Abstract
The bone marrow microenvironment contains cellular niches that maintain the pool of hematopoietic stem and progenitor cells and support hematopoietic maturation. Malignant hematopoietic cells also co-opt normal cellular interactions in order to promote their own growth and evade therapy. In vivo systems to study human hematopoiesis have mostly been achieved through transplantation into immunodeficient mouse models. However, incomplete cross-compatibility between the murine stroma and transplanted human hematopoietic cells limit the rate of engraftment and the study of relevant interactions. To supplement in vivo xenotransplantation models, complementary strategies have recently been developed, including the use of three-dimensional human bone marrow organoids in vivo, generated from bone marrow stromal cells seeded onto osteo-inductive scaffolds, as well as the use of ex vivo bioreactor models. These topics were the focus of the Spring 2020 International Society for Experimental Hematology New Investigator webinar. We review here the latest advances in generating humanized hematopoietic organoids and how they allow for the study of novel microenvironmental interactions.
Keywords: Ossicle, bioreactor, hematopoiesis, hematopoietic stem cell, niche
Introduction
Hematopoietic stem and progenitor cells (HSPCs), primarily residing within the bone marrow (BM), sustain life-long hematopoiesis1–4. The BM microenvironment is composed of a range of cell types – from mesenchymal stromal cells (MSCs) and endothelium to various mature hematopoietic cell types – and provide various supportive extrinsic cues5, 6. The accumulation of somatic mutations within HSPCs is well described as driver of a range of hematological disorders such as myelodysplastic syndromes (MDS), myeloproliferative neoplasms (MPNs), and leukemias7–12. However, there is increasing recognition for the interplay between mutant HSPCs and altered BM stroma in the development and progression of hematological diseases13.
While hematopoietic niches have long been studied in model organisms, particularly mouse and zebrafish, understanding the microenvironmental interactions specific to human hematopoiesis and leukemogenesis are essential to the development of new therapeutic strategies. Replicating the human BM microenvironment in vitro is challenging and, correspondingly, supporting primary HSPCs in culture has also remained difficult14. Additionally, how purified populations of HSPCs grow ex vivo may differ to their activities within the context of their native cellular niche. The development of mutant and genetically engineered mouse strains expressing human hematopoietic factors has greatly improved the engraftment of human HSPCs15, but uncertainties remain regarding the ability of these models to replicate all relevant microenvironmental interactions.
Recent advances in humanized BM organoids provide an approach to grow human hematopoietic cells in a conspecific microenvironment for better engraftment and experimental interrogation of cellular interactions3, 16, 17. Engineered hematopoietic environments were the focus of the Spring 2020 International Society for Experimental Hematology New Investigator Committee Webinar. This webinar (also available here: https://www.youtube.com/watch?v=R3XSTALtmjs&feature=youtu.be) included presentations from Drs. Dominique Bonnet and Paul Bourgine, who discussed humanized ossicle and bioreactor technologies, respectively, and was moderated by Dr. François Mercier. In this Perspective, we introduce traditional xenotransplantation assays and then discuss recent progress in the humanization of the hematopoietic niche using ectopic ossicle and bioreactor technologies.
Traditional xenotransplantation assays
The transplantation of human hematopoietic cells into immunodeficient mice has been pivotal in defining the nature of human hematopoiesis and leukemia including phenotyping human HSPCs and hematopoietic hierarchies18–20, providing evidence for leukemic stem cells and leukemic evolution12, 15, 21, 22, and evaluating new therapeutic strategies15, 23, 24. However, there remain limitations to xenotransplantation. In particular, individual cases of acute myeloid leukemia (AML) and MDS display variable engraftment25, as the ability of hematological malignancies to engraft is usually characteristic of cases with an aggressive clinical course. It has also been observed that clonally heterogeneous mixtures of leukemic cells do not always engraft representatively26.
Since the development of the first NOD/Scid immunodeficient mouse model18, 21, various derivatives of this mouse strain have been developed to further improve human hematopoiesis in mice15, 23. This includes the NOD/Scid/IL2Rg-KO (NSG) mice, Kit-mutant NSG mice (NSG-W41 and NBSGW), and human cytokine-expressing NSG mice (e.g., the NSG-S and MISTRG) strains27–32. Expression of human cytokines has improved the development of human innate immune cells in mice compared to earlier models (e.g. NOD/Scid), thereby considerably improving the engraftment of different patient samples in these xenotransplantation settings15. The generation of murine recipient strains that are even more permissive for human hematopoiesis, native or diseased, is pursued through the suppression of innate immune responses or humanization of additional ligands (and is reviewed in detail elsewhere33, 34). However, all these models rely on the engraftment of human HSPCs within mouse hematopoietic organs, particularly the mouse bone marrow and spleen. Although these microenvironments can support the long-term engraftment of human cells, species differences mean these microenvironments are not fully analogous to human. For example, numerous ligand-receptor pairs are yet to be humanized in these mouse models. For the study of these specific interactions in human hematopoiesis, generating human hematopoietic niches de novo offers flexible, complementary strategies.
In vivo ossicle models
Over the last decade, several groups including Dr. Bonnet’s have developed methods to model the human BM microenvironment in mice (Figure 1A) using subcutaneous humanized ossicles35–41. These protocols often involve the seeding of human MSCs onto a 3D scaffold composed of extracellular matrix, then subcutaneously implanting the scaffold into NSG mice. However, other methods such as those characterized by the Majeti laboratory36, 37 subcutaneously inject MSCs mixed in an extracellular matrix gel to generate the ossicle (Figure 1A). Within the mouse, these humanized ossicles become colonized with mouse endothelium and mouse hematopoietic cells38. The microenvironment within the ossicle allows engraftment of injected human HSPCs and supports human HSPC expansion and differentiation. Highlighting that much remains to be learned about the human HSPC-niche interactions, unknown differences between MSC donors cause major variations in the composition and engraftment of human hematopoietic lineages in these models38.
Figure 1: Schematic summary of ossicle and bioreactor technologies.
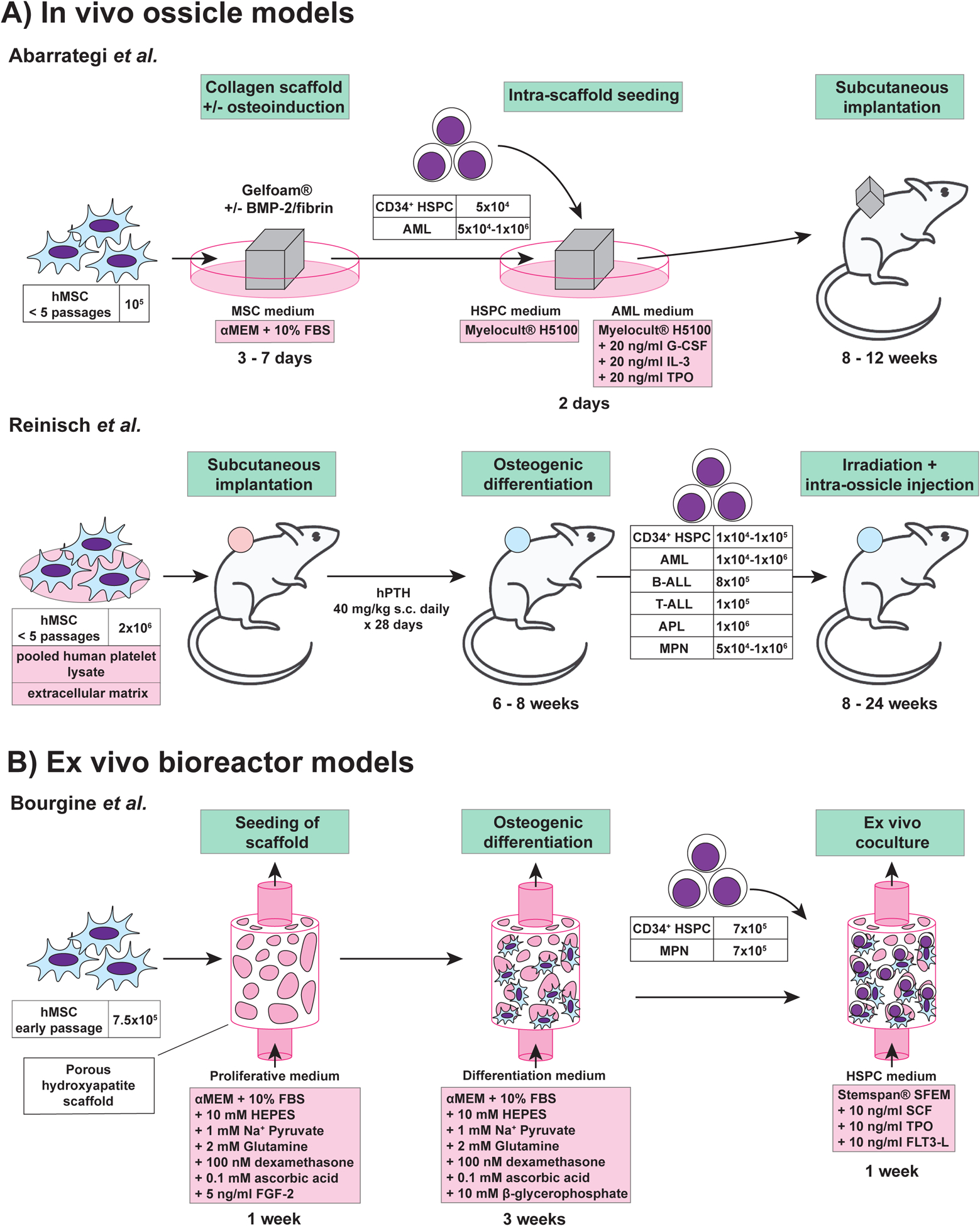
Approaches for engineering tri-dimensional hematopoietic environments. A. In vivo ossicle models. B. Ex vivo bioreactor model. ALL: acute lymphoblastic leukemia; AML: acute myeloid leukemia; APL: acute promyelocytic leukemia; BMP-2: bone morphogenetic protein; FBS: fetal bovine serum; FGF-2: fibroblast growth factor-2; FLT3-L: Fms-related tyrosine kinase 3 ligand; G-CSF: granulocyte colony stimulating factor; HSPC: hematopoietic stem and progenitor cells; hMSC: human mesenchymal stromal cells; hPTH: human parathyroid hormone; IL-3: interleukin-3; MPN: myeloproliferative neoplasm; SCF: stem cell factor; TPO: thrombopoietin.
Various improvements to this basic ossicle formation protocol have been made. For example, to further improve endochondral ossification within the scaffold, the Bonnet lab incorporated bone morphogenic protein 2 (BMP-2), an osteo-inductive signal, into the collagen scaffold38. Similarly, Bourgine and colleagues primed MSCs for cartilage differentiation via transgenic expression of stromal-derived factor 1 alpha (SDF1α, also known as CXCL12), with the aim of mimicking the endochondral ossification pathway seen during embryonic bone development41. These methods also highlight the potential to genetically modify the MSCs to study the role of specific signaling pathways in these humanized hematopoietic microenvironments. Additionally, the Bonnet lab showed that seeding the scaffold with human endothelial cells can lead to formation of human vasculature within the ossicle, further humanizing this BM niche38.
One goal of ossicle technology is to overcome the variable engraftment of hematological malignancies in mice25. A comparison between the engraftment of samples derived from patients with AML, using traditional xenotransplantation assays and ossicle transplantation assays, found that ossicles were superior38,36. In particular, humanized ossicles allowed the engraftment of AML samples that engrafted poorly otherwise38. This highlights the utility of the ossicle system and its advantages over traditional xenotransplantation assays. Ossicle technology is now being expanded to other diseases, such as MDS, in which patient samples poorly engraft in traditional murine xenotransplantation and are therefore difficult to study in vivo42. To date, MSCs have been collected from healthy donors, but there is also great interest to generate ossicles from patient derived MSCs in the future. Using ossicle models, it should be possible to dissect the cellular and molecular interactions within the human BM niche in health and disease, and ultimately help to identify new therapeutic targets for disease intervention.
Ex vivo bioreactors models
While the development of humanized ossicles represents a unique opportunity to study hematologic diseases in vivo43, some limitations remain (Table 1). For example, established in vivo models remain chimeric, since vasculature as well as nerve fibers are of murine origin. This also holds true for some circulating cytokines, which are not always conserved between mouse and human, such as GM-CSF44. Thus, additional techniques are needed to facilitate the study of human HSPCs and hematopoietic malignancies, including engineered in vitro BM systems45. These methods can be used for HSPC expansion, drug screening or disease modeling. Several models have been established, from 2D cultures to static 3D scaffold systems up to dynamic 3D setups, which implement the component of perfusion.
Table 1:
Comparison of ossicle and bioreactor technologies
| Model | Major Pros | Major Cons |
|---|---|---|
| In vivo ossicle models | Support functional human HSCs long-term as well as various myeloid malignancies; offers an in vivo model of the human bone marrow niche | Requires use of immunodeficient animals; ossicles can become chimeric with infiltration of mouse cells; batch-to-batch variability between donor MSCs |
| Ex vivo bioreactor technology | Fully defined 3D microenvironment that can be easily modulated real-time; amenable to time course analysis, perturbations, etc. | Current bioreactor technology cannot currently stably support HSCs long-term; requires constant perfusion |
Bourgine and colleagues recently engineered a dynamic human 3D in vitro BM niche43 (Figure 1B). This bioreactor consists of four components: (I) a porous ceramic material, which mimics the bone structure, (II) human MSCs, (III) human HSPCs, and (IV) perfusion of serum-free medium. Briefly, MSCs were allowed to colonize the ceramic scaffold for one week, and osteogenesis primed for three weeks. After this initial period of engineered niche (eN) formation, CD34+ HSPCs and recombinant growth factors (SCF, TPO, Flt3-L) were added. Time-course analysis identified gradual seeding of hematopoietic cell populations within the eN after addition of HSPCs. In comparison to the control condition, where only the ceramic scaffold was used, the eN setting promoted the expansion of phenotypic HSPCs. Functional analysis revealed in vitro maintenance of stem cells. However, stem cell performance was still reduced compared with freshly isolated human HSPCs. Within the eN, formation of extracellular matrix (ECM) and osteocalcin at the scaffold surface was observed, characterizing the eN as osteoblastic. Interestingly, a functional compartmentalization could be observed, which mimicked the biological cellular distribution of normal BM. The human HSCs were preferentially located in the ECM close to MSCs, while more committed progenitors were also detected in the supernatant. The presented model exhibits characteristic features of a human osteoblastic BM niche and can be used to study the effects of different extrinsic factors on HSCs or for disease modeling.
In a proof-of-concept experiment, Dr. Bourgine showed that overexpression of SDF1α in human MSCs led to increased maintenance and quiescence of cultured human HSCs43. In addition, niche injury was modeled in vitro by application of the DNA-damaging compound bleomycin. Bleomycin led to a decrease in HSPC numbers in the ECM compartment of the bioreactor and an increase in the number of cycling cells. These proof-of-principle approaches demonstrated that such bioreactor systems can be used to analyze the influences of different biological and chemical factors on the MSC and HSPC compartments. Current applications of this bioreactor technology include the ex vivo maintenance and study of HSPCs derived from patients suffering from MPNs. This approach will potentially allow for mechanistic analyses and drug screenings to be performed using patient-derived MPN HSPCs ex vivo, investigations that have been challenging to date.
Together, these studies demonstrate that complex biological environments such as the human BM can be engineered in vitro, allowing for improved functional maintenance of HSPCs. The platform can be used to investigate the role of different factors and disease development. In the future, it will be of interest to study the long-term culture of HSPCs using bioreactor systems. Recently, long-term culture of mouse HSPCs has been improved considerably46, 47. Refined compositions of culture medium in combination with engineered BM niches could facilitate culture and expansion of HSPCs. In addition to the analysis of HSPCs derived from patients suffering from hematological diseases, it will be also important to have a closer look at matched patient-derived MSCs. Such studies could reveal additional mechanistic interactions within the diseased niche and enable the identification of potential niche-specific therapeutic targets.
Conclusions
The development of new models to study human hematopoiesis and leukemogenesis and the bone marrow microenvironment, including in vivo ossicle models and ex vivo bioreactors, are providing new biological and biomedical insights. In the recent ISEH webinar, Drs. Bonnet and Bourgine presented how humanized BM organoids can be used to improve the engraftment of hard-to-transplant patient samples, characterize the effect of microenvironmental perturbations on HSPCs, and expand human hematopoietic progenitor cells ex vivo. We expect that these applications are only the start for these powerful technologies. Further optimization and characterization of these engineered microenvironments will undoubtedly yield new fundamental and practical insights in the regulation of normal and aberrant hematopoiesis.
Highlights:
A review of the use of ossicle models to model human hematopoiesis
Discusses the use of bioreactors to grow human HSPCs ex vivo
Acknowledgments:
We thank ISEH staff and colleagues on the ISEH New Investigator Committee for their support.
References:
- 1.Eaves CJ. Hematopoietic stem cells: concepts, definitions, and the new reality. Blood. April 2015;125(17):2605–13. doi: 10.1182/blood-2014-12-570200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wilkinson AC, Igarashi KJ, Nakauchi H. Haematopoietic stem cell self-renewal in vivo and ex vivo. Nat Rev Genet. May 2020;doi: 10.1038/s41576-020-0241-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Loughran S, Haas S, Wilkinson A, Klein A, Brand M. Lineage commitment of hematopoietic stem cells and progenitors: insights from recent single cell and lineage tracing technologies. Experimental Hematology. 2020;doi: 10.1016/j.exphem.2020.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Orkin SH, Zon LI. Hematopoiesis: An evolving paradigm for stem cell biology. Cell. February 22 2008;132(4):631–644. doi:Doi 10.1016/J.Cell.2008.01.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Morrison SJ, Scadden DT. The bone marrow niche for haematopoietic stem cells. Nature. January 2014;505(7483):327–34. doi: 10.1038/nature12984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pinho S, Frenette PS. Haematopoietic stem cell activity and interactions with the niche. Nat Rev Mol Cell Biol. May 2019;20(5):303–320. doi: 10.1038/s41580-019-0103-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jan M, Ebert BL, Jaiswal S. Clonal hematopoiesis. Semin Hematol. January 2017;54(1):43–50. doi: 10.1053/j.seminhematol.2016.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bowman RL, Busque L, Levine RL. Clonal Hematopoiesis and Evolution to Hematopoietic Malignancies. Cell Stem Cell. February 2018;22(2):157–170. doi: 10.1016/j.stem.2018.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Luis TC, Wilkinson AC, Beerman I, Jaiswal S, Shlush LI. Biological implications of clonal hematopoiesis. Exp Hematol. 09 2019;77:1–5. doi: 10.1016/j.exphem.2019.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Park SJ, Bejar R. Clonal hematopoiesis in cancer. Exp Hematol. 03 2020;83:105–112. doi: 10.1016/j.exphem.2020.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Steensma DP, Ebert BL. Clonal hematopoiesis as a model for premalignant changes during aging. Exp Hematol. March 2020;83:48–56. doi: 10.1016/j.exphem.2019.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Corces-Zimmerman MR, Majeti R. Pre-leukemic evolution of hematopoietic stem cells: the importance of early mutations in leukemogenesis. Leukemia. December 2014;28(12):2276–82. doi: 10.1038/leu.2014.211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Méndez-Ferrer S, Bonnet D, Steensma DP, et al. Bone marrow niches in haematological malignancies. Nat Rev Cancer. 05 2020;20(5):285–298. doi: 10.1038/s41568-020-0245-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wilkinson AC, Nakauchi H. Stabilizing hematopoietic stem cells in vitro. Current Opinion in Genetics & Development. 2020;64:1–5. doi: 10.1016/j.gde.2020.05.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goyama S, Wunderlich M, Mulloy JC. Xenograft models for normal and malignant stem cells. Blood. April 2015;125(17):2630–40. doi: 10.1182/blood-2014-11-570218 [DOI] [PubMed] [Google Scholar]
- 16.Gundry MC, Dever DP, Yudovich D, et al. Technical considerations for the use of CRISPR/Cas9 in hematology research. Exp Hematol. 10 2017;54:4–11. doi: 10.1016/j.exphem.2017.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carrelha J, Lin D, Rodriguez-Fraticelli A, et al. Single-cell lineage tracing approaches in hematology research – technical considerations. Experimental Hematology. 2020;doi: 10.1016/j.exphem.2020.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kamel-Reid S, Dick JE. Engraftment of immune-deficient mice with human hematopoietic stem cells. Science. December 1988;242(4886):1706–9. doi: 10.1126/science.2904703 [DOI] [PubMed] [Google Scholar]
- 19.Notta F, Doulatov S, Laurenti E, Poeppl A, Jurisica I, Dick JE. Isolation of single human hematopoietic stem cells capable of long-term multilineage engraftment. Science. July 2011;333(6039):218–21. doi: 10.1126/science.1201219 [DOI] [PubMed] [Google Scholar]
- 20.Notta F, Zandi S, Takayama N, et al. Distinct routes of lineage development reshape the human blood hierarchy across ontogeny. Science. January 2016;351(6269):aab2116. doi: 10.1126/science.aab2116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bonnet D, Dick JE. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat Med. July 1997;3(7):730–7. doi: 10.1038/nm0797-730 [DOI] [PubMed] [Google Scholar]
- 22.Kreso A, Dick JE. Evolution of the cancer stem cell model. Cell Stem Cell. March 2014;14(3):275–91. doi: 10.1016/j.stem.2014.02.006 [DOI] [PubMed] [Google Scholar]
- 23.Abarrategi A, Mian SA, Passaro D, Rouault-Pierre K, Grey W, Bonnet D. Modeling the human bone marrow niche in mice: From host bone marrow engraftment to bioengineering approaches. J Exp Med. 03 2018;215(3):729–743. doi: 10.1084/jem.20172139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Milan T, Canaj H, Villeneuve C, et al. Pediatric leukemia: Moving toward more accurate models. Exp Hematol. 06 2019;74:1–12. doi: 10.1016/j.exphem.2019.05.003 [DOI] [PubMed] [Google Scholar]
- 25.Griessinger E, Vargaftig J, Horswell S, Taussig DC, Gribben J, Bonnet D. Acute myeloid leukemia xenograft success prediction: Saving time. Exp Hematol. 03 2018;59:66–71.e4. doi: 10.1016/j.exphem.2017.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Klco JM, Spencer DH, Miller CA, et al. Functional heterogeneity of genetically defined subclones in acute myeloid leukemia. Cancer Cell. March 2014;25(3):379–92. doi: 10.1016/j.ccr.2014.01.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wunderlich M, Chou FS, Link KA, et al. AML xenograft efficiency is significantly improved in NOD/SCID-IL2RG mice constitutively expressing human SCF, GM-CSF and IL-3. Leukemia. October 2010;24(10):1785–8. doi: 10.1038/leu.2010.158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rongvaux A, Willinger T, Martinek J, et al. Development and function of human innate immune cells in a humanized mouse model. Nat Biotechnol. April 2014;32(4):364–72. doi: 10.1038/nbt.2858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Das R, Strowig T, Verma R, et al. Microenvironment-dependent growth of preneoplastic and malignant plasma cells in humanized mice. Nat Med. 11 2016;22(11):1351–1357. doi: 10.1038/nm.4202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ellegast JM, Rauch PJ, Kovtonyuk LV, et al. inv(16) and NPM1mut AMLs engraft human cytokine knock-in mice. Blood. 10 2016;128(17):2130–2134. doi: 10.1182/blood-2015-12-689356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cosgun KN, Rahmig S, Mende N, et al. Kit regulates HSC engraftment across the human-mouse species barrier. Cell Stem Cell. August 2014;15(2):227–38. doi: 10.1016/j.stem.2014.06.001 [DOI] [PubMed] [Google Scholar]
- 32.McIntosh BE, Brown ME, Duffin BM, et al. Nonirradiated NOD,B6.SCID Il2rγ−/− Kit(W41/W41) (NBSGW) mice support multilineage engraftment of human hematopoietic cells. Stem Cell Reports. February 2015;4(2):171–80. doi: 10.1016/j.stemcr.2014.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saito Y, Shultz LD, Ishikawa F. Understanding Normal and Malignant Human Hematopoiesis Using Next-Generation Humanized Mice. Trends Immunol. 08 2020;41(8):706–720. doi: 10.1016/j.it.2020.06.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gbyli R, Song Y, Halene S. Humanized mice as preclinical models for myeloid malignancies. Biochem Pharmacol. 04 2020;174:113794. doi: 10.1016/j.bcp.2020.113794 [DOI] [PubMed] [Google Scholar]
- 35.Vaiselbuh SR, Edelman M, Lipton JM, Liu JM. Ectopic human mesenchymal stem cell-coated scaffolds in NOD/SCID mice: an in vivo model of the leukemia niche. Tissue Eng Part C Methods. December 2010;16(6):1523–31. doi: 10.1089/ten.tec.2010.0179 [DOI] [PubMed] [Google Scholar]
- 36.Reinisch A, Thomas D, Corces MR, et al. A humanized bone marrow ossicle xenotransplantation model enables improved engraftment of healthy and leukemic human hematopoietic cells. Nat Med. 07 2016;22(7):812–21. doi: 10.1038/nm.4103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reinisch A, Hernandez DC, Schallmoser K, Majeti R. Generation and use of a humanized bone-marrow-ossicle niche for hematopoietic xenotransplantation into mice. Nat Protoc. October 2017;12(10):2169–2188. doi: 10.1038/nprot.2017.088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Abarrategi A, Foster K, Hamilton A, et al. Versatile humanized niche model enables study of normal and malignant human hematopoiesis. J Clin Invest. February 2017;127(2):543–548. doi: 10.1172/JCI89364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Passaro D, Abarrategi A, Foster K, Ariza-McNaughton L, Bonnet D. Bioengineering of Humanized Bone Marrow Microenvironments in Mouse and Their Visualization by Live Imaging. J Vis Exp. 08 2017;(126)doi: 10.3791/55914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fritsch K, Pigeot S, Feng X, et al. Engineered humanized bone organs maintain human hematopoiesis in vivo. Exp Hematol. 05 2018;61:45–51.e5. doi: 10.1016/j.exphem.2018.01.004 [DOI] [PubMed] [Google Scholar]
- 41.Bourgine PE, Fritsch K, Pigeot S, et al. Fate Distribution and Regulatory Role of Human Mesenchymal Stromal Cells in Engineered Hematopoietic Bone Organs. iScience. September 2019;19:504–513. doi: 10.1016/j.isci.2019.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Côme C, Balhuizen A, Bonnet D, Porse BT. Myelodysplastic syndrome patient-derived xenografts: from no options to many. Haematologica. April 2020;105(4):864–869. doi: 10.3324/haematol.2019.233320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bourgine PE, Klein T, Paczulla AM, et al. In vitro biomimetic engineering of a human hematopoietic niche with functional properties. Proc Natl Acad Sci U S A. 06 2018;115(25):E5688–E5695. doi: 10.1073/pnas.1805440115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dupard SJ, Grigoryan A, Farhat S, Coutu DL, Bourgine PE. Development of Humanized Ossicles: Bridging the Hematopoietic Gap. Trends Mol Med. June 2020;26(6):552–569. doi: 10.1016/j.molmed.2020.01.016 [DOI] [PubMed] [Google Scholar]
- 45.Bourgine PE, Martin I, Schroeder T. Engineering Human Bone Marrow Proxies. Cell Stem Cell. 03 2018;22(3):298–301. doi: 10.1016/j.stem.2018.01.002 [DOI] [PubMed] [Google Scholar]
- 46.Wilkinson AC, Ishida R, Kikuchi M, et al. Long-term ex vivo haematopoietic-stem-cell expansion allows nonconditioned transplantation. Nature. May 2019;571(7763):117–121. doi: 10.1038/s41586-019-1244-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wilkinson AC, Ishida R, Nakauchi H, Yamazaki S. Long-term ex vivo expansion of mouse hematopoietic stem cells. Nat Protoc. January 2020;doi: 10.1038/s41596-019-0263-2 [DOI] [PMC free article] [PubMed] [Google Scholar]


