Veglia et al. characterize the heterogeneity of polymorphonuclear neutrophils (PMNs) in the tumor microenvironment, spleen, and peripheral blood. The identification of neutrophil populations with potent immune suppressive activity opens selective targeting opportunities.
Abstract
In this study, using single-cell RNA-seq, cell mass spectrometry, flow cytometry, and functional analysis, we characterized the heterogeneity of polymorphonuclear neutrophils (PMNs) in cancer. We describe three populations of PMNs in tumor-bearing mice: classical PMNs, polymorphonuclear myeloid-derived suppressor cells (PMN-MDSCs), and activated PMN-MDSCs with potent immune suppressive activity. In spleens of mice, PMN-MDSCs gradually replaced PMNs during tumor progression. Activated PMN-MDSCs were found only in tumors, where they were present at the very early stages of the disease. These populations of PMNs in mice could be separated based on the expression of CD14. In peripheral blood of cancer patients, we identified two distinct populations of PMNs with characteristics of classical PMNs and PMN-MDSCs. The gene signature of tumor PMN-MDSCs was similar to that in mouse activated PMN-MDSCs and was closely associated with negative clinical outcome in cancer patients. Thus, we provide evidence that PMN-MDSCs are a distinct population of PMNs with unique features and potential for selective targeting opportunities.
Introduction
Polymorphonuclear neutrophils (PMNs) are the most abundant innate immune cells, with a well-defined role in protection of the host from different pathogens and in tissue remodeling. In recent years, their role in regulation of adaptive immunity has emerged. It is especially evident in cancer. PMNs are important components of the tumor microenvironment (TME; Coffelt et al., 2016; Shaul et al., 2020). PMNs in cancer are functionally diverse (Mishalian et al., 2017; Ng et al., 2019). PMNs have been implicated in antitumor activity (Fridlender et al., 2009; Granot et al., 2011; Singhal et al., 2016). However, a large body of evidence indicates that their potent role in negative regulation of immune responses in cancer and their presence in cancer patients is associated with poor prognosis and therapeutic outcomes (Zhou et al., 2018). Immune suppressive activity of PMNs is attributed to polymorphonuclear myeloid-derived suppressor cells (PMN-MDSCs), a population of pathologically activated PMNs. These cells suppress the functions of T lymphocytes, B lymphocytes, and natural killer cells and also promote tumor progression and metastasis via nonimmune mechanisms (Condamine et al., 2015b; Veglia et al., 2018). The question is how to separate PMNs with neutral or antitumor activity from those with protumorigenic activity. Can these two populations of cells coexist in the same tumor hosts? If yes, what is the relationship between these cells? Answers to these questions are important not only for better understanding of the biology of the cells, but also for the development of selective targeting of PMN-MDSCs.
Until now, separation of PMN-MDSCs from other PMNs in the same mouse was not possible due to the absence of specific markers. Therefore, all PMNs in tumor-bearing (TB) mice are considered PMN-MDSCs, based on the fact that immune suppressive activity was found only in PMN-MDSCs from TB mice, but not in PMNs from tumor-free mice. Thus, the nature of PMN diversity in cancer and the specific contribution of different populations of cells to regulation of immune function and tumor progression has remained unclear.
In cancer patients, PMN-MDSCs can be separated from other PMNs by gradient centrifugation. This allows for identification of a distinct transcriptomic profile of PMN-MDSCs and identification of LOX-1 as a marker of human PMN-MDSCs (Condamine et al., 2016). Recent studies confirmed the clinical relevance of this marker (Kim et al., 2019; Kumar et al., 2017; Si et al., 2019). However, one marker may not fully identify the population of the cells, and the coexistence of different groups of PMNs in cancer patients has remained unclear. A number of studies of the tumor landscape demonstrated a strong negative prognostic value of PMNs in tumor tissues (Gentles et al., 2015; Zhang et al., 2020). However, what population of PMNs contributes to this phenomenon remained unknown.
Recent studies established a gene signature of PMNs and PMN-MDSCs at the single-cell level from transcriptomes obtained from sequenced Gr1+ cells in mice and CD45+ cells in humans. Using single-cell RNA sequencing (scRNA-seq), it has been confirmed that PMNs isolated from mice with spontaneous mammary tumors showed a gene signature that was different from PMNs from control mice. Importantly, the MDSC state was largely conserved between mice and humans (Alshetaiwi et al., 2020). Although it helps to characterize MDSCs, it does not capture the PMN repertoire. The biggest challenge is identification of the PMN profile in tumor tissues. Low transcriptional activity of these cells may contribute to the lack of critical information. To overcome these limitations and obtain a comprehensive understanding of the heterogeneity of PMNs in cancer, we used scRNA-seq in a pure population of PMNs followed by cell mass spectrometry, flow cytometry, and functional analysis. We identified molecular, phenotypic, and functional characteristics of PMN-MDSCs and their relations to other PMNs. We determined that populations of classical PMNs and PMN-MDSCs coexist in the same TB hosts and identified potential marker of mouse PMN-MDSCs.
Results
PMN-MDSCs represent two distinct populations of neutrophils in TB mice
To assess the heterogeneity of PMNs in cancer, we employed scRNA-seq. Since transcriptional activity of PMNs is much lower than most other cells in TME, RNA-seq of unseparated cells in TME usually results in underrepresentation of transcripts associated with PMNs. Therefore, we sorted CD45+Ly6G+Ly6Clow PMNs (Fig. 1 A) and processed for scRNA-seq using the 10x Genomics Chromium platform. We evaluated PMNs from spleens of tumor-free mice (n = 3), as well as from spleens (n = 3) and tumors (n = 4) of Lewis lung carcinoma (LLC) TB mice. In total, 66,854 single cells expressing at least 400 genes (each gene with at least one count) were considered for further analysis. The scran (Lun et al., 2016) and scater (McCarthy et al., 2017) packages were used to normalize the raw counts to control cell-specific biases. These normalized counts were then supplied to Seurat package (v3.1.1; Stuart et al., 2019) to perform batch correction, clustering, and marker identification for the predicted cell clusters.
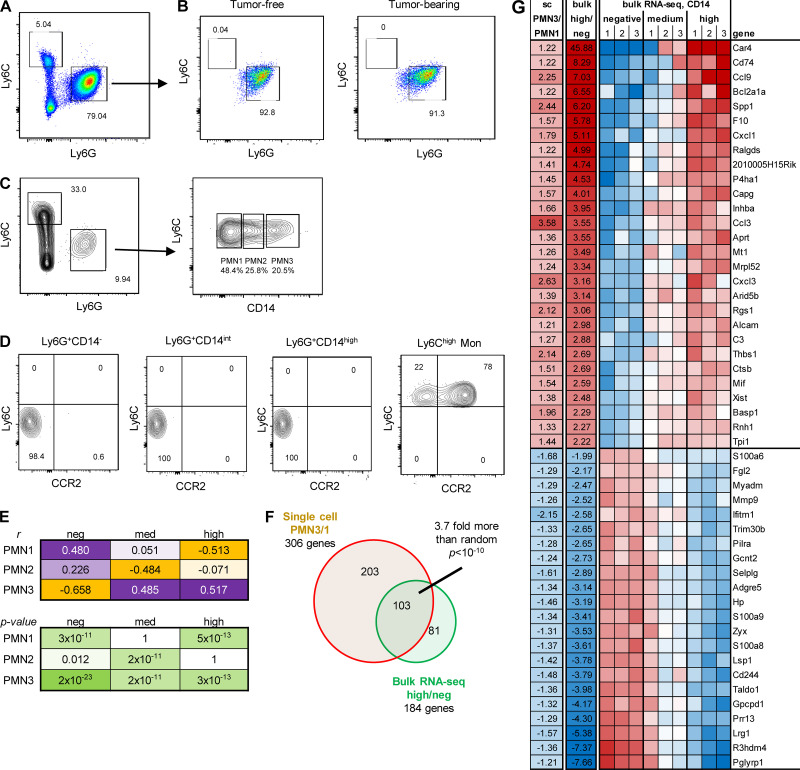
Figure 1.
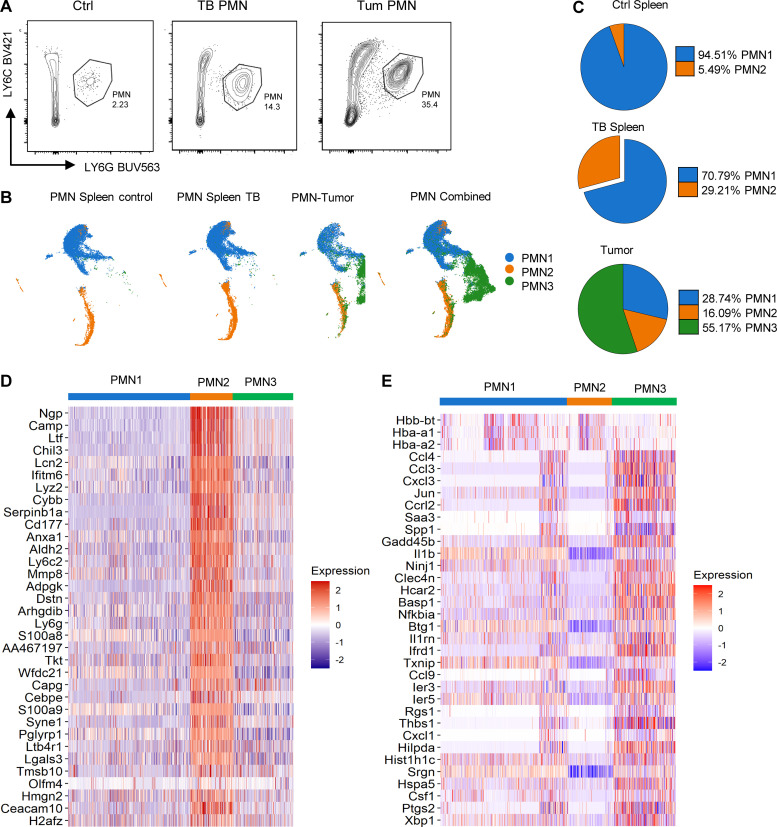
PMN clusters in TB mice. (A) Example of gating of PMNs. (B) Unsupervised clustering of PMNs. (C) Distribution of three PMN populations in spleen and tumors. (D) Expression of genes associated with immune suppression in different clusters of PMNs. (E) Expression of genes associated with cell activation and inflammation in different PMN clusters. Experiments were performed three times with spleen PMNs, three times with spleen PMNs from TB mice, and four times with tumor PMNs.
To determine cell identity, we matched our dataset with two separate public mouse datasets (Heng et al., 2008; Benayoun et al., 2019) using singleR package (v1.0.0; Aran et al., 2019). Based on transcriptomic results, we projected them into two dimensions with Uniform Manifold Approximation and Projection (UMAP; Becht et al., 2018). A PMN-specific signature was identified in 92.29% of all cells analyzed. A macrophage signature was found in 1.29%, monocytes in 1.96%, and B cells in 3.18% of all cells. The remaining cell types were either undetectable or <0.3%. Cells with PMN signature were selected for further analysis. Unbiased, graph-based clustering identified three main clusters of PMNs with different representation in naive and TB mice, referenced as PMN1, PMN2, and PMN3. Spleen PMNs from control and TB mice contained only two clusters (PMN1 and PMN2), whereas tumor-infiltrating PMNs had all three clusters (Fig. 1 B). Almost all (95%) of control spleen PMNs were represented by the PMN1 cluster. In contrast, 30% of PMNs in spleen from TB mice were PMN2 cells. In tumors, the PMN1 cluster represented <30% of PMNs, with the majority of PMNs belonging to the PMN3 cluster (Fig. 1 C). When compared with PMN1, the PMN2 cluster demonstrated enrichment for genes associated with immature neutrophils and PMN-MDSC–associated genes (Ngp, Ltf, Cd177, Anxa1, Mmp8, S100a8, S100a9, Cebpe, Ltb4r1, and Cybb; Fig. 1 D), suggesting that PMN2 represents the population of PMN-MDSCs. PMN3 also had higher expression of those genes than in PMN1; however, surprisingly, their expression was lower than in PMN2 (Fig. 1 D). Instead, the PMN3 cluster was enriched for transcripts of chemokines and chemokine receptors as well as genes associated with cell activation, inflammation, and ER stress (Ccl4, Ccl3, Cxcl2, Cxcl3, Spp1, Il1b, Nfkbia, Socs3, Mif, Klf6, Atf3, Ptgs2, and Xbp1; Fig. 1 E). In ingenuity pathway analysis (IPA) of differentially expressed genes, the PMN3 cluster demonstrated upregulation of multiple pathways associated with IL-6, HMGB1, TNFR1, IL-1, TLR signaling, nitric oxide (NO), and ROS production, confirming the activated state of these cells (Fig. S1). Thus, it appears that the PMN1 cluster may represent classical PMNs; PMN2, PMN-MDSCs; and PMN3, “activated” PMN-MDSCs.
Figure S1.
Pathways significantly changed in different populations of PMNs.
We hypothesized that PMN2 may be a transient stage to more potently activated PMN3 cells. Using computational trajectory analysis, we assessed the relationship between the three populations of PMNs. Scran normalized data were input into Monocle3 (Cao et al., 2019), and batch correction, scaling, clustering, trajectory prediction, and gene identification were performed. PMN1 was used as the initial stage (starting cells) for the trajectory, assuming that the pseudotime reflects a direction of possible transition from PMN1 classical cells to PMN2 and PMN3 types. Trajectory analysis revealed that PMN1 cells can transform to either PMN2 or PMN3 by taking different paths, and PMN2 are not transformed to PMN3 (Fig. 2). Thus, PMN2 is not a transient state between PMN1 and PMN3 but may reflect different pathways of PMN-MDSC activation in different microenvironments.
Figure 2.
Clustering of PMN cells and their trajectory by Monocle3. Classical PMN cells (PMN1) were used as initial stage for the trajectory. The forked pseudotime trajectory suggests that transformation of PMN1 into PMN3 does not involve a transient PMN2 state.
CD14 is a marker of different populations of PMNs in TB mice
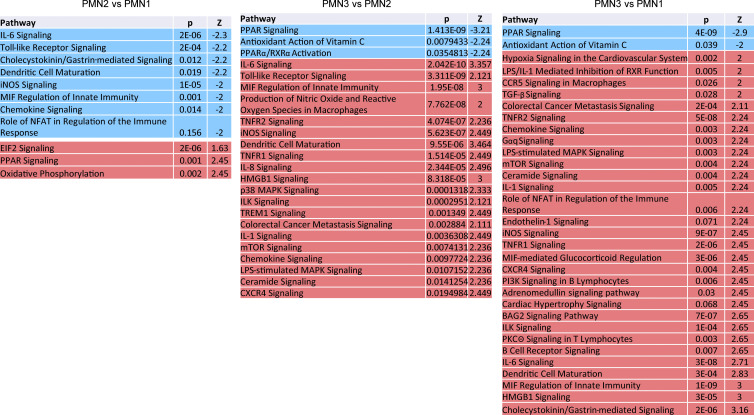
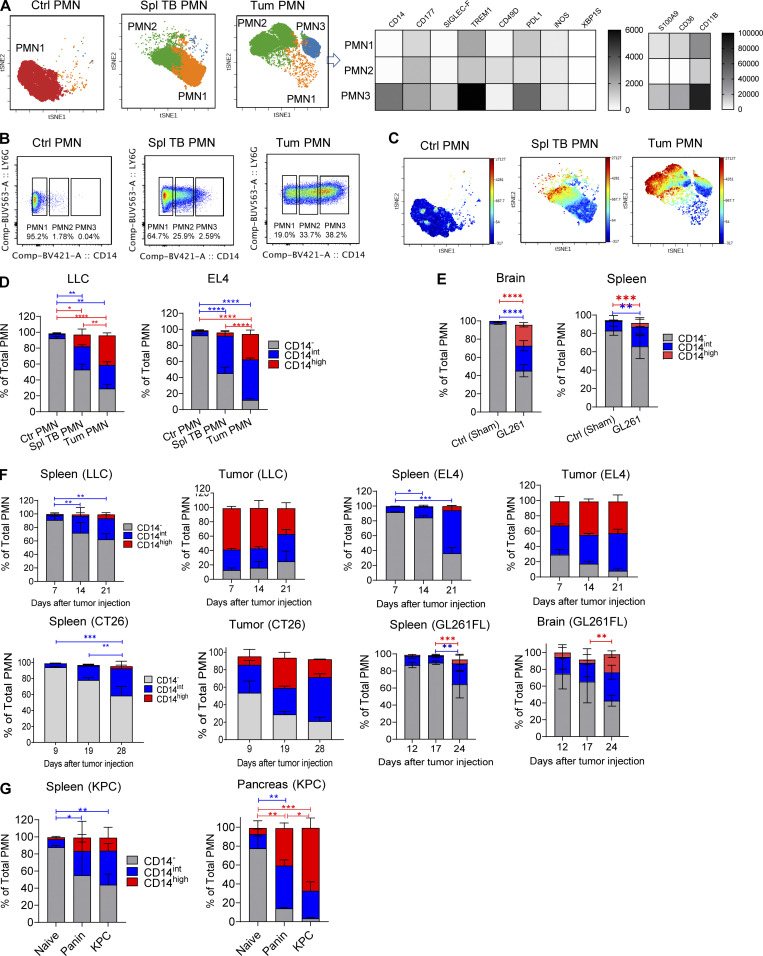
To better characterize the PMN3 population, we evaluated the expression of transmembrane receptors. PMN3 cells had increased expression of several transcripts that encoded proteins with known association with PMNs (Fig. S2 A). Interestingly, the PMN2 and PMN3 populations had increased expression of Cd14, which is a marker of monocytic cells and is expressed at much lower levels in granulocytes. To independently test the hypothesis of the coexistence of different populations of PMNs, we employed flow cytometry and flow self-organizing maps (FlowSOM). FlowSOM is an algorithm that allows for processing and analysis of high-dimensional single-cell data for the identification of novel subpopulations having specific combinations of markers. We designed a fluorescence antibody panel that included markers needed for identification of neutrophils (LY6G, LY6C, CD11B, CD45, SIGLEC-F, and CD177), functional intracellular proteins associated with PMN-MDSCs (induced NO synthase [iNOS], S100A9, sXBPS1, and CD36), and surface markers identified by scRNA-seq as associated with PMN2 and PMN3 clusters (CD14, TREM1, and CD49d). By combining flow cytometry and FlowSOM, we found that spleens of control mice had just one main cluster of PMNs, whereas PMNs from spleens of TB mice had two clusters, and tumor PMNs had three clusters (Fig. 3 A), which were similar to the clusters identified by scRNA-seq. In tumors, expression of several proteins associated with PMN-MDSCs was higher in the putative PMN3 population than in the two others (Fig. 3 A). When evaluated separately, no differences were observed in the expression of TREM1, SIGLEC-F, CD49D, PD-L1, and CD84 between PMNs from naive and TB mice (Fig. S2 B).
Figure S2.
Expression of cell surface markers on PMN-MDSCs from spleen and tumor. (A) Fold change of cell surface marker expression in PMN3 compared with PMN1 or PMN2. (B) Flow cytometric analysis of the expression of Siglec-F, CD49D, PD-L1, TREM1, and CD84 on PMNs from spleen and tumor. Dot plot representative of 5–8 mice analyzed. (C) Frequency in live cells and absolute number of the populations of PMNs from spleen and tumor of LLC- and EL4-bearing mice on days 7, 14, and 21 (five mice per group). (D) Frequency in live cells and absolute number of CD14−, CD14int, CD14high PMNs from spleen and pancreas of naive, Panin, and KPC mice (five mice per group). In C and D, mean and SD are shown. P values were calculated by ANOVA test with corrections for multiple comparisons. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
Figure 3.
CD14 expression delineates three populations of PMNs in TB mice. (A) FlowSOM clusters overlaid on a viSNE plot of PMN (left). Five samples (five mice) per group from five independent experiments were concatenated before applying viSNE and FlowSOM. Heatmap generated using raw median value of each marker expressed in each cluster identified (right). Spl, spleen; Tum, tumor. (B) Flow cytometric analysis of the expression of CD14 on tumor-associated PMNs. Typical result of five experiments is shown. (C) Expression of CD14 in FlowSOM clusters overlaid on viSNE plot. Typical example of five experiments is shown. (D) Frequency of CD14−, CD14int, CD14high PMNs in spleens of control mice and spleens and tumors of LLC and EL4 TB mice (n = 5–8 mice per group, from three independent experiments). Data are presented as mean ± SD. For comparisons between groups, ANOVA with correction for multiple comparisons was used. *, P < 0.05; **, P < 0.01; ****, P < 0.0001. (E) GL261FL cells or vehicle were injected intracranially into mice. Frequency of PMN subsets in control and TB mice, analyzed by flow cytometry. Mean and SD are shown (n = 3). Experiments were reproduced twice. **; P < 0.01; ***, P < 0.001; ****, P < 0.0001 in two-sided Student’s t test. (F) Kinetics of expansion of PMNs in spleen and tumors of indicated tumor models (n = 5 mice per group, from five independent experiments). *, P < 0.05; **, P < 0.01; ***, P < 0.001. (G) Expansion of PMNs in spleen and pancreatic tumors of KPC mice (n = 5 mice per group from three experiments). Data are presented as mean ± SD. For comparisons between groups, ANOVA with correction for multiple comparisons was used. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
Expression of CD14 demonstrated a different pattern. It was barely detectable in PMNs from spleens of tumor-free mice and was almost exclusively expressed in PMNs from TB mice (Fig. 3 B). The expression of CD14 was much higher in tumor-infiltrating PMNs than in spleen PMNs. Based on the expression of CD14, we defined three subsets of PMNs in spleen and tumor tissues of TB mice: negative (CD14−), intermediately positive (CD14int), and highly positive (CD14high; Fig. 3 B). t-Distributed stochastic neighbor embedding (t-SNE) analysis based on the expression of CD14 revealed segregation of PMNs on three populations based on CD14 expression (Fig. 3 C). In control spleens, CD14− PMNs represented >95% of all PMNs. By 4 wk after tumor injections, in spleens of LLC and EL4 TB mice, the CD14int population increased markedly to almost 40% of total PMNs. In tumor tissues, CD14int and CD14high PMNs were the dominant populations, representing >75% of all PMNs. CD14high cells represented >30% of all PMNs (Fig. 3 D). Similar analysis was performed in an orthotopic model of brain cancer (GL261FL). Accumulation of CD14int PMNs was observed in spleens of mice on day 24 after tumor inoculation. In brain tissue of TB mice, there was marked increase of both CD14int and CD14high PMNs (Fig. 2 E). Using transplantable s.c. tumor models (LLC, EL4, and CT26), we found that, among PMNs, CD14− cells were gradually replaced with CD14int cells during tumor progression. A very small population of CD14high cells became visible in spleens by day 21, when tumors became bulky with 1.5 cm in diameter (Fig. 3 F). In contrast, in tumor tissues, CD14int and CD14high populations of PMNs represented >50% of PMNs from the very early stages of tumor development, and their proportion was not substantially increased with tumor progression. Almost half of PMNs in tumors were CD14high cells (Fig. 3 F). In the spleens of mice with brain tumors (GL261FL), we observed the presence of a very small population of CD14high PMNs only at a late time point (day 24). CD14int PMNs were present at early time points (days 12 and 17) and were significantly increased at a late stage (day 24). In brain tumors, CD14int and CD14high PMNs were present at early time points, representing 60% of all PMNs (Fig. 3 F).
The association of different populations of PMNs with tumor stage was evaluated in a genetically engineered model of spontaneous pancreatic cancer (KPC). This model allows for distinction of relatively early stages of pancreatic cancer, pancreatic intraepithelial neoplasia (PanIN), and fully developed pancreatic ductal adenocarcinoma (PDAC; Hingorani et al., 2005; Kaplan-Lefko et al., 2003). Similar to the data from transplantable tumor models, there was a stage-dependent replacement of CD14− PMNs with CD14int and CD14high PMNs in spleen. In tumors, CD14int and CD14high PMNs represented vast majority of PMNs, even at PanIN stage. With progression of tumors to PDAC stage, CD14high PMNs became predominant population of PMNs (Fig. 2 G). When the absolute number of PMNs was evaluated, it was evident that tumor progression was associated with expansion of all three populations of PMNs in spleens, including CD14− cells. In contrast, in tumor tissues of LLC TB mice, the increase in CD14− PMNs during tumor progression was very small, and it was undetectable in EL4 and KPC tumors (Fig. S2, C and D).
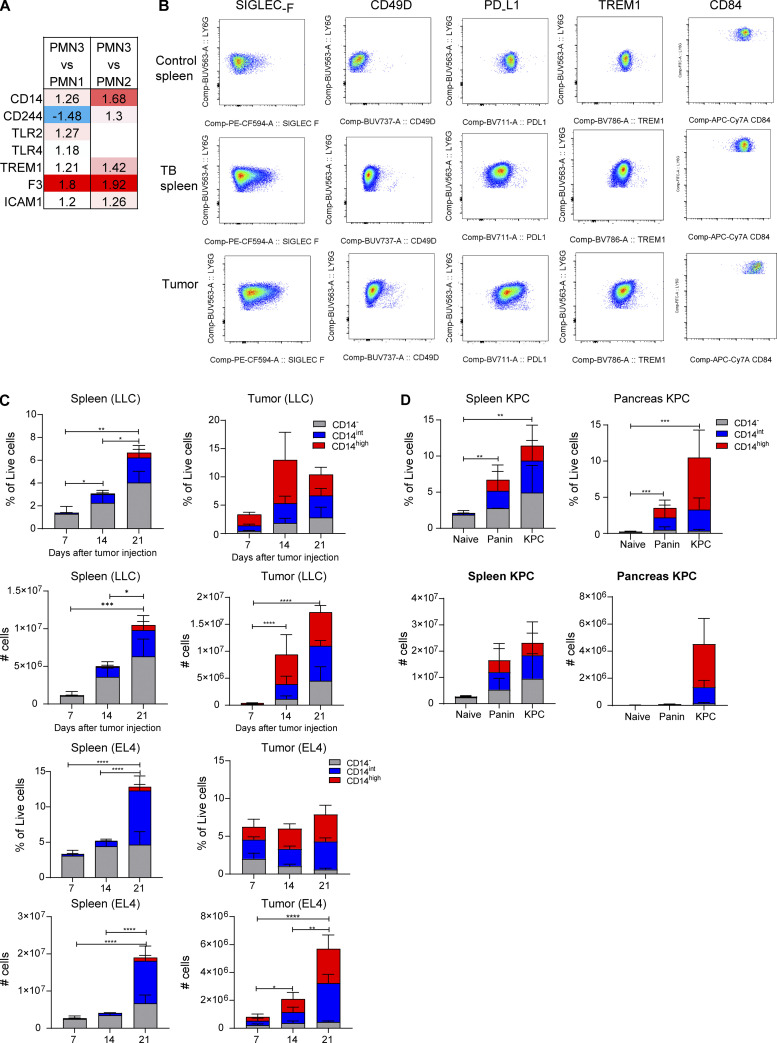
Since CD14 is a recognized marker of monocytic cells, we tested the hypothesis that PMN-MDSCs may acquire phenotype of monocytic cells. PMNs and PMN-MDSCs were isolated from bone marrow (BM) of tumor-free and GL261FL TB mice and cultured with GM-CSF for 24 h. We found no presence of monocytic cells after the culture (Fig. 4, A and B). Next, we assessed the possible presence of M-MDSCs within the populations of tumor CD14−, CD14int, and CD14high PMNs (Fig. 4 C). No presence of Ly6ChighLy6G−CCR2+ M-MDSCs was found in any PMN population (Fig. 4 D).
Figure 4.
Characteristics of PMNs with different expression of CD14. PMNs were sorted from BM obtained from control and GL261FL TB mice and cultured for 24 h in the presence of GM-CSF. (A) Representative gating strategy used for sorting. (B) Frequency of monocytic cells differentiated from PMNs isolated from tumor-free or TB mice. Experiments were performed twice with the same results. (C and D) Myeloid cells were isolated from GL26FL tumors on day 24 after tumor injections. (C) Gating strategy used for the identification of PMNs and their subsets in brain tumor tissue. (D) M-MDSC (Ly6ChighLy6G−CCR2+) present in different populations of PMNs. Ly6Chigh M-MDSC were used as positive controls of staining. Experiments were performed three times with the same results. (E) Correlation between three main single-cell clusters and three CD14 level groups from bulk RNA-seq. Correlation was done on 185 genes that passed FDR <5% threshold in both single-cell and bulk RNA-seq. (F) Overlap between single-cell PMN3/1 clusters and bulk RNA-seq high/negative CD14 cells. (G) Top 50 genes overlapped between single-cell and bulk RNA-seq in comparison between PMN3 versus PMN1 and high versus low CD14.
Three populations of tumor PMNs (CD14−, CD14int, and CD14high) were sorted, and bulk RNA-seq analysis was performed. We compared expression profiles of these three PMN populations with three clusters obtained by scRNA-seq analysis (PMN1–3). We found that CD14− PMNs were the most similar to the PMN1 group, and CD14high were the most similar to the PMN3 group (Fig. 4 E). There was a significant overlap of genes that were different between PMN3 versus PMN1 and genes different between cells with high versus low CD14 (P < 10−10 by Fisher’s exact test; Fig. 4 F); the top 50 such genes are shown in Fig. 4 G. Thus, tumor progression is associated with accumulation of a CD14high population of PMNs in spleens. Inside the tumors, these cells were dominant from the early stages of tumor growth.
Functional characterization of the populations of PMNs
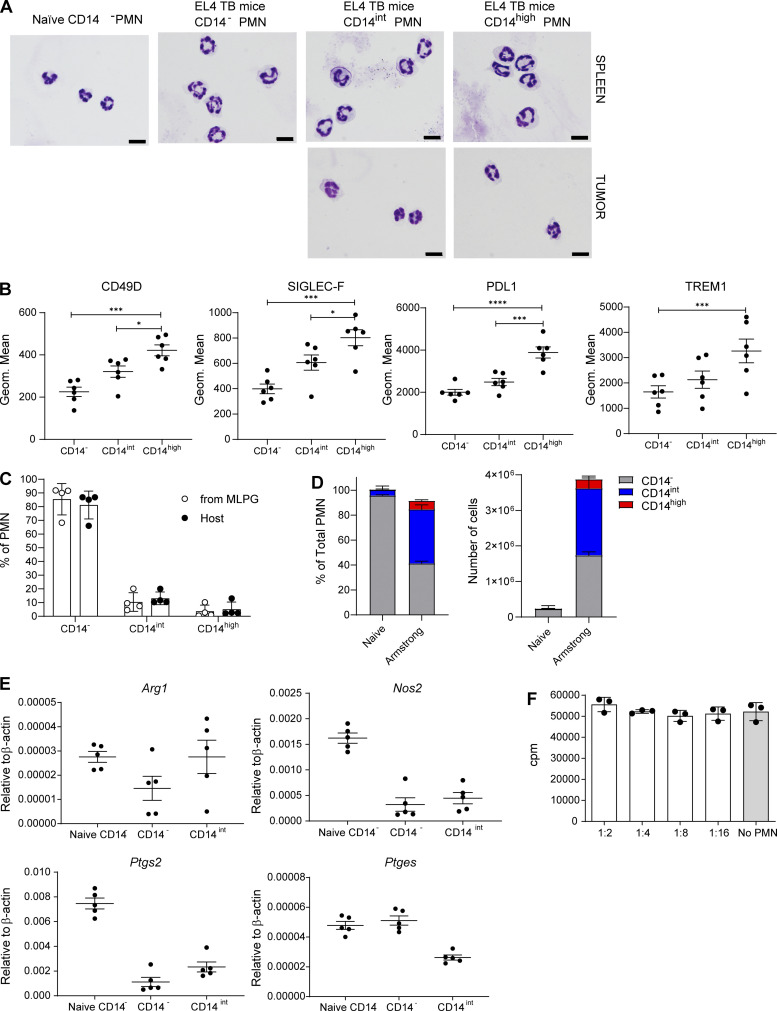
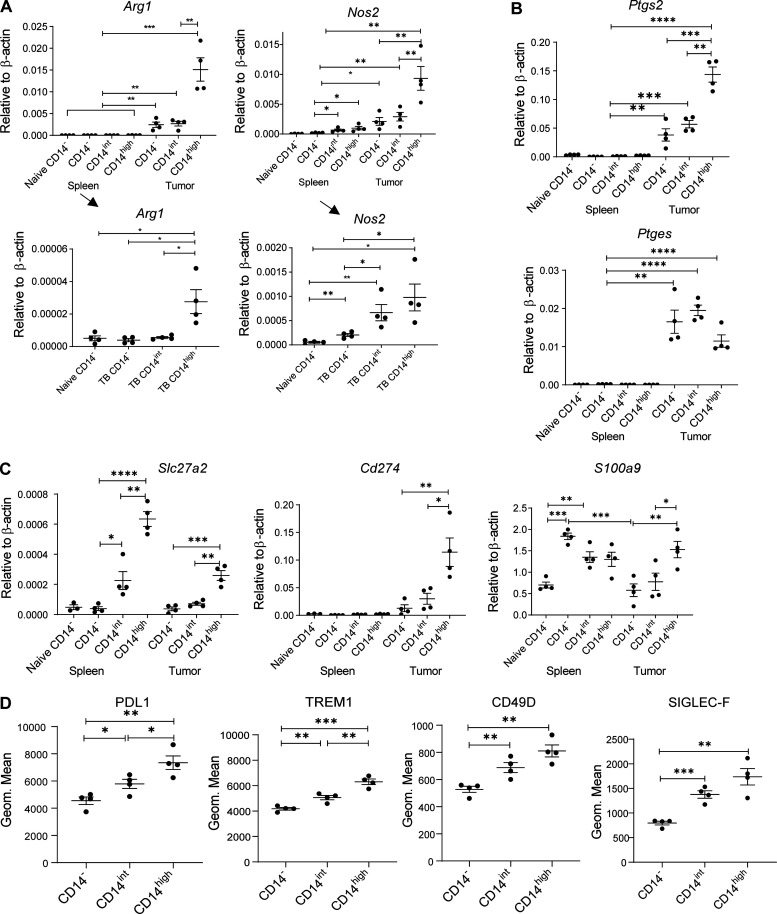
Next, we sought to identify functional characteristics of populations of PMNs in TB mice. PMNs were sorted based on the expression of CD14 marker. No clear morphological differences between populations of PMNs were evident (Fig. S3 A). Arginase1 (encoded by Arg1) and NO (produced by inducible NO synthase, Nos2) are major contributors to MDSC immune suppressive activity (Veglia et al., 2018). We observed markedly higher expression of Arg1 in tumor PMNs as compared to spleen PMNs. CD14high tumor PMNs had substantially higher expression than other populations of tumor PMNs. High Nos2 expression was associated exclusively with the CD14high population of tumor PMNs (Fig. 5 A). Expression of Arg1 and Nos2 in spleen PMNs was substantially lower than in tumor PMNs; however, it was also associated primarily with CD14high PMNs (Fig. 5 A). PGE2 production is another major mechanism of PMN-MDSC activity (He et al., 2018; Veglia et al., 2019). PGE2 synthesis is controlled by two enzymes, COX2 (encoded by Ptgs2) and PTGES (encoded by Ptges). We found substantially higher expression of Ptgs2 in tumor CD14high PMNs than in any other PMN population. Tumor CD14int PMNs but not CD14− had higher expression of Ptgs2 than control CD14− PMNs. High Ptges expression was associated with all three populations of tumor PMNs (Fig. 5 B). We have previously identified fatty acid transport protein 2 (FATP2) encoded by Slc27a2 as selectively upregulated in PMN-MDSCs and involved in their suppressive activity (Veglia et al., 2019). We found that the expression of Slc27a2 was higher in CD14int PMNs than in CD14− PMNs. Expression of this gene in CD14high PMNs in spleens and tumors was markedly higher than in CD14− and CD14int PMNs (Fig. 5 C). Expression of Cd274 that encodes PD-L1 was dramatically higher in tumor CD14high PMNs than in other populations of PMNs. No differences in the expression of S100a9 were seen between the populations of spleen PMNs. In contrast, tumor CD14high PMNs expressed substantially higher S100a9 than tumor CD14− PMNs (Fig. 5 C). We also assessed the expression of some surface proteins associated with PMN-MDSC. In EL4 TB mice, spleen CD14int PMNs expressed markedly higher CD49D, SIGLEC-F, PD-L1, and TREM1 proteins than CD14− PMNs. The expression of these proteins was even higher in CD14high PMNs (Fig. 5 D). Similar results were obtained in LLC TB mice (Fig. S3 B).
Figure S3.
Morphology and phenotype of the populations of PMNs and CD14 expression and the function of the populations of PMNs in an acute infection model. (A) Giemsa staining of indicated populations of PMNs in LLC TB mice. Scale bars = 10 µm. (B) Expression of indicated molecules by flow cytometry in the populations of from tumors of LLC TB mice; 6 mice per group were analyzed. Frequency of CD14−, CD14int, and CD14high PMN differentiated from MLPG (n = 4). Mean and SD are shown. P values were calculated in unpaired two-sided Student’s t test. (D) Frequency of the populations of PMNs in spleens of mice 7 d after infection with Armstrong strain of LCMV; n = 5 mice per group. (E) Expression of arg1, nos2, ptgs2, and ptges by qPCR in control spleen CD14− PMNs, as well as CD14− and CD14int PMNs isolated from spleen of LCMV-infected mice; n = 5 mice per group. (F) T cell proliferation in the presence of PMNs isolated from spleens of LCMV-infected mice (n = 3 mice per group). Mean and SEM are shown. *, P < 0.05; ****, P < 0.0001.
Figure 5.
Expression of genes related to PMN-MDSC activity in different populations of PMNs. (A–C) Expression of indicated genes by qPCR in different populations of PMNs isolated from spleen and tumors of LLC TB mice. Four experiments were performed. (D) Expression of selected markers by flow cytometry in the populations of PMNs from tumors of LLC TB mice (n = 4 per group). All data are presented as mean ± SD. P values were calculated in ANOVA with correction for multiple comparisons. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
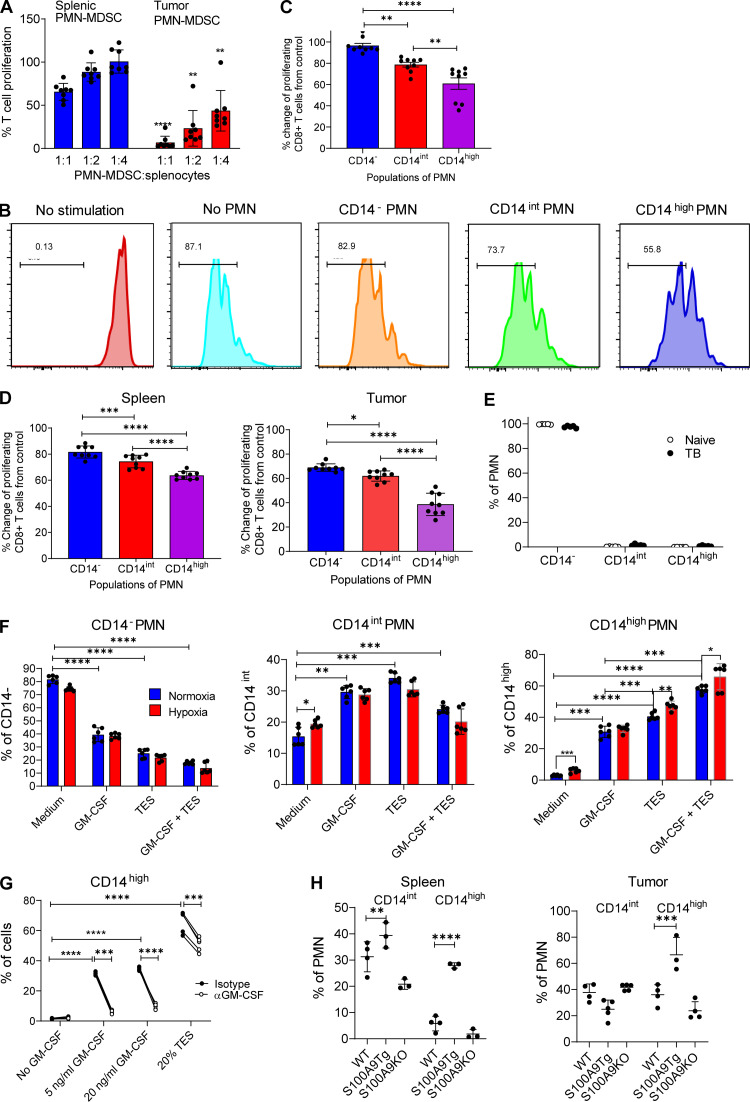
Our data demonstrated markedly higher presence of CD14high PMNs in tumors than in spleens. If CD14high PMNs represented more potent PMN-MDSCs, then tumor PMNs would have more suppressive activity than spleen PMN-MDSCs. We tested this concept in side-by-side experiments using pull-down with magnetic beads of Ly6G+ PMNs from spleens and tumors and determined that on a per-cell basis, tumor PMNs were much more suppressive than spleen PMNs in LLC TB mice (Fig. 6 A). Next, we assessed the ability of different populations of PMNs in tumors to suppress T cell function. FACS sorting of PMN-MDSCs impaired their functional activity because of the high sensitivity of these cells to sorting. However, it was unavoidable due to the requirements to use several surface markers on the same cell. PMNs were sorted from tumors and assessed in antigen-specific suppression assays. CD14− PMNs had no detectable suppressive activity. CD14int PMNs were weakly suppressive, whereas CD14high PMNs demonstrated more potent suppression (Fig. 6, B and C). In another tumor model (EL4 lymphoma), we compared side-by-side the suppressive activity of different populations of PMNs in spleens and tumors. Tumor CD14high PMNs had more potent activity than spleen CD14high PMNs. However, in both, spleen and tumor CD14high PMNs showed substantially higher suppressive activity than CD14int PMNs. CD14int PMNs were more suppressive than CD14− cells, which had little suppressive activity (Fig. 6 D). Together with the gene expression data, these functional data indicated that CD14high cells are potently suppressive cells and probably major contributors to the suppressive activity associated with PMN-MDSCs.
Figure 6.
Suppressive activity of PMN in TB mice. (A) Antigen-specific suppressive activity of PMN-MDSCs isolated from spleens and tumors of LLC TB mice (n = 8 mice per group). Changes from T cell proliferation in the absence of PMNs (100%) are shown. (B) Representative flow cytometric analysis of proliferation of CD8+ T cells upon stimulation with cognate antigen with or without PMNs isolated from tumors of LLC TB mice. (C) Proliferation of antigen-specific T cells in the presence of different populations of PMNs isolated from tumors of LLC TB mice. Changes from T cell proliferation in the absence of PMNs (100%) are shown. n = 9 mice per group. (D) Suppressive activity of PMNs isolated from spleen and tumors of EL-4 TB mice. PMNs were sorted from spleens and tumors of EL-4 TB mice 3 wk after tumor inoculation. PMNs were added to splenocytes from PMEL mice at a 1:1 ratio in the presence of cognate peptide. Proliferation of T cells was measured as dilution of Cell Trace by flow cytometry (n = 9). (E) Frequency of PMN populations in BM of naive (n = 4) and TB (n = 4) mice. (F) Frequency of PMN populations after stimulation of BM-derived PMNs with TES in normal and hypoxic conditions (n = 6). (G) PMNs were isolated using magnetic beads from BM of naive mice and were cultured with different concentrations of GM-CSF or 20% TES, with or without GM-CSF neutralizing antibody. Frequency of PMNs was analyzed after 24 h by flow cytometry (n = 4). (H) Frequency of populations of PMNs in spleen and tumors of LLC TB WT, S100A9Tg, or S100A9KO mice (n = 3). All data in the figure are presented as mean ± SD. For comparisons between groups, one-way ANOVA with correction for multiple comparisons was used. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
We recently identified monocyte-like precursors of PMN-MDSCs (MLPGs) responsible for the generation of a substantial proportion of PMN-MDSCs in TB mice (Mastio et al., 2019). Since CD14 is a monocytic differentiation marker, we asked whether there is a specific differentiation pathway of CD14int/high PMN-MDSCs from MLPGs. MLPGs (CD11b+Ly6G−Ly6ChighCD117+) were isolated from BM of LLC-bearing CD45.2+ mice and transferred into sublethally irradiated naive CD45.1 mice. After 3 d, splenic PMNs (CD11b+Ly6CintLy6G+) were analyzed. Consistent with previous observations (Mastio et al., 2019), >90% of MLPG-derived cells were PMNs. Most of PMNs were CD14−, with only a small proportion of cells expressing CD14 marker. No differences between MLPG-derived and host PMNs was observed (Fig. S3 C). PMNs in BM contained very few CD14+ PMNs, and no differences were observed between naive and TB mice (Fig. 6 E). Taken together, these results strongly suggest that the expression of CD14 on tumor PMN-MDSCs was not the result of specific pathways of differentiation in BM, but probably the effect of local factors in the microenvironment. To test this hypothesis, we isolated PMNs from BM of naive mice and cultured them for 24 h in the presence of tumor explant supernatants (TESs) and (or) hypoxia (0.5% O2). GM-CSF and TESs caused a very prominent increase in the proportions of CD14int and CD14high PMNs. TESs caused a stronger increase of CD14high PMNs than GM-CSF. Hypoxia alone caused an increase in the presence of CD14int and CD14high PMNs (Fig. 6 F). To assess the role of GM-CSF in induction of CD14high PMNs by TESs, we isolated PMNs from BM of naive mice using magnetic beads and cultured them for 24 h with GM-CSF or TESs in the presence of GM-CSF blocking antibody. As expected, neutralizing GM-CSF antibody completely abrogated the effect of GM-CSF on accumulation of CD14high PMNs. GM-CSF antibody reduced the effect of TESs on CD14high accumulation (Fig. 6 G). However, it did not abrogate this effect. These data indicate that TES-derived GM-CSF contributes to regulation of CD14 expression on PMNs, but other tumor-derived factors also contribute to this phenomenon.
Upregulation of S100A8/A9 protein is one of the hallmarks of PMN-MDSCs (Bronte et al., 2016). Our data showed association of S100a9 expression with CD14high PMN-MDSC in tumors (Fig. 5 C). We asked whether S100A8/A9 could influence the expression of CD14 on PMNs. To test this hypothesis, we evaluated PMNs in S100A9 transgenic (Tg) mice that overexpress S100a9 and in S100A9 knockout (KO) mice (Cheng et al., 2008; Manitz et al., 2003; Ortiz et al., 2015). LLC was established s.c., and spleens and tumors were evaluated 3 wk after tumor inoculation. PMNs from S100A9Tg LLC TB mice had a markedly higher presence of CD14high PMNs in both spleens and tumors than WT mice (Fig. 6 H). The proportion of CD14int and CD14high PMNs in S100A9KO mice was the same as in WT mice. These results suggest that S100A9 might contribute to upregulation of CD14 expression on the surface of PMNs.
We investigated whether CD14 expression can also define populations of PMNs in a model of acute infection. To this end, we used acute lymphocytic choriomeningitis virus (LCMV) infection (Armstrong). Spleens were collected on day 7 after infection. Infection was associated with the expansion of PMNs in spleens and an increase in the presence of CD14int PMNs but not CD14high cells (Fig. S3 D). CD14int PMNs in LCMV-infected mice did not have upregulation of the major genes associated with PMN-MDSC activity (Fig. S3 E) and lacked suppressive activity (Fig. S3 F). Thus, our data indicated that during tumor progression, two distinct populations of PMN-MDSCs gradually replaced classical PMNs in peripheral lymphoid organs. In tumor sites, potently suppressive CD14high PMN-MDSCs were present at very early stages of tumor growth.
PMN-MDSC gene signature in tumors is associated with negative clinical outcome in cancer patients
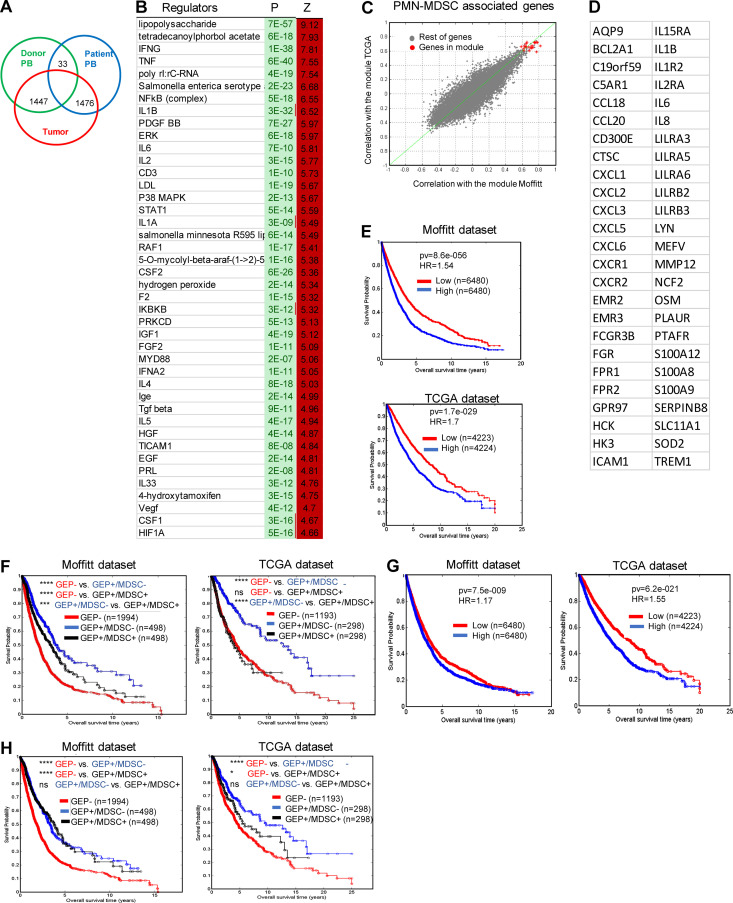
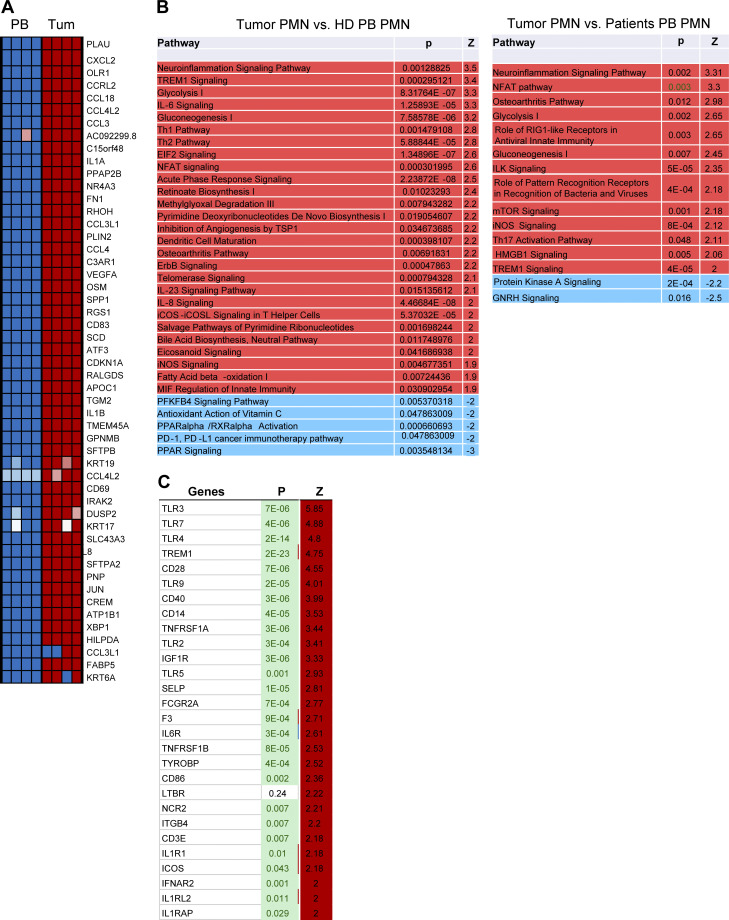
We compared the gene expression profile (GEP) of the total population of PMNs (CD45+CD14−CD15+CD66b+) sorted from tumors of cancer patients and compared it with peripheral blood PMNs with the same phenotype from heathy donors and the same cancer patients. Consistent with previous observations (Condamine et al., 2016), peripheral blood PMNs from healthy donors and cancer patients demonstrated a similar GEP, which probably reflects the relatively small proportion of PMN-MDSCs among the total PMN population. Only 33 genes were differentially expressed (false discovery rate [FDR] <5%). In contrast, the GEP of tumor PMNs was quite distinct, with >1,400 genes differentially expressed (Fig. 7 A). The top 50 changed genes are shown in Fig. S4 A. There was a substantial overlap in genes overexpressed in tumor PMNs with the previously identified signature of blood PMN-MDSCs (Condamine et al., 2016). In tumor PMNs, IPA revealed enrichment of pathways associated with inflammatory response, iNOS signaling, fatty acid β oxidation, and ER stress response (Fig. S4 B). This was confirmed by the analysis of gene regulators. Among increased regulators were NF-κB complex, IL1, IFNγ, IL6, HGF, STAT1, ATF4, CEBPB, and thapsigargin (Fig. 7 B). We also compared the expression of transmembrane receptors in PMNs from tumor and peripheral blood of cancer patients. TLR3, 7, 4, CD14, TREM1, and CD40 were among those upregulated in tumor PMNs (Fig. S4 C). Mouse and human genes associated with PMN-MDSCs are presented in Table 1.
Figure 7.
GEP of human tumor PMNs. (A) The number of differentially expressed genes (FDR <5%) between PMNs isolated from peripheral blood and tumors. (B) Gene regulators enriched in tumor PMNs in comparison with peripheral blood PMNs from the same patients. (C and D) Genes found to be coexpressed with PMN-MDSC signature, with correlation coefficient >0.6 after analysis of TCGA and Moffitt datasets. (E) Association of PMN-MDSC signature with survival of patients in Moffitt and TCGA databases. (F) Association of PMN-MDSC signature with survival of patients with GEP of T cell infiltrated tumors. (G) Association of PMN signature with survival of patients. (H) Association of PMN signature with survival of patients with GEP of T cell infiltrated tumors. Number of samples analyzed, P values, and HRs are shown on graphs or presented as *, P < 0.05; ***, P < 0.001; ****, P < 0.0001.
Figure S4.
Gene expression in cancer patient PMNs. (A) Top 50 changed genes in tumor PMNs compared with peripheral blood PMNs in the same patients. (B) IPA of tumor PMNs compared with peripheral blood (PB) healthy donor (HD) or patient PMNs. (C) Changes in the expression of genes encoding transmembrane receptors in tumor PMNs compared with peripheral blood PMNs from the same patient.
Table 1. Genes associated with mouse and human PMN-MDSC.
| Mouse | Human | ||
|---|---|---|---|
| PMN-MDSC | Activated PMN-MDSC | PMN-MDSC | |
| Ngp | Ccl4 | AQP9 | LILRA5 |
| Camp | Ccl3 | BCL2A1 | LILRA6 |
| Ltf | Cxcl3 | C19orf59 | LIRRB2 |
| Chil3 | Jun | C5AR1 | LILRB3 |
| Lcn2 | Ccrl2 | CCL18 | LYN |
| Iftm6 | Saa3 | CCL20 | MEFV |
| Lyz2 | Spp1 | CD300E | MMP12 |
| Cybb | Gad45b | CTSC | NCF2 |
| Serpinb1a | Il1b | CXCL1 | OSM |
| Cd177 | Ninj1 | CXCL2 | PLAUR |
| Anxa1 | Clec4n | CXCL3 | PTAFR |
| Aldh2 | Hcar2 | CXCL5 | S100A12 |
| Mmp8 | Basp1 | CXCL6 | S100A8 |
| Adpgk | Nfkbia | CXCR1 | S100A9 |
| Dstn | Btg1 | CXCR2 | SERPINB8 |
| Arhgdib | Il1rn | EMR2 | SLC11A1 |
| S100a8 | Ifrd1 | EMR3 | SOD2 |
| AA467197 | Txnip | FCGR3B | TREM1 |
| Tkt | Ccl9 | FGR | |
| Wfdc21 | Ier3 | FPR1 | |
| Capg | Ier5 | FPR2 | |
| Cebpe | Rs1 | GPR97 | |
| S100a9 | Thbs1 | HCK | |
| Syn1 | Cxcl1 | HK3 | |
| Pglyrp1 | Hilpda | ICAM1 | |
| Ltb4r1 | Hist1h1c | IL15RA | |
| Lgals3 | Srgn | IL1B | |
| Tmsb10 | Hspa5 | IL1R2 | |
| Olfm4 | Csf1 | IL2RA | |
| Hmgn2 | Pgts2 | IL6 | |
| Ceacam10 | Xbp1 | IL8 | |
| H2afz | LILRA3 | ||
Genes shared between human and mouse PMN-MDSCs are shown in red.
We asked whether the gene signature of tumor PMN-MDSCs was associated with clinical outcome across different types of cancer. A gene expression fingerprint developed based on all PMN-associated genes has been projected into two large cohorts of GEPs from human tumor samples (The Cancer Genome Atlas [TCGA] and Moffitt datasets) and analyzed for coherence and consistency. The average coexpression of the genes in the fingerprint was found to be 0.6 in TCGA and 0.65 in Moffitt, indicating highly coherent expression. A metaprofile corresponding to the average of the genes in the fingerprint was then used for further analysis. 50 genes were found to be coexpressed with tumor PMN-MDSC signature, with a correlation coefficient >0.6 (Fig. 7, C and D). Patients with high level of expression of PMN-MDSC signature had substantially shorter survival than patients with lower expression of that signature in both Moffitt and TCGA databases. The hazard ratio (HR) for the Moffitt dataset was 1.54, and for the TCGA dataset, 1.7 (Fig. 7 E). We further explored the prognostic value of PMN-MDSCs in tumor types with GEP of T cell–infiltrated tumors as it has been defined previously (Ayers et al., 2018). This GEP was shown to be associated with better clinical outcome (Ayers et al., 2017). In patients with GEP of T cell–infiltrated tumors, high level of PMN-MDSC signature was associated with markedly shorter survival than low PMN-MDSC signature (Fig. 7 F). These data suggest that tumor PMN-MDSCs are associated with negative clinical outcome, even in patients with strong T cell infiltration (based on GEP). We asked if the signature of classical PMNs would also be associated with negative clinical outcome. We used the gene signature of LOX-1− PMNs identified in the peripheral blood of cancer patients (Condamine et al., 2016). In the Moffitt dataset, PMN signature was not associated with negative clinical outcome (HR = 1.17), whereas in the TCGA dataset, this association of markedly lower than in PMN-MDSCs (HR = 1.55; Fig. 7 G). In the Moffitt dataset, PMN signature was not associated with shorter survival in patients with strong T cell infiltration. In the TCGA dataset, this association was markedly smaller than in the PMN-MDSC signature (Fig. 7 H). Thus, tumor PMN-MDSCs, but not PMNs, are strongly associated with negative clinical outcome, even in patients with tumors infiltrated by T cells.
Identification of distinct populations of PMNs in cancer patients
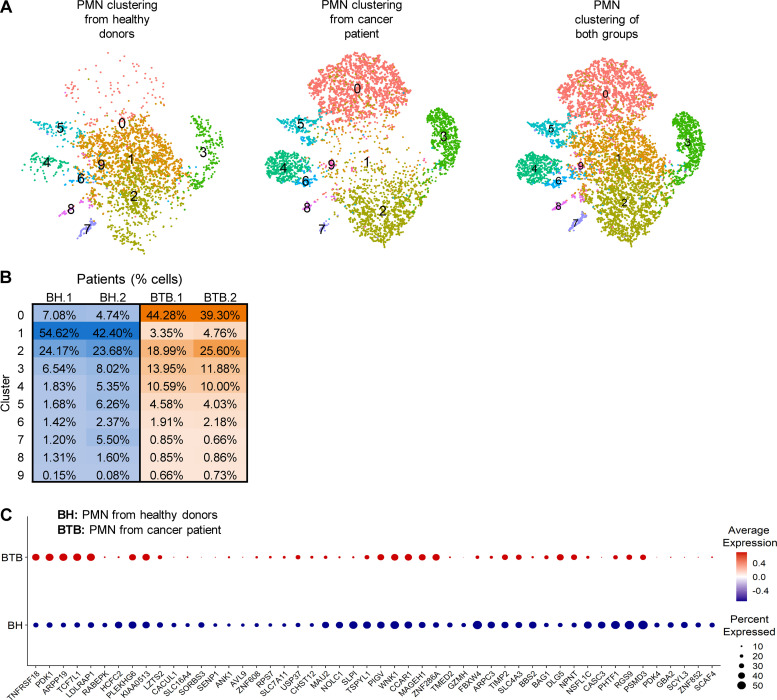
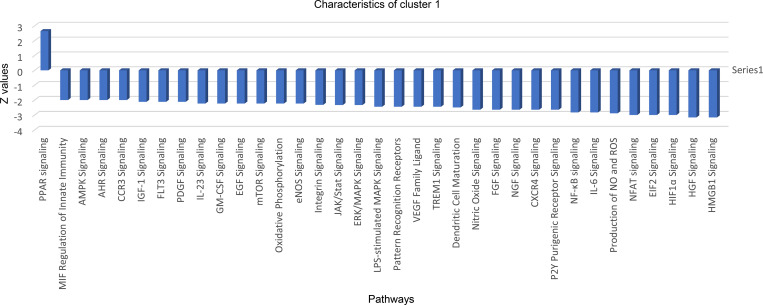
To evaluate the heterogeneity of PMNs in cancer patients, we sorted CD45+CD14−CD15+CD66b+ PMNs from the peripheral blood of healthy donors and cancer patients and performed scRNA-seq. 10 distinct clusters could be defined in PMNs (Fig. 8 A). However, only four of them represented >5% of cells. Cluster 1 included 50% of all PMNs in healthy donors, but <5% in cancer patients (Fig. 8 A). Pathway analysis revealed that this cluster had markedly lower expression of genes associated with HMGB1 and HIF1α signaling, TREM1 signaling, MAPK signaling, IL-6, ER stress, JAK/STAT signaling inflammation, and NO and ROS production than other clusters (Fig. S5). Thus, this cluster of PMNs had a GEP similar to that of classical nonactivated PMNs. Cluster 2 represented 20–25% of PMNs in both healthy donors and cancer patients. Pathway analysis did not reveal significant differences between these two clusters, which may suggest that clusters 1 and 2 could represent classical nonactivated PMNs. In contrast, cluster 0 represented <8% of PMNs in healthy individuals and 40% of PMNs in cancer patients (Fig. 8 B). When individual genes were compared between PMNs from healthy donors and cancer patients, several genes associated with PMN-MDSC signature (LDLRAP1, PRDM15, TIMP2, NPNT, DLG5, and BAG1) were upregulated in patient PMNs (Fig. 8 C).
Figure 8.
Clustering of peripheral blood human PMNs. (A) t-SNE plots of cells clustering (n = 10 clusters) of PMNs from healthy donors and cancer patients. (B) Percentage distribution of PMN cells among different clusters. (C) Dot plot of PMN-MDSC–associated genes that are upregulated in PMN from cancer patients. The red and blue colors indicate up- and downregulated expression, respectively. BH, PMN from healthy donors; BTB, PMN from cancer patient.
Figure S5.
Pathways changed in cluster 1 in patient PMNs. AHR, aryl hydrocarbon receptor (AHR); AMPK, AMP-activated protein kinase; EGF, epidermal growth factor; eNOS, endothelial NOS; FGF, fibrobalst growth factor; HGF, hepatocyte growth factor; mTOR, mechanistic target of rapamcyin; NGF, nerve growth factor; PDGF, platelet derived growth factor; PPAR, peroxisome proliferator-activated receptors; VEGF, vascular endothelial growth factor.
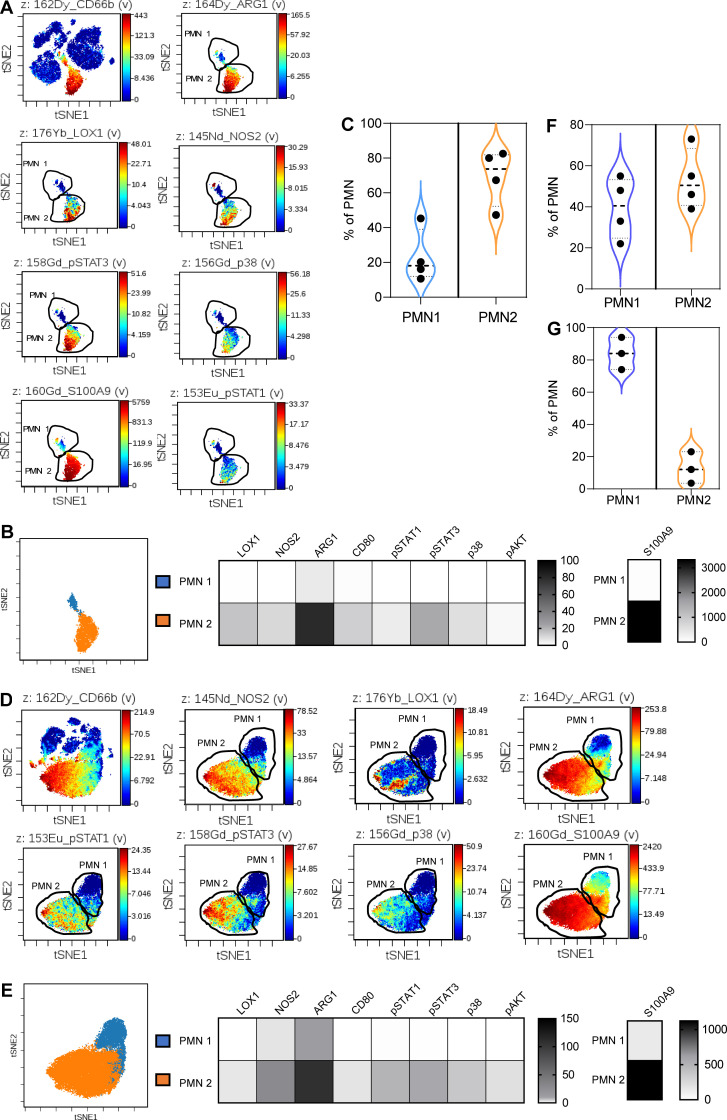
These results suggested the existence of at least two major populations of PMNs in blood of cancer patients. However, detailed characterization of these populations based on scRNA-seq was very difficult due to low representation of the transcripts in these cells. Therefore, we used cytometry by time of flight (CyTOF). We assembled a cocktail of antibodies that allowed for identification of different functional and phenotypical markers known to be associated with PMNs in healthy donors and PMN-MDSCs in cancer patients (Table S1). Samples were collected from tumor tissues and peripheral blood of cancer patients and healthy donors. PMNs were gated based on the expression of CD15 and CD66b markers (Fig. 9 A). Two distinct populations of PMNs were identified in tumors (Fig. 9, A and B). One population (PMN1) with phenotype CD11b+CD15+CD16+CD66bhighArg1+/−Lox1−pSTAT3−S100A9+, resembled classical PMNs, and the other one (PMN2) had phenotype CD11bhighCD15highCD66bhighCD33highArg1highNos2highS100A9highLox1high pSTAT3highp38+pSTAT1+, typical for PMN-MDSCs (Fig. 9 B). The PMN2 population represented a substantial majority of PMNs in tumors (70%; Fig. 9 C). In peripheral blood of cancer patients, the same two populations of PMNs were evident (Fig. 9, D and E). The proportions of PMN1 and PMN2 were equally represented among PMNs in peripheral blood (Fig. 9 F). In contrast, in peripheral blood of healthy donors, the population of PMN1 represented most PMNs (>80%; Fig. 9 G). Because mouse data demonstrated a specific role of CD14 in defining populations of PMNs, we evaluated the expression of CD14 on human PMNs. However, no detectable expression was observed in PMNs from healthy donors or cancer patients.
Figure 9.
Analysis of populations of PMNs in cancer patients by CyTOF. (A) Representative viSNE analysis of PMNs from tumors. CD66b+CD15+ PMNs were gated and the analysis of markers was performed within total population of PMNs. (B) Heatmap generated using raw median values of selected markers and proportion of PMN1 and PMN2 among all PMNs (n = 4). (C) The proportion of PMN1 and PMN2 populations among PMNs from tumors (n = 4). (D) Representative viSNE analysis of PMNs from peripheral blood of cancer patients. Analysis was performed as described in Fig. 9 A. (E) Heatmap generated using raw median values of selected markers and proportion of PMN1 and PMN2 among all PMNs (n = 4). (F) The proportion of PMN1 and PMN2 populations among PMNs from peripheral blood of cancer patients (n = 4). (G) The proportion of PMN1 and PMN2 populations among PMNs from peripheral blood of healthy donors (n = 4).
Discussion
In this study, we identified three distinct populations of PMNs in mice. PMN1 represented almost all PMNs in spleens of tumor-free mice. During tumor progression, it was gradually replaced by the PMN2 population, which had a GEP similar to that described for PMN-MDSCs (Youn et al., 2012). It appears that PMN1 is likely a population of nonactivated classical PMNs, as opposed to PMN2, which had a GEP of PMN-MDSCs. In tumors, a third population of PMNs was most prominent. These cells appeared at the very early stages of tumor development and remained the most abundant population of PMNs in tumors. Although PMN3 cells had a gene profile similar to PMN-MDSCs, they had high expression of genes associated with chemotaxis, typically expressed by more mature PMNs. These cells also showed activation of pathways of inflammation, NO, ROS, mechanistic target of rapamycin, TREM1, and HMGB1 signaling. All these pathways have been associated with suppressive properties of MDSCs (Condamine et al., 2015a; Gabrilovich, 2017). These results indicate that in TME, PMN-MDSCs are represented by the distinct group of cells with much stronger activation and potentially more suppressive features than PMN-MDSCs in peripheral lymphoid organs. This population coexists not only with the small group of classical PMNs, but also with less activated and less suppressive PMN-MDSCs. Contrary to our initial expectations, computational trajectory prediction analysis demonstrated that these two populations of PMNs had distinct trajectories, suggesting that they were not directly linked.
scRNA-seq cannot address the question of functional diversity of these cells. Therefore, we attempted to identify surface molecules that could be associated with these populations. A number of surface molecules were differentially expressed on PMNs and PMN-MDSCs. However, only one, CD14, showed a pattern of expression remarkably similar to three populations of PMNs defined by RNA-seq: CD14−, CD14int, and CD14high. CD14 is strongly positive in monocytes and most tissue macrophages. Myeloblasts and other granulocytic precursors do not express CD14, but neutrophils may be weakly positive. CD14 is a glycosylphosphatidylinositol-anchored receptor, which together with TLR4 and MD-2 forms the multireceptor complex that recognizes LPS on the cell membrane (Jerala, 2007; Lloyd and Kubes, 2006; McAvoy et al., 2011; Yoshino et al., 2000). Downstream signaling from that complex involves NF-κB, src family kinase, and PLCγ 2 activation. The latter hydrolyzes membrane phosphatidylinositol bisphosphate, resulting in generation of inositol-triphosphate and diacylglycerol. This leads to activation of NFAT transcription factors (Zanoni et al., 2009). Activation of the NFAT pathway in dendritic cells can result in upregulation of PGE2 (Zanoni et al., 2012) via the transcription of Ptges (Zanoni and Granucci, 2012). Remarkably, PMN3 showed upregulation of LPS-mediated MAPK signaling, dendritic cell maturation signaling, TLR signaling, and NFAT signaling. These pathways were not upregulated in the PMN2 population. This confirmed that CD14 expression is associated mostly with PMN3.
CD14− PMNs represented almost 95% of PMNs in spleens of tumor-free mice, suggesting that CD14− PMNs may represent classical PMNs. Analysis of the expression of genes directly implicated in suppressive activity of PMN-MDSCs (arg1, nos2, ptgs2, and slc27a2) demonstrated that CD14− PMNs from TB mice had either the same or only slightly elevated expression of these genes as compared with CD14− PMNs from tumor-free mice. These cells in tumors lacked T cell suppressive activity. CD14int PMNs had modest upregulation of immune-suppressive genes and suppressive activity. In contrast, CD14high PMNs had strong upregulation of immune suppressive genes and potent suppressive activity. CD14high PMNs also expressed higher levels of cd274 (encoding PDL1), PDL1, and SIGLEC F that recently was found to be associated with PMN-MDSCs (Engblom et al., 2017). Increased expression of slc27a2 and cd36 suggest that CD14high PMNs may have a dysregulated lipid metabolism. Previous study implicated slc27a2 and its protein FATP2 in regulation of suppressive activity of PMN-MDSCs via production of PGE2 (Veglia et al., 2019). Moreover, CD36 has also been involved in the regulation of functions of MDSC in cancer (Al-Khami et al., 2017).
CD14high PMNs were almost exclusively present in TME. Our data indicate that these cells did not differentiate from dedicated precursors in BM but activated in situ in the response to different soluble factors present in TESs (including GM-CSF) and hypoxia. Interestingly, the hypoxia-associated pathway was upregulated in the PMN3 population, further supporting the hypothesis that CD14high and PMN3 represent the same population of PMNs. To our surprise, the population of CD14high PMNs was present in tumors at early stages and did not change with time, suggesting that regulation of immune responses by activated PMN-MDSCs may take place very early in tumor development.
Several years ago, CD14 was implicated as a TLR4 coreceptor in the S100A8/A9-induced cytokine response (He et al., 2016). S100A8/A9 is one of the hallmarks of PMN-MDSC (Bronte et al., 2016). Our data show that PMN2 and PMN3 populations had higher expression of s100a8 and s100a9 than in PMN1. It has been described that S100A9-deficient PMNs lose suppressive activity (He et al., 2018; Ortiz et al., 2015). Since S100A9 released by PMNs binds to CD14/TLR4 complex, high expression of CD14 on PMN-MDSCs may facilitate their suppressive activity. Recently, CD84 has been identified as marker of MDSCs in spleens of TB mice (Alshetaiwi et al., 2020) We did not find differences in CD84 expression in PMNs from spleen and tumors. Apparently, in our tumor models, TME did not impact CD84 expression.
In humans, substantial differences between tumor and peripheral blood PMNs were identified in bulk RNA-seq. PMNs from peripheral blood of healthy donors and cancer patients demonstrated very similar transcriptomes, with only a few genes differentially expressed. This was consistent with a previous report (Condamine et al., 2016). However, PMNs in tumors were markedly different and expressed profiles with increased expression of chemokine receptors, S100A8, A9, TREM1, and other genes associated with PMN-MDSCs, as well as upregulation of ER stress, mammalian target of rapamycin, and inflammatory pathways. Interestingly, tumor PMNs also had upregulation of NFAT pathway downstream of CD14. Thus, tumor PMNs in cancer patients had similar transcriptomic features with mouse PMN3 population. Similar to mouse PMN-MDSCs, human tumor PMNs had upregulation of CD14. Gene signatures based on tumor PMNs had very strong negative predictive values in patients with activated T cell response, underscoring the important role of these cells in regulation of tumor progression and possible response to immunotherapy.
The concept of coexistence of classical PMNs and PMN-MDSCs in peripheral blood was confirmed in cancer patients using scRNA-seq. We identified several major clusters of PMNs. Two clusters representing >70% of all PMNs in healthy donors and only 20–25% in cancer patients had a transcriptome of classical PMNs, whereas one cluster with <8% representation among PMNs from healthy donors and 40% in cancer patients had some features of PMN-MDSC. We confirmed this concept by using multiparameter CyTOF analysis, where we used multiple surface and intracellular molecules identified previously or during this study as associated with PMN-MDSCs, to better define the populations of PMNs in peripheral blood and tumors. We observed two distinct populations of PMNs. One population had typical features of PMN-MDSCs, with high expression of ARG1, iNOS, pSTAT3, LOX-1, and S100A9, whereas the other could be characterized as classical PMNs. Since CD14 expression was associated with tumor PMNs, we expected to identify a specific role of CD14 in human PMNs. Expression of CD14 was associated with hybrid PMNs that were found in tissue of early-stage lung cancer patients (Singhal et al., 2016). However, unlike mouse PMNs, CD14 was not expressed on human PMNs.
Thus, this study demonstrated coexistence of the populations of classical PMNs and immune suppressive PMN-MDSC in mice and humans and identified the marker of PMN-MDSC in mice, CD14. Although in spleens, CD14int PMN-MDSCs only gradually replace classical PMNs, in tumors, activated and potently suppressive CD14high PMN-MDSCs appear very early in tumor development. In humans, the gene signature of tumor PMNs is strongly associated with negative clinical outcome in patients with signature of T cell infiltration and suggests novel targeting opportunities.
Materials and methods
Human samples
Samples of peripheral blood and tumor tissues were collected from patients at the Helen F. Graham Cancer Center and the University of Pennsylvania. The study was approved by the Institutional Review Board (IRB) of the Christiana Care Health System at the Helen F. Graham Cancer Center and the Wistar IRB. All patients signed IRB-approved consent forms. Samples were collected at Helen F. Graham Cancer Center from six patients with previously untreated stage II–IV non–small cell lung cancer. Blood from healthy volunteers was collected at the Wistar Institute. All patients and donors signed IRB-approved consent forms.
Mouse models
Animal experiments were approved by the Wistar Institute Animal Care and Use Committee and the H. Lee Moffitt Cancer Center Animal Care and Use Committee. C57BL/6 and C57BL/6-Ly5.1 mice (female, 6–8 wk old) were obtained from Charles River; OT-I TCR-Tg mice (C57Bl/6-Tg(TCRaTCRb)1100mjb; female, 6–8 wk old) were purchased from The Jackson Laboratory. KrasLSL.G12D/+; p53R172H/+; PdxCretg/+ (or KPC) mice were obtained from Robert H. Vonderheide at University of Pennsylvania. S100A9Tg and S100A9 KO mice were described previously (Cheng et al., 2008; Kato et al., 1998; Manitz et al., 2003; Meyer et al., 2011; Ortiz et al., 2015).
Tumor cell lines
EL4 (lymphoma) and LLC were from ATCC. All cells were maintained in DMEM supplemented with 10% FBS (Sigma-Aldrich) at 37°C, 5% CO2. Tumor cells were injected s.c. at 5 × 105 cells per mouse and formed tumors with a 1.5-cm diameter within 2–3 wk of injection. Tumor cell lines were tested for mycoplasma contamination by using the Universal Mycoplasma detection kit (ATCC) every 3 mo. GL261 cells were obtained from Dr. Rintaro Hashizume (Northwestern University, Chicago, IL).
Orthotopic glioma cell injection
Mice were intracranially injected with 40,000 GL261 cells expressing luciferase into the right frontal cortex (1 mm caudal, 1.5 mm lateral from bregma, and 2 mm deep) using a stereotactic apparatus. Mice were monitored daily for neurological symptoms, lethargy, and hunched posture that would qualify as signs of tumor burden.
RNA-seq
RNA-seq data were aligned using bowtie229 against mm10 version of the mouse genome, and RSEM v1.2.12 software was used to estimate raw read counts using Ensemble v84 gene information. DESeq231 was used to estimate the significance of differential expression between sample groups. Overall gene expression changes were considered significant if they passed the FDR <5% threshold. Significant genes affected at least twofold were analyzed for enrichment of upstream regulators using Qiagen’s Ingenuity Pathway Analysis software (upstream analysis option). Only regulators with significantly enriched P < 0.005 targets (at least 20) with significantly predicted activation states (activation z-score, |Z| > 2) were considered.
Single-cell library preparation and scRNA-seq data processing
Single-cell libraries were prepared with isolated PBMCs using Chromium Single Cell 3′ v2 Reagent Kits. The Cell Ranger Software Suite (v3.1.0) was used to process mouse and human PMN scRNA-seq data with command cell ranger count. For mice PMN samples, cellranger count was executed with refdata-cellranger-mm10-3.0.0 transcriptome to map reads on the mouse genome (mm10) using STAR (v2.5.2b; Dobin et al., 2013), and unique molecular identifiers (UMIs) were counted. The human PMN samples were processed in a similar fashion with refdata-cellranger-GRCh38-3.0.0 transcriptome for mapping reads on the human genome (GRCh38/hg38), and UMIs were counted. The outputs of cellranger count were loaded in R-statistical environment using the Read10X function. The low-quality cells with few expressed genes (n < 400) were removed. After filtering, a total of 66,854 cells expressing at least 400 genes were selected for the following analysis.
The UMIs of 66,854 cells were supplied to scran package (v1.14.5; 27122128) to group cells into clusters of similar expression using the quickCluster function. Normalization within each cluster was performed to compute size factors of each cell, and the factors were rescaled by normalization between clusters using the computeSumFactors function. The logNormCounts function from scater R package (28088763) was used to normalize expression values, which controls cell-specific biases. These normalized expression values were supplied to Seurat package (v3.1.1; Stuart et al., 2019) to perform batch correction, cell clustering, and marker identification. Briefly, the FindVariableFeatures function was used to find highly variable genes (n = 2,000), followed by anchor detection and sample integration using FindIntegrationAnchors and IntegrateData functions, respectively, to control the batch effect among the PMN samples. The scaling was performed on integrated assay data using ScaleData function. The principal components (PCs) were estimated by RunPCA function. The first 30 PCs were used for cell clustering using the FindNeighbors and FindClusters functions, with resolution 0.6 and UMAP visualization (using RunUMAP function). Differentially expressed genes among cell clusters were identified using the FindAllMarkers function with the following criteria: only.pos = false, min.pct = 0.25, and thresh.use = 0.25. Gene set enrichment analysis was accomplished using Qiagen’s Ingenuity Pathway Analysis software using “Pathways,” “Networks,” “Regulator Effects,” and “Diseases & Functions” on the genes found significant (FDR<5%) between PMN3 versus PMN1, PMN3 versus PMN2, PMN3 versus PMN1 and PMN2, PMN2 versus PMN1, and PMN2 and PMN3 versus PMN1 comparisons. Sequences were deposited to GEO, accession no. GSE163834.
Pseudotime analysis
Monocle 3 (v3.0.2.0; 30787437) was used to estimate a pseudotemporal path of three populations of PMNs cells. The normalized counts of cells clustered in PMN1, PMN2, and PMN3 by Seurat analysis were loaded to Monocle3 using the new_cell_data_set function to create a monocle object. This object was preprocessed using preprocess_cds function (with 30 PCs and 0.6 resolution) and batch corrected by align_cds function. The cells were ordered in pseudotime along a trajectory using reduce_Dimension with the UMAP method and order_cells functions. PMN1 population was selected as the starting point (initial stage). We identified genes that are differentially expressed in different subsets of PMN cells using graph_test function. These genes were then grouped into modules (PMN1-, PMN2-, PMN3-, and other-coregulated genes) using find_gene_modules function. This function runs UMAP on the genes and then groups them into modules using Louvain community analysis. The aggregate expression values were used to draw a heatmap of genes modules that are specific to PMN1, PMN2, PMN3, and others using pheatmap function. As described above, gene set enrichment analysis was done using IPA software on the significant genes (FDR <0.05) that are specific to PMN1, PMN2, and PMN3.
Overlap bulk RNA-seq with scRNA-seq data
Bulk RNA-seq data were aligned using bowtie2 (Langmead and Salzberg, 2012) algorithm against hg19 human genome version, and RSEM v1.2.12 software (Li and Dewey, 2011) was used to estimate read counts and RPKM values using gene information from Ensemble transcriptome version GRCh37.p13. Raw counts were normalized and used to estimate significance of differential expression difference between two experimental groups using DESeq2 (Love et al., 2014). Overall gene expression changes were considered significant if they passed the FDR <5% threshold.
Normalized bulk RNA-seq counts were z-score transformed, and genes that passed FDR <5% in both experiments were taken for analysis. Comparison between average z-scores of three bulk RNA-seq groups and z-scores of three single-cell clusters was performed using Pearson correlation, and nominal P values were corrected for multiple testing using Bonferroni procedure. Significance of overlap was estimated by hypergeometric test, using the 2,000 most variable detected genes used for clustering differential expression analysis in a single-cell experiment as a reference set.
Data availability
Raw UMI counts for single-cell RNA-seq are publicly available at GEO, accession no. GSE163834.
Isolation of mouse and human neutrophils
For mouse neutrophils (PMNs), single-cell suspensions were prepared from BM and spleen, followed by red blood cell removal using ammonium chloride lysis buffer. Single-cell suspensions from tumor tissues were prepared using Mouse Tumor Dissociation Kit according to the manufacturer’s recommendation (Miltenyi). Then cells were stained and sorted on BD FACS Aria (BD Biosciences) using the following gate strategy: CD45+CD11b+Ly6G+Ly6Clo. In some experiments, we also included CD14 marker. Human PMNs were isolated, and whole blood was enriched for PMNs using MACSxpress Neutrophil Isolation Kit (Miltenyi) following the manufacturer’s instructions. PMNs form tumor tissue using a modified method for disaggregation of fresh human lung tumors that preserves the phenotype and function of the immune cells as described in (Quatromoni et al., 2014). Briefly, tumor fragments were incubated in a 50-ml centrifuge tube with Hyclone Leibovitz L-15 medium (25 ml/0.5 g tissue) supplemented with 2% FBS and containing collagen I (170 mg/liter; 45–60 U/ml), collagen II (56 mg/liter; 15–20 U/ml), collagen IV (170 mg/liter; 45–60 U/ml), DNase I (25 mg/liter; 50 K U/ml; 0.002%), and elastase (25 mg/liter; 0.075 U/ml; 0.002%). Cells were placed on a shaker and incubated for 60 min at 37°C. After 30 min of incubation, tumor particles were pipetted vigorously by use of a 10-ml pipette to enhance disaggregation and incubated for further 30 min at 37°C. Then cells were passed through a 70-µM nylon cell strainer. Cells were stained and FACS sorted on BD FACS Aria using the following gate strategy: CD45+CD15+CD66b+CD14−. PMNs were used for further analysis.
Flow cytometry
For surface staining, 0.5 × 106 cells were incubated with a cocktail of surface antibodies, FC-block (BD Biosciences), and Aqua Fixable viability dye at room temperature (RT) for 20 min. The list of antibodies is provided in Table S2. After washing, cells were fixed and permeabilized with Fixation and Permeabilization Buffers (BD Biosciences) for 15 min at RT, washed twice with wash buffer (BD Biosciences), and incubated with a cocktail of intracellular antibody at RT for 30 min. Cells were run on either BD FACSymphony (BD Biosciences) or BD LSRII (BD Biosciences), and data were analyzed by FlowJo (Tristar).
CyTOF antibody conjugation and staining
Antibodies were labeled using the X8 antibody labeling kit as per the manufacturer’s protocol (Fluidigm). Single-cell suspensions (1 × 106 cells) were incubated with monoisotopic cisplatin-194Pt (Fluidigm) at RT for 5 min, washed with 1× PBS, incubated with Fc receptor–blocking solution to each tube, and incubated for 10 min at RT, without washing off Fc receptor–blocking solution. The antibody was added in Maxpar Cell Staining Buffer (Fluidigm) at RT for 30 min. After washing with PBS, the cells were fixed with 1× Maxpar Fix I Buffer (Fluidigm) for 30 min at RT, washed twice with Perm-S Buffer (Fluidigm), and then incubated with an intracellular antibody cocktail in Perm-S Buffer for 30 min at RT. The cells were washed and incubated overnight with Cell-ID Intercalator-Ir diluted in Maxpar Fix and Perm Buffer (Fluidigm). After another wash with 0.5% BSA in PBS, the cells were filtered, washed twice with 0.1% BSA, and run in a Helios mass cytometer (Fluidigm) at the University of Pennsylvania. The mass cytometry data were normalized to Equation 4-element calibration bead signal (Fluidigm) in 100-s-interval windows using normalization software version 2 (Fluidigm). Data were analyzed using Cytobank software. A list of reagents is provided in Table S3.
Processing and analyzing high-dimensional single-cell data
For flow cytometry datasets, cell debris, doublets, and dead cells, exclusion and biexponential transformation were run in FlowJo. PMN (CD45+CD11b+LY6G+LY6Clo) from four to five different mice were concatenated together and exported as a single FCS file and uploaded to the Cytobank software. Data were transformed by arcsinh, and dimension reduction visualization was performed using the vi stochastic neighbors embedding (viSNE) algorithm (Amir et al., 2013). The FlowSOM algorithm (Van Gassen et al., 2015) was applied and overlaid on viSNE to automatically and objectively identify cell populations based on the expression of markers selected for the analysis. Metaclusters with similar expression of these markers were combined, using automatic gate cluster functionality in Cytobank. For marker expression visualization in heatmaps, the raw median value of each marker in each cluster was generated using Cytobank and plotted using GraphPad Prism 8. For CyTOF human datasets, individual samples were uploaded in Cytobank and manually gated using Cytobank to eliminate cell debris, dead cells, and doublets, and for the identification of CD45+ cisplatin-negative cells. Dimension reduction visualization was performed using viSNE, viSNE plots were generated for all CD45+ cells, and PMN subsets were analyzed.
Suppression assay
Single-cell suspensions from spleens and tumors were prepared as described above. Cells were stained and sorted on BD FACS Aria (BD Biosciences). PMN-MDSCs (CD45+CD11b+Ly6G+Ly6CloCD14−/int/high) were plated in U-bottom 96-well plates (three replicates) in RPMI with 10% FBS and cocultured at different ratios with splenocytes from Pmel Tg mice in the presence of cognate peptides: murine gp100 peptide (EGSRNQDWL; 0.1 µg/ml). After 48 h, cells were incubated with [3H]thymidine (PerkinElmer) for 16–18 h. Proliferation was measured by using TopCount NXT instrument (PerkinElmer). For the CellTrace assay, Pmel splenocytes were labeled with CellTrace Far Red for 20 min at 37°C and cocultured with PMN-MDSCs and mgp100 peptide.
Quantitative real-time PCR (qPCR)
RNA was extracted using a Total RNA Kit according to the manufacturer’s instructions. DNase digestion was performed, and cDNA was generated with a High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems). qPCR was performed using Power SYBR Green PCR Master Mix (Applied Biosystems) in 96- or 384-well plates. Plates were read with ABI 7900 (Applied Biosystems). A list of primers is provided in Table S4.
MLPG differentiation in vivo
MLPGs were isolated from BM of LLC-TB C57BL/6 mice. Cells were incubated with Fc block followed by 20-min incubation with Aqua Fixable viability dye and antibodies cocktail. MLPGs were isolated through cell sorting using a MoFlo Astrios EQ (Beckman Coulter) cell sorter as Aqua−CD11b+Ly6G−Ly6ChighCD117+ cells. Isolated MLPGs were transferred by i.v. injection to sublethally irradiated (450 rad) C57BL/6-Ly5.1. After 3 d, CD14 expression was analyzed on endogenous splenic PMNs (CD45.1+) or MLPG-derived PMNs (CD45.2+).
PMN in vitro treatment
PMNs were isolated from BM of naive mice using anti-Ly-6G MicroBeads (Miltenyi Biotec), plated in 1 ml of complete medium at 106/ml with 20 ng/ml GM-CSF (Peprotech) and 20% (vol/vol) TES, and cultured for 24 h in normoxic or hypoxic (0.5% O2) conditions using a hypoxic chamber (BioSpherix). Mouse TES was generated as previously described (Mastio et al., 2019). After 24 h, CD14 expression was assessed by flow cytometry on live PMNs (Aqua−Ly6G+). In some experiments, anti-mouse GM-CSF neutralizing antibody (clone MP1-22E9; BioXcell) was added to the culture for 18–24 h.
Statistical analyses
After testing for normal distribution of data, statistical analyses were performed using two-tailed Student’s t test and Prism 5 software (GraphPad Software). All data are presented as mean ± SD, and P values <0.05 were considered significant. One-way ANOVA test with correction for multiple comparisons (Kruskal–Wallis or Tukey test) was used in experiments with more than two groups.
Online supplemental material
Fig. S1 shows pathways significantly changed in different populations of PMN. Fig. S2 shows expression of cell surface markers on PMN-MDSC from spleen and tumor. Fig. S3 presents morphology and phenotype of the populations of PMNs and CD14 expression and the function of the populations of PMNs in an acute infection model. Fig. S4 shows gene expression in cancer patient PMNs. Fig. S5 shows pathways changed in cluster 1 in patient PMNs. Table S1 lists antibodies used in CyTOF. Table S2 lists antibodies used in flow cytometry. Table S3 lists reagents used in the study. Table S4 shows primers used in qPCR.
Supplementary Material
lists antibodies used in CyTOF.
lists antibodies used in flow cytometry.
lists reagents used in this study.
shows primers used in qPCR.
Acknowledgments
This work was supported by a pilot grant from the University of Pennsylvania and flow cytometry and animal cores of the Wistar Institute (P30 CA010815). This work was also supported in part by the Flow Cytometry Core at the H. Lee Moffitt Cancer Center and Research Institute (P30-CA076292).
Author contributions: F. Veglia designed and conducted experiments, analyzed data, and contributed to writing the manuscript; A. Hashimoto conducted experiments, analyzed data, and contributed to writing the manuscript; H. Dweep, A. Kossenkov, and A. Loboda performed bioinformatic analysis; A. De Leo, E. Sanseviero, E. Tcyganov, J. Patel, V. Bhargava, P. Wilkinson, and D. Smirnov performed experiments and data analysis; C. Mulligan, B. Nam, G. Masters, S. Singhal, and E.B. Eruslanov provided materials; E.B. Eruslanov, M.A. Sepulveda, R. Cristescu, and Y. Nefedova participated in data analysis and reviewed manuscript; D.I. Gabrilovich designed experiments, analyzed data, wrote the manuscript, and acquired funding.
References
- Al-Khami, A.A., Zheng L., Del Valle L., Hossain F., Wyczechowska D., Zabaleta J., Sanchez M.D., Dean M.J., Rodriguez P.C., and Ochoa A.C.. 2017. Exogenous lipid uptake induces metabolic and functional reprogramming of tumor-associated myeloid-derived suppressor cells. OncoImmunology. 6:e1344804. 10.1080/2162402X.2017.1344804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alshetaiwi, H., Pervolarakis N., McIntyre L.L., Ma D., Nguyen Q., Rath J.A., Nee K., Hernandez G., Evans K., Torosian L., et al. 2020. Defining the emergence of myeloid-derived suppressor cells in breast cancer using single-cell transcriptomics. Sci. Immunol. 5:eaay6017. 10.1126/sciimmunol.aay6017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amir, E.D., Davis K.L., Tadmor M.D., Simonds E.F., Levine J.H., Bendall S.C., Shenfeld D.K., Krishnaswamy S., Nolan G.P., and Pe’er D.. 2013. viSNE enables visualization of high dimensional single-cell data and reveals phenotypic heterogeneity of leukemia. Nat. Biotechnol. 31(6):545–552. 10.1038/nbt.2594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aran, D., Looney A.P., Liu L., Wu E., Fong V., Hsu A., Chak S., Naikawadi R.P., Wolters P.J., Abate A.R., et al. 2019. Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nat. Immunol. 20:163–172. 10.1038/s41590-018-0276-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayers, M., Nebozhyn M., Cristescu R., McClanahan T.K., Perini R., Rubin E., Cheng J.D., Kaufman D.R., and Loboda A.. 2018. Molecular Profiling of Cohorts of Tumor Samples to Guide Clinical Development of Pembrolizumab as Monotherapy. Clin. Cancer Res. 25(5):1564–1573. 10.1158/1078-0432.CCR-18-1316 [DOI] [PubMed] [Google Scholar]
- Ayers, M., Lunceford J., Nebozhyn M., Murphy E., Loboda A., Kaufman D.R., Albright A., Cheng J.D., Kang S.P., Shankaran V., et al. 2017. IFN-γ–related mRNA profile predicts clinical response to PD-1 blockade. J. Clin. Invest. 127(8):2930–2940. 10.1172/JCI91190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becht, E., McInnes L., Healy J., Dutertre C.A., Kwok I.W.H., Ng L.G., Ginhoux F., and Newell E.W.. 2018. Dimensionality reduction for visualizing single-cell data using UMAP. Nat. Biotechnol. [DOI] [PubMed] [Google Scholar]
- Benayoun, B.A., Pollina E.A., Singh P.P., Mahmoudi S., Harel I., Casey K.M., Dulken B.W., Kundaje A., and Brunet A.. 2019. Remodeling of epigenome and transcriptome landscapes with aging in mice reveals widespread induction of inflammatory responses. Genome Res. 29:697–709. 10.1101/gr.240093.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bronte, V., Brandau S., Chen S.-H., Colombo M.P., Frey A.B., Greten T.F., Mandruzzato S., Murray P.J., Ochoa A., Ostrand-Rosenberg S., et al. 2016. Recommendations for myeloid-derived suppressor cell nomenclature and characterization standards. Nat. Commun. 7:12150. 10.1038/ncomms12150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, J., Spielmann M., Qiu X., Huang X., Ibrahim D.M., Hill A.J., Zhang F., Mundlos S., Christiansen L., Steemers F.J., et al. 2019. The single-cell transcriptional landscape of mammalian organogenesis. Nature. 566:496–502. 10.1038/s41586-019-0969-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, P., Corzo C.A., Luetteke N., Yu B., Nagaraj S., Bui M.M., Ortiz M., Nacken W., Sorg C., Vogl T., et al. 2008. Inhibition of dendritic cell differentiation and accumulation of myeloid-derived suppressor cells in cancer is regulated by S100A9 protein. J. Exp. Med. 205:2235–2249. 10.1084/jem.20080132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffelt, S.B., Wellenstein M.D., and de Visser K.E.. 2016. Neutrophils in cancer: neutral no more. Nat. Rev. Cancer. 16:431–446. 10.1038/nrc.2016.52 [DOI] [PubMed] [Google Scholar]
- Condamine, T., Dominguez G.A., Youn J.I., Kossenkov A.V., Mony S., Alicea-Torres K., Tcyganov E., Hashimoto A., Nefedova Y., Lin C., et al. 2016. Lectin-type oxidized LDL receptor-1 distinguishes population of human polymorphonuclear myeloid-derived suppressor cells in cancer patients. Sci. Immunol. 1:aaf8943. 10.1126/sciimmunol.aaf8943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condamine, T., Mastio J., and Gabrilovich D.I.. 2015a. Transcriptional regulation of myeloid-derived suppressor cells. J. Leukoc. Biol. 98:913–922. 10.1189/jlb.4RI0515-204R [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condamine, T., Ramachandran I., Youn J.I., and Gabrilovich D.I.. 2015b. Regulation of tumor metastasis by myeloid-derived suppressor cells. Annu. Rev. Med. 66:97–110. 10.1146/annurev-med-051013-052304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin, A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., and Gingeras T.R.. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 29:15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engblom, C., Pfirschke C., Zilionis R., Da Silva Martins J., Bos S.A., Courties G., Rickelt S., Severe N., Baryawno N., Faget J., et al. 2017. Osteoblasts remotely supply lung tumors with cancer-promoting SiglecFhigh neutrophils. Science. 358:eaal5081. 10.1126/science.aal5081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridlender, Z.G., Sun J., Kim S., Kapoor V., Cheng G., Ling L., Worthen G.S., and Albelda S.M.. 2009. Polarization of tumor-associated neutrophil phenotype by TGF-beta: “N1” versus “N2” TAN. Cancer Cell. 16:183–194. 10.1016/j.ccr.2009.06.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabrilovich, D.I. 2017. Myeloid-Derived Suppressor Cells. Cancer Immunol. Res. 5:3–8. 10.1158/2326-6066.CIR-16-0297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentles, A.J., Newman A.M., Liu C.L., Bratman S.V., Feng W., Kim D., Nair V.S., Xu Y., Khuong A., Hoang C.D., et al. 2015. The prognostic landscape of genes and infiltrating immune cells across human cancers. Nat. Med. 21:938–945. 10.1038/nm.3909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granot, Z., Henke E., Comen E.A., King T.A., Norton L., and Benezra R.. 2011. Tumor entrained neutrophils inhibit seeding in the premetastatic lung. Cancer Cell. 20:300–314. 10.1016/j.ccr.2011.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, Y.M., Li X., Perego M., Nefedova Y., Kossenkov A.V., Jensen E.A., Kagan V., Liu Y.F., Fu S.Y., Ye Q.J., et al. 2018. Transitory presence of myeloid-derived suppressor cells in neonates is critical for control of inflammation. Nat. Med. 24:224–231. 10.1038/nm.4467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, Z., Riva M., Björk P., Swärd K., Mörgelin M., Leanderson T., and Ivars F.. 2016. CD14 Is a Co-Receptor for TLR4 in the S100A9-Induced Pro-Inflammatory Response in Monocytes. PLoS One. 11:e0156377. 10.1371/journal.pone.0156377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heng, T.S., and Painter M.W.. Immunological Genome Project Consortium . 2008. The Immunological Genome Project: networks of gene expression in immune cells. Nat. Immunol. 9:1091–1094. 10.1038/ni1008-1091 [DOI] [PubMed] [Google Scholar]
- Hingorani, S.R., Wang L., Multani A.S., Combs C., Deramaudt T.B., Hruban R.H., Rustgi A.K., Chang S., and Tuveson D.A.. 2005. Trp53R172H and KrasG12D cooperate to promote chromosomal instability and widely metastatic pancreatic ductal adenocarcinoma in mice. Cancer Cell. 7:469–483. 10.1016/j.ccr.2005.04.023 [DOI] [PubMed] [Google Scholar]
- Jerala, R. 2007. Structural biology of the LPS recognition. Int. J. Med. Microbiol. 297:353–363. 10.1016/j.ijmm.2007.04.001 [DOI] [PubMed] [Google Scholar]
- Kaplan-Lefko, P.J., Chen T.M., Ittmann M.M., Barrios R.J., Ayala G.E., Huss W.J., Maddison L.A., Foster B.A., and Greenberg N.M.. 2003. Pathobiology of autochthonous prostate cancer in a pre-clinical transgenic mouse model. Prostate. 55:219–237. 10.1002/pros.10215 [DOI] [PubMed] [Google Scholar]
- Kato, M., Takahashi M., Akhand A.A., Liu W., Dai Y., Shimizu S., Iwamoto T., Suzuki H., and Nakashima I.. 1998. Transgenic mouse model for skin malignant melanoma. Oncogene. 17:1885–1888. 10.1038/sj.onc.1202077 [DOI] [PubMed] [Google Scholar]
- Kim, H.R., Park S.M., Seo S.U., Jung I., Yoon H.I., Gabrilovich D.I., Cho B.C., Seong S.Y., Ha S.J., and Youn J.I.. 2019. The Ratio of Peripheral Regulatory T Cells to Lox-1+ Polymorphonuclear Myeloid-derived Suppressor Cells Predicts the Early Response to Anti-PD-1 Therapy in Patients with Non-Small Cell Lung Cancer. Am. J. Respir. Crit. Care Med. 199:243–246. 10.1164/rccm.201808-1502LE [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, V., Donthireddy L., Marvel D., Condamine T., Wang F., Lavilla-Alonso S., Hashimoto A., Vonteddu P., Behera R., Goins M.A., et al. 2017. Cancer-Associated Fibroblasts Neutralize the Anti-tumor Effect of CSF1 Receptor Blockade by Inducing PMN-MDSC Infiltration of Tumors. Cancer Cell. 32:654–668.e5. 10.1016/j.ccell.2017.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead, B., and Salzberg S.L.. 2012. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 9:357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, B., and Dewey C.N.. 2011. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 12:323. 10.1186/1471-2105-12-323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd, K.L., and Kubes P.. 2006. GPI-linked endothelial CD14 contributes to the detection of LPS. Am. J. Physiol. Heart Circ. Physiol. 291:H473–H481. 10.1152/ajpheart.01234.2005 [DOI] [PubMed] [Google Scholar]
- Love, M.I., Huber W., and Anders S.. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550. 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lun, A.T., Bach K., and Marioni J.C.. 2016. Pooling across cells to normalize single-cell RNA sequencing data with many zero counts. Genome Biol. 17:75. 10.1186/s13059-016-0947-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manitz, M.P., Horst B., Seeliger S., Strey A., Skryabin B.V., Gunzer M., Frings W., Schönlau F., Roth J., Sorg C., and Nacken W.. 2003. Loss of S100A9 (MRP14) results in reduced interleukin-8-induced CD11b surface expression, a polarized microfilament system, and diminished responsiveness to chemoattractants in vitro. Mol. Cell. Biol. 23:1034–1043. 10.1128/MCB.23.3.1034-1043.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastio, J., Condamine T., Dominguez G., Kossenkov A.V., Donthireddy L., Veglia F., Lin C., Wang F., Fu S., Zhou J., et al. 2019. Identification of monocyte-like precursors of granulocytes in cancer as a mechanism for accumulation of PMN-MDSCs. J. Exp. Med. 216:2150–2169. 10.1084/jem.20181952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAvoy, E.F., McDonald B., Parsons S.A., Wong C.H., Landmann R., and Kubes P.. 2011. The role of CD14 in neutrophil recruitment within the liver microcirculation during endotoxemia. J. Immunol. 186:2592–2601. 10.4049/jimmunol.1002248 [DOI] [PubMed] [Google Scholar]
- McCarthy, D.J., Campbell K.R., Lun A.T., and Wills Q.F.. 2017. Scater: pre-processing, quality control, normalization and visualization of single-cell RNA-seq data in R. Bioinformatics. 33:1179–1186. 10.1093/bioinformatics/btw777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer, C., Sevko A., Ramacher M., Bazhin A.V., Falk C.S., Osen W., Borrello I., Kato M., Schadendorf D., Baniyash M., and Umansky V.. 2011. Chronic inflammation promotes myeloid-derived suppressor cell activation blocking antitumor immunity in transgenic mouse melanoma model. Proc. Natl. Acad. Sci. USA. 108:17111–17116. 10.1073/pnas.1108121108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishalian, I., Granot Z., and Fridlender Z.G.. 2017. The diversity of circulating neutrophils in cancer. Immunobiology. 222:82–88. 10.1016/j.imbio.2016.02.001 [DOI] [PubMed] [Google Scholar]
- Ng, L.G., Ostuni R., and Hidalgo A.. 2019. Heterogeneity of neutrophils. Nat. Rev. Immunol. 19:255–265. 10.1038/s41577-019-0141-8 [DOI] [PubMed] [Google Scholar]
- Ortiz, M.L., Kumar V., Martner A., Mony S., Donthireddy L., Condamine T., Seykora J., Knight S.C., Malietzis G., Lee G.H., et al. 2015. Immature myeloid cells directly contribute to skin tumor development by recruiting IL-17-producing CD4+ T cells. J. Exp. Med. 212:351–367. 10.1084/jem.20140835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quatromoni, J.G., Singhal S., Bhojnagarwala P., Hancock W.W., Albelda S.M., and Eruslanov E.. 2014. An optimized disaggregation method for human lung tumors that preserves the phenotype and function of the immune cells. J. Leukoc. Biol. 97(1):201–209. 10.1189/jlb.5TA0814-373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaul, M.E., Eyal O., Guglietta S., Aloni P., Zlotnik A., Forkosh E., Levy L., Weber L.M., Levin Y., Pomerantz A., et al. 2020. Circulating neutrophil subsets in advanced lung cancer patients exhibit unique immune signature and relate to prognosis. FASEB J. 34:4204–4218. 10.1096/fj.201902467R [DOI] [PubMed] [Google Scholar]
- Si, Y., Merz S.F., Jansen P., Wang B., Bruderek K., Altenhoff P., Mattheis S., Lang S., Gunzer M., Klode J., et al. 2019. Multidimensional imaging provides evidence for down-regulation of T cell effector function by MDSC in human cancer tissue. Sci. Immunol. 4:eaaw9159. 10.1126/sciimmunol.aaw9159 [DOI] [PubMed] [Google Scholar]
- Singhal, S., Bhojnagarwala P.S., O’Brien S., Moon E.K., Garfall A.L., Rao A.S., Quatromoni J.G., Stephen T.L., Litzky L., Deshpande C., et al. 2016. Origin and Role of a Subset of Tumor-Associated Neutrophils with Antigen-Presenting Cell Features in Early-Stage Human Lung Cancer. Cancer Cell. 30:120–135. 10.1016/j.ccell.2016.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart, T., Butler A., Hoffman P., Hafemeister C., Papalexi E., Mauck W.M. III, Hao Y., Stoeckius M., Smibert P., and Satija R.. 2019. Comprehensive Integration of Single-Cell Data. Cell. 177:1888–1902.e21. 10.1016/j.cell.2019.05.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Gassen, S., Callebaut B., Van Helden M.J., Lambrecht B.N., Demeester P., Dhaene T., and Saeys Y.. 2015. FlowSOM: Using self-organizing maps for visualization and interpretation of cytometry data. Cytometry A. 87:636–645. 10.1002/cyto.a.22625 [DOI] [PubMed] [Google Scholar]
- Veglia, F., Perego M., and Gabrilovich D.. 2018. Myeloid-derived suppressor cells coming of age. Nat. Immunol. 19:108–119. 10.1038/s41590-017-0022-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veglia, F., Tyurin V.A., Blasi M., De Leo A., Kossenkov A.V., Donthireddy L., To T.K.J., Schug Z., Basu S., Wang F., et al. 2019. Fatty acid transport protein 2 reprograms neutrophils in cancer. Nature. 569:73–78. 10.1038/s41586-019-1118-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshino, T., Miyake K., Ichimura K., Mannami T., Ohara N., Hamazaki S., and Akagi T.. 2000. Increased incidence of follicular lymphoma in the duodenum. Am. J. Surg. Pathol. 24:688–693. 10.1097/00000478-200005000-00007 [DOI] [PubMed] [Google Scholar]
- Youn, J.I., Collazo M., Shalova I.N., Biswas S.K., and Gabrilovich D.I.. 2012. Characterization of the nature of granulocytic myeloid-derived suppressor cells in tumor-bearing mice. J. Leukoc. Biol. 91:167–181. 10.1189/jlb.0311177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanoni, I., and Granucci F.. 2012. Regulation and dysregulation of innate immunity by NFAT signaling downstream of pattern recognition receptors (PRRs). Eur. J. Immunol. 42:1924–1931. 10.1002/eji.201242580 [DOI] [PubMed] [Google Scholar]
- Zanoni, I., Ostuni R., Capuano G., Collini M., Caccia M., Ronchi A.E., Rocchetti M., Mingozzi F., Foti M., Chirico G., et al. 2009. CD14 regulates the dendritic cell life cycle after LPS exposure through NFAT activation. Nature. 460:264–268. 10.1038/nature08118 [DOI] [PubMed] [Google Scholar]
- Zanoni, I., Ostuni R., Barresi S., Di Gioia M., Broggi A., Costa B., Marzi R., and Granucci F.. 2012. CD14 and NFAT mediate lipopolysaccharide-induced skin edema formation in mice. J. Clin. Invest. 122:1747–1757. 10.1172/JCI60688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, W.H., Wang W.Q., Gao H.L., Xu S.S., Li S., Li T.J., Han X., Xu H.X., Li H., Jiang W., et al. 2020. Tumor-Infiltrating Neutrophils Predict Poor Survival of Non-Functional Pancreatic Neuroendocrine Tumor. J. Clin. Endocrinol. Metab. 105:2217–2228. 10.1210/clinem/dgaa196 [DOI] [PubMed] [Google Scholar]
- Zhou, J., Nefedova Y., Lei A., and Gabrilovich D.. 2018. Neutrophils and PMN-MDSC: Their biological role and interaction with stromal cells. Semin. Immunol. 35:19–28. 10.1016/j.smim.2017.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
lists antibodies used in CyTOF.
lists antibodies used in flow cytometry.
lists reagents used in this study.
shows primers used in qPCR.
Data Availability Statement
Raw UMI counts for single-cell RNA-seq are publicly available at GEO, accession no. GSE163834.