Abstract
Introduction
Most of the approximately 60 genes that if mutated cause steroid-resistant nephrotic syndrome (SRNS) are highly expressed in the glomerular podocyte, rendering SRNS a “podocytopathy.”
Methods
We performed whole-exome sequencing (WES) in 1200 nephrotic syndrome (NS) patients.
Results
We discovered homozygous truncating and homozygous missense mutation in SYNPO2 (synaptopodin-2) (p.Lys1124∗ and p.Ala1134Thr) in 2 patients with childhood-onset NS. We found SYNPO2 expression in both podocytes and mesangial cells; however, notably, immunofluorescence staining of adult human and rat kidney cryosections indicated that SYNPO2 is localized mainly in mesangial cells. Subcellular localization studies reveal that in these cells SYNPO2 partially co-localizes with α-actinin and filamin A−containing F-actin filaments. Upon transfection in mesangial cells or podocytes, EGFP-SYNPO2 co-localized with α-actinin-4, which gene is mutated in autosomal dominant SRNS in humans. SYNPO2 overexpression increases mesangial cell migration rate (MMR), whereas shRNA knockdown reduces MMR. Decreased MMR was rescued by transfection of wild-type mouse Synpo2 cDNA but only partially by cDNA representing mutations from the NS patients. The increased mesangial cell migration rate (MMR) by SYNPO2 overexpression was inhibited by ARP complex inhibitor CK666. SYNPO2 shRNA knockdown in podocytes decreased active Rac1, which was rescued by transfection of wild-type SYNPO2 cDNA but not by cDNA representing any of the 2 mutant variants.
Conclusion
We show that SYNPO2 variants may lead to Rac1-ARP3 dysregulation, and may play a role in the pathogenesis of nephrotic syndrome.
Keywords: monogenic kidney disease, nephrotic syndrome, SYNPO2
Nephrotic syndrome (NS) is characterized by persistent proteinuria, caused by disruption of the glomerular filtration barrier.1 It is the second most frequent cause of chronic kidney disease (CKD) before the age of 25 years. Its incidence of 1.15 to 16.9 per 100,000 children varies by ethnicity and region.2,3 Nephrotic syndrome is classified by its response to steroid treatment into steroid-sensitive nephrotic syndrome (SSNS) and steroid-resistant nephrotic syndrome (SRNS), with clinical overlap, as some individuals with SSNS may later develop SRNS. Steroid-sensitive nephrotic syndrome constitutes 80% of all childhood NS. Steroid-resistant nephrotic syndrome is associated with increased risk of progression to end-stage renal disease.2
The identification of monogenic causes of SSNS and SRNS has revealed 60 single-gene etiologies3,4 that have generated deep insight into the pathogenesis of SRNS, such as dysregulation of the slit glomerular membrane and dysfunction of actin remodeling causing podocyte dysfunction. These disease genes are predominantly expressed in the glomerular podocyte, and mutations in these genes will cause defects in podocyte structure and function,5,6 leading to the definition of SRNS as a “podocytopathy.” However, of these monogenic causes of SRNS,4 6 (TRPC6, SCARB2, KANK2, ITSN1, KANK4, and EMP2) are more specifically expressed in mesangial cells rather than podocytes when evaluating data on ScRNA sequencing7 (Supplementary Figure S1).
The podocyte has a unique cytoskeletal architecture that relies on actin cytoskeleton to stabilize the glomerular capillaries and to participate in glomerular barrier function via cell–cell junction and cell–matrix proteins.8 Support for the pivotal role of podocyte actin cytoskeleton remodeling in the pathogenesis of NS came from the discovery in patients with SRNS of monogenic mutations in actin cytoskeleton−related genes, such as ACTN4 (encoding α-actinin-4),9 MYO1E (encoding myosin IE),10 INF2 (encoding inverted formin-2),11 ARHGDIA (encoding Rho GDP dissociation inhibitor α),12 and AVIL (encoding advillin).13 Mutations in the above genes cause profound changes in the podocyte actin cytoskeleton. Extensive research has shown that actin cytoskeletal dynamics are modulated by the Rho-like small GTPases, RhoA/Rac1/ Cdc42, although the mechanism is not fully understood.14
Mesangial cells are smooth muscle−like cells that maintain the structural integrity of the glomerular microvascular bed and mesangial matrix homeostasis in communication with podocytes.15 In PDGFB- or PDGFBR-deficient mice, glomeruli are lacking mesangial cells and appear as balloon-like structures.16 Mice with homozygous deletion of Itgb8 show a renal glomerular phenotype that features endothelial cell apoptosis,17 whereas mesangial cell−specific conditional Itgb8 knockout mice show glomerular capillary microaneurysms and delayed recovery after injury.18 Mesangial cells and their matrix form the central stalk of the glomerulus and are part of a functional unit interacting closely with endothelial cells and podocytes.19 These 3 cell types each play critical roles during capillary tuft development, known as cellular cross-talk of VEGF/VEGFR between podocyte and endothelial cells, PDGFB/PDGFBRB between endothelial cells and mesangial cells.16Alterations in 1 cell type can produce changes in the others.
To identify additional monogenic causes of NS that might help to better understand its pathogenesis, we applied homozygosity mapping (HM) and whole-exome sequencing (WES) to our cohort of 1200 families with SRNS and discovered recessive mutations in the gene SYNPO2 in 2 unrelated families as a likely novel monogenic cause of SRNS. We demonstrate that SYNPO2 is more strongly expressed in glomerular mesangial cells in vivo than in podocytes, and that the synaptopodin-2 (SYNPO2) protein co-localizes with F-actin and α-actinin-4, which, if mutated, cause autosomal dominant SRNS. We demonstrate that cell migration defects upon SYNPO2 knockdown are rescued by re-expression of SYNPO2, but not by mutants detected in SRNS patients. We delineate a novel pathogenic axis for NS caused by SYNPO2 mutations that includes loss of Rac1 activation and defects of actin remodeling along the Rac1-ARP2/3 pathway. Targeting the Rac1-ARP3 pathway may offer a therapeutic approach to SRNS.
Materials and Methods
For information on materials and methods used, please see Supplementary Materials and Methods, available online.
Results
Recessive Mutations of SYNPO2 Cause SRNS
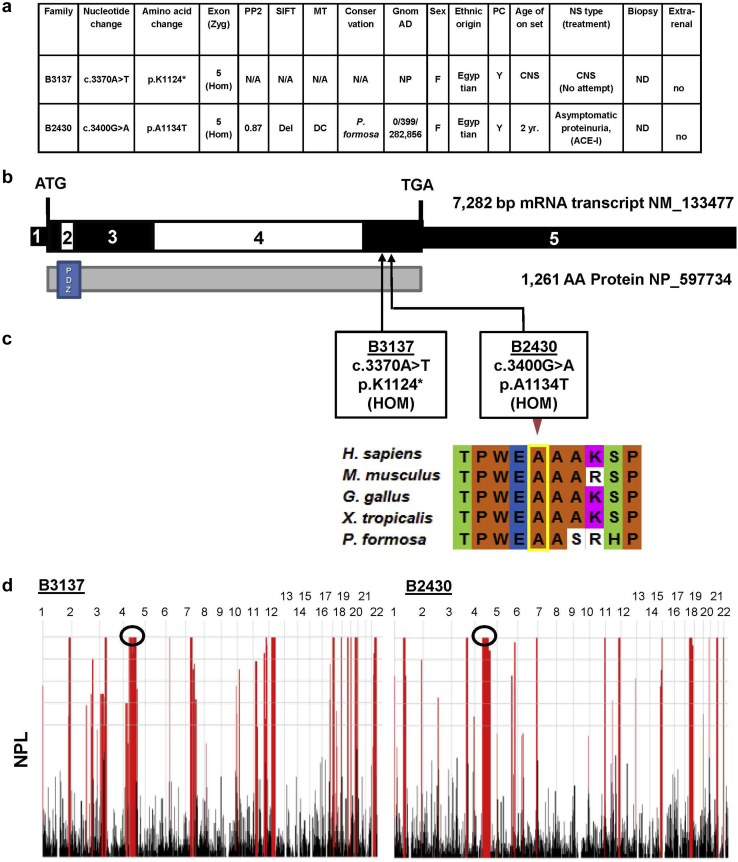
To elucidate the molecular pathogenesis of SRNS, we applied homozygosity mapping and WES of our cohort of approximately 1200 individuals with SRNS. Two consanguineous families were identified with homozygous SYNPO2 mutations (Figure 1a and Supplementary Figure S2). Subject B3137, a girl of Egyptian descendants, had congenital-onset nephrotic syndrome. By WES, we identified a homozygous SYNPO2 truncating mutation (c.3370A>T, p.Lys1124∗) (Figure 1). This variant was never reported in either a homozygous or heterozygous state in the control genome database gnomAD. No treatment was attempted.
Figure 1.
SYNPO2 mutations identified in 2 families in nephrotic syndrome (NS). (a) Summary of genetic and phenotypic data for 2 patients with SYNPO2 mutations. ACE-I, angiotensin-converting enzyme inhibitor; CNS, congenital nephrotic syndrome; DC, disease causing; Del, deleterious; F, female; gnomAD, Genome Aggregation database; Hom, homozygous; M, male; MT, mutation taster; N/A, not applicable; ND, not done; NP, not present in control variant database; PC, parental consanguinity; PP2, PolyPhen-2 prediction score; SIFT, “Sorting Tolerant from Intolerant” prediction score; Zyg, Zygosity. (b) Exon structure (upper bar) and protein domain content (lower bar) structures of SYNPO2 are shown with arrows indicating positions of mutations in patients (B3137 and B2430) with NS or persistent proteinuria. (c) Evolutionary conservation of amino acid position A1134 in SYNPO2 protein across evolution. (d) Homozygosity mapping data across the genome was generated using nonparametric LOD scores (NPL scores) based on WES variant data using Homozygosity Mapper for individuals B3137 and B2430. Black circles demonstrate the NPL peak regions, in which SYNPO2 mutations were positioned.
Subject B2430, a 2-year old girl, was diagnosed with persistent proteinuria, around 300 to 500 mg/d. She had no edema and received angiotensin-converting enzyme inhibitor treatment. By WES, we detected a homozygous missense mutation (c.3400G>A, p.A1134T). This homozygous variant was never reported in the gnomAD database. Ala1134 is conserved to Poecillia formosa (Figure 1). The mutation yielded strong in silico prediction scores for being deleterious (Figure 1a).
SYNPO2 mRNA Expression in Kidney and Mesangial Cells
Since the SYNPO2 gene has several transcripts (Supplementary Figure S4), it was important to show that the exon containing the mutations is expressed in the (human) kidney. First, we analyzed the expression data available from the Human Protein Atlas at https://v15.proteinatlas.org/. The p.Lys1124∗ and p.Ala1134Thr mutations are located in human exon 5, which is alternatively spliced and expressed tissue specifically. Using reverse transcription−polymerase chain reaction experiments with primers covering all different human or rat exons, we could confirm expression of the mutation-containing exon in the human kidney (Supplementary Figure S4), and in rat kidney mesangial and smooth muscle cells (Supplementary Figure S5). In Xenopus, we validated expression enrichment around the proximal pronephrous/glomus region with no labeling by a control probe (Supplementary Figure S4).
SYNPO2 Shows Strongest Expression in Glomeruli Mesangial Cells
We checked published databases to evaluate which glomerular cell types show the highest mRNA expression levels of SYNPO2, and found that expression is almost exclusively detected in mesangial cells and less so in podocytes, endothelial cells, or other cell types (Supplementary Figure S3).
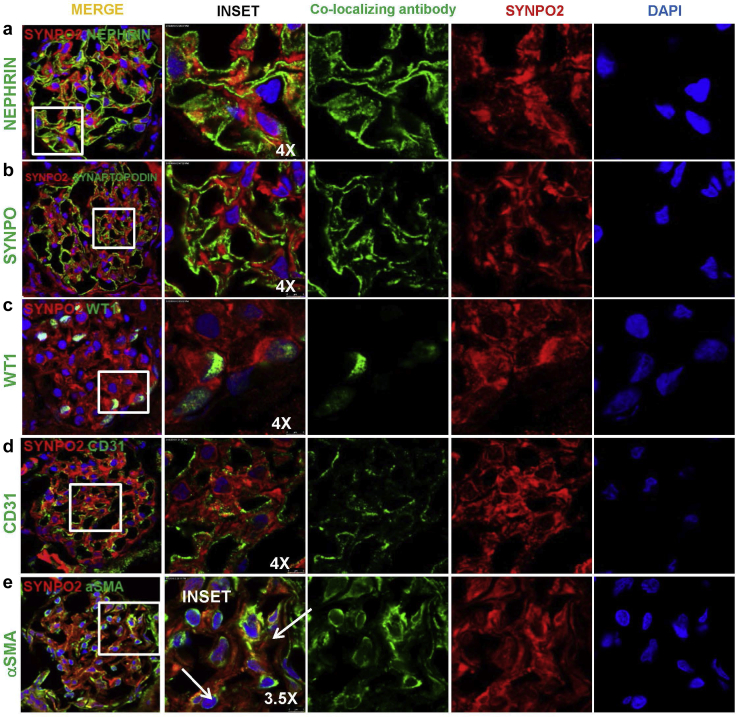
We tested endogenous SYNPO2 expression in renal glomeruli using commercially available and well-established home-made anti-SYNPO2 antibodies20,21 (Supplementary Figure S8). We established further SYNPO2 antibodies (#1 and #2) identified tagged SYNPO2 protein by immunoblotting and immunofluorescence (Supplementary Figures S6 and S7). We found signal produced by this antibody clearly reduced in SYNPO2 CRISPR podocytes by immunofluorescence (Supplementary Figure S6 and S7), thus specifically recognizing SYNPO2. Using antibody #1, we performed SYNPO2 co-staining with characteristic marker of glomerular cells, WT1 and Nephrin (podocytes), CD31 (endothelial cells), and aSMA (mesangial cells) in rat kidney frozen sections. We detected SYNPO2 in rat glomeruli by immunofluorescence, partially co-localizing with α−smooth muscle actin (Figure 2). SYNPO2 was not co-localized with the podocyte or endothelial cell markers (Figure 2). We also tested SYNPO2 localization with the mouse monoclonal antibody “HH9” in rat and human frozen kidney sections. The SYNPO2 signal localized mainly to the mesangial matrix (Supplementary Figure S8). Importantly, HH9 monoclonal and M2 polyclonal antibodies resulted in an identical staining pattern in human kidney cryosections. It is the same pattern observed in human FFPE kidney sections, where immunohistochemistry also localized SYNPO2 to mesangial matrix (Supplementary Figure S8). These results show that SYNPO2 expression is highest in mesangial cells. This was consistent with single-cell RNA sequencing data from mouse7 and human (http://humphreyslab.com/SingleCell) (Supplementary Figure S3). We thus discovered the unusual situation that a potential monogenic cause of NS (i.e., SYNPO2 mutation) might result from a cellular dysfunction affecting mesangial cells more than podocytes.
Figure 2.
SYNPO2 immunofluorescence stain with antibody (Ab#1 Abcam, ab50192) in rat glomeruli. Adult rat kidney sections were stained with SYNPO2, and co-stained with cell type marker antibodies against nephrin (podocyte slit membrane), SYNPO (podocyte cytoplasm), WT1 (podocyte nucleus), CD31 (endothelial cell), and aSMA (mesangial cells). SYNPO2 staining is detected in rat glomeruli by immunofluorescence on kidney frozen sections but is not co-localizing with nephrin, SYNAPTOPODIN, WT1, or CD31. However, SYNPO2 partially co-localizes with aSMA, a mesangial cell marker (white arrows). DAPI stains nuclear (blue). Bar = 5 μm.
SYNPO2 Co-localizes With F-Actin and Induces Formation of Distinct F-Actin Networks
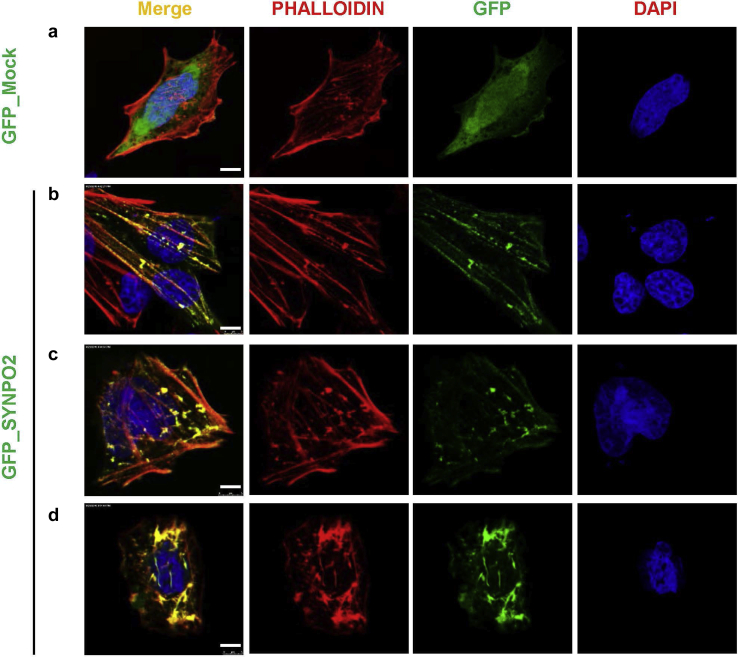
To further determine its subcellular localization and potential cellular function, we overexpressed green fluorescent protein (GFP)-tagged SYNPO2 in a rat mesangial cell line and co-stained with phalloidin (Figure 3).22 SYNPO2 overexpression induced 2 distinct F-actin patterns and differentially co-localized with these networks. Co-staining for F-Actin with phalloidin showed long and well-organized actin bundles frequently oriented in parallel along axis of the cell (Figure 3b and 3c). The alternative pattern seen upon GFP-SYNPO2 overexpression showed perinuclear circles and bipolar fusiform morphology with thick irregular actin bundles (Figure 3d).
Figure 3.
SYNPO2 co-localizes with F-actin networks in 2 distinct patterns in mesangial cells. (a) Rat mesangial cells (RMCs) transfected with green fluorescent protein (GFP) mock negative control were almost devoid of large actin fibers in the cell body but displayed strong F-actin staining around the cell periphery. (b, c) Transfection of RMCs with GFP_SYNPO2. SYNPO2 induces 2 distinct F-actin patterns, co-localizing with these networks. F-actin staining shows long and well-organized actin bundles, frequently oriented parallel along the axis of the cell. (d) The alternative pattern shows perinuclear and bipolar fusiform morphology with thick actin irregular bundles. Bar = 5 μm.
SYNPO2 Interacts With ACTN1 and ACTN4, Whereas Mutants Partially Abrogate This Interaction
We analyzed the expression of SYNPO2 in a rat mesangial cells with specific antibodies, and compared the staining pattern to that of α-actinin and filamin, 2 binding partners of SYNPO2. Indeed, SYNPO2 was found to be at least partially co-localized with both F-actin−binding proteins (Supplementary Figure S9).
We previously showed that the variant of SYNPO2 expressed in skeletal muscle interacts with ACTN2.20 Because data from http://humphreyslab.com/SingleCell indicate that mesangial cells express ACTN1 and ACTN4, we investigated whether SYNPO2 also interacts with these α-actinin isoforms. We hypothesized that SYNPO2 might also interact with ACTN4, an actin-binding protein that is known to cause monogenic SRNS if mutated. We found that, upon overexpression in mesangial cells, ACTN4 co-localized with GFP-SYNPO2, but not with GFP-Mock (Supplementary Figure S9), in a pattern similar to the pattern that we found with SYNPO2−F-actin co-localization (Figure 3). ACTN4 co-localized with SYNPO2 in a pattern of bundles parallel to the long axis of the cell and in a perinuclear meshwork (Supplementary Figure S9).
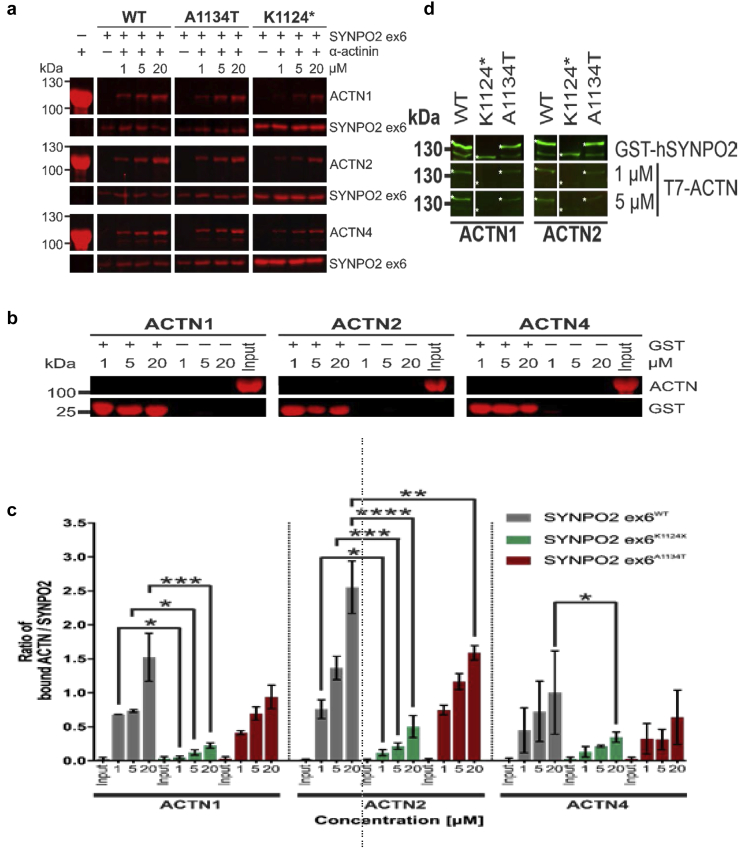
Pulldown assays confirmed direct interaction of SYNPO2 with ACTN2, and also revealed interaction of SYNPO2 with ACTN1 and ACTN4 (Figure 4). Differences in the amount of bound α-actinin indicated that ACTN2 bound strongest, whereas ACTN4 showed the weakest interaction. Because SYNPO2 contains several independent ACTN-binding sites,20 we analyzed binding of only the part of the protein that is encoded by exon 5 (starting with amino acid 1085) alone using GST pull down assays, and found that this part of the protein contains yet another α-actinin binding site (Figure 4a). We analyzed the effects of both mutations on the capacity of the carboxy terminus of the SYNPO2 variants to bind α-actinins. Both p.Lys1124∗ and p.Ala1134Thr SYNPO2 showed reduced binding. These experiments revealed that this part of the protein contained another independent binding site for all α-actinins tested and that the binding strength of the SYNPO2 mutants is considerably reduced when compared to that of wild-type SYNPO2 (Figure 4c). Because of the large size of full-length SYNPO2, we also analyzed a truncated version that lacks the amino-terminal 394 amino acids (the start of the main isoform expressed in skeletal muscles20) in our interaction assays (Figure 4d). Western blot overlay experiments in which the SYNPO2 variants were run on a gel, blotted to nitrocellulose, and overlaid with the different α-actinin isoforms confirmed these findings: ACTN1 and ACTN2 bound strongest to wild-type SYNPO2, whereas interaction to p.Ala1134Thr and p.Lys1124∗ SYNPO2 was slightly and strongly reduced, respectively (Figure 4d). In this assay, ACTN4 binding was probably too weak to show binding to any of the SYNPO2 variants. We speculate that the mislocalization of ACTN (Figure 3) resulted from the reduced binding to mutant SYNPO2.
Figure 4.
Mutations in SYNPO2 reduce binding to α-actinin. (a) Pulldown experiments. Different quantities of α-actinin (ACTN) were added to GST-SYNPO2 ex6 (aminoacids 1085−1261) variants bound to GSH-beads. Gel fractions show bound ACTN1, 2, and 4 (upper panels), as well as GSH-bound SYNPO2 (lower panels). Left panels show α-actinin for quality control. Concentrations (μM) correspond to amount of actinin input. (b) Control pulldown experiments. No nonspecific binding of α-actinin to GSH-beads (−) or GST-coupled beads (+) could be observed. (c) Quantification of the experiment shown in (a). Ratios of α-actinin (ACTN) to SYNPO2 bands were calculated to quantify bound α-actinin-1, α-actinin-2, and α-actinin-4. ACTN1, ACTN2, and ACTN4 show significantly impaired binding to SYNPO2K1124∗ mutants compared to wild-type SYNPO2. The A1134T-mutation significantly impairs binding of α-actinin2 at higher concentrations. Bars represent standard error of n = 3. ∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001; ∗∗∗∗P < 0.0001. (d) Western blot overlay of α-actinin1, 2 to immobilized SYNPO2. Purified WT or mutant SYNPO2 (amino acids 396−1261) are indicated with asterisks. In comparison to wild type, binding of α-actinin1, and 2 is impaired for both SYNPO2 mutants.
Expression of Mutant SYNPO2 Affects Actin Cytoskeleton Formation
To further analyze the effects of the mutations on the actin cytoskeleton, we transfected PtK2 cells (Potorous tridactylis kidney cells) with the different SYNPO2 variants. Wild-type GFP- SYNPO2 bound mainly to stress fibers in a periodic fashion, whereas the localization of α-actinin was not affected (indicated by arrows). By contrast, GFP-SYNPO2 A1134T and GFP_SYNPO2 K1124∗ mainly co-localized with α actinin in granula and showed partial (A1134T) or hardly any (K1124∗) α actinin left in stress fibers (indicated by arrowheads). (Supplementary Figure S10).
Wild-Type SYNPO2, but Not Mutants, Rescues Rac1 Activation
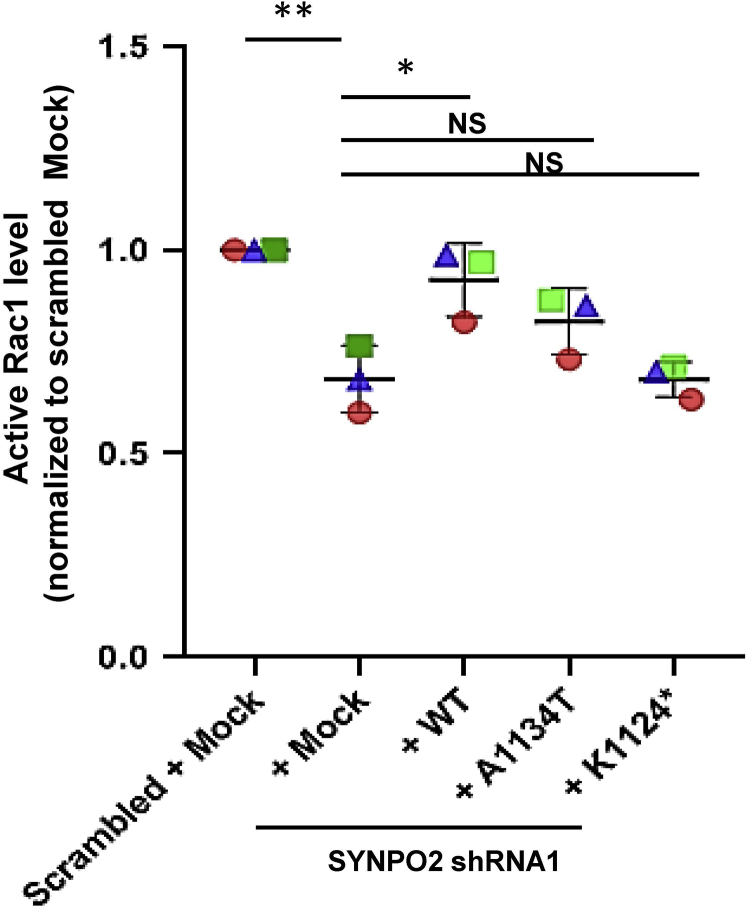
The small Rho-like GTPases, RhoA, Rac1, and Cdc42 play a pivotal role in podocyte cell shape change and in the pathogenesis of monogenic forms of SRNS.23 To examine whether pathogenic mechanisms of SYNPO2 loss of function are connected to altered Rho/Rac/CDC42 activation with consecutive defects of actin remodeling, we performed Rho/Rac/CDC42 G-LISA. We have generated shRNA-induced SYNPO2-knockdown podocyte cell line and confirmed protein knockdown by western blot (Supplementary Figure S12 and S13). We found that upon SYNPO2 knockdown, active Rac1 was significantly reduced compared to scrambled shRNA-negative control cells. This reduction was rescued by wild-type Synpo2 cDNA transfection, Synpo2 cDNA constructs representing the missense mutation of Ala1134Thr and Lys1124∗ failed to rescue (Figure 5).
Figure 5.
Synpo2 rescued active Rac1 in SYNPO2 knockdown cell line, but not by mutants from nephrotic syndrome (NS) patients. Active levels of Rac1 were measured by Rac1 G-LISA assay. shRNA-mediated knockdown of SYNPO2 in human podocytes (+ Mock) reduced active Rac1. Overexpression of wild-type Synpo2 cDNA (+ WT) rescued this effect, but the Synpo2 cDNA constructs reflecting mutations in NS patients, failed to rescue reduction of active Rac1 (+A1134T, +K1124∗). P values calculated by one-way analysis of variance. ∗P < 0.05, ∗∗P < 0.01. NS, nonsignificant.
There was no significant difference in active Rac1 level upon SYNPO2 overexpression compared to that in mock overexpression (Supplementary Figure S11). There was also no significant difference in CDC42 activation upon SYNPO2 overexpression and knockdown cells (Supplementary Figure S11), in either of RhoA (data not shown). The results demonstrate that SYNPO2 mutations may interfere with podocyte migration through lack of Rac1 activation.
Wild-Type Synpo2, but Not Mutants From NS Patients, Rescues Mesangial Cell Migration Rate
Because of the role that we discovered for SYNPO2 in mesangial cells and actin cytoskeleton remodeling, we hypothesized that SYNPO2 might regulate mesangial cell migration rate (MMR), which is an established intermediate phenotype of numerous SRNS disease genes.
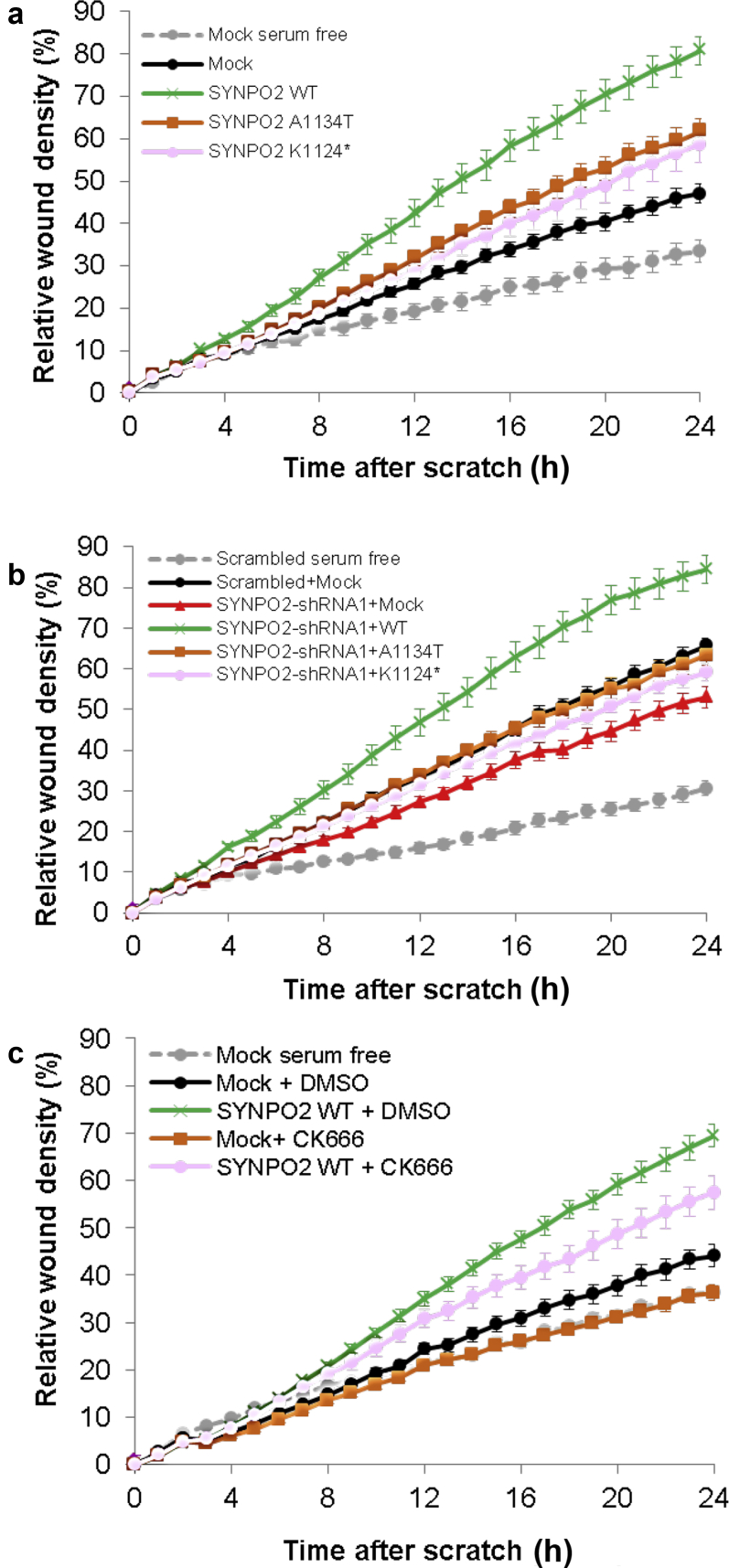
We observed that overexpression of SYNPO2 WT strongly increased MMR, whereas human SYNPO2 cDNA constructs representing both mutants from NS patients showed a milder increase in migration rate compared to WT SYNPO2 (Figure 6a). Mesangial cell migration rate was reduced by SYNPO2 shRNA knockdown. This reduction was rescued by transient expression of wild-type Synpo2. By contrast, mouse Synpo2 cDNA constructs reflecting mutations in NS patients only partially rescued MMR (Figure 6b).
Figure 6.
Wild-type but not mutant SYNPO2 increases migration rate and acts upstream of ARP2/3. (a) In a human mesangial cell line expressing Mock negative control, MMR is increased following serum addition (black) relative to serum-free conditions (gray). All subsequent experiments were performed in serum replete conditions. Overexpression of SYNPO2 WT (green) strongly increased MMR compared to Mock (black). Human SYNPO2 cDNA constructs representing mutants from NS patients (p.A1134T and p. K1124∗) partially rescued MMR compared to Mock (orange and pink). (B) In a human mesangial cell line expressing scrambled shRNA, MMR is increased following serum addition (black) relative to serum-free conditions (gray). All subsequent experiments were performed in serum-replete conditions. Knockdown of SYNPO2 (red) showed reduced MMR compared with scrambled shRNA with Mock overexpression (black). Migration was rescued by overexpression of WT Synpo2 construct (green). Mouse Synpo2 cDNA constructs reflecting mutations in NS patients only partially rescued the MMR (orange and pink). (c) ARP3 complex is an effector for mesangial cell migration downstream of SYNPO2. In a human mesangial cell line expressing Mock negative control, MMR is increased following serum addition (black) with dimethylsulfoxide (DMSO) control relative to serum-free conditions (gray). All subsequent experiments were performed in serum-replete conditions. Overexpression of SYNPO2 WT (green) with DMSO control strongly increased MMR compared to Mock (black) with DMSO control. However, the increased migration (green) was reduced with ARP3 inhibitor CK666 (pink). In Mock transfected cells, migration was also reduced with CK666 (orange) compared to DMSO (black). CK666 = 50 μM.
ARP3 Is Required to Enhance Cell Migration Mediated by SYNPO2 Overexpression
We previously showed that SYNPO2 induces lamellipodia formation through ARP2/3 complex.24 We therefore tested whether the ARP3 inhibitor CK666 could reduce migration induced by SYNPO2.
We found that the human mesangial cell migration rate was increased by SYNPO2 wild-type overexpression, but this was inhibited in the presence of ARP complex inhibitor CK666 (Figure 6c). In Mock-transfected cells, migration was also reduced with CK666 treatment.
Discussion
In this study, we identified recessive mutations in SYNPO2 as a potential novel single-gene cause of SRNS. We delineate that SYNPO2 mutations result in defective mesangial cell migration rate and impaired Rac1 activation upstream of ARP2/3 complex formation. SYNPO2 belongs to the synaptopodin family and is a paralogue of synaptopodin (SYNPO).25 SYNPO, a specific marker for podocytes,26 is an actin-binding protein that plays an important role in the regulation of the actin cytoskeleton through RhoA.27,28 SYNPO2 is also an actin binding protein, known to induce distinct actin bundling patterns.29 However, little is known about its role in kidney physiology and renal pathogenesis. We have shown that SYNPO2 localizes to the cytoplasm of rat and human glomerular mesangial cells more than in podocytes. A recent report by Chung et al.30 suggested that Synpo2 expression may be largely enhanced in smooth muscle cells rather than mesangial cells; however, our findings are consistent with most of the recently published single cell RNA sequencing data7 (http://humphreyslab.com/SingleCell). Furthermore, we confirm that SYNPO2 co-localizes with F-actin and has F-actin bundling activity, and both mutant variants showed decreased actin bundling activity.
α-Actinin-4 (ACTN4) is an actin-binding protein that interacts with actin filaments, linking transmembrane proteins to the actin cytoskeleton by forming focal adhesions with other proteins including integrins, vinculin, zyxin, and paxillin.31,32 ACTN4 is expressed in human glomeruli, and mutations in ACTN4 cause focal segmental glomerulosclerosis.9 Mice deficient in Actn4 have severe proteinuria and glomerular disease.33 It was previously shown that SYNPO2 interacts with α-actinin-2.20 Here we show that SYNPO2 interacts with α-actinin-1 and α-actinin-4, the isoforms that are expressed in glomerular mesangial cells, which suggests that the pathogenic mechanism of SYNPO2 loss of function may be α-actinin related. This notion is strongly suggested by our finding that SYNPO2 mutants show a disrupted interaction with α-actinin, probably leading to pathologic cytoskeleton dysregulation.
SYNPO2 plays an important role in the metastasis of breast cancer via cell migration.34, 35, 36 Moreover, SYNPO2 phosphorylation by interaction of integrin-linked kinase (ILK) leads to suppression of cell growth and motility in prostate cancer cells.37 Furthermore SYNPO2 can promote lamellipodia formation.24 In this study, we demonstrate that SYNPO2 loss of function leads to migration defects in mesangial cell via an ARP3-related pathway. ARP2/3 complex mediates F-actin nucleation, thereby enhancing F-actin branching and promoting lamellipodia protrusion.38 We therefore speculate that a possible mechanism of SYNPO2 loss of function is related to decreased ARP3 activation, leading to less lamelipodia formation and migration defects.
The Rho-like small GTPases RhoA, Rac1, and Cdc42 are regulators of the actin cytoskeleton.39 Podocytes depend on a highly dynamic and tightly regulated actin cytoskeleton to generate and maintain their actin-based foot processes and the slit membrane of the renal glomerular filter.8 Dysregulation of these Rho-like small GTPases has been observed in several monogenic forms of human SRNS40, 41, 42, 43 and murine models of SRNS.44 Rho/Rac1/CDC42 are also important in maintaining mesangial cell function, as well as in the pathogenesis of kidney disease. Rac1 was shown to be required for normal mesangial cell morphology and thrombospondin-1 expression,45 and RhoA activation leads to matrix upregulation in mesangial cells.46 We observed that knockdown of SYNPO2 in mesangial cells and immortalized human podocytes decreased the activity state of Rac1, which was rescued by wild-type SYNPO2 but not by mutants detected in patients. This suggests that SYNPO2 mutations may exert their pathogenic effect via dysregulation of Rac1 signaling. In context of our finding that SYNPO2 overexpression resulted in increased migration that was inhibited by the ARP3 inhibitor CK666, we speculate that SYNPO2 mutations interfere with cell migration through the Rac1−ARP2/3 pathway. However, a RAC1-independent pathway may also be involved, as active RAC1 was not increased upon SYNPO2 overexpression. The specific cellular and subcellular localization pattern of SYNPO2 could be further characterized in the future by additional experiments in mesangial cells that use phalloidin staining in cells expressing wild-type or mutant SYNPO2 and by endogenous immunoprecipitation experiments of SYNPO2 and α-actinin-4.
One of the limitations in our study is the low number of families found with variants in SYNPO2 in our cohort, which is below the ACMG threshold defining the discovery of a gene in 3 or more families as strong supporting evidence.47 In addition, 1 of the identified variants (rs137992021) appears to be a common single-nucleotide polymorphism in the African population, with a frequency in gnomAD of 1.5% (although no homozygous variants were reported). Therefore, we suggest SYNPO2 as a candidate gene for SRNS. A more definite conclusion regarding the causality of SYNPO2 in SRNS will emerge from future studies such as the generation of Synpo2 knockout mouse model.
We delineate novel pathogenic axis for NS with loss-of-function mutations in SYNPO2, leading to both podocytic and mesangial cell dysfunction through Rac1−ARP3 dysregulation. It is of note that the pathogenesis of this potential monogenic form of NS might be mediated mostly by mesangial cell defects, as SYNPO2 is more profoundly expressed in mesangial cells. However, a contribution of podocytic SYNPO2 loss of function to the pathogenesis of SRNS cannot be excluded, even though its expression level in podocytes is low.
Disclosure
FH is a co-founder of Goldfinch Biopharma Inc. All the other authors declare no competing interests.
Acknowledgments
We are grateful to the families and study individuals for their contribution. FH is the William E. Harmon Professor of Pediatrics. This research is supported by a grant from the National Institutes of Health to FH (5R01DK076683-13) and by the “EPT” program of Shanghai Children’s Medical Center to YM. AJM was supported by an NIH Training Grant (T32DK-007726), the 2017 Post-doctoral Fellowship Grant from the Harvard Stem Cell Institute, and the American Society of Nephrology Lipps Research Program 2018 Polycystic Kidney Disease Foundation Jared J. Grantham Research Fellowship. FB was supported by a fellowship grant (404527522) from the German Research Foundation (DFG). ACO is supported by the National Institutes of Health F32 Ruth L. Kirschstein Postdoctoral Individual National Research Service Award (DK122766).
Author Contributions
YM, RS, VK, FB, TMK, NM, MN, ACO-W, AJM, TH, KD, WZ, SS, and FH performed genetic analysis and functional studies. YM and RS performed cDNA cloning, IFs, migration assay, and G-LISA.
KL, MA, PFMvdV, and DOF performed GST-Pulldown assay, RT-PCR, transfections, and IF in kidney. NNM and RD performed lamellipodia formation. JM and MK performed in situ hybridizaion experiments in Xenopus. HMF recruited patients and gathered detailed clinical information for the study. SM and RPL performed WES. FH conceived of and directed the project. YM, RS, and FH wrote the paper, which was critically reviewed by all authors. HMF, JAK, SED, LAE, HSA, and MA-S contributed patient data.
Footnotes
Supplementary Methods
Figure S1. Single cell RNA sequencing results from mouse glomerulus from Muller data. mRNA expression (Z-score) of 60 known SRNS genes.
Figure S2. Sanger sequencing confirms biallelic SYNPO2 mutations in 2 families with nephrotic syndrome.
Figure S3. SYNPO2 expression in kidney.
Figure S4. SYNPO2 isoform expression in the human kidney and other tissues (A-D), and expression pattern in situ hybridization in Xenopus (E).
Figure S5. Extent of the SYNPO2 antigen used to generate for the SYNPO2 antibody used in study.
Figure S6. Characterization of SYNPO2 antibody (Ab#1, Abcam, ab50192) specificity.
Figure S7. SYNPO2 antibody (Ab#2, Sigma, HPA030665, Ab#3, Prosci, 6051) characterization.
Figure S8. SYNPO2 staining of human kidney section.
Figure S9. SYNPO2 co-localizes with aactinin and filamin A and partially co-localizes with ACTN4 in rat mesangial cells (RMC).
Figure S10. Transient transfections of PtK2 cells with eGFP-fusion proteins of wild-type and mutant variants of SYNPO2.
Figure S11. G-LISA for active Rac1 and CDC42 with SYNPO2.
Figure S12. SYNPO2 knockdown in human podocytes and human mesangial cells.
Figure S13. Western blot from lysates of stable SYNPO2-shRNA and scrambled shRNA expressing human immortalized podocytes co-transfected for rescue constructs.
Supplementary Material
Supplementary Methods
Figure S1. Single cell RNA sequencing results from mouse glomerulus from Muller data. mRNA expression (Z-score) of 60 known SRNS genes.
Figure S2. Sanger sequencing confirms biallelic SYNPO2 mutations in 2 families with nephrotic syndrome.
Figure S3. SYNPO2 expression in kidney.
Figure S4. SYNPO2 isoform expression in the human kidney and other tissues (A-D), and expression pattern in situ hybridization in Xenopus (E).
Figure S5. Extent of the SYNPO2 antigen used to generate for the SYNPO2 antibody used in study.
Figure S6. Characterization of SYNPO2 antibody (Ab#1, Abcam, ab50192) specificity.
Figure S7. SYNPO2 antibody (Ab#2, Sigma, HPA030665, Ab#3, Prosci, 6051) characterization.
Figure S8. SYNPO2 staining of human kidney section.
Figure S9. SYNPO2 co-localizes with aactinin and filamin A and partially co-localizes with ACTN4 in rat mesangial cells (RMC).
Figure S10. Transient transfections of PtK2 cells with eGFP-fusion proteins of wild-type and mutant variants of SYNPO2.
Figure S11. GLISA for active Rac1 and CDC42 with SYNPO2.
Figure S12. SYNPO2 knockdown in human podocytes and human mesangial cells.
Figure S13. Western blot from lysates of stable SYNPO2-shRNA and scrambled shRNA expressing human immortalized podocytes co-transfected for rescue constructs.
References
- 1.Wiggins R.C. The spectrum of podocytopathies: a unifying view of glomerular diseases. Kidney Int. 2007;71:1205–1214. doi: 10.1038/sj.ki.5002222. [DOI] [PubMed] [Google Scholar]
- 2.Trautmann A., Schnaidt S., Lipska-Zietkiewicz B.S. Long-term outcome of steroid-resistant nephrotic syndrome in children. J Am Soc Nephrol. 2017;28:3055–3065. doi: 10.1681/ASN.2016101121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sadowski C.E., Lovric S., Ashraf S. A single-gene cause in 29.5% of cases of steroid-resistant nephrotic syndrome. J Am Soc Nephrol. 2015;26:1279–1289. doi: 10.1681/ASN.2014050489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Warejko J.K., Tan W., Daga A. Whole exome sequencing of patients with steroid-resistant nephrotic syndrome. Clin J Am Soc Nephrol. 2018;13:53–62. doi: 10.2215/CJN.04120417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Park J., Shrestha R., Qiu C. Single-cell transcriptomics of the mouse kidney reveals potential cellular targets of kidney disease. Science. 2018;360:758–763. doi: 10.1126/science.aar2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wharram B.L., Goyal M., Wiggins J.E. Podocyte depletion causes glomerulosclerosis: diphtheria toxin-induced podocyte depletion in rats expressing human diphtheria toxin receptor transgene. J Am Soc Nephrol. 2005;16:2941–2952. doi: 10.1681/ASN.2005010055. [DOI] [PubMed] [Google Scholar]
- 7.Karaiskos N., Rahmatollahi M., Boltengagen A. A single-cell transcriptome atlas of the mouse glomerulus. J Am Soc Nephrol. 2018;29:2060–2068. doi: 10.1681/ASN.2018030238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Faul C., Asanuma K., Yanagida-Asanuma E. Actin up: regulation of podocyte structure and function by components of the actin cytoskeleton. Trends Cell Biol. 2007;17:428–437. doi: 10.1016/j.tcb.2007.06.006. [DOI] [PubMed] [Google Scholar]
- 9.Kaplan J.M., Kim S.H., North K.N. Mutations in ACTN4, encoding alphaactinin-4, cause familial focal segmental glomerulosclerosis. Nat Genet. 2000;24:251–256. doi: 10.1038/73456. [DOI] [PubMed] [Google Scholar]
- 10.Mele C., Iatropoulos P., Donadelli R. MYO1E mutations and childhood familial focal segmental glomerulosclerosis. N Engl J Med. 2011;365:295–306. doi: 10.1056/NEJMoa1101273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brown E.J., Schlondorff J.S., Becker D.J. Mutations in the formin gene INF2 cause focal segmental glomerulosclerosis. Nat Genet. 2010;42:72–76. doi: 10.1038/ng.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gee H.Y., Saisawat P., Ashraf S. ARHGDIA mutations cause nephrotic syndrome via defective RHO GTPase signaling. J Clin Invest. 2013;123:3243–3253. doi: 10.1172/JCI69134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rao J., Ashraf S., Tan W. Advillin acts upstream of phospholipase C 1 in steroid-resistant nephrotic syndrome. J Clin Invest. 2017;127:4257–4269. doi: 10.1172/JCI94138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Perico L., Conti S., Benigni A., Remuzzi G. Podocyte-actin dynamics in health and disease. Nat Rev Nephrol. 2016;12:692–710. doi: 10.1038/nrneph.2016.127. [DOI] [PubMed] [Google Scholar]
- 15.Schlondorff D., Banas B. The mesangial cell revisited: no cell is an island. J Am Soc Nephrol. 2009;20:1179–1187. doi: 10.1681/ASN.2008050549. [DOI] [PubMed] [Google Scholar]
- 16.Vaughan M.R., Quaggin S.E. How do mesangial and endothelial cells form the glomerular tuft? J Am Soc Nephrol. 2008;19:24–33. doi: 10.1681/ASN.2007040471. [DOI] [PubMed] [Google Scholar]
- 17.Khan S., Lakhe-Reddy S., McCarty J.H. Mesangial cell integrin alphavbeta8 provides glomerular endothelial cell cytoprotection by sequestering TGF-beta and regulating PECAM-1. Am J Pathol. 2011;178:609–620. doi: 10.1016/j.ajpath.2010.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lakhe-Reddy S., Li V., Arnold T.D. Mesangial cell alphavbeta8-integrin regulates glomerular capillary integrity and repair. Am J Physiol Renal Physiol. 2014;306:F1400–F1409. doi: 10.1152/ajprenal.00624.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lennon R., Hosawi S. Glomerular cell crosstalk. Curr Opin Nephrol Hypertens. 2016;25:187–193. doi: 10.1097/MNH.0000000000000221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Linnemann A., van der Ven P.F., Vakeel P. The sarcomeric Z-disc component myopodin is a multiadapter protein that interacts with filamin and alphaactinin. Eur J Cell Biol. 2010;89:681–692. doi: 10.1016/j.ejcb.2010.04.004. [DOI] [PubMed] [Google Scholar]
- 21.Ulbricht A., Eppler F.J., Tapia V.E. Cellular mechanotransduction relies on tension-induced and chaperone-assisted autophagy. Curr Biol. 2013;23:430–435. doi: 10.1016/j.cub.2013.01.064. [DOI] [PubMed] [Google Scholar]
- 22.Kai F., Duncan R. Prostate cancer cell migration induced by myopodin isoforms is associated with formation of morphologically and biochemically distinct actin networks. FASEB J. 2013;27:5046–5058. doi: 10.1096/fj.13-231571. [DOI] [PubMed] [Google Scholar]
- 23.Kistler A.D., Altintas M.M., Reiser J. Podocyte GTPases regulate kidney filter dynamics. Kidney Int. 2012;81:1053–1055. doi: 10.1038/ki.2012.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kai F., Fawcett J.P., Duncan R. Synaptopodin-2 induces assembly of peripheral actin bundles and immature focal adhesions to promote lamellipodia formation and prostate cancer cell migration. Oncotarget. 2015;6:11162–11174. doi: 10.18632/oncotarget.3578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chalovich J.M., Schroeter M.M. Synaptopodin family of natively unfolded, actin binding proteins: physical properties and potential biological functions. Biophys Rev. 2010;2:181–189. doi: 10.1007/s12551-010-0040-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mundel P., Heid H.W., Mundel T.M. Synaptopodin: an actin-associated protein in telencephalic dendrites and renal podocytes. J Cell Biol. 1997;139:193–204. doi: 10.1083/jcb.139.1.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Asanuma K., Yanagida-Asanuma E., Faul C. Synaptopodin orchestrates actin organization and cell motility via regulation of RhoA signalling. Nat Cell Biol. 2006;8:485–491. doi: 10.1038/ncb1400. [DOI] [PubMed] [Google Scholar]
- 28.Faul C., Donnelly M., Merscher-Gomez S. The actin cytoskeleton of kidney podocytes is a direct target of the antiproteinuric effect of cyclosporine A. Nat Med. 2008;14:931–938. doi: 10.1038/nm.1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Linnemann A., Vakeel P., Bezerra E. Myopodin is an F-actin bundling protein with multiple independent actin-binding regions. J Muscle Res Cell Motil. 2013;34:61–69. doi: 10.1007/s10974-012-9334-5. [DOI] [PubMed] [Google Scholar]
- 30.Chung J.-J., Goldstein L., Chen Y.-J.J. Single-cell transcriptome profiling of the kidney glomerulus identifies key cell types and reactions to injury. J Am Soc Nephrol. 2020;31:2341–2354. doi: 10.1681/ASN.2020020220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sjoblom B., Salmazo A., Djinovic-Carugo K. Alphaactinin structure and regulation. Cell Mol Life Sci. 2008;65:2688–2701. doi: 10.1007/s00018-008-8080-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weins A., Schlondorff J.S., Nakamura F. Disease-associated mutant alphaactinin-4 reveals a mechanism for regulating its F-actin-binding affinity. Proc Natl Acad Sci U S A. 2007;104:16080–16085. doi: 10.1073/pnas.0702451104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kos C.H., Le T.C., Sinha S. Mice deficient in alphaactinin-4 have severe glomerular disease. J Clin Invest. 2003;111:1683–1690. doi: 10.1172/JCI17988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lin F., Yu Y.P., Woods J. Myopodin, a synaptopodin homologue, is frequently deleted in invasive prostate cancers. Am J Pathol. 2001;159:1603–1612. doi: 10.1016/S0002-9440(10)63006-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu Y.P., Luo J.H. Myopodin-mediated suppression of prostate cancer cell migration involves interaction with zyxin. Cancer Res. 2006;66:7414–7419. doi: 10.1158/0008-5472.CAN-06-0227. [DOI] [PubMed] [Google Scholar]
- 36.Kai F., Tanner K., King C., Duncan R. Myopodin isoforms alter the chemokinetic response of PC3 cells in response to different migration stimuli via differential effects on Rho-ROCK signaling pathways. Carcinogenesis. 2012;33:2100–2107. doi: 10.1093/carcin/bgs268. [DOI] [PubMed] [Google Scholar]
- 37.Yu Y.P., Luo J.H. Phosphorylation and interaction of myopodin by integrin-link kinase lead to suppression of cell growth and motility in prostate cancer cells. Oncogene. 2011;30:4855–4863. doi: 10.1038/onc.2011.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Krause M., Gautreau A. Steering cell migration: lamellipodium dynamics and the regulation of directional persistence. Nat Rev Mol Cell Biol. 2014;15:577–590. doi: 10.1038/nrm3861. [DOI] [PubMed] [Google Scholar]
- 39.Etienne-Manneville S., Hall A. Rho GTPases in cell biology. Nature. 2002;420:629–635. doi: 10.1038/nature01148. [DOI] [PubMed] [Google Scholar]
- 40.Primary nephrotic syndrome in children: clinical significance of histopathologic variants of minimal change and of diffuse mesangial hypercellularity. A report of the International Study of Kidney Disease in Children. Kidney Int. 1981;20:765–771. doi: 10.1038/ki.1981.209. [DOI] [PubMed] [Google Scholar]
- 41.Boyer O., Nevo F., Plaisier E. INF2 mutations in Charcot-Marie-Tooth disease with glomerulopathy. N Engl J Med. 2011;365:2377–2388. doi: 10.1056/NEJMoa1109122. [DOI] [PubMed] [Google Scholar]
- 42.Gee H.Y., Zhang F., Ashraf S. KANK deficiency leads to podocyte dysfunction and nephrotic syndrome. J Clin Invest. 2015;125:2375–2384. doi: 10.1172/JCI79504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gee H.Y., Ashraf S., Wan X. Mutations in EMP2 cause childhood-onset nephrotic syndrome. Am J Hum Genet. 2014;94:884–890. doi: 10.1016/j.ajhg.2014.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Scott R.P., Hawley S.P., Ruston J. Podocyte-specific loss of Cdc42 leads to congenital nephropathy. J Am Soc Nephrol. 2012;23:1149–1154. doi: 10.1681/ASN.2011121206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Giehl K., Graness A., Goppelt-Struebe M. The small GTPase Rac-1 is a regulator of mesangial cell morphology and thrombospondin-1 expression. Am J Physiol Renal Physiol. 2008;294:F407–F413. doi: 10.1152/ajprenal.00093.2007. [DOI] [PubMed] [Google Scholar]
- 46.Wu S.Z., Peng F.F., Li J.L. Akt and RhoA activation in response to high glucose require caveolin-1 phosphorylation in mesangial cells. Am J Physiol Renal Physiol. 2014;306:F1308–F1317. doi: 10.1152/ajprenal.00447.2013. [DOI] [PubMed] [Google Scholar]
- 47.Richards S., Aziz N., Bale S. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.