Abstract
We report the performance of a variety of commercially available SARS-CoV-2 PCR kits, used in several different sites across Ireland to determine if Ct values across platforms are comparable. We also investigate whether a Ct value, a surrogate for calculated viral loads in the absence of viral culture of > 34 can be used to exclude SARS-CoV-2 infection and its complications. We found a variation in Ct values from different assays for the same calculated viral load; this should be taken into consideration for result interpretation.
Keywords: SARS-CoV-2, RT-PCR, Cycle threshold
The interpretation of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) real-time (RT)-PCR tests presents multiple conundrums with respect to viral load to cause infection and be infectious, age, clinical phase (presymptomatic, symptomatic, asymptomatic, resolution, re-infection or persistent positivity), testing purposes (diagnostic or surveillance), trending of previous test results if available and use of test result (e.g. for infection prevention control or occupational health purposes). In recent months, various publications have suggested that the use of cycle threshold (Ct) values as surrogate for calculated viral load, may help in the management of patients [1-4].
In this study, we investigate if Ct values obtained by a variety of commercially available SARS-CoV-2 PCR kits, used in several different sites across Ireland, are comparable across platforms. We also explore whether a Ct value of > 34 [3], in the absence of viral culture, can be used to exclude SARS-CoV-2 infection.
Ethical statement
No patient data or specimens were used in this study, therefore, ethical approval was not required.
PCR platform comparison
In April 2020, Quality Control for Molecular Diagnostics (QCMD) produced a SARS-CoV-2 external quality assessment (EQA) panel [5]. The panel contained eight samples of which five were positive for SARS-CoV-2. Laboratories participating in the EQA were given the panel of samples without the respective information on positivity or negativity. Each laboratory processed the samples as if the provided material was viral transport media (VTM) from a SARS-CoV-2 inoculated swab, and tested the EQA panel according to their RT-PCR of choice and own laboratory procedures, then returned a ‘Detected’ or ‘Not detected’ result to QCMD. Following submission of results from all participants, in June 2020, QCMD provided a report to all participants, now detailing the digital (d)PCR log10 copies/mL of SARS-CoV-2 in the samples for reference purpose. The dPCR log10 copies/mL for the five SARS-CoV-2 positive samples were 4.3, 3.3, 4.3, 5.3, and 2.3 respectively.
In this study, we analyse the results of 16 participating clinical diagnostic laboratories across Ireland in more detail, using in particular the Ct values that they obtained with their RT-PCR assays for each of the five positive samples. For each laboratory, the Ct values for the five samples and the corresponding dPCR log10 copies/mL were employed to produce standard curves for each assay (or for each assay target). When several laboratories used a common assay, this allowed to assess the performance of the same assay across the platforms. Moreover, when laboratories used different assays, it was possible to compare outputs across assays. Six for 16 laboratories submitted data on more than one assay. In total nine assays with in total 15 gene targets were analysed (Table 1).
Table 1. Real-time PCR assays considered in the study, Ireland, June 2020 (n = 9 assays).
| Assay | Number of laboratories | Number of gene targets | Genes targeted | Total datasets generated |
|---|---|---|---|---|
| GeneXpert (Cepheid, Sunnyvale, California, United States) | 6 | 2 | E gene N gene |
12 |
| Logix Smart (Co-Diagnostics, Inc, Salt Lake City, Utah, United States) | 2 | 1 | RdRp gene | 2 |
| Cobas 4800 (Roche Diagnostics, Basel, Switzerland) | 2 | 2 | ORF1a/b E gene |
4 |
| RealStar (Altona DiagnosticsGmbH, Hamburg, Germany) | 1 | 1 | E gene | 1 |
| genesig (Primerdesign, Southampton, Hants, United Kingdom) | 4 | 1 | ORF1a/b | 4 |
| RespiBio (Serosep, Limerick, Ireland) | 2 | 1 | RdRp gene | 2 |
| VIASURE (CerTest Biotec, Zaragoza, Spain) | 3 | 2 | ORF1a/b N gene |
3 (2 genes combined) |
| Abbott RealTime SARS-CoV-2 (Abbott Park, Illinois, United States) | 1 | 2 | RdRp gene N gene |
1 (2 genes combined) |
| Allplex SARS-CoV-2 (Seegene, Seoul, South Korea) | 1 | 3 | RdRp gene N gene E gene |
3 |
E: envelope; N: nucleocapsid; ORF: open reading frame; RdRp: RNA-dependent RNA polymerase; SARS-CoV-2: severe acute respiratory syndrome coronavirus 2.
There were a number of different RNA extraction systems, either prior to the RT-PCR or incorporated within this procedure (no external extraction for GeneXpert (Cepheid)). Each of these systems used a different amount of sample (200 μL to 750 μL), with a varying portion of the total recovered RNA as subsequent PCR template. A standard curve was created in Microsoft Excel for each assay (one for each gene target). The standard curve was created by plotting the Ct value for each of the five samples against the dPCR log10 copies/mL provided by QCMD. R2 values of all the assays ranged from 0.9497 to 0.9997, with a mean of 0.9885. From each standard curve, estimated viral load (log10 copies per mL) was extrapolated (using the equation of the individual standard curve) for each Ct value. The Abbott RealTime SARS-CoV-2 assay reports cycle number (CN) values, which are not equivalent to Ct values and thus are not directly comparable [6] and was therefore excluded from further analysis.
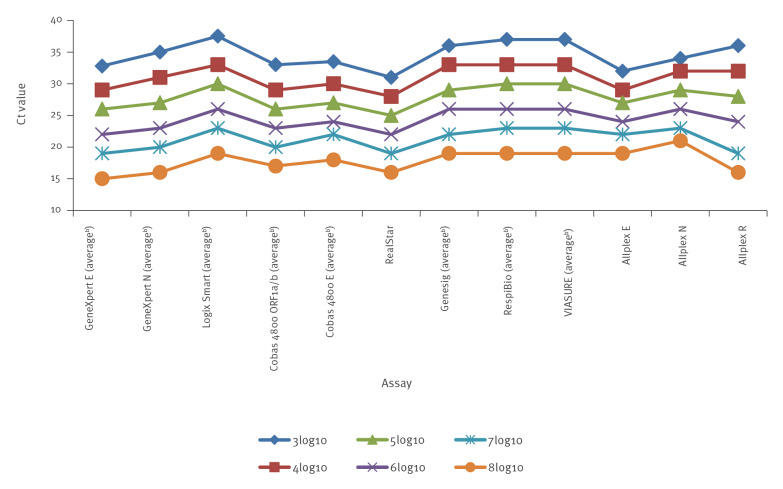
Mean Ct values for all assays and standard deviations were calculated across a range of log10 copies per mL values (Table 2 and 3). Where only one laboratory tested an assay, absolute values were used for comparison. Correlation of Ct results between the same assays used across different sites was good for all assays (mean: 1.6; standard deviation: 0–5.1) (Figure, Table 2). Data from all assays correlated with the internationally recognised 3.3 cycle difference for every 1log10 copies/mL change in viral load. However, there was a wide variation in Ct values for different assays for the same viral loads, 6.5 cycle difference (31–37.5) at 3log10 down to 4 cycle difference (22–26) at 6log10. But the range difference in Ct values between assays was stable across all log values (Figure). These data demonstrate that reporting Ct values per se can be misleading and is non comparative between different assays, unless the Ct value is correlated with the calculated viral load for the particular assay used and also reported.
Table 2. Average and standard deviation of Ct values at calculated SARS-CoV-2 viral loads of 3log10 to 8log10 copies/mL, Ireland, June 2020 (n = 8 assays)a .
| Assay | Average Ct @ 3log10 (SD) |
Average Ct @ 4log10 (SD) |
Average Ct @ 5log10 (SD) |
Average Ct @ 6log10 (SD) |
Average Ct @ 7log10 (SD) |
Average Ct @ 8log10 (SD) |
|
|---|---|---|---|---|---|---|---|
| GeneXpert | E | 32.8 (1.32) | 29 (0.57) | 26 (0.63) | 22 (0.72) | 19 (0.79) | 15 (0.89) |
| N | 35 (1.34) | 31 (0.27) | 27 (0.21) | 23 (0.20) | 20 (0.22) | 16 (0.28) | |
| Logix Smart | RdRp | 37.5 (0.70) | 33 (0.29) | 30 (0.14) | 26 (0.05) | 23 (0.20) | 19 (0.40) |
| Cobas 4800 | ORF1a/b | 33 (0.19) | 29 (0.02) | 26 (0.03) | 23 (0.13) | 20 (0.2) | 17 (0.27) |
| E gene | 33.5 (0.7) | 30 (0.05) | 27 (0.03) | 24 (0.10) | 22 (0.15) | 18 (0.25) | |
| RealStarb | E | 31 (NA) | 28 (NA) | 25 (NA) | 22 (NA) | 19 (NA) | 16 (NA) |
| genesig | ORF1a/b | 36 (1.63) | 33 (0.43) | 29 (0.72) | 26 (0.97) | 22 (1.31) | 19 (1.59) |
| RespiBio | RdRp | 37 (1.41) | 33 (0.03) | 30 (0.22) | 26 (0.54) | 23 (0.79) | 19 (1.12) |
| VIASURE | ORF1a/b N (combined) | 37 (3.78) | 33 (0.96) | 30 (0.87) | 26 (0.99) | 23 (1.21) | 19 (1.6) |
| Allplex SARS-CoV-2b | E | 32 (NA) | 29 (NA) | 27 (NA) | 24 (NA) | 22 (NA) | 19 (NA) |
| N | 34 (NA) | 32 (NA) | 29 (NA) | 26 (NA) | 23 (NA) | 21 (NA) | |
| RdRp | 36 (NA) | 32 (NA) | 28 (NA) | 24 (NA) | 19 (NA) | 16 (NA) | |
Ct: cycle threshold; E: envelope; NA: not applicable; N: nucleocapsid; ORF: open reading frame; RdRp: RNA-dependent RNA polymerase; SARS-CoV-2: severe acute respiratory syndrome coronavirus 2; SD: standard deviation.
a While a total of nine SARS-CoV-2 real-time PCR assays were used in the study-participating laboratories, the Abbott RealTime SARS-CoV-2 assay reports cycle number values, which are not equivalent to Ct values and thus are not directly comparable. This assay was therefore excluded from the analysis.
b Absolute values for Altona and Seegene targets as only data from one laboratory.
Table 3. Average SARS-CoV-2 viral loads of log10 copies/mL and standard deviation at selected Ct values, Ireland, June 2020 (n = 8 assays)a .
| Ct | Average log10 (standard deviation) (copies/mL) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GeneXpert (Cepheid) |
Logix Smart (Co-Diagnostics Inc) |
Cobas 4800 (Roche) |
RealStar (Altona Diagnostics)b |
genesig (Primerdesign) |
RespiBio (Serosep) |
VIASURE (CerTest Biotec) |
Allplex SARS-CoV-2 (Seegene) |
|||||
| E | N | RdPd | E | ORF1a/b | E | ORF1a/b | RdPd | ORF1a/b, N (combined) | Eb | Nb | RdRpb | |
| 30 | 3.84 (0.54) | 4.23 (0.27) | 4.99 (0.19) | 3.63 (0.07) | 4.10 (0.11) | 3.50 (NA) | 4.77 (0.56) | 5.02 (0.14) | 4.95 (0.88) | 3.77 (NA) | 4.64 (NA) | 4.48 (NA) |
| 31 | 3.55 (0.53) | 3.97 (0.28) | 4.70 (0.24) | 3.30 (0.09) | 3.76 (0.13) | 3.19 (NA) | 4.48 (0.49) | 4.74 (0.05) | 4.67 (0.92) | 3.37 (NA) | 4.27 (NA) | 4.24 (NA) |
| 32 | 3.26 (0.52) | 3.70 (0.30) | 4.42 (0.29) | 2.97 (0.12) | 3.41 (0.16) | 2.87 (NA) | 4.19 (0.43) | 4.46 (0.03) | 4.38 (0.96) | 2.98 (NA) | 3.90 (NA) | 4.01 (NA) |
| 33 | 2.97 (0.50) | 3.43 (0.32) | 4.14 (0.34) | 2.64 (0.14) | 3.07 (0.19) | 2.55 (NA) | 3.91 (0.38) | 4.17 (0.11) | 4.10 (1.02) | 2.58 (NA) | 3.54 (NA) | 3.77 (NA) |
| 34 | 2.69 (0.50) | 3.17 (0.34) | 3.86 (0.39) | 2.31 (0.17) | 2.72 (0.22) | 2.24 (NA) | 3.62 (0.35) | 3.89 (0.19) | 3.82 (1.09) | 2.19 (NA) | 3.17 (NA) | 3.53 (NA) |
| 35 | 2.40 (0.49) | 2.90 (0.36) | 3.58 (0.44) | 1.98 (0.19) | 2.38 (0.24) | 1.92 (NA) | 3.33 (0.34) | 3.61 (0.27) | 3.53 (1.17) | 1.79 (NA) | 2.81 (NA) | 3.30 (NA) |
| 36 | 2.11 (0.49) | 2.63 (0.38) | 3.30 (0.49) | 1.65 (0.22) | 2.04 (0.27) | 1.61 (NA) | 3.05 (0.35) | 3.32 (0.36) | 3.25 (1.26) | 1.40 (NA) | 2.44 (NA) | 3.06 (NA) |
| 37 | 1.83 (0.54) | 2.37 (0.27) | 3.02 (0.19) | 1.32 (0.07) | 1.69 (0.11) | 1.29 (NA) | 2.76 (0.56) | 3.04 (0.14) | 2.97 (0.88) | 1.00 (NA) | 2.08 (NA) | 2.83 (NA) |
Ct: cycle threshold; E: envelope; NA: not applicable; N: nucleocapsid; ORF: open reading frame; RdRp: RNA-dependent RNA polymerase; SARS-CoV-2: severe acute respiratory syndrome coronavirus 2.
a While a total of nine SARS-CoV-2 real-time PCR assays were used in the study-participating laboratories, the Abbott RealTime SARS-CoV-2 assay reports cycle number values, which are not equivalent to Ct values and thus are not directly comparable. This assay was therefore excluded from the analysis.
b Absolute values for Altona and Seegene targets as only data from one laboratory.
Figure.
Average Ct values at SARS-CoV-2 viral loads of 3log10 to 8log10 copies/mL, Ireland, June 2020 (n = 8 assays)a
Ct: cycle threshold; E: envelope; N: nucleocapsid; ORF: open reading frame; R: RNA-dependent RNA polymerase; SARS-CoV-2: severe acute respiratory syndrome coronavirus 2.
a While a total of nine SARS-CoV-2 real-time PCR assays were used in the study-participating laboratories, the Abbott RealTime SARS-CoV-2 assay reports cycle number values, which are not equivalent to Ct values and thus are not directly comparable. This assay was therefore excluded from the analysis.
b When more than one laboratory used an assay (or targets in the assay), the average Cts obtained across the laboratories are presented for each assay (or assay target).
Average Ct values at calculated viral loads of 3log10 to 8log10 copies/mL.
Discussion
Inferred viral loads
Tom et al. 2020 [3] noted that the issue of high Ct values can be problematic for clinicians, especially when there are less than 100 copies of the virus present, as this could reflect presymptomatic, early infection, late infection, persistent positivity or nonviable virus. From our data even at a Ct as high as 37, six of eight assays had at least one gene target correlating to calculated viral load of ≥ 100 copies/mL, although our study did not include viral culture, nor infer cultureability. The Ct cut off of 34 described by La Scola et al. [7] at which they propose patients can be discharged from isolation may need to be considered cautiously, as our data show that a Ct value of 34 has a range of calculated viral loads from 2.19log10 to 3.89log10 (equivalent to approximately 150 – > 7000 viral copies per mL (Table 3).
Our National coronavirus disease (COVID-19) guidelines [1,2] also describe the difficulty of interpreting positive ‘high’ (> 30) Ct PCR results from asymptomatic individuals, they too suggest that a Ct of 34 equates to < 100 copies/mL, however, our data indicate that the Ct value could be up to 38 for a calculated viral load of 100 copies/mL (data not shown).
A number of different RNA extraction systems were used by the participating laboratories, the effect this has on the results is unquantifiable. While it is a limitation of this study, our results support the view of Chik-Yan et al. [8] who state that differences in Ct values may be due to differences in specimen source or preparation or differences in cycling parameters and reagents, even though there is no significant difference in sensitivity. Indeed, for our data there was good correlation in Ct results using the same assay at different sites. Another limitation of the current study is that it investigated a small number of laboratories, with only one to six laboratories using the same assay (giving either absolute Ct values or averages based on small number of replicates). A larger study in the future would be useful to support these results.
Consensus on the correlation between Ct value and disease severity has not been reached, Sang Hyun Ra et al. [9] found that there was no significant difference in mean Ct values from symptomatic or asymptomatic cases, whereas Salvatore et al. [4] reported higher Ct values in asymptomatic individuals. Likewise Prubelli et al. [10] identified an increase in Ct values that correlated with a decrease in severe cases in Italy. While diagnostic test results play a role in identification and aid management of infected individuals, it is imperative to have a thorough understanding of the performance characteristics of individual PCR assays to aid the accurate interpretation of results [11].
Cycle threshold values in patient management
Using Ct values to influence patient management is complex and must be done with caution. Including the Ct value on positive results may be confusing and misleading [12]. With no clearly defined infectious dose for SARS-CoV-2, viral culture not available routinely and Ct values differing by up to 6.5 cycles between platforms, one would question the value of the routine use of reporting Ct values for patient management. This is particularly challenging in a community testing setting, where frequently there is no accompanying patient clinical information to aid interpretation. However, this may be ameliorated in an acute hospital testing environment with access to patient clinical data. In this setting, analysis of Ct trends (either rising or decreasing) from repeat testing using the same assay may give more insight to an individual’s disease progression or resolution.
To aid the clinical interpretative value of COVID-19 PCR results, we agree with Tom et al. that binary reporting (‘Detected’ or ‘Not detected’) could be enhanced by the additional reporting of Ct value ranges in ‘high’, ‘medium’, ‘low’ categories . However, our data infer that such ranges if used, should be based on calculated viral loads of each assay used, not on absolute Ct values. The viral loads for ‘high’, ‘medium’ and ‘low’ categories would need to be defined. This reporting would be more accurate and informative to aid clinical and public health decisions, particularly when considered in the context of individual clinical data.
Acknowledgements
We would like to thank all contributing laboratories; The Coombe Women & Infants University Hospital, University Hospital Waterford, Our Lady's Hospital Navan, PHL-HSE-Dublin, Cork University Hospital, University Hospital Limerick, Midland Regional Hospital Portlaoise, University Hospital Kerry, Children's Health Ireland (CHI) at Crumlin, UCD National Virus Reference Laboratory, Letterkenny University Hospital, University Hospital Galway (UHG), The Meath Foundation Tallaght University Hospital, Tallaght, Midland Regional Hospital at Tullamore, The Mater Misericordiae University Hospital and Rotunda Hospital.
Conflict of interest: None declared.
Authors’ contributions: Anne Carroll: study design, data collection, data analysis and manuscript preparation. Eleanor McNamara: study design, critically revised manuscript.
References
- 1.Health Service Executive (HSE). HSE 2020, Proposal for the management of weak positive (high Ct value) PCR results in the setting of mass testing of asymptomatic individuals for SARS-CoV-2, V1.1 08.10.2020. Ireland: HSE; Dec 2020. Available from: https://www.hpsc.ie/az/respiratory/coronavirus/novelcoronavirus/guidance/outbreakmanagementguidance/PCR%20weak%20results%20guidance.pdf
- 2.Health Service Executive (HSE) Health Protection Surveillance Centre. (HPSC). Guidance on the management of weak positive (high Ct value) PCR results in the setting of testing individuals for SARS-CoV-2 V1.2 22.12.2020. Ireland: HSE HPSC. Available from: https://www.hpsc.ie/a-z/respiratory/coronavirus/novelcoronavirus/guidance/outbreakmanagementguidance/PCR%20weak%20results%20guidance.pdf
- 3. Tom MR, Mina MJ. To Interpret the SARS-CoV-2 Test, Consider the Cycle Threshold Value. Clin Infect Dis. 2020;71(16):2252-4. 10.1093/cid/ciaa619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Salvatore PP, Dawson P, Wadhwa A, Rabold EM, Buono S, Dietrich EA, et al. Epidemiological Correlates of Polymerase Chain Reaction Cycle Threshold Values in the Detection of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2). Clin Infect Dis. 2020;ciaa1469. 10.1093/cid/ciaa1469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Quality Control for Molecular Diagnostics (QCMD). [Accessed 11 Feb 2021]. Available from: http://www.qcmd.org/
- 6. Marais G, Naidoo M, Hsiao NY, Valley-Omar Z, Smuts H, Hardie D. The implementation of a rapid sample preparation method for the detection of SARS-CoV-2 in a diagnostic laboratory in South Africa. PLoS One. 2020;15(10):e0241029. 10.1371/journal.pone.0241029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. La Scola B, Le Bideau M, Andreani J, Hoang VT, Grimaldier C, Colson P, et al. Viral RNA load as determined by cell culture as a management tool for discharge of SARS-CoV-2 patients from infectious disease wards. Eur J Clin Microbiol Infect Dis. 2020;39(6):1059-61. 10.1007/s10096-020-03913-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yip CC, Sridhar S, Cheng AK, Leung KH, Choi GK, Chen JH, et al. Evaluation of the commercially available LightMix® Modular E-gene kit using clinical and proficiency testing specimens for SARS-CoV-2 detection. J Clin Virol. 2020;129:104476. 10.1016/j.jcv.2020.104476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ra SH, Lim JS, Kim GU, Kim MJ, Jung J, Kim SH. Upper respiratory viral load in asymptomatic individuals and mildly symptomatic patients with SARS-CoV-2 infection. Thorax. 2021;76(1):61-3. 10.1136/thoraxjnl-2020-215042 [DOI] [PubMed] [Google Scholar]
- 10. Piubelli C, Deiana M, Pomari E, Silva R, Bisoffi Z, Formenti F, et al. Overall decrease in SARS-CoV-2 viral load and reduction in clinical burden: the experience of a hospital in northern Italy. Clin Microbiol Infect. 2021;27(1):131.e1-3. 10.1016/j.cmi.2020.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Buchan BW, Hoff JS, Gmehlin CG, Perez A, Faron ML, Munoz-Price LS, et al. Distribution of SARS-CoV-2 PCR Cycle Threshold Values Provide Practical Insight Into Overall and Target-Specific Sensitivity Among Symptomatic Patients. Am J Clin Pathol. 2020;154(4):479-85. 10.1093/ajcp/aqaa133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Binnicker MJ. Challenges and Controversies to Testing for COVID-19. J Clin Microbiol. 2020;58(11):e01695-20. 10.1128/JCM.01695-20 [DOI] [PMC free article] [PubMed] [Google Scholar]