Abstract
Metaviridae is a family of retrotransposons and reverse-transcribing viruses with long terminal repeats belonging to the order Ortervirales. Members of the genera Errantivirus and Metavirus include, respectively, Saccharomyces cerevisiae Ty3 virus and its Gypsy-like relatives in drosophilids. This is a summary of the International Committee on Taxonomy of Viruses (ICTV) Report on the family Metaviridae, which is available at ictv.global/report/metaviridae.
Keywords: Metaviridae, ICTV, taxononmy, retrotransposon
Virion
The morphology of virus-like particles (VLPs) is poorly characterized; a high-resolution structure is only available for Saccharomyces cerevisiae Ty3 virus (Table 1, Fig. 1). Saccharomyces cerevisiae Ty3 VLPs are icosahedral (T=9) and built from a capsid protein, which is homologous to the corresponding protein of retroviruses and other members of the order Ortervirales [1–3]. Most VLPs do not appear to be infectious extracellularly, although Drosophila melanogaster Gypsy virus does generate infective VLPs [4]. By analogy with members of the family Retroviridae, VLPs are thought to contain two copies of the viral RNA genome, complexed with the nucleocapsid protein, cellular tRNAs involved in the priming of reverse transcription, reverse transcriptase and integrase. Virions of Drosophila melanogaster Gypsy virus also contain the envelope glycoprotein responsible for recognition and binding of VLPs to host cells and fusion of the viral and cellular membranes [5].
Table 1.
Characteristics of members of the family Metaviridae
|
Typical member: |
Saccharomyces cerevisiae Ty3 virus (M34549), species Saccharomyces cerevisiae Ty3 virus, genus Metavirus |
|---|---|
|
Virion |
Virions are icosahedral (T=9) and might be enveloped |
|
Genome |
Two identical copies of linear single-stranded, positive-sense RNA |
|
Replication |
Replication by reverse-transcription primed with a host-encoded tRNA |
|
Translation |
Genomic RNA is translated into one or more polyproteins |
|
Host range |
Fungi, plants and animals |
|
Taxonomy |
Realm Riboviria, kingdom Pararnavirae, phylum Artverviricota, class Revtraviricetes, order Ortervirales, family Metaviridae; the genera Errantivirus and Metavirus include >30 species |
Fig. 1.
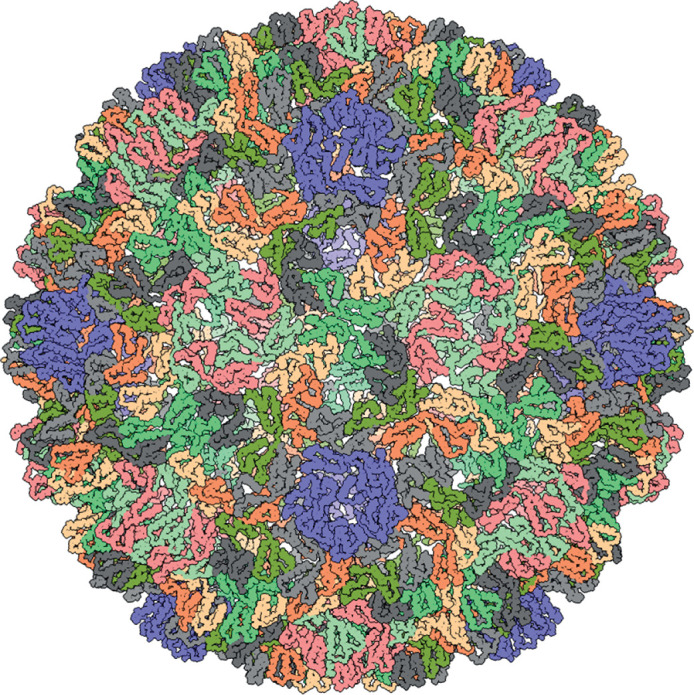
Three-dimensional reconstruction of a VLP of Saccharomyces cerevisiae Ty3. Diameter about 47 nm. PDB id: 6R24 [1].
Genome
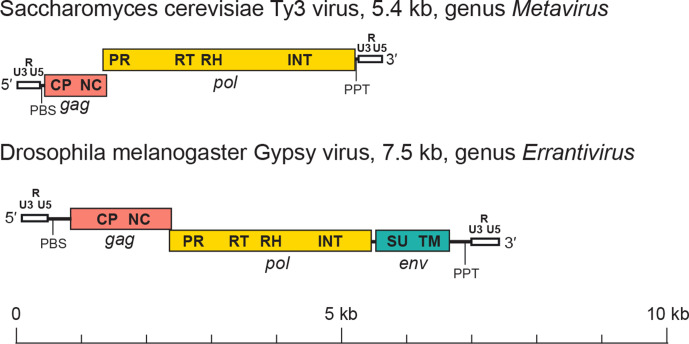
The genome of a canonical member of the Metaviridae has an internal region that may range from 3 kb to > 15 kb flanked by two homologous non-coding sequences called long terminal repeats (LTRs) (Fig. 2). A canonical LTR has three subregions, U3-R-U5, which are analogous to those of retroviruses. ‘U3’ (200–1200 nt) contains the promoters; ‘R’ is repeated on each end of the transcript; and ‘U5’ (75–250 nt) constitutes the first portion of the reverse-transcribed genome. The internal region is delimited by two small motifs: an 18 nt sequence downstream of the 5′-LTR (the primer binding site, PBS) and about ten A/G residues located upstream of the 3′-LTR (the polypurine tract, PPT). The internal region may have one (gag-pol), two (gag and pol) or three (gag, pol and env) genes. The gag gene encodes domains for the capsid and the nucleocapsid proteins, whereas the pol gene includes domains for the protease, reverse transcriptase, ribonuclease H and integrase proteins. Where present, envelope (Env) polyproteins contain transmembrane and surface domains and are encoded downstream of the integrase domain [6, 7].
Fig. 2.
Metavirus genome structure. LTRs (white) are labelled with the U3, R and U5 regions. Other labels are PBS (primer binding site), PPT (polypurine tract), gag (pink) with its capsid (CP) and nucleocapsid (NC) domains, pol (yellow) with its protease (PR), reverse transcriptase (RT), ribonuclease H (RH) and integrase (INT) domains, and env (blue-green) with its surface (SU) and transmembrane (TM) domains.
Replication
Members of the family Metaviridae replicate via reverse transcription within intracellular VLPs. The cellular tRNA packaged in the VLP anneals to the RNA genome in the PBS region complementary to the 3′-end of that tRNA and is used by reverse transcriptase as an initiation primer. The proviral cDNA is imported into the nucleus, followed by integration into a chromosomal target site by the integrase.
Taxonomy
Current taxonomy: ictv.global/taxonomy. The genera Errantivirus and Metavirus differ by the presence or absence of the env gene, respectively. However, this criterion is inconsistent with current knowledge of the evolutionary history of the family Metaviridae. While the genus Errantivirus constitutes a monophyletic clade, members of the genus Metavirus, form distinct clades showing polyphyletic relationships with each other. Furthermore, not all errantiviruses have env genes and, conversely, certain members of the genus Metavirus have env genes. A revision of the Metaviridae is required.
Resources
Full ICTV Report on the family Metaviridae: ictv.global/report/metaviridae
Gypsy Database (GyDB) devoted to viruses and mobile genetic elements: http://gydb.org
Funding information
Beatriz Soriano was supported by the pre-doctoral research fellowship from Industrial Doctorates of MINECO (Grant 659 DI-17–09134). Production of this Profile, the ICTV Report, and associated resources was funded by a grant from the Wellcome Trust (WT108418AIA).
Acknowledgements
Members of the ICTV Report Consortium are Stuart G. Siddell, Andrew J. Davison, Elliot J. Lefkowitz, Sead Sabanadzovic, Peter Simmonds, Donald B. Smith, Richard J. Orton and Balázs Harrach.
Conflicts of interest
The authors declare that there are no conflicts of interest
Footnotes
Abbreviations: LTR, long terminal repeat; PBS, primer binding site; PPT, polypurine tract; VLP, virus-like particle.
References
- 1.Dodonova SO, Prinz S, Bilanchone V, Sandmeyer S, Briggs JAG. Structure of the Ty3/Gypsy retrotransposon capsid and the evolution of retroviruses. Proc Natl Acad Sci USA. 2019;116:10048–10057. doi: 10.1073/pnas.1900931116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Krupovic M, Koonin EV. Homologous capsid proteins testify to the common ancestry of retroviruses, caulimoviruses, pseudoviruses, and metaviruses. J Virol. 2017;91:e00210-17. doi: 10.1128/JVI.00210-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Krupovic M, Blomberg J, Coffin JM, Dasgupta I, Fan H, et al. Ortervirales: new virus order unifying five families of reverse-transcribing viruses. J Virol. 2018;92:e00515-18. doi: 10.1128/JVI.00515-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim A, Terzian C, Santamaria P, Pélisson A, Purd'homme N, et al. Retroviruses in invertebrates: the gypsy retrotransposon is apparently an infectious retrovirus of Drosophila melanogaster . Proc Natl Acad Sci USA. 1994;91:1285–1289. doi: 10.1073/pnas.91.4.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Song SU, Gerasimova T, Kurkulos M, Boeke JD, Corces VG. An env-like protein encoded by a Drosophila retroelement: evidence that gypsy is an infectious retrovirus. Genes Dev. 1994;8:2046–2057. doi: 10.1101/gad.8.17.2046. [DOI] [PubMed] [Google Scholar]
- 6.Wright DA, Voytas DF. Athila4 of Arabidopsis and Calypso of soybean define a lineage of endogenous plant retroviruses. Genome Res. 2002;12:122–131. doi: 10.1101/gr.196001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pantazidis A, Labrador M, Fontdevila A. The retrotransposon Osvaldo from Drosophila buzzatii displays all structural features of a functional retrovirus. Mol Biol Evol. 1999;16:909–921. doi: 10.1093/oxfordjournals.molbev.a026180. [DOI] [PubMed] [Google Scholar]