Abstract
Introduction
Steroid-resistant nephrotic syndrome (SRNS) is the second most common cause of chronic kidney disease during childhood. Identification of 63 monogenic human genes has delineated 12 distinct pathogenic pathways.
Methods
Here, we generated 2 independent sets of nephrotic syndrome (NS) candidate genes to augment the discovery of additional monogenic causes based on whole-exome sequencing (WES) data from 1382 families with NS.
Results
We first identified 63 known monogenic causes of NS in mice from public databases and scientific publications, and 12 of these genes overlapped with the 63 known human monogenic SRNS genes. Second, we used a set of 64 genes that are regulated by the transcription factor Wilms tumor 1 (WT1), which causes SRNS if mutated. Thirteen of these WT1-regulated genes overlapped with human or murine NS genes. Finally, we overlapped these lists of murine and WT1 candidate genes with our list of 120 candidate genes generated from WES in 1382 NS families, to identify novel candidate genes for monogenic human SRNS. Using this approach, we identified 7 overlapping genes, of which 3 genes were shared by all datasets, including SYNPO. We show that loss-of-function of SYNPO leads to decreased CDC42 activity and reduced podocyte migration rate, both of which are rescued by overexpression of wild-type complementary DNA (cDNA), but not by cDNA representing the patient mutation.
Conclusion
Thus, we identified 3 novel candidate genes for human SRNS using 3 independent, nonoverlapping hypotheses, and generated functional evidence for SYNPO as a novel potential monogenic cause of NS.
Keywords: pediatric nephrology, proteinuria, recessive disease, whole-exome sequencing
Graphical abstract
NS is a kidney disease defined by proteinuria with resulting hypoalbuminemia, frequently causing edema and hyperlipidemia.1,2 In the clinical setting, “steroid-sensitive” versus “steroid-resistant” NS (SSNS vs. SRNS) can be distinguished based on the patient’s response to standard steroid therapy.3 SRNS with the histological correlate of focal segmental glomerulosclerosis (FSGS) invariably leads to end-stage renal failure. SRNS is the second most common cause of chronic kidney disease in childhood, with a reported incidence of 1.15 to 16.9 per 100,000 children.4,5 Over the past years, 63 genes have been identified as causing monogenic forms of SRNS (recessive or dominant) in humans with an onset <25 years.6, 7, 8, 9 Characterization of the cellular function of these 63 genes helped delineate 12 distinct pathogenic pathways of SRNS/FSGS.6, 7, 8, 9, 10, 11 As these genes are mainly expressed in podocytes, gene identification has helped to reveal a central role of the podocyte in the pathogenesis of SRNS.6, 7, 8, 9, 10 Monogenic causation accounts for 11.0% to 29.5% of patients with SRNS with onset before 25 years,9,10,12,13 leaving up to 70% of cases in that age group genetically unsolved. Thus, we surmised that novel genes still remain to be discovered.
Most SRNS genes identified in recent years were discovered by WES. Thus, WES, as an unbiased approach, still represents the most efficient method to identify potential novel causes of SRNS in humans. However, as childhood-onset SRNS represents a rare disease with a limited number of patients worldwide, and as WES analysis often reveals multiple variants, an independent or “orthogonal” dataset of candidate genes may facilitate the WES filtering process. Here, we generated 2 datasets of candidate genes for SRNS. After validating both sets against a list of known human NS genes, we overlapped these with our list of unique candidate genes from WES in 1382 families with NS. Specifically, we generated the following lists of known or candidate genes: (i) 63 known human NS genes were used for validation, (ii) 63 known monogenic causes of murine NS were retrieved from the literature, (iii) 64 genes regulated by the transcription factor WT1 were used.14,15
By validating the mouse and WT1-derived candidate datasets, we found that 12 of the murine NS genes and 5 of the WT1 downstream targets overlapped with the validation set of known human NS genes. We then overlapped the mouse and WT1-derived candidate datasets with our unique list of 120 NS candidate genes (iv).9,10,13 This approach allowed us to identify 3 potential novel candidate genes (ITGB8, SEMA3G, Buerger, 2020, personal communication, functional studies in progress, synaptopodin [SYNPO]) for human NS.
Within this set of 3 novel candidate genes, we identified a homozygous mutation in the gene SYNPO in a family with steroid-dependent NS. We show that loss-of-function of SYNPO leads to decreased CDC42 activity and reduced podocyte migration rate, which were each rescued by overexpression of wild-type cDNA, but not by cDNA representing the patient mutation.
Thus, we identified 3 novel candidate genes for human SRNS using 3 independent nonoverlapping hypotheses, generating functional evidence for SYNPO as a potential novel candidate gene for human NS.
Methods
Research Subjects
This study was approved by the institutional review board of Boston Children’s Hospital. We obtained blood samples and pedigrees following informed consent from families with NS. The diagnoses of NS were based on published clinical criteria and renal biopsies criteria evaluated by renal pathologists.16 Clinical data were obtained using a standardized questionnaire (http://www.renalgenes.org).
WES and Mutation Calling
WES was performed in 1382 NS families from worldwide sources, as previously described,17 using Agilent SureSelect or Illumina Nextera human exome capture arrays (Thermo Fisher Scientific, Waltham, MA) with next generation sequencing on an Illumina (San Diego, CA) platform. In total we examined 1382 families (1624 individuals) with at least 1 affected individual with NS (1198 families had only 1 affected individual, 142 families had 2 affected individuals, 33 families had 3 affected individuals, 6 families had 4 affected individuals, 1 family had each 5 or 6 affected individuals, and 1 family had 8 affected individuals; families with multiple affected individuals mostly represented sibling cases, some of them also included affected parents, uncles, aunts, or cousins). Trio/duo evaluation was performed in case DNA of unaffected parents was available (trios: 320, duos [maternal or paternal DNA available]: 138). Sequence reads were mapped against the human reference genome (NCBI build 37/hg19) using CLC Genomics Workbench (version 6.5.1) (CLC bio, Aarhus, Denmark). Genetic location information is according to the February 2009 Human Genome Browser data, hg19 assembly (http://www.genome.ucsc.edu). Downstream processing of aligned BAM files were done using Picard and samtools60, and SNV calling was done using Genome Analysis Tool Kit 5. Mutation calling was performed in line with proposed guidelines,18 and the criteria were used as previously described.13,17,19, 20, 21, 22, 23, 24 Briefly, after alignment to the human reference genome, variants were filtered for most likely deleterious variants. The variants included were rare in the population with mean allele frequency <1% in dbSNP147 and with only 0–1 homozygotes in the adult genome database gnomAD. Synonymous and intronic variants that were not located within splice-site regions were excluded.
First, WES data were analyzed for 1 of the 63 known NS genes. In a subcohort of families we have identified a known monogenic NS gene in up to 29.5% of the cases; this information is not included in this study but is published elsewhere.9,10,12,13 Unsolved cases were then subjected to an unbiased WES analysis. Subsequently, variant severity was stratified based on protein impact (truncating frameshift or nonsense mutations, essential or extended splice-site mutations, and missense mutations). Splice-site mutations were assessed by in silico tools MaxEnt and NNSPLICE splice-site mutation prediction scores,20,21,25 as well as conservation across human splice sites as described previously.17 Missense mutations were assessed based on SIFT, MutationTaster, and PolyPhen 2.0 conservation prediction scores22, 23, 24 and evolutionary conservation based on manually derived multiple sequence alignments.
Homozygosity Mapping
Homozygosity mapping was performed based on WES data. In brief, aligned BAM files were processed using Picard and SAMtools as described by other groups.26 Single nucleotide variant calling was performed using Genome Analysis Tool Kit.27 The resulting VCF files were used to generate homozygosity mapping data and visual outputs using the program Homozygosity Mapper.28 In case an individual had a relevant region of homozygosity (>60 MBp), we prioritized regions of homozygosity mapping in our evaluation of WES by initially analyzing variants identified in regions of homozygosity as published recently.29 Approximately 35% of the families, in which we identified 1 of the 120 candidate genes, had relevant homozygosity (>60 MBp). The 120 candidate genes each represented the strongest variants identified by this unbiased approach.
Generating a Candidate Gene Set of Monogenic Mouse Models of NS
Stringent criteria were used to generate a set of known monogenic mouse models of NS (Supplementary Figure S1) Two approaches were combined: (i) An “MGI Human-Mouse” Disease Connection search (http://www.informatics.jax.org) was performed by filtering for the terms: “nephrotic syndrome,” “proteinuria,” “foot process effacement,” or “glomerulonephritis.” Mouse models with nephritis rather than NS, hypertension models or autoimmune disease–related causes were excluded. (ii) A PubMed search (https://www.ncbi.nlm.nih.gov/pubmed/) was performed by searching for the following terms: “mouse model” AND “nephrotic”, “mice” AND “nephrotic”, “mice” AND “foot process effacement”, “mice” AND “proteinuria” AND “gene”. The same exclusion criteria were applied. In this way, 111 monogenic mouse models of nephrotic syndrome/proteinuria were identified (Supplementary Figure S1). Of these, only 75 models were based on global knockout, 36 tissue-specific mouse models were excluded, as they did not generate a hypothesis of a monogenic NS phenotype. Twelve of the 75 mouse NS genes were excluded from the study, as the murine NS phenotype was reported after the human NS gene was published in the literature, leaving us with 63 genes (Supplementary Tables S1 S2, and S3).
cDNA Cloning
Human SYNPO cDNA clones were purchased (Genscript, Piscataway, NJ; OHu26207). Human SYNPO full-length protein (GenBank accession NP_ 001159680) is encoded by GenBank accession NM_001166208. Mutagenesis was performed using the QuikChange II XL Site-Directed Mutagenesis Kit (Agilent Technologies, Santa Clara, CA). Expression constructs (pRK5-N-Myc, pCDNA6.2-N-GFP) were produced using LR Clonase (Invitrogen, Thermo Fisher Scientific) following the manufacturer’s instructions. Clones were validated prior to usage (Supplementary Figures S2 and S3).
Cell Lines
Experiments were done in HEK293T cells (ATCC, Manassas, VA) or immortalized human podocytes (gift from Moin Saleem, University of Bristol, Bristol, UK). Podocytes were cultured as previously described.30
Short Hairpin RNA Transduction
Short hairpin RNA (shRNA) targeting human SYNPO was subcloned into pSIREN RetroQ for retroviral transduction using HEK293T cells 48 hours after transduction puromycin was added to the medium for selection of transduced cells at a final concentration of 2.4 μg/ml.
Reagents and Antibodies
The following primary antibodies were used: mouse monoclonal anti-SYNPO (G1D4; PROGEN, Heidelberg, Germany), rabbit anti-SYNPO (PA5–56997; Thermo Fisher Scientific), mouse horseradish peroxidase–linked anti-beta actin (ab20272; Abcam, Cambridge, UK). Donkey anti-mouse, anti-rabbit Alexa 488– and Alexa 594–conjugated secondary antibodies; 4′,6-diamidino-2-phenylindole (DAPI) staining reagents; Phalloidin-Alexa 594 were obtained from Invitrogen (Thermo Fisher Scientific). Horseradish peroxidase–labeled secondary antibodies were purchased from Santa Cruz Biotechnology (Dallas, TX).
Immunofluorescence and Confocal Laser Scanning Microscopy
For overexpression studies, human podocytes were transiently transfected using Lipofectamine 2000 (Thermo Fisher Scientific) according to the manufacturer’s instructions. Experiments were performed 24 to 48 hours after transfection. Cells were fixed for 15 minutes using 4% paraformaldehyde and permeabilized with 0.5% Triton-X 100. After blocking, sections were incubated overnight at 4°C with primary antibody. The cells were incubated in secondary antibodies for 60 minutes at room temperature followed by mounting in hardening medium with 4′,6-diamidino-2-phenylindole (DAPI). Confocal imaging was performed using the Leica (Wetzlar, Germany) SP5X system with an upright DM6000 microscope, images were processed with the Leica AF software suite.
G-LISA Activation Assays
Cells were transfected with wild-type or Mock constructs using Lipofectamine-2000. Transfected cells were incubated in RPMI medium with 10% fetal bovine serum for 24 hours and then in serum-free medium for 24 hours. RAC1 or CDC42 activity was determined using a G-LISA Activation Assay Biochem Kit (Cytoskeleton, Denver, CO), according to the manufacturer’s instructions.
Podocyte Migration Assay
Podocyte migration assay was performed using Incucyte ZOOM System (Essen Bioscience, Ann Arbor, MI) according to the manufacturer’s instructions.31 SEM is presented.
Statistical Analysis
Results are presented as SEM or mean ± SD for the indicated number of experiments. Statistical analysis was performed with 1-way analysis of variance using GraphPad (La Jolla, CA) Prism. P < 0.05 was considered statistically significant.
Results
Three Orthogonal Approaches to Generate Human NS Candidate Genes
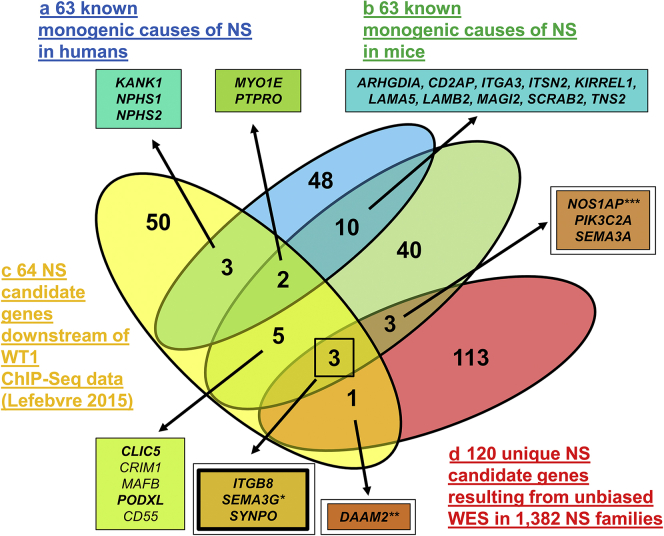
To identify novel candidate genes for human monogenic NS, we generated 3 candidate gene lists that were considered “orthogonal” (i.e., independent) with regard to their biological and functional candidate hypothesis: (i) the first approach comprised a list of 63 published monogenic mouse models of NS or proteinuria (Figure 1, part B, Supplementary Figure S1, Supplementary Tables S1 and S2). Stringent search criteria were applied and 63 genes were identified (Supplementary Figure S1). To generate this list, 36 proteinuric mouse models based on conditional gene knockout were excluded. Furthermore, 12 mouse models with an equivalent human phenotype that were generated after the human NS gene was found were excluded (Supplementary Table S3). (ii) Second, we used a published list of 64 candidate genes based on the hypothesis that podocytic genes, downstream of the master regulator WT1, are potential causes of NS2 (Figure 1, part C, Supplementary Figure S4). (iii) Finally, we overlapped both candidate gene sets (mouse and WT1) with a list of 120 candidate genes that we generated by unbiased WES evaluation in a cohort of 1,382 families with NS (Figure 1, part D).
Figure 1.
Venn diagram of 3 independent approaches to generate candidate genes for monogenic nephrotic syndrome (NS) in humans. (a) Overview of 63 known monogenic causes of NS in humans (blue oval) that serve as a positive control. Twelve of the 63 genes (19%) overlap with 63 genes of known mouse model for NS/proteinuria (green oval) and 5 of the 63 genes (8%) overlap with 64 Wilms Tumor 1 (WT1) downstream targets.14,15 (b) Overview of 63 known monogenic mouse models of NS/proteinuria (green oval) (Supplementary Table S1, Supplementary Figure S1). Twelve of the 63 mouse NS genes (19%) overlap with 63 known monogenic causes of human NS. Ten of the 63 mouse NS genes (16%) overlap with WT1 downstream targets (yellow oval), and 6 of the 63 mouse NS genes (10%) genes overlap with unique NS candidate genes resulting from unbiased whole exome sequencing (WES) (red oval). (c) Overview of 64 NS candidate genes (yellow oval) based on WT1 chromatin immunoprecipitation sequencing (ChIP-Seq) data on E18.5 mice published by Lefebvre et al.14 (Supplementary Figure S4). Five of the 64 genes (8%, KANK1, NPHS1, NPHS2, MYO1E, PTPRO) overlap with 63 known monogenic causes of NS in humans that served as a positive control. Ten of 64 genes (15.6%, MYO1E, PTPRO, CLIC5, CRIM1, MAFB, PODXL, CD55, ITGB8, SEMA3G, SYNPO) overlap with known monogenic mouse models of NS (green oval). Four of 64 genes (7.8%, ITGB8, SEMA3G, SYNPO, DAAM2) overlap with 120 unique NS candidate genes generated by us from unbiased WES (red oval). (d) Overview of 120 unique NS candidate genes generated from unbiased WES in 1382 NS families from a worldwide cohort (red oval). Four of the 120 genes (3.3%, ITGB8, SEMA3G, SYNPO, DAAM2) overlap with 64 WT1 downstream genes (yellow oval) and 6 of the 120 genes (5%, ITGB8, SEMA3G, SYNPO, NOS1AP, PIK3C2A, SEMA3A) overlap with mouse NS genes (green). Three of the 120 genes (2.5%, ITGB8, SEMA3G, SYNPO) overlap with both WT1 and mouse NS candidate genes. Bold font indicates that a family with a potentially causative mutation in this gene was detected. Double frame boxes indicate the 7 strongest candidate genes. ∗Buerger, personal communication, 2020; ∗∗Schneider32; ∗∗∗Majmundar and Buerger.33
Murine NS Genes
To test the validity of the mouse list, we examined it for overlap with the published 63 known human NS genes (Figure 1, parts A and B, blue: human NS, green: mouse NS).6, 7, 8, 9, 10 Twelve of the 63 NS mouse genes (19%) overlapped with the 63 known human NS genes (Figure 1, Supplementary Table S2) (MYO1E, PTPRO, ARHGDIA, CD2AP, ITGA3, ITSN2, KIRREL1, LAMA5, LAMB2, MAGI2, SCARB2, TNS2). This finding validated the mouse candidate list as suitable for the identification of novel potential monogenic human NS genes.
WT1 Downstream Targets in Podocytes Based on Chromatin Immunoprecipitation Sequencing in E18.5 Mouse Kidneys
WT1 is a master regulator for podocyte development and maintenance and is highly expressed in mature podocytes.34 Mutations in WT1 have been found to lead to different glomerular diseases, including isolated NS,35,36 Denys-Drash Syndrome,37 or Frasier Syndrome.38,39 WT1 encodes a tumor suppressor protein and putative splicing cofactor, based on a zinc finger structure that has multiple key functions in kidney development, although exact mechanisms remain elusive.40, 41, 42, 43 Recently, different WT1-related studies using chromatin immunoprecipitation sequencing, exon array, and bioinformatical analyses have shown a specific role for WT1 in regulating the podocyte-specific transcriptome.1,2,44,45 We therefore hypothesized that genes downstream of WT1 represent promising NS candidate genes and generated a second candidate list for this study (Figure 1, part C, WT1 genes: yellow Supplementary Figure S4) based on the following publications: (i) Motamedi et al.15 performed in vivo chromatin immunoprecipitation sequencing analyses on E18.5 kidneys. In this work, 36,512 WT1-associated regions were identified, which were reduced to 5547 or 2940 peaks, by setting an irreproducibility discovery rate cutoff of ≤0.1 or 0.01, respectively (Supplementary Figure S4).15 (ii) To identify podocyte-specific genes that are potentially regulated by WT1, Lefebvre et al.14 then evaluated these WT1 chromatin immunoprecipitation sequencing–associated genes, with a list of genes that represented the “Top200” genes highly expressed in podocytes (Supplementary Figure S4).1,46 Sixty-four of the 192 podocytic genes were associated with WT1-bound regions (irreproducibility discovery rate ≤ 0.01) and harbored a WT1 motif, within 2.5 kb of the genes’ transcriptional start site.1,46
Thus, 64 podocytic genes were found to be potentially regulated by WT1 according to the criteria employed by Lefebvre et al.14 We therefore tested the hypothesis that some of these genes represent potential WT1-derived candidates for novel human NS genes (Figure 1, part C, blue: human NS, yellow: WT1 genes). To validate this candidate gene approach “a posteriori,” we overlapped the WT1 genes with human NS genes. Interestingly, 5 of the 64 WT1-derived genes (8%) overlapped with the 63 known human NS genes (Figure 1) (KANK1, NPHS1, NPHS2, MYO1E, PTPRO), of which 2 genes (MYO1E, PTPRO) overlapped with both the human and murine NS lists (Figure 1). This overlap demonstrated that the known human NS genes are enriched 26-fold in the WT1-derived list compared to the approximately 20,000 genes of the entire human genome (Supplementary Figure S5A). After validating the WT1 list against the human NS genes, we then compared the WT1 genes with the candidate list of murine monogenic NS genes (Figure 1). Here we found that 10 of the 64 WT1 candidate genes (15.6%) overlapped with the dataset of 63 mouse NS genes (MYO1E, PTPRO, CLIC5, CRIM1, MAFB, PODXL, CD55, ITGB8, SEMA3G, SYNPO). Hence, monogenic murine NS genes were enriched 50-fold within the WT1 candidate dataset compared to all genes of the entire human genome (Supplementary Figure S5B). We therefore concluded that due to this enrichment, the remaining 59 of the 64 WT1-derived genes, that have not been reported to be a human NS gene, may represent novel candidate genes for monogenic NS in humans.
Unique NS Candidate Genes Resulting From Unbiased WES in 1382 NS Families
Over the past 12 years, our laboratory has performed WES analyses in 1382 families in a worldwide cohort with NS. In a subcohort of families, we have identified a known monogenic NS gene in up to 29.5% of cases.9,10,12,13 Cases unsolved for known NS genes were then subjected to unbiased WES analysis. Using this approach, we have discovered, functionally characterized, and published 25 novel NS genes.47, 48, 49, 50, 51, 52, 53, 54, 55, 56, 57, 58, 59, 60 Mutation calling was performed in line with proposed unbiased guidelines.18 In addition to identifying known NS genes, we generated potential novel candidate genes for human monogenic NS in 120 families in which only one strong candidate gene resulted from WES evaluation (Figure 1d, red: WES candidates; also see Figure 1 of Waireko et al.9). The following criteria were used11,19: Only rare, biallelic (recessive) variants with a mean allele frequency <1% and 0–1 homozygote allele carriers in the control genome/exome database gnomAD were included. Nonsynonymous variants and/or variants located within splice sites were analyzed. Subsequently, variant severity was stratified by the following: splice-site mutations were assessed by in silico prediction scores.20,21,25 Missense mutations were assessed based on SIFT, MutationTaster, and PolyPhen 2.0 prediction scores,22, 23, 24 and evolutionary conservation was evaluated based on manually derived multiple sequence alignments.
Having validated the mouse (Figure 1b, green) and WT1 (Figure 1c, yellow) candidate gene lists against the human (Figure 1a, blue) NS known genes, we then hypothesized that any genes from the mouse or WT1 candidate list that overlap with a WES-derived candidate gene may generate a strong candidate of our 120 WES candidates. We found that 6 of the WES genes overlapped with the mouse candidate list (ITGB8, 1 family; SEMA3G, 3 families, Buerger, 2020, personal communication, functional studies in progress; SYNPO, 1 family; NOS1AP, 2 families, Majmundar and Buerger33; PIK3C2A, 2 families; SEMA3A, 1 family), whereas 4 genes overlapped with the WT1-derived candidate dataset (ITGB8, 1 family; SEMA3G, 3 families, Buerger, 2020, personal communication, functional studies in progress; SYNPO, DAAM2, 4 families, Schneider32). Interestingly, 3 genes were included in all 3 of the candidate gene lists (ITGB8, 1 family; SEMA3G, 3 families, Buerger, 2020, personal communication, functional studies in progress; SYNPO, 1 family) (Figure 1, Supplementary Table S4).
Overlap Between Candidate Gene Sets
Seven genes from our WES-derived candidate dataset overlapped with either the mouse or the WT1 candidate gene lists (Figure 1, Supplementary Table S5). Interestingly, for 3 of these genes, we have identified different families with multiple, likely causative mutations (NOS1AP, Majmundar and Buerger33; DAAM2, Schneider32; SEMA3G, Buerger, 2020, personal communication, functional studies in progress) (Figure 1). Furthermore, we discovered families with NS with mutations in the following genes: PIK3C2A, SEMA3A, and ITGB8 (Figure 1, Supplementary Table S5).
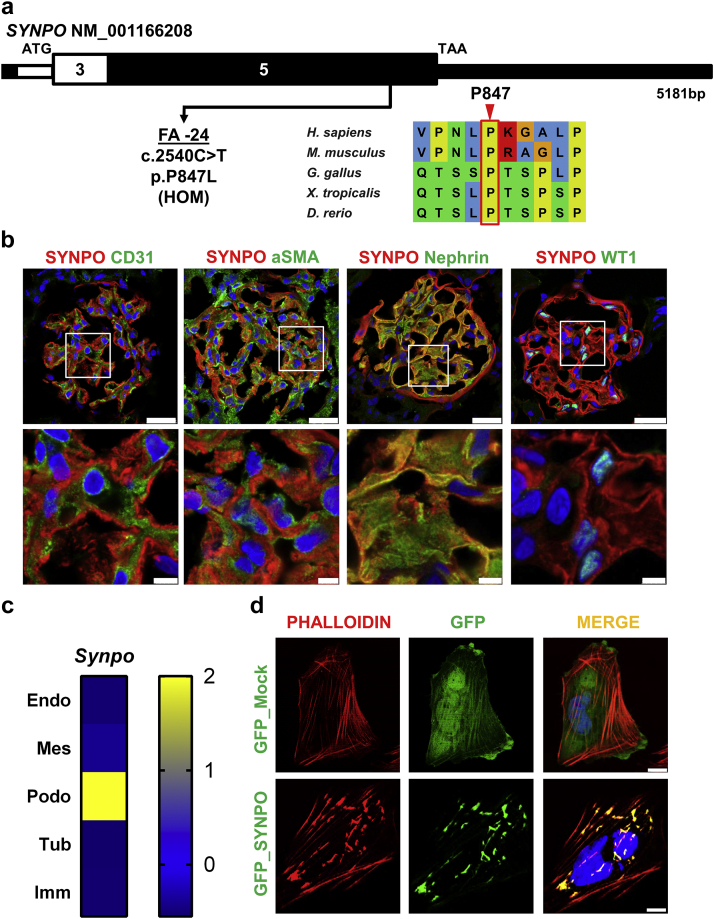
In addition, we identified a consanguineous family (FA) harboring a homozygous missense mutation in the gene SYNPO (c.2540C>T, p.P847L) (Figure 2a, Supplementary Table S4). The patient was from Saudi Arabian ethnicity, consanguineous descent, and had an onset of NS at the age of 4 years (Supplementary Table S4). The here-identified variant was absent from the control database gnomAD, and the mutation yielded strong in silico prediction scores (Supplementary Table S4). The proline at position 847 has been evolutionarily conserved since Danio rerio (Figure 2a). The SYNPO mutation was present homozygously in the affected patient FA-24, whereas the healthy parents and siblings were heterozygous carriers of the mutation (Supplementary Figure S6). The patient was reported to initially respond to glucocorticoid treatment but remained steroid-dependent. No renal biopsy was performed. The patient continued to have proteinuria, with slow progression of renal disease despite ongoing steroid therapy. Extrarenal symptoms included muscular dystrophy caused by a homozygous LAMA2 mutation (c.2096G>T, p.R699M). The patient’s brother also suffered from this disease, but not from NS. Hypotonia developed at the age of 40 days. The patient has dysmorphic features with plagiocephaly and scoliosis and is reported to be wheelchair-bound.
Figure 2.
Biallelic SYNPO mutation identified in family FA with nephrotic syndrome. (a) Exon structure of SYNPO is shown with arrow indicating position of mutation of patient FA-24 with nephrotic syndrome (NS). Position of start codon (ATG) and stop codon (TAA) are indicated. Exon numbers are marked on a black or white background. Evolutionary conservation is shown for sequence surrounding amino acid position P847 in SYNPO protein. (b) Immunofluorescence staining for Synpo and colocalization in rat glomeruli is shown for different cell type marker proteins: costaining with antibodies against CD31 (endothelial cells), αSMA (mesangial cells), nephrin (podocytes), and Wilms Tumor 1 (WT1) (podocyte nuclei). Insets are shown enlarged in the lower row. Note that there is a strong colocalization of Synpo staining only with nephrin, which labels podocyte slit membrane structures, thereby demonstrating Synpo localization to podocytes but not endothelial or mesangial cells. Bars = 5, 2.5 μm. (c) Single-cell type-specific average expression of Synpo. Data were modified from Karaiskos et al.63 The heat map is based on Z-scores. Endo, endothelial cell; Mes, mesangial cell; Podo, podocyte; Tub, tubule cell; Imm, immune cell. (d) Upper row: Transfection of a human podocyte cell line30 with negative control GFP_Mock (green). Cells were almost devoid of large actin fibers in perinuclear cytoplasm but displayed strong F-actin staining (red) within the cell periphery. Lower row: upon transfection with GFP_SYNPO wild-type cDNA (green), SYNPO colocalizes (yellow) with F-actin (red) in thick irregular cytoplasmic perinuclear actin clusters. Bar = 10 μm.
SYNPO Expression in Podocytes
To further characterize the deleteriousness of the patient-derived recessive SYNPO mutation, we performed functional in vitro studies. SYNPO encodes a linear proline-rich protein, highly present in neuronal dendrites and in foot processes of podocytes.61,62 Here, we confirm that Synpo colocalizes with nephrin, which, if mutated, causes NS, and which is a marker for podocytes and the glomerular slit membrane (Figure 2b). There was no colocalization with an endothelial cell marker (CD31) or with a mesangial cell marker (αSMA) in rat glomeruli (Figure 2b). Single-cell RNA sequencing databases also show a strong podocytic expression63 (Figure 2c). Thus, we confirm that Synpo is predominantly expressed in podocytes. SYNPO has been shown to be important for podocyte function and has been extensively studied as an actin-related protein in human podocytes.61,64 To determine its subcellular localization on overexpression in cultured human podocytes, we expressed GFP_SYNPO cDNA in a human podocyte cell line and tested for colocalization with phalloidin. On overexpression, SYNPO induced the formation of F-actin networks in podocytes and colocalized with F-actin in thick irregular or parallel arrangements of actin bundles in the perinuclear cytoplasm (Figure 2d, lower row), whereas cells that were transfected with an empty GFP vector did not harbor large actin fiber aggregates in the perinuclear region, but displayed strong F-actin staining within the cell periphery (Figure 2d, upper row).
Wild-Type SYNPO, But Not Mutant, Rescues CDC42 Activation
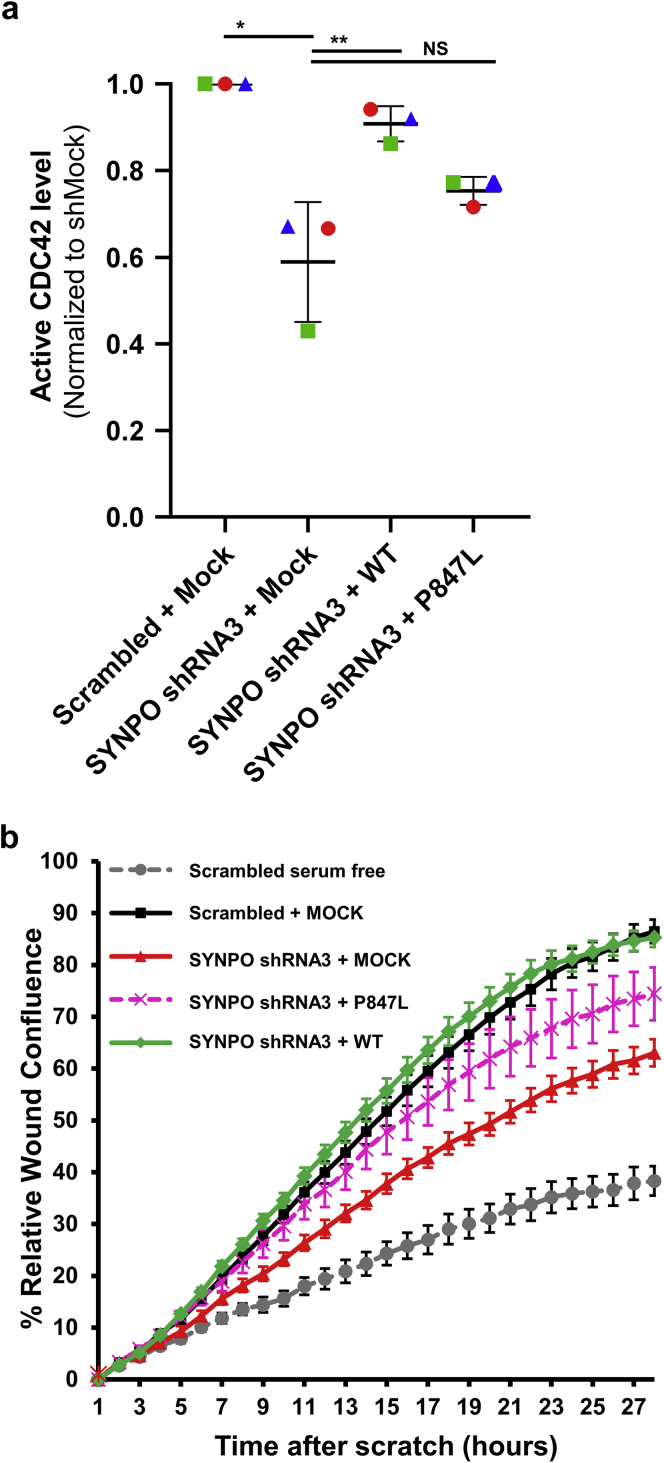
The small Rho-like GTPases (RHOA, RAC1, and CDC42) play a pivotal role in podocyte morphogenesis, migration, and the pathogenesis of monogenic forms of NS.60 SYNPO has been shown to induce stress fibers by competitively blocking Smurf1-mediated ubiquitination of RHOA.65 To examine whether the potential pathogenic mechanism of SYNPO loss-of-function in patient FA-24 is connected to altered RAC1/CDC42 activation, we performed G-LISA assays in cultured human podocytes. In shRNA-mediated SYNPO knockdown podocytes (Supplementary Figure S2), CDC42 activation was significantly reduced compared with scrambled shRNA cells (Figure 3a). The reduction of CDC42 activity was rescued by overexpressing murine Synpo wild-type cDNA. The Synpo cDNA construct reflecting the mutation of the NS patient FA-24 failed to rescue CDC42 activity (Figure 3a). There was no difference in the activation of RAC1 (Supplementary Figure S7).
Figure 3.
CDC42 activity and podocyte migration rate (PMR) is reduced by SYNPO knockdown and rescued by transfection with wild-type Synpo, but not by complementary DNA (cDNA), reflecting the mutation of patient with nephrotic syndrome (NS) FA-24. (a) Active levels of CDC42 were measured by CDC42 G-LISA assay in a human podocyte cell line. Short hairpin RNA (shRNA)-mediated knockdown of SYNPO and overexpression of empty vector negative control (+ Mock) reduced active CDC42. Overexpression of mouse wild-type Synpo cDNA (+ WT) rescued this effect. A Synpo cDNA construct reflecting the mutation from NS patient FA-24 (+ P847L) failed to rescue the phenotype. P-values calculated by 1-way analysis of variance. ∗P < 0.01, ∗∗P < 0.01. NS, nonsignificant. (b) In a human podocyte cell line expressing scrambled shRNA, PMR is increased following serum addition (black) relative to serum-deplete conditions (gray). All subsequent experiments were performed in presence of serum. Knockdown of SYNPO (red) showed reduced PMR compared with scrambled shRNA (black). Migration was rescued by overexpression of wild-type Synpo construct (green). However, mouse Synpo cDNA constructs reflecting the mutation (p.P847L) of NS patient FA-24 only partially rescued PMR (pink).
Wild-Type Synpo, But Not the Patient Variant, Rescues Podocyte Migration
Because wild-type Synpo, but not the patient variant, rescued CDC42 activity upon SYNPO knockdown in cultured human podocytes (Figure 3a), we hypothesized that SYNPO may also regulate the podocyte migration rate (PMR), a well-established intermediate phenotype for SRNS disease genes.52,53,57 Using the IncuCyte ZOOM live cell imaging system, we found that knockdown of SYNPO reduced PMR compared with scrambled shRNA (Figure 3b). Migration phenotype was rescued by transient overexpression of murine wild-type Synpo cDNA. In contrast, a Synpo cDNA construct reflecting the mutation found in the NS patient failed to rescue the PMR (Figure 3b), suggesting deleteriousness of the mutation of NS family FA.
Discussion
In this study, we generated 2 independent lists of NS candidate genes to augment the discovery of additional monogenic causes of human NS from WES data generated in 1382 families with SRNS. A set of 63 known monogenic causes of NS in mice and another set of 64 genes described to be regulated by the transcription factor WT114,15 were identified. We then validated the murine and WT1 candidate gene sets by overlapping them with known human NS genes, used as a positive control toward our hypothesis: that the 2 sets of candidate genes may be relevant for the discovery of novel human SRNS genes. Finally, we compared the mouse and WT1 candidate genes (Figure 1) with our unique NS WES-derived candidate list. Hereby, we identified novel candidate genes as potential causes for monogenic human NS, including a SYNPO mutation in a family with steroid-dependent NS. We demonstrate that lack of SYNPO reduced CDC42 activity and PMR in cultured human podocytes, and show that wild-type, but not cDNA representing the patient mutation, was able to rescue the phenotype. This demonstrated the deleteriousness of this mutation in assays relevant for NS pathogenesis. Thus, we conclude that SYNPO may be a novel candidate gene for human NS.
SYNPO mutations have previously been discussed as potential monogenic causes of NS in humans. That assumption is supported by the following publications: (i) Dai et al.66 published 2 cases with heterozygous mutations in the SYNPO promoter region in patients with NS; however, no homozygous mutations in SYNPO in patients with NS have been reported. (ii) It has been shown that patients with FSGS,67, 68, 69 HIV-associated nephropathy,70 and IgA nephropathy71 have reduced expression of SYNPO in the glomerulus. (iii) Srivastava et al.68 observed a statistical difference of SYNPO expression between patients with FSGS and minimal change disease. (iv) Synpo-deficient mice display impaired recovery from protamine sulfate–induced foot process effacement and lipopolysaccharide-induced NS.72 Just recently, it was also shown that Synpo-deficient mice demonstrated increased susceptibility to Adriamycin nephropathy.73 (v) Digenic heterozygosity for Cd2ap and Synpo was sufficient to induce FSGS in mice.74 (vi) SYNPO was identified as an inhibitor of the Cdc42:IRSp53:Mena signaling, and it was suggested to play a role in a potential antiproteinuric signaling pathway,75 thus confirming SYNPO as a strong candidate gene for monogenic NS, if mutated.
SYNPO has also been well-studied in podocytes,61,62 and has an established role in actin cytoskeleton regulation.65,72 In this context, gene silencing of SYNPO reduced the PMR, whereas the expression of SYNPO led to the induction of stress fibers.65 We here showed that knockdown of SYNPO in human podocytes leads to an impaired PMR and reduced CDC42 activity (Figure 3a and b). Of note, only wild-type Synpo, but not cDNA representing the patient’s mutation, was able to rescue the phenotype, demonstrating the deleteriousness of the patient’s homozygous allele. This is consistent with findings that implicated dysregulation of Rho-like small GTPases as a role in disease mechanism in several monogenic forms of human and murine SRNS.51,53,54,60 We therefore suggest that the SYNPO mutation may partly exert its pathogenic effect via dysregulation of CDC42 signaling.
However, it should be emphasized that SYNPO, as of now, can be considered a candidate gene only for monogenic human NS. Identification of additional mutations in SYNPO in other families with NS and functional characterization is essential to substantiate its candidate status. Therefore, the validity of the here-identified mutation must be considered with caution, despite the loss-of-function data we generated.
Monogenic mouse disease genes have successfully been used in the past to identify novel human disease genes.76, 77, 78 Here, we show that 12 of the 63 known mouse NS candidate genes overlap with the 63 known human genes that cause NS if mutated (Figure 1). Before generating the 63 mouse candidate gene list, we had excluded 12 mouse NS genes that were published after the human NS phenotype was discovered, to maintain an unbiased approach (Supplementary Table S3). This emphasizes that the remaining mouse genes, without an associated known human NS phenotype, might be promising candidates for this disease. The fact that equivalent human mutations have not been discovered yet may be partially explained by the fact that childhood-onset SRNS is a rare disease, with a limited number of patients sequenced to date. Furthermore, human and mouse pathophysiology can differ, and some genes may lead to variable disease phenotypes depending on the organism.
In summary, we identified 3 novel candidate genes for human SRNS using 3 independent nonoverlapping hypotheses, based on monogenic mouse models of NS, WT1 downstream targets and unbiased WES, thereby generating further evidence for SYNPO as a potential novel cause of human NS. We demonstrate loss-of-function for the patient mutation and suggest that SYNPO mutations may lead to disease through CDC42 dysregulation.
Disclosure
FH is a co-founder and Scientific Advisory Board member of Goldfinch Biopharma Inc. All the other authors declared no competing interests.
Acknowledgments
We are grateful to the families and study individuals for their contribution. FH is the William E. Harmon Professor of Pediatrics. This research is supported by grants from the National Institutes of Health (NIH) to FH (5R01DK076683–13), ACO-W (F32 DK122766), and AJM (T32-DK007726). AJM is supported by the 2017 Harvard Stem Cell Institute Fellowship Grant and the 2018 Jared J. Grantham Research Fellowship from the American Society of Nephrology Ben J. Lipps Research Fellowship Program. YM was supported by the “EPT” program of Shanghai Children’s Medical Center. VK and FB are supported by the Deutsche Forschungsgemeinschaft (VK-403877094/KL3224/2-1 and FB-404527522). EW was supported by the Leopoldina Fellowship Program, Deutsche Akademie der Naturforscher Leopoldina – Nationale Akademie der Wissenschaften (German National Academy of Sciences Leopoldina - grant LPDS 2015–07). Sequencing and analysis were provided by Yale Center for Mendelian Genomics (NIH UM1 HG006504) and the Broad Institute of MIT and Harvard Center for Mendelian Genomics (CMG), which was funded by the National Human Genome Research Institute, the National Eye Institute, and the National Heart, Lung, and Blood Institute grant UM1 HG008900. No part of this manuscript has been previously published.
Author Contributions
VK and FH designed the study. YM performed cell biological experiments. YM, VK, and FH analyzed the data. VK, YM, RS, FB, ACO-W, KD, TMK, MN, AJM, NM, EW, WT, and SS performed WES analysis and Sanger sequencing. HLR, SM, and RPL generated sequence data and provided analysis support. VK, YM, and FH drafted the paper. All authors revised the manuscript and approved of the final version.
TRANSLATIONAL STATEMENT
Here, we identified 3 novel candidate genes for human steroid-resistant nephrotic syndrome using 3 independent non-overlapping hypotheses, based on monogenic mouse models, WT1 downstream targets, and unbiased whole-exome sequencing. We generated further evidence for SYNPO as a potential novel cause of human nephrotic syndrome. Identification of additional families with mutations in these genes will further corroborate their candidate status and may help delineate additional insights into pathomechanisms of steroid-resistant nephrotic syndrome.
Footnotes
Figure S1. Workflow for searching for mouse candidate genes via MGI and PubMed.
Figure S2. (A) SYNPO knockdown in human podocytes. (B) Immuno blot from lysates of stable scrambled shRNA negative control and SYNPO-shRNA3 expression in human immortalized podocytes co-transfected for rescue construct of wild-type versus p.P847L SYNPO constructs.
Figure S3. SYNPO antibody and cDNA clone characterization.
Figure S4. Workflow for filtering criteria from WT1 ChIP Seq data on E18.5 kidneys (2 replicates) published by Motamedi et al.2 and Lefebvre et al.1
Figure S5. Fold enrichment resulting from overlapping WT1 downstream targets1 when being overlapped with human and murine NS genes as positive controls.
Figure S6. Family pedigree and Sanger sequencing traces of SYNPO mutation.
Figure S7. G-LISA for active RAC1 in human podocyte cell lines overexpression SYNPO cDNA constructs or in stable scrambled shRNA negative control vs. SYNPO-shRNA3 podocytes.
Table S1. List of 63 genes that lead to proteinuria/nephrotic syndrome in mice in case of a global knockout. Genes that are underlined have a published human nephrotic syndrome phenotype.
Table S2. Overview of 12 human nephrotic syndrome or phenocopy genes that had a nephrotic mouse with a global knockout published before the human equivalent was reported.
Table S3. Overview of 12 global knockout mice that were excluded in this study as the equivalent human NS phenotype was published before the mouse model was reported.
Table S4. Recessive SYNPO mutation in 1 family (FA) with nephrotic syndrome.
Table S5. Summary of families with 7 candidate genes.
Supplementary Material
References
- 1.Hildebrandt F. Genetic kidney diseases. Lancet. 2010;375:1287–1295. doi: 10.1016/S0140-6736(10)60236-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wiggins R.C. The spectrum of podocytopathies: a unifying view of glomerular diseases. Kidney Int. 2007;71:1205–1214. doi: 10.1038/sj.ki.5002222. [DOI] [PubMed] [Google Scholar]
- 3.Koide K., Sano M. [Glucocorticoid therapy in renal diseases--its indication and therapeutic schedule] Nihon Rinsho. 1994;52:728–733. [PubMed] [Google Scholar]
- 4.Noone D.G., Iijima K., Parekh R. Idiopathic nephrotic syndrome in children. Lancet. 2018;392:61–74. doi: 10.1016/S0140-6736(18)30536-1. [DOI] [PubMed] [Google Scholar]
- 5.Tryggvason K., Patrakka J., Wartiovaara J. Hereditary proteinuria syndromes and mechanisms of proteinuria. N Engl J Med. 2006;354:1387–1401. doi: 10.1056/NEJMra052131. [DOI] [PubMed] [Google Scholar]
- 6.Arrondel C., Missoury S., Snoek R. Defects in t(6)A tRNA modification due to GON7 and YRDC mutations lead to Galloway-Mowat syndrome. Nat Commun. 2019;10:3967. doi: 10.1038/s41467-019-11951-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lovric S., Ashraf S., Tan W. Genetic testing in steroid-resistant nephrotic syndrome: when and how? Nephrol Dial Transplant. 2016;31:1802–1813. doi: 10.1093/ndt/gfv355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Somlo S., Mundel P. Getting a foothold in nephrotic syndrome. Nat Genet. 2000;24:333–335. doi: 10.1038/74139. [DOI] [PubMed] [Google Scholar]
- 9.Warejko J.K., Tan W., Daga A. Whole exome sequencing of patients with steroid-resistant nephrotic syndrome. Clin J Am Soc Nephrol. 2018;13:53–62. doi: 10.2215/CJN.04120417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sadowski C.E., Lovric S., Ashraf S. A single-gene cause in 29.5% of cases of steroid-resistant nephrotic syndrome. J Am Soc Nephrol. 2015;26:1279–1289. doi: 10.1681/ASN.2014050489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vivante A., Hildebrandt F. Exploring the genetic basis of early-onset chronic kidney disease. Nat Rev Nephrol. 2016;12:133–146. doi: 10.1038/nrneph.2015.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Connaughton D.M., Kennedy C., Shril S. Monogenic causes of chronic kidney disease in adults. Kidney Int. 2019;95:914–928. doi: 10.1016/j.kint.2018.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tan W., Lovric S., Ashraf S. Analysis of 24 genes reveals a monogenic cause in 11.1% of cases with steroid-resistant nephrotic syndrome at a single center. Pediatr Nephrol. 2018;33:305–314. doi: 10.1007/s00467-017-3801-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lefebvre J., Clarkson M., Massa F. Alternatively spliced isoforms of WT1 control podocyte-specific gene expression. Kidney Int. 2015;88:321–331. doi: 10.1038/ki.2015.140. [DOI] [PubMed] [Google Scholar]
- 15.Motamedi F.J., Badro D.A., Clarkson M. WT1 controls antagonistic FGF and BMP-pSMAD pathways in early renal progenitors. Nat Commun. 2014;5:4444. doi: 10.1038/ncomms5444. [DOI] [PubMed] [Google Scholar]
- 16.Primary nephrotic syndrome in children: clinical significance of histopathologic variants of minimal change and of diffuse mesangial hypercellularity. A Report of the International Study of Kidney Disease in Children. Kidney Int. 1981;20:765–771. doi: 10.1038/ki.1981.209. [DOI] [PubMed] [Google Scholar]
- 17.Braun D.A., Sadowski C.E., Kohl S. Mutations in nuclear pore genes NUP93, NUP205 and XPO5 cause steroid-resistant nephrotic syndrome. Nat Genet. 2016;48:457–465. doi: 10.1038/ng.3512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.MacArthur D.G., Manolio T.A., Dimmock D.P. Guidelines for investigating causality of sequence variants in human disease. Nature. 2014;508:469–476. doi: 10.1038/nature13127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van der Ven A.T., Connaughton D.M., Ityel H. Whole-exome sequencing identifies causative mutations in families with congenital anomalies of the kidney and urinary tract. J Am Soc Nephrol. 2018;29:2348–2361. doi: 10.1681/ASN.2017121265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reese M.G., Eeckman F.H., Kulp D. Improved splice site detection in Genie. J Comput Biol. 1997;4:311–323. doi: 10.1089/cmb.1997.4.311. [DOI] [PubMed] [Google Scholar]
- 21.Yeo G., Burge C.B. Maximum entropy modeling of short sequence motifs with applications to RNA splicing signals. J Comput Biol. 2004;11:377–394. doi: 10.1089/1066527041410418. [DOI] [PubMed] [Google Scholar]
- 22.Adzhubei I.A., Schmidt S., Peshkin L. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schwarz J.M., Cooper D.N., Schuelke M. MutationTaster2:mutation prediction for the deep-sequencing age. Nat Methods. 2014;11:361–362. doi: 10.1038/nmeth.2890. [DOI] [PubMed] [Google Scholar]
- 24.Sim N.L., Kumar P., Hu J. SIFT web server: predicting effects of amino acid substitutions on proteins. Nucleic Acids Res. 2012;40:W452–W457. doi: 10.1093/nar/gks539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sibley C.R., Blazquez L., Ule J. Lessons from non-canonical splicing. Nat Rev Genet. 2016;17:407–421. doi: 10.1038/nrg.2016.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li H., Handsaker B., Wysoker A. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Van der Auwera G.A., Carneiro M.O., Hartl C. From FastQ data to high confidence variant calls:the Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinformatics. 2013;43:11–33. doi: 10.1002/0471250953.bi1110s43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Seelow D., Schuelke M., Hildebrandt F. HomozygosityMapper--an interactive approach to homozygosity mapping. Nucleic Acids Res. 2009;37:W593–W599. doi: 10.1093/nar/gkp369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hildebrandt F., Heeringa S.F., Ruschendorf F. A systematic approach to mapping recessive disease genes in individuals from outbred populations. PLoS Genet. 2009;5 doi: 10.1371/journal.pgen.1000353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saleem M.A., O'Hare M.J., Reiser J. A conditionally immortalized human podocyte cell line demonstrating nephrin and podocin expression. J Am Soc Nephrol. 2002;13:630–638. doi: 10.1681/ASN.V133630. [DOI] [PubMed] [Google Scholar]
- 31.Price S.T., Beckham T.H., Cheng J.C. Sphingosine 1-phosphate receptor 2 regulates the migration, proliferation, and differentiation of mesenchymal stem cells. Int J Stem Cell Res Ther. 2015;2:014. doi: 10.23937/2469-570x/1410014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schneider R., Deutsch K., Hoeprich G.J. DAAM2 variants cause nephrotic syndrome via actin dysregulation. Am J Hum Genet. 2020;107:1113–1128. doi: 10.1016/j.ajhg.2020.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Majmundar A.J., Buerger F., Forbes T.A. Recessive NOS1AP variants impair actin remodeling and cause glomerulopathy in humans and mice. Sci Adv. 2020;7 doi: 10.1126/sciadv.abe1386. eabe1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gebeshuber C.A., Kornauth C., Dong L. Focal segmental glomerulosclerosis is induced by microRNA-193a and its downregulation of WT1. Nat Med. 2013;19:481–487. doi: 10.1038/nm.3142. [DOI] [PubMed] [Google Scholar]
- 35.Jeanpierre C., Denamur E., Henry I. Identification of constitutional WT1 mutations, in patients with isolated diffuse mesangial sclerosis, and analysis of genotype/phenotype correlations by use of a computerized mutation database. Am J Hum Genet. 1998;62:824–833. doi: 10.1086/301806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chernin G., Vega-Warner V., Schoeb D.S. Genotype/phenotype correlation in nephrotic syndrome caused by WT1 mutations. Clin J Am Soc Nephrol. 2010;5:1655–1662. doi: 10.2215/CJN.09351209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pelletier J., Bruening W., Kashtan C.E. Germline mutations in the Wilms' tumor suppressor gene are associated with abnormal urogenital development in Denys-Drash syndrome. Cell. 1991;67:437–447. doi: 10.1016/0092-8674(91)90194-4. [DOI] [PubMed] [Google Scholar]
- 38.Barbaux S., Niaudet P., Gubler M.C. Donor splice-site mutations in WT1 are responsible for Frasier syndrome. Nat Genet. 1997;17:467–470. doi: 10.1038/ng1297-467. [DOI] [PubMed] [Google Scholar]
- 39.Klamt B., Koziell A., Poulat F. Frasier syndrome is caused by defective alternative splicing of WT1 leading to an altered ratio of WT1 +/-KTS splice isoforms. Hum Mol Genet. 1998;7:709–714. doi: 10.1093/hmg/7.4.709. [DOI] [PubMed] [Google Scholar]
- 40.Drummond I.A., Rupprecht H.D., Rohwer-Nutter P. DNA recognition by splicing variants of the Wilms' tumor suppressor, WT1. Mol Cell Biol. 1994;14:3800–3809. doi: 10.1128/mcb.14.6.3800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guo J.K., Menke A.L., Gubler M.C. WT1 is a key regulator of podocyte function:reduced expression levels cause crescentic glomerulonephritis and mesangial sclerosis. Hum Mol Genet. 2002;11:651–659. doi: 10.1093/hmg/11.6.651. [DOI] [PubMed] [Google Scholar]
- 42.Moore A.W., McInnes L., Kreidberg J. YAC complementation shows a requirement for Wt1 in the development of epicardium, adrenal gland and throughout nephrogenesis. Development. 1999;126:1845–1857. doi: 10.1242/dev.126.9.1845. [DOI] [PubMed] [Google Scholar]
- 43.Hohenstein P., Hastie N.D. The many facets of the Wilms' tumour gene, WT1. Hum Mol Genet. 2006;15(Spec No. 2):R196–R201. doi: 10.1093/hmg/ddl196. [DOI] [PubMed] [Google Scholar]
- 44.Dong L., Pietsch S., Tan Z. Integration of cistromic and transcriptomic analyses identifies Nphs2, Mafb, and Magi2 as Wilms' Tumor 1 target genes in podocyte differentiation and maintenance. J Am Soc Nephrol. 2015;26:2118–2128. doi: 10.1681/ASN.2014080819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kann M., Ettou S., Jung Y.L. Genome-wide analysis of Wilms' Tumor 1-controlled gene expression in podocytes reveals key regulatory mechanisms. J Am Soc Nephrol. 2015;26:2097–2104. doi: 10.1681/ASN.2014090940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brunskill E.W., Georgas K., Rumballe B. Defining the molecular character of the developing and adult kidney podocyte. PLoS One. 2011;6 doi: 10.1371/journal.pone.0024640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ashraf S., Gee H.Y., Woerner S. ADCK4 mutations promote steroid-resistant nephrotic syndrome through CoQ10 biosynthesis disruption. J Clin Invest. 2013;123:5179–5189. doi: 10.1172/JCI69000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Braun D.A., Lovric S., Schapiro D. Mutations in multiple components of the nuclear pore complex cause nephrotic syndrome. J Clin Invest. 2018;128:4313–4328. doi: 10.1172/JCI98688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Braun D.A., Rao J., Mollet G. Mutations in KEOPS-complex genes cause nephrotic syndrome with primary microcephaly. Nat Genet. 2017;49:1529–1538. doi: 10.1038/ng.3933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ebarasi L., Ashraf S., Bierzynska A. Defects of CRB2 cause steroid-resistant nephrotic syndrome. Am J Hum Genet. 2015;96:153–161. doi: 10.1016/j.ajhg.2014.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gee H.Y., Ashraf S., Wan X. Mutations in EMP2 cause childhood-onset nephrotic syndrome. Am J Hum Genet. 2014;94:884–890. doi: 10.1016/j.ajhg.2014.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gee H.Y., Sadowski C.E., Aggarwal P.K. FAT1 mutations cause a glomerulotubular nephropathy. Nat Commun. 2016;7:10822. doi: 10.1038/ncomms10822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gee H.Y., Saisawat P., Ashraf S. ARHGDIA mutations cause nephrotic syndrome via defective RHO GTPase signaling. J Clin Invest. 2013;123:3243–3253. doi: 10.1172/JCI69134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gee H.Y., Zhang F., Ashraf S. KANK deficiency leads to podocyte dysfunction and nephrotic syndrome. J Clin Invest. 2015;125:2375–2384. doi: 10.1172/JCI79504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Heeringa S.F., Chernin G., Chaki M. COQ6 mutations in human patients produce nephrotic syndrome with sensorineural deafness. J Clin Invest. 2011;121:2013–2024. doi: 10.1172/JCI45693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hinkes B., Wiggins R.C., Gbadegesin R. Positional cloning uncovers mutations in PLCE1 responsible for a nephrotic syndrome variant that may be reversible. Nat Genet. 2006;38:1397–1405. doi: 10.1038/ng1918. [DOI] [PubMed] [Google Scholar]
- 57.Lovric S., Goncalves S., Gee H.Y. Mutations in sphingosine-1-phosphate lyase cause nephrosis with ichthyosis and adrenal insufficiency. J Clin Invest. 2017;127:912–928. doi: 10.1172/JCI89626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ovunc B., Otto E.A., Vega-Warner V. Exome sequencing reveals cubilin mutation as a single-gene cause of proteinuria. J Am Soc Nephrol. 2011;22:1815–1820. doi: 10.1681/ASN.2011040337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rao J., Ashraf S., Tan W. Advillin acts upstream of phospholipase C 1 in steroid-resistant nephrotic syndrome. J Clin Invest. 2017;127:4257–4269. doi: 10.1172/JCI94138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ashraf S., Kudo H., Rao J. Mutations in six nephrosis genes delineate a pathogenic pathway amenable to treatment. Nat Commun. 2018;9:1960. doi: 10.1038/s41467-018-04193-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mundel P., Heid H.W., Mundel T.M. Synaptopodin:an actin-associated protein in telencephalic dendrites and renal podocytes. J Cell Biol. 1997;139:193–204. doi: 10.1083/jcb.139.1.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mundel P. [Synaptopodin:an actin-associated protein of telencephalic dendrites and of podocytes in the kidney glomerulus] Ann Anat. 1998;180:391–392. [PubMed] [Google Scholar]
- 63.Karaiskos N., Rahmatollahi M., Boltengagen A. A single-cell transcriptome atlas of the mouse glomerulus. J Am Soc Nephrol. 2018;29:2060–2068. doi: 10.1681/ASN.2018030238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yu S.M., Nissaisorakarn P., Husain I. Proteinuric kidney diseases: a podocyte's slit diaphragm and cytoskeleton approach. Front Med (Lausanne) 2018;5:221. doi: 10.3389/fmed.2018.00221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Asanuma K., Yanagida-Asanuma E., Faul C. Synaptopodin orchestrates actin organization and cell motility via regulation of RhoA signalling. Nat Cell Biol. 2006;8:485–491. doi: 10.1038/ncb1400. [DOI] [PubMed] [Google Scholar]
- 66.Dai S., Wang Z., Pan X. Functional analysis of promoter mutations in the ACTN4 and SYNPO genes in focal segmental glomerulosclerosis. Nephrol Dial Transplant. 2010;25:824–835. doi: 10.1093/ndt/gfp394. [DOI] [PubMed] [Google Scholar]
- 67.Hirakawa M., Tsuruya K., Yotsueda H. Expression of synaptopodin and GLEPP1 as markers of steroid responsiveness in primary focal segmental glomerulosclerosis. Life Sci. 2006;79:757–763. doi: 10.1016/j.lfs.2006.02.031. [DOI] [PubMed] [Google Scholar]
- 68.Srivastava T., Garola R.E., Whiting J.M. Synaptopodin expression in idiopathic nephrotic syndrome of childhood. Kidney Int. 2001;59:118–125. doi: 10.1046/j.1523-1755.2001.00472.x. [DOI] [PubMed] [Google Scholar]
- 69.Wagrowska-Danilewicz M., Danilewicz M. [Synaptopodin immunoexpression in steroid-responsive and steroid-resistant minimal change disease and focal segmental glomerulosclerosis] Nefrologia. 2007;27:710–715. [PubMed] [Google Scholar]
- 70.Barisoni L., Kriz W., Mundel P. The dysregulated podocyte phenotype:a novel concept in the pathogenesis of collapsing idiopathic focal segmental glomerulosclerosis and HIV-associated nephropathy. J Am Soc Nephrol. 1999;10:51–61. doi: 10.1681/ASN.V10151. [DOI] [PubMed] [Google Scholar]
- 71.Hill G.S., Karoui K.E., Karras A. Focal segmental glomerulosclerosis plays a major role in the progression of IgA nephropathy. I. Immunohistochemical studies. Kidney Int. 2011;79:635–642. doi: 10.1038/ki.2010.466. [DOI] [PubMed] [Google Scholar]
- 72.Asanuma K., Kim K., Oh J. Synaptopodin regulates the actin-bundling activity of alpha-actinin in an isoform-specific manner. J Clin Invest. 2005;115:1188–1198. doi: 10.1172/JCI23371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ning L., Suleiman H.Y., Miner J.H. Synaptopodin is dispensable for normal podocyte homeostasis but is protective in the context of acute podocyte injury. J Am Soc Nephrol. 2020;31:2815–2832. doi: 10.1681/ASN.2020050572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Huber T.B., Kwoh C., Wu H. Bigenic mouse models of focal segmental glomerulosclerosis involving pairwise interaction of CD2AP, Fyn, and synaptopodin. J Clin Invest. 2006;116:1337–1345. doi: 10.1172/JCI27400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yanagida-Asanuma E., Asanuma K., Kim K. Synaptopodin protects against proteinuria by disrupting Cdc42:IRSp53:Mena signaling complexes in kidney podocytes. Am J Pathol. 2007;171:415–427. doi: 10.2353/ajpath.2007.070075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chen C.K., Mungall C.J., Gkoutos G.V. MouseFinder: candidate disease genes from mouse phenotype data. Hum Mutat. 2012;33:858–866. doi: 10.1002/humu.22051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kohl S., Hwang D.Y., Dworschak G.C. Mild recessive mutations in six Fraser syndrome-related genes cause isolated congenital anomalies of the kidney and urinary tract. J Am Soc Nephrol. 2014;25:1917–1922. doi: 10.1681/ASN.2013101103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Schofield P.N., Hoehndorf R., Gkoutos G.V. Mouse genetic and phenotypic resources for human genetics. Hum Mutat. 2012;33:826–836. doi: 10.1002/humu.22077. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.