This research identifies the interactors of TORC1 by genome-wide BiFC screens and reveals that a spatially distinct portion of TORC1 interacts with RNA-binding proteins. As a component of mRNP complexes, TORC1 phosphorylates Scd6 to negatively regulate Scd6 activity in translational repression and P-body accumulation.
Abstract
The target of rapamycin complex 1 (TORC1) is mainly localized to the vacuolar membrane and regulates eukaryotic cell growth in response to nutrient availability. To obtain deeper insights into the functional roles of TORC1, we performed a genome-wide analysis of the TORC1 interactome in yeast using the bimolecular fluorescence complementation (BiFC) assay. We found that while most of the BiFC signals are observed at the vacuolar membrane, a fraction of them are detected at cytoplasmic messenger ribonucleoprotein (mRNP) granules. Moreover, mRNA-binding proteins are enriched in the TORC1 interactome, suggesting a functional relationship between TORC1 and mRNA metabolism. We show that a portion of TORC1 is consistently associated with mRNP complexes and interacts with a specific subset of mRNAs. We also demonstrate that TORC1 directly targets a translational repressor Scd6 and that the activity of Scd6 is inhibited by TORC1-dependent phosphorylation. Collectively, our data suggest that TORC1 plays a novel role in posttranscriptional regulation by controlling the activity of Scd6.
Introduction
The target of rapamycin (TOR) was first identified in budding yeast during screening for mutants that have resistance to rapamycin, an antifungal antibiotic (Heitman et al., 1991). TOR proteins belong to the phosphatidylinositol kinase–related kinase family and are well conserved in eukaryotic cells from yeast to mammalian cells (Cafferkey et al., 1993; Helliwell et al., 1994). Yeast has two TOR proteins, Tor1 and Tor2, whereas higher eukaryotic cells possess a single TOR protein. TOR complex 1 (TORC1) and TORC2 are two distinct complexes that contain TOR proteins (Loewith et al., 2002; Reinke et al., 2004). TORC1, which is sensitive to rapamycin, comprises Kog1, Lst8, and Tco89 and either Tor1 or Tor2. TORC1 controls cell growth by regulating the cell cycle, ribosome biogenesis, translation, and autophagy (Loewith et al., 2002; Wullschleger et al., 2006). On the other hand, rapamycin-insensitive TORC2 consists of Avo1, Avo2, Lst8, Bit61, and only Tor2. TORC2 serves as an essential regulator of plasma membrane homeostasis and cell wall integrity (Roelants et al., 2017).
The TORC1 signaling pathway plays a crucial role in the integration of extracellular stimuli regulating cell growth. TORC1 is activated by nutrients, especially amino acids, and inhibited by various stresses, including heat stress, osmotic stress, and glucose starvation (Wullschleger et al., 2006). TORC1 is mainly localized to the vacuolar membrane and regulated by the EGO (exit from rapamycin-induced growth arrest) complex, which consists of Gtr1, Gtr2, Ego1, Ego2, and Ego3 (Binda et al., 2009; Dubouloz et al., 2005; Powis et al., 2015). Ego1 is the vacuolar membrane–anchoring subunit of the EGO complex, and Gtr1/Gtr2 activates TORC1 in response to amino acid availability (Dubouloz et al., 2005; Kim et al., 2008). The best-characterized substrate of TORC1 is Sch9, which regulates ribosome biogenesis, translation initiation, and entry into the G0 phase (Urban et al., 2007). Given that Sch9 is concentrated at the vacuolar membrane, TORC1 has been suggested to be activated at the vacuolar membrane (Binda et al., 2009; Urban et al., 2007). These regulatory mechanisms have been further confirmed in mammalian cells wherein mammalian TORC1 (mTORC1) is localized and regulated at the lysosomal membrane by RAGULATOR and Rag GTPases (Sancak et al., 2010). However, TORC1 and mTORC1 are also detected at other subcellular sites, such as stress granules, the endoplasmic reticulum, the plasma membrane, and the nucleus, raising the possibility that TORC1 has unknown targets or functions at subcellular locations other than the vacuolar/lysosomal membrane (Bridges et al., 2012; Reinke et al., 2004; Takahara and Maeda, 2012; Thedieck et al., 2013; Wedaman et al., 2003; Wippich et al., 2013).
Interactome analysis of a protein provides insight into its function. Various techniques for high-throughput protein–protein interaction (PPI) analysis have been developed and have expanded the knowledge of cellular processes over the past decades. The bimolecular fluorescence complementation (BiFC) assay is a method for analyzing PPIs that uses fluorescent proteins as reporters. In the BiFC assay, two nonfluorescent fragments of a fluorescent protein are fused to two proteins of interest (Hu et al., 2002; Kerppola, 2006). The interaction between two target proteins brings the fragments of the fluorescent protein into close proximity and facilitates reconstitution of a functional fluorescent protein that can be directly visualized by fluorescence microscopy. The BiFC assay is a useful tool for detecting binary PPIs in the cellular context with minimal disturbance and has been successfully used in genome-wide analyses of the small ubiquitin-related modifier interactome and protein homomerization in yeast (Kim et al., 2019; Sung et al., 2013)
In this study, we performed a genome-wide BiFC screen for the TORC1 interactome to gain a deeper and broader understanding of the functional roles of TORC1. Interestingly, we found that some interactors of TORC1 are localized to cytoplasmic messenger RNP (mRNP) granules and that mRNA-binding proteins are enriched in the TORC1 interactome, suggesting a functional relationship between TORC1 and mRNA metabolism. We show that nonvacuolar TORC1 interacts with specific mRNAs and that three mRNA-binding proteins in the TORC1 interactome—Scd6, Ssd1, and Whi3—are phosphorylated by TORC1. Notably, the activity of Scd6, an mRNA-binding protein that represses translation initiation, is inhibited upon phosphorylation by TORC1. Taken together, we suggest that TORC1 plays a novel role in posttranscriptional regulation.
Results
Genome-wide BiFC screens identify novel interactors of TORC1
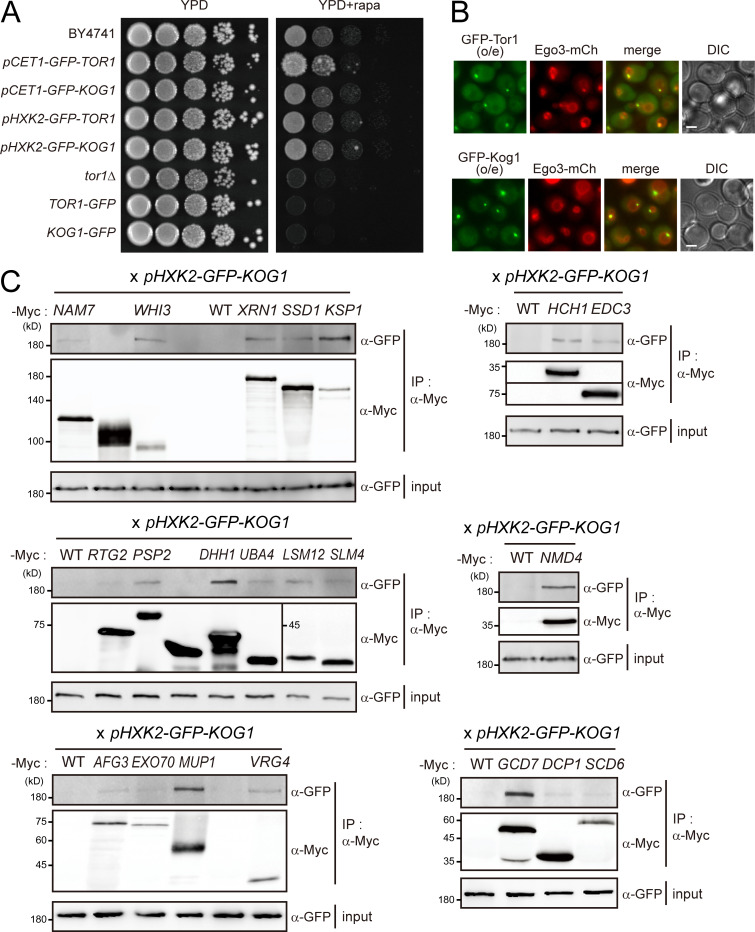
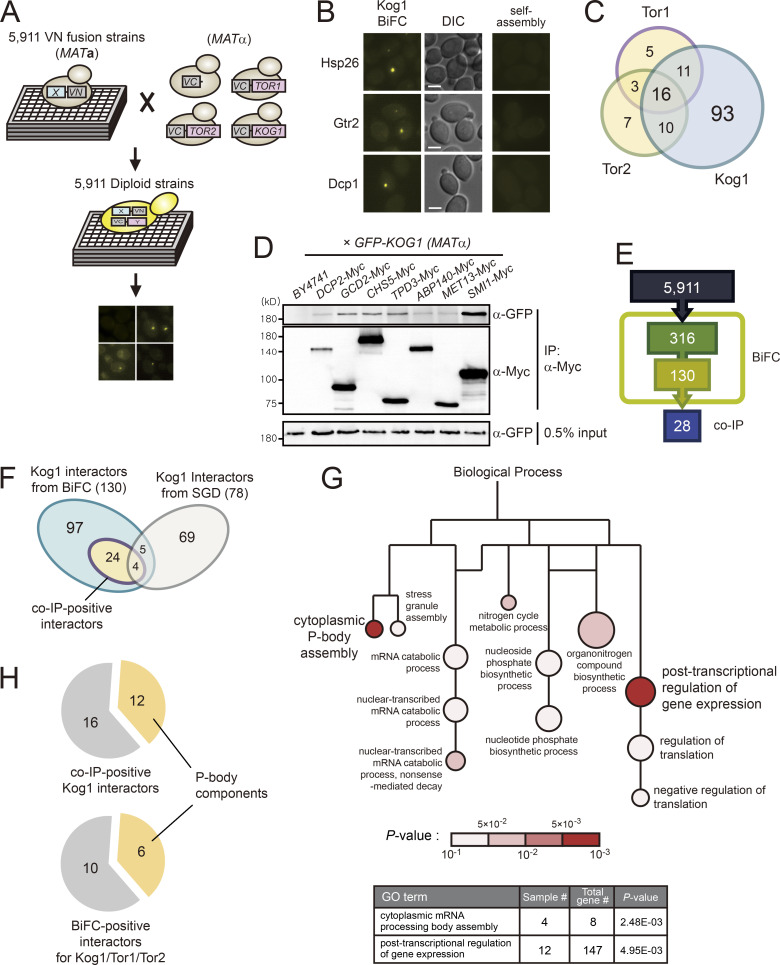
To systematically analyze the TORC1 interactome, we performed a genome-wide BiFC screen using a collection of 5,911 MATα strains expressing full-length proteins tagged with the N-terminal fragment of Venus (VN) from their own native promoters (Sung et al., 2013). Because C-terminal tagging of TORC1 subunits caused severe defects in their activity (Fig. S1 A), we constructed MATα strains in which the C-terminal fragment of Venus (VC) was tagged to the N-terminus of Tor1, Tor2, or Kog1. The endogenous expression levels of Tor1, Tor2, or Kog1 were too low to produce BiFC signals, thus we expressed VC-tagged Tor1, Tor2, or Kog1 under the control of the HXK2 promoter. These VC-tagged strains were mated with each strain of the VN fusion library, and the resultant diploid cells were analyzed by fluorescence microscopy (Fig. 1 A). From these genome-wide BiFC screens, we initially identified 96, 73, and 316 positive signals for Tor1, Tor2, and Kog1, respectively. All the BiFC signals appeared as cytoplasmic foci (Fig. 1 B). These signal patterns were similar to those of GFP-tagged Tor1 and Kog1 expressed under the HXK2 promoter; the GFP signals for both Tor1 and Kog1 appeared as bright cytoplasmic foci that mostly overlapped with the vacuolar membrane (Fig. S1 B). To eliminate potential false-positive BiFC signals arising from the self-assembly of VN and VC, we performed a BiFC screen with a MATα strain expressing VC without fusion to a protein. During this self-assembly analysis, diploid strains that showed the BiFC signals above the background level were regarded as false positives. By excluding these false positives, we finally identified 35, 36, and 130 proteins as putative interactors of Tor1, Tor2, and Kog1, respectively, with 16 proteins overlapping in all three BiFC screens (Fig. 1 C; and Table S1, Table S2, and Table S3). The interactors of Tor1 and Tor2 partially overlapped, and most of the Tor1 and Tor2 interactors overlapped with the Kog1 interactors. Given that Kog1 is a defining component of TORC1 (Loewith et al., 2002), we assumed that the 130 Kog1 interactors could represent the majority of the TORC1 interactome, and, thus, further analysis of the TORC1 interactome was performed with these 130 Kog1 interactors.
Figure S1.
Interactome analysis of TORC1. (A) Impaired TORC1 activity by C-terminally tagging to Tor1 or Kog1. (B) Localization of overexpressed TORC1 at the vacuolar membrane. o/e indicates overexpression under the HXK2 promoter. Scale bars, 1 µm. (C) CoIP assay of Kog1 interactors. C-terminally Myc-tagged proteins were immunoprecipitated, and coimmunoprecipitated GFP-Kog1 was detected. GFP-Kog1 was expressed under the HXK2 promoter. The coIP assay for seven interactors is shown in Fig. 1 D.
Figure 1.
Genome-wide BiFC screens identify numerous novel interactors of TORC1. (A) Scheme of BiFC screens for the TORC1 interactome. (B) Representative BiFC images of Kog1 interactors. DIC, direct interference contrast. Scale bars, 2 µm. (C) Venn diagram depicting the overlap among the interactors of Kog1, Tor1, and Tor2. (D) Examples of coIP assays for 130 Kog1 interactors. GFP-Kog1 was expressed under the HXK2 promoter, and Myc-tagged proteins were expressed under their native promoters. (E) The number of Kog1 interactors that were identified by BiFC screens and coIP assays. (F) Comparison of Kog1 interactors identified in this study with the known interactors of TORC1 listed in the Saccharomyces Genome Database (SGD). (G) GO analysis of 130 Kog1 interactors. GO analysis for biological process was performed using the PANTHER classification system. Lines indicate hierarchical relationships, and sizes of circles are proportional to the number of genes included in the GO terms. (H) Enrichment of P-body components in TORC1 interactors.
To validate the 130 Kog1 interactors identified by the BiFC assay, we performed coimmunoprecipitation (coIP) assay with Kog1 interactors (Fig. 1 D). A total of 130 MATα strains expressing C-terminally Myc-tagged interactors of Kog1 were constructed by an epitope-switching strategy (Sung et al., 2008) and mated with a MATα strain expressing N-terminally GFP-tagged Kog1 under the HXK2 promoter. Upon examination of the resultant diploid strains by coIP assay, we observed that 28 out of the 130 proteins were coimmunoprecipitated with Kog1 (Fig. 1, D and E; Fig. S1 C; and Table S4). According to the Saccharomyces Genome Database (http://www.yeastgenome.org, as of July 9, 2019), 78 proteins are known to physically interact with Kog1. Among the 130 Kog1 interactors that were identified by the BiFC assay, 9 overlapped with the 78 interactors of Kog1 reported in previous datasets (Fig. 1 F and Table S5). Notably, 24 out of the 28 coIP-positive Kog1 interactors were included among the 121 proteins newly defined by the BiFC assay in this study. These results suggest that a genome-wide BiFC screen is a useful tool for studying PPIs in a high-throughput format and that our dataset is highly complementary to the existing datasets.
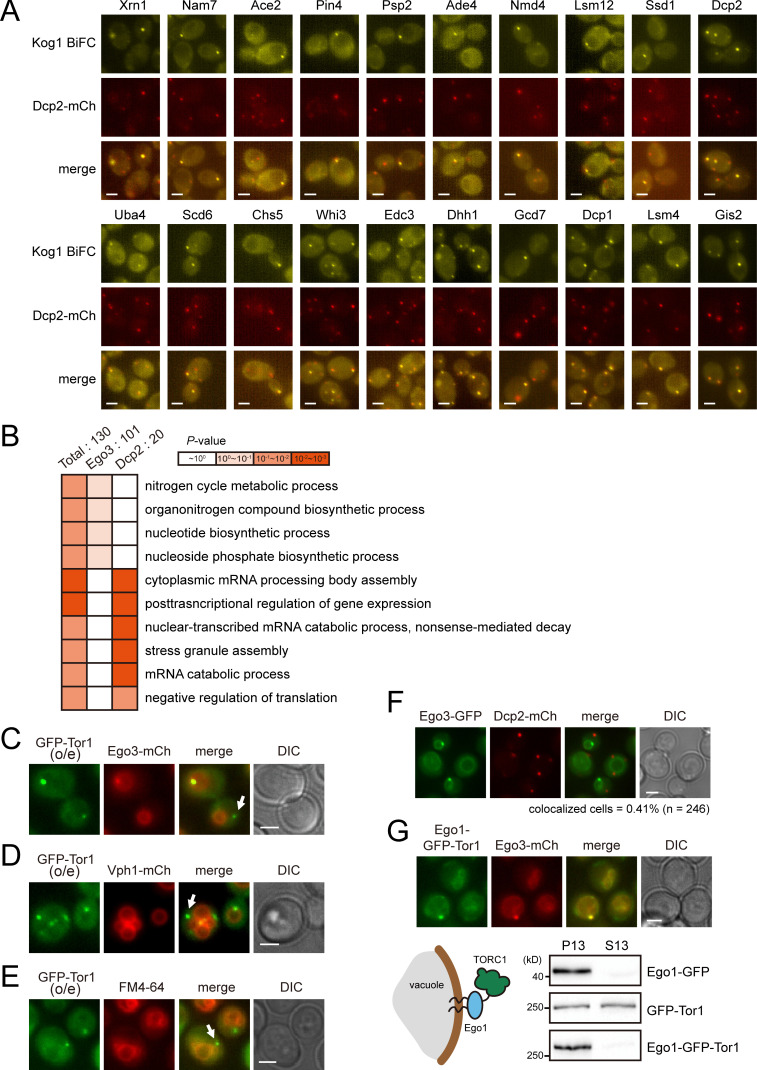
Gene Ontology (GO) analysis of 130 Kog1 interactors suggested the involvement of TORC1 in posttranscriptional regulation of mRNAs. The GO biological process terms “cytoplasmic mRNA processing body (P-body) assembly” and “posttranscriptional regulation of gene expression” were significantly overrepresented among the 130 Kog1 interactors (Fig. 1 G and Table S6). Notably, among the 28 coIP-positive Kog1 interactors, 12 are known to be located at P-bodies (Fig. 1 H and Table S4). In addition, among the 16 proteins that overlapped in all three BiFC screens, 6 (Dcp1, Dcp2, Gis2, Nam7, Psp2, and Xrn1) are known P-body components. These results raise the possibility that TORC1 may be involved in the posttranscriptional regulation of mRNAs.
TORC1 interacts with two distinct groups
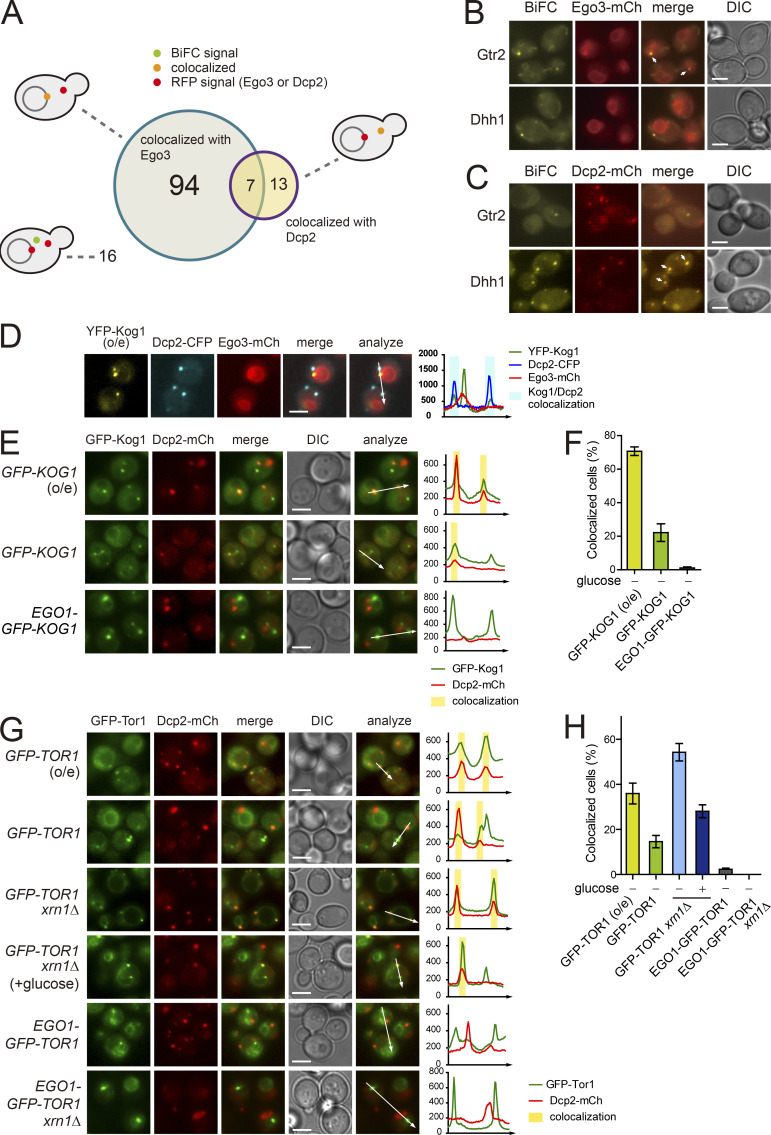
TORC1 is predominantly localized to the vacuolar membrane and activated by the EGO complex (Dubouloz et al., 2005). Consistent with this, it has been shown that TORC1 is highly colocalized with the EGO complex (Binda et al., 2009). The BiFC assay has the advantage of providing information on not only the occurrence but also the subcellular localization of PPIs. To investigate whether the interactions between TORC1 and Kog1 interactors occur at the vacuolar membrane, we examined the colocalization of BiFC signals with Ego3, a subunit of the EGO complex. Among the 130 BiFC signals of the Kog1 interactors, 101 were colocalized with Ego3, while 29 were not (Fig. 2, A and B; and Table S1). Interestingly, 10 out of the 29 proteins that were not colocalized with Ego3 were P-body components. P-bodies are mRNP granules that are visualized as punctate signals in the cytoplasm under various stress conditions such as glucose starvation, heat stress, and osmotic stress (Cowart et al., 2010; Teixeira et al., 2005). We next monitored the colocalization of BiFC signals with the P-body component Dcp2 following the induction of P-bodies by glucose depletion. 20 out of the 130 BiFC signals were colocalized with Dcp2 under glucose starvation conditions (Fig. 2, A and C; Fig. S2 A; and Table S1). Among these 20 BiFC signals, 7 were also colocalized with Ego3. GO analysis of these two groups within the 130 identified Kog1 interactors (i.e., the 101 proteins colocalizing with Ego3 and the 20 proteins colocalizing with Dcp2) showed that they are not only spatially but also functionally distinct (Fig. S2 B). The 20 proteins whose BiFC signals were colocalized with Dcp2 were enriched with those having a role in the posttranscriptional regulation pathway, whereas the 101 proteins whose BiFC signals were localized to the vacuolar membrane were enriched with those involved in metabolic processes.
Figure 2.
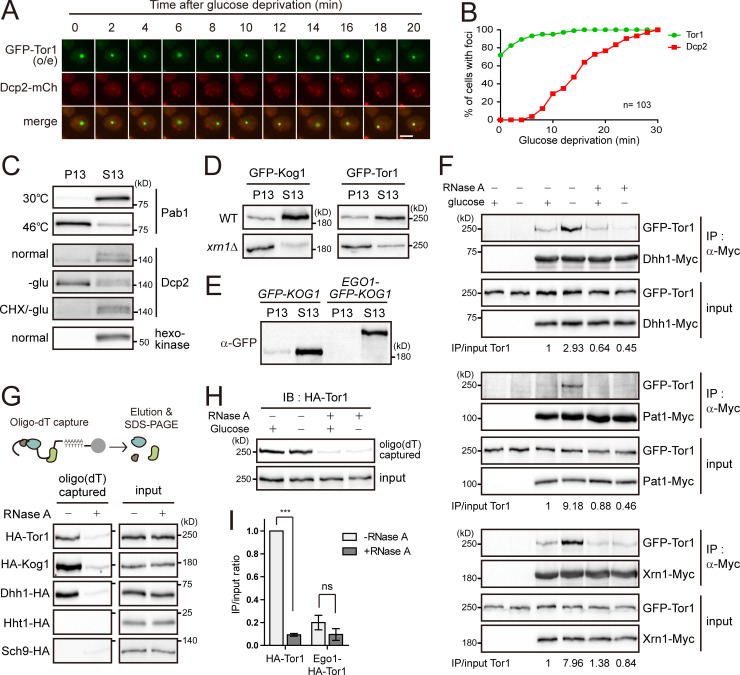
A portion of TORC1 localizes to P-bodies. (A) Venn diagram showing the numbers of the BiFC signals for Kog1 interactors that were colocalized with Ego3 or Dcp2. (B) Examples of the BiFC signals that were colocalized (Gtr2) or not colocalized (Dhh1) with Ego3. Arrows indicate colocalized signals. (C) Examples of the BiFC signals that were colocalized (Dhh1) or not colocalized (Gtr2) with Dcp2. Cells were starved of glucose for 30 min. Arrows indicate colocalized signals. (D) Three-color colocalization analysis of TORC1 (YFP-Kog1), P-bodies (Dcp2-CFP), and the vacuolar membrane (Ego3-mCherry). Fluorescent signals were visualized after 30 min of glucose starvation. (E) Colocalization between Kog1 and P-bodies. Cells expressing GFP-Kog1 under the HXK2 or CET1 promoter and Ego1-GFP-Kog1 under the CET1 promoter were starved of glucose for 30 min to induce P-body formation. (F) Quantification of the ratio of cells showing the colocalization of Kog1 with P-bodies. (G) Colocalization between Tor1 and P-bodies. Cells were starved of glucose for 30 min to induce P-body formation, except “GFP-TOR1 xrn1Δ (+glucose)” cells, which were observed under normal growth conditions. (H) Quantification of the ratio of cells showing the colocalization of Tor1 with P-bodies. Error bars represent SD, and values represent the average of three independent experiments (n > 200) in F and H. Scale bars, 2 µm for B–E and G. Fluorescence intensity profiles along the arrows were obtained using ImageJ software and shown on the right in D, E, and G. o/e indicates overexpression under the HXK2 promoter.
Figure S2.
Foci accumulation of the BiFC signals for the TORC1 interactome. (A) 20 proteins that have interaction with Kog1 in P-bodies. MATα VN fusion strains were mated with a MATα strain expressing VC-KOG1 and DCP2-mCherry. Cells were starved of glucose for 30 min before fluorescence microscopy. (B) GO analysis of Kog1 interactors colocalizing with Ego3 and those colocalizing with Dcp2. GO analysis for biological process was performed using the PANTHER classification system. (C–E) Detection of a portion of Tor1 foci outside the vacuolar membrane. GFP-Tor1 was expressed under the HXK2 promoter. Fluorescence signals were detected under normal growth conditions. Arrows indicate Tor1 foci that were not localized to the vacuolar membrane. (F) Different localization of Ego3 and P-bodies. Cells were starved of glucose for 30 min before fluorescence microscopy. (G) Anchorage of TORC1 to the vacuolar membrane by conjugating Ego1 to Tor1. Top: Colocalization between Ego1-conjugated Tor1 and Ego3. Fluorescence signals were detected under normal growth conditions. Bottom: Fractionation of Tor1. Subcellular fractionation was performed by centrifugation at 13,000×g. Western blotting was performed with anti-GFP antibody. Scale bars, 2 µm.
A portion of TORC1 is colocalized with P-bodies
Several studies have found that TORC1 accumulates in the perivacuolar foci or the limiting membrane of the vacuole, wherein TORC1 colocalizes with the EGO complex (Binda et al., 2009; Sturgill et al., 2008; Urban et al., 2007). We also observed that the TORC1 foci were mainly colocalized with Ego3 (Fig. S1 B). However, we observed that a small fraction of TORC1 foci were not colocalized with Ego3 or other vacuolar markers (Fig. S2, C–E), suggesting that TORC1 can accumulate in different types of foci from the perivacuolar foci. From the three-color colocalization assay of Kog1, Dcp2, and Ego3 under glucose starvation, we found that most of the TORC1 foci that did not overlap with Ego3 colocalized with P-bodies (Fig. 2 D). When we overexpressed Kog1 under the HXK2 promoter, Kog1 colocalized with P-bodies in 71% of cells under glucose starvation conditions (Fig. 2, E and F). We also observed the colocalization of Kog1 with P-bodies in 22% of cells when Kog1 was expressed under the CET1 promoter, which has a similar strength to the KOG1 promoter (Fig. S3, A and B). Tor1 was also colocalized with P-bodies whether it was expressed under the HXK2 promoter or the CET1 promoter (Fig. 2, G and H). It has been reported that P-bodies are increased in number and size in the xrn1Δ mutant due to the accumulation of decapped mRNAs (Sheth and Parker, 2003). Notably, deletion of XRN1 increased the colocalization of Tor1 with Dcp2 in up to 54% of cells under glucose starvation conditions and 28% of cells under normal conditions, even though Tor1 was not overexpressed. These results suggest that the colocalization of TORC1 with P-bodies is not an artifact caused by overexpression of TORC1 but an intrinsic feature of TORC1.
Figure S3.
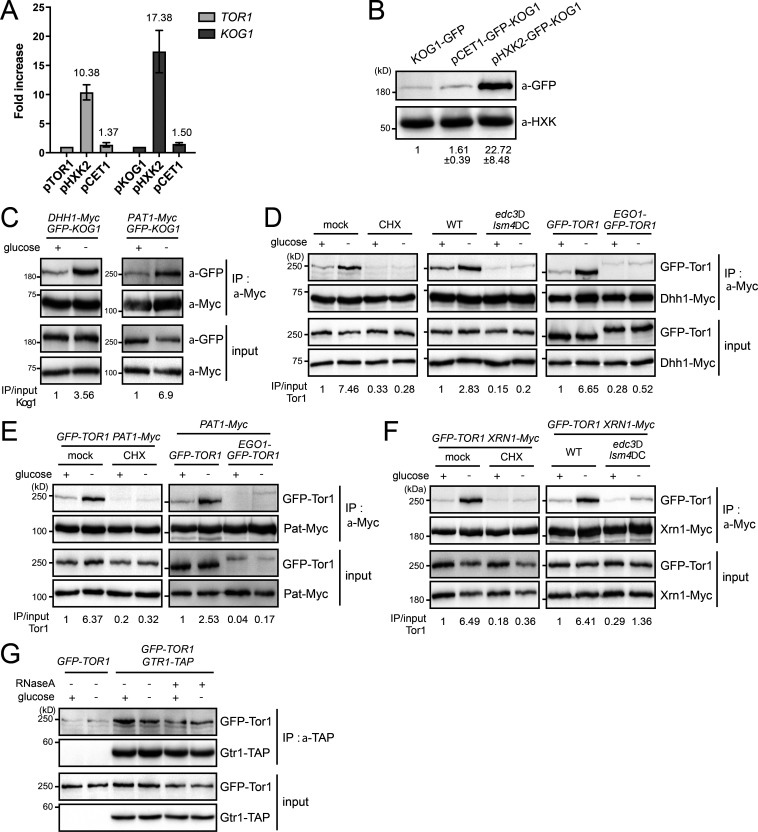
RNA mediates the interaction between TORC1 and P-body components under glucose starvation (A) Comparison of promoter strengths. The mRNA expression levels of TOR1 and KOG1 under different promoters were compared. The mRNA levels of TOR1 and KOG1 were normalized against those of ACT1. Numbers indicate fold increases compared with TOR1 and KOG1 expressed from their own promoters. Error bars represent SD. (B) Protein expression levels of Kog1 under different promoters. The relative ratio of Kog1 to hexokinase was calculated, and the mean ± SD of triplicate experiments is shown below each lane. (C–F) Interaction between TORC1 and P-body components. Immunoprecipitation was performed with anti-c-Myc antibody, and coimmunoprecipitated GFP-Kog1 or GFP-Tor1 was detected by Western blotting with anti-GFP antibody. The relative ratio of coimmunoprecipitated Kog1-GFP or Tor1-GFP to input proteins is shown below each lane. (C) Interaction between Kog1 and P-body components under glucose starvation. (D) Interaction between Tor1 and a P-body component Dhh1 in cells with cycloheximide treatment, edc3Δ lsm4ΔC double mutation, or Ego1-conjugated Tor1. For cycloheximide treatment, cells were pretreated with cycloheximide 10 min before glucose starvation. (E) Interaction between Tor1 and a P-body component Pat1 in cells with cycloheximide treatment or Ego1-conjugated Tor1. (F) Interaction between Tor1 and a P-body component Xrn1 in cells with cycloheximide treatment or edc3Δ lsm4ΔC double mutation. (G) Interaction between Tor1 and Gtr1 under glucose starvation or RNase treatment.
Consistent with the fact that P-bodies are cytoplasmic foci, P-bodies rarely colocalized with the vacuolar membrane (Fig. S2 F). To further confirm that TORC1 foci colocalizing with P-bodies are nonvacuolar, we anchored TORC1 to the vacuolar membrane by conjugating Ego1 to TORC1 as previously described (Takahara and Maeda, 2012) and monitored its colocalization with P-bodies. Remarkably, Ego1-conjugated TORC1 did not colocalize with P-bodies (Fig. 2, E and F; and Fig. S2 G), and deletion of XRN1 did not increase the colocalization of Ego1-conjugated TORC1 with Dcp2 (Fig. 2, G and H). Taken together, these results suggest that a portion of TORC1 is colocalized with P-bodies, apart from the vacuolar membrane, under certain conditions that induce P-bodies.
TORC1 is a component of mRNP complexes
The interactions between mRNAs and proteins induce the aggregation of mRNP complexes even under normal conditions, leading to liquid–liquid phase separation and the formation of mRNA granules (Brangwynne, 2013). A previous study showed that P-bodies are formed by the recruitment of P-body components to preexisting mRNA granules (Lui et al., 2014). To investigate what mediates the colocalization of TORC1 with P-bodies, we took time-lapse images of the colocalization between Tor1 and Dcp2 after glucose starvation (Fig. 3 A). When we analyzed 103 cells that exhibited the colocalization of Tor1 foci with P-bodies at 30 min of glucose starvation, ∼75% of the cells showed Tor1 foci from the outset of glucose starvation, while cells with Dcp2 accumulating in P-bodies began to be observed only after 6 min of glucose starvation (Fig. 3 B). This result suggests that a portion of TORC1 localizes to preexisting mRNA granules that can recruit P-body components under glucose starvation.
Figure 3.
TORC1 has interaction with mRNAs. (A) Time-course analysis of Tor1 foci and P-body formation. Images of cells were taken every 2 min after glucose deprivation. Scale bar, 2 µm. (B) Kinetics of Tor1 foci and P-body formation under glucose starvation. Images of Tor1 foci and P-bodies were taken every 2 min after glucose deprivation. Among 245 cells examined, 103 cells exhibited the colocalization of Tor1 foci and Dcp2 at 30 min of glucose starvation. For these 103 cells, we analyzed the percentage of cells showing Tor1 or Dcp2 accumulation in the foci at each time point. (C) Fractionation of stress granules or P-bodies under stress conditions. Cells expressing Pab1-TAP or Dcp2-TAP were treated with heat stress for 10 min or glucose starvation (-glu) for 30 min or pretreated with cycloheximide (CHX) for 10 min. (D) Fractionation of TORC1. GFP-Kog1 and GFP-Tor1 were expressed under the CET1 promoter. (E) Fractionation of Kog1. GFP-Kog1 and Ego1-GFP-Kog1 were expressed under the CET1 promoter. (F) RNA-mediated interaction between Tor1 and P-body components under glucose starvation conditions. GFP-Tor1 was expressed under the CET1 promoter. Cells were starved of glucose for 30 min. Cell lysates were treated with 50 µg/ml RNase A and incubated for 10 min at 37°C before immunoprecipitation (IP). The relative ratio of coimmunoprecipitated GFP-Tor1 to input GFP-Tor1 is shown below each lane. (G) Interaction between TORC1 and mRNAs. mRNAs from cells expressing the indicated proteins were precipitated by using oligo(dT) cellulose, and copurified proteins were detected. RNase A was treated for 10 min at 37°C before the addition of oligo(dT) cellulose. (H) Glucose-independent interaction between TORC1 and mRNAs. Cell lysates extracted from nonstarved or glucose-starved cells were incubated with oligo(dT) cellulose. IB, immunoblot. (I) Localization-dependent interaction between TORC1 and mRNAs. mRNAs from cells expressing HA-Tor1 or Ego1-HA-Tor1 were precipitated by using oligo(dT) cellulose, and copurified proteins were detected. The relative ratios of oligo(dT)-captured proteins to input proteins are represented as bars. Values represent the average of three independent experiments. Error bars represent SD. P values were determined by Student’s t test (***, P < 0.001; ns, not significant [P > 0.05]).
It has been reported that nonmembranous structures such as P-bodies, stress granules, and protein aggregates can be isolated by fractionation in the presence of detergent (Hubstenberger et al., 2017; Jain et al., 2016; Wallace et al., 2015). We also observed that the induction of stress granules by heat stress or P-bodies by glucose starvation increased the accumulation of Pab1 or Dcp2 in the pellet (P13) fraction obtained from centrifugation at 13,000×g for 10 min (Fig. 3 C), suggesting that the accumulation of proteins in mRNP granules can be monitored by their presence in the P13 fraction. Given this result, we examined the accumulation of TORC1 in mRNA granules by fractionation. A portion of TORC1 was detected in the P13 fraction under normal growth conditions, and the amount of TORC1 in the P13 fraction was increased in the xrn1Δ mutant, in which TORC1 was highly colocalized with P-bodies (Fig. 3 D). These results suggest that TORC1 in the P13 fraction represents the accumulation of TORC1 in mRNP granules. Consistent with the colocalization data (Fig. 2, E–H), the vacuole-anchored TORC1 was only detected in the supernatant (S13) fraction (Fig. 3 E), indicating that precipitation in the P13 fraction is mediated by nonvacuolar TORC1.
The increase in the colocalization between TORC1 and P-bodies under glucose starvation conditions led us to ask whether their interactions are also increased by glucose depletion. Indeed, the interactions between TORC1 and P-body components, such as Dhh1, Pat1, and Xrn1, were increased under glucose starvation conditions (Figs. 3 F and S3 C). However, cells with cycloheximide treatment or the edc3Δ lsm4ΔC double mutation, wherein P-bodies are not induced (Buchan et al., 2008), did not exhibit increased interactions between TORC1 and P-body components under glucose starvation conditions (Fig. S3, D–F). The vacuole-anchored TORC1 also did not show increased interactions with P-body components under glucose starvation conditions. These observations suggest that the colocalization between TORC1 and P-bodies is required for their interactions. Strikingly, RNase treatment abolished the increase in the interactions of TORC1 with P-body components under glucose starvation conditions (Fig. 3 F), indicating that RNA mediates the interactions between TORC1 and P-body components. On the other hand, the interaction between Tor1 and Gtr1, a subunit of the EGO complex, was not affected by glucose deprivation or RNase treatment (Fig. S3 G). Taken together, these results suggest that RNA mediates the interactions of TORC1 with P-body components under glucose starvation conditions.
Given that localization at mRNA granules and RNA-mediated interactions are common features of mRNA-binding proteins, we examined the possibility of interaction between TORC1 and mRNAs by using a native oligo(dT) pull-down assay (Fig. 3 G). In this assay, poly(A)+ mRNAs were captured by oligo(dT)-cellulose after mock or RNase treatment, and copurified proteins were analyzed. Interestingly, both Tor1 and Kog1 were copurified with mRNAs similarly to Dhh1, a known mRNA-binding protein, even when they were not overexpressed. As expected, the negative control histone H3 (Hht1) and Sch9 were not copurified with mRNAs, confirming the specificity of this assay. Although the oligo(dT) pull-down assay could not demonstrate whether TORC1 directly binds to mRNAs, this result suggests that TORC1 is associated with mRNAs as a component of mRNP complexes.
While RNA-mediated interactions between Tor1 and P-body components were increased under glucose starvation, the TORC1–mRNA interaction was not affected by glucose deprivation (Fig. 3 H). This result suggests that under glucose starvation, P-body components are recruited to mRNAs that have already been associated with TORC1. This result is also consistent with the aforementioned time-course colocalization data showing that P-body components are recruited to preexisting TORC1 foci under glucose starvation (Fig. 3, A and B). Next, to examine whether the interaction between TORC1 and mRNAs is dependent on TORC1 location, we performed an oligo(dT) pull-down assay with the vacuole-anchored Tor1. In contrast to unconjugated Tor1, Ego1-conjugated Tor1 was rarely detected in the oligo(dT) precipitate (Fig. 3 I). This result indicates that interactions between TORC1 and mRNAs occur at a distinct location from the vacuolar membrane.
TORC1 phosphorylates Scd6
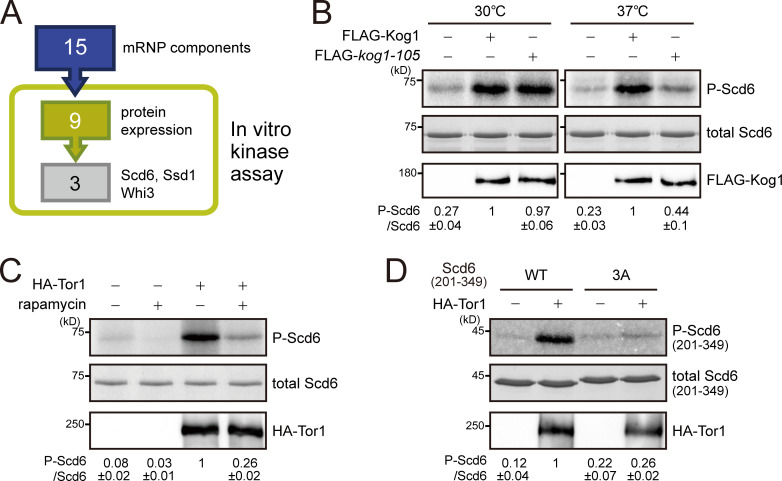
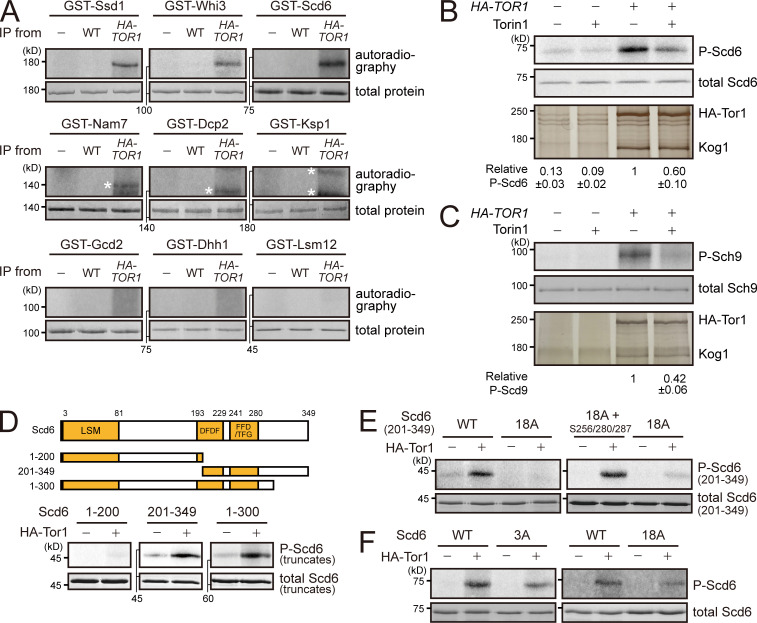
TORC1 is a Ser/Thr kinase complex, but none of the known substrates of TORC1 have been reported to associate with mRNAs. Thus, we investigated the possibility that TORC1 might target novel substrates among the components of mRNP complexes. In our coIP experiments, we identified 15 mRNP components that interact with TORC1 (Table S4). We performed an in vitro kinase assay of TORC1 with these proteins to examine whether they can be phosphorylated by TORC1. Among the nine proteins that were successfully purified from Escherichia coli, Scd6, Ssd1, and Whi3 were phosphorylated by immunoprecipitated TORC1 (Figs. 4 A and S4 A). Because the BiFC signals of Ssd1 and Whi3 were also detected at the vacuolar membrane (Table S1), we focused on Scd6, whose BiFC signal was colocalized with Dcp2, but not with Ego3. We further confirmed whether phosphorylation of Scd6 is mediated by TORC1 by an in vitro kinase assay using a temperature-sensitive mutant of KOG1, kog1-105 (Nakashima et al., 2008). TORC1 purified from cells expressing the kog1-105 mutant at the nonpermissive temperature (37°C) could not phosphorylate Scd6, whereas TORC1 from cells expressing wild-type KOG1 could (Fig. 4 B). Moreover, TORC1 purified from rapamycin-treated cells reduced phosphorylation of Scd6 (Fig. 4 C). We also used Torin1, an ATP-competitive inhibitor of TORC1 (Liu et al., 2012; Tanigawa and Maeda, 2017), during the in vitro kinase reaction. The addition of 2 µM Torin1 (50% inhibitory concentration = 500 nM) reduced phosphorylation of Scd6 as well as Sch9 (Fig. S4, B and C). On the basis of these results, we conclude that Scd6 is phosphorylated by TORC1.
Figure 4.
TORC1 phosphorylates Scd6. (A) In vitro kinase assay screen for 15 mRNA-binding proteins that interact with TORC1. (B) In vitro kinase assay using a temperature-sensitive mutant of Kog1, kog1-105. Kog1 was immunoprecipitated from lysates of cells and incubated with E. coli–purified Scd6. The relative ratio of phosphorylated Scd6 to total Scd6 was calculated, and the mean ± SD of triplicate experiments is shown below each lane. (C) In vitro kinase assay under rapamycin treatment. Tor1 was immunoprecipitated from cell lysates that were extracted from mock or rapamycin-treated cells. (D) In vitro kinase assay with the C-terminal fragment of Scd6 (amino acids 201–349). 3A indicates a mutant of Scd6(201–349) in which S256, S280, and S287 residues were replaced with Ala.
Figure S4.
TORC1 phosphorylates mRNA-binding proteins. (A) In vitro kinase assay for nine mRNP complex components purified from E. coli. Asterisks denote nonspecific signals. (B) Inhibition of in vitro phosphorylation of Scd6 by Torin1. Tor1 was immunoprecipitated by anti-HA−conjugated Dynabeads protein G from cell lysates. Immunoprecipitated proteins were silver stained. The relative ratio of phosphorylated Scd6 to total Scd6 was calculated, and the mean ± SD of triplicate experiments is shown below each lane. (C) Inhibition of in vitro phosphorylation of Sch9 by Torin1. The relative ratio of phosphorylated Sch9 to total Sch9 was calculated, and the mean ± SD of triplicate experiments is shown below each lane. (D) In vitro kinase assay with truncated Scd6. DFDF, DFDF domain (PROSITE #PS51512).; FFD, FFD box (PROSITE #PS51513); LSM, Sm-like ribonucleoproteins domain (SUPERFAMILY #SSF50182); TFG, TFG box (PROSITE #PS51536). (E) Contribution of the S256, S280, and S287 residues in TORC1-mediated phosphorylation of Scd6(201–349). (F) Phosphorylation of full-length Scd6-3A and Scd6-18A mutants by TORC1.
To identify the residues of Scd6 that are phosphorylated by TORC1, we performed an in vitro kinase assay with truncated Scd6. While the N-terminal fragment of Scd6, comprising amino acids 1–200, was hardly phosphorylated by TORC1, Scd6 truncations comprising amino acids 1–300 and 201–349 were strongly phosphorylated by TORC1 (Fig. S4 D), suggesting that the middle region of Scd6 (amino acids 201–300) is enriched with the target residues of TORC1. Indeed, a mutant of Scd6(201–349) in which all 18 Ser/Thr residues in the region of amino acids 201–300 were replaced with Ala was poorly phosphorylated by TORC1 (Fig. S4 E, left). By using Scd6 with point mutations at various combinations of Ser/Thr residues, we found that phosphorylation of Scd6 by TORC1 was considerably decreased when the S256, S280, and S287 residues were replaced with Ala (Fig. 4 D). Furthermore, when we restored these three residues in the 18A mutant of Scd6(201–349), phosphorylation by TORC1 was recovered (Fig. S4 E, right). These results suggest that S256, S280, and S287 are the target residues of TORC1. However, full-length Scd6-3A (Scd6[S256A/S280A/S287A]) and Scd6-18A were still phosphorylated by TORC1 (Fig. S4 F), implying that residues other than S256, S280, and S287 are also responsible for TORC1-mediated phosphorylation of Scd6.
mRNP complex–localized TORC1 down-regulates Scd6 activity
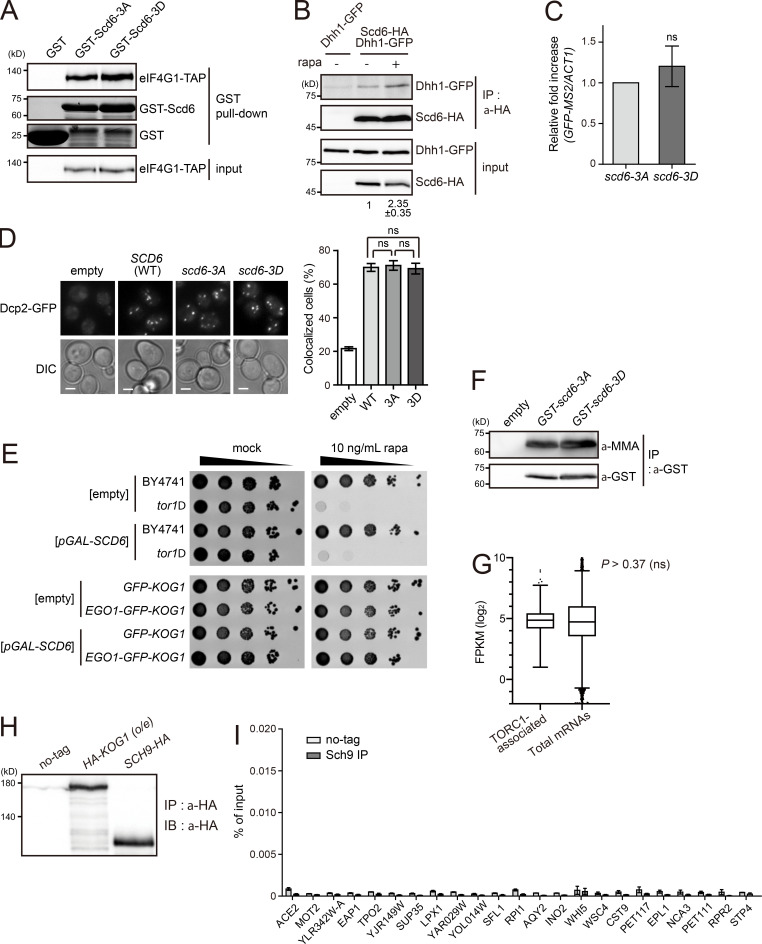
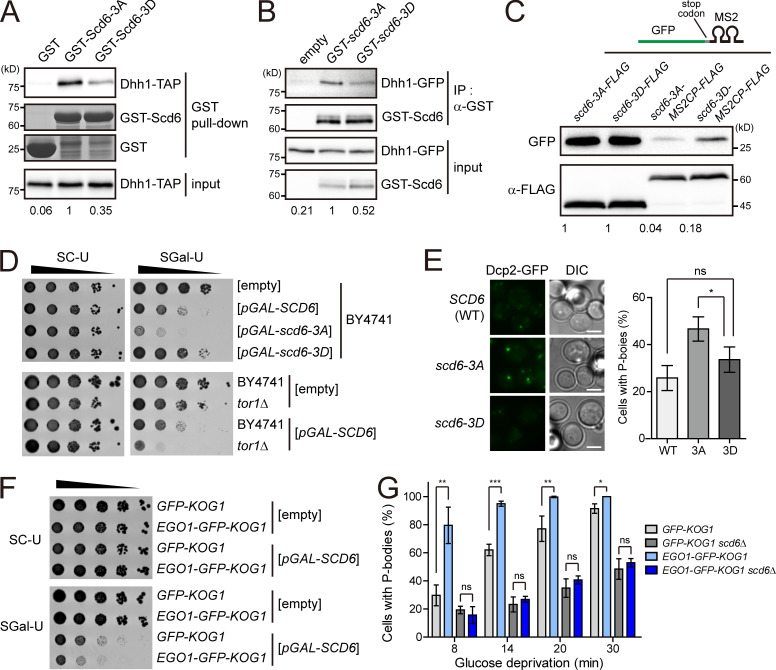
Scd6 is an RGG motif protein that inhibits translation initiation. The interaction between Scd6 and eukaryotic translation initiation factor (eIF) 4G prevents the recruitment of the 43S preinitiation complex, leading to translational repression at the initiation step (Rajyaguru et al., 2012). We examined whether TORC1-mediated phosphorylation of Scd6 affects its interaction with eIF4G using Scd6-3A and a phosphomimetic form of Scd6, Scd6-3D (Scd6[S256D/S280D/S287D]). However, we observed little, if any, difference between Scd6-3A and Scd6-3D in the interaction with eIF4G (Fig. S5 A). Thus, we tried to find another factor that could regulate Scd6-mediated translational repression. Recently, it has been reported that Dhh1 has a direct interaction with Scd6 and is a target of Scd6 in translational repression (Zeidan et al., 2018). When we examined the effects of TORC1-mediated phosphorylation of Scd6 on its interaction with Dhh1, Scd6-3D exhibited a decrease in the interaction with Dhh1 in both the in vitro GST pull-down assay (Fig. 5 A) and the in vivo coIP assay (Fig. 5 B). Furthermore, acute inhibition of TORC1 increased the Dhh1–Scd6 interaction in vivo (Fig. S5 B), suggesting that this interaction is regulated by TORC1-mediated phosphorylation. Given that the interaction of Scd6 with Dhh1 is believed to mediate translational repression, we next investigated whether the phosphorylation status of Scd6 affects the translational efficiency of Scd6-binding mRNAs. To do this, we tethered Scd6 mutants to GFP mRNAs using the MS2-CP system (Zeidan et al., 2018) and measured the translational efficiency of GFP mRNAs. The translational efficiency of GFP mRNAs tethered with Scd6-3A was much lower than that of GFP mRNAs tethered with Scd6-3D (Fig. 5 C). There was no significant difference in the turnover of GFP mRNAs tethered with Scd6-3A or Scd6-3D (Fig. S5 C). Taken together, these results suggest that TORC1-dependent phosphorylation of Scd6 on the S256/S280/S287 residues down-regulates the activity of Scd6 in translational repression by reducing its affinity to Dhh1.
Figure S5.
Phenotypes of scd6-3A and scd6-3D mutants. (A) Interaction of Scd6 mutants with eIF4G. E. coli–purified Scd6 mutants were incubated with lysates from cells expressing TIF4631-TAP. (B) Increased interaction between Scd6 and Dhh1 under TORC1 inhibition by rapamycin (rapa). The relative ratio of coimmunoprecipitated Dhh1-GFP to input Dhh1-GFP was calculated, and the mean ± SD of duplicate experiments is shown below each lane. (C) Effects of tethering of Scd6 mutants on the stability of GFP mRNAs. GFP-MS2 mRNAs were expressed under the ADH1 promoter, and Scd6 mutants were expressed under the TDH3 promoter. The mRNA levels of GFP-MS2 were normalized against to those of ACT1. P values were determined by Student’s t test (ns, P > 0.05). (D) P-body formation in cells overexpressing Scd6 mutants. Cells harboring p426GAL-SCD6 vectors were grown in 2% raffinose-containing media and Scd6 was induced by the addition of 2% galactose for 1.5 h. Scale bars, 2 µm. Right: Quantification of the ratio of cells with P-bodies. More than 200 cells were counted for each experiment. Values represent the average of three independent experiments. P values were determined by Student’s t test (ns, P > 0.05). (E) Effect of the vacuolar membrane anchoring of TORC1 on its activity. Cells were spotted in 10-fold serial dilutions on SC-Ura plates in the absence or presence of 10 ng/ml rapamycin. (F) Arginine methylation of Scd6 mutants. Scd6 was purified from cells expressing GST-Scd6-3A or GST-Scd6-3D, and arginine methylation was detected by anti-mono and dimethyl arginine (MMA) antibody (Abcam; ab412). (G) Expression levels of TORC1-associated mRNAs and total mRNAs. The whiskers represent the 5th and the 95th percentiles. P value was determined by Mann-Whitney U test. (H) Western blotting of Kog1 and Sch9 showing that the amount of immunoprecipitated Sch9 was not significantly different from that of immunoprecipitated Kog1 used in Fig. 6 B. (I) Enrichment of all tested mRNAs in the Sch9 immunoprecipitates. The amounts of immunoprecipitated mRNAs were divided by the amounts of input mRNAs and are represented as percentage of input. Values represent the average of three independent experiments. All error bars represent SD.
Figure 5.
TORC1 regulates the activity of Scd6 via phosphorylation. (A) In vitro GST pull-down assay with Scd6 mutants. E. coli–purified GST or GST-Scd6 mutants were incubated with lysates of cells expressing Dhh1-TAP. Dhh1-TAP was detected by immunoblotting. GST and GST-Scd6 mutants were visualized by Coomassie blue staining. (B) CoIP assay for the interaction between Dhh1 and Scd6. Dhh1-GFP coimmunoprecipitated with GST-Scd6 mutants was detected by Western blotting. The relative ratio of pulled-down or coimmunoprecipitated Dhh1-TAP to input Dhh1-TAP is shown below each lane. (C) Translational repression of the GFP reporter by tethering of Scd6 mutants. GFP-MS2 mRNAs were expressed under the ADH1 promoter and Scd6-3A/3D-FLAG or Scd6-3A/3D-MS2CP-FLAG were expressed under the TDH3 promoter. The relative ratio of GFP to FLAG-tagged protein is shown below each lane. (D) TORC1-dependent regulation of growth defect caused by Scd6 overexpression. Wild-type (BY4741) or tor1Δ cells harboring the indicated plasmids were spotted in 10-fold serial dilutions on SC-Ura (SC-U) and SGal-Ura (SGal-U) plates. (E) P-body formation in cells with Scd6 mutants under glucose starvation. Left: P-body formation was observed by fluorescence microscopy. scd6-3A and scd6-3D indicate cells expressing Scd6-3A and Scd6-3D, respectively. Cells were starved of glucose for 10 min. Scale bars, 2 µm. Right: Quantification of the ratio of cells with P-bodies (n > 200). (F) The effect of the vacuole-anchored TORC1 on growth defect caused by Scd6 overexpression. Cells expressing GFP-Kog1 or Ego1-GFP-Kog1 were transformed with the indicated plasmids and spotted in 10-fold serial dilutions on SC-Ura and SGal-Ura plates. (G) Increase in P-body formation in cells with the vacuole-anchored TORC1. Time-course images of cells expressing Dcp2-mCherry were taken at the indicated times after glucose starvation, and the ratio of cells with P-bodies was calculated using ImageJ software. Values represent the average of three independent experiments. Error bars represent SD. P values were determined by Student’s t test (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, P > 0.05).
Several phenotypes related to Scd6 have been reported. First, overexpression of Scd6 induces P-bodies and stress granules under nonstress conditions (Lien et al., 2016; Rajyaguru et al., 2012). Second, Scd6 stimulates the formation of P-bodies under glucose starvation conditions (Rajyaguru et al., 2012). Third, overexpression of Scd6 causes growth defects, similar to other translational repressors such as Dhh1 and Pat1, presumably due to increased translational repression activity (Coller and Parker, 2005; Nissan et al., 2010). To determine the role of TORC1-mediated phosphorylation of Scd6, we examined the effects of overexpression of Scd6-3A and Scd6-3D on cell growth. As we expected from the above translational efficiency analysis, the growth defect caused by overexpression of Scd6 was affected by the mutation of the S256/S280/S287 residues. Cells overexpressing Scd6-3A exhibited more severe growth defects than cells overexpressing wild-type Scd6, whereas overexpression of Scd6-3D rescued the growth defects (Fig. 5 D, top). Consistent with our finding that TORC1 phosphorylates the S256/S280/S287 residues, deletion of TOR1 enhanced the growth defects caused by Scd6 overexpression (Fig. 5 D, bottom). These results suggest that TORC1-dependent phosphorylation of Scd6 on the S256/S280/S287 residues inhibits the activity of Scd6 as a translational repressor. In addition, P-body formation under glucose starvation conditions was accelerated in cells expressing Scd6-3A compared with cells expressing wild-type Scd6 or Scd6-3D (Fig. 5 E), although overexpression of Scd6 induced P-body formation regardless of its phosphorylation status under normal conditions (Fig. S5 D). It is presumable that accelerated P-body formation in cells with Scd6-3A may be due to its enhanced interaction with Dhh1.
Next, we asked whether Scd6 is regulated by a portion of TORC1 that is associated with mRNAs in mRNP complexes. Interestingly, upon overexpression of Scd6, cells with the vacuole-anchored TORC1 exhibited more severe growth defects than cells with free TORC1 (Fig. 5 F), while a rapamycin sensitivity assay showed that the overall TORC1 activity of cells with the vacuole-anchored TORC1 was similar to that of cells with free TORC1 (Fig. S5 E). This result suggests that the activity of Scd6 is inhibited by nonvacuolar TORC1. We also examined P-body formation under glucose starvation conditions, which is known to be stimulated by Scd6 (Rajyaguru et al., 2012), in cells with the vacuole-anchored TORC1. Consistent with the above result, P-body formation was significantly increased in cells with the vacuole-anchored TORC1 at the early time points of glucose starvation (Fig. 5 G). Deletion of SCD6 abolished the difference in P-body formation between cells with the vacuole-anchored TORC1 and cells with free TORC1, indicating that the increase in P-body formation in cells with the vacuole-anchored TORC1 is mediated by Scd6. Overall, these results suggest that mRNP complex–localized TORC1 down-regulates the activity of Scd6 as a translational repressor.
RIP-seq identifies specific mRNAs that are associated with TORC1
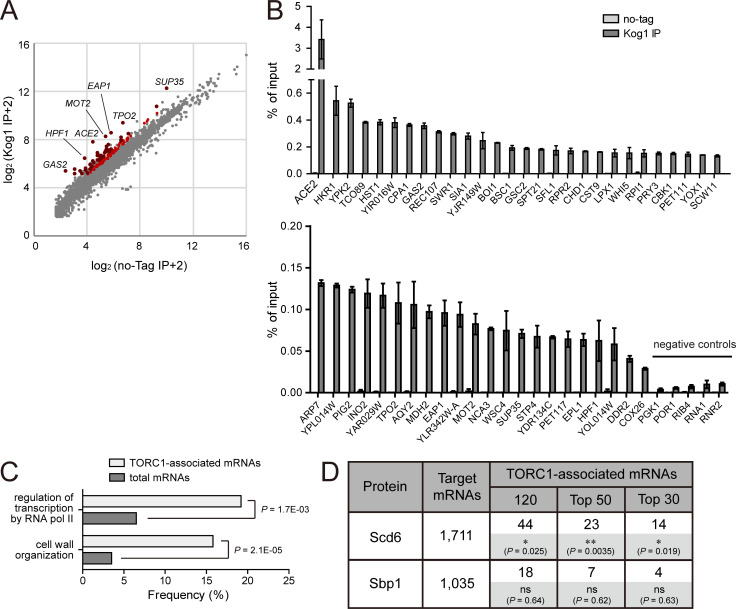
To identify mRNAs that interact with TORC1, we performed native RNA immunoprecipitation followed by deep sequencing (RIP-seq). Immunoprecipitation was performed with anti-HA antibody in HA-KOG1 cells that express both Tor1 and HA-Kog1 under the HXK2 promoter, and coimmunoprecipitated RNAs were analyzed by next-generation sequencing. From this analysis, we identified 120 genes whose mRNAs were enriched in the immunoprecipitates of HA-KOG1 cells compared with those of no-tag control cells (Fig. 6 A and Table S7 A). There was no bias in the expression levels of the 120 TORC1-associated mRNAs (Fig. S5 G) and none of these enrichments was caused by differential expression of the mRNAs in HA-KOG1 cells (Table S7 B). To further confirm the RIP-seq results, the 50 most enriched mRNAs in the Kog1 immunoprecipitates were analyzed by quantitative real-time PCR. All of the tested mRNAs were significantly enriched in Kog1 immunoprecipitates compared with the no-tag control, while five mRNAs randomly selected from RIP-seq–negative genes were not enriched (Fig. 6 B). We also analyzed the specificity of the enrichment of these mRNAs using the immunoprecipitates of cells expressing Sch9-HA. None of these mRNAs were enriched in the Sch9 immunoprecipitates (Fig. S5, H and I), indicating that these mRNAs interact with TORC1 in a specific manner.
Figure 6.
TORC1-associated mRNAs significantly overlap with Scd6-interacting mRNAs. (A) RIP-seq analysis of Kog1. Reads per kilobase million value of each gene obtained from cells expressing HA-Kog1 (y axis) was plotted against that obtained from wild-type cells (x axis). Red dots indicate 120 mRNAs that were enriched in Kog1 immunoprecipitates, and dark red dots among them indicate the 50 most enriched mRNAs confirmed by quantitative real-time PCR. (B) RIP/quantitative real-time PCR assay for the 50 most enriched mRNAs among TORC1-associated mRNAs and five negative controls. The amounts of immunoprecipitated mRNAs are represented as percentage of input. Values represent the average of three independent experiments. Error bars represent SD. (C) GO analysis of 120 TORC1-associated mRNAs. GO analysis for biological process was performed using the GO term finder (Boyle et al., 2004). (D) Comparison of TORC1-associated mRNAs with those interacting with Scd6 and Sbp1. The numbers of overlapped mRNAs are represented. P values were determined by Fisher’s exact test (*, P < 0.05; **, P < 0.01; ns, P > 0.05).
To characterize TORC1-associated mRNAs, we performed GO analysis for the 120 genes whose mRNAs were enriched in the Kog1 immunoprecipitates. Two GO biological process terms, “cell wall organization” and “regulation of transcription by RNA pol II,” were significantly enriched among the 120 genes compared with the whole genome (Fig. 6 C). Given that TORC1 phosphorylates Scd6 and mRNP complex–localized TORC1 regulates the activity of Scd6, we next investigated the relationships between the target mRNAs of TORC1 and Scd6. A previous study identified 1,711 mRNAs that interact with Scd6 (Tsvetanova et al., 2010). We compared the list of 120 TORC1-associated mRNAs with that of Scd6 target mRNAs. Notably, Scd6 target mRNAs were significantly enriched in TORC1-associated mRNAs, and the significance was further increased for the 50 and 30 most enriched mRNAs in the Kog1 immunoprecipitates (Fig. 6 D). As a control, we also compared TORC1-associated mRNAs with the target mRNAs of Sbp1, which is another RGG motif protein (Mitchell et al., 2013; Rajyaguru et al., 2012). However, we could not observe any significant overlap between the target mRNAs of TORC1 and those of Sbp1. Taken together, it is probable that TORC1 may regulate the translation of its associated mRNAs through phosphorylation of Scd6.
Discussion
In this study, we identified interactors of TORC1 by using the BiFC assay. With the advantages of the BiFC assay, we identified numerous novel interactors, some of which were verified by the coIP assay. Our dataset includes weak or transient interactions that have not been detected in other high-throughput PPI experiments. The enrichment of mRNA-binding proteins in the TORC1 interactome prompted us to investigate the relationship between TORC1 and the posttranscriptional regulation of mRNAs. We show that nonvacuolar TORC1 interacts with a specific subset of mRNAs and is localized to mRNP granules. We also provide evidence that the RGG motif protein Scd6 is a novel substrate of TORC1. TORC1-dependent phosphorylation of Scd6 seems to repress the activity of Scd6, and this regulation is mediated by nonvacuolar TORC1. Given that TORC1-associated mRNAs significantly overlap with Scd6-associated mRNAs, it is presumable that TORC1–Scd6 signaling may regulate the translation of a specific subset of mRNAs.
Although the vacuolar membrane is known to be the main site of TORC1 activation, there are several reports showing that TORC1 also localizes to other sites. Nuclear-localized TORC1 regulates the transcription of 35S rRNA (Li et al., 2006), and a small fraction of TORC1 is detected on the plasma membrane (Reinke et al., 2004; Wedaman et al., 2003). Perivacuolar foci of TORC1 have recently been suggested to be endosomes and nonvesicular aggregates of TORC1 (TORC1 organized in inhibited domains), which are induced in different situations (Hatakeyama et al., 2019; Prouteau et al., 2017). Notably, the localization of TORC1 to stress granules has been observed in yeast as well as mammalian cells (Takahara and Maeda, 2012; Thedieck et al., 2013). TORC1 is sequestered into stress granules in response to heat stress, and the mTORC1 component raptor is recruited to stress granules upon arsenite stress. In both cases, the sequestration of TORC1/mTORC1 to stress granules is regarded as a transient mechanism of TORC1/mTORC1 inhibition. However, in this study, we demonstrate that a portion of TORC1 is consistently associated with mRNP complexes and suggest that TORC1 may play a role in the posttranscriptional regulation of specific mRNAs.
mTORC1 directly phosphorylates LARP1, which binds 5′ terminal oligopyrimidine mRNAs, such as ribosomal protein mRNAs, and enhances the translation of LARP1-associated mRNAs (Hong et al., 2017; Meyuhas and Kahan, 2015; Tcherkezian et al., 2014). Given that TORC1 phosphorylates the mRNA-binding protein Scd6, which regulates translation, the mTORC1–LARP1 pathway seems similar to the TORC1–Scd6 pathway identified in this study. However, because yeast does not have a 5′ terminal oligopyrimidine sequence and Scd6 does not have functional or structural homology with LARP1, we do not believe that these two pathways are homologous. In Saccharomyces cerevisiae, Slf1 and Sro9 are two homologues of LARP1 (Bousquet-Antonelli and Deragon, 2009). In the initial BiFC screen, Sro9 showed positive BiFC signals with both Tor1 and Kog1, but it was eliminated from the TORC1 interactome during the self-assembly analysis. Nevertheless, it is still possible that the BiFC signals between Sro9 and TORC1 were produced by true interactions. It would be interesting to investigate whether Sro9 is a target of TORC1 and whether the mTORC1–LARP1 pathway is related to Sro9 in yeast.
In this study, we detected the in vitro phosphorylation of Ssd1 and Whi3, in addition to Scd6, by TORC1. Ssd1 is an RNA-binding protein that is mainly associated with mRNAs encoding cell wall proteins (Jansen et al., 2009). Cbk1 is the only known kinase of Ssd1 that inhibits its translational repressor activity and its association with P-bodies (Jansen et al., 2009; Kurischko and Broach, 2017). Ssd1 has also been linked to both TORC1 and TORC2 based on genetic interactions (Cardon et al., 2012; Reinke et al., 2004). Notably, the rapamycin hypersensitivity of tor1Δ and tco89Δ mutants was rescued by overexpression of Ssd1, suggesting a functional role of Ssd1 in the TORC1 pathway. Although the precise mechanisms have not yet been elucidated, our results suggest the possibility that Ssd1 is a downstream effector of TORC1.
Recently, it has been reported that arginine methylation regulates the activity of Scd6. When arginine methylation is impaired, Scd6 cannot accumulate in P-bodies properly and its interaction with eIF4G is decreased, resulting in the reduction of its translational repressor activity (Lien et al., 2016; Poornima et al., 2016). Given that S287 is proximal to R288, which is one of the methylated residues, it can be assumed that the mutation of S287 might affect the activity of Scd6 by interfering with methylation on R288. To check this possibility, we analyzed arginine methylation of Scd6 mutants using anti-mono and dimethyl arginine antibody. However, we could not detect any difference in arginine methylation between Scd6-3A and Scd6-3D (Fig. S5 F). The phenotypes of cells expressing Scd6-3A are also different from those of methylation-deficient cells; the methylation-deficient mutation of Scd6 does not affect P-body formation under glucose starvation (Lien et al., 2016), whereas Scd6-3A does (Fig. 5 E). In addition, the interaction between Scd6 and eIF4G is not altered in cells expressing Scd6-3A or Scd6-3D (Fig. S5 A). It has been suggested that the arginine methylation of Scd6 is not required for the cooperative function of Scd6 and Dhh1 in the formation of P-bodies under glucose starvation conditions. Given these facts, we suggest that phosphorylation on the S256/S280/S287 residues is another regulatory mechanism of Scd6 and that the function of this phosphorylation is related to the recruitment of Dhh1. Further investigation into the relationship between the phosphorylation and arginine methylation of Scd6 will increase our knowledge about the precise molecular mechanisms of Scd6 in translational regulation.
Materials and methods
Yeast strains, plasmids, and culture conditions
S. cerevisiae strains, plasmids, and oligonucleotide primers used in this study are listed in Table S8. Yeast cells were grown at 30°C in YPD (1% yeast extract, 2% peptone, and 2% glucose) or synthetic complete (SC) medium as indicated. For glucose starvation, cells grown to mid-log phase (OD600 = 0.7∼1.0) were transferred to YP medium (1% yeast extract and 2% peptone) or SC medium lacking glucose and cultured for the indicated times. For cycloheximide treatment, cells grown to mid-log phase were treated with 100 µg/ml of cycloheximide and cultured for the indicated times.
Genome-wide BiFC screen
VC-tagged MATα strains (VC-TOR1, VC-TOR2, and VC-KOG1) were generated by PCR-based gene targeting procedure (Sung and Huh, 2007). Each VC-tagged strain was mated with 5,911 MATα strains expressing full-length proteins tagged with VN (Sung et al., 2013). The resulting diploid cells were incubated in SC medium to mid-log phase and microscopically analyzed in 96-well glass-bottomed microplates (Matrical Bioscience, MGP096). Microscopy was performed on a Nikon eclipse E1 microscope with a Plan Fluor 100×/1.30 NA oil immersion objective.
CoIP assay
Yeast cells grown to mid-log phase were harvested and disrupted with glass beads in lysis buffer (50 mM Tris-Cl, pH 7.5, 150 mM NaCl, and 0.15% NP-40) containing 1 mM phenylmethylsulphonyl fluoride (PMSF) and protease inhibitors. Lysates were clarified by centrifugation at 3,300×g for 10 min at 4°C and then diluted to a concentration of ∼5 mg/ml. For RNase A treatment, 50 µg/ml RNase A was added to the cell lysates and incubated for 10 min at 37°C. After taking input samples, diluted extracts were incubated with anti-c-Myc antibody (Santa Cruz Biotechnology; SC-40)-conjugated protein A-Sepharose beads (GE Healthcare; 17513801) for Myc-tagged proteins or IgG-Sepharose (GE Healthcare; 17096901) for TAP-tagged proteins. After incubation for 2 h, beads were washed five times with lysis buffer and boiled with SDS sample buffer. Immunoprecipitated samples and 0.5% input samples were subjected to SDS-PAGE. Immunoblots were performed with HRP-conjugated rabbit anti-mouse IgG antibody (Sigma-Aldrich; A9044) for TAP-tagged proteins, HRP-conjugated anti-GFP antibody (Santa Cruz Biotechnology; SC-9996 HRP) for GFP-tagged proteins, and anti–c-Myc antibody (Santa Cruz Biotechnology; SC-40 HRP) for Myc-tagged proteins.
Live-cell imaging and image analysis
Images for BiFC screens were taken by a Nikon eclipse E1 microscope as described above. All images except Fig. 1 B, Fig. 2, B and C, and Fig. S2 A were taken by a DeltaVision System (Applied Precision) using a 100× objective lens. Cells were grown in SC medium and starved for glucose when indicated. Cells were transferred to a coverglass-bottom dish (SPL; 100350), and time-lapse microscopy of cells was performed in a 30°C environmental chamber of the DeltaVision System as previously described (Lim et al., 2020). For counting P-bodies or colocalized foci, 10 images were taken with 2 µm spacing. The acquired images were analyzed with ImageJ software.
Fractionation
To separate nonmembranous structures from soluble proteins (Fig. 3, C–E), 50 OD600 of yeast cells grown to mid-log phase were harvested and disrupted with glass beads in 1 ml of 1% Triton X-100 lysis buffer (50 mM Tris-Cl, pH 7.5, 150 mM NaCl, and 1% Triton X-100) containing 1 mM PMSF and protease inhibitors. Lysed cells were clarified by centrifugation at 2,000×g for 5 min at 4°C, and then crude cell extracts were centrifuged at 13,000×g for 10 min at 4°C. Supernatants were taken for S13 samples. Pellets were briefly washed with PBS and resuspended in 200 µl SDS sample buffer for P13 samples.
Subcellular fractionation of membrane proteins (Fig. S2 G, lower) was conducted as previously described (Kaiser et al., 2002) with minor modifications. Briefly, 30 OD600 of yeast cells grown to mid-log phase were converted to spheroplasts by incubation with 100 U zymolyase in the presence of 3,3′-dithiodipropionic acid. The resulting spheroplasts were resuspended in lysis buffer (50 mM Tris-Cl, pH 7.5, 0.2 M sorbitol, and 1 mM EDTA) and disrupted with up-and-down strokes of Dounce homogenizer. Cell lysates were clarified by centrifugation at 500×g for 5 min at 4°C and then fractionated by centrifugation at 13,000×g for 10 min at 4°C. Supernatants were used for S13 fraction samples, and pellets were used for P13 fraction after resuspension with lysis buffer containing 1% NP-40.
Native oligo(dT) pull-down assay
Cell extracts were prepared from cells grown to mid-log phase in YPD medium by bead beating in lysis buffer (50 mM Tris-Cl, pH 7.5, 150 mM NaCl, 1% NP-40, and 1 mM EDTA) containing 1 mM PMSF, protease inhibitors, and RNasin (Promega; N2111). Cell extracts were incubated with or without 50 µg/ml RNase A for 10 min at 37°C and then incubated with oligo(dT)25 Cellulose Beads (New England Biolabs; S2408) for 1 h at 4°C to precipitate mRNA–protein complexes. Oligo(dT)-captured proteins were eluted by heating with SDS sample buffer and analyzed by Western blotting.
In vitro kinase assay
Cells were grown in YPD medium at 30°C to OD600 0.4–0.6 and treated with 3 mM dithiobis[succinimidyl propionate] cross-linker to preserve TORC1. For nonpermissive temperature conditions, cells were moved to 37°C and incubated for 2 h. For rapamycin treatment, 200 ng/ml rapamycin was added to cell culture and incubated for 30 min at 30°C. Cells were harvested and lysed with glass beads in lysis buffer (50 mM Tris-Cl, pH 7.5, 150 mM NaCl, and 1% NP-40) containing protease inhibitors and phosphatase inhibitors. After centrifugation at 6,000×g for 10 min, cell lysates were incubated with anti-Flag M2 affinity gel (Sigma-Aldrich; F2426) for Flag-tagged proteins or anti-HA antibody (Santa Cruz Biotechnology; SC-7392)–conjugated Dynabeads protein G (Invitrogen; 10003D) for HA-tagged proteins. SCD6 was cloned into the pGEX4T-1 plasmid, and GST-Scd6 was purified from E. coli by GST affinity purification. Kinase reactions were performed in kinase buffer (25 mM Hepes, pH 7.5, 10 mM MgCl2, and 50 µM ATP). Immunoprecipitated TORC1 and purified GST-Scd6 were incubated with 4 µCi [γ-32P]-ATP for 15 min at 30°C. The reactions were terminated by the addition of SDS-PAGE sample buffer followed by heating at 95°C for 10 min. Autoradiography signals were detected by BAS-2500 (Bio-Rad), and Western blotting was performed with anti-Flag antibody (Rockland; 600–401-383) or anti-HA antibody (Santa Cruz Biotechnology; SC-7392 HRP).
RIP-seq
Yeast cells grown to mid-log phase in YPD medium were lysed by bead beating in lysis buffer (50 mM Tris-Cl, pH 7.5, 150 mM NaCl, 1% NP-40, and 1 mM EDTA) containing 1 mM PMSF, protease inhibitors, and RNasin. Cell extracts were clarified by centrifugation at 3,300×g for 10 min at 4°C and then diluted to a concentration of ∼5 mg/ml. Anti-HA agarose beads (Sigma-Aldrich; A2095) were added to cell extracts and incubated with gentle rocking for 2 h at 4°C. RNAs from input and immunoprecipitated samples were extracted by TRIzol. cDNA libraries were constructed using TruSeq Stranded Total RNA with Ribo-Zero kit (Illumina) following the manufacturer’s instruction. Next-generation sequencing was performed using Illumina HiSeq2000.
Quantitative real-time PCR
Cell lysates were prepared as described above in RNA immunoprecipitation. 3% of cell lysates were taken for input samples, and the remaining lysates were incubated with anti-c-Myc antibody-conjugated Dynabeads protein G. RNAs were prepared as described above and cDNAs were synthesized using random hexamer (IDT) and M-MLV Reverse transcription (MBiotech; 19500). The amount of mRNAs of interest was analyzed by the QuantStudio 3 Real-Time PCR system with the SensiFAST SYBR Lo-ROX kit (Bioline; BIO-94005).
Statistical analysis
GraphPad Prism software was used to perform statistical analysis for Fig. 2, F and H, Fig. 3 I, Fig. 5, E and G, Fig. 6 B, and Fig. S5, C, D, and I. The data represent mean ± SD of triplicate experiments, and P values were determined by unpaired, two-tailed t tests. For quantification of cells with colocalization or P-bodies in Fig. 2, F and H, Fig. 5, E and G, and Fig. S5 D, more than 200 cells were counted for individual experiments. P values in Fig. 6 D were determined by Fisher’s exact test.
Online supplemental material
Fig. S1 shows the analysis of coIP-positive Kog1 interactors. Fig. S2 shows the accumulation of TORC1 in cytoplasmic foci distinct from the vacuolar membrane. Fig. S3 shows the comparison of promoter strengths and that RNA mediates the interaction between TORC1 and P-body components under glucose starvation. Fig. S4 shows phosphorylation of mRNA-binding proteins by TORC1. Fig. S5 shows the phenotypes of cells expressing Scd6-3A and Scd6-3D mutants and that Sch9 does not interact with TORC1-associated mRNAs. Table S1 lists 130 BiFC-positive interactors of Kog1. Table S2 lists 35 BiFC-positive interactors of Tor1. Table S3 lists 36 BiFC-positive interactors of Tor2. Table S4 lists 28 coIP-positive interactors of Kog1. Table S5 lists known physical interactors of Kog1. Table S6 shows GO analysis of 130 BiFC-positive Kog1 interactors. Table S7 lists 120 TORC1-associated mRNAs. Table S8 lists yeast strains, plasmids, and oligonucleotide primers used in this study.
Supplementary Material
lists 130 BiFC-positive interactors of Kog1.
lists 35 BiFC-positive interactors of Tor1.
lists 36 BiFC-positive interactors of Tor2.
lists 28 coIP-positive interactors of Kog1.
lists known physical interactors of Kog1.
shows GO analysis of 130 BiFC-positive Kog1 interactors.
lists 120 TORC1-associated mRNAs. Table S7 A shows fold enrichment of genes in Kog1 immunoprecipitates. Reads per kilobase million values were determined by a RIP-seq experiment. TOR1, KOG1, and genes with reads per kilobase million <1 were eliminated. Table S7 B shows differentially expressed genes in HA-KOG1 cells that were used in RIP-seq experiments. Fold increase values were determined by duplicated RNA sequencing.
lists yeast strains, plasmids, and oligonucleotide primers used in this study.
Acknowledgments
We thank Jeffrey E. Gerst (Weizmann Institute of Science, Rehovot, Israel) for generously providing the plasmids for m-TAG system, Yoshiaki Kamada (National Institute for Basic Biology, Okazaki, Japan) for yeast strains for the temperature-sensitive mutant of KOG1, and Seung Bum Park for technical assistance in the three-color colocalization experiment. We also thank Ankur Jain for supporting the completion of the revision and the members of our laboratory for helpful discussions.
This work was supported by the National Research Foundation of Korea (grants 2015R1A2A1A01007871 and 2018R1A2B2009169), which is funded by the Ministry of Education, Science and Technology, Republic of Korea.
The authors declare no competing financial interests.
Author contributions: Conceptualization: Y. Chang and W.-K. Huh; Investigation: Y. Chang and G. Lim; Formal analysis: Y. Chang and G. Lim; Visualization: Y. Chang and W.-K. Huh; Writing – original draft: Y. Chang and W.-K. Huh; Writing – review and editing: all authors; Supervision, W.-K. Huh.
References
- Binda, M., Péli-Gulli M.-P., Bonfils G., Panchaud N., Urban J., Sturgill T.W., Loewith R., and De Virgilio C.. 2009. The Vam6 GEF controls TORC1 by activating the EGO complex. Mol. Cell. 35:563–573. 10.1016/j.molcel.2009.06.033 [DOI] [PubMed] [Google Scholar]
- Bousquet-Antonelli, C., and Deragon J.M.. 2009. A comprehensive analysis of the La-motif protein superfamily. RNA. 15:750–764. 10.1261/rna.1478709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle, E.I., Weng S., Gollub J., Jin H., Botstein D., Cherry J.M., and Sherlock G.. 2004. GO:TermFinder--open source software for accessing Gene Ontology information and finding significantly enriched Gene Ontology terms associated with a list of genes. Bioinformatics. 20:3710–3715. 10.1093/bioinformatics/bth456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brangwynne, C.P. 2013. Phase transitions and size scaling of membrane-less organelles. J. Cell Biol. 203:875–881. 10.1083/jcb.201308087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridges, D., Ma J.-T., Park S., Inoki K., Weisman L.S., and Saltiel A.R.. 2012. Phosphatidylinositol 3,5-bisphosphate plays a role in the activation and subcellular localization of mechanistic target of rapamycin 1. Mol. Biol. Cell. 23:2955–2962. 10.1091/mbc.e11-12-1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan, J.R., Muhlrad D., and Parker R.. 2008. P bodies promote stress granule assembly in Saccharomyces cerevisiae. J. Cell Biol. 183:441–455. 10.1083/jcb.200807043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cafferkey, R., Young P.R., McLaughlin M.M., Bergsma D.J., Koltin Y., Sathe G.M., Faucette L., Eng W.K., Johnson R.K., and Livi G.P.. 1993. Dominant missense mutations in a novel yeast protein related to mammalian phosphatidylinositol 3-kinase and VPS34 abrogate rapamycin cytotoxicity. Mol. Cell. Biol. 13:6012–6023. 10.1128/MCB.13.10.6012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardon, C.M., Beck T., Hall M.N., and Rutter J.. 2012. PAS kinase promotes cell survival and growth through activation of Rho1. Sci. Signal. 5:ra9. 10.1126/scisignal.2002435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coller, J., and Parker R.. 2005. General translational repression by activators of mRNA decapping. Cell. 122:875–886. 10.1016/j.cell.2005.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowart, L.A., Gandy J.L., Tholanikunnel B., and Hannun Y.A.. 2010. Sphingolipids mediate formation of mRNA processing bodies during the heat-stress response of Saccharomyces cerevisiae. Biochem. J. 431:31–38. 10.1042/BJ20100307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubouloz, F., Deloche O., Wanke V., Cameroni E., and De Virgilio C.. 2005. The TOR and EGO protein complexes orchestrate microautophagy in yeast. Mol. Cell. 19:15–26. 10.1016/j.molcel.2005.05.020 [DOI] [PubMed] [Google Scholar]
- Hatakeyama, R., Péli-Gulli M.P., Hu Z., Jaquenoud M., Garcia Osuna G.M., Sardu A., Dengjel J., and De Virgilio C.. 2019. Spatially Distinct Pools of TORC1 Balance Protein Homeostasis. Mol. Cell. 73:325–338.e8. 10.1016/j.molcel.2018.10.040 [DOI] [PubMed] [Google Scholar]
- Heitman, J., Movva N.R., and Hall M.N.. 1991. Targets for cell cycle arrest by the immunosuppressant rapamycin in yeast. Science. 253:905–909. 10.1126/science.1715094 [DOI] [PubMed] [Google Scholar]
- Helliwell, S.B., Wagner P., Kunz J., Deuter-Reinhard M., Henriquez R., and Hall M.N.. 1994. TOR1 and TOR2 are structurally and functionally similar but not identical phosphatidylinositol kinase homologues in yeast. Mol. Biol. Cell. 5:105–118. 10.1091/mbc.5.1.105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong, S., Freeberg M.A., Han T., Kamath A., Yao Y., Fukuda T., Suzuki T., Kim J.K., and Inoki K.. 2017. LARP1 functions as a molecular switch for mTORC1-mediated translation of an essential class of mRNAs. eLife. 6:e25237. 10.7554/eLife.25237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, C.-D., Chinenov Y., and Kerppola T.K.. 2002. Visualization of interactions among bZIP and Rel family proteins in living cells using bimolecular fluorescence complementation. Mol. Cell. 9:789–798. 10.1016/S1097-2765(02)00496-3 [DOI] [PubMed] [Google Scholar]
- Hubstenberger, A., Courel M., Bénard M., Souquere S., Ernoult-Lange M., Chouaib R., Yi Z., Morlot J.B., Munier A., Fradet M., et al. 2017. P-Body Purification Reveals the Condensation of Repressed mRNA Regulons. Mol. Cell. 68:144–157.e5. 10.1016/j.molcel.2017.09.003 [DOI] [PubMed] [Google Scholar]
- Jain, S., Wheeler J.R., Walters R.W., Agrawal A., Barsic A., and Parker R.. 2016. ATPase-Modulated Stress Granules Contain a Diverse Proteome and Substructure. Cell. 164:487–498. 10.1016/j.cell.2015.12.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen, J.M., Wanless A.G., Seidel C.W., and Weiss E.L.. 2009. Cbk1 regulation of the RNA-binding protein Ssd1 integrates cell fate with translational control. Curr. Biol. 19:2114–2120. 10.1016/j.cub.2009.10.071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser, C.A., Chen E.J., and Losko S.. 2002. Subcellular fractionation of secretory organelles. Methods Enzymol. 351:325–338. 10.1016/S0076-6879(02)51855-3 [DOI] [PubMed] [Google Scholar]
- Kerppola, T.K. 2006. Design and implementation of bimolecular fluorescence complementation (BiFC) assays for the visualization of protein interactions in living cells. Nat. Protoc. 1:1278–1286. 10.1038/nprot.2006.201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, E., Goraksha-Hicks P., Li L., Neufeld T.P., and Guan K.-L.. 2008. Regulation of TORC1 by Rag GTPases in nutrient response. Nat. Cell Biol. 10:935–945. 10.1038/ncb1753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, Y., Jung J.P., Pack C.G., and Huh W.K.. 2019. Global analysis of protein homomerization in Saccharomyces cerevisiae. Genome Res. 29:135–145. 10.1101/gr.231860.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurischko, C., and Broach J.R.. 2017. Phosphorylation and nuclear transit modulate the balance between normal function and terminal aggregation of the yeast RNA-binding protein Ssd1. Mol. Biol. Cell. 28:3057–3069. 10.1091/mbc.e17-02-0100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H., Tsang C.K., Watkins M., Bertram P.G., and Zheng X.F.S.. 2006. Nutrient regulates Tor1 nuclear localization and association with rDNA promoter. Nature. 442:1058–1061. 10.1038/nature05020 [DOI] [PubMed] [Google Scholar]
- Lien, P.T., Izumikawa K., Muroi K., Irie K., Suda Y., and Irie K.. 2016. Analysis of the Physiological Activities of Scd6 through Its Interaction with Hmt1. PLoS One. 11:e0164773. 10.1371/journal.pone.0164773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim, G., Chang Y., and Huh W.K.. 2020. Phosphoregulation of Rad51/Rad52 by CDK1 functions as a molecular switch for cell cycle-specific activation of homologous recombination. Sci. Adv. 6:eaay2669. 10.1126/sciadv.aay2669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Q., Ren T., Fresques T., Oppliger W., Niles B.J., Hur W., Sabatini D.M., Hall M.N., Powers T., and Gray N.S.. 2012. Selective ATP-competitive inhibitors of TOR suppress rapamycin-insensitive function of TORC2 in Saccharomyces cerevisiae. ACS Chem. Biol. 7:982–987. 10.1021/cb300058v [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewith, R., Jacinto E., Wullschleger S., Lorberg A., Crespo J.L., Bonenfant D., Oppliger W., Jenoe P., and Hall M.N.. 2002. Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. Mol. Cell. 10:457–468. 10.1016/S1097-2765(02)00636-6 [DOI] [PubMed] [Google Scholar]
- Lui, J., Castelli L.M., Pizzinga M., Simpson C.E., Hoyle N.P., Bailey K.L., Campbell S.G., and Ashe M.P.. 2014. Granules harboring translationally active mRNAs provide a platform for P-body formation following stress. Cell Rep. 9:944–954. 10.1016/j.celrep.2014.09.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyuhas, O., and Kahan T.. 2015. The race to decipher the top secrets of TOP mRNAs. Biochim. Biophys. Acta. 1849:801–811. 10.1016/j.bbagrm.2014.08.015 [DOI] [PubMed] [Google Scholar]
- Mitchell, S.F., Jain S., She M., and Parker R.. 2013. Global analysis of yeast mRNPs. Nat. Struct. Mol. Biol. 20:127–133. 10.1038/nsmb.2468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima, A., Maruki Y., Imamura Y., Kondo C., Kawamata T., Kawanishi I., Takata H., Matsuura A., Lee K.S., Kikkawa U., et al. 2008. The yeast Tor signaling pathway is involved in G2/M transition via polo-kinase. PLoS One. 3:e2223. 10.1371/journal.pone.0002223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nissan, T., Rajyaguru P., She M., Song H., and Parker R.. 2010. Decapping activators in Saccharomyces cerevisiae act by multiple mechanisms. Mol. Cell. 39:773–783. 10.1016/j.molcel.2010.08.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poornima, G., Shah S., Vignesh V., Parker R., and Rajyaguru P.I.. 2016. Arginine methylation promotes translation repression activity of eIF4G-binding protein, Scd6. Nucleic Acids Res. 44:9358–9368. 10.1093/nar/gkw762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powis, K., Zhang T., Panchaud N., Wang R., De Virgilio C., and Ding J.. 2015. Crystal structure of the Ego1-Ego2-Ego3 complex and its role in promoting Rag GTPase-dependent TORC1 signaling. Cell Res. 25:1043–1059. 10.1038/cr.2015.86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prouteau, M., Desfosses A., Sieben C., Bourgoint C., Lydia Mozaffari N., Demurtas D., Mitra A.K., Guichard P., Manley S., and Loewith R.. 2017. TORC1 organized in inhibited domains (TOROIDs) regulate TORC1 activity. Nature. 550:265–269. 10.1038/nature24021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajyaguru, P., She M., and Parker R.. 2012. Scd6 targets eIF4G to repress translation: RGG motif proteins as a class of eIF4G-binding proteins. Mol. Cell. 45:244–254. 10.1016/j.molcel.2011.11.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinke, A., Anderson S., McCaffery J.M., Yates J. III, Aronova S., Chu S., Fairclough S., Iverson C., Wedaman K.P., and Powers T.. 2004. TOR complex 1 includes a novel component, Tco89p (YPL180w), and cooperates with Ssd1p to maintain cellular integrity in Saccharomyces cerevisiae. J. Biol. Chem. 279:14752–14762. 10.1074/jbc.M313062200 [DOI] [PubMed] [Google Scholar]
- Roelants, F.M., Leskoske K.L., Martinez Marshall M.N., Locke M.N., and Thorner J.. 2017. The TORC2-Dependent Signaling Network in the Yeast Saccharomyces cerevisiae. Biomolecules. 7:7. 10.3390/biom7030066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancak, Y., Bar-Peled L., Zoncu R., Markhard A.L., Nada S., and Sabatini D.M.. 2010. Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell. 141:290–303. 10.1016/j.cell.2010.02.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheth, U., and Parker R.. 2003. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science. 300:805–808. 10.1126/science.1082320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturgill, T.W., Cohen A., Diefenbacher M., Trautwein M., Martin D.E., and Hall M.N.. 2008. TOR1 and TOR2 have distinct locations in live cells. Eukaryot. Cell. 7:1819–1830. 10.1128/EC.00088-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung, M.K., and Huh W.K.. 2007. Bimolecular fluorescence complementation analysis system for in vivo detection of protein-protein interaction in Saccharomyces cerevisiae. Yeast. 24:767–775. 10.1002/yea.1504 [DOI] [PubMed] [Google Scholar]
- Sung, M.-K., Ha C.W., and Huh W.-K.. 2008. A vector system for efficient and economical switching of C-terminal epitope tags in Saccharomyces cerevisiae. Yeast. 25:301–311. 10.1002/yea.1588 [DOI] [PubMed] [Google Scholar]
- Sung, M.-K., Lim G., Yi D.-G., Chang Y.J., Yang E.B., Lee K., and Huh W.-K.. 2013. Genome-wide bimolecular fluorescence complementation analysis of SUMO interactome in yeast. Genome Res. 23:736–746. 10.1101/gr.148346.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahara, T., and Maeda T.. 2012. Transient sequestration of TORC1 into stress granules during heat stress. Mol. Cell. 47:242–252. 10.1016/j.molcel.2012.05.019 [DOI] [PubMed] [Google Scholar]
- Tanigawa, M., and Maeda T.. 2017. An In Vitro TORC1 Kinase Assay That Recapitulates the Gtr-Independent Glutamine-Responsive TORC1 Activation Mechanism on Yeast Vacuoles. Mol. Cell. Biol. 37:e00075-17. 10.1128/MCB.00075-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tcherkezian, J., Cargnello M., Romeo Y., Huttlin E.L., Lavoie G., Gygi S.P., and Roux P.P.. 2014. Proteomic analysis of cap-dependent translation identifies LARP1 as a key regulator of 5'TOP mRNA translation. Genes Dev. 28:357–371. 10.1101/gad.231407.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira, D., Sheth U., Valencia-Sanchez M.A., Brengues M., and Parker R.. 2005. Processing bodies require RNA for assembly and contain nontranslating mRNAs. RNA. 11:371–382. 10.1261/rna.7258505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thedieck, K., Holzwarth B., Prentzell M.T., Boehlke C., Kläsener K., Ruf S., Sonntag A.G., Maerz L., Grellscheid S.-N., Kremmer E., et al. 2013. Inhibition of mTORC1 by astrin and stress granules prevents apoptosis in cancer cells. Cell. 154:859–874. 10.1016/j.cell.2013.07.031 [DOI] [PubMed] [Google Scholar]
- Tsvetanova, N.G., Klass D.M., Salzman J., and Brown P.O.. 2010. Proteome-wide search reveals unexpected RNA-binding proteins in Saccharomyces cerevisiae. PLoS One. 5:e12671. 10.1371/journal.pone.0012671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban, J., Soulard A., Huber A., Lippman S., Mukhopadhyay D., Deloche O., Wanke V., Anrather D., Ammerer G., Riezman H., et al. 2007. Sch9 is a major target of TORC1 in Saccharomyces cerevisiae. Mol. Cell. 26:663–674. 10.1016/j.molcel.2007.04.020 [DOI] [PubMed] [Google Scholar]
- Wallace, E.W., Kear-Scott J.L., Pilipenko E.V., Schwartz M.H., Laskowski P.R., Rojek A.E., Katanski C.D., Riback J.A., Dion M.F., Franks A.M., et al. 2015. Reversible, Specific, Active Aggregates of Endogenous Proteins Assemble upon Heat Stress. Cell. 162:1286–1298. 10.1016/j.cell.2015.08.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wedaman, K.P., Reinke A., Anderson S., Yates J. III, McCaffery J.M., and Powers T.. 2003. Tor kinases are in distinct membrane-associated protein complexes in Saccharomyces cerevisiae. Mol. Biol. Cell. 14:1204–1220. 10.1091/mbc.e02-09-0609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wippich, F., Bodenmiller B., Trajkovska M.G., Wanka S., Aebersold R., and Pelkmans L.. 2013. Dual specificity kinase DYRK3 couples stress granule condensation/dissolution to mTORC1 signaling. Cell. 152:791–805. 10.1016/j.cell.2013.01.033 [DOI] [PubMed] [Google Scholar]
- Wullschleger, S., Loewith R., and Hall M.N.. 2006. TOR signaling in growth and metabolism. Cell. 124:471–484. 10.1016/j.cell.2006.01.016 [DOI] [PubMed] [Google Scholar]
- Zeidan, Q., He F., Zhang F., Zhang H., Jacobson A., and Hinnebusch A.G.. 2018. Conserved mRNA-granule component Scd6 targets Dhh1 to repress translation initiation and activates Dcp2-mediated mRNA decay in vivo. PLoS Genet. 14:e1007806. 10.1371/journal.pgen.1007806 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
lists 130 BiFC-positive interactors of Kog1.
lists 35 BiFC-positive interactors of Tor1.
lists 36 BiFC-positive interactors of Tor2.
lists 28 coIP-positive interactors of Kog1.
lists known physical interactors of Kog1.
shows GO analysis of 130 BiFC-positive Kog1 interactors.
lists 120 TORC1-associated mRNAs. Table S7 A shows fold enrichment of genes in Kog1 immunoprecipitates. Reads per kilobase million values were determined by a RIP-seq experiment. TOR1, KOG1, and genes with reads per kilobase million <1 were eliminated. Table S7 B shows differentially expressed genes in HA-KOG1 cells that were used in RIP-seq experiments. Fold increase values were determined by duplicated RNA sequencing.
lists yeast strains, plasmids, and oligonucleotide primers used in this study.