Summary
Dysregulated immune cell responses have been linked to the severity of coronavirus disease 2019 (COVID-19), but the specific viral factors of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) were currently unknown. Herein, we reveal that the Immunoglobulin-like fold ectodomain of the viral protein SARS-CoV-2 ORF7a interacts with high efficiency to CD14+ monocytes in human peripheral blood, compared to pathogenic protein SARS-CoV ORF7a. The crystal structure of SARS-CoV-2 ORF7a at 2.2 Å resolution reveals three remarkable changes on the amphipathic side of the four-stranded β-sheet, implying a potential functional interface of the viral protein. Importantly, SARS-CoV-2 ORF7a coincubation with CD14+ monocytes ex vivo triggered a decrease in HLA-DR/DP/DQ expression levels and upregulated significant production of proinflammatory cytokines, including IL-6, IL-1β, IL-8, and TNF-α. Our work demonstrates that SARS-CoV-2 ORF7a is an immunomodulating factor for immune cell binding and triggers dramatic inflammatory responses, providing promising therapeutic drug targets for pandemic COVID-19.
Subject areas: Immunology, Virology
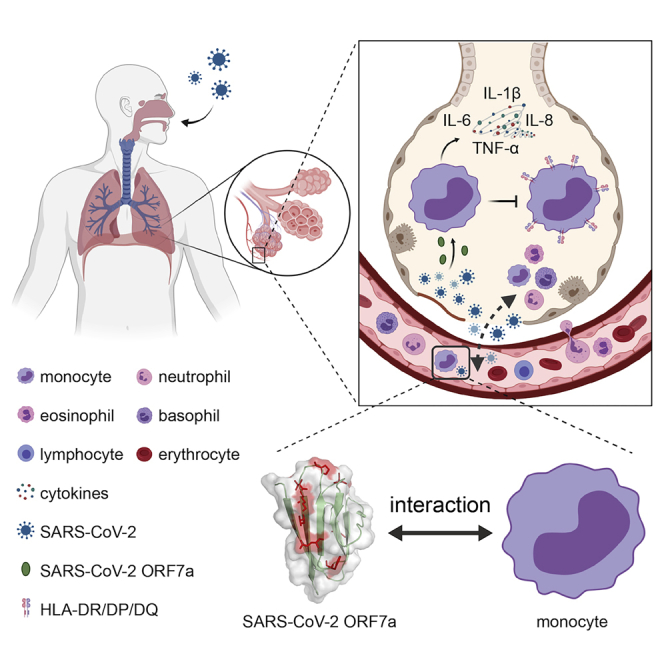
Graphical abstract
Highlights
-
•
Ig-like SARS-CoV-2 ORF7a efficiently interacts with CD14+ monocytes from PBMCs
-
•
SARS-CoV-2 ORF7a structure shows distinct binding pattern to specific immune cells
-
•
SARS-CoV-2 ORF7a triggers the antigen-presenting suppression of CD14+ monocytes
-
•
SARS-CoV-2 ORF7a is involved in cytokine storm propagation of patients with COVID-19
Immunology; Virology
Introduction
The pandemic COVID-19 outbreak, caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has resulted in a total of 41,570,883 confirmed cases and 1,134,940 deaths all over the world, as of October 23, 2020 (World Health Organization, 2020). The worldwide spread of SARS-CoV-2 infections has led to disastrous consequences for public health (Guan et al., 2020). It is estimated that COVID-19 is twenty-fold more deadly than seasonal influenza virus infections based on mid-April reported cases (Faust and Del Rio, 2020). People with COVID-19 have typical symptoms of fever, dry cough, myalgia, shortness of breath, and pneumonia. Critically ill patients with COVID-19 often develop acute respiratory distress syndrome (ARDS) (Chen et al., 2020b; Huang et al., 2020).
Monocytes are innate immune cells that participate in the complex functional processes of inflammatory responses, including phagocytosis, antigen presentation, and cytokine secretion, within the circulation or after their migration into tissues and lymphoid organs (Jakubzick et al., 2017). Several studies have characterized monocytes in the immunopathology of patients with COVID-19. A recent study found that the percentage of CD14+CD16+ inflammatory monocytes in the peripheral blood was higher in the 33 hospitalized patients diagnosed with COVID-19, especially in intensive care unit patients (Zhou et al., 2020). Additionally, a significant increase in IL-6 production of inflammatory CD14+CD16+ monocytes was noted in patients with mild and severe COVID-19, suggesting that monocytes are key contributors to cytokine storms in COVID-19 (Zhou et al., 2020). Similarly, several researchers found a substantial decrease in circulating monocytes and specific enrichment of monocytes in severe and critical COVID-19 patient tissues (Sanchez-Cerrillo et al., 2020). An observational study of 54 individuals infected with SARS-CoV-2 found evidence of profound immune dysregulation mediated by monocyte dysfunction in all these patients (Giamarellos-Bourboulis et al., 2020). Recent studies based on single-cell transcriptome technology reported several unique interferon response-enriched monocyte clusters that correlated with the severity of the disease (Arunachalam et al., 2020; Schulte-Schrepping et al., 2020; Silvin et al., 2020). These results suggest a link between activated monocytes in tissues and the pathogenesis of severe COVID-19.
Immunoglobulin-like (Ig-like) domain-containing proteins play vital roles in mediating macromolecular interactions in the immune system. Indeed, viruses can evolve a series of countermeasures featuring Ig-like domains to evade or hijack the host immune system, delaying or attenuating the host immune response. Previous data suggest that the SARS-CoV Ig-like viral protein ORF7a interacts with immune cells, as mediated by its interaction with integrin LFA-1 (Hanel et al., 2006; Hanel and Willbold, 2007; Shimaoka et al., 2003). SARS-CoV-2 ORF7a, similar to SARS-CoV ORF7a, is predicted to be a member of the Ig-like domain superfamily (Huang et al., 2006; Ito et al., 2005; Nelson et al., 2005; Schaecher et al., 2007; Tan et al., 2020). Although numerous structural studies have been performed for some other SARS-CoV-2 viral proteins (i.e., spike protein and nucleocapsid protein) (Kang et al., 2020; Lan et al., 2020; Walls et al., 2020; Wrapp et al., 2020; Yan et al., 2020), little is known about the structural or functional aspects of the Ig-like viral protein SARS-CoV-2 ORF7a, especially related to its potential ability to bind to immune cells.
Herein, we discovered high binding efficiency of SARS-CoV-2 ORF7a to human monocytes via viral protein-based flow cytometry and cell surface marker analysis but did not find similar results for SARS-CoV ORF7a. We then solved the crystal structure of SARS-CoV-2 ORF7a at 2.2 Å resolution. Structure-based superimposition indicates that the residue variations on the potential intramolecular interfaces are probably critical for its binding capability. Furthermore, we sought to identify the biological significance of SARS-CoV-2 ORF7a–leukocyte interactions and subsequent immune responses, which may provide potential therapeutic targets for pandemic COVID-19.
Results
The binding capability of Ig-like fold protein SARS-CoV-2 ORF7a to human leukocytes
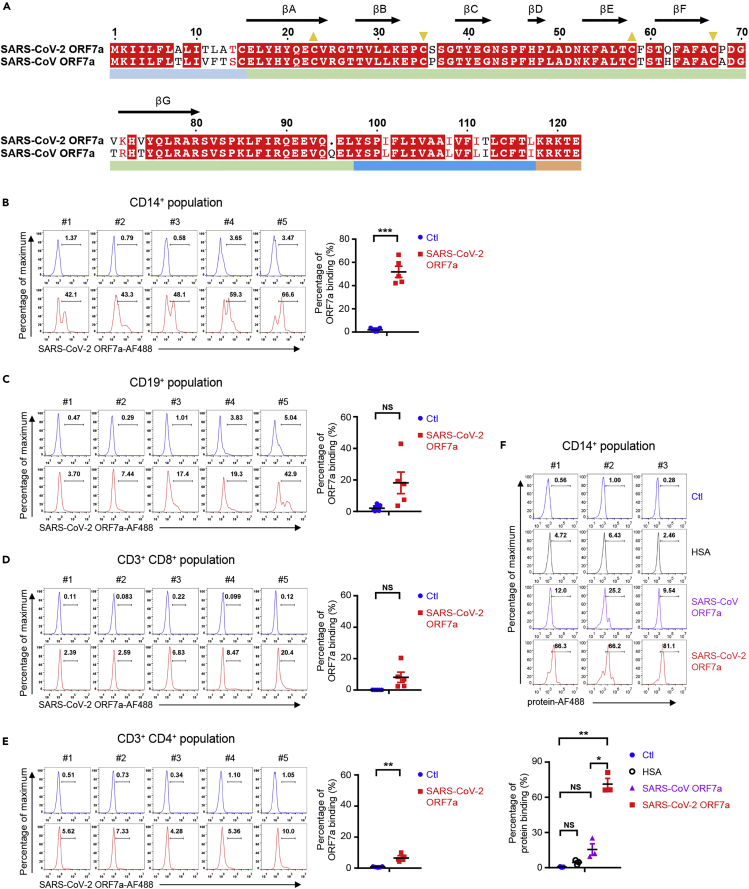
Similar to SARS-CoV ORF7a, SARS-CoV-2 ORF7a is a type-I transmembrane protein with 121 amino acid residues, consisting of an N-terminal signaling region (residues 1–15), an Ig-like ectodomain (residues 16–96), a hydrophobic transmembrane domain (residues 97–116), and a typical ER retention motif (residues 117–121) (Figure S1A). The immunoglobulin domain has been found in many other proteins with diverse biological functions, such as intercellular adhesion, identification, or binding. However, it is rarely recognized among the human pathogenic coronavirus proteins except as an accessory protein. The sequence alignment of SARS-CoV-2 ORF7a with SARS-CoV ORF7a displays a conserved Ig-like β-sandwich fold with seven β-strands (Figure 1A). Most of the sequence variations are distributed on the ectodomain (8 residue variations, as shown in Figure 1A), implying potentially diverse protein functions.
Figure 1.
Sequence alignment and human PBMC binding assay of SARS-CoV-2 ORF7a
(A) Sequence alignment of SARS-CoV-2 ORF7a (UniProt: A0A6B9VSS2) with SARS-CoV ORF7a (UniProt: P59635). Yellow triangles in the same direction indicate the paired cysteines composing disulfide bonds. The arrows with labels denote the β-strand regions of the structure. The color bars below the alignment denote the corresponding domains.
(B–E) Human PBMCs from healthy donors (n = 5) were incubated with or without Alexa Fluor 488-coupled SARS-CoV-2 ORF7a protein (SARS-CoV-2 ORF7a-AF488) for 2 hr, and then the binding ability was analyzed via flow cytometry. Various immune cells were gated by specific antibodies, e.g., CD14+ monocytes (B), CD19+ B cells (C), CD3+ CD8+ T cells (D), and CD3+CD4+ T cells (E).
(F) Human PBMCs from healthy donors (n = 3) were incubated with Alexa Fluor 488-coupled human serum albumin (HSA), SARS-CoV ORF7a, or SARS-CoV-2 ORF7a protein for 2 hr, and then the binding ability of the CD14+ monocytes was analyzed via flow cytometry. All data were analyzed using paired two-tailed Student's t-tests, and the error bars show the means ± s.e.m. (∗, p < 0.05; ∗∗, p < 0.01; ∗∗∗, p < 0.001; NS, nonsignificant).
See also Figure S1.
To investigate whether the SARS-CoV-2 ORF7a ectodomain is involved in the interaction of SARS-CoV-2 with immune cells, human peripheral blood mononuclear cells (PBMCs) from healthy donors (without SARS-CoV-2 infection history) were incubated with fluorescence-labeled SARS-CoV-2 ORF7a ectodomain protein (residues 14–96) and then analyzed via flow cytometry. The SARS-CoV-2 ORF7a coupled with fluorescent dye bound to CD14+ monocytes from human PBMCs with the highest efficiency (Figure 1B), while much weaker signals were detected on CD19+ B cells, CD8+ T cells, and CD4+ T cells (Figures 1C–1E). Intriguingly, SARS-CoV ORF7a showed a significantly lower binding ability to CD14+ monocytes comparing to SARS-CoV-2 ORF7a (Figure 1F), reflecting a difference in their direct interactions with CD14+ monocytes.
Crystal structure of SARS-CoV-2 ORF7a
To explore the mechanism of the differential binding capabilities of SARS-CoV-2 ORF7a and SARS-CoV ORF7a to human CD14+ monocytes, we solved the structure of SARS-CoV-2 ORF7a at 2.2 Å resolution using X-ray crystallography technology. Briefly, residues 14–96 of the SARS-CoV-2 ORF7a protein were constructed, expressed, and purified as described in the denatured refolding protocol (Transparent Methods). The structure was determined by molecular replacement using the SARS-CoV ORF7a structure (PDB: 1XAK) as the search model (Nelson et al., 2005). The final structure, with a visible electron density spanning residues 14–82, was refined to R factor and R-free values of 0.201 and 0.246, respectively. All the statistics for the data collection, phasing, and refinement are presented in Table S1.
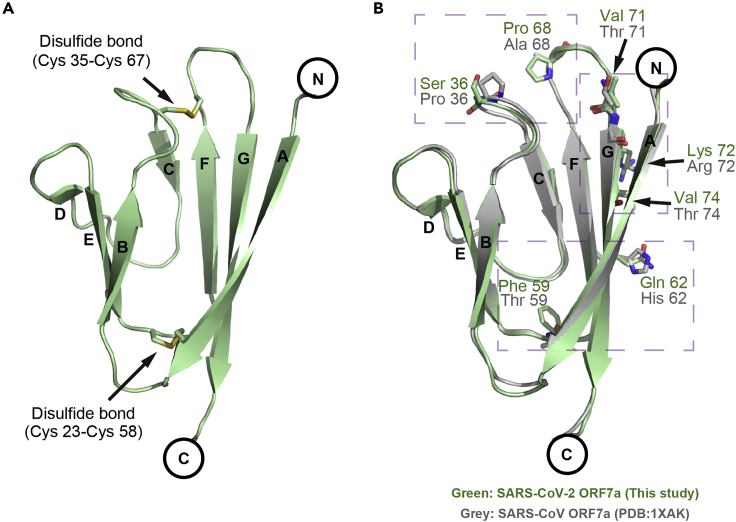
The overall structure of the SARS-CoV-2 ORF7a consists of seven β-strands divided into two tightly packed β-sheets (Figures 2A and S1B). Strands A, G, F, and C form the larger 4-stranded β-sheet, while strands B, E, and D compose the smaller 3-stranded β-sheet. Two disulfide bonds (formed by residues Cys23-Cys58 and Cys35-Cys67) stabilize the β-sheet structure “sandwich” by connecting strand A to the EF loop and the BC loop to the FG loop (Figures 2A and S1B). The SARS-CoV-2 ORF7a structure is similar to SARS-CoV ORF7a with a root-mean-square deviation (RMSD) of 0.48 Å as determined by secondary structure matching superimposition of 68 aligned residues in Coot software (Emsley and Cowtan, 2004). Compared with SARS-CoV ORF7a, the varied residues were distributed on the amphipathic side of the larger 4-stranded β-sheet (strands A, G, F, and C) or the loop regions, with none located on the 3-stranded β-sheet (strands B, E, or D) (Figure 2B), implying that the key residues are located on the major functional interface.
Figure 2.
Structural overview and alignment of the SARS-CoV-2 ORF7a
(A) Ribbon representation of a SARS-CoV-2 ORF7a molecule. Disulfide bonds are represented with stick models. The β-strands are assigned in alphabetical order from the N-terminus to the C-terminus.
(B) Structural alignment between SARS-CoV-2 ORF7a (this study) and SARS-CoV ORF7a (PDB: 1XAK). The diverse residues are shown as sticks. The residues of SARS-CoV-2 ORF7a are colored green, whereas the residues of SARS-CoV ORF7a (PDB: 1XAK) are colored gray. The purple dashed boxes indicate the diverse residues for the subsequent detailed view.
See also Table S1.
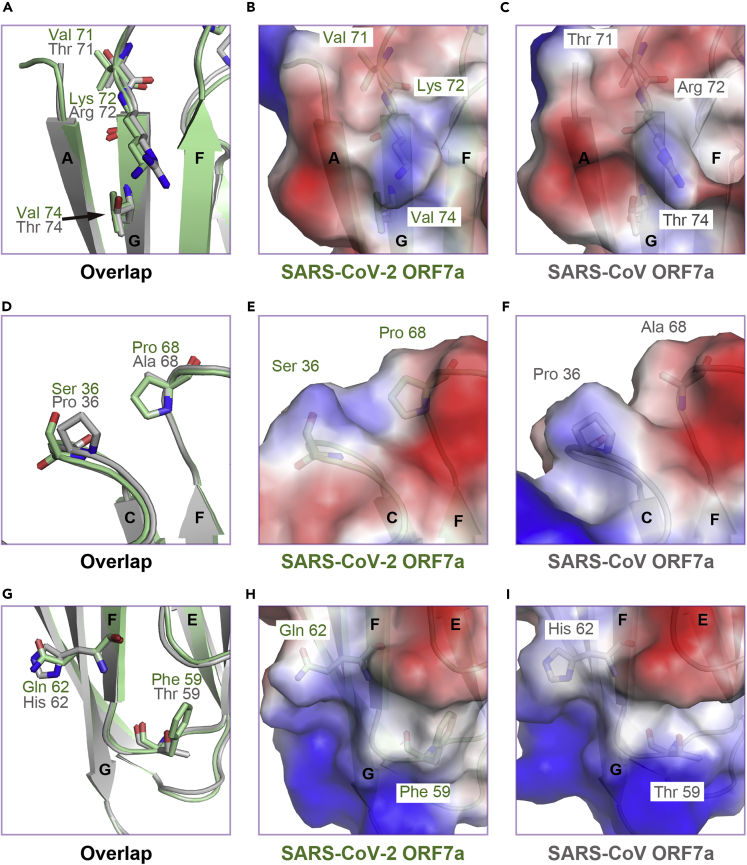
Three major differences were highlighted upon superimposition (Figure 2B). First, SARS-CoV-2 ORF7a residues Val71, Lys72, and Val74 composed a nonpolar network on β-strand G in the 4-stranded β-sheet, whereas the equivalent residues of SARS-CoV ORF7a comprised a relatively polar surface (Figures 3A–3C). Second, the SARS-CoV-2 ORF7a residue Ser36 in the BC loop and Pro68 in the FG loop displayed a neutral and flat electrostatic potential surface compared with the equivalent residues in SARS-CoV ORF7a (Figures 3D–3F). Last, the SARS-CoV-2 ORF7a residue Gln62 on β-strand F in the 4-stranded β-sheet increased the polarity of the surface compared with SARS-CoV ORF7a, and Phe59 in the EF loop increased the hydrophobicity of the surface compared with that of SARS-CoV ORF7a (Figures 3G–3I). In general, varying the residues resulted in different electrostatic potentials on the surface (Figures S1C–S1E), although the overall structures of SARS-CoV-2 ORF7a and SARS-CoV ORF7a display a similar Ig-like fold (Hanel et al., 2006; Nelson et al., 2005).
Figure 3.
Distinct features of diverse residues between SARS-CoV-2 ORF7a and SARS-CoV ORF7a
(A) Superimposition of diverse residues on strand βG, which are depicted as ball-and-stick models.
(B and C) Semitransparency electrostatic potential mapped onto the accessible molecular surface of SARS-CoV-2 ORF7a (B) and SARS-CoV ORF7a (C). Blue denotes a net positive charge (+5.6 kT), and red denotes a negative charge (−5.6 kT). The feature residues are shown as transparency ball-and-stick models.
(D–F) Superimposition of diverse residues on the BC loop and FG loop.
(G–I) Superimposition of diverse residues on strand βF and the EF loop.
See also Figure S1.
A distinct binding pattern of the SARS-CoV-2 ORF7a ectodomain to human leukocytes
To identify the proteins with structural similarity to the SARS-CoV-2 ORF7a, we next performed network service DALI searching of the PDB database (Holm, 2020). Although the folds with the greatest similarity are adhesion-related molecules, the sequence identity of SARS-CoV-2 ORF7a to that of structurally similar proteins is low (Figure S2A). Among the top 3 structures with Z-scores greater than 6.1, the identical structures are SARS-CoV ORF7a (Hanel et al., 2006) (PDB: 1YO4, residues 1–86, Z score of 10.9, RMSD of 1.3 Å for 69 aligned residues, with 87% sequence identity), a bacterial adhesin MpAFP RIII-2 domain (Guo et al., 2017) (PDB: 5K8G, residues 1–511, Z score of 6.2, RMSD of 2.5 Å for 65 aligned residues, with 11% sequence identity), and an intercellular adhesion molecule with an ICAM3 D1 domain (Song et al., 2005) (PDB: 1T0P, residues 1–86, Z score of 6.1, RMSD of 2.5 Å for 65 aligned residues, with 8% sequence identity) (Table S2).
The MpAFP protein-mediated adherence of Marinomonas primoryensis to ice occurs by binding to Ca2+, and the RIII region of MpAFP is a Ca2+-stabilized Y-shaped ligand-binding site. The ICAM3 protein is constitutively expressed on resting leukocytes and mediates firm leukocyte-endothelial cell adherence through divalent cation-dependent interactions with integrin in different cellular settings. Consistent with our results, previous evidence has suggested that the ICAM adhesion family protein (i.e., ICAM1 (PDB: 1MQ8) (Shimaoka et al., 2003)) was the best hit for the closely related viral protein SARS-CoV ORF7a when DALI online homological structural proteins were searched (Hanel et al., 2006; Nelson et al., 2005). ICAM-integrin interactions regulate leukocyte trafficking and infiltration, which are important host defense mechanisms against infectious agents.
To further validate the similarity of these proteins, we compared their topological and tertiary structures. SARS-CoV-2 ORF7a shows a unique topological structure with the C-F-G-A and B-E-D sheets compared to the D-E-B-A and (A′)-G-F-C sheets of ICAM3 or the F-G-A and B-E-D-C sheets of MpAFP (Figures S2B–S2D). Although the DALI results suggest that the ICAM3 D1 domain has a score similar to that of the MpAFP RIII-2 domain in SARS-CoV-2 ORF7a, suggesting structural similarity, the superimposition results are quite different. The orientation of the β-sheets in SARS-CoV-2 ORF7a and the ICAM3 D1 domain is different (Figure S2E), whereas strands F-G-A and B-E-D in the MpAFP RIII-2 domain are perfectly aligned with their SARS-CoV-2 ORF7a counterparts (Figures S2D and S2F). Also, we compared SARS-CoV-2 ORF7a to ICAM1, ICAM2, and IL-1R, respectively (Figure S3), which were found to be the most similar to SARS-CoV ORF7a (Hanel et al., 2006). Despite ICAM2 and ICAM3 both belong to the ICAM adhesion family, there are still minor differences between them. The main difference between ICAM2 and ICAM3 is the strand βC and the BC loop (Figures S2E and S3B), leading to different DALI searching results between SARS-CoV-2 ORF7a and SARS-CoV ORF7a. Collectively, these superimposition results suggest that SARS-CoV-2 ORF7a may play a vital role in recognizing macromolecules on the cellular surface, as the three most-similar structures are all adhesion molecules.
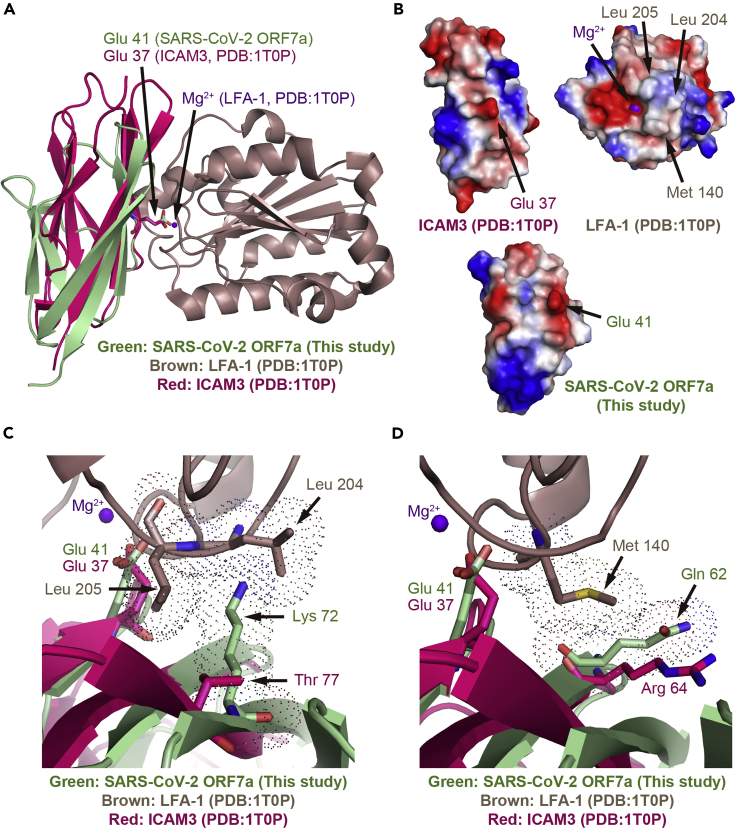
To explore the potential roles of diverse residues in the SARS-CoV-2 ORF7a ectodomain, we next examined the structural superimpositions between SARS-CoV-2 ORF7a and the complex structure of ICAM3 D1–LFA-1. Previous studies have indicated that SARS-CoV ORF7a interacted with the integrin LFA-1 in leukocytes (Hanel and Willbold, 2007). Therefore, we aligned the key residues Glu41 or Glu37 between these two structures (Figure 4A). As shown in Figures S1C, S1D, and 4A, the varied residues were located on the same surface facing toward the LFA-1 interaction sites. LFA-1 binding is mediated by Mg2+ via the recognition of Glu37 of ICAM3 in the ICAM3–LFA-1 complex (Figures 4A and 4B). The varied residues Lys72 and Gln62 of SARS-CoV-2 ORF7a show significant clashes to the counterparts Leu204 and Met140 of LFA-1 at the interaction surface, respectively (Figures 4C and 4D). These results suggest that the interaction pattern of SARS-CoV-2 ORF7a to leukocytes may not be similar to the ICAM3–LFA-1 complex.
Figure 4.
Structural superimposition and electrostatic surface comparison of SARS-CoV-2 ORF7a to the crystal structure of ICAM3–LFA-1 complex
(A) Structural superimposition of SARS-CoV-2 ORF7a with the ICAM3 in the crystal structure of the ICAM3–LFA-1 complex.
(B) Electrostatic surface comparison of SARS-CoV-2 ORF7a with the crystal structure of ICAM3–LFA-1 complex. The ICAM3–LFA-1 complex is split with the interaction interfaces from a front view perspective. The electrostatic surface of SARS-CoV-2 ORF7a was generated based on its structural superimposition to ICAM3.
(C) View of the detailed interactions of the varied residues (Lys72) in SARS-CoV-2 ORF7a with the nonpolar residues (Leu204 and Leu205) in LFA-1.
(D) Detailed interaction view of the varied residues (Gln62) in SARS-CoV-2 ORF7a with the nonpolar residues (Met140) in LFA-1. Dot representation indicates steric occupation of the residues. LFA-1 is in brown, ICAM3 is in red, and Mg2+ ions are in purple.
See also Figures S2–S6 and Table S2.
To test the hypothesis suggesting structural dissimilarities in LFA-1 interactions, we carried out several in vitro protein–protein interaction (PPI) analyses using the recombinant active or inactive form of the LFA-1 αL I-domain interacting with the SARS-CoV-2 or SARS-CoV ORF7a ectodomain. We performed surface plasmon resonance analysis (Figure S4), protein pull-down assays (Figure S5A), biolayer interferometry assays (Figures S5B and S5C), and size exclusive chromatography analysis (Figures S5D–S5G). All of the results suggested little interaction between LFA-1 and SARS-CoV-2 ORF7a, indicating that the interaction of SARS-CoV-2 ORF7a with immune cells is independent of LFA-1.
SARS-CoV-2 ORF7a triggers immune responses by human monocytes
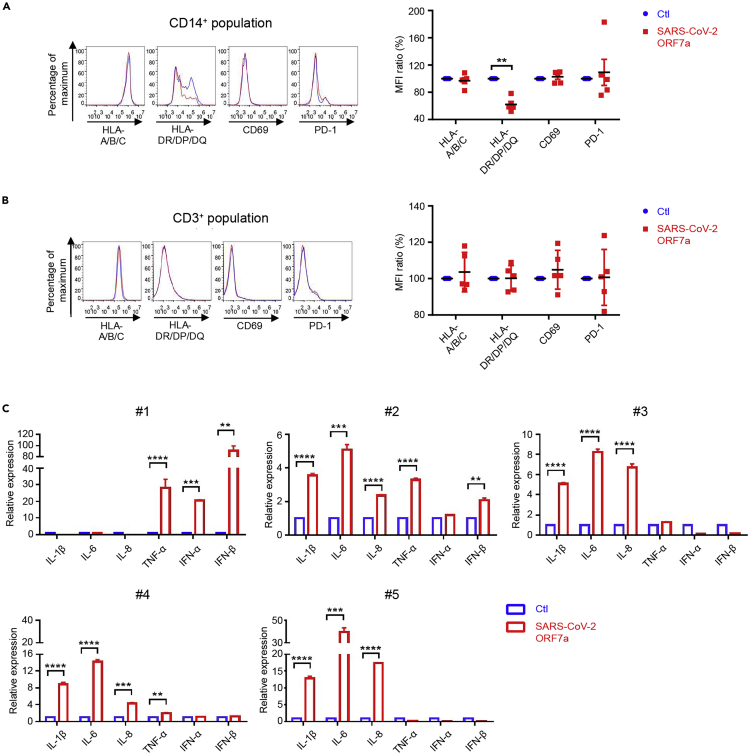
To investigate the biological significance of the SARS-CoV-2 ORF7a protein binding to immune cells, the ORF7a protein ectodomain was incubated with human PBMCs from healthy donors for 24 hr, and the expression levels of the surface molecules on the PBMCs were then analyzed by flow cytometry. As shown in Figure 5A, the antigen-presenting-related cell surface molecules HLA-DR/DP/DQ on CD14+ monocytes were significantly decreased, by approximately 30%, after SARS-CoV-2 ORF7a incubation, whereas HLA-A/B/C expression was unchanged. Furthermore, neither the lymphocyte activation marker CD69 nor the exhaustion marker PD-1 significantly changed upon SARS-CoV-2 ORF7a incubation. Additionally, none of these molecules were altered on the CD3+ T cells (Figure 5B). Moreover, the expression of various cytokines was tested by quantitative real-time polymerase chain reaction (qRT-PCR) after SARS-CoV-2 ORF7a incubation overnight. Intriguingly, SARS-CoV-2 ORF7a coincubation with monocytes triggered the significant upregulation of the proinflammatory cytokines IL-6, IL-1β, IL-8, and TNF-α (Figure 5C), the most abundant cytokines propagated during cytokine storms in SARS-CoV-2 infection (Chen et al., 2020a; Huang et al., 2020; Qin et al., 2020). The PBMCs from different donors showed diverse proinflammatory cytokine profiles. These outcomes are consistent with the previous reports, which demonstrated that SARS-CoV-2 infection triggered distinct proinflammatory parameters in different patients (Giamarellos-Bourboulis et al., 2020; Huang et al., 2020; Silvin et al., 2020). Taken together, the data suggest that the SARS-CoV-2 ORF7a ectodomain directly interacts with human immune cells, leading to decreased antigen-presenting ability and inducing the dramatic expression of proinflammatory cytokines by human CD14+ monocytes.
Figure 5.
SARS-CoV-2 ORF7a triggers immune responses
(A and B) Human PBMCs from healthy donors (n = 5) were incubated with or without purified SARS-CoV-2 ORF7a protein for 24 hr, and the expression of the different cell surface molecules was analyzed via flow cytometry. Blue lines represent the staining signal of the negative control group, and red lines denote the SARS-CoV-2 ORF7a protein treatment group.
(C) Monocytes isolated from healthy individual donor PBMCs were incubated with or without purified SARS-CoV-2 ORF7a protein for 24 hr, and differential gene expression was analyzed via qRT-PCR. All data were analyzed using paired two-tailed Student's t-tests, and the error bars show the means ± s.e.m. (∗∗, p < 0.01; ∗∗∗, p < 0.001; ∗∗∗∗, p < 0.0001). The flow cytometry pictures are representative of one donor in (A) and (B).
See also Table S3.
Discussion
With a report of 72,314 cases from the Chinese Center for Disease Control and Prevention, the fatality rate of patients in critical condition was exceptionally high at 49% (Epidemiology Working Group for NCIP Epidemic Response, Chinese Center for Disease Control and Prevention, 2020). The excessive inflammatory response to SARS-CoV-2 is thought to also be a preeminent cause of disease severity and mortality in patients with COVID-19 (Mehta et al., 2020). It is associated with high levels of cytokines, profound lymphopenia, and substantial monocyte or macrophage infiltration into the lungs, heart, spleen, lymph nodes, and kidney, as observed during postmortem analysis (Xu et al., 2020). These results imply that the aberrant inflammatory immune response is important for disease progression, but the host factor or viral protein that mediates this process remains to be elucidated.
From the published literature (Huang et al., 2006), Huang et al. found that SARS-CoV ORF7a was a viral structural protein incorporated into the viral particles. They analyzed the supernatant of SARS-CoV infected Caco2 cells via sucrose gradient centrifugation, and found that SARS-CoV ORF7a was present in the fractions containing S, N, and M structural protein. To further confirm the presence of SARS-CoV ORF7a, they purified the viral particles by anti-spike monoclonal antibodies. The subsequent western blotting assays with specific anti-7a antibodies suggested that SARS-CoV ORF7a was a novel structural protein in viral particles. Herein, SARS-CoV-2 ORF7a and SARS-CoV ORF7a share an identical ER retention motif (Figure 1A, the C-terminal KRKTE sequence), which suggests SARS-CoV-2 ORF7a may be incorporated in the viral envelope as SARS-CoV ORF7a. However, it is unclear about the functions of SARS-CoV-2 ORF7a as a potential structural protein.
For the first time, to our knowledge, we demonstrate that SARS-CoV-2 ORF7a ectodomain binds to CD14+ monocytes in human peripheral blood with high efficiency (Figures 1B and 1F), whereas SARS-CoV ORF7a ectodomain shows a significantly lower binding ability to monocytes (Figure 1F). Monocytes are pivotal for the immunopathology of COVID-19, as shown in hospitalized patients with COVID-19 and postmortem samples (Giamarellos-Bourboulis et al., 2020; Sanchez-Cerrillo et al., 2020; Zhou et al., 2020). By using single-cell RNA sequencing, a recent study found that proinflammatory monocyte-derived macrophages were significantly enriched in bronchoalveolar lavage fluid from patients with severe COVID-19 compared to patients with mild disease and healthy controls (Liao et al., 2020). As the lung is the major site for SARS-CoV-2 replication and virus production, the direct interactions between SARS-CoV-2 ORF7a and monocytes indicate that SARS-CoV-2 ORF7a is probably a key viral factor that contributes to the recruitment of monocytes to infected lungs during COVID-19.
We further solved the crystal structure of SARS-CoV-2 accessory protein ORF7a ectodomain, a COVID-19 factor, to explore the mechanism of the different binding capabilities of SARS-CoV-2 ORF7a and SARS-CoV ORF7a to human monocytes. Considering the structure alignment results, we discovered that SARS-CoV-2 ORF7a is structurally similar to SARS-CoV ORF7a (PDB: 1XAK). All of these varied residues are mainly distributed at the amphipathic side of the larger 4-stranded β-sheet, which may be the potential functional interface. This structure is very likely to be basis of the molecular mechanism underlying the different pathological processes of SARS strains. However, the structural alignment results between SARS-CoV-2 ORF7a with ICAM proteins, the similar protein family to SARS-CoV ORF7a, suggest that the interaction pattern of SARS-CoV-2 ORF7a to leukocytes may not be similar to the ICAM–LFA-1 complex. Indeed, four kinds of in vitro PPI assays support this observation. All of the assays demonstrate the interactions of the SARS-CoV-2 ORF7a to leukocytes are independent of LFA-1.
To further investigate the consequence of the viral protein bound to monocytes, we next explored the immune response of human monocytes ex vivo. SARS-CoV-2 ORF7a binding to CD14+ monocytes led to a significant decrease in the HLA-DR/DP/DQ molecules on CD14+ monocytes, which suggests that ORF7a may suppress the antigen-presenting ability of these monocytes (Figure 5A). It will be interesting to functionally characterize how SARS-CoV-2 ORF7a modulates the antigen-presenting ability of immune cells in future studies. Consistent with our results, a recent study on COVID-19 patients with severe respiratory failure suggests that severe respiratory failure is associated with a major decrease in HLA-DR on CD14+ monocytes as a result of immune dysregulation of hyperinflammatory responses (Giamarellos-Bourboulis et al., 2020). Other recent works also confirmed the accumulation of CD14+HLA-DRLow monocytes in patients with severe COVID-19 via single-cell RNA sequencing and single-cell proteomics (Arunachalam et al., 2020; Schulte-Schrepping et al., 2020; Silvin et al., 2020). Impressively, SARS-CoV-2 ORF7a coincubation with monocytes triggers the significant upregulation of multiple proinflammatory cytokines, indicating that ORF7a might be a key factor in cytokine storm progression in COVID-19 (Guan et al., 2020; Huang et al., 2020; Qin et al., 2020). Taken together, we presume that the potential immunomodulatory ability of SARS-CoV-2 ORF7a on human monocytes serves as a viral immune escape strategy. To be noted, it may not be the soluble ectodomain of ORF7a functions as circulating and interacting with monocytes, but the ORF7a at the viral particle surface potentially does. Although the specific molecules that bind to SARS-CoV-2 ORF7a require further investigation, our work has identified the novel roles of the viral ORF7a protein in the immunopathology of COVID-19, and SARS-CoV-2 ORF7a may be a promising therapeutic target for pandemic COVID-19.
Limitations of the study
This study demonstrated that SARS-CoV-2 ORF7a was essential for the binding of SARS-CoV-2 to human CD14+ monocytes. Although we indicated that SARS-CoV-2 ORF7a triggers the antigen-presenting suppression of CD14+ monocytes and is involved in cytokine storm propagation of patients with COVID-19, we could not specify the direct binding molecule.
Resource availability
Lead contact
The lead contact is Shoudeng Chen (chenshd5@mail.sysu.edu.cn).
Materials availability
Materials generated in this study are available from the lead contact with a completed Materials Transfer Agreement.
Data and code availability
The accession number for the structure reported in this paper is Protein Data Bank (PDB): 7CI3.
Methods
All methods can be found in the accompanying Transparent Methods supplemental file.
Acknowledgments
This work was supported by the National Key R&D Program of China (2019YFA0110300, 2020YFA0509400) granted to J.C.; the Special Fund for Scientific and Technological Innovation Strategy of Guangdong Province of China (2018B030306029) and COVID-19 Emerging Prevention Products, Research Special Fund of Zhuhai City (ZH22036302200016PWC) granted to S.C.; the National Natural Science Foundation of China (82071745), the Fundamental Research Funds for the Central Universities (19ykzd36) and the Science and Technology Program of Guangzhou (202002030069) and a grant from MOE Key Laboratory of Gene Function and Regulation granted to J.C. This study was approved by The Medical Ethics Committee of The Fifth Affiliated Hospital, Sun Yat-sen University (2020-K195-1). We thank the staff of the BL18U1 beamline at the Shanghai Synchrotron Radiation Facility (SSRF) for their help with X-ray diffraction data collection. The graphical abstract was created with the help of BioRender.com.
Author contributions
S.C. and J.C. contributed to the conception of the study. S.C., J.C., Ziliang Z., C.H., and Zhechong Z. generated the outline of the article, designed the experiments, drew the figures, and wrote the manuscript. Ziliang Z. and Zhechong Z. performed the protein purification and crystallization experiments, PPIs in vitro, and the biochemical tests. S.C. and Ziliang Z. performed the structural determination and validation. C.H. carried out the cellular experiments. Z.H. prepared the materials for the experiments. L.S. and X.R. assisted in collecting the peripheral blood samples from The Health Management Center, The Fifth Affiliated Hospital, Sun Yat-sen University and Guangzhou Blood Center, respectively. F.X., S.K., X.C., Q.C., and S.H. helped to analyze the experimental data.
Declaration of interests
The authors declare no competing interests.
Published: March 19, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2021.102187.
Contributor Information
Jun Chen, Email: chenjun23@mail.sysu.edu.cn.
Shoudeng Chen, Email: chenshd5@mail.sysu.edu.cn.
Supplemental information
References
- Arunachalam P.S., Wimmers F., Mok C.K.P., Perera R., Scott M., Hagan T., Sigal N., Feng Y., Bristow L., Tak-Yin Tsang O. Systems biological assessment of immunity to mild versus severe COVID-19 infection in humans. Science. 2020;369:1210–1220. doi: 10.1126/science.abc6261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G., Wu D., Guo W., Cao Y., Huang D., Wang H., Wang T., Zhang X., Chen H., Yu H. Clinical and immunological features of severe and moderate coronavirus disease 2019. J. Clin. Invest. 2020;130:2620–2629. doi: 10.1172/JCI137244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen N., Zhou M., Dong X., Qu J., Gong F., Han Y., Qiu Y., Wang J., Liu Y., Wei Y. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. The Lancet. 2020;395:507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P., Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D. Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Epidemiology Working Group for NCIP Epidemic Response, Chinese Center for Disease Control and Prevention The epidemiological characteristics of an outbreak of 2019 novel coronavirus diseases (COVID-19) in China. Zhonghua. Liu. Xing. Bing. Xue. Za. Zhi. 2020;41:145–151. doi: 10.3760/cma.j.issn.0254-6450.2020.02.003. [DOI] [PubMed] [Google Scholar]
- Faust J.S., Del Rio C. Assessment of deaths from COVID-19 and from seasonal influenza. JAMA Intern. Med. 2020;180:1045–1046. doi: 10.1001/jamainternmed.2020.2306. [DOI] [PubMed] [Google Scholar]
- Giamarellos-Bourboulis E.J., Netea M.G., Rovina N., Akinosoglou K., Antoniadou A., Antonakos N., Damoraki G., Gkavogianni T., Adami M.E., Katsaounou P. Complex immune dysregulation in COVID-19 patients with severe respiratory failure. Cell Host Microbe. 2020;27:992–1000. doi: 10.1016/j.chom.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan W., Ni Z., Hu Y., Liang W., Ou C., He J., Liu L., Shan H., Lei C., Hui D.S.C. Clinical characteristics of coronavirus disease 2019 in China. N. Engl. J. Med. 2020;382:1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo S.Q., Stevens C.A., Vance T.D.R., Olijve L.L.C., Graham L.A., Campbell R.L., Yazdi S.R., Escobedo C., Bar-Dolev M., Yashunsky V. Structure of a 1.5-MDa adhesin that binds its Antarctic bacterium to diatoms and ice. Sci. Adv. 2017;3:e1701440. doi: 10.1126/sciadv.1701440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanel K., Stangler T., Stoldt M., Willbold D. Solution structure of the X4 protein coded by the SARS related coronavirus reveals an immunoglobulin like fold and suggests a binding activity to integrin I domains. J. Biomed. Sci. 2006;13:281–293. doi: 10.1007/s11373-005-9043-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanel K., Willbold D. SARS-CoV accessory protein 7a directly interacts with human LFA-1. Biol. Chem. 2007;388:1325–1332. doi: 10.1515/BC.2007.157. [DOI] [PubMed] [Google Scholar]
- Holm L. DALI and the persistence of protein shape. Protein Sci. 2020;29:128–140. doi: 10.1002/pro.3749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Ito N., Tseng C.T.K., Makino S. Severe acute respiratory syndrome coronavirus 7a accessory protein is a viral structural protein. J. Virol. 2006;80:7287–7294. doi: 10.1128/JVI.00414-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito N., Mossel E.C., Narayanan K., Popov V.L., Huang C., Inoue T., Peters C.J., Makino S. Severe acute respiratory syndrome coronavirus 3a protein is a viral structural protein. J. Virol. 2005;79:3182–3186. doi: 10.1128/JVI.79.5.3182-3186.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakubzick C.V., Randolph G.J., Henson P.M. Monocyte differentiation and antigen-presenting functions. Nat. Rev. Immunol. 2017;17:349–362. doi: 10.1038/nri.2017.28. [DOI] [PubMed] [Google Scholar]
- Kang S., Yang M., Hong Z., Zhang L., Huang Z., Chen X., He S., Zhou Z., Zhou Z., Chen Q. Crystal structure of SARS-CoV-2 nucleocapsid protein RNA binding domain reveals potential unique drug targeting sites. Acta Pharm. Sin. B. 2020;10:1228–1238. doi: 10.1016/j.apsb.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan J., Ge J.W., Yu J.F., Shan S.S., Zhou H., Fan S.L., Zhang Q., Shi X.L., Wang Q.S., Zhang L.Q. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- Liao M., Liu Y., Yuan J., Wen Y., Xu G., Zhao J., Cheng L., Li J., Wang X., Wang F. Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19. Nat. Med. 2020;26:842–844. doi: 10.1038/s41591-020-0901-9. [DOI] [PubMed] [Google Scholar]
- Mehta P., McAuley D.F., Brown M., Sanchez E., Tattersall R.S., Manson J.J., Hlh Across Speciality Collaboration U.K. COVID-19: consider cytokine storm syndromes and immunosuppression. The Lancet. 2020;395:1033–1034. doi: 10.1016/S0140-6736(20)30628-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson C.A., Pekosz A., Lee C.A., Diamond M.S., Fremont D.H. Structure and intracellular targeting of the SARS-coronavirus ORF7a accessory protein. Structure. 2005;13:75–85. doi: 10.1016/j.str.2004.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin C., Zhou L., Hu Z., Zhang S., Yang S., Tao Y., Xie C., Ma K., Shang K., Wang W. Dysregulation of immune response in patients with COVID-19 in Wuhan, China. Clin. Infect. Dis. 2020;71:762–768. doi: 10.1093/cid/ciaa248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Cerrillo I., Landete P., Aldave B., Sanchez-Alonso S., Sanchez-Azofra A., Marcos-Jimenez A., Avalos E., Alcaraz-Serna A., de los Santos I., Mateu-Albero T. Differential redistribution of activated monocyte and dendritic cell subsets to the lung associates with severity of COVID-19. medRxiv. 2020 doi: 10.1101/2020.05.13.20100925. [DOI] [Google Scholar]
- Schaecher S.R., Mackenzie J.M., Pekosz A. The ORF7b protein of severe acute respiratory syndrome coronavirus (SARS-CoV) is expressed in virus-infected cells and incorporated into SARS-CoV particles. J. Virol. 2007;81:718–731. doi: 10.1128/JVI.01691-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulte-Schrepping J., Reusch N., Paclik D., Baßler K., Schlickeiser S., Zhang B., Kramer B., Krammer T., Brumhard S., Bonaguro L. Severe COVID-19 is marked by a dysregulated myeloid cell compartment. Cell. 2020;182:1419–1440.e23. doi: 10.1016/j.cell.2020.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimaoka M., Xiao T., Liu J.H., Yang Y., Dong Y., Jun C.D., McCormack A., Zhang R., Joachimiak A., Takagi J. Structures of the alpha L I domain and its complex with ICAM-1 reveal a shape-shifting pathway for integrin regulation. Cell. 2003;112:99–111. doi: 10.1016/s0092-8674(02)01257-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silvin A., Chapuis N., Dunsmore G., Goubet A.-G., Dubuisson A., Derosa L., Almire C., Hénon C., Kosmider O., Droin N. Elevated calprotectin and abnormal myeloid cell subsets discriminate severe from mild COVID-19. Cell. 2020;182:1401–1418.e18. doi: 10.1016/j.cell.2020.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song G., Yang Y.T., Liu J.H., Casasnovas J.M., Shimaoka M., Springer T.A., Wang J.H. An atomic resolution view of ICAM recognition in a complex between the binding domains of ICAM-3 and integrin alpha(L)beta(2) Proc. Natl. Acad. Sci. U S A. 2005;102:3366–3371. doi: 10.1073/pnas.0500200102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Y., Schneider T., Leong M., Aravind L., Zhang D. Novel immunoglobulin domain proteins provide insights into evolution and pathogenesis of SARS-CoV-2-related viruses. mBio. 2020;11 doi: 10.1128/mBio.00760-20. e00760-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181:281–292. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization Weekly Update on COVID-19 - 23 October 2020. 2020. https://www.who.int/publications/m/item/weekly-update-on-covid-19---23-october
- Wrapp D., Wang N.S., Corbett K.S., Goldsmith J.A., Hsieh C.L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z., Shi L., Wang Y., Zhang J., Huang L., Zhang C., Liu S., Zhao P., Liu H., Zhu L. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir. Med. 2020;8:420–422. doi: 10.1016/S2213-2600(20)30076-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan R.H., Zhang Y.Y., Li Y.N., Xia L., Guo Y.Y., Zhou Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;367:1444–1448. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Fu B., Zheng X., Wang D., Zhao C., Qi Y., Sun R., Tian Z., Xu X., Wei H. Pathogenic T-cells and inflammatory monocytes incite inflammatory storms in severe COVID-19 patients. Natl. Sci. Rev. 2020;7:998–1002. doi: 10.1093/nsr/nwaa041. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The accession number for the structure reported in this paper is Protein Data Bank (PDB): 7CI3.