell.2020.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kaneko N., et al. Loss of Bcl-6-expressing T follicular helper cells and germinal centers in COVID-19. Cell. 2020;183:143–157.e13. doi: 10.1016/j.cell.2020.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Long Q.-X., et al. Clinical and immunological assessment of asymptomatic SARS-CoV-2 infections. Nat. Med. 2020;26:1200–1204. doi: 10.1038/s41591-020-0965-6. [DOI] [PubMed] [Google Scholar]
- 62.Seow J., et al. Longitudinal evaluation and decline of antibody responses in SARS-CoV-2 infection. medRxiv. 2020 doi: 10.1101/2020.07.09.20148429. Published online July 11, 2020. [DOI] [Google Scholar]
- 63.Chen Y., et al. Quick COVID-19 Healers sustain anti-SARS-CoV-2 antibody production. Cell. 2020;183:1496–1507.e16. doi: 10.1016/j.cell.2020.10.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tang F., et al. Lack of peripheral memory B cell responses in recovered patients with severe acute respiratory syndrome: a six-year follow-up study. J. Immunol. 2011;186:7264–7268. doi: 10.4049/jimmunol.0903490. [DOI] [PubMed] [Google Scholar]
- 65.Zhou F., et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395:1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Youm Y.H., et al. The Nlrp3 inflammasome promotes age-related thymic demise and immunosenescence. Cell Rep. 2012;1:56–68. doi: 10.1016/j.celrep.2011.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Qian F., et al. Impaired interferon signaling in dendritic cells from older donors infected in vitro with West Nile virus. J. Infect. Dis. 2011;203:1415–1424. doi: 10.1093/infdis/jir048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Agrawal A. Mechanisms and implications of age-associated impaired innate interferon secretion by dendritic cells: a mini-review. Gerontology. 2013;59:421–426. doi: 10.1159/000350536. [DOI] [PubMed] [Google Scholar]
- 69.Channappanavar R., et al. IFN-I response timing relative to virus replication determines MERS coronavirus infection outcomes. J. Clin. Invest. 2019;129:3625–3639. doi: 10.1172/JCI126363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wilson E.B., et al. Blockade of chronic type I interferon signaling to control persistent LCMV infection. Science. 2013;340:202–207. doi: 10.1126/science.1235208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Teijaro J.R., et al. Persistent LCMV infection is controlled by blockade of type I interferon signaling. Science. 2013;340:207–211. doi: 10.1126/science.1235214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhao J., et al. Evasion by stealth: inefficient immune activation underlies poor T cell response and severe disease in SARS-CoV-infected mice. PLoS Pathog. 2009;5 doi: 10.1371/journal.ppat.1000636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bolles M., et al. A double-inactivated severe acute respiratory syndrome coronavirus vaccine provides incomplete protection in mice and induces increased eosinophilic proinflammatory pulmonary response upon challenge. J. Virol. 2011;85:12201–12215. doi: 10.1128/JVI.06048-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Agrawal A., et al. Role of dendritic cells in inflammation and loss of tolerance in the elderly. Front. Immunol. 2017;8:896. doi: 10.3389/fimmu.2017.00896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Toapanta F.R., Ross T.M. Impaired immune responses in the lungs of aged mice following influenza infection. Respir. Res. 2009;10:112. doi: 10.1186/1465-9921-10-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hartwig S.M., et al. Depletion of alveolar macrophages ameliorates virus-induced disease following a pulmonary coronavirus infection. PloS one. 2014;9 doi: 10.1371/journal.pone.0090720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhao J., et al. Age-related increases in PGD 2 expression impair respiratory DC migration, resulting in diminished T cell responses upon respiratory virus infection in mice. J. Clin. Invest. 2011;121:4921–4930. doi: 10.1172/JCI59777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Westover J.B., et al. Vascular leak and hypercytokinemia associated with severe fever with thrombocytopenia syndrome virus infection in mice. Pathogens. 2019;8:158. doi: 10.3390/pathogens8040158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Iyer S.S., Cheng G. Role of interleukin 10 transcriptional regulation in inflammation and autoimmune disease. Critical Reviews™ in. Immunology. 2012;32:23–63. doi: 10.1615/critrevimmunol.v32.i1.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Jiang L., et al. COVID-19 and multisystem inflammatory syndrome in children and adolescents. Lancet Infect. Dis. 2020;20:e276–e288. doi: 10.1016/S1473-3099(20)30651-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Liu Q., et al. The cytokine storm of severe influenza and development of immunomodulatory therapy. Cell. Mol. Immunol. 2016;13:3–10. doi: 10.1038/cmi.2015.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhang Y., et al. Analysis of serum cytokines in patients with severe acute respiratory syndrome. Infect. Immun. 2004;72:4410–4415. doi: 10.1128/IAI.72.8.4410-4415.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Reghunathan R., et al. Expression profile of immune response genes in patients with severe acute respiratory syndrome. BMC Immunol. 2005;6:1–11. doi: 10.1186/1471-2172-6-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Liu J., et al. Longitudinal characteristics of lymphocyte responses and cytokine profiles in the peripheral blood of SARS-CoV-2 infected patients. EBioMedicine. 2020;55 doi: 10.1016/j.ebiom.2020.102763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Moore J.B., June C.H. Cytokine release syndrome in severe COVID-19. Science. 2020;368:473–474. doi: 10.1126/science.abb8925. [DOI] [PubMed] [Google Scholar]
- 86.Cookson B.T., Brennan M.A. Pro-inflammatory programmed cell death. Trends Microbiol. 2001;9:113–114. doi: 10.1016/s0966-842x(00)01936-3. [DOI] [PubMed] [Google Scholar]
- 87.Li S., et al. SARS-CoV-2 triggers inflammatory responses and cell death through caspase-8 activation. Signal Transduct. Target. Ther. 2020;5:1–10. doi: 10.1038/s41392-020-00334-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yang M. Cell pyroptosis, a potential pathogenic mechanism of 2019-nCoV infection. SSRN. 2020 doi: 10.2139/ssrn.3527420. Published online January 29, 2020. [DOI] [Google Scholar]
- 89.Velazquez-Salinas L., et al. The role of interleukin 6 during viral infections. Front. Microbiol. 2019;10:1057. doi: 10.3389/fmicb.2019.01057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Cho J.-H., et al. Calcineurin-dependent negative regulation of CD94/NKG2A expression on naive CD8+ T cells. Blood J. Am. Soc. Hematol. 2011;118:116–128. doi: 10.1182/blood-2010-11-317396. [DOI] [PubMed] [Google Scholar]
- 91.Kim K.D., et al. Adaptive immune cells temper initial innate responses. Nat. Med. 2007;13:1248–1252. doi: 10.1038/nm1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lokugamage K.G., et al. SARS-CoV-2 is sensitive to type I interferon pretreatment. bioRxiv. 2020 doi: 10.1101/2020.03.07.982264. Published online April 9, 2020. [DOI] [Google Scholar]
- 93.Broggi A., et al. Type III interferons disrupt the lung epithelial barrier upon viral recognition. Science. 2020;369:706–712. doi: 10.1126/science.abc3545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Higgins P., et al. Intranasal interferon as protection against experimental respiratory coronavirus infection in volunteers. Antimicrob. Agents Chemother. 1983;24:713–715. doi: 10.1128/aac.24.5.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Gao L., et al. A randomized controlled trial of low-dose recombinant human interferons α-2b nasal spray to prevent acute viral respiratory infections in military recruits. Vaccine. 2010;28:4445–4451. doi: 10.1016/j.vaccine.2010.03.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Sallard E., et al. Type 1 interferons as a potential treatment against COVID-19. Antivir. Res. 2020;178 doi: 10.1016/j.antiviral.2020.104791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Furlow B. COVACTA trial raises questions about tocilizumab's benefit in COVID-19. Lancet Rheumatol. 2020;2:e592. doi: 10.1016/S2665-9913(20)30313-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Poland G.A., et al. SARS-CoV-2 immunity: review and applications to phase 3 vaccine candidates. Lancet. 2020;396:P1595–P1606. doi: 10.1016/S0140-6736(20)32137-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Grigoryan L., Pulendran B. The immunology of SARS-CoV-2 infections and vaccines. Semin. Immunol. 2020;50 doi: 10.1016/j.smim.2020.101422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Tang J.W., et al. Emergence of a new SARS-CoV-2 variant in the UK. J. Infect. 2020 doi: 10.1016/j.jinf.2020.12.024. Published online December 28, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wadman M. Public needs to prep for vaccine side effects. Science. 2020;370:1022. doi: 10.1126/science.370.6520.1022. [DOI] [PubMed] [Google Scholar]
- 102.Van Hoecke L., et al. The opposing effect of type I IFN on the T cell response by non-modified mRNA-lipoplex vaccines is determined by the route of administration. Mol. Ther. Nucleic Acids. 2020;22:373–381. doi: 10.1016/j.omtn.2020.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]