The molecular mechanisms that control the early events of adipogenesis are not yet fully elucidated. Audano et al. show that Zc3h10 is a proadipogenic factor essential to achieve protein synthesis repression and for proper remodeling of filamentous actin to provide a functional population of mitochondria to support the adipogenic program.
Abstract
The commitment of mesenchymal stem cells to preadipocytes is stimulated by hormonal induction. Preadipocytes induced to differentiate repress protein synthesis, remodel their cytoskeleton, and increase mitochondrial function to support anabolic pathways. These changes enable differentiation into mature adipocytes. Our understanding of the factors that coordinately regulate the early events of adipocyte differentiation remains incomplete. Here, by using multipronged approaches, we have identified zinc finger CCCH-type containing 10 (Zc3h10) as a critical regulator of the early stages of adipogenesis. Zc3h10 depletion in preadipocytes resulted in increased protein translation and impaired filamentous (F)-actin remodeling, with the latter detrimental effect leading to mitochondrial and metabolic dysfunction. These defects negatively affected differentiation to mature adipocytes. In contrast, Zc3h10 overexpression yielded mature adipocytes with remarkably increased lipid droplet size. Overall, our study establishes Zc3h10 as a fundamental proadipogenic transcription factor that represses protein synthesis and promotes F-actin/mitochondria dynamics to ensure proper energy metabolism and favor lipid accumulation.
Introduction
Adipocytes originate from pluripotent mesenchymal stem cells (MSCs). In mammals, MSCs localize to the vascular stroma of adipose tissue and to bone marrow. MSCs become restricted to the adipocyte lineage by means of specific chemical/physical cues that promote their commitment to preadipocytes that then give rise to lipid-loaded mature adipocytes (Tang and Lane, 2012).
Besides the upregulation of adipogenic transcription factors (Lefterova et al., 2014; Siersbæk et al., 2017; Ghaben and Scherer, 2019), the inhibition of translation is associated with the generation of mature adipocytes. Translation activity is reduced to arrest cell growth and proliferation. In adipose-derived MSCs, translation regulation occurs mainly at the initiation step and involves the modulation of the eukaryotic translation initiation factor (eIF) 4F complex (Marcon et al., 2017). This complex is composed of three nonidentical subunits—the ATP-dependent RNA helicase eIF4A, the cap-binding protein eIF4E, and the scaffold protein eIF4G—that bind to the 5′ cap of mRNAs to promote eukaryotic translation initiation (Aitken and Lorsch, 2012; Merrick, 2015). Activation of this complex occurs mainly through the phosphorylation of repressor eIF4E-binding proteins (4E-BPs; Jackson et al., 2010). Indeed, hypophosphorylated 4E-BP1 avidly binds to eIF4E, resulting in cap-dependent mRNA translation inhibition (Gingras et al., 1999); however, hyperphosphorylated 4E-BP1 releases eIF4E, leading to increased translation activity. The amount of hypophosphorylated 4E-BP1 is increased during the first days of adipogenic differentiation, resulting in the downregulation of protein synthesis (Marcon et al., 2017).
Cytoskeletal and mitochondrial remodeling are also fundamental prerequisite steps for the morphologic transitions from MSCs to preadipocytes to mature adipocytes (Spiegelman and Farmer, 1982). During the course of the adipogenic program, MSCs are transformed into mature adipocytes that are occupied by large lipid droplets (Feng et al., 2010), a change that involves the disruption of filamentous (F)-actin, a key cytoskeletal component, via the downregulation of Ras homologue family member A (RhoA)/Rho-associated protein kinase (ROCK) signaling (Hansson et al., 2019). In addition to these actin-dependent morphologic changes, mitochondria in preadipocytes also form interconnected networks that are dynamically remodeled to sustain anabolic pathways to produce lipid-filled mature adipocytes (Wilson-Fritch et al., 2003; Spinelli and Haigis, 2018; De Pauw et al., 2009). Despite knowing that translation, cytoskeletal remodeling, and mitochondrial dynamics/function are critical for the proper development of MSCs into adipocytes, the molecular mechanisms underlying the coordinated regulation of these processes and the metabolic activation that occurs during the white adipogenic program have yet to be fully elucidated.
Previously, we showed that zinc finger CCCH-type containing 10 (Zc3h10) is a mitochondrial regulator required during myogenesis (Audano et al., 2018); however, other molecular mechanisms and aspects of general cell biology that are regulated by Zc3h10 during the white adipogenic program remain unknown.
In this work, by using gain- and loss-of-function approaches integrated with transcriptomics, metabolomics, and functional validation analyses, we have identified Zc3h10 as an early regulator of the white adipogenic program. We have also elucidated a transcriptional program governed by Zc3h10 that represses protein translation and promotes cytoskeletal remodeling that determines mitochondrial function, metabolic activation, and, ultimately, lipid loading.
Results
Zc3h10 regulates the white adipogenic program
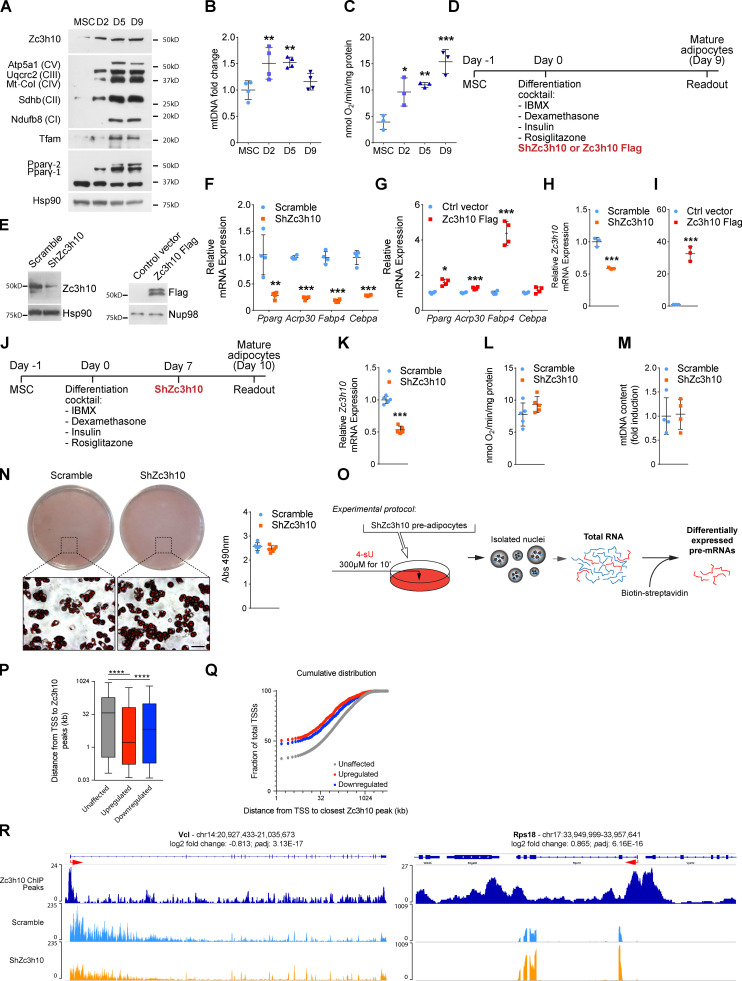
To assess whether Zc3h10 affects the white adipogenic program, we first investigated its expression during CH310T1/2 MSC differentiation into white adipocytes. Zc3h10 protein levels were higher during the early phase of hormonal induction and preceded protein levels of the bona fide hormone-stimulated adipogenic factor, peroxisome proliferator–activated receptor γ2 (Tontonoz et al., 1994), and mitochondrial transcription factor A (Fig. S1 A). Notably, the increase in Zc3h10 protein levels also paralleled the increased expression of different subunits belonging to the oxidative phosphorylation (Fig. S1 A), as well as increased mitochondrial DNA content and basal oxygen consumption rates (Fig. S1, B and C).
Figure S1.
Zc3h10 expression increases in the early phases of the adipogenic program and promotes adipogenesis. (A) Western blot of Zc3h10, oxidative phosphorylation subunits (Atp5a1, Uqcrc2, Mt-CoI, Sdhb, Ndufb8), Tfam, and peroxisome proliferator-activated receptor γ (Pparγ) at different time points during adipogenesis. (B and C) Mitochondrial DNA (B; n = 4) and oxygen consumption levels (C; n = 3) were measured in proliferating MSCs and at 2, 5, and 9 d after differentiation induction; statistical analysis was performed by using one-way ANOVA followed by Dunnett’s post-hoc test; *P < 0.05, **P < 0.01, ***P < 0.001 vs. MSCs. (D) Representation of the experimental protocol followed for the infection and differentiation of C3H10T1/2 and primary MSCs. (E) Validation of Zc3h10 expression levels by Western blot in Scramble, ShZc3h10 (left), control vector, and Zc3h10-Flag (right) C3H10T1/2 preadipocytes (36 h). (F and G) Gene expression analysis of the adipogenic markers Pparg, Adipoq, Fabp4, and Cebpa, in Scramble and ShZc3h10 (F) and control (Ctrl; G) vector and Zc3h10 Flag mature adipocytes (day 9). n = 4; statistical analysis was performed using unpaired parametric Student’s t tests; *P < 0.05, **P < 0.01, ***P < 0.001 vs. Scramble or Ctrl vector, respectively. (H and I) Relative mRNA expression of Zc3h10 in primary stromal vascular cells (SVCs) exposed to Scramble or ShRNA against Zc3h10 (H) and control vector or Zc3h10-Flag (I), and then differentiated to adipocytes for 8 d. n = 3; statistical analysis was performed using unpaired parametric Student’s t tests; ***P < 0.001 vs. Scramble or control vector, respectively. (J) Representation of the experimental protocol followed for the differentiation and infection of C3H10T1/2 mature adipocytes. (K–N) Relative Zc3h10 mRNA levels (K), basal cell respiration (L), relative mitochondrial DNA (mtDNA) content (M), and lipid accumulation (Oil-Red-O staining; N) in Scramble and ShZc3h10 mature adipocytes generated following the protocol in J. (O) Schematic representation of the experimental protocol for pre-mRNA labeling, isolation, and analysis in Scramble and ShZc3h10 preadipocytes. Before nuclei isolation, cells were exposed to 300 µM 4-sU for 10 min. Total RNA was isolated and quantified, and then 50 µg of total RNA were used to isolate 4-sU–labeled pre-mRNAs. (P) Box plot representing the distances (kb) from TSSs to Zc3h10 binding sites on chromatin in unaffected, upregulated (red), and downregulated (blue) pre-mRNAs of ShZc3h10 preadipocytes (36 h) compared with Scramble control. Statistical analysis was performed using Mann-Whitney (nonparametric) two-tailed tests; ****P < 0.0001. (Q) Cumulative distribution of Zc3h10 binding sites on chromatin in unaffected, upregulated (red), and downregulated (blue) pre-mRNAs of ShZc3h10 compared with Scramble control preadipocytes (36 h after differentiation). (R) Zc3h10 chromatin immunoprecipitation sequencing peaks (dark blue track) and signals from 4-sU-seq analysis for Vcl and Rps18 as examples of downregulated and upregulated genes, respectively, in Zc3h10-silenced preadipocytes 36 h after differentiation induction. The light blue track indicates the pre-mRNA reads for Scramble and the orange track shows the pre-mRNA reads for ShZc3h10 preadipocytes. Log2 fold change and adjusted P value (padj) are shown on top of the graph. IBMX, 3-isobutyl-1-methylxanthine.
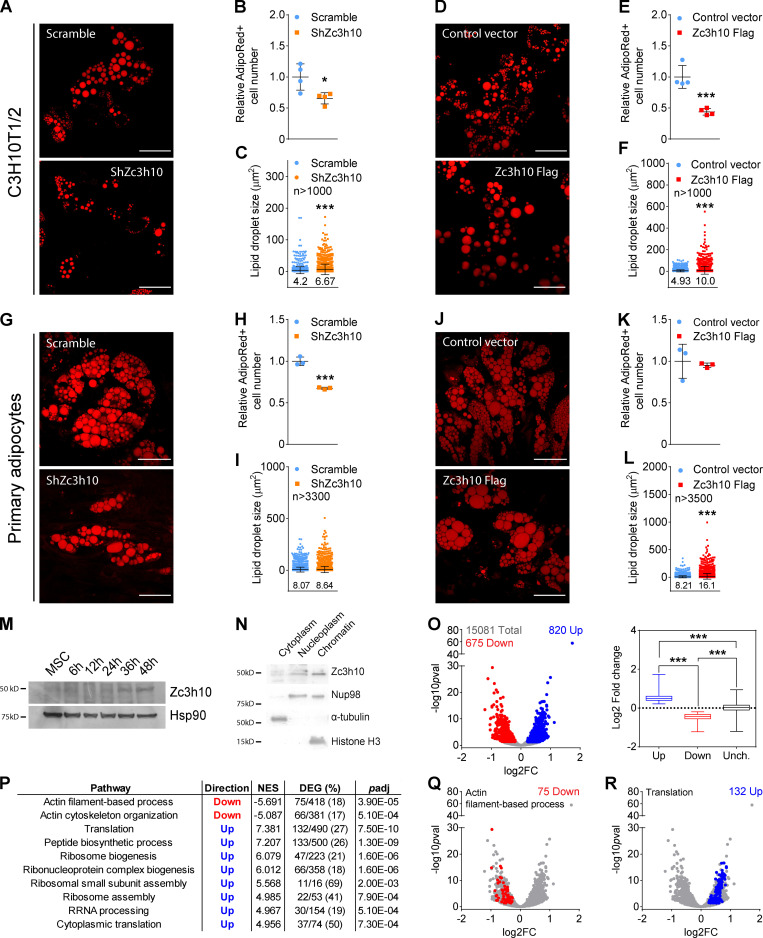
Next, we performed loss- and gain-of-function studies to elucidate the role of Zc3h10 in this process. We infected MSCs at the induction of differentiation by using an adenovirus carrying an shRNA against Zc3h10 (Fig. S1 D). Following knockdown (Fig. S1 E, left), adipocyte numbers were shown to be significantly reduced (Fig. 1, A and B), while lipid droplet sizes were slightly increased in terminally differentiated adipocytes (Fig. 1 C). These effects were accompanied by the decreased expression of classical markers of adipogenesis (Fig. S1 F). In our gain-of-function studies, overexpression of Zc3h10 (Fig. S1 E, right) resulted in fewer lipid-containing cells on day 9 of adipogenesis (Fig. 1, D and E), characterized by dramatically increased lipid droplet size (Fig. 1 F) and the upregulated expression of adipogenic genes (Fig. S1 G). Most of these findings were recapitulated in primary mouse stromal vascular cells differentiated to mature adipocytes (Fig. 1, G–L; and Fig. S1, H and I), suggesting that Zc3h10 plays an important role in the early stages of adipocyte differentiation.
Figure 1.
Zc3h10 controls white adipogenesis and pre-mRNA transcription in preadipocytes. (A and D) Lipid droplets were labeled by using AdipoRed dye to evaluate lipid accumulation in mature adipocytes (day 9). Scale bar: 50 µm. (B and E) Relative quantification of AdipoRed-positive (AdipoRed+) cells in mature adipocytes (day 9). n = 4; unpaired parametric Student’s t test; *P < 0.05 vs. Scramble; ***P < 0.001 vs. Control. (C and F) Lipid droplet size adipocytes differentiated for 9 d. n > 1,000; unpaired and nonparametric Mann-Whitney test; ***P < 0.001 vs. Scramble/Control. (G and J) Primary stromal vascular cell (SVC) differentiation was evaluated in Scramble and ShZc3h10 mature adipocytes (day 8) by using AdipoRed dye. Scale bar: 50 µm. (H and K) AdipoRed+ cell counts in Scramble and ShZc3h10 primary adipocytes (day 8). n = 3; unpaired parametric Student’s t test; ***P < 0.001 vs. Scramble. (I and L) Lipid droplet size in primary mature adipocytes (day 8). n > 3,300 ShZc3h10; n > 3,500 Zc3h10 Flag; unpaired nonparametric Mann-Whitney test; ***P < 0.001 vs. Control. (M) Western blot of Zc3h10 in whole-cell lysates during the first 48 h of differentiation. (N) Western blot of Zc3h10, Nup98 (nuclear marker), α-tubulin (cytoplasmic marker), and histone H3 (chromatin marker) in the indicated cell fractions 36 h after differentiation induction. (O) Volcano and box plots of downregulated (red), upregulated (blue), and unchanged (gray) pre-mRNAs in ShZc3h10 preadipocytes at 36 h. Scramble: n = 2; ShZc3h10: n = 3; volcano plot: false discovery rate (FDR) < 0.05; box plot: Kruskal-Wallis test of rank with Dunn’s post-hoc test; ***P < 0.001 vs. Scramble. (P) GAUGE analysis results of significantly down- and upregulated pre-mRNA clusters in ShZc3h10 36-h-differentiated preadipocytes. Table indicates pathway name, trend, normalized enrichment score (NES), number of differentially expressed genes (DEGs) along with their percentage of the total genes in the cluster, and adjusted P value (padj). (Q) Volcano plot of the actin filament–based process cluster genes. Red dots represent DEGs that were significantly downregulated (FDR < 0.05) in ShZc3h10 cells 36 h after differentiation induction. (R) Volcano plot of the translation gene cluster. Genes that were significantly upregulated (FDR < 0.05) in ShZc3h10 preadipocytes 36 h after differentiation induction are shown in blue.
We then silenced Zc3h10 at day 7 of differentiation (Fig. S1 J) and found that these cells showed unchanged basal respiration, mitochondrial DNA content, and lipid accumulation (Fig. S1, K–N). These results indicate that the effects of the loss of Zc3h10 on lipid accumulation and mitochondrial function depend on the stage of differentiation. More specifically, Zc3h10 is more important for regulating processes that occur at the beginning of the adipocyte differentiation program.
Zc3h10 controls nascent mRNA levels of genes that regulate actin filament–based processes and translation pathways in preadipocytes
We next sought to unveil the molecular mechanism by which Zc3h10 controls adipogenesis. A more detailed analysis of Zc3h10 expression showed that protein levels were higher 36 h after exposure to adipogenic cocktail (preadipocytes, Fig. 1 M). Moreover, Zc3h10 was enriched in the chromatin fraction, which is consistent with its transcription factor activity (Yi et al., 2019; Fig. 1 N). Based on these results, to capture changes in actively transcribed mRNAs, we investigated the effects of Zc3h10 depletion on nascent/premRNA synthesis in preadipocytes (Fig. S1 O; Rabani et al., 2011). 4-Thiouridine (4-sU) labeling of nascent RNAs, coupled with next-generation sequencing (4-sU-seq), revealed significant transcriptomic differences in Zc3h10-silenced preadipocytes, with 820 upregulated and 675 downregulated pre-mRNAs (adjusted P value ≤ 0.05; Fig. 1 O) observed in the absence of Zc3h10. Subsequent generically applicable gene-set enrichment (GAUGE) pathway analysis showed that Zc3h10 depletion resulted in the downregulation of pre-mRNAs related to actin filament–based cytoskeleton organization and the upregulation of pre-mRNAs related to translation and ribosome biogenesis (Fig. 1, P–R).
We next queried published Zc3h10 chromatin immunoprecipitation coupled to sequencing data in brown adipocytes (Yi et al., 2019) and found that most of the statistically downregulated or upregulated genes from our 4-sU-seq were actually bound by Zc3h10 within 1 kb of the gene transcription start site (TSS; Fig. S1, P and Q; and Table S1). For instance, pre-mRNAs for vinculin (Vcl), a member of the actin filament–based process cluster, were downregulated, while pre-mRNAs for ribosomal protein S18 (Rps18), a gene belonging to the translation pathway, were upregulated in the absence of Zc3h10. Of note, Zc3h10 binds around the corresponding TSS of each of these genes (Fig. S1 R). Together, these data indicate that Zc3h10 promotes the expression of actin filament–based process genes and blunts the expression of those genes involved in peptide biosynthetic processes.
Zc3h10 governs F-actin morphology to determine mitochondrial dynamics
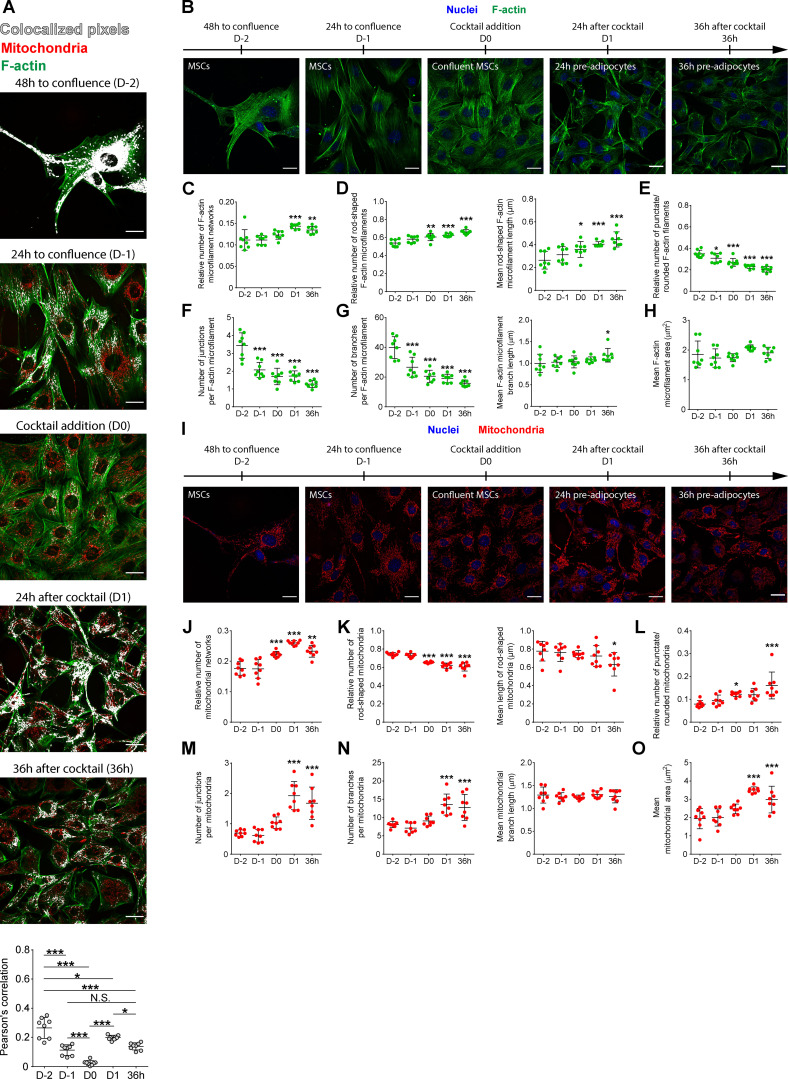
The main pathways negatively affected by Zc3h10 depletion in preadipocytes were actin filament–based processes and cytoskeleton organization (Fig. 1, P and Q). F-actin enables cell migration, force generation, and cell shape remodeling (Halder et al., 2012), and defines the functional mitochondrial network; however, the latter data are so far restricted to HeLa and COS-7 cells and human epidermal keratinocytes (Moore et al., 2016). We first sought to examine the extent to which Zc3h10 may be involved in these processes by characterizing F-actin and mitochondrial dynamics during the differentiation of unmodified MSCs after hormonal induction. As shown in Fig. S2, F-actin/mitochondria contacts in MSCs declined over time from day −2 to day 0 (Fig. S2 A). This drop was due to cell confluence (Fig. S2 B), leading to F-actin stress fiber breakdown (decreased numbers of punctate/rounded filaments, junctions, and branches; increased number and length of rod-shaped filaments; Fig. S2, C–G), as previously reported (Nobusue et al., 2014). These effects occurred without affecting the F-actin area (Fig. S2 H). During the same timeframe, we observed increased mitochondrial fission, as evidenced by increased numbers of mitochondrial networks and punctuate/rounded mitochondria, along with decreased numbers of rod-shaped mitochondria (Fig. S2, I–L). After hormonal stimulation of the adipogenic program (day 0 to day 1), F-actin/mitochondria contacts and stress fiber breakdown continued to increase (Fig. S2, A–G). In addition, F-actin reorganization toward the cell periphery started at day 1 and became more evident at 36 h (Fig. S2 B), while, at the same time, the contacts detected between F-actin and mitochondria decreased (Fig. S2 A). This phenotype was associated with increases in mitochondrial junctions, branches, and area (Fig. S2, M–O). Together, our findings suggest that there are two distinct phases of F-actin/mitochondria interaction during the transition from MSCs to preadipocytes. In the first phase (before day 0), the reduction of contacts between F-actin and mitochondria promotes mitochondrial fission; however, in the second phase (from day 0), there is a priming and transient interaction that promotes mitochondrial biogenesis and fusion.
Figure S2.
Morphology of F-actin, mitochondrial networks, and their colocalization during the early stages of adipogenesis. (A) Representative images and quantitative analysis of mitochondria/F-actin colocalization in the early phases of adipogenesis (from proliferating MSCs to 36-h-differentiated preadipocytes). Red: mitochondria; green: F-actin; white: colocalized pixels. n = 8; statistical analysis was performed by using one-way ANOVA followed by Tukey’s post-hoc test; *P < 0.05, ***P < 0.001. Scale bar: 25 µm. (B–H) Representative images (B) and quantitative analysis of F-actin networks (C–H) during the early phases of C3H10T1/2 differentiation to adipocytes (from proliferating MSCs to 36-h-differentiated preadipocytes). Green: F-actin; blue: nuclei. n = 8; statistical analysis was performed by using one-way ANOVA followed by Dunnett’s post-hoc test; *P < 0.05, **P < 0.01, ***P < 0.01 vs. D-2. Scale bar: 25 µm. (I–O) Representative images (I) and quantitative analysis of mitochondrial networks (J–O) during the early phases of adipogenesis (from proliferating MSCs to 36-h-differentiated preadipocytes). Red: mitochondria; blue: nuclei. n = 8; statistical analysis was performed by using one-way ANOVA followed by Dunnett’s post-hoc test; *P < 0.05, **P < 0.01, ***P < 0.01 vs. D-2. Scale bar: 25 µm.
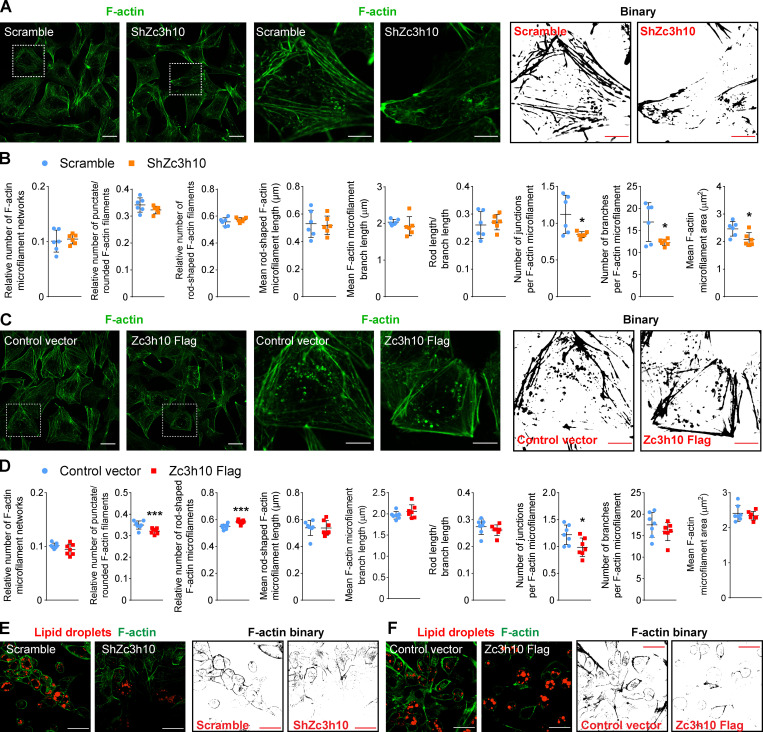
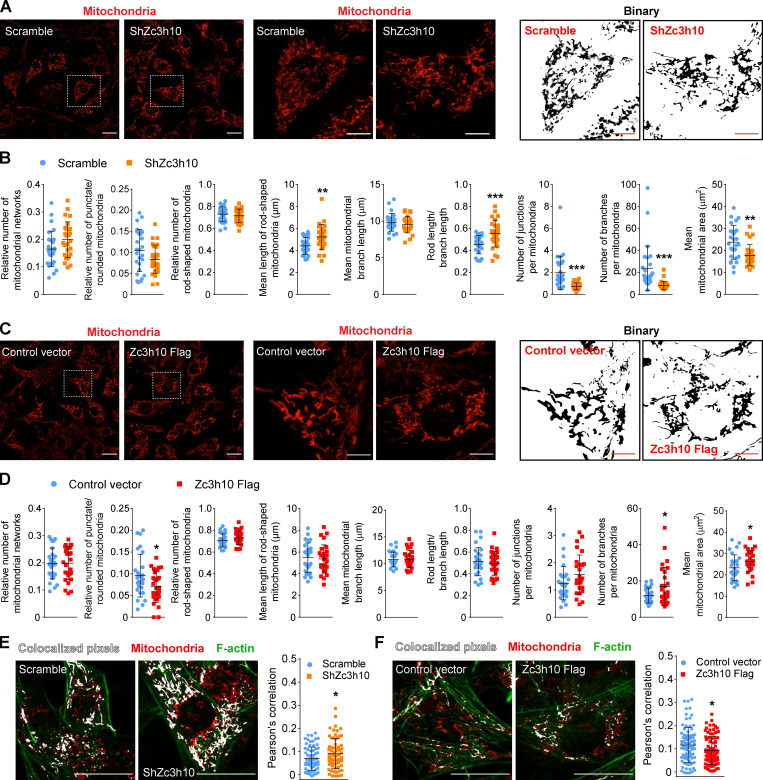
In this context, Zc3h10 depletion led to decreased F-actin junctions, branches, and area, suggesting accelerated F-actin breakdown (Fig. 2, A and B). Instead, Zc3h10 overexpression enhanced the priming of F-actin reorganization toward the cell periphery (Fig. 2 C), as evidenced by decreased punctate/rounded filaments, increased rod-shaped filaments, and unchanged filament length (Fig. 2 D). These latter results were also accompanied by decreased F-actin junctions, while branches and filament area were unchanged (Fig. 2 D). Strikingly, at later stages of the adipogenic program (day 5 after the induction of differentiation), the decreased F-actin organization due to Zc3h10 depletion was associated with reduced lipid accumulation, while Zc3h10-overexpressing preadipocytes showed peculiar cortical F-actin rings with larger lipid droplets (Fig. 2, E and F). Collectively, these results show that Zc3h10 controls the F-actin network at the transcriptional level to ensure proper cytoskeletal remodeling in the early phases of adipogenesis.
Figure 2.
The Zc3h10 transcriptomic signature is associated with F-actin dynamics in preadipocytes. (A and C) Left: Representative confocal images of F-actin detected by using the SiR-actin probe in preadipocytes (36 h). Middle and right: Enlarged images (middle) and relative binary images (right) are also shown. Scale bar: 50 µm (left), 10 µm (middle and right). (B and D) Quantitative analysis of the F-actin networks preadipocytes (36 h). n = 6 ShZc3h10; n = 7 Zc3h10-Flag; unpaired parametric Student’s t tests; *P < 0.05 vs. Scramble (B); *P < 0.05 (D); ***P < 0.001 vs. Control. (E and F) Representative confocal images of F-actin (green) and lipid droplets (red), as well as F-actin binary images of preadipocytes on day 5 after differentiation. Scale bar: 50 µm.
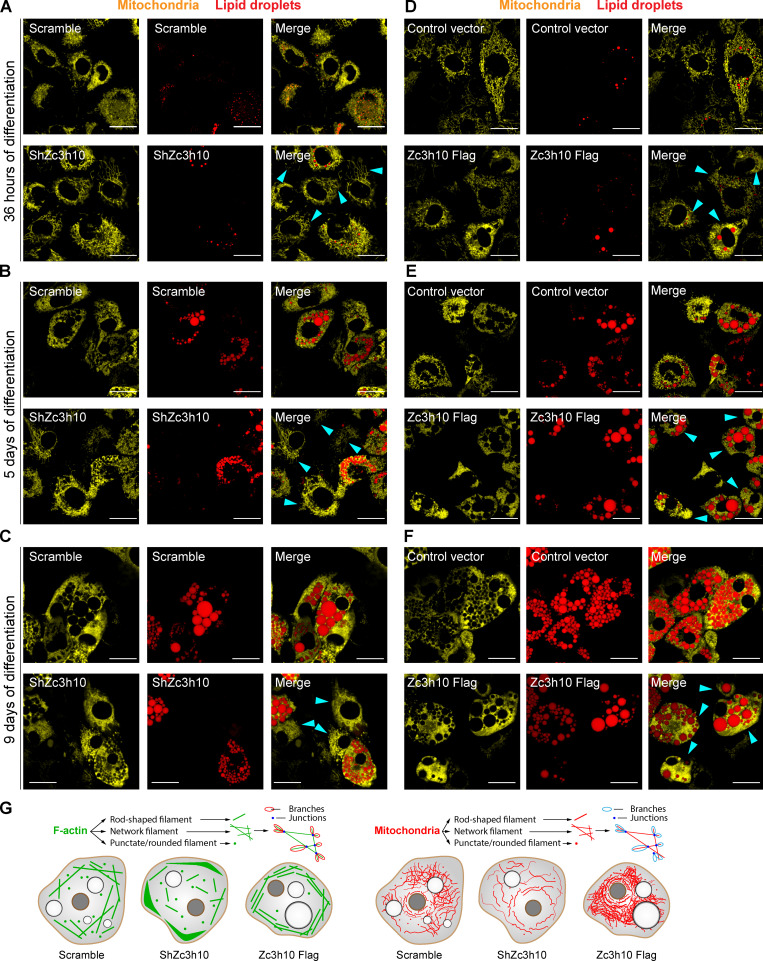
Zc3h10-depleted cells also showed a different mitochondrial pattern (Fig. 3 A). Quantitative morphologic analysis of preadipocytes at 36 h postdifferentiation initiation revealed that Zc3h10 silencing increased the length of rod-shaped mitochondria and the ratio of rod length to branch length (Fig. 3 B). Of note, the numbers of junctions and branches, as well as the mitochondrial area, were also significantly reduced in cells lacking Zc3h10 (Fig. 3 B). The priming activity of Zc3h10 on mitochondrial structure was also confirmed with a gain-of-function approach. Zc3h10-overexpressing preadipocytes exhibited different mitochondrial morphologies after 36 h of differentiation relative to control vector–infected cells (Fig. 3 C) which consisted of a significant decrease in the number of punctate/rounded mitochondria, an increased number of branches, and greater mitochondrial area (Fig. 3 D). Strikingly, our results showed that Zc3h10 depletion in preadipocytes led to significant increases in F-actin/mitochondria interaction (Fig. 3 E). In contrast, overexpression of Zc3h10 reduced these interactions (Fig. 3 F). Furthermore, knockdown of Zc3h10 consistently altered mitochondrial morphology during the adipogenic program, which was linked to a progressive reduction in lipid accumulation (Fig. S3, A–C). In contrast, Zc3h10 overexpression led to increased mitochondrial fusion and lipid accumulation (Fig. S3, D–F). Overall, these results indicate that Zc3h10 favors the breakdown of F-actin stress fibers and peripheral reorganization, ultimately increasing the remodeling of the mitochondrial network and its complexity (Fig. S3 G).
Figure 3.
Zc3h10-mediated F-actin remodeling determines mitochondrial network complexity. (A and C) Left: Representative confocal images of mitochondria detected by the Mitotracker Red CM-H2XROS probe preadipocytes (36 h). Middle and right: Enlarged images (middle) and relative binary (right) are also shown. Scale bar: 50 µm (left), 10 µm (middle and right). (B and D) Quantitative analysis of mitochondrial networks in preadipocytes (36 h). n = 27 cells in ShZc3h10; n = 28 cells in Zc3h10-Flag; unpaired parametric Student’s t tests; **P < 0.01, ***P < 0.001 vs. Scramble, *P < 0.05 vs. Control vector. (E and F) Colocalization analysis of mitochondria (red) and F-actin (green) in 36-h-differentiated cells. n = 68 cells in ShZc3h10; n = 85 cells in Zc3h10-Flag; unpaired parametric Student’s t test; *P < 0.05 vs. Scramble/Control. Scale bar: 25 µm.
Figure S3.
Mitochondrial networks are regulated by Zc3h10 during adipogenesis and are associated with lipid accumulation. (A–C) Representative images of lipid droplets (red) and mitochondria (yellow) in Scramble and ShZc3h10 cells differentiated for 36 h (A), 5 d (B), and 9 d (C). Cyan arrows indicate major alterations of the mitochondrial network. Scale bar: 25 µm. (D–F) Representative images of lipid droplets (red) and mitochondria (yellow) in control vector and Zc3h10-Flag overexpressing cells differentiated for 36 h (D), 5 d (E), and 9 d (F). Cyan arrows indicate major differences in the mitochondrial network. Scale bar: 25 µm. (G) Schematic of the F-actin and mitochondrial analysis with a pictorial summary of the main findings.
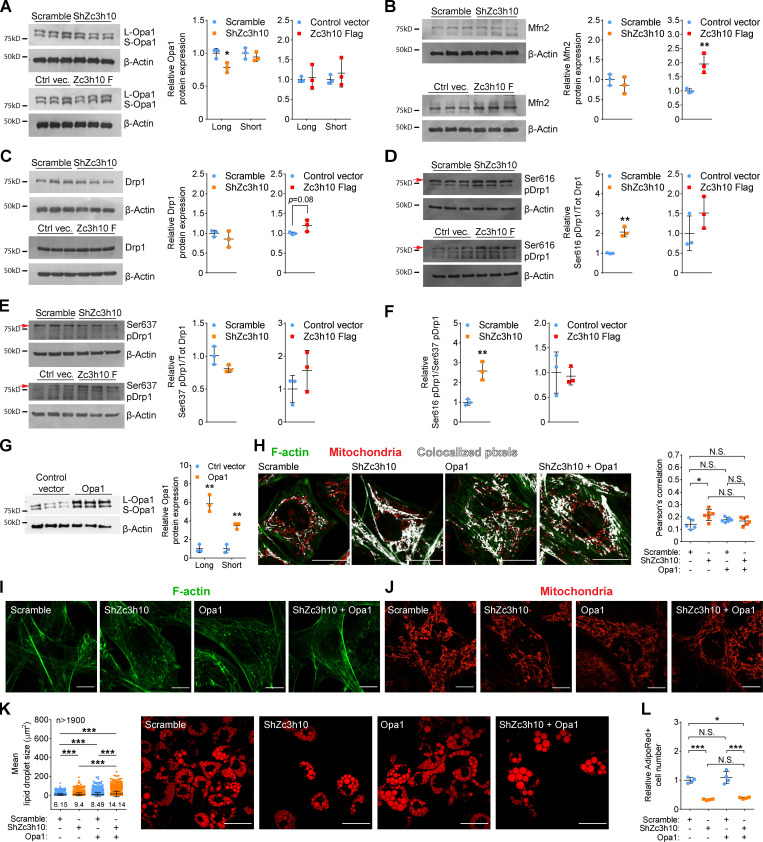
To further assess the impact of Zc3h10 on preadipocyte mitochondrial morphology, we measured the expression levels of proteins involved in mitochondrial dynamics. We found that long-optic atrophy 1 (Opa1, associated to mitochondrial fusion; Pernas and Scorrano, 2016; Griparic et al., 2007; Tondera et al., 2009; Ehses et al., 2009; Head et al., 2009) levels were decreased with Zc3h10 knockdown, but were unaffected by Zc3h10 overexpression (Fig. 4 A). Instead, mitofusin 2 (Chen et al., 2003; Rambold et al., 2011; Cipolat et al., 2004; Giacomello et al., 2020) protein levels were increased with Zc3h10 overexpression, but were unaffected with Zc3h10 knockdown (Fig. 4 B). We also examined the expression and activation levels of the mitochondrial fission protein dynamin-related protein 1 (Drp1). Drp1 translocation to mitochondria is regulated by post-translational modifications (Alaimo et al., 2014; Smirnova et al., 2001; Chen et al., 2003; Cipolat et al., 2004); phosphorylation on Ser616 (pSer616) by cyclin-dependent kinase 1/cyclin B or cyclin-dependent kinase 5 favors mitochondrial fission during mitosis (Taguchi et al., 2007; Liesa et al., 2009; Giacomello et al., 2020). Conversely, Drp1 pSer637 by protein kinase A (PKA) promotes the release of Drp1 from mitochondria, resulting in the inhibition of mitochondrial fission (Kashatus et al., 2011; Wang et al., 2012; Giacomello et al., 2020). Hence, the Drp1 pSer616/pSer637 ratio represents an index of the capacity of Drp1-mediated mitochondrial dynamics. While we did not observe significant increases in Drp1 expression in either preadipocyte model, we did see increased Drp1 pSer616 in Zc3h10-depleted cells (Fig. 4, C and D). Notably, the pSer616/pSer637 Drp1 ratio was also significantly increased in Zc3h10-silenced cells, suggesting increased mitochondrial fission (Fig. 4, D–F). Taken together, these data confirm that Zc3h10 favors mitochondrial network mass and complexity by enhancing mitochondrial fusion and counteracting fission.
Figure 4.
Zc3h10 impacts mitochondrial dynamics and function. (A–E) Representative Western blots of Opa1 (A), mitofusin 2 (Mfn2; B), total Drp1 (C), Drp1 pSer616 (D), and pSer637 (E) with the relative quantifications in preadipocytes (36 h). n = 3; unpaired parametric Student’s t tests; *P < 0.05 vs. Scramble (A); **P < 0.01 vs. Control (B); **P < 0.01 vs. Scramble (D). The β-actin used for the phosphorylation status of Drp1 (D and E) is identical to that used to assess the phosphorylation status of 4E-BP1 reported in Fig. 8, D and E since the samples were evaluated on the same membranes. (F). pSer616/pSer637 Drp1 ratio in preadipocytes (36 h). n = 3; unpaired parametric Student’s t tests; **P < 0.01 vs. Scramble. (G) Overexpression levels of Opa1 in preadipocytes (36 h) with relative quantifications of long and short isoforms. n = 3; unpaired parametric Student’s t tests; **P < 0.01 vs. Control vector (vec). (H) Representative confocal images of preadipocytes (36 h) for colocalization analysis of mitochondria and F-actin. n = 5 for Scramble and n = 6 for the other conditions; one-way ANOVA followed by Tukey’s post-hoc test; *P < 0.05. Scale bar: 25 µm. (I and J) Representative confocal images of F-actin (I) and mitochondria (J) in preadipocytes (36 h) expressing the indicated molecules. Scale bar: 25 µm. (K) Lipid droplet size in mature adipocytes (9 d) overexpressing Opa1; n > 1,900 lipid droplets; statistical analysis was performed by one-way ANOVA followed by Tukey’s post-hoc test; ***P < 0.001. Scale bar: 25 µm. (L) Relative quantification of AdipoRed+ cells in mature adipocytes (9 d). n = 4; one-way ANOVA followed by Tukey’s post-hoc test; *P < 0.05; ***P < 0.001.
Next, we performed genetic experiments to evaluate whether promoting mitochondrial fusion is sufficient to overcome defects in F-actin remodeling and to promote lipid droplet formation in Zc3h10-silenced cells. Overexpression of the cristae-shaping Opa1 in Zc3h10-depleted preadipocytes normalized the F-actin/mitochondria interactions by promoting mitochondrial fusion that was associated with F-actin reorganization to the cell periphery (Fig. 4, G–J). Opa1 overexpression also increased lipid droplet size; however, the number of AdipoRed-positive cells was not rescued (Fig. 4, K and L). Overall, these findings indicate that Zc3h10 controls the early phases of adipogenesis by modulating F-actin/mitochondria interactions, enabling the proper mitochondrial fusion and biogenesis to support lipid accumulation.
Zc3h10 controls mitochondrial function and metabolism
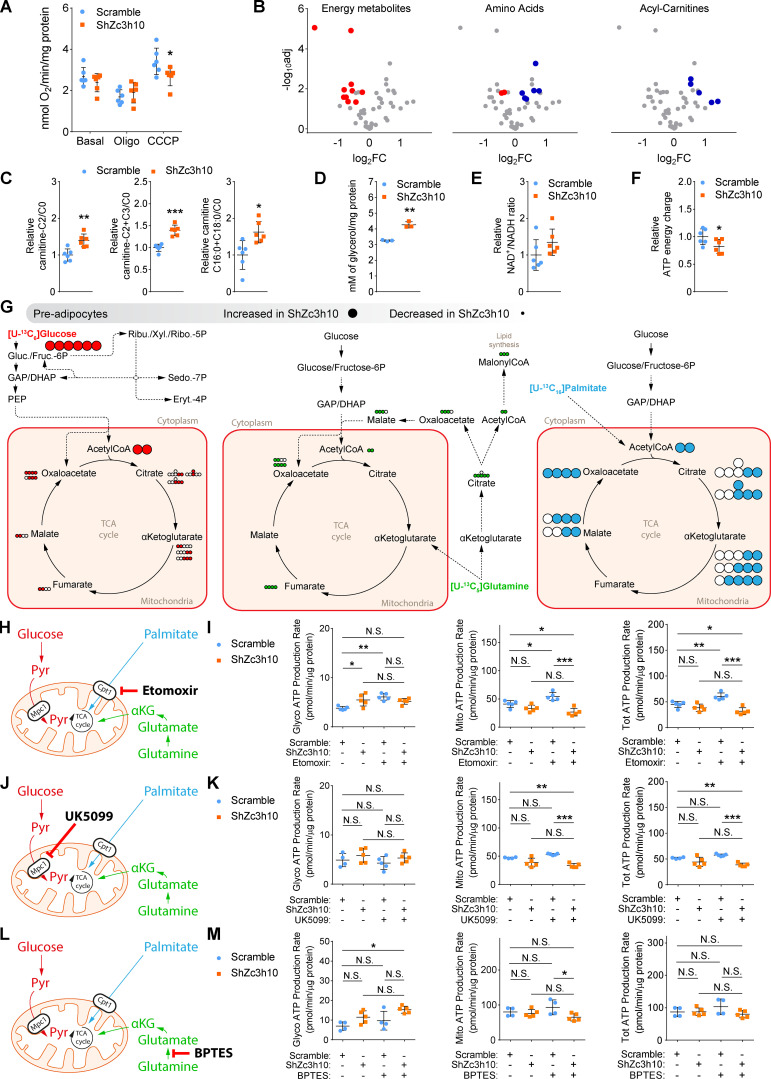
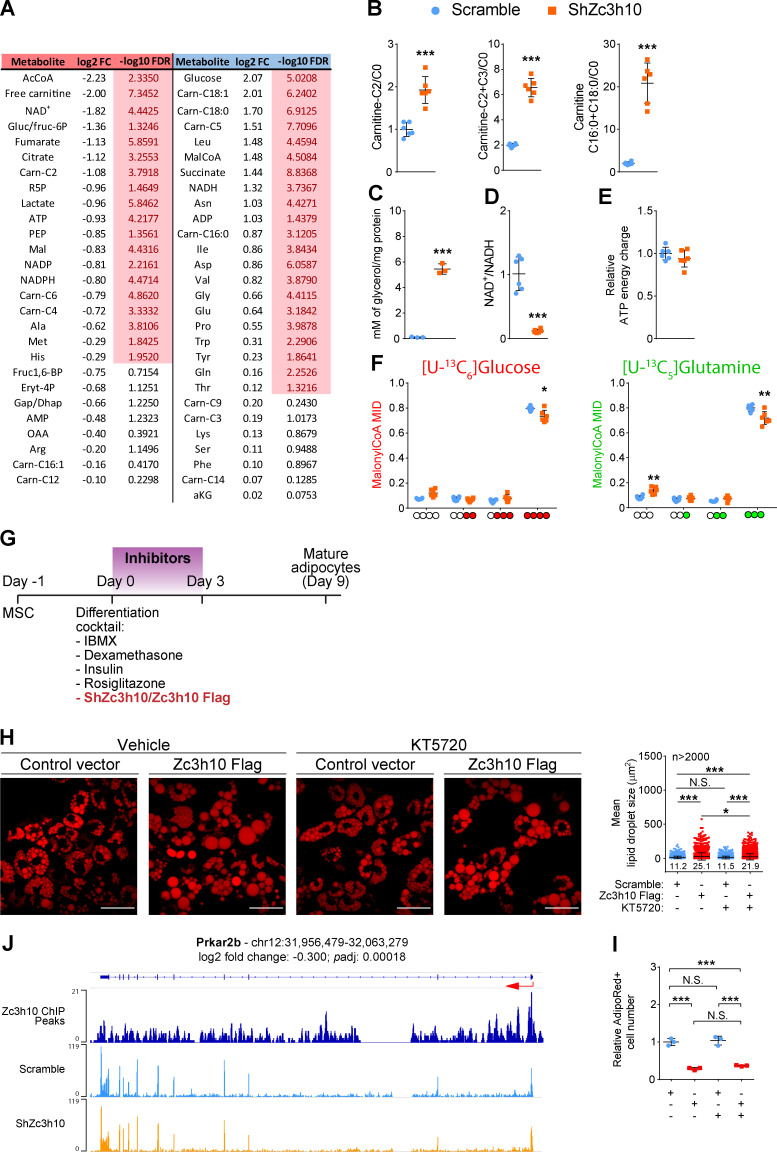
To provide evidence that Zc3h10-dependent mitochondrial fusion is a key feature to sustain mitochondrial activities and metabolism, we next applied functional and metabolomic analyses. Zc3h10-silenced preadipocytes showed no differences in basal and uncoupled respiration; however, we observed reduced electron transport chain activity when cells were challenged with the mitochondrial uncoupler carbonyl cyanide m-chlorophenyl hydrazine (CCCP; Fig. 5 A). Conversely, metabolomic analysis revealed decreased levels of some glycolysis intermediates, as well as energy and tricarboxylic acid (TCxA) cycle metabolites, while various amino acids and acylcarnitines were increased in cells lacking Zc3h10 (Figs. 5 B and S4 A). Furthermore, carnitine C2/C0 and C2+C3/C0 ratios, along with the carnitine palmitoyl transferase 1a (Cpt1a) activity index (C16:0+C18:0/C0), were consistently increased in Zc3h10-depleted cells (Fig. 5 C). These results were accompanied by increased glycerol release (as a marker of triacylglycerol breakdown; Fig. 5 D), a trend toward an increased NAD+/NADH ratio (Fig. 5 E), and reduced ATP energy charge in Zc3h10-silenced preadipocytes (Fig. 5 F).
Figure 5.
Zc3h10 orchestrates mitochondrial energy metabolism in preadipocytes. (A) Basal, uncoupled (Oligo), and maximal uncoupled respiration (CCCP) in preadipocytes (3 d). n = 6; unpaired parametric Student’s t tests; *P < 0.05 vs. Scramble. (B) Volcano plots representing significantly decreased (red dots), increased (blue dots), and unchanged (gray dots) energy metabolites, amino acids, and acyl-carnitines in preadipocytes (3 d). n = 6; unpaired parametric multiple Student’s t tests followed by false discovery rate (FDR) correction; FDR < 0.05 vs. Scramble. (C) C2 acyl-carnitine/free carnitine (C0; left), C2+C3 acyl-carnitines/C0 (middle), and C16:0+C18:0 acyl-carnitines/C0 (right) ratios in preadipocytes (3 d). n = 6; unpaired parametric Student’s t tests; *P < 0.05, **P < 0.01, ***P < 0.001 vs. Scramble. (D) Glycerol quantification in the media of preadipocytes (3 d). Glycerol concentrations were normalized to cellular protein content. n = 3; unpaired parametric Student’s t test; **P < 0.01 vs. Scramble. (E and F) NAD+/NADH ratio (E) and ATP energy charge (F) in preadipocytes (3 d). n = 6; unpaired parametric Student’s t tests; *P < 0.05 vs. Scramble. (G) Schematic representation of metabolic flux analyses in preadipocytes (3 d). Red, green, and light blue dots represent labeled carbons originating from [U-13C6]glucose, [U-13C5]glutamine, and [U-13C16]palmitate, respectively. Big and small dots represent increased and decreased labeling, respectively, of the indicated isotopomer in ShZc3h10 compared with Scramble cells. (H, J, and L) Schematic representation of etomoxir (H), UK5099 (J), and BPTES (L) mechanism of action. (I, K, and M) Glycolytic (left), mitochondrial (middle), and total ATP levels (right) calculated by the Seahorse XFp Real-Time induced ATP Rate Assay in preadipocytes (3 d) treated with 4 µM etomoxir (I), 2 µM UK5099 (K), and 3 µM BPTES (M). n = 5; one-way ANOVA followed by Tukey’s post-hoc test; *P < 0.05, **P < 0.01, ***P < 0.001. αKG, α-ketoglutarate.
Figure S4.
Zc3h10 regulates metabolite levels and metabolic flux in the TCxA cycle of preadipocytes. (A) Targeted metabolomics results in preadipocytes (3 d) indicating log2 fold change and −log10 adjusted P value for each metabolite. Red and blue legends indicate down- and upregulated molecules, respectively, while significantly changed metabolites are highlighted in pink. n = 6; statistical analysis was performed by using unpaired parametric multiple Student’s t tests followed by false discovery rate (FDR) correction; FDR < 0.05 vs. Scramble. (B) [U-13C6]Glucose-derived isotopomers in preadipocytes (3 d). Red dots indicate labeled carbons. n = 6; statistical analysis was performed by using unpaired parametric Student’s t tests; *P < 0.05, **P < 0.01, ***P < 0.01 vs. Scramble. (C) mRNA expression levels of the indicated genes in Scramble and ShZc3h10 in preadipocytes (3 d). n = 6; statistical analysis was performed by using unpaired parametric Student’s t tests; ***P < 0.01 vs. Scramble. (D) [U-13C5]Glutamine-derived isotopomers in preadipocytes (3 d). Green dots indicate labeled carbons. n = 6; statistical analysis was performed by using unpaired parametric Student’s t tests; *P < 0.05, ***P < 0.01 vs. Scramble. (E) [U-13C16]Palmitate-derived isotopomers in preadipocytes (3 d). Light blue dots indicate labeled carbons. n = 6; statistical analysis was performed by using unpaired parametric Student’s t tests; *P < 0.05, **P < 0.01, ***P < 0.01 vs. Scramble. αKG, α-ketoglutarate. MID, mass isotopomer distribution.
To disentangle the flux of carbons associated with glucose, glutamine, and palmitate utilization, we performed metabolic flux analysis. We found increased levels of glucose-derived 13Carbon in M6 glucose-6P/fructose-6P and decreased levels of labeled carbon in TCxA cycle isotopomers, such as M2 and M3 citrate, M2 and M3 α-ketoglutarate, and M2 fumarate, in Zc3h10-depleted preadipocytes (Fig. 5 G and S4 B). These results were consistent with reduced isocitrate dehydrogenase 3α (Idh3a) and malate dehydrogenase 2 (Mdh2) mRNA levels in the absence of Zc3h10 (Fig. S4 C). Zc3h10-silenced cells also displayed increased glucose-derived M2 acetyl-CoA (AcCoA) levels and decreased M2 malate, as well as M3 and M4 oxaloacetate (OAA) levels, which was consistent with reduced expression of citrate synthase (Fig. 5 G; and Fig. S4, B and C). Furthermore, glutamine utilization through oxidative metabolism was impaired in cells lacking Zc3h10, as shown by decreased M4 fumarate and M4 OAA. Reductive glutamine metabolism was also affected, as evidenced by reduced M2 AcCoA, M5 citrate, M3 and M2 OAA, and M3 MalonylCoA levels (Figs. 5 G and S4 D), which was consistent with the decreased expressions of ATP citrate lyase (Acly) and pyruvate carboxylase (Pcx; Fig. S4 C). Next, Zc3h10-depleted cells and the relative controls were exposed to [U-13C16]palmitate. Palmitate-derived 13Carbon incorporation was increased in M2 AcCoA and in different TCxA cycle metabolites, such as M2 and M4 citrate; M2, M3, and M4 α-ketoglutarate; M2 and M3 malate; and M4 OAA, in Zc3h10-depleted cells (Figs. 5 G and S4 E). These data indicate that fatty acids were preferentially used by Zc3h10-silenced cells.
We also performed metabolic stress-based experiments by using Seahorse technology to investigate the contribution of different metabolic fuels to ATP production. We exposed Zc3h10-silenced preadipocytes to Etomoxir (an inhibitor of fatty acid oxidation; Fig. 5 H), UK5099 (mitochondrial pyruvate carrier inhibitor; Fig. 5 J), and BPTES (an inhibitor of glutaminase I; Fig. 5 L). ATP rate assays confirmed no differences in total ATP levels in untreated cells (Fig. 5, I, K, and M, total ATP in the absence of metabolic inhibitors, right); however, Zc3h10-depleted preadipocytes showed significant decreases in mitochondrial ATP production rates when challenged with inhibitors (Fig. 5, I, K, and M, mitochondrial ATP, middle), with these effects being maintained on total ATP levels only with Etomoxir and UK5099 treatments (Fig. 5, I, K, and M, total ATP with metabolic inhibitors, right). Notably, Zc3h10-depleted cells coped with mitochondrial ATP impairment by glycolysis when glutaminolysis was inhibited (Fig. 5 M, glycolysis ATP, left). These results disclose the inability of Zc3h10-silenced cells to adapt to metabolic stress, mainly due to glucose and fatty acid oxidation inhibition.
Next, we sought to determine whether losing Zc3h10 at the beginning of adipogenesis resulted in changes in metabolism in mature adipocytes. Zc3h10-silenced mature adipocytes (Fig. S1 D) showed a more altered metabolic profile than silenced preadipocytes (Fig. S5 A), characterized by increased C2/C0 and C2+C3/C0 carnitine ratios, Cpt1a activity index, and glycerol release (Fig. S5, B and C), along with a decreased NAD+/NADH ratio (Fig. S5 D) and unchanged energy charge (Fig. S5 E). Metabolic flux analysis confirmed that Zc3h10-depleted mature adipocytes experienced reduced glucose and glutamine metabolism contributing to lower levels of the lipogenic metabolite MalonylCoA (Fig. S5 F). Overall, our results show that knockdown of Zc3h10 in pre- and mature adipocytes leads to significant rewiring of metabolic pathways fueling the TCxA cycle, favoring fatty acid oxidation at the expense of glucose and glutamine metabolism.
Figure S5.
Zc3h10 tunes energy metabolism in mature adipocytes and its function partially relies on PKA activity in preadipocytes. (A) Targeted metabolomics results in mature adipocytes (9 d) indicating log2 fold change and −log10 adjusted P value for each metabolite. Red and blue legends indicate down- and upregulated molecules, respectively, while significantly changed metabolites are highlighted in pink. n = 6; statistical analysis was performed using unpaired parametric multiple Student’s t test followed by false discovery rate (FDR) correction; FDR < 0.05 vs. Scramble. (B) C2 acyl-carnitine/free carnitine (C0; left), C2+C3 acyl-carnitines/C0 (middle), and C16:0+C18:0 acyl-carnitines/C0 (right) ratios in Scramble and ShZc3h10 in mature adipocytes (9 d). n = 6; statistical analysis was performed by using unpaired parametric Student’s t tests; ***P < 0.001 vs. Scramble. (C) Glycerol quantification in the media of Scramble and ShZc3h10 in mature adipocytes (9 d). Glycerol concentrations were normalized to cellular protein content. n = 3; statistical analysis was performed by using an unpaired parametric Student’s t test; **P < 0.01 vs. Scramble. (D) NAD+/NADH ratio in Scramble and ShZc3h10 in mature adipocytes (9 d). n = 6; statistical analysis was performed by using an unpaired parametric Student’s t test; ***P < 0.001 vs. Scramble. (E) ATP energy charge in Scramble and ShZc3h10 in mature adipocytes (9 d). n = 6; statistical analysis was performed by using an unpaired parametric Student’s t test. (F) [U-13C6]Glucose- and [U-13C5]glutamine-derived MalonylCoA in Scramble and ShZc3h10 in mature adipocytes (9 d). n = 6; statistical analysis was performed by using unpaired parametric Student’s t tests; *P < 0.05, **P < 0.01 vs. Scramble. MID, mass isotopomer distribution. (G) Representation of the experimental protocol used for preadipocyte treatment with PKA and Rho GTPase pathway inhibitors (e.g., KT5720, Y26732, ML141, and NSC23766). (H) Lipid droplet images and relative quantification in vehicle or KT5720 treated Scramble and ShZc3h10 in mature adipocytes (9 d). n > 2,000; statistical analysis was performed by using one-way ANOVA followed by Tukey’s post-hoc test; *P < 0.05, ***P < 0.01. Scale bar: 25 µm. (I) AdipoRed+ cell counts by cytofluorimetry in Scramble and ShZc3h10 in mature adipocytes (9 d) treated with vehicle or KT5720; n = 3; statistical analysis was performed by using one-way ANOVA followed by Tukey’s post-hoc test; ***P < 0.001. (J) Zc3h10 chromatin immunoprecipitation (ChIP) sequencing peaks (dark blue track) and signals from 4-sU-seq analysis for Prkar2b, a subunit of PKA. The light blue track indicates the pre-mRNA reads for Scramble cells, and the orange track shows the pre-mRNA reads for ShZc3h10 preadipocytes. Log2 fold change and adjusted P value (padj) are shown on top of the graph. IBMX, 3-isobutyl-1-methylxanthine.
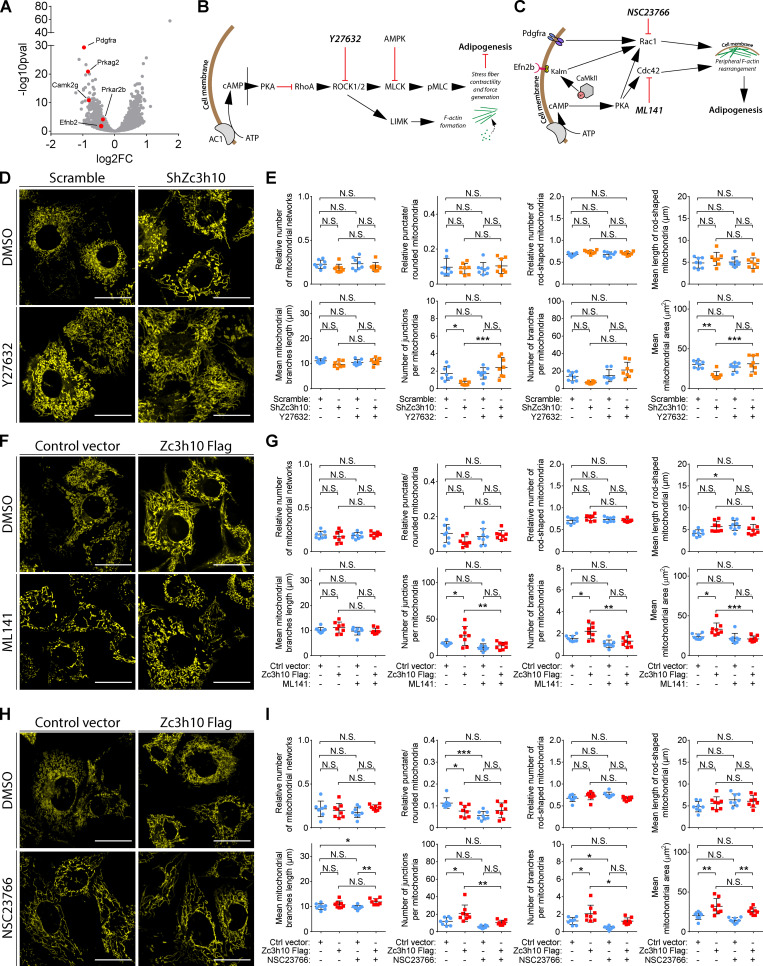
Zc3h10 favors Rho GTPases to stimulate F-actin remodeling, mitochondrial function, and lipid accumulation
PKA activation is one of the main and early pathways that regulate F-actin remodeling during the adipogenic program by affecting the activity of Rho GTPases (Yu et al., 2013; Cristancho and Lazar, 2011; Howe, 2004). The three most common members of this family are cell division control protein 42 homologue (Cdc42), Ras-related C3 botulinum toxin substrate 1 (Rac1), and RhoA (Heasman and Ridley, 2008; Hall, 2012; Jaffe and Hall, 2005). Inhibition of Rho/ROCK, a downstream effector of RhoA, favors adipogenesis (Noguchi et al., 2007; Romani et al., 2019; Nobusue et al., 2014; Halder et al., 2012). Rac1 and Cdc42 activation regulates cell cycle progression (Lamarche et al., 1996) and dorsal F-actin organization to promote terminal adipogenesis (Liu et al., 2005; Cristancho and Lazar, 2011). In addition, PKA activates Rac1 and Cdc42 and inhibits RhoA pathways (Bachmann et al., 2013; O’Connor and Mercurio, 2001; Price et al., 1998; Howe, 2004). Hence, we tested whether Zc3h10 requires PKA activation to favor lipid accumulation (Fig. S5 G). PKA inhibition (KT5720) reduced lipid droplet size in Zc3h10-overexpressing cells, but the number of adipocytes was unchanged (Fig. S5, H and I). Notably, Zc3h10 silencing resulted in decreased expression of pre-mRNA for the regulatory subunit of PKA, Prkar2b, which appears to be a direct target gene of Zc3h10 (Fig. S5 J). Taken together, these results indicate that Zc3h10 partially requires PKA signaling during adipogenesis.
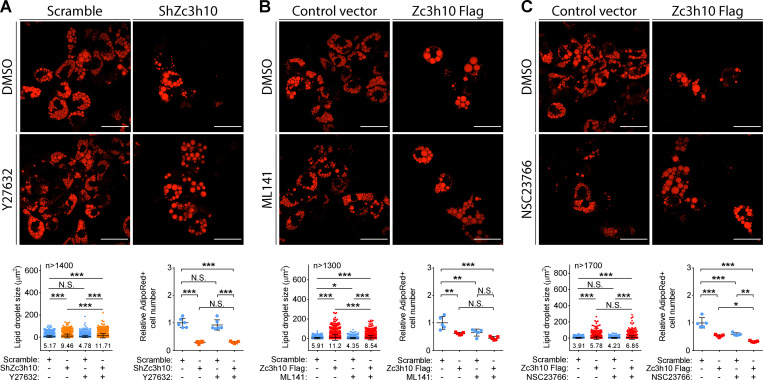
Of note, 4-sU-seq results demonstrated that Zc3h10 silencing inhibited the expression of target genes involved in cytoskeletal remodeling by Rho GTPase activities, such as PDGF factor receptor α (Pdgfra), protein kinase AMP-activated noncatalytic subunit γ 2 (Prkag2), Ca2+/CaM-dependent protein kinase II subunit Camk2g, and ephrin B2 (Efnb2; Fig. 6 A and Table S1; Feng et al., 2011; Salt and Hardie, 2017; Rangamani et al., 2016; Murai and Pasquale, 2003). Therefore, we inhibited the three Rho GTPases to better define the molecular pathways regulated by Zc3h10 that control F-actin remodeling and, consequently, mitochondrial function (Fig. 6, B and C; and Fig. S5 G). ROCK inhibition (Y27632) rescued the number of junctions per mitochondria, mitochondrial area, and increased lipid droplet size without affecting the number of lipid-positive cells in Zc3h10-depleted preadipocytes (Fig. 6, D and E; and Fig. 7 A). The Cdc42 inhibitor ML141 reduced the number of mitochondrial junctions and branches, as well as the mitochondrial area and lipid droplet size in Zc3h10-overexpressing cells (Fig. 6, F and G; and Fig. 7 B). Rac1 inhibition (NSC23766) reduced the number of mitochondrial junctions and branches, as well as reduced the number of adipocytes, resulting in blunted lipid accumulation in Zc3h10-overexpressing cells (Fig. 6, H and I; and Fig. 7 C). Together, these findings indicate that Zc3h10 activates a transcriptional program that sets the basis for the downstream functionality of pathways involving PKA and Rho GTPases that can then execute F-actin remodeling and determine mitochondrial dynamics, ultimately enabling the white adipogenic program.
Figure 6.
ROCK, Cdc42, and Rac1 mediate Zc3h10 activity on mitochondrial networks. (A) Volcano plot of differentially expressed pre-mRNAs in ShZc3h10 and Scramble cells (36 h after differentiation induction). Red dots indicate labeled downregulated genes in ShZc3h10 preadipocytes regulating F-actin organization by Rho GTPases activity. Scramble: n = 2; ShZc3h10: n = 3; false discovery rate (FDR) < 0.05. (B) Schematic representation of the RhoA/ROCK pathway and the mechanism of action of Y27632. PKA inhibits RhoA, leading to reduced contractility of F-actin stress fibers and, ultimately, induces adipogenesis. (C) Schematic representation of the Rac1/Cdc42 pathway and the mechanisms of action of ML141 and NSC23766. PKA induces both Rac1 and Cdc42 activities, leading to peripheral F-actin remodeling in preadipocytes that favors adipogenesis. (D, F, and H) Representative confocal images of mitochondria in preadipocytes (36 h) that were exposed to 10 µM Y27632 (D), 10 µM ML141 (F), and 50 µM NSC23766 (H), and stained with Mitotracker Deep Red FM probe. Scale bar: 10 µm. (E, G, and I) Mitochondrial network analysis of the cells treated with 10 µM Y27632 (E), 10 µM ML141 (G), and 50 µM NSC23766 (I). n = 8 cells; one-way ANOVA followed by Tukey’s post-hoc test; *P < 0.05, **P < 0.01, ***P < 0.001.
Figure 7.
Rho GTPase activity is required for adipogenesis. (A) Confocal images of lipid droplets labeled with AdipoRed in mature adipocytes exposed to DMSO or 10 µM Y27632 (top). Quantifications of lipid droplet size (bottom left) and number of AdipoRed+ cells (bottom right) are shown. Lipid droplet size: n > 1,400; AdipoRed+ cell count: n = 6; one-way ANOVA followed by Tukey’s post-hoc test; *P < 0.05, **P < 0.01, ***P < 0.001. (B) Confocal images of lipid droplets labeled with AdipoRed in mature adipocytes exposed to DMSO or 10 µM ML141. Quantifications of lipid droplet size (bottom left) and number of AdipoRed+ cells (bottom right) are shown. Lipid droplet size: n > 1,300; AdipoRed+ cell count: n = 5; one-way ANOVA followed by Tukey’s post-hoc test; *P < 0.05, **P < 0.01, ***P < 0.001. (C) Confocal images of lipid droplets labeled with AdipoRed in mature adipocytes exposed to DMSO or 50 µM NSC23766. Quantifications of lipid droplet size (bottom left) and number of AdipoRed+ cells (bottom right) are shown. Lipid droplet size: n > 1,700; AdipoRed+ cell count: n = 5; one-way ANOVA followed by Tukey’s post-hoc test; *P < 0.05, **P < 0.01, ***P < 0.001.
A Zc3h10 transcriptomic signature is associated with reduced translation activity
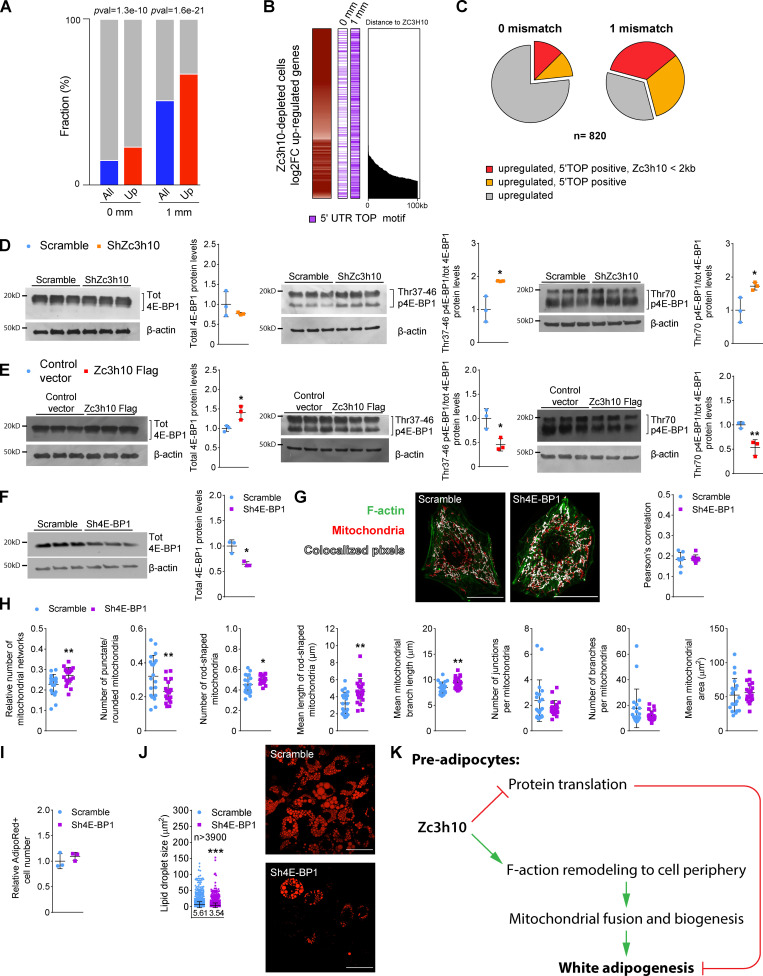
During the early phases of the adipogenic program, preadipocytes show reduced translational activity (Marcon et al., 2017; Tsukiyama-Kohara et al., 2001; Blackshear et al., 1997). Indeed, we found 132 pre-mRNAs of the translation pathway that were upregulated in Zc3h10-silenced preadipocytes (Fig. 1, P and R). Therefore, we investigated whether these transcriptional changes were associated with increased translation activity. As functional markers of the early stages of translation initiation, we evaluated the enrichment of the 5′ UTR terminal oligopyrimidine (5′TOP) motif (Jefferies et al., 1994) in upregulated genes and the phosphorylation status of 4E-BP1 that regulates the assembly of the preinitiation complex. To gather information regarding whether upregulated genes in Zc3h10-depleted cells are significantly enriched in transcripts harboring a 5′TOP motif, we annotated all the mm10 RefSeq 5′ UTRs and transcripts with both a perfect match and at most one mismatch to the CUYUYYC motif (Jensen et al., 2020). Of 820, 191 (23.3%) transcripts upregulated in Zc3h10-silenced cells had a perfect TOP motif in their 5′ UTR overlap, which increased to 547 (66.7%) transcripts when considering RNAs with TOP motifs containing at most one mismatch. Hypergeometric test (Fisher’s exact test) for both sets of transcripts indicated that the overlap was significantly enriched in upregulated mRNAs of Zc3h10-depleted cells (P value = 1.27e-10; and P value = 1.6e-21 respectively; Fig. 8 A). In addition, 5′TOP-positive transcripts were associated with Zc3h10 binding, with more than 50% of these genes being within 2 kb of a Zc3h10 chip-seq peak published by Yi et al. (2019; Fig. 8, B and C; and Table S1).
Figure 8.
Protein synthesis regulated by Zc3h10 affects the accumulation of lipids. (A) Enrichment of 0 mismatch (mm) and 1 mm 5′ UTR TOP motif enriched transcripts in Zc3h10-silenced preadipocytes compared with all upregulated genes. Statistical analysis was performed by using hypergeometric tests (Fisher’s exact test). (B) 5′TOP motif distribution in genes upregulated in Zc3h10-silenced cells and its distance (kb) from Zc3h10 binding sites. (C) Enrichment of 5′TOP motifs with and without associated (within 2 kb) Zc3h10 binding sites in genes that were upregulated in Zc3h10-silenced cells. (D and E) Western blot of total, pThr37-46, and pThr70 4E-BP1 with relative quantifications in ShZc3h10 (D) and Zc3h10-Flag (E) preadipocytes differentiated for 36 h. n = 3; unpaired parametric Student’s t tests; *P < 0.05 vs. Scramble (D); *P < 0.05 (E), **P < 0.01 vs. Control. (F) Western blot of 4E-BP1 in Scramble and Sh4E-BP1 preadipocytes (36 h), with relative quantification. n = 3; unpaired parametric Student’s t test; *P < 0.05 vs. Scramble. (G) Analysis of F-actin/mitochondria localization in Scramble and Sh4E-BP1 preadipocytes (36 h). n = 12 cells; scale bar: 25 µm. (H) Mitochondrial morphologic analysis of Scramble and Sh4E-BP1 preadipocytes (36 h); n = 20 cells; unpaired parametric Student’s t tests; *P < 0.05, **P < 0.01 vs. Scramble. (I) AdipoRed+ cell count by cytofluorimetry in Scramble and Sh4E-BP1 mature adipocytes (9 d). n = 3. (J) Lipid droplet size in Scramble and Sh4E-BP1 mature adipocytes differentiated for 9 d. n > 3,900; unpaired nonparametric Mann-Whitney test; ***P < 0.001 vs. Scramble. (K) Schematic representation of the role of Zc3h10 in adipogenesis. Zc3h10 represses protein translation and, by promoting the expression and functions of F-actin regulators (PKA, Cdc42, and Rac1), favors F-actin reorganization to the cell periphery in preadipocytes, fundamental processes for adipogenesis.
Translational control of 5′TOP mRNAs relies, at least in part, on the regulation of eIF4E by 4E-BPs (Hsieh et al., 2012; Thoreen et al., 2012). The hierarchical phosphate addition to 4E-BP1 requires pThr37 and pThr46, followed by pThr70 and, ultimately, pSer65 to inhibit 4E-BP1 activity and favor translation (Gingras et al., 1999, 2001; Proud, 2007). Zc3h10-depleted cells showed no difference in total 4E-BP1 levels; however, pThr37, -46, and -70 were significantly increased (Fig. 8 D). Conversely, Zc3h10-overexpressing cells showed hypophosphorylation of the same amino acid residues, despite a significant increase in total 4E-BP1 levels (Fig. 8 E).
4E-BP1–silenced preadipocytes (Fig. 8 F) showed no differences in F-actin/mitochondria interactions (Fig. 8 G) or in F-actin morphology (data not shown). Instead, the numbers of mitochondrial networks and rod-shaped mitochondria, as well as mitochondrial length and branch length, were increased, while punctate/rounded mitochondria were decreased. Notably, the number of mitochondrial branches and junctions in addition to mitochondrial area (indexes of fusion) were unaffected, meaning that 4E-BP1 knockdown was unable to mimic the effects of Zc3h10 depletion on F-actin/mitochondria dynamics (Fig. 8 H). Finally, while the number of mature adipocytes was unaffected, lipid droplet size was significantly reduced in 4E-BP1–silenced cells (Fig. 8, I and J). These results indicate that Zc3h10 negatively controls the expression of specific genes involved in protein translation pathways, a key step in adipogenesis, as well as the F-actin/mitochondria dynamics necessary to promote lipid loading.
Discussion
A hormonal regimen is widely used to commit MSCs to preadipocytes, which then differentiate into lipid-filled mature white adipocytes (Tang and Lane, 2012). The early phases of this multistep process require the reduction of translation to arrest cell proliferation (Marcon et al., 2017, 2019; Tsukiyama-Kohara et al., 2001), cytoskeletal remodeling that involves F-actin reorganization (Spiegelman and Farmer, 1982; Hansson et al., 2019), and an increase in mitochondrial network complexity (Tharp et al., 2018; Wilson-Fritch et al., 2003; De Pauw et al., 2009). However, knowledge of the molecular mechanisms that control these events in a coordinated fashion still remains incomplete. In this study, we used gain- and loss-of-function approaches to unravel the biochemical and molecular functions of Zc3h10 during the white adipogenic program. By using integrated multidisciplinary approaches, we demonstrated that Zc3h10 resides in the chromatin fraction and is required in the early stages of adipogenesis to determine proper terminal adipocyte differentiation. Depletion of Zc3h10 in mouse preadipocytes resulted in reduced lipid accumulation, while Zc3h10 overexpression before its physiologic expression increased lipid droplet size, though it reduced the number of mature adipocytes. These results highlight the importance of the fine-tuned and time-controlled expression of Zc3h10 during the white adipogenic program.
In the early stages of the adipogenic program, Zc3h10 controls the expression of genes belonging to the translation and actin cytoskeleton organization pathways. Functional validation experiments demonstrated that Zc3h10 depletion in preadipocytes was associated with translation initiation, as evidenced by the enrichment of the 5′TOP motif in upregulated genes and the hyperphosphorylated status of the repressor of translation 4E-BP1. 4E-BP1 is mainly controlled by phosphorylation mediated by the mammalian target of rapamycin (mTOR) pathway (Siddiqui and Sonenberg, 2015). During the white adipogenic program, the total protein levels of mTOR in mature adipocytes are higher than those observed in preadipocytes (Kim and Chen, 2004). Moreover, mTOR phosphorylation at Ser2448 rises as a proxy of mTOR activation concomitantly with the strongly increased expression of two well-known downstream targets of mTOR: ribosomal protein S6 kinase 1 and 4E-BP1. Consistent with our results, mTOR activation is detected at the preadipocyte stage to relieve translation suppression and allow the protein synthesis required to form mature adipocytes (Kim and Chen, 2004). Of note, 4E-BP1 knockdown in preadipocytes is associated with a significant decrease of lipid accumulation, suggesting a crucial role for reduced protein translation in the positive regulation of adipogenesis.
Our investigations into the early phases of adipogenesis indicate a key role for Zc3h10 in orchestrating F-actin remodeling to determine a discrete subpopulation of functional and fused mitochondria to provide energy and anabolic substrates for the buildup of lipids. PKA and downstream Rho GTPases are the main controllers of cytoskeleton remodeling, cell shape, and organelle development (Heasman and Ridley, 2008; Hall, 2012; Jaffe and Hall, 2005). Our work using chemical inhibitors of either PKA or Rho GTPases in Zc3h10 knockdown or Zc3h10-overexpressing cells supports the notion that Zc3h10 activates a transcriptional program that enables the activities of PKA/Rho GTPases and results in F-actin remodeling to trigger mitochondrial fusion and network complexity. All these early events controlled by Zc3h10 allow for the formation of mature adipocytes. This concept is supported by our analyses conducted at the late stage of adipogenesis that showed that Zc3h10-depleted mature adipocytes experience mitochondrial dysfunction, leading to fatty acid utilization instead of accumulation, ultimately resulting in an impaired adipogenic program.
In closing, we have defined a physiologic role for Zc3h10 as an adipogenic transcription factor that, through the coordinated control of the cytoskeletal structure, mitochondrial function/dynamics, and metabolism in preadipocytes, sets the basis for the white adipogenic program (Fig. 8 K).
Materials and methods
Cell lines and animal models
C3H10T1/2 murine MSCs (ATCC CCL-226) were maintained in growth medium made of high-glucose DMEM supplemented with 10% FBS (vol/vol; Euroclone), 1% glutamine (vol/vol; Thermo Fisher Scientific), and 1% pen/strep (vol/vol; Thermo Fisher Scientific) at 37°C in a 5% CO2 and 90% humidity atmosphere. C3H10T1/2 cells were differentiated to adipocytes 24 h after reaching complete confluence. Growth medium was then switched to differentiation medium composed of high-glucose DMEM, 10% FBS (vol/vol), 1% glutamine (vol/vol), 1% pen/strep (vol/vol), 5 µg/ml insulin (Sigma-Aldrich), 2 µg/ml dexamethasone (Sigma-Aldrich), 0.5 mM 3-isobutyl-1-methylxanthine (Sigma-Aldrich), and 5 µM rosiglitazone (Cayman Chemicals). After 3 d, preadipocytes were maintained in growth medium supplemented with 5 µg/ml insulin for 6 d until completely differentiated. Rho GTPase and PKA inhibitors were used at the following concentrations: 10 µM Y27632 (Tocris, 1254), 10 µM ML141 (Tocris, 4266), 50 µM NSC23766 (Tocris, 2161), and 1 µM KT5720 (Tocris, 1288).
Primary stromal vascular cells were isolated from inguinal and epididymal white adipose tissue of C57BL/6J male mice. Tissues were minced with a sterile blade to a very fine consistency and then incubated in Hepes solution (150 mM NaCl, 62.5 mM KCl, 6.2 mM D-glucose, 2% BSA [wt/vol], 1.2 mM CaCl2, pH 7.6) containing 1 mg/ml type II collagenase (Sigma Life Science, C6885) for 1 h at 37°C with shaking (115 rpm). The digested fat solution was then poured through a 100-µm cell strainer and centrifuged at 200 × g for 10 min at RT to remove the collagenase solution and mature adipocytes. Cells were then incubated with erythrocyte lysis buffer (155 mM NH4Cl, 5.7 mM K2HPO4, 0.1 mM EDTA) for 5 min, and the suspension was filtered through a 100-µm cell strainer and centrifuged as above. The supernatant was discarded and the pellet containing stromal vascular cells was resuspended in 10 ml of plating medium (DMEM/F12; Thermo Fisher Scientific) supplemented with 10% FBS (vol/vol; Euroclone), 1% glutamine (vol/vol; Thermo Fisher Scientific), 1% pen/strep (vol/vol; Thermo Fisher Scientific), and 1% Na-pyruvate (vol/vol; Thermo Fisher Scientific). To remove endothelial cell clumps, the cell suspension was filtered through a 40-µm cell strainer. 24 h after confluency, plating medium was then switched to differentiation medium composed of DMEM/F12, 10% FBS (vol/vol), 1% glutamine (vol/vol), 1% pen/strep (vol/vol), 1% Na-pyruvate (vol/vol), 10 µg/ml insulin (Sigma-Aldrich), 1 µM dexamethasone (Sigma-Aldrich), 0.5 mM 3-isobutyl-1-methylxanthine (Sigma-Aldrich) and 0.5 µM rosiglitazone (Cayman Chemicals). After 48 h, preadipocytes were maintained in DMEM/F12, 10% FBS (vol/vol), 1% glutamine (vol/vol), 1% pen/strep (vol/vol), 1% Na-pyruvate (vol/vol), and 1 µg/ml insulin for 6 d until completely differentiated. All experiments were conducted following the ARRIVE guidelines and regulations of the European Community (EU Directive 2010/663/EU, Official Journal of the European Union L 276/33, 20/10/2010) as well as local regulations (Italian Legislative Decree n. 26—04/03/2014) for the care and use of laboratory animals. The Italian Ministry of Health approved the animal protocols of this study (ministerial decree no. 579/2015-PR).
Adenoviral transduction for gene overexpression and downregulation
Zc3h10 overexpression in C3H10T1/2 MSCs was performed by adding adenovirus (Human Adenovirus Type 5 dE1/dE3) containing a validated adenoviral vector to express mouse Zc3h10 cDNA fused to a Flag tag at the C terminus (Zc3h10-Flag) controlled by a cytomegalovirus promoter to confluent cells in differentiation medium. Cells were incubated with virus for 3 d or 36 h, where indicated. As a control, we used the same construct expressing GFP under the control of a cytomegalovirus promoter (Vector Biolabs, 1768). Zc3h10-Flag and GFP were transduced with an MOI of 25. Zc3h10 and 4E-BP1 targeted silencing in C3H10T1/2 MSCs was performed as described above by using a virus harboring a validated adenoviral construct with a specific shRNA sequence under the control of the U6 promoter (Vector Biolabs, Zc3h10 ShADV-276549 and 4E-BP1 ShADV-258238). The control vector contained a scrambled sequence (Scramble; Vector Biolabs, 1122). To silence Zc3h10 and 4E-BP1, cells were infected with 50 and 100 MOI, respectively. Opa1 overexpression was performed by using retroviral infection of proliferating MSCs as previously described (Cipolat et al., 2004). Briefly, for the generation of retrovirus supernatants, Platinum-E packaging cells were seeded 1 d before transfection. Cells were transfected the next day at 70% confluency by using polyethylenimine (Polysciences). Supernatant was collected 24, 48, and 72 h after transfection and filtered with a 0.45-mm polyvinylidene difluoride filter (Millipore). Polybrene (4 µg/ml) was added to the viral supernatants and used to infect.
Cytoplasm-nucleoplasm-chromatin separation
36 h after differentiation induction, C3H10T1/2 MSCs were collected for cytoplasm, nucleoplasm, and chromatin protein extraction. All centrifugations were performed at 4°C and samples were kept on ice throughout the procedure. Cells were rinsed with ice-cold PBS, suspended in 300 µl of cytoplasmic lysis buffer (10 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.15% NP-40 [vol/vol]), and incubated for 5 min on ice. Lysates were layered on top of 750 µl of a chilled sucrose cushion (24% [wt/vol] of sucrose in cytoplasmic lysis buffer without NP-40) and centrifuged at 18,000 × g for 10 min. The supernatant contained the cytoplasmic protein fraction. Nuclei pellets were resuspended in 300 µl of cold glycerol buffer (20 mM Tris-HCl, pH 7.9, 75 mM NaCl, 0.5 mM EDTA, 0.85 mM DTT, 50% glycerol [vol/vol]), and then 300 µl of nuclei lysis buffer (20 mM Hepes, pH 7.6, 7.5 mM MgCl2, 0.2 mM EDTA, 300 mM NaCl, 1 M urea, 1% NP-40 [vol/vol], 0.1 mM DTT) was added. Samples were incubated on ice for 1 min and were then centrifuged at 18,000 × g for 2 min. The supernatant contained the nucleoplasmic protein fraction. The chromatin pellet was resuspended in 50 µl PBS and was used for chromatin-associated protein analysis. Proteins were then equally loaded for SDS-PAGE analysis.
Total DNA and mitochondrial DNA quantification
Total genomic and mitochondrial DNA were isolated by using a kit to recover genomic DNA from tissues (Macherey-Nagel, 740952/250). Briefly, cells were lysed in 200 µl of Buffer T1, and then DNA was isolated following the manufacturer’s instructions. Samples were eluted in 100 µl of DNase- and RNase-free water and quantified by UV spectrophotometry (NanoDrop 1000 Spectrophotometer; Thermo Fisher Scientific). Mitochondrial DNA content was evaluated by assessing mt-Co2 and 36B4 content by quantitative PCR as examples of mitochondrial and nuclear-encoded genes, respectively. Primers and probes were obtained from Eurofins Genomics MWG-Operon and sequences are available upon request.
Gene expression analysis
Total RNA was obtained from C3H10T1/2 cells by using a commercial kit (NucleoSpin RNA extraction kit, Macherey-Nagel, 740955.250). Briefly, cells were washed in ice-cold PBS and lysed in 350 µl of lysis buffer (RA1) supplemented with 1% (vol/vol) β-mercaptoethanol. Total RNA was then isolated following the manufacturer’s instructions and eluted from columns with 60 µl of RNase-free water. Total RNA amounts were quantified by UV spectrophotometry (NanoDrop 1000 Spectrophotometer, Thermo Fisher Scientific). Samples were then diluted to 10 ng/µl and used for mRNA quantification. RNA was quantitated by quantitative RT-PCR (qRT-PCR) using iScript One Step for Probes (Bio-Rad, 1725141) and iTaq Universal SYBR Green One-Step Kit for quantitative PCR (Bio-Rad, 1725151), following the manufacturer’s instructions. The qRT-PCR protocol consisted of 40 cycles of amplification, each including a denaturation step at 95°C for 15 s and an annealing/extension step at 60°C for 60 s. The oligonucleotides used for qRT-PCR were obtained from Eurofins MWG Operon (Ebersberg, Germany). qRT-PCR primer sequences are available upon request.
Oil-Red-O staining
Cells were washed with PBS and incubated with 2 ml of 10% formalin at RT. After 30 min, formalin was removed and cells were washed with distilled water. After removing the water, 2 ml of 60% isopropanol were added to cover the samples for 5 min. After isopropanol removal, samples were incubated with Oil-Red-O working solution (prepared following the manufacturer’s instructions; Lonza, 0–0625) for 5 min. Cells were then washed with water until all the excess Oil-Red-O was removed. Images were acquired by using brightfield microscopy. After image acquisition, the Oil-Red-O stain was extracted by adding 1 ml of 100% isopropanol to the samples for 5 min with gentle rocking. 100 µl of each sample was then transferred to a 96-well plate and quantified at 492 nm.
RNA synthesis assay
4-sU (Sigma Aldrich, T4509)–labeled RNA isolation was performed as previously described (Rabani et al., 2011). Briefly, 300 µM 4-sU was added for 10 min to label confluent 36-h differentiated Scramble- and ShZc3h10-treated preadipocytes. Total RNA extraction was performed, as described above, and 1/50 of the total RNA was saved as input. The labeled RNA was isolated and processed as described (Austenaa et al., 2015). Briefly, the nascent 4-sU–labeled RNA was extracted from 50 µg of total TRIzol-isolated RNA, conjugated to N-[6-(biotinamido)hexyl]-3'-(2'-pyridyldithio)propionamide (biotin-HPDP; Abcam), and precipitated with 50 µl streptavidin (MyOne Streptavidin T1, Invitrogen). 4-sU RNA samples were eluted in 20 µl of RNase-free water and analyzed by next-generation sequencing. Isolated nascent 4-sU–labeled RNA (20–30 ng) was used for cDNA library synthesis by using the QIAseq Stranded Total RNA Lib Kit (Qiagen, 180745) with no ribosomal depletion or polyA selection. Libraries were sequenced on an Illumina NextSeq 500.
Computational analysis of RNA synthesis and chromatin immunoprecipitation
Nascent RNA sequencing reads were aligned to the mm10 genome assembly (GRCM38/mm10 Dec. 2011), together with transcript annotation from RefSeq (ncbiRefSeqCurated 2017/11/16) using topHat2 (TopHat v2.1.1; Kim et al., 2013) with parameters “–max-multihits 2–b2-very-sensitive”. Multimapped reads were removed by selecting for reads with “NH:i:” flag set to 1. Reads mapped within “blacklist” regions (Amemiya et al., 2019) were removed by using standard bedtools operations (bedtools v2.29.2). An appropriate scaling factor was obtained by using a method described previously (Gualdrini et al., 2016). Briefly, a set of invariant genes was identified as those having differences across samples following a Log-normal distribution. Genes within 1σ of the mean difference of such a distribution were selected as invariant and were used to compute normalization-scaling factors. Differential gene expression analysis was performed by using DESeq2 (Love et al., 2014) by comparing Zc3h10 knockdown cells against control and selecting those genes with an associated adjusted P value of ≤0.05.
Chromatin immunoprecipitation data for Zc3h10 were retrieved from GEO (accession no. GSE139029). Paired-end reads were mapped in pair-end mode to the mm10 genome assembly (GRCM38/mm10 Dec. 2011) by using bowtie2 (version 2.3.5.1; Langmead and Salzberg, 2012) with parameters “-X 1000 -k 2 -t”. Reads mapped and in proper pairs were selected by using basic samtools operations (samtools version 1.9). For the RNa sequencing, sample reads within blacklist regions were excluded and duplicates were filtered by using the samtools markdup tool. Zc3h10 peaks were then identified by using MACS2 (version 2.2.6; Zhang et al., 2008) with parameters “–format=BAMPE -B -g mm–nomodel–extsize 300–nolambda–call-summits”. Zc3h10 peaks with an associated qval ≤ −log10(10−10) were selected. Distances to RefSeq genes’ TSSs were computed in R with the library “GenomicRanges” (R version 3.6.2 2019–12-12).
Western blot
Protein relative quantification analyses were performed by separating cell lysates by SDS-PAGE. Cells were rinsed in ice-cold PBS and lysed in RIPA buffer. Protein concentrations were measured by using BCA methods (Euroclone). Proper protein amounts were than loaded onto a 12.5% SDS-PAGE gel. After the gel was run, proteins were transferred to a nitrocellulose membrane and blocked in 0.1% TBS-Tween 20 and 5% (wt/vol) BSA for 1 h at RT. Membranes were then incubated overnight at 4°C with primary antibodies suspended in 0.1% TBS-Tween 20 and 3% BSA. After extensive washing, membranes were incubated with HRP-conjugated secondary antibodies for 1 h at RT. After washing, membranes were finally incubated with ECL substrate to detect bands. Primary and secondary antibodies were diluted as follows: Zc3h10 1:1,000 (Aviva Systems Biology, ARP60671_P050), mitochondrial transcription factor A 1:500 (Aviva Systems Biology, ARP31400_P050), oxidative phosphorylation cocktail 1:2,000 (Abcam, ab110413), Hsp90 1:1,000 (Santa Cruz Biotechnology, sc-13119), β-actin 1:5,000 (Sigma-Aldrich, A5441), histone H3 1:1,000 (Millipore, 06–755), α-tubulin 1:1,000 (Sigma-Aldrich, T9026), Flag-M2 1:5,000 (Sigma-Aldrich, F3165), peroxisome proliferator-activated receptor γ 1:1,000 (Cell Signaling Technology, 2443S), Nup98 1:1,000 (Cell Signaling Technology, 2597s), 4E-BP1 1:1,000 (Cell Signaling Technology, 9644), phospho-4E-BP1 (Thr37/46) 1:1,000 (Cell Signaling Technology, 2855), phospho-4E-BP1 (Thr70) 1:1,000 (Cell Signaling Technology, 9455), DRP1 1:1,000 (Cell Signaling Technology, 14647), phospho-DRP1 (Ser637) 1:1,000 (Cell Signaling Technology, 6319), phospho-DRP1 (Ser616) 1:1,000 (Cell Signaling Technology, 4494), OPA1 1:1,000 (Cell Signaling Technology, 80471), mitofusin-2 1:1,000 (Cell Signaling Technology, 9482), anti-mouse IgG, HRP-linked (Cell Signaling Technology, 7076) and anti-rabbit IgG, HRP-linked (Cell Signaling Technology, 7074). Blots were quantified by using ImageJ software. Relative protein levels were shown as ratiometric quantifications relative to the loading control or to total protein levels for phosphorylated proteins.
Metabolite extraction and liquid chromatography/tandem mass spectrometry analysis
Cells were grown in six-well plates for 36 h or 9 d, harvested in 250 µl of ice-cold methanol/acetonitrile 1:1 containing 1 ng/µl [U-13C6]glucose (internal standard, Sigma-Aldrich, 389374) and 1 ng/µl [U-13C5]glutamine (internal standard, Sigma-Aldrich, 605166), and spun at 20,000 × g for 5 min at 4°C. Supernatants were then passed through a regenerated cellulose filter, dried, and resuspended in 100 µl methanol for subsequent analysis. Amino acid quantification was performed through previous derivatization. Briefly, 50 µl of 5% phenyl isothiocyanate in 31.5% ethanol and 31.5% pyridine in water were added to 10 µl of each sample. Mixtures were then incubated with phenyl isothiocyanate solution for 20 min at RT, dried under N2 flow, and suspended in 100 µl of 5 mM ammonium acetate in methanol/H2O 1:1.
The identities of all metabolites were confirmed by using pure standards. Quantification of different metabolites was performed with a liquid chromatography/tandem mass spectrometry method using a C18 column (Biocrates) for amino acids, a Pursuit XRs Ultra 2.8 Diphenyl (Varian Medical Systems) for acyl-carnitine, and a cyano-phase LUNA column (50 mm × 4.6 mm, 5 µm; Phenomenex) for metabolites, respectively. Methanolic samples were analyzed by 10- and 3-min runs in positive (amino acids and acyl-carnitine, respectively) and 5-min runs in negative (all other metabolites) ion mode with a 35-multiple-reaction monitoring (MRM) transition in positive ion mode and a 30-MRM transition in negative ion mode, respectively. The mobile phases for the amino acid analysis were phase A: 0.2% formic acid in water; and phase B: 0.2% formic acid in acetonitrile. The gradient was T0 100% A, T5.5 min 5% A, T7 min 100% A with a flow rate of 500 µl/min. The mobile phase for the acyl-carnitine analysis was 0.1% formic acid in methanol under isocratic conditions with a flow rate of 300 µl/min. The mobile phase for negative ion mode analysis (all other metabolites) was phase A: water; and B: 2 mM ammonium acetate in methanol. The gradient was 90% B for all analyses with a flow rate of 500 µl/min.
Metabolomic data were acquired on an API-4000 triple quadrupole mass spectrometer (AB Sciex) coupled with an HPLC system (Agilent Technologies), a CTC PAL HTS autosampler (PAL System), and a Sciex Triple Quad 3500 (AB Sciex) with an HPLC system (AB Sciex) and a built-in autosampler (AB Sciex). MultiQuant software (version 3.0.2) was used for data analysis and peak review of chromatograms.
Metabolomic data were normalized by defining (relative metabolite area) as
where xn represents the peak areas of metabolite n for samples a, b, …, z, and represents the sum of peak areas of metabolite n for samples a, b, …, z.
Relative metabolite area () was then divided by the sum of relative metabolite areas analyzed in each sample to obtain the relative metabolite abundance (m), as
where represents the sum of relative metabolite areas 1, 2, …, n for sample a. Internal standards were used to control instrument sensitivity. ATP energy charge was calculated as
For metabolic tracing analyses, 2- and 8-d differentiated cells were exposed for 24 h to 1 mM [U-13C6]glucose (Sigma-Aldrich, 389374), 2 mM [U-13C5]glutamine (Sigma-Aldrich, 605166), or 100 µM [U-13C16]palmitate (Sigma-Aldrich, 605573). Metabolite quantification was performed as described above by increasing the MRM transitions in negative ion mode to 139 to analyze the different isotopomers.
Oxygen consumption measurements
Oxygen consumption rate analysis on whole cells was performed by using a Clark-type oxygen electrode (Hansatech, DW1 electrode chamber). C3H10T1/2 cells were rinsed in prewarmed PBS (37°C) and suspended in coupled respiration buffer (2% fatty acid–free BSA [wt/vol], 1 mM Na-pyruvate, 25 mM D-glucose, and 40 ng/ml digitonin in PBS) or electron flow buffer (2% fatty acid–free BSA [wt/vol], 10 mM Na-pyruvate, 2 mM malate, 4 µM CCCP, and 40 ng/ml digitonin in PBS). Samples were then transferred to the electrode chamber for the oxygen consumption rate measurement. After measuring basal respiration, uncoupled and maximal respiration were evaluated by adding 12.5 µM oligomycin and 5 µM CCCP, respectively. Complex I, II, and IV activities were evaluated through the electron flow protocol. Once transferred into the chamber, complex I activity was evaluated. After 20 µM rotenone and 1 mM succinate addition, we assessed complex II activity. We then added 150 µM antimycin A and 40 mM/1.6 mM ascorbate/N, N, N′, N′-tetramethyl-p-phenylenediamine to measure complex IV activity. All sample values were normalized to total protein content.
ATP production
Measurements of glycolytic and mitochondrial real-time ATP production in Scramble and ShZc3h10 cells were performed by using a Seahorse XFp real-time induced ATP Rate Assay Kit (Agilent Technologies, 103591–100) and an XFe24 Seahorse following the manufacturer’s instructions. To test weather metabolic pathways fueled ATP production, Scramble and ShZc3h10 preadipocytes were separately treated with 4 µM etomoxir, 2 µM UK5099, or 3 µM BPTES. 1.5 µM oligomycin and 0.5 µM rotenone/antimycin A were then serially injected. Data were analyzed by using the Seahorse XFp Real-Time ATP Rate Assay Report Generator and normalized to amounts of total protein.
Cytofluorimetric analyses
Adipocyte counts were performed as previously described (Higuchi et al., 2013). Cells were grown in 12-well plates and exposed to AdipoRed assay reagent (1:200, Lonza, PT-7009) for 15 min in the dark at 37°C. Successively, cells were washed and then harvested in 300 µl of ice-cold PBS. Cells were analyzed a with cytofluorimeter (Novocyte 3000, Acea, Agilent Technologies) at 485 nm excitation and 572 nm emission. Unstained cells were used as a negative control.
Confocal microscopy
Cells were cultured on Nunc glass-based dishes (27 mm; Thermo Fisher Scientific, 150682) and analyzed at different stages of differentiation. For staining, cell medium was replaced with staining medium—high-glucose DMEM without phenol red, without FBS, and supplemented with 1% glutamine and 1% pen/strep—containing different probes used for staining F-actin, mitochondria, lipid droplets, and nuclei. For F-actin staining, we incubated live cells with SiR-actin probe (λ Ex/Em: 652/674 nm) and verapamil (1:2,000 and 1:1,000, respectively) for 1 h at 37°C, 5% CO2 in the dark (Tebu-Bio, SC001). Mitochondria in live cells were stained with Mitotracker Red CM-H2XROS (100nM, λ Ex/Em: 579/599 nm) or with Mitotracker Deep Red FM (500 nM, λ Ex/Em: 644/665 nm) for 30 min at 37°C, 5% CO2 in the dark (Thermo Fisher Scientific, M7513, and Cell Signaling Technology, 8778). For lipid droplets, live cells were incubated with AdipoRed assay reagent (1:200, λ Ex/Em: 550/600 nm, Lonza, PT-7009) for 30 min at 37°C in the dark. Cells were washed in dye-free staining medium and images were acquired by confocal microscopy using an LSM 900 (Zeiss).
Confocal microscopy image processing and analysis
F-actin and mitochondrial morphologies were analyzed by adapting the protocol described by Valente et al. (2017). Briefly, mitochondria and F-actin images were analyzed by using Fiji software. Images were smoothed and binarized to analyze the area of single particles. Binary images were then skeletonized, and each skeleton was quantified to obtain the numbers of networks, junctions, and branches, as well as the length of each particle. Skeletonized particle quantification was performed with no elimination of loops and end points.
For lipid droplet size, we binarized and applied a watershed algorithm to the images. Lipid droplet areas were then obtained—with a minimal decimal value of 0.01—and the values were plotted in histograms.
Statistical analysis
All data are expressed as mean ± SD. The n reported in figures indicates the number of biologic replicates. P values were calculated from the indicated n independent experiments according to Vaux et al. (2012). Statistical analyses were performed with specific Student’s t tests, Mann-Whitney test, or one-way ANOVA followed by Dunnett’s or Tukey’s posttest using GraphPad Prism (version 6.0).
Data and software availability
The data discussed in this manuscript has been deposited in NCBI's Gene Expression Omnibus and is accessible through the GEO Series accession no. GSE147941 for the C3H10T1/2 4-sU-seq dataset.
Generally applicable gene-set enrichment (GAUGE) analysis was performed to evaluate significantly enriched gene clusters in C3H10T1/2 preadipocytes and mature adipocytes (Ge et al., 2018). Image quantifications were performed using ImageJ (https://imagej.nih.gov/ij/). Metabolomic mass spectrometric raw data were analyzed with MultiQuantsoftware (version 3.0.2; https://sciex.com/products/software/multiquant-software).
Online supplemental material
Fig. S1 shows results from additional experiments supplementing Fig. 1. Fig. S2 shows results from additional experiments supplementing Figs. 2 and 3. Fig. S3 shows results from additional experiments showing the effect of Zc3h10 on mitochondrial network and lipid accumulation during adipogenesis. Figs. S4 and S5 show results from additional experiments supplementing Figs. 5, 6, and 7. Table S1 lists information for downregulated or upregulated genes from 4-sU-seq analysis bound by Zc3h10 within 1 kb of the gene transcription start site and those harboring a 5′TOP motif.
Supplementary Material
lists information for downregulated or upregulated genes from 4-sU-seq analysis bound by Zc3h10 within 1 kb of the gene transcription start site and those harboring a 5′TOP motif.
Acknowledgments
We thank F. Giavarini for his valuable help with HPLC and mass spectrometry. We thank Dr. Enrique Saez (The Scripps Research Institute, La Jolla, CA) for critically reading our manuscript and for the helpful comments. We are indebted to Dr. Marta Marchesi, Dr. Daniela Rossi, and Ms. E. Desiderio Pinto for administrative assistance. We are extremely grateful to Amy Sullivan (Obrizus Communications) for her contribution to the editing of this manuscript.
This work is supported by a Giovanni Armenise-Harvard Foundation career development grant, Cariplo Foundation grant 2014-0991 (N. Mitro), the European Foundation for the Study of Diabetes/Lilly European Diabetes Research Program 2015 and Intramural Transition Grant (N. Mitro), and by a grant from Ministry of Education, University and Research (MIUR) Progetto Eccellenza (2018-2022).
The authors declare no competing financial interests.
Author contributions: N. Mitro, M. Audano, S. Pedretti, and E. De Fabiani developed the study, conceived the experiments, analyzed data, and wrote the manuscript. M. Audano, S. Pedretti, and S. Ligorio performed the majority of the biochemical, biologic, and molecular biology experiments. C. Bean prepared Opa1-overexpressing virus and analyzed data. D. Caruso, M. Audano, and S. Pedretti performed and analyzed data from the metabolomic and fluxomic experiments. S. Ghisletti, M. Russo, and S. Polletti performed the 4-sU-seq experiment. F. Gualdrini analyzed the data from the 4-sU-seq experiment and reanalyzed chromatin immunoprecipitation sequencing data. E. De Fabiani, D. Caruso, and M. Crestani analyzed data and participated in the interpretation of results. All authors read and edited the manuscript.
References
- Aitken, C.E., and Lorsch J.R.. 2012. A mechanistic overview of translation initiation in eukaryotes. Nat. Struct. Mol. Biol. 19:568–576. 10.1038/nsmb.2303 [DOI] [PubMed] [Google Scholar]
- Alaimo, A., Gorojod R.M., Beauquis J., Muñoz M.J., Saravia F., and Kotler M.L.. 2014. Deregulation of mitochondria-shaping proteins Opa-1 and Drp-1 in manganese-induced apoptosis. PLoS One. 9:e91848. 10.1371/journal.pone.0091848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amemiya, H.M., Kundaje A., and Boyle A.P.. 2019. The ENCODE Blacklist: Identification of Problematic Regions of the Genome. Sci. Rep. 9:9354. 10.1038/s41598-019-45839-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audano, M., Pedretti S., Cermenati G., Brioschi E., Diaferia G.R., Ghisletti S., Cuomo A., Bonaldi T., Salerno F., Mora M., et al. 2018. Zc3h10 is a novel mitochondrial regulator. EMBO Rep. 19:e45531. 10.15252/embr.201745531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austenaa, L.M., Barozzi I., Simonatto M., Masella S., Della Chiara G., Ghisletti S., Curina A., de Wit E., Bouwman B.A., de Pretis S., et al. 2015. Transcription of Mammalian cis-Regulatory Elements Is Restrained by Actively Enforced Early Termination. Mol. Cell. 60:460–474. 10.1016/j.molcel.2015.09.018 [DOI] [PubMed] [Google Scholar]
- Bachmann, V.A., Riml A., Huber R.G., Baillie G.S., Liedl K.R., Valovka T., and Stefan E.. 2013. Reciprocal regulation of PKA and Rac signaling. Proc. Natl. Acad. Sci. USA. 110:8531–8536. 10.1073/pnas.1215902110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackshear, P.J., Stumpo D.J., Carballo E., and Lawrence J.C. Jr. 1997. Disruption of the gene encoding the mitogen-regulated translational modulator PHAS-I in mice. J. Biol. Chem. 272:31510–31514. 10.1074/jbc.272.50.31510 [DOI] [PubMed] [Google Scholar]
- Chen, H., Detmer S.A., Ewald A.J., Griffin E.E., Fraser S.E., and Chan D.C.. 2003. Mitofusins Mfn1 and Mfn2 coordinately regulate mitochondrial fusion and are essential for embryonic development. J. Cell Biol. 160:189–200. 10.1083/jcb.200211046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cipolat, S., Martins de Brito O., Dal Zilio B., and Scorrano L.. 2004. OPA1 requires mitofusin 1 to promote mitochondrial fusion. Proc. Natl. Acad. Sci. USA. 101:15927–15932. 10.1073/pnas.0407043101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cristancho, A.G., and Lazar M.A.. 2011. Forming functional fat: a growing understanding of adipocyte differentiation. Nat. Rev. Mol. Cell Biol. 12:722–734. 10.1038/nrm3198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Pauw, A., Tejerina S., Raes M., Keijer J., and Arnould T.. 2009. Mitochondrial (dys)function in adipocyte (de)differentiation and systemic metabolic alterations. Am. J. Pathol. 175:927–939. 10.2353/ajpath.2009.081155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehses, S., Raschke I., Mancuso G., Bernacchia A., Geimer S., Tondera D., Martinou J.C., Westermann B., Rugarli E.I., and Langer T.. 2009. Regulation of OPA1 processing and mitochondrial fusion by m-AAA protease isoenzymes and OMA1. J. Cell Biol. 187:1023–1036. 10.1083/jcb.200906084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng, T., Szabo E., Dziak E., and Opas M.. 2010. Cytoskeletal disassembly and cell rounding promotes adipogenesis from ES cells. Stem Cell Rev. Rep. 6:74–85. 10.1007/s12015-010-9115-8 [DOI] [PubMed] [Google Scholar]
- Feng, H., Hu B., Liu K.W., Li Y., Lu X., Cheng T., Yiin J.J., Lu S., Keezer S., Fenton T., et al. 2011. Activation of Rac1 by Src-dependent phosphorylation of Dock180(Y1811) mediates PDGFRα-stimulated glioma tumorigenesis in mice and humans. J. Clin. Invest. 121:4670–4684. 10.1172/JCI58559 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Ge, S.X., Son E.W., and Yao R.. 2018. iDEP: an integrated web application for differential expression and pathway analysis of RNA-Seq data. BMC Bioinformatics. 19:534. 10.1186/s12859-018-2486-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghaben, A.L., and Scherer P.E.. 2019. Adipogenesis and metabolic health. Nat. Rev. Mol. Cell Biol. 20:242–258. 10.1038/s41580-018-0093-z [DOI] [PubMed] [Google Scholar]
- Giacomello, M., Pyakurel A., Glytsou C., and Scorrano L.. 2020. The cell biology of mitochondrial membrane dynamics. Nat. Rev. Mol. Cell Biol. 21:204–224. 10.1038/s41580-020-0210-7 [DOI] [PubMed] [Google Scholar]
- Gingras, A.C., Gygi S.P., Raught B., Polakiewicz R.D., Abraham R.T., Hoekstra M.F., Aebersold R., and Sonenberg N.. 1999. Regulation of 4E-BP1 phosphorylation: a novel two-step mechanism. Genes Dev. 13:1422–1437. 10.1101/gad.13.11.1422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingras, A.C., Raught B., Gygi S.P., Niedzwiecka A., Miron M., Burley S.K., Polakiewicz R.D., Wyslouch-Cieszynska A., Aebersold R., and Sonenberg N.. 2001. Hierarchical phosphorylation of the translation inhibitor 4E-BP1. Genes Dev. 15:2852–2864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griparic, L., Kanazawa T., and van der Bliek A.M.. 2007. Regulation of the mitochondrial dynamin-like protein Opa1 by proteolytic cleavage. J. Cell Biol. 178:757–764. 10.1083/jcb.200704112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gualdrini, F., Esnault C., Horswell S., Stewart A., Matthews N., and Treisman R.. 2016. SRF Co-factors Control the Balance between Cell Proliferation and Contractility. Mol. Cell. 64:1048–1061. 10.1016/j.molcel.2016.10.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halder, G., Dupont S., and Piccolo S.. 2012. Transduction of mechanical and cytoskeletal cues by YAP and TAZ. Nat. Rev. Mol. Cell Biol. 13:591–600. 10.1038/nrm3416 [DOI] [PubMed] [Google Scholar]
- Hall, A. 2012. Rho family GTPases. Biochem. Soc. Trans. 40:1378–1382. 10.1042/BST20120103 [DOI] [PubMed] [Google Scholar]
- Hansson, B., Morén B., Fryklund C., Vliex L., Wasserstrom S., Albinsson S., Berger K., and Stenkula K.G.. 2019. Adipose cell size changes are associated with a drastic actin remodeling. Sci. Rep. 9:12941. 10.1038/s41598-019-49418-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Head, B., Griparic L., Amiri M., Gandre-Babbe S., and van der Bliek A.M.. 2009. Inducible proteolytic inactivation of OPA1 mediated by the OMA1 protease in mammalian cells. J. Cell Biol. 187:959–966. 10.1083/jcb.200906083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heasman, S.J., and Ridley A.J.. 2008. Mammalian Rho GTPases: new insights into their functions from in vivo studies. Nat. Rev. Mol. Cell Biol. 9:690–701. 10.1038/nrm2476 [DOI] [PubMed] [Google Scholar]
- Higuchi, M., Dusting G.J., Peshavariya H., Jiang F., Hsiao S.T.F., Chan E.C., and Liu G.S.. 2013. Differentiation of human adipose-derived stem cells into fat involves reactive oxygen species and Forkhead box O1 mediated upregulation of antioxidant enzymes. Stem Cells Dev. 22:878–888. 10.1089/scd.2012.0306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe, A.K. 2004. Regulation of actin-based cell migration by cAMP/PKA. Biochim. Biophys. Acta. 1692:159–174. 10.1016/j.bbamcr.2004.03.005 [DOI] [PubMed] [Google Scholar]
- Hsieh, A.C., Liu Y., Edlind M.P., Ingolia N.T., Janes M.R., Sher A., Shi E.Y., Stumpf C.R., Christensen C., Bonham M.J., et al. 2012. The translational landscape of mTOR signalling steers cancer initiation and metastasis. Nature. 485:55–61. 10.1038/nature10912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson, R.J., Hellen C.U.T., and Pestova T.V.. 2010. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat. Rev. Mol. Cell Biol. 11:113–127. 10.1038/nrm2838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffe, A.B., and Hall A.. 2005. Rho GTPases: biochemistry and biology. Annu. Rev. Cell Dev. Biol. 21:247–269. 10.1146/annurev.cellbio.21.020604.150721 [DOI] [PubMed] [Google Scholar]
- Jefferies, H.B.J., Reinhard C., Kozma S.C., and Thomas G.. 1994. Rapamycin selectively represses translation of the “polypyrimidine tract” mRNA family. Proc. Natl. Acad. Sci. USA. 91:4441–4445. 10.1073/pnas.91.10.4441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen, K.B., Dredge B.K., Toubia J., Jin X., Iadevaia V., Goodall G.J., and Proud C.G.. 2020. capCLIP: a new tool to probe protein synthesis in human cells through capture and identification of the eIF4E-mRNA interactome. bioRxiv. doi: (Preprint posted April 18, 2020). 10.1101/2020.04.18.047571 [DOI] [PMC free article] [PubMed]
- Kashatus, D.F., Lim K.H., Brady D.C., Pershing N.L.K., Cox A.D., and Counter C.M.. 2011. RALA and RALBP1 regulate mitochondrial fission at mitosis. Nat. Cell Biol. 13:1108–1115. 10.1038/ncb2310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, J.E., and Chen J.. 2004. regulation of peroxisome proliferator-activated receptor-γ activity by mammalian target of rapamycin and amino acids in adipogenesis. Diabetes. 53:2748–2756. 10.2337/diabetes.53.11.2748 [DOI] [PubMed] [Google Scholar]
- Kim, D., Pertea G., Trapnell C., Pimentel H., Kelley R., and Salzberg S.L.. 2013. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14:R36. 10.1186/gb-2013-14-4-r36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamarche, N., Tapon N., Stowers L., Burbelo P.D., Aspenström P., Bridges T., Chant J., and Hall A.. 1996. Rac and Cdc42 induce actin polymerization and G1 cell cycle progression independently of p65PAK and the JNK/SAPK MAP kinase cascade. Cell. 87:519–529. 10.1016/S0092-8674(00)81371-9 [DOI] [PubMed] [Google Scholar]
- Langmead, B., and Salzberg S.L.. 2012. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 9:357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefterova, M.I., Haakonsson A.K., Lazar M.A., and Mandrup S.. 2014. PPARγ and the global map of adipogenesis and beyond. Trends Endocrinol. Metab. 25:293–302. 10.1016/j.tem.2014.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liesa, M., Palacín M., and Zorzano A.. 2009. Mitochondrial dynamics in mammalian health and disease. Physiol. Rev. 89:799–845. 10.1152/physrev.00030.2008 [DOI] [PubMed] [Google Scholar]
- Liu, J., DeYoung S.M., Zhang M., Zhang M., Cheng A., and Saltiel A.R.. 2005. Changes in integrin expression during adipocyte differentiation. Cell Metab. 2:165–177. 10.1016/j.cmet.2005.08.006 [DOI] [PubMed] [Google Scholar]
- Love, M.I., Huber W., and Anders S.. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550. 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcon, B.H., Holetz F.B., Eastman G., Origa-Alves A.C., Amorós M.A., de Aguiar A.M., Rebelatto C.K., Brofman P.R.S., Sotelo-Silveira J., and Dallagiovanna B.. 2017. Downregulation of the protein synthesis machinery is a major regulatory event during early adipogenic differentiation of human adipose-derived stromal cells. Stem Cell Res. (Amst.). 25:191–201. 10.1016/j.scr.2017.10.027 [DOI] [PubMed] [Google Scholar]
- Marcon, B.H., Shigunov P., Spangenberg L., Pereira I.T., de Aguiar A.M., Amorín R., Rebelatto C.K., Correa A., and Dallagiovanna B.. 2019. Cell cycle genes are downregulated after adipogenic triggering in human adipose tissue-derived stem cells by regulation of mRNA abundance. Sci. Rep. 9:5611. 10.1038/s41598-019-42005-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrick, W.C. 2015. eIF4F: a retrospective. J. Biol. Chem. 290:24091–24099. 10.1074/jbc.R115.675280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore, A.S., Wong Y.C., Simpson C.L., and Holzbaur E.L.F.. 2016. Dynamic actin cycling through mitochondrial subpopulations locally regulates the fission-fusion balance within mitochondrial networks. Nat. Commun. 7:12886. 10.1038/ncomms12886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murai, K.K., and Pasquale E.B.. 2003. 'Eph’ective signaling: forward, reverse and crosstalk. J. Cell Sci. 116:2823–2832. 10.1242/jcs.00625 [DOI] [PubMed] [Google Scholar]
- Nobusue, H., Onishi N., Shimizu T., Sugihara E., Oki Y., Sumikawa Y., Chiyoda T., Akashi K., Saya H., and Kano K.. 2014. Regulation of MKL1 via actin cytoskeleton dynamics drives adipocyte differentiation. Nat. Commun. 5:3368. 10.1038/ncomms4368 [DOI] [PubMed] [Google Scholar]
- Noguchi, M., Hosoda K., Fujikura J., Fujimoto M., Iwakura H., Tomita T., Ishii T., Arai N., Hirata M., Ebihara K., et al. 2007. Genetic and pharmacological inhibition of Rho-associated kinase II enhances adipogenesis. J. Biol. Chem. 282:29574–29583. 10.1074/jbc.M705972200 [DOI] [PubMed] [Google Scholar]
- O’Connor, K.L., and Mercurio A.M.. 2001. Protein kinase A regulates Rac and is required for the growth factor-stimulated migration of carcinoma cells. J. Biol. Chem. 276:47895–47900. 10.1074/jbc.M107235200 [DOI] [PubMed] [Google Scholar]
- Pernas, L., and Scorrano L.. 2016. Mito-Morphosis: Mitochondrial Fusion, Fission, and Cristae Remodeling as Key Mediators of Cellular Function. Annu. Rev. Physiol. 78:505–531. 10.1146/annurev-physiol-021115-105011 [DOI] [PubMed] [Google Scholar]
- Price, L.S., Leng J., Schwartz M.A., and Bokoch G.M.. 1998. Activation of Rac and Cdc42 by integrins mediates cell spreading. Mol. Biol. Cell. 9:1863–1871. 10.1091/mbc.9.7.1863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proud, C.G. 2007. Signalling to translation: how signal transduction pathways control the protein synthetic machinery. Biochem. J. 403:217–234. 10.1042/BJ20070024 [DOI] [PubMed] [Google Scholar]
- Rabani, M., Levin J.Z., Fan L., Adiconis X., Raychowdhury R., Garber M., Gnirke A., Nusbaum C., Hacohen N., Friedman N., et al. 2011. Metabolic labeling of RNA uncovers principles of RNA production and degradation dynamics in mammalian cells. Nat. Biotechnol. 29:436–442. 10.1038/nbt.1861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambold, A.S., Kostelecky B., Elia N., and Lippincott-Schwartz J.. 2011. Tubular network formation protects mitochondria from autophagosomal degradation during nutrient starvation. Proc. Natl. Acad. Sci. USA. 108:10190–10195. 10.1073/pnas.1107402108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangamani, P., Levy M.G., Khan S., and Oster G.. 2016. Paradoxical signaling regulates structural plasticity in dendritic spines. Proc. Natl. Acad. Sci. USA. 113:E5298–E5307. 10.1073/pnas.1610391113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romani, P., Brian I., Santinon G., Pocaterra A., Audano M., Pedretti S., Mathieu S., Forcato M., Bicciato S., Manneville J.-B.J.-B.B., et al. 2019. Extracellular matrix mechanical cues regulate lipid metabolism through Lipin-1 and SREBP. Nat. Cell Biol. 21:338–347. 10.1038/s41556-018-0270-5 [DOI] [PubMed] [Google Scholar]
- Salt, I.P., and Hardie D.G.. 2017. AMP-Activated Protein Kinase: An Ubiquitous Signaling Pathway With Key Roles in the Cardiovascular System. Circ. Res. 120:1825–1841. 10.1161/CIRCRESAHA.117.309633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiqui, N., and Sonenberg N.. 2015. Signalling to eIF4E in cancer. Biochem. Soc. Trans. 43:763–772. 10.1042/BST20150126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siersbæk, R., Madsen J.G.S., Javierre B.M., Nielsen R., Bagge E.K., Cairns J., Wingett S.W., Traynor S., Spivakov M., Fraser P., and Mandrup S.. 2017. Dynamic Rewiring of Promoter-Anchored Chromatin Loops during Adipocyte Differentiation. Mol. Cell. 66:420–435.e5. 10.1016/j.molcel.2017.04.010 [DOI] [PubMed] [Google Scholar]
- Smirnova, E., Griparic L., Shurland D.L., and van der Bliek A.M.. 2001. Dynamin-related protein Drp1 is required for mitochondrial division in mammalian cells. Mol. Biol. Cell. 12:2245–2256. 10.1091/mbc.12.8.2245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiegelman, B.M., and Farmer S.R.. 1982. Decreases in tubulin and actin gene expression prior to morphological differentiation of 3T3 adipocytes. Cell. 29:53–60. 10.1016/0092-8674(82)90089-7 [DOI] [PubMed] [Google Scholar]
- Spinelli, J.B., and Haigis M.C.. 2018. The multifaceted contributions of mitochondria to cellular metabolism. Nat. Cell Biol. 20:745–754. 10.1038/s41556-018-0124-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taguchi, N., Ishihara N., Jofuku A., Oka T., and Mihara K.. 2007. Mitotic phosphorylation of dynamin-related GTPase Drp1 participates in mitochondrial fission. J. Biol. Chem. 282:11521–11529. 10.1074/jbc.M607279200 [DOI] [PubMed] [Google Scholar]
- Tang, Q.Q., and Lane M.D.. 2012. Adipogenesis: from stem cell to adipocyte. Annu. Rev. Biochem. 81:715–736. 10.1146/annurev-biochem-052110-115718 [DOI] [PubMed] [Google Scholar]
- Tharp, K.M., Kang M.S., Timblin G.A., Dempersmier J., Dempsey G.E., Zushin P.H., Benavides J., Choi C., Li C.X., Jha A.K., et al. 2018. Actomyosin-Mediated Tension Orchestrates Uncoupled Respiration in Adipose Tissues. Cell Metab. 27:602–615.e4. 10.1016/j.cmet.2018.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoreen, C.C., Chantranupong L., Keys H.R., Wang T., Gray N.S., and Sabatini D.M.. 2012. A unifying model for mTORC1-mediated regulation of mRNA translation. Nature. 485:109–113. 10.1038/nature11083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tondera, D., Grandemange S., Jourdain A., Karbowski M., Mattenberger Y., Herzig S., Da Cruz S., Clerc P., Raschke I., Merkwirth C., et al. 2009. SLP-2 is required for stress-induced mitochondrial hyperfusion. EMBO J. 28:1589–1600. 10.1038/emboj.2009.89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tontonoz, P., Hu E., and Spiegelman B.M.. 1994. Stimulation of adipogenesis in fibroblasts by PPAR γ 2, a lipid-activated transcription factor. Cell. 79:1147–1156. 10.1016/0092-8674(94)90006-X [DOI] [PubMed] [Google Scholar]
- Tsukiyama-Kohara, K., Poulin F., Kohara M., DeMaria C.T., Cheng A., Wu Z., Gingras A.C., Katsume A., Elchebly M., Spiegelman B.M., et al. 2001. Adipose tissue reduction in mice lacking the translational inhibitor 4E-BP1. Nat. Med. 7:1128–1132. 10.1038/nm1001-1128 [DOI] [PubMed] [Google Scholar]
- Valente, A.J., Maddalena L.A., Robb E.L., Moradi F., and Stuart J.A.. 2017. A simple ImageJ macro tool for analyzing mitochondrial network morphology in mammalian cell culture. Acta Histochem. 119:315–326. 10.1016/j.acthis.2017.03.001 [DOI] [PubMed] [Google Scholar]
- Vaux, D.L., Fidler F., and Cumming G.. 2012. Replicates and repeats--what is the difference and is it significant? A brief discussion of statistics and experimental design. EMBO Rep. 13:291–296. 10.1038/embor.2012.36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Z., Jiang H., Chen S., Du F., and Wang X.. 2012. The mitochondrial phosphatase PGAM5 functions at the convergence point of multiple necrotic death pathways. Cell. 148:228–243. 10.1016/j.cell.2011.11.030 [DOI] [PubMed] [Google Scholar]
- Wilson-Fritch, L., Burkart A., Bell G., Mendelson K., Leszyk J., Nicoloro S., Czech M., and Corvera S.. 2003. Mitochondrial biogenesis and remodeling during adipogenesis and in response to the insulin sensitizer rosiglitazone. Mol. Cell. Biol. 23:1085–1094. 10.1128/MCB.23.3.1085-1094.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi, D., Dempersmier J.M., Nguyen H.P., Viscarra J.A., Dinh J., Tabuchi C., Wang Y., and Sul H.S.. 2019. Zc3h10 Acts as a Transcription Factor and Is Phosphorylated to Activate the Thermogenic Program. Cell Rep. 29:2621–2633.e4. 10.1016/j.celrep.2019.10.099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, F.X., Zhang Y., Park H.W., Jewell J.L., Chen Q., Deng Y., Pan D., Taylor S.S., Lai Z.C., and Guan K.L.. 2013. Protein kinase A activates the Hippo pathway to modulate cell proliferation and differentiation. Genes Dev. 27:1223–1232. 10.1101/gad.219402.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W., and Liu X.S.. 2008. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9:R137. 10.1186/gb-2008-9-9-r137 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
lists information for downregulated or upregulated genes from 4-sU-seq analysis bound by Zc3h10 within 1 kb of the gene transcription start site and those harboring a 5′TOP motif.
Data Availability Statement
The data discussed in this manuscript has been deposited in NCBI's Gene Expression Omnibus and is accessible through the GEO Series accession no. GSE147941 for the C3H10T1/2 4-sU-seq dataset.
Generally applicable gene-set enrichment (GAUGE) analysis was performed to evaluate significantly enriched gene clusters in C3H10T1/2 preadipocytes and mature adipocytes (Ge et al., 2018). Image quantifications were performed using ImageJ (https://imagej.nih.gov/ij/). Metabolomic mass spectrometric raw data were analyzed with MultiQuantsoftware (version 3.0.2; https://sciex.com/products/software/multiquant-software).