The incidence, clinical features, and severity of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection among immunosuppressed patients with glomerular disease are not well described. During the initial outbreak of SARS-CoV-2 infection in the United Kingdom, because polymerase chain reaction (PCR) testing for SARS-CoV-2 was widely available in hospital settings but not in the community, patients with mild disease generally did not receive diagnostic testing. Thus, the identification of coronavirus disease 2019 (COVID-19) based on PCR testing alone underestimates infection rates and may bias the interpretation of clinical outcomes, representing only those patients who have severe disease.
Assessing the true rates of COVID-19 within patient populations requires a combination of PCR testing at the time of disease presentation and subsequent serologic testing for SARS-CoV-2 infection. There are a large number of assays available to detect antibodies to SARS-CoV-2,S1–S8 although data on their performance in immunosuppressed patient populations are limited.
The aims of this study were to estimate the incidence of SARS-CoV-2 infection within a large cohort of patients with glomerular disease, to report their clinical features and outcomes, and to describe our experience in using 2 different serologic assays to identify antibodies to SARS-CoV-2 in these patients.
We screened 493 patients, showing that 7.5% had serological evidence of previous SARS-CoV-2 infection. Although the burden of immunosuppression was high, the majority of patients (60%) had mild disease. Overall case fatality was 7%. Thus, immunosuppression does not universally portend poor outcome from COVID-19 in this patient group. We compared 2 different serological assays and propose that receptor binding domain-based immuoassays may be preferable to detect seroconversion to SARS-CoV-2 in immunosuppressed patients.
Results
Patients With PCR-proven SARS-CoV-2 Infection Had Severe Disease and Poor Outcomes
We identified 10 patients within our clinic cohort of approximately 1500 patients who had PCR-proven COVID-19 (Table 1). Nine were receiving immunosuppression at the time of diagnosis, 7 with 2 or more agents (Supplementary Figure S1A). Most patients (70%) were B cell deplete after recent rituximab treatment and/or on corticosteroids. Eight patients required hospital admission, and 3 died (Supplementary Figure S1B and C). Of the 3 patients who died, 1 was admitted to intensive care, and the other 2 died on a general ward.
Table 1.
Patient characteristics according to severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection identified by swab testing or serologic screening
| Variable | PCR-proven SARS-Cov-2 |
Serologic screening for SARS-CoV-2 |
||
|---|---|---|---|---|
| PCR Positive (%) (n = 10) |
Abbott positive (%) (n = 31) |
Abbott indeterminate/RBD positive (%) (n = 7) |
Abbott indeterminate/RBD negative (%) (n = 18) |
|
| Sex | ||||
| Male | 4 (40) | 13 (42) | 4 (57) | 6 (33) |
| Female | 6 (60) | 18 (58) | 3 (43) | 12 (67) |
| Mean age (range), yr | 49 (19–72) | 49 (18–78) | 48 (26–65) | 49 (24–80) |
| Underlying diagnosis | ||||
| SLE | 3 (30) | 10 (33) | 2 (29) | 9 (50) |
| AAV | 3 (30) | 1 (3) | 1 (14) | 3 (17) |
| MN | 2 (20) | 6 (20) | 1 (14) | 1 (6) |
| IgAN | 1 (10) | 3 (10) | 0 | 3 (17) |
| MCD | 0 | 1 (3) | 0 | 1 (6) |
| FSGS | 1 (10) | 3 (10) | 1 (14) | 1 (6) |
| Anti-GBM | 0 | 2 (7) | 0) | 0 |
| Other | 0 | 5 (16) | 2 (29) | 0 |
| Immunosuppression | ||||
| Yes | 9 (90) | 22 (71) | 3 (43) | 13 (72) |
| ≥2 agents | 7 (70) | 4 (13) | 2 (29) | 7 (39) |
| Rituximab | 7 (70) | 3 (10) | 1 (14) | 5 (28) |
| CyP | 3 (30) | 1 (3) | 0 | 0 |
| Corticosteroids | 7 (70) | 9 (29) | 2 (29) | 6 (34) |
| Tacrolimus | 1 (10) | 3 (10) | 2 (29) | 2 (11) |
| MMF | 0 | 3 (10) | 2 (29) | 4 (22) |
| Azathioprine | 0 | 4 (13) | 0 | 4 (22) |
| MTX | 2 (20) | 3 (10) | 0 | 1 (6) |
| Symptomatic, yes | 9 (90) | 25 (81) | 6 (86) | 2 (11) |
| Fever | 8 (80) | 18 (58) | 3 (43) | 0 |
| Cough | 6 (60) | 13 (42) | 1 (14) | 1 (6) |
| Dyspnea | 6 (60) | 8 (26) | 2 (29) | 0 |
| Myalgia | 3 (30) | 6 (19) | 4 (57) | 0 |
| Fatigue | 2 (20) | 10 (32) | 3 (43) | 1 (6) |
| Diarrhea | 2 (20) | 5 (16) | 0 | 0 |
| Nausea and vomiting | 1 (10) | 3 (10) | 0 | 0 |
| Anosmia | 1 (10) | 12 (39) | 2 (29) | 1 (6) |
| Disease severity | ||||
| Asymptomatic | 1 (10) | 6 (19) | 1 (14) | 16 (89) |
| Mild | 1 (10) | 19 (61) | 6 (86) | 2 (11) |
| Moderate | 5 (50) | 6 (19) | 0 | 0 |
| Severe | 0 | 0 | 0 | 0 |
| Fatal | 3 (30) | 0 | 0 | 0 |
| Mean (range) SARS-CoV-2 IgG titer, RLUa | 3.79 (0.01–7.44) | 4.22 (1.40–9.10) | 0.87 (0.48–1.21) | 0.51 (0.26–1.15) |
AAV, antineutrophil cytoplasmic antibody–associated vasculitis; CyP, cyclophosphamide; FSGS, focal segmental glomerulosclerosis; GBM, glomerular basement membrane disease; IgAN, IgA nephropathy; MCD, minimal change disease; MMF, mycophenolate mofetil; MN, membranous nephropathy; MTX, methotrexate; PCR, polymerase chain reaction; RBD, receptor binding domain; RLU, relative light unit; SLE, systemic lupus erythematosus.
Measured using Abbott SARS-CoV-2 IgG assay.
Immunosuppressed Patients Seroconvert After PCR-proven SARS-CoV-2 Infection
All 7 patients who survived received subsequent serology testing using the Abbott (Abbott Park, IL) SARS-CoV-2 IgG assay. The mean interval between the PCR result and serologic testing was 86 days (range, 55–145 days). Five patients tested positive, and 2 tested negative (mean IgG titer = 3.90 relative light units). One of the 2 negative patients retested with the S-protein/receptor binding domain (RBD) dual antigen binding assay (DABA) assay (using the same serum samples) was positive. Thus, 6 of the 7 surviving patients seroconverted after PCR-proven SARS-CoV-2 infection.
Serologic Screening Identifies Additional Patients With Previously Undiagnosed SARS-CoV-2 Infection With Milder Disease
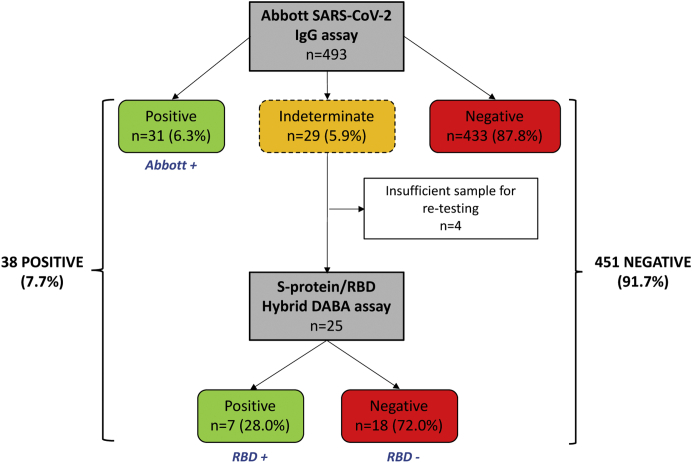
A total of 493 patients were screened for antibodies to SARS-CoV-2 using the N-protein–based Abbott IgG assay. Thirty-one patients (6.3%) had positive serology (IgG titer > 1.4 relative light units), and 29 (5.9%) had indeterminate serology (IgG titer = 0.25–1.3 relative light units). Indeterminate samples were reprocessed using the S-protein/RBD DABA assay, identifying a further 7 patients with positive serology (Figure 1). Hence, a total of 38 of 493 patients (7.7%) in our cohort had serologic evidence of previous SARS-CoV-2 infection.
Figure 1.
A flow diagram of serologic screening. A total of 493 patients attending their planned clinic appointments received serologic testing for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) during the study period. The flow diagram indicates the number of patients who tested positive, indeterminate, or negative using the Abbott SARS-CoV-2 IgG assay. Serum samples with an indeterminate test result on the Abbott assay were reprocessed using the S-protein/receptor binding domain (RBD) hybrid dual antigen binding assay (DABA) assay and the number of positive and negative results from this second assay are indicated. The 3 serologic groups that underwent subsequent comparative analysis are indicated (positive on Abbott assay [Abbott+], indeterminate on Abbott but positive on S-protein/RBD DABA assay [RBD+], and indeterminate on Abbott and negative on S-protein/RBD DABA assay [RBD−]).
Patients were analyzed in 3 groups according to their serology status (Figure 1): positive on Abbott assay (Abbott+), indeterminate on Abbott but positive on S-protein/RBD DABA assay (RBD+), and indeterminate on Abbott and negative on S-protein/RBD DABA assay (RBD−). Five of the 7 patients with PCR-proven COVID-19 who received subsequent serology testing for SARS-CoV-2 were positive on the Abbott assay and are included in the Abbott+ group in this section. Surprisingly, the remaining 2 patients were negative (rather than indeterminate) on the Abbott assay (although 1 was subsequently positive on the RBD assay) and were not included in the analysis of these 3 serologic groups.
Most patients in the Abbott+ and RBD+ groups reported prior COVID-19 symptoms, whereas the RBD− group was largely asymptomatic (Table 1). The majority of symptomatic patients reported an onset of symptoms within a month before or after the UK lockdown on 23 March 2020 (Supplementary Figure S2A). Fever, cough, anosmia, and fatigue were the most commonly reported symptoms (Supplementary Figure S2B). The overall disease severity was notably milder compared with the PCR-positive group (Table 1). A greater proportion of patients in the Abbott+ group had moderate disease compared with the RBD+ group. Conversely, patients with moderate disease developed higher IgG titers to SARS-CoV-2 (Abbott) compared with patients with mild or asymptomatic disease (Supplementary Figure S3A).
IgG titers to SARS-Cov-2 (Abbott) were significantly higher in the RBD+ compared with the RBD− group (Supplementary Figure S3B). The absence of positive serology on either immunoassay combined with the low incidence of COVID-19 symptoms in the RBD− group suggest that these patients did not have prior infection with SARS-CoV-2.
The Abbott+ and RBD+ groups were compared to determine factors that may have contributed to the difference in performance of the Abbott IgG assay. More patients in the RBD+ group were receiving immunosuppression with multiple agents at the time of testing (Table 1). However, there was no difference in patient age, male-to-female ratio, or the time interval between symptom onset and serologic testing (Supplementary Figure S4).
Overall Severity of COVID-19 Cases Identified Through PCR or Serologic Testing
Using a combination of PCR and serologic testing, we identified 43 patients who had COVID-19. This overall analysis included the 38 patients with positive SARS-CoV-2 serology (which includes the 5 PCR-positive patients who had positive serology using the Abbott assay) combined with the remaining 5 PCR-positive patients. Nineteen patients (44%) were male and 24 (56%) female, and their mean age was 49 years. Thirty-one patients (72%) were receiving immunosuppression, 11 (26%) with 2 or more agents.
With respect to overall disease severity, 6 (14%) were asymptomatic, 26 (60%) had mild disease that did not require hospital admission, 8 (19%) required hospital admission but were subsequently discharged, and a further 3 patients (7%) were admitted to the hospital and died during admission.
Discussion
In our clinic cohort of patients with glomerular disease, only 10 patients had PCR-proven SARS-CoV-2 infection. Eighty percent required hospital admission, and 3 patients died (30%).
Systematic serology screening for SARS-CoV-2 on 493 patients (performed over a 9-week period after the peak of cases in the United Kingdom) identified additional, previously undiagnosed cases of COVID-19. Thirty-eight patients (7.7%) had serologic evidence of previous SARS-CoV-2 infection. The majority reported prior symptoms compatible with COVID-19, but disease severity was notably milder compared with the PCR-proven group; only 6 of 38 patients (15.8%) required hospital admission compared with 8 of 10 (80%) in the PCR-proven group. As of December 2020, all of the patients in our clinic cohort of approximately 1500 have either attended an outpatient appointment or have been contacted by phone. We have not identified any patients who have died of COVID-19 at home or any additional cases of COVID-19 that have been diagnosed and managed at hospitals outside of our area.
Thus, the identification of COVID-19 infection based on PCR testing alone underestimated the SARS-CoV-2 infection rate in our cohort. Moreover, analysis of disease severity and clinical outcome in either PCR-proven or serologically proven groups in isolation is subject to selection bias because patients receiving PCR testing were more likely to be hospitalized, and serology testing excludes patients who died from COVID-19. A combined analysis of all cases (identified either by PCR or serologic testing) showed that the majority of patients had mild or asymptomatic disease, 19% required hospital admission (but subsequently recovered and were discharged), and the overall case fatality rate was 7% (substantially lower than the 30% mortality reported for the PCR-proven group). This provides a more accurate analysis of COVID-19 severity in immunosuppressed patients with glomerular disease and is somewhat reassuring because the use of immunosuppressive treatment does not appear to universally portend poor outcome in COVID-19 (the overall burden of immunosuppression was high). Nevertheless, the overall disease severity from SARS-CoV-2 infection in this patient group was higher than that reported for the general population. No epidemiologic studies examining COVID-19 disease severity in the general population in London have been published for direct comparison. However, a recently published, large Icelandic epidemiologic study reported that of 1797 patients identified with SARS-CoV-2 infection (with a similar male-to-female ratio and average age to our patient cohort), 5.6% were hospitalized, 1.5% required admission to the intensive therapy unit, and 0.6% died.1 Further work is now needed to understand the role of different immunosuppressive treatments, the underlying disease for which they are prescribed, and comorbidities in determining COVID-19 outcome.
Seroprevalence was 7.7% in our cohort, which was lower than that reported in London over the course of the study period (10%–15%)2 but higher than expected in a vulnerable cohort of patients who were advised to take strict “shielding” measures during the pandemic. However, we note that that all but 1 of the PCR- or serologically proven infections occurred within 4 weeks of this recommendation and the introduction of the national lockdown (Supplementary Figure S2A). The overall rate of infection was also significantly lower than in the hemodialysis population at our center (36%), who were similarly vulnerable but unable to “shield” because of their need to attend in-center hemodialysis.3 This suggests that shielding recommendations, although not completely effective, did impact the rates of infection in our patient group.
Importantly, 6 of the 7 surviving patients with PCR-confirmed infection (86%) showed seroconversion, suggesting that immunosuppressed patients with moderate disease are able to seroconvert to SARS-CoV-2 infection, even when B cell deplete at the time of COVID-19 (57% of surviving patients).
The S-protein/RBD DABA assay identified seroconversion to SARS-CoV-2 in 28% of patients testing indeterminate on the Abbott assay. The difference in performance between these 2 assays may be due to differences in the immunogenicity of the N protein compared with the S-protein RBD.4, 5, 6 Notably, patients with asymptomatic or mild disease had a weaker serologic response compared with those with moderate disease, confirming previous reports.4,7, 8, 9 Thus, an RBD antigen-based assay may be more sensitive for assessing seroconversion in immunosuppressed patients, particularly for those who have mild disease or those on multiple immunosuppressive agents.
We believe this is the first study to perform systematic serologic screening on a large cohort of patients with glomerular disease, providing a representative analysis of COVID-19 disease characteristics and severity in this immunosuppressed population and demonstrating, somewhat reassuringly, that many patients had mild illness. We compared the performance of 2 different serologic tests for SARS-CoV-2 and propose that an RBD antigen-based immunoassay is preferred in immunosuppressed patients. This should be considered in future studies performing serologic screening in these patients and in clinical practice.
Disclosure
All the authors declared no competing interests.
Acknowledgments
This research is supported by the National Institute for Health Research Imperial Biomedical Research Centre based at Imperial College Healthcare NHS Trust and Imperial College London and funded in part by a UK Research Institute Medical Research Council Grant (MC_PC_19078) and the Auchi Renal Research Fund of Imperial Health Charity at Imperial College London. TT-S is funded by a Medical Research Council grant MR/M003159/1. The authors thank the West London Kidney Patient Association, all of the patients and staff in the Specialist Glomerulonephritis Clinic at Imperial College Healthcare NHS Trust (in particular, Ms Fabiana Costa, Mr Rafael Santiago, Ms Dejin Yang, and Dr Eva Santos), and the staff within the North West London Pathology laboratories.
Footnotes
Supplementary Methods.
Figure S1. Clinical characteristics of COVID-19 and outcomes in patients with PCR-proven disease.
Figure S2. COVID-19 symptoms and timing of onset.
Figure S3. SARS-CoV-2 IgG titers measured by the Abbott assay.
Figure S4. The effect of age, sex, and testing interval on the performance of Abbott IgG SARS-CoV-2 assay.
Supplementary References
Supplementary Material
Supplementary Methods.
Figure S1. Clinical characteristics of COVID-19 and outcomes in patients with PCR-proven disease.
Figure S2. COVID-19 symptoms and timing of onset.
Figure S3. SARS-CoV-2 IgG titers measured by the Abbott assay.
Figure S4. The effect of age, sex, and testing interval on the performance of Abbott IgG SARS-CoV-2 assay.
Supplementary References
References
- 1.Eythorsson E., Helgason D., Ingvarsson R.F. Clinical spectrum of coronavirus disease 2019 in Iceland: population based cohort study. BMJ. 2020;371:m4529. doi: 10.1136/bmj.m4529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Public Health England Weekly COVID-19 Surveillance Report (Week 29) https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/901803/Weekly_COVID19_Surveillance_Report_week_29_FINAL.pdf Available at: In: ENGLAND, P. H. (Ed.), 2020.
- 3.Clarke C., Prendecki M., Dhutia A. High prevalence of asymptomatic COVID-19 infection in hemodialysis patients detected using serologic screening. J Am Soc Nephrol. 2020;31:1969–1975. doi: 10.1681/ASN.2020060827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Faustini S.E., Jossi S.E., Perez-Toledo M. Detection of antibodies to the SARS-CoV-2 spike glycoprotein in both serum and saliva enhances detection of infection [e-pub ahead of print]. medRxiv. [DOI]
- 5.Johnson M., Wagstaffe H.R., Gilmour K.C. Evaluation of a novel multiplexed assay for determining IgG levels and functional activity to SARS-CoV-2. J Clin Virol. 2020;130:104572. doi: 10.1016/j.jcv.2020.104572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rosadas C., Randell P., Khan M. Testing for responses to the wrong SARS-CoV-2 antigen? Lancet. 2020;396:e23. doi: 10.1016/S0140-6736(20)31830-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burgess S., Ponsford M.J., Gill D. Are we underestimating seroprevalence of SARS-CoV-2? BMJ. 2020;370:m3364. doi: 10.1136/bmj.m3364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sekine T., Perez-Potti A., Rivera-Ballesteros O. Robust T cell immunity in convalescent individuals with asymptomatic or mild COVID-19. Cell. 2020;183:158–168. doi: 10.1016/j.cell.2020.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Long Q.X., Tang X.J., Shi Q.L. Clinical and immunological assessment of asymptomatic SARS-CoV-2 infections. Nat Med. 2020;26:1200–1204. doi: 10.1038/s41591-020-0965-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.