Abstract
Background
Fostemsavir is a prodrug of a first-in-class HIV-1 attachment inhibitor, temsavir, that binds to gp120 and blocks attachment to the host-cell CD4 receptor, preventing entry and infection of the target cell. Previous studies using a limited number of clinical isolates showed that there was intrinsic variability in their susceptibility to temsavir.
Objectives
Here, an analysis was performed using all clinical isolates analysed in the Monogram Biosciences PhenoSense® Entry assay as part of the development programme.
Methods
In total, 1337 individual envelopes encompassing 20 different HIV-1 subtypes were examined for their susceptibility to temsavir. However, only seven subtypes (B, C, F1, A, [B, F1], BF and A1) were present more than five times, with subtype B (881 isolates) and subtype C (156 isolates) having the largest numbers.
Results
As expected, variability in susceptibility was observed within all subtypes. However, for the great majority of these viruses, temsavir was highly potent, with most viruses exhibiting IC50s <10 nM. One exception was CRF01_AE viruses, where all five isolates exhibited IC50s >100 nM. For the 607 isolates where tropism data were available, geometric mean temsavir IC50 values were remarkably similar for CCR5-, CXCR4- and dual mixed-tropic envelopes from infected individuals.
Conclusions
These data show that HIV-1 viruses from most subtypes are highly susceptible to temsavir and that temsavir susceptibility is independent of tropism.
Introduction
Fostemsavir (FTR, GSK3684934; formerly known as BMS-663068) is a prodrug of a HIV-1 gp120-directed attachment inhibitor, temsavir (TMR, GSK2616713, formerly BMS-626529). It has recently been approved in the USA under the brand name RUKOBIA, which, in combination with other antiretroviral(s), is indicated for the treatment of HIV-1 infection in heavily treatment-experienced adults with MDR HIV-1 infection failing their current antiretroviral regimen due to resistance, intolerance or safety considerations.1,2 Structural studies of temsavir bound into the gp140 SOSIP envelope,3 modelling and functional studies4–6 have suggested a mechanism of action whereby temsavir binds within gp120 and interacts with the β20–β21 bridging sheet near the CD4-binding pocket within the gp120 subunit of gp160.3 The temsavir-bound gp120 is then believed to be held in the ‘closed’ state 1 conformation,3,6,7 which precludes binding of gp120 to the CD4 cell-surface receptor. Therefore, temsavir binds directly to the gp120 subunit within the HIV-1 envelope glycoprotein gp160 complex and prevents the initial interaction between the virus and cellular CD4 receptors, thereby preventing viral attachment to cellular CD4 and entry into host cells. This mechanism is similar to the one observed for a class of broadly neutralizing monoclonal antibodies (bnAbs) that target gp120 and keep the envelope in the ‘closed’ conformation. 8
Much like bnAbs, the susceptibility of viral envelopes to temsavir is highly context dependent.9–12 Susceptibilities within and between subtypes can range from very low picomolar to IC50s >10 μM. Some of this heterogeneity is due to amino acid polymorphisms found at four sites surrounding the binding site of temsavir within gp160. Thus, certain natural polymorphisms at amino acid positions S375, M426, M434 and M475 can reduce susceptibility to temsavir in a context-dependent manner. However, in BRIGHTE, a Phase 3 study (205888) of fostemsavir in highly treatment-experienced individuals, gp120 polymorphisms of interest, temsavir IC50 and HIV-1 subtype did not reliably predict virological outcome at Day 8 of fostemsavir functional monotherapy and did not impact durability of response (HIV-1 RNA <40 copies/mL) to fostemsavir added to optimized background therapy (OBT) through Week 96 of therapy.13 In addition, emergence of substitutions at these polymorphic amino acid positions during treatment can occur inconsistently around the time of virological failure.13,14 Thus, the susceptibility of viral envelopes to temsavir is highly context dependent.
Previously, variability of susceptibility to temsavir within different subtypes was analysed using clinical samples tested in PBMCs.15 This analysis is a slow, highly variable and labour-intensive method. Presently, a method used to examine entry inhibitor susceptibility of clinical envelopes is the PhenoSense® Entry assay performed at Monogram Biosciences (San Francisco, CA, USA).16 During the development phases for fostemsavir, this assay was used exclusively to obtain susceptibility information on temsavir. In an attempt to characterize the overall susceptibility of temsavir to clinical envelopes, the PhenoSense® Entry assay data available for baseline (untreated) samples performed during the clinical development of temsavir, first at Bristol-Myers Squibb and then at ViiV Healthcare, were evaluated. In all, 1337 clinical envelopes from 20 different HIV-1 subtypes have been analysed in this assay. These envelopes included all those examined as part of the Phase 2a (GSK 206267) and Phase 2b (GSK 205889) clinical studies, as well as the ongoing Phase 3 BRIGHTE study in heavily treatment-experienced individuals (GSK 205888). The results show that although there is a broad range of susceptibility in most subtypes, the majority of viruses within any subtype are highly susceptible to temsavir (IC50s <10 nM), with no effect of viral tropism on susceptibility.
Methods
PhenoSense® Entry assay
The PhenoSense® Entry HIV-1 assay was performed at Monogram Biosciences, with samples generated in their labs.16 The original PhenoSense® assay was developed to evaluate PIs and reverse transcriptase inhibitors17 and, for the PhenoSense® Entry assay, it has been modified to amplify and test pseudotyped viruses with envelopes from the clinical isolates. The data obtained from Monogram Biosciences were evaluated both as an IC50 and as a fold change compared with a control envelope. As the control envelope in the assay was changed during the course of these studies, only the IC50 results are discussed in this report.
Origin of samples
The data presented here are from all HIV-1-infected individuals in the clinical studies for fostemsavir (GSK 206267, GSK 205889 and BRIGHTE, GSK 205888; n = 1123), clinical samples from the Bristol-Myers Squibb collection or collaborators (n = 156) and anonymous isolates chosen by Monogram Biosciences (n = 58). The clinical samples used in this research were obtained from HIV-1-infected clinical trial participants from global centres that took part in clinical development studies. For the clinical samples from the fostemsavir studies, either the screening or baseline sample (not both) was used in this analysis, with baseline samples as the default, if available. The samples from GSK 206267 (n = 46), GSK 205889 (n = 467) and BRIGHTE (n = 610) included all screening samples that were successful in the PhenoSense® Entry assay, regardless of whether they were eventually enrolled in the study. No on-treatment samples were included in the analyses. The clinical studies were conducted in accordance with International Conference of Harmonization (‘ICH’) principles and were approved by independent Ethics Committees and Institutional Review Boards. Prior to the conduct of any study procedures, all participants consented to study participation and to the research use of their biological samples.
The determinations of virus subtype and tropism were performed at Monogram Biosciences. Virus tropism was determined in the BRIGHTE study using the Trofile® assay,18 using the sample generated for the PhenoSense® Entry assay.
Results
Epidemiology of samples based upon genotype
Table 1 illustrates the subtypes of the envelopes examined in the PhenoSense® Entry assay through December 2019. A total of 1337 independent isolates have been examined thus far in this assay. These included viruses from all participants screened and successfully phenotyped in the Phase 2a, 2b and 3 studies for fostemsavir [Study 206267 (AI438-006), Study 205889 (AI438-011) and BRIGHTE], plus an additional 214 clinical samples run during preclinical analyses. A total of 881 of these samples were from subtype B-infected individuals, 156 samples were from subtype C-infected individuals, 43 samples were from subtype A-infected individuals and 48 samples were identified as being from the F1 subtype. In addition, 28 envelopes from individuals with complex genotypes (designated Complex) could not be definitively assigned to a single subtype and the subtype of 84 envelopes could not be determined (designated NA or NR).
Table 1.
Characterization of isolates examined in the PhenoSense® Entry assay
| Subtype | Number of isolates | Subtype | Number of isolates |
|---|---|---|---|
| B | 881 | B, D | 4 |
| C | 156 | G | 4 |
| NAa | 58 | D, F1 | 3 |
| F1 | 48 | F | 3 |
| A | 43 | A, B | 2 |
| B, F1 | 29 | A, G | 2 |
| Complex | 28 | A, AE | 1 |
| NRb | 26 | A, C | 1 |
| BF | 19 | B, C | 1 |
| A1 | 17 | B, G | 1 |
| CRF01_AE | 5 | B, J | 1 |
| AG | 4 |
Not available.
Not recorded.
Susceptibility of envelopes to temsavir based upon subtype
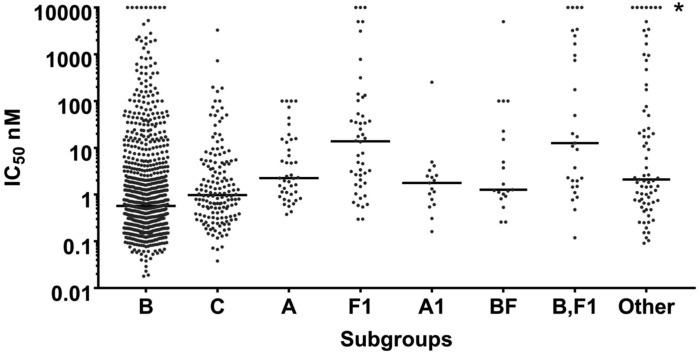
Figure 1 illustrates the susceptibilities of the different envelopes based upon their HIV-1 subtype. Known subtypes with more than five samples were analysed separately. Envelopes with genotypes from subtypes with less than or equal to five independent samples were included in the ‘Other’ column, along with envelopes where subtypes were not assigned (NA, NR and Complex). In all cases, a wide range of susceptibilities to temsavir was observed, with most subtypes having IC50s ranging from the low picomolar to >5 μM.
Figure 1.
Range of temsavir susceptibility observed within subtypes. Viruses with more than five isolates are grouped by subtype, while the remaining isolates are grouped into ‘Other’, along with isolates for which a subtype could not be determined. The geometric means of each group are shown by the horizontal lines and are included in Table 2. Symbols in line with the asterisk represent envelopes without a defined IC50, but which have an IC50 above the highest concentration tested.
The data from Figure 1 were analysed by a different approach in Table 2. The numbers and percentages within several ranges of IC50s were evaluated and geometric means of the totals were calculated. In addition, median IC50 and the 90th percentile IC50 concentration levels were determined (Table 2). The median IC50 for all 1337 isolates was 0.8 nM, while the 90th percentile IC50 was 75.4 nM. Subtypes A1 (17 isolates), C (156 isolates) and B (881 isolates) showed the greatest susceptibility to temsavir, with 90th percentile IC50s of 4.3, 22.9 and 47.6 nM, respectively. For subtype F1 isolates, the median IC50 was 13.8 nM and the 90th percentile IC50 was 892.9 nM. The BF subtype also showed a less robust susceptibility, with a 90th percentile IC50 of 190.4 nM. All five of the CRF01_AE strains tested exhibited IC50s >100 nM.
Table 2.
Susceptibility of envelopes to temsavir based on the indicated IC50s
| Subtype | # | IC50 <1 nM (%) | IC50 >1 to <10 nM (%) | IC50 >10 to <100 nM (%) | IC50 >100 nM (%) | Geometric mean IC50 (nM)a | Median IC50 (nM)a | 90th percentile IC50 (nM)a |
|---|---|---|---|---|---|---|---|---|
| All | 1337 | 719 (53.8) | 352 (26.3) | 146 (10.9) | 120 (9.0) | 1.7 | 0.8 | 75.4 |
| B | 881 | 544 (61.7) | 196 (22.2) | 85 (9.6) | 56 (6.4) | 1.2 | 0.6 | 47.6 |
| C | 156 | 78 (50.0) | 56 (35.9) | 15 (9.6) | 7 (4.5) | 1.5 | 1.0 | 22.9 |
| A1 | 17 | 5 (29.4) | 11 (64.7) | 0 (0) | 1 (5.9) | 1.8 | 1.8 | 4.3 |
| A | 43 | 10 (23.3) | 21 (48.8) | 8 (18.6) | 4 (9.3) | 3.8 | 2.3 | 57.7 |
| BF | 19 | 5 (26.3) | 8 (42.1) | 2 (10.5) | 4 (21.1) | 4.6 | 1.3 | 190.4 |
| F1 | 48 | 8 (16.7) | 15 (31.3) | 13 (27.1) | 12 (25.0) | 17.9 | 13.8 | 892.9 |
| B, F1 | 29 | 4 (13.8) | 9 (31.0) | 5 (17.2) | 11 (37.9) | 38.4 | 12.7 | 6925.3 |
| CRF01_AE | 5 | 0 (0) | 0 (0) | 0 (0) | 5 (100) | – | – | – |
May include isolates with >MAX IC50 (MAX = 5000 nM or >100 nM IC50).
The geometric mean values also indicated that the great majority of viruses (excluding CRF01_AE) were highly susceptible to temsavir (Table 2). The geometric mean value for all 1337 viruses was 1.7 nM, which is similar to the geometric mean for B, C and A1 subtypes, with the A and BF geometric means being only slightly higher (3.8 and 4.6 nM, respectively). The geometric mean IC50s for F1 and [B, F1] subtypes were higher, at 17.9 and 38.4 nM, respectively.
Temsavir activity is tropism independent
In an earlier study, the tropism effect on temsavir susceptibility was evaluated.9 In that study, the geometric mean IC50 values for a set of 11 CXCR4-tropic and 12 CCR5-tropic envelopes from a dual-infected individual were nearly identical. In order to substantiate that observation, all viruses with tropism results from the BRIGHTE study (GSK 205888) were grouped and evaluated for susceptibility to temsavir (Table 3). In total, 607 virus isolates had both tropism and measured IC50s. These included 43 CXCR4-tropic envelopes, 225 CCR5-tropic envelopes and 339 dual mixed-tropic envelopes. The percentages of viral envelopes within each range of susceptibilities were strikingly similar between the three different tropism classes. Envelopes with IC50s ≤1 nM were 48.8% of the CXCR4-tropic population, 48.4% of the CCR5-tropic population and 54.6% of the dual mixed-tropic population. Perhaps most importantly, the percentage of envelopes with IC50s in the Monogram Biosciences PhenoSense® Entry assay >100 nM was 11.6% for CXCR4-tropic envelopes, 12.9% for CCR5-tropic envelopes and 10.3% for dual mixed-tropic envelopes. Accordingly, the geometric mean IC50s for the three tropism classes were almost identical, as dual mixed-tropic envelopes had a geometric mean of 1.89 nM, CCR5-tropic envelopes had a geometric mean of 2.77 nM and CXCR4-tropic envelopes had a geometric mean of 2.94 nM. Thus, this analysis provides a strong indication that susceptibility to temsavir (or fostemsavir) is tropism independent.
Table 3.
Susceptibility of envelopes to temsavir based upon envelope tropism
| Tropism | # | IC50 ≤1 nM (%) | IC50 >1 to ≤10 nM (%) | IC50 >10 to ≤100 nM (%) | IC50 >100 nM (%) | Geometric mean (nM)a |
|---|---|---|---|---|---|---|
| All | 607 | 315 (51.9) | 148 (24.4) | 75 (12.4) | 69 (11.4) | 2.25 |
| CXCR4 | 43 | 21 (48.8) | 9 (20.9) | 8 (18.6) | 5 (11.6) | 2.94 |
| CCR5 | 225 | 109 (48.4) | 54 (24.0) | 33 (14.7) | 29 (12.9) | 2.77 |
| Dual mixed | 339 | 185 (54.6) | 85 (25.1) | 34 (10.0) | 35 (10.3) | 1.89 |
May include isolates with >MAX IC50 (MAX = 5000 nM).
Discussion
Fostemsavir is a novel prodrug of temsavir that—in turn—inhibits attachment of gp120 to host-cell CD4 receptors, inhibiting infection and leaving host cells intact.5,19 Temsavir has been shown to bind within the CD4-binding pocket, locking the envelope into a pre-fusion (state 1) conformation that is unable to bind to the CD4 receptor.3,4,6 This is similar to what is observed with the class of CD4 binding site bnAbs.4,8 Fostemsavir has recently been approved for use in a subpopulation of HIV-infected patients with the greatest unmet medical need, the highly treatment-experienced population.1,2 Previous studies on the susceptibility of clinical isolates in PBMCs to temsavir suggested that viruses within most subtypes exhibit a wide range of susceptibilities.15 In an effort to better understand the variability in susceptibility observed in global clinical envelopes, an analysis was performed on all isolates thus far examined in the PhenoSense® Entry assay.16
The data show that each subtype, and the population as a whole, exhibits a wide range of susceptibilities to temsavir. This is similar to what was observed with a smaller number of clinical isolates examined in PBMCs15 and is in some part due to the presence of polymorphisms at amino acid positions 375, 426, 434 and 475 that are known to alter the susceptibility of envelopes to temsavir.9,14 However, this analysis also shows that, despite the wide range observed in the phenotyping assay, a large majority of the viruses were highly susceptible to temsavir. For instance, 53.8% of the 1337 isolates exhibited IC50s <1 nM, while 80.1% exhibited IC50s <10 nM. Of the 1337 viruses, only 9.0% had IC50s >100 nM (Table 2), with only 3.8% having IC50s >1000 nM. This analysis is skewed by the preponderance of subtype B viruses and lack of certain subtypes. Thus, it is important that each subtype be analysed individually as illustrated in Table 2 for subtypes with more than five isolates (excluding cohorts labelled as NA, NR or Complex, where subtypes were unknown). Analysis of individual subtypes shows that subtypes B, C, A1 and A show a similar pattern of high susceptibility to temsavir; geometric mean IC50s for the B, C, A1 and A subtypes ranged from 1.2 to 3.8 nM. Even the less susceptible subtypes (BF, F1 and [B, F1]) exhibited relatively low median IC50s between 1.3 and 13.8 nM. Susceptibility in the clinic is more complicated, as plasma levels and protein-binding properties of the drug must be considered, with the trough (Cmin) value being an important factor for efficacy. The relatively high plasma trough (Cmin; 478 ng/mL or ∼1.01 μM)1 and Cmax (1770 ng/mL or ∼3.74 μM)1 values of temsavir observed in clinical trials along with moderate protein-binding properties (88.4% protein bound)1 suggest that the great majority of virus isolates would be susceptible to fostemsavir treatment, which was illustrated in the BRIGHTE study.
One potential exception are viruses from the CRF01_AE subtype, as envelopes from this subtype tend to exhibit high IC50s of temsavir due to existing polymorphisms. Only 5 CRF01_AE viruses exist as part of the 1337 virus cohort and all 5 exhibited IC50s >100 nM. Previously, it has been shown that CRF01_AE envelopes were associated with higher IC50s of temsavir due to the prevalence of amino acid polymorphisms at S375H and M475I in this subtype.9,20 In the BRIGHTE study, two participants with subtype CRF01_AE virus at screening in the randomized cohort were enrolled. One participant (temsavir IC50 >5000 nM and gp120 substitutions S375H and M475I at baseline) did not respond to fostemsavir at Day 8, while a second participant (temsavir IC50 = 222.859 and gp120 substitution S375N at baseline) received placebo during functional monotherapy.1,21 However, both participants had an excellent response on fostemsavir-based therapy through 96 weeks, achieving and maintaining virological suppression (HIV-1 RNA <40 copies/mL) by that timepoint, as well as exhibiting increases in baseline CD4 count of 262 and 490 cells/mm3. Notably, each of these participants had relatively low Day 1 viral loads (∼1000 and ∼14 000 copies/mL).
Additional polymorphisms at S375 (S375I/M//N/T), M426 (M426L/P) and M434 (M434I/K) have been shown to display the potential to affect susceptibility to temsavir in vitro.14,21 However, these polymorphisms are highly context dependent with respect to temsavir susceptibility and the presence of a known polymorphism does not preclude an envelope from being susceptible to temsavir and responding well to fostemsavir treatment.13,14 Results from the BRIGHTE study suggest that higher baseline temsavir IC50 has been associated with a trend towards reduced virological response to fostemsavir monotherapy; however, baseline factors, including the presence of temsavir-relevant gp160 substitutions, temsavir IC50 and subtype, were not reliably predictive of response to short-term fostemsavir functional monotherapy and did not impact durability of response to fostemsavir plus OBT through 96 weeks of therapy.13
Previously, data from a single infected individual with a dual mixed-tropic envelope phenotype suggested that susceptibility to temsavir was not affected by tropism.9 In that study, separate envelopes isolated from this one individual were examined in a cell–cell fusion assay for susceptibility to temsavir. Although variability was seen for both the CCR5- and CXCR4-tropic envelopes, the mean IC50s for each tropism cohort were nearly identical. In this 1337 isolate cohort, tropism information was available for 607 isolates, with 55.8%, 37.1% and 7.1% of the envelopes encoding envelopes from dual mixed-tropic infected individuals, CCR5-tropic infected individuals and CXCR4-tropic infected individuals, respectively (Table 3). The preponderance of dual mixed-tropic infected individuals presumably reflects the fact that this is a highly treatment-experienced population. Within this 607 isolate subset, the different tropism classes exhibited a remarkably similar pattern of susceptibility to temsavir, with susceptibility within each defined IC50 range differing by only a few percentage points and the geometric mean values being almost identical. Given that tropism maps to changes in the V3 loop22,23 and the amino acids known to affect temsavir susceptibility map to constant regions,9 this finding is not surprising, but confirms that viral tropism does not affect temsavir susceptibility.
This analysis of clinical samples tested for susceptibility to temsavir in the PhenoSense® Entry assay supports previous work that showed wide variability in susceptibility to the agent within subtypes and throughout the population. However, the data do show that a great majority of viruses (especially from subtypes B, C, A and A1) are highly susceptible to temsavir. This correlates well with recently published data from a small study of 24 patients with MDR HIV-1 enrolled in the PRESTIGIO Registry,24 while this present analysis encompassed 1337 independent samples. Also, analysis of this data set strongly shows that temsavir susceptibility is tropism independent and thus fostemsavir could be a useful alternative in the highly treatment-experienced HIV-infected subpopulation.
Acknowledgements
We would like to thank all the patients who participated in the clinical studies and their families, as well as Bristol-Myers Squibb and the scientists/clinicians who originated studies on fostemsavir and temsavir.
Funding
All funding for this work originated from internal sources, either from Bristol-Myers Squibb or ViiV Healthcare.
Transparency declarations
All authors are employees of ViiV Healthcare or GlaxoSmithKline, which markets fostemsavir, and as such receive salary, grants of GSK stock and additional benefits from the companies.
References
- 1.RUKOBIA. 2020. https://www.accessdata.fda.gov/drugsatfda_docs/label/2020/212950s000lbl.pdf.
- 2. Kozal M, Aberg J, Pialoux G. et al. Fostemsavir in adults with multidrug-resistant HIV-1 infection. N Engl J Med 2020; 382: 1232–43. [DOI] [PubMed] [Google Scholar]
- 3. Pancera M, Lai YT, Bylund T. et al. Crystal structures of trimeric HIV envelope with entry inhibitors BMS-378806 and BMS-626529. Nat Chem Biol 2017; 13: 1115–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Munro JB, Gorman J, Ma X. et al. Conformational dynamics of single HIV-1 envelope trimers on the surface of native virions. Science 2014; 346: 759–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ho HT, Fan L, Nowicka-Sans B. et al. Envelope conformational changes induced by human immunodeficiency virus type 1 attachment inhibitors prevent CD4 binding and downstream entry events. J Virol 2006; 80: 4017–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Langley DR, Kimura SR, Sivaprakasam P. et al. Homology models of the HIV-1 attachment inhibitor BMS-626529 bound to gp120 suggest a unique mechanism of action. Proteins 2015; 83: 331–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Herschhorn A, Ma X, Gu C. et al. Release of gp120 restraints leads to an entry-competent intermediate state of the HIV-1 envelope glycoproteins. mBio 2016; 7: e01598-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sok D, Burton DR.. Recent progress in broadly neutralizing antibodies to HIV. Nat Immunol 2018; 19: 1179–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhou N, Nowicka-Sans B, McAuliffe B. et al. Genotypic correlates of susceptibility to HIV-1 attachment inhibitor BMS-626529, the active agent of the prodrug BMS-663068. J Antimicrob Chemother 2014; 69: 573–81. [DOI] [PubMed] [Google Scholar]
- 10. Zhou N, Nowicka-Sans B, Zhang S. et al. In vivo patterns of resistance to the HIV attachment inhibitor BMS-488043. Antimicrob Agents Chemother 2011; 55: 729–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Malherbe DC, Sanders RW, van Gils MJ. et al. HIV-1 envelope glycoprotein resistance to monoclonal antibody 2G12 is subject-specific and context-dependent in macaques and humans. PLoS One 2013; 8: e75277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chen W, Dimitrov DS.. Monoclonal antibody-based candidate therapeutics against HIV type 1. AIDS Res Hum Retroviruses 2012; 28: 425–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lataillade M, Lalezari JP, Kozal M. et al. Safety and efficacy of the HIV-1 attachment inhibitor prodrug fostemsavir in heavily treatment-experienced individuals: week 96 results of the phase 3 BRIGHTE study. Lancet HIV 2020; 7: e740–51. [DOI] [PubMed] [Google Scholar]
- 14. Lataillade M, Zhou N, Joshi SR. et al. Viral drug resistance through 48 weeks, in a phase 2b, randomized, controlled trial of the HIV-1 attachment inhibitor prodrug, fostemsavir. J Acquir Immune Defic Syndr 2018; 77: 299–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Nowicka-Sans B, Gong YF, McAuliffe B. et al. In vitro antiviral characteristics of HIV-1 attachment inhibitor BMS-626529, the active component of the prodrug BMS-663068. Antimicrob Agents Chemother 2012; 56: 3498–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Monogram Biosciences. PhenoSense® Entry. 2020. https://www.monogrambio.com/resources/phenotyping/phenosense-entry.
- 17. Petropoulos CJ, Parkin NT, Limoli KL. et al. A novel phenotypic drug susceptibility assay for human immunodeficiency virus type 1. Antimicrob Agents Chemother 2000; 44: 920–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Monogram Biosciences. Trofile® 2020. https://www.monogrambio.com/resources/tropism-assays/trofile.
- 19. Meanwell NA, Krystal MR, Nowicka-Sans B. et al. Inhibitors of HIV-1 attachment: the discovery and development of temsavir and its prodrug fostemsavir. J Med Chem 2018; 61: 62–80. [DOI] [PubMed] [Google Scholar]
- 20. Schader SM, Colby-Germinario SP, Quashie PK. et al. HIV gp120 H375 is unique to HIV-1 subtype CRF01_AE and confers strong resistance to the entry inhibitor BMS-599793, a candidate microbicide drug. Antimicrob Agents Chemother 2012; 56: 4257–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ackerman P, Llamoso C, Pierce A. et al. Baseline and emergent genotypic and phenotypic results in HIV-1-infected, heavily treatment-experienced (HTE) participants meeting protocol-defined virologic failure (PDVF) criteria through week 96 in the fostemsavir (FTR) phase 3 BRIGHTE study. Seventeenth European AIDS Conference, Basel, Switzerland, 2019. Abstract PE17/6. [Google Scholar]
- 22. Poveda E, Alcami J, Paredes R. et al. Genotypic determination of HIV tropism - clinical and methodological recommendations to guide the therapeutic use of CCR5 antagonists. AIDS Rev 2010; 12: 135–48. [PubMed] [Google Scholar]
- 23. Swenson LC, Daumer M, Paredes R.. Next-generation sequencing to assess HIV tropism. Curr Opin HIV AIDS 2012; 7: 478–85. [DOI] [PubMed] [Google Scholar]
- 24. Saladini F, Giannini A, Giammarino F. et al. In vitro susceptibility to fostemsavir is not affected by long-term exposure to antiviral therapy in MDR HIV-1-infected patients. J Antimicrob Chemother 2020; 75: 2547–53. [DOI] [PubMed] [Google Scholar]