Summary
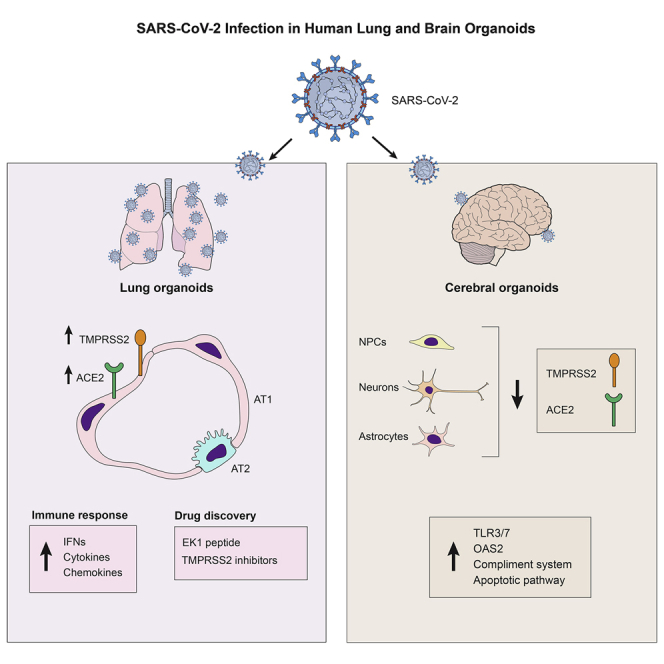
COVID-19 is a transmissible respiratory disease caused by a novel coronavirus, SARS-CoV-2, and has become a global health emergency. There is an urgent need for robust and practical in vitro model systems to investigate viral pathogenesis. Here, we generated human induced pluripotent stem cell (iPSC)-derived lung organoids (LORGs), cerebral organoids (CORGs), neural progenitor cells (NPCs), neurons, and astrocytes. LORGs containing epithelial cells, alveolar types 1 and 2, highly express ACE2 and TMPRSS2 and are permissive to SARS-CoV-2 infection. SARS-CoV-2 infection induces interferons, cytokines, and chemokines and activates critical inflammasome pathway genes. Spike protein inhibitor, EK1 peptide, and TMPRSS2 inhibitors (camostat/nafamostat) block viral entry in LORGs. Conversely, CORGs, NPCs, astrocytes, and neurons express low levels of ACE2 and TMPRSS2 and correspondingly are not highly permissive to SARS-CoV-2 infection. Infection in neuronal cells activates TLR3/7, OAS2, complement system, and apoptotic genes. These findings will aid in understanding COVID-19 pathogenesis and facilitate drug discovery.
Key words: iPSCs, SARS-CoV-2, lungs organoids, cerebral organoids, host-pathogen interactions, ACE2, TMPRSS2
Graphical abstract
Highlights
-
•
Lung organoids express ACE2/TMPRSS2 and are permissive to SARS-CoV-2 infection
-
•
SARS-CoV-2 induces IFNs and cytokines/chemokines, and activates inflammasome pathway
-
•
SARS-CoV-2 infection in neurons activates TLR3/7, OAS2, and apoptotic genes
-
•
Spike protein and TMPRSS2 inhibitors reduce SARS-CoV-2 infection in organoids
In this report, Rana and colleagues show that human iPSC-derived lung and brain organoids express ACE2/TMPRSS2 and are permissive to SARS-CoV-2 infection inducing specific pathways in lung and brain cells. Spike protein inhibitor, EK1 peptide, and TMPRSS2 inhibitors (camostat/nafamostat) reduced SARS-CoV-2 infection in lung organoids, providing a model system for drug development.
Introduction
Coronavirus disease 2019 (COVID-19), a transmissible respiratory disease caused by a novel severe acute respiratory syndrome coronavirus, SARS-CoV-2, has become a global health emergency since its outbreak in 2019 (Lu et al., 2020). There are currently no approved drugs for this disease, and there is an urgent need for developing therapeutic strategies. In addition, there is a lack of robust and practical in vitro model systems to investigate the pathophysiology of COVID-19. The etiologic agent of COVID-19, SARS-CoV-2, is an enveloped positive-sense single-stranded RNA virus. SARS-CoV-2 uses angiotensin-converting enzyme 2 (ACE2) as a host cell receptor and transmembrane serine protease 2 (TMPRSS2) to cleave its spike protein for entry into the host cell (Hoffmann et al., 2020). Symptoms of COVID-19 vary in severity and manifest in many ways involving multiple human organ systems. Due to the permissiveness of viral infection in various tissues, SARS-CoV-2 could lead to multiorgan failure and death (Puelles et al., 2020). Our current understanding of COVID-19 disease is based on clinical outcomes and studies using cancer cell lines or transgenic mouse models expressing ACE2 receptor for viral entry, which have limitations in recapitulating the human physiology to decipher fundamental molecular mechanisms regulating host-pathogen interactions, viral replication kinetics, and tropism. Thus, there is an urgent need for robust and practical in vitro model systems to investigate the pathophysiology of SARS-CoV-2 infection.
Induced pluripotent stem cells (iPSCs) or multipotent adult tissue stem cells can be differentiated to generate three-dimensional (3D) complex organoid structures containing multiple cell types and assemblies that have characteristics of spatial organization and function of a tissue (Benito-Kwiecinski and Lancaster, 2019). Organoids have shown great promise in modeling human diseases, investigating host-pathogen interactions, and drug screening. In earlier studies, we and others have utilized embryonic stem cell/iPSC-derived human neural progenitor cells (NPCs) and cerebral organoids (CORGs) to understand Zika virus-associated neuronal injury (Dang et al., 2016; Dutta and Clevers, 2017; Muffat et al., 2018; Xu et al., 2016). Recently, we reported glial cell diversity and methamphetamine-induced neuroinflammation in human CORGs by using single-cell RNA sequencing (Dang et al., 2020). Several studies using stem cell-derived organoids have started to provide further insights into SARS-CoV-2 infection of various cell types and host responses, and the discovery of new drug candidates (Han et al., 2020; Jacob et al., 2020; Katsura et al., 2020; Lamers et al., 2020; Mykytyn et al., 2021; Pellegrini et al., 2020; Ramani et al., 2020; Yang et al., 2020). Interestingly, SARS-CoV-2 productively infects brain choroid plexus epithelium, leading to cell death and functional deficits (Jacob et al., 2020; Pellegrini et al., 2020).
To investigate the tissue-specific SARS-CoV-2 infection and host responses in lungs and brain, we generated 3D human LORGs and CORGs, and brain cell types such as NPCs, neurons, and astrocytes. By using a SARS-CoV-2 pseudovirus and USA-WA1/2020 SARS-CoV-2, we determined viral tropism, host responses, and viral inhibition by specific TMPRSS2 drugs and a spike-protein-binding peptide EK1.
Results
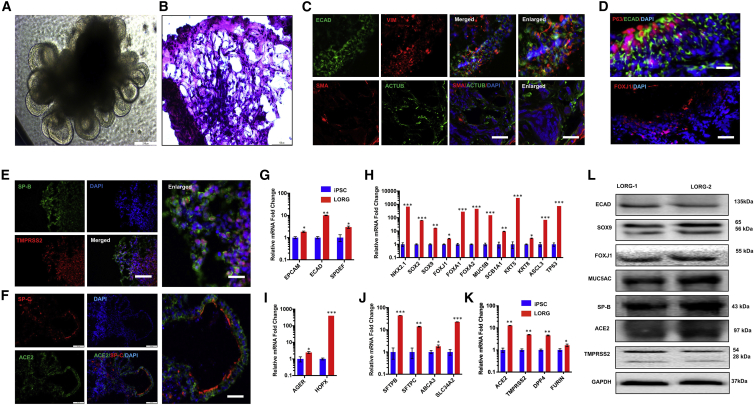
To develop a model system for investigating the human lung pathophysiology of SARS-CoV-2 infection, we differentiated stepwise human iPSCs into lung organoids (LORGs) as described previously, with minor modification (Leibel et al., 2019; Miller et al., 2019). First, we differentiated human iPSCs into definitive endoderm (DE) followed by specification into anterior foregut endoderm/spheroids (AFE), lung progenitor cells), and finally LORGs (Figures 1A, S1A, and S1B). During differentiation, we characterized the authenticity of each step of differentiation into specific cell types. AFE in LORG medium by day 12 generated an epithelial-like structure and as the organoids progressively grow, mesenchymal populations became visible, surrounding epithelial structures (Figures 1A and S1B). Next, AFE differentiation was confirmed by immunolabeling of AFE markers such as NK2 homeobox 1 (NKX2.1+) cells and western blotting of NKX2.1 to confirm their differentiation into AFE cells (Figures S1C and S1D). In addition, AFE also contains progenitor cells such as SOX2+ cells (Figure S1D). By day 60 (60D) in culture, human LORGs have prominent epithelial structure as visualized by hematoxylin and eosin (H&E) staining (Figure 1B). During the development of 3D LORG, we detected expression of both proximal and distal regions of lung by SOX2 and SOX9, respectively (Figure S1D). These proximal and distal regions of lung contain epithelial cells specializing in diverse functions such as secretory, basal, and ciliated cells in the airways, and type 1 and 2 cells lining the alveoli (Barkauskas et al., 2017). SOX9-expressing progenitors play a role in development of the entire lung epithelium and are involved in formation of airway and alveolar cell types in lungs (Ostrin et al., 2018). LORG epithelial cells (ECAD+) were surrounded by mesenchymal cells as shown by vimentin (VIM) (Figure 1C). LORGs also showed mesenchymal cells on immunolabeling with smooth muscle markers smooth muscle actin and acetylated tubulin (Figure 1C). Furthermore, LORGs exhibited pseudostratified epithelial structures with P63+ basal-like cells, lining the basal surfaces and population of ciliated cells such as FOXJ1+ cells (Figure 1D). Importantly, alveolar cell markers SFTPB and SFTPC showed co-labeling with SARS-CoV-2 receptor ACE2 and protease TMPRSS2 (Figures 1E, 1F, and S1G). In addition, LORGs exhibited expression of alveolar type 1 markers AGER and HOPX (Figures S1E and S1F). Furthermore, quantitative gene expression analysis of 60D differentiated LORGs showed the presence of diverse cell types including progenitor cells markers (SOX2 and NKX2.1), proximal lung epithelial cell genes (SOX9, FOXJ1, FOXA1, FOXA2, SCB1A1, MUC5B, KRT5, KRT8, ASCL3, and TP63), lung epithelial cell markers (EPCAM, ECAD, and SPDEF), alveolar type 1 cells (AGER and HOPX), and alveolar type 2 cells (SFTPB, SFTPC, ABCA3, and SLC34A2) as compared with iPSCs (Figures 1G–1J). Altogether, these results show that we have obtained bona fide iPSC-derived LORGs containing multiple cell types including alveolar types 1 and 2.
Figure 1.
Human iPSC differentiation into 3D lung organoids for modeling SARS-CoV-2 infection
(A) Representative phase-contrast image of lung organoids (LORGs) at 60 days (60D). Scale bar, 200 μm.
(B) H&E staining of 60D LORG showing alveolar-like morphology. Scale bars, 100 μm.
(C) Confocal images showing labeling for cells surrounding epithelial structures (ECAD, green) co-labeled with mesenchymal-cell-type marker vimentin (VIM, red). LORGs also show mesenchymal cells by immunolabeling with smooth muscle marker, smooth muscle actin (SMA, red), and acetylated tubulin (ACTUB, green). Scale bars, 100 μm; for enlarged images, 50 μm.
(D) Epithelial structures (ECAD, green) showing basal-like cells (P63, red) on basolateral surface and ciliated cells (FOXJ1, red) on the luminal side of the epithelium. Scale bars, 100 μm.
(E and F) Representative confocal images showing labeling for alveolar type 2 cells (SP-B, green and SP-C, red) co-labeled with TMPRSS2 (E, red) or SARS-CoV-2 receptor ACE2 (F, green). Scale bars, 100 μm; for enlarged images, 50 μm.
(G) LORGs express lung epithelial cell markers EPCAM, ECAD, and SPDEF. Mean ± SEM of n = 3 organoids cultured in different wells. ∗p < 0.05, ∗∗p < 0.01 by Student's t test.
(H) Expression analysis of proximal lung epithelial cell markers (SOX9, FOXJ1, FOXA1, FOXA2, SCB1A1, MUC5B, KRT5, KRT8, ASCL3, and TP63) and progenitor cell genes (NKX2.1 and SOX2) expressed in developing lungs. Mean ± SEM of n = 3 organoids cultured in different wells. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 by Student's t test.
(I) Analysis of lung alveolar type 1 cell genes AGER and HOPX. Mean ± SEM of n = 3 organoids cultured in different wells. ∗p < 0.05, ∗∗∗p < 0.001.
(J) Expression analysis of expression of lung alveolar type 2 cell genes SFTPB, SFTPC, ABCA3, and SLC34A2. Mean ± SEM of n = 3 organoids cultured in different wells. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
(K) qRT-PCR analysis of 60D LORG shows expression of ACE2, TMPRSS2, DPP4, and Furin. Data presented as mean ± SEM of n = 3 organoids cultured in different wells. ∗p < 0.05, ∗∗p < 0.01.
(L) Western blot analysis of 60D organoid showing protein levels for ECAD, SOX9, FOXJ1, MUC5AC, and alveolar type 2 cells (surfactant protein B [SP-B]), SARS-CoV-2 receptor (ACE2), and protease (TMPRSS2). n = 2 organoids.
Since SARS-CoV-2 uses ACE2 receptor and the TMPRSS2 to cleave its S protein for entry into the cell, we quantified ACE2 and TMPRSS2 mRNA expression in iPSCs and LORGs. In addition, Hoffmann et al. (2020) showed that furin protease cleavage of spike protein at the S1/S2 site was essential for spike protein-mediated cell-cell fusion and entry into human lung cells. We also included analysis of expression of dipeptidylpeptidase 4 (DPP4), a functional receptor for MERS-CoV. We analyzed the expression of host proteases and found a significant increase in expression of all these genes in LORGs as compared with iPSCs, with increase in ACE2/PLASMIN being the highest, >10 fold (Figures 1K and S1H). To confirm cell-specific expression of ACE2 and TMPRSS2, we performed immunostaining of LORGs, whereby our results showed that alveolar type 2 cells (SP-B and SP-C) expressed ACE2 and TMPRSS2 (Figures 1E, 1F, and S1G). Furthermore, western blot analysis of 60D organoids showed strong protein expression of ACE2 and TMPRSS2 in alveolar type 2 cells (Figure 1L). Overall, these results demonstrate that LORGs express SARS-CoV-2 receptor and co-factors required for viral entry.
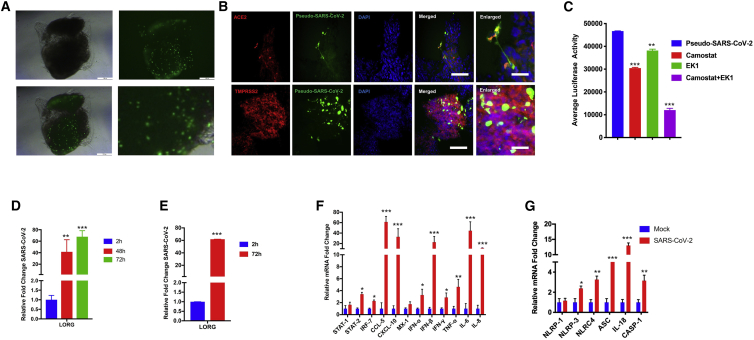
To determine whether LORGs were permissive to SARS-CoV-2 infection, we constructed SARS-CoV-2 pseudovirus (GFP or luciferase) based on VSV to investigate the viral entry process (Wang et al., 2020). We inoculated LORGs with SARS-CoV-2 pseudovirus (multiplicity of infection [MOI] = 2) and analyzed for viral entry and infection at 24 h post infection based on EGFP signals (Figure 2A). Bright-field imaging of the whole LORG merged with robust EGFP signals as well as sections of LORG showed integration of EGFP, indicating successful incorporation of SARS-CoV-2 into organoids (Figure 2A). Furthermore, SARS-CoV-2 showed co-labeling with ACE2 and TMPRSS2, indicating virus binding with the receptor and internalization in the cell by TMPRSS2 proteases in LORG (Figure 2B). To quantify virus infection, we infected LORGs with SARS-CoV-2 pseudovirus expressing luciferase at MOI = 2 for 24 h. We observed a robust infection of organoids as quantified by luciferase activity (Figure 2C).
Figure 2.
SARS-CoV-2 robustly replicates in human LORGs and induces expression of genes involved in innate immunity and inflammation
(A) Phase-contrast and fluorescence imaging shows infection of SARS-CoV-2-GFP pseudovirus at 24 h post infection and MOI = 2. Scale bars, 200 μm.
(B) Fluorescence image showing that SARS-CoV-2 GFP pseudovirus infects ACE2 and TMPRSS2+ cells of the LORG. Scale bars, 100 μm; for enlarged images, 50 μm.
(C) Pseudo-SARS-CoV-2-luciferase pseudovirus infection is blocked by viral entry inhibitors. Bar graph shows the luciferase activity in the presence and absence of TMPRSS2 inhibitor (camostat mesylate) and a fusion inhibitor EK1 peptide to LORG infected with SARS-CoV-2 pseudovirus. Mean ± SEM of n = 3 organoids cultured in different wells. ∗∗∗p < 0.001 by Student’s t test.
(D and E) Lung organoids were infected with SARS-CoV-2 USA-WA1/2020 virus at MOI = 2, and viral RNAs from supernatant (D) and cellular (E) fractions were quantified at indicated times of infection. Mean ± SEM of n = 2 organoids cultured and infected in different wells. ∗∗p < 0.01, ∗∗∗p < 0.001 by Student's t test.
(F and G) LORGs were infected as above in (E), and gene expression of indicated genes was quantified by qRT-PCR at 72 h post infection. Bar graph shows expression of immune response genes and cytokines (F) and inflammasome markers (G). Mean ± SEM of n = 2 organoids cultured and infected in different wells. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 by Student's t test.
To define the S-mediated and TMPRSS2-dependent SARS-CoV-2 infection of LORGs and to assess the utility of LORGs to test drugs that could inhibit viral infection, we treated LORGs with a fusion inhibition peptide, EK1, targeting the HR1 domain of the spike protein and TMPRSS2 inhibitor camostat, then quantified the luciferase activity. We showed that camostat or EK1 peptide inhibited viral infection, confirming that pseudovirus entry is mediated by the spike protein and is dependent on TMPRSS2 function (Figure 2C). As expected, combination of camostat and EK1 peptide robustly inhibited viral infection in LORGs (Figure 2C). Next, we compared these results with a lung epithelial cell line, Calu-3, and obtained similar results whereby EK1 peptide or TMPRSS2 inhibitors, camostat and nafamostat, repressed viral infection, which was further reduced when both classes of inhibitors were combined (Figure S1I). We then tested the inhibitory activities of nafamostat in Calu-3 cells infected with a patient-derived isolate of SARS-CoV-2, whereby our results showed that nafamostat significantly reduced SARS-CoV-2 infection (Figures S1J–S1K) as observed in LORGs. Together, these results show that iPSC-derived LORGs provide a 3D lung model system to evaluate spike protein-mediated viral infection and drug efficacy to inhibit SARS-CoV-2.
To determine the utility of the LORG model system for understanding the pathogenesis of SARS-CoV-2 infection, we used a patient-derived isolate, SARS-CoV-2 USA-WA1/2020, for LORG infection. LORGs were infected with SARS-CoV-2 USA-WA1/2020, and infection rates were quantified by evaluating viral RNA of the infected cells and supernatants. We found that the viral strain productively infected LORGs and the infection rate was increased in a time-dependent manner (Figures 2D and 2E). To investigate physiological effects of virus infection, we examined the host response genes and we found that expression of a number of immune-regulatory genes including STAT1, STAT2, interferons (IFNs), and chemokines were upregulated in LORGs (Figure 2F). Our results showed that CCL5, CXCL10, IFN-β, interleukin-6 (IL-6), and IL-8 were all upregulated >10-fold, and these findings are consistent with the data obtained from SARS-CoV-2 infection of lung epithelial cells (Blanco-Melo et al., 2020) and COVID-19 patients relative to healthy donors (Daamen et al., 2020). Interestingly, our results also indicate that SARS-CoV-2 infection activates the inflammasome pathway because the key genes of this pathway including NLRP3, ASC, IL-18, and caspase-1 were upregulated (Figure 2G). Together, our results demonstrate that SARS-CoV-2 robustly replicates in human LORGs and provide a model system to study the mechanisms of viral pathogenesis as well as to test new therapeutics.
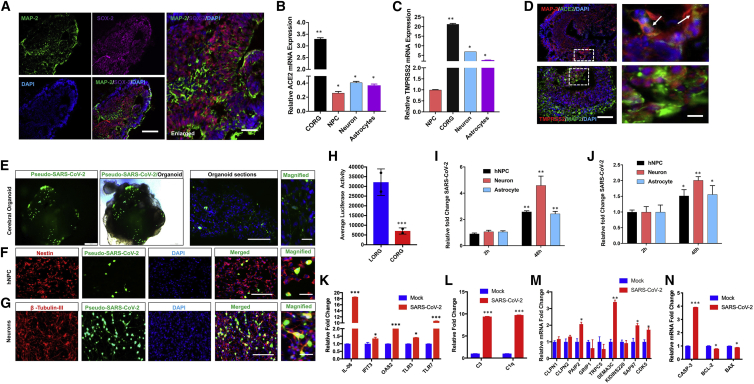
Clinical case reports describe a wide range of neurological and neuropsychiatric manifestations in severe COVID-19 (Helms et al., 2020). However, many questions about COVID-19 and neurological manifestations remain. (1) Does SARS-CoV-2 infect brain? (2) Which cell types in brain are infected and support viral replication? (3) What are the host responses to infection and how does the virus cause neuronal injury and observed phenotypes? To address these questions in a human-relevant in vitro model, we generated 3D CORGs and investigated SARS-CoV-2 infection, which was based on the brain organoid we used to investigate Zika virus infection and methamphetamine-associated neurotoxicity (Dang et al., 2016, 2020). To understand the neurotrophic potential of the SARS-CoV-2 virus, we differentiated human iPSCs into CORG, which showed presence of various cell types, such as NPCs, neurons, and glial cells characterized by their respective markers SOX-2, MAP-2, and GFAP (Figures 3A and S2A–S2D). In addition, we differentiated iPSCs into NPCs, neurons, and astrocytes, as confirmed by their characteristic markers (Figures S2B–S2D). We also quantified the mRNA expression of neural progenitor genes (Nestin and SOX2, Figure S2B), neuronal gene (MAP2, Figure S2C), and glial genes (S100β and GFAP, Figure S2D) in 3D CORG and in 2D NPCs, neurons, and astrocytes.
Figure 3.
SARS-CoV-2 infects human cerebral organoids and activates expression of innate immunity and neurodegeneration genes in neurons
(A) Representative immunofluorescence image of an 80-day-old organoid shows various cell types including neurons (MAP-2, green), neuronal progenitor (SOX-2, magenta), and nuclear stain (DAPI, blue); merged images are also shown. Scale bars, 100 μm.
(B and C) qRT-PCR analysis shows expression of ACE2 (B) and TMPRSS2 (C) in CORGs, NPCs, neurons, and astrocytes. Mean ± SEM of n = 3 independent organoids and n = 3 independent experiments from NPCs, neurons, and astrocytes. ∗p < 0.05, ∗∗p < 0.01 by Student's t test.
(D) Confocal imaging of CORGs showing immunostaining of neuronal marker (MAP-2, red) co-labeled with SARS-CoV-2 receptor (ACE2, green) or TMPRSS2 (red) co-labeled with MAP-2 (green). White boxes were enlarged (right), and arrows show co-labeling of MAP-2/ACE2 and MAP-2/TMPRSS2 in magnified images. Scale bars, 100 μm; for enlarged images, 25 μm.
(E–G) Organoids, NPCs, and neurons were infected with pseudo-SARS-CoV-2-GFP virus at MOI = 2 and analyzed after 24 h of infection. (E) Immunofluorescence and phase-contrast images of an organoid showing SARS-CoV-2 (green). Scale bars, 200 μm. (F) Immunofluorescence confocal images of NPCs stained with antibody against Nestin (red), SARS-CoV-2 (green), and nuclear stain DAPI (blue). Scale bars, 100 μm (merged) and 25 μm (magnified). (G) Immunofluorescence confocal images of NPC differentiated neurons stained with antibody against β-tubulin-III (red), SARS-CoV-2 (green), and nuclear stain DAPI (blue). Scale bars, 100 μm (merged) and 25 μm (magnified).
(H) Organoids were infected with pseudo-SARS-CoV-2-luciferase virus at MOI = 2, and luciferase activities were measured after 24 h of infection. Mean ± SEM of n = 3 independent organoids. ∗∗∗p < 0.001 by Student's t test.
(I and J) NPCs, neurons, and astrocytes were infected with SARS-CoV-2 USA-WA1/2020 virus at MOI = 2, and viral RNAs from supernatant (I) and cellular (J) fractions were quantified at indicated times of infection. Mean ± SEM of n = 3 independent organoids. ∗∗∗p < 0.001, ∗∗p < 0.001, ∗p < 0.05 by Student's t test.
(K–N) Neurons were infected as above in (I) and (J), and gene expression of indicated genes was quantified by qRT-PCR at 48 h post infection. Bar graph shows expression of innate immune response genes (K), C3 and C1q complement genes (L), synaptic function of neurons (M), and apoptotic pathway genes (N). Mean ± SEM of n = 3 independent organoids. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 by Student's t test.
See also Figure S2.
Next, to determine the potential of organoids and various cell types in brain to support SARS-CoV-2 infection, we determined ACE2 and TMPRSS2 expression in 3D CORGs as well as in differentiated NPCs, neurons, and astrocytes. qRT-PCR results showed that ACE2 and TMPRSS2 are expressed in CORGs and in all three cell types analyzed including NPCs, neurons, and astrocytes (Figures 3B and 3C). In addition, we analyzed the expression of host factors involved in entry and infection of SARS-CoV-2 such as FURIN, PLASMIN, CTSL1, NRP1, and DPP4 and found upregulation of these genes in CORGs and astrocytes compared with NPCs (Figure S2E). We performed immunostaining to examine the expression of ACE2 and TMPRSS2 in organoids. ACE2/TMPRSS2 expression was readily detected with neuronal marker MAP2 (Figure 3D) and very low TMPRSS2 expression with glial cells GFAP (Figures S2F–S2I). These results show that CORGs and three cell types analyzed express ACE2 and TMPRSS2, and, interestingly, ACE2 and TMPRSS2 expression levels were the highest in CORGs followed by neurons and astrocytes.
To determine whether CORGs were permissive to SARS-CoV-2 infection, we used SARS-CoV-2 pseudovirus (GFP or luciferase) as described above for LORGs. We inoculated CORGs with SARS-CoV-2 pseudovirus (MOI = 2) and at 24 h post infection and analyzed them for viral entry and infection based on EGFP signals (Figure 3E). Bright-field imaging of the whole CORG merged with robust EGFP signals as well as sections of CORG, indicating successful infection of SARS-CoV-2 (Figure 3E). Next, to examine the cell-specific SARS-CoV-2 pseudovirus infection, we infected neurons and NPCs with virus and carried out GFP analysis. Our results showed that infection was more efficient in neurons than in NPCs (Figures 3F and 3G), an observation that correlates with the levels of ACE2 and TMPRSS2 expression in neurons and NPCs (Figures 3B and 3C). So far, our results indicate that both LORGs and CORGs can be infected with SARS-CoV-2 GFP pseudovirus. Since major clinical complications in COVID-19 patients are lung related, we wondered about the efficiency of viral infection in LORGs and CORGs. To address this question, we infected both CORGs and LORGs with SARS-CoV-2-luciferase pseudovirus at MOI = 2, and luciferase activities were measured after 24 h of infection. Our quantification showed that viral infection was ~6-fold higher in LORGs in comparison with CORGs (Figure 3H).
To determine the cell-type-specific infectious capabilities of SAR-CoV-2 and to probe specific gene expression leading to observed clinical manifestations in COVID-19 patients, we used a patient-derived isolate, SARS-CoV-2 USA-WA1/2020, and infected NPCs, neurons, and astrocytes. All three cell types were infected under identical conditions, MOI = 2 for 48 h, and viral replication was quantified by qRT-PCR in both supernatant and cellular fractions (Figures 3I and 3J). Consistent with the pseudovirus infection results (Figures 3F and 3G), we found that the virus infected neurons more efficiently than NPCs and astrocytes. In addition, we evaluated whether these three cell types support replication of SARS-CoV-2 virus by comparing the viral RNA collected from cells at 2 h post infection. Our results indicate that all three cell types supported viral replication over time, with the amount of viral RNA being highest in neurons when compared with NPCs and astrocytes (Figures 3I and 3J).
Since virus replication levels were the highest in neurons, we analyzed host responses to viral infection in neurons. We selected genes specific to pathways that could potentially be involved in observed neurological complications in COVID-19 patients. Gene expressions from neurons infected with USA-WA1/2020 for 48 h were analyzed by qRT-PCR. Our analyses showed that SARS-COV-2 infection upregulated genes involved in innate immunity including IL6, IFIT3, OAS2, TLR3, and TLR7, complement system, calpain pathways, CDK5, apoptosis, and RIPK1/3-regulated necroptosis pathways (Figures 3K, 3L, 3M, and S2J). Apoptotic gene caspase-3 was upregulated while the antiapoptotic genes, BCL2 and BAX, were downregulated in infected neurons (Figure 3N). Altogether, our results demonstrate that SARS-CoV-2 infects human CORGs and regulates specific gene expressions in neurons involved in neuronal functions. We also show that iPSC-derived organoids can be employed to study tissue-specific differences in supporting viral infection as well as inducing cell-specific host responses.
Discussion
The SARS family of viruses, including SARS-CoV-2, targets the human respiratory system and leads to adverse effects, as observed during the COVID-19 pandemic. However, the range of human tissues permissive to SARS-CoV-2 infection is wide and includes the heart, brain, liver, and gastrointestinal system. To understand viral tropism and investigate host-pathogen biology, we utilized iPSC-derived lung and brain organoids to investigate SARS-CoV-2 infection and host responses. We found that alveolar epithelial cells highly expressed ACE2 and TMPRSS2 (Figures 1E, 1F, and 1K) and were permissive to SARS-CoV-2 infection, and supported efficient replication kinetics (Figures 2D and 2E). In addition, host factors such as neuropilin-1 (NRP1), cathepsin L (CTSL1), plasmin, and kallikrein-related peptidase 13 (KLK13) were expressed in both LORG and CORG model systems (Figures 1K, S1H, and S2E). In response to infection, most of the key genes in innate immunity, cytokine/chemokines, and inflammasome were upregulated in LORGs (Figures 2F and 2G). For example, immunity-related genes include STAT1/2, IRF7, CCL5, CXCL10, TNFα, IL-6, IL-8, and IFN, which corroborate with the patient-derived results indicating production of a cytokine storm leading to severe lung injury. In addition, we found that SARS-CoV-2 infection of LORGs activated genes involved in inflammasome pathways (NLRP3, NLRC4, ASC, IL-18, and caspase-1), suggesting that inflammasome activation causes pulmonary inflammation and cell death in lungs.
There is growing evidence of neurological complications in people with COVID-19 (Helms et al., 2020; Kremer et al., 2020; Mao et al., 2020). However, the SARS-CoV-2 permissiveness of brain and specific cell types is not well established. Our results show that CORGs express both ACE2 and TMPRSS2 and support virus infection. Corresponding to the lower expression of ACE2 and TMPRSS2 in CORGs, we observed lower viral infection rates in CORGs as compared with LORGs (Figures 3B, 3C, and 3H). Similarly, neurons expressed more ACE2 and TMPRSS2 than other cell types and were more permissive to viral infection than astrocytes and NPCs (Figures 3B, 3C, 3F, 3G, 3I, and 3J). Consistent with our results, two groups reported that SARS-CoV-2 does not infect neurons but efficiently infects choroid plexus epithelium, leading to cell damage and death (Jacob et al., 2020; Pellegrini et al., 2020). Interestingly, SARS-CoV-2 infection of neurons exhibited stimulation of gene expression different from LORGs because we found TLRs, OAS, and IL-6 but no IFNs (Figures 3K, 3L, and S2J). Furthermore, Zika, which is also a positive strand virus akin to SARS-CoV-2, also activates TLR3 to cause neuronal injury in NPCs (Dang et al., 2016). In addition, complement system gene expression was strongly enhanced during SARS-CoV-2 infection, suggesting a potential mechanism for COVID-19-related symptoms whereby the complement system is activated in response to injury, infection, and neurodegenerative diseases and triggers synaptic loss and neuronal cell death. Due to enhanced inflammatory response, neurons undergo apoptotic cell death as suggested by enhanced expression of caspase-3 and downregulation of antiapoptotic genes BCL2 and BAX.
Finally, we showed that viral entry was spike protein-mediated and that this was blocked by a specific inhibitor of spike protein, EK1 peptide, and TMPRSS2 inhibitors camostat and nafamostat in LORGs (Figure 2C), Calu-3 cells (Figures S1I–S1K). These results are in agreement with previous studies showing the activities of EK1 peptide (Xia et al., 2020) and TMPRSS2 inhibitors (Hoffmann et al., 2020) in repressing SARS-CoV-2, further validating the utility of stem cell-derived organoids to identify new therapeutics. Altogether, the described organoid model studies could facilitate the speed of drug discovery and provide useful knowledge about the mechanism of action and potential side effects of new drugs.
Experimental procedures
All studies were performed in accordance with the approved IRB protocols by the University of California, San Diego. Human iPSC cells were differentiated into LORGs and CORGs by following the previously established methods (Leibel et al., 2019; Miller et al., 2019) with minor modifications. In brief, ~75%–85% confluent undifferentiated human iPSC colonies were treated with 1 mg/mL dispase solution for 5–10 min at 37°C, washing cells gently with DMEM/F12 and scraping the colonies with a cell scraper in mTeSRTM1 medium. Next, iPSC colonies were dissociated into smaller colonies using serological pipette, and 0.5 mL of the smaller human iPSC clumps were plated in 24-well Matrigel-coated plates. When cells reached to 50%–75% confluence, stepwise differentiation was started into DE AFE, and LORGs. CORGs were prepared as described previously (Dang et al., 2016, 2020). Twenty 80-day organoids were transferred to bioreactors (125 mL) containing a magnetic shaft and stirring speed was maintained at 50–60 rpm. A detailed description of stem cell differentiation, immunofluorescence characterization, qRT-PCR, western blotting, viral infection, and statistical analysis is provided in Supplemental experimental procedures.
Author contributions
S.K.T. designed and performed experiments, analyzed the data, and wrote the manuscript draft; S.W. designed and performed experiments; D.S. provided resources; A.C. gave advice in experimental plans and performed experiments; T.M.R. conceived the overall project and experimental design, and participated in data analyses, interpretations, and manuscript writing.
Declaration of interests
T.M.R. is a founder of ViRx Pharmaceuticals and has an equity interest in the company. The terms of this arrangement have been reviewed and approved by the University of California, San Diego in accordance with its conflict of interest policies.
Acknowledgments
We thank F. Furnari for the iPSC lines and members of the Rana lab for helpful discussions and advice. The following reagent was deposited by the Centers for Disease Control and Prevention and obtained through BEI Resources, NIAID, NIH: SARS-Related Coronavirus 2, Isolate USA-WA1/2020, NR-52281. This work was supported by a Career Award for Medical Scientists from the Burroughs Wellcome Fund USA and a grant from the National Institutes of Health USA (K08 AI130381) to AFC, John and Mary Tu Foundation USA, and in part by grants from the National Institutes of Health USA (CA177322, DA039562, DA049524, and AI125103).
Published: March 9, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.stemcr.2021.02.005.
Supplemental information
References
- Barkauskas C.E., Chung M.I., Fioret B., Gao X., Katsura H., Hogan B.L. Lung organoids: current uses and future promise. Development. 2017;144:986–997. doi: 10.1242/dev.140103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benito-Kwiecinski S., Lancaster M.A. Brain organoids: human neurodevelopment in a dish. Cold Spring Harb. Perspect. Biol. 2019;12:a035709. doi: 10.1101/cshperspect.a035709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco-Melo D., Nilsson-Payant B.E., Liu W.C., Uhl S., Hoagland D., Moller R., Jordan T.X., Oishi K., Panis M., Sachs D. Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell. 2020;181:1036–1045 e1039. doi: 10.1016/j.cell.2020.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daamen A.R., Bachali P., Owen K.A., Kingsmore K.M., Hubbard E.L., Labonte A.C., Robl R., Shrotri S., Grammer A.C., Lipsky P.E. Comprehensive transcriptomic analysis of COVID-19 blood, lung, and airway. bioRxiv. 2020 doi: 10.1101/2020.05.28.121889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang J., Tiwari S.K., Agrawal K., Hui H., Qin Y., Rana T.M. Glial cell diversity and methamphetamine-induced neuroinflammation in human cerebral organoids. Mol. Psychiatry. 2020 doi: 10.1038/s41380-020-0676-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang J., Tiwari S.K., Lichinchi G., Qin Y., Patil V.S., Eroshkin A.M., Rana T.M. Zika virus depletes neural progenitors in human cerebral organoids through activation of the innate immune receptor TLR3. Cell Stem Cell. 2016;19:258–265. doi: 10.1016/j.stem.2016.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta D., Clevers H. Organoid culture systems to study host-pathogen interactions. Curr. Opin. Immunol. 2017;48:15–22. doi: 10.1016/j.coi.2017.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y., Duan X., Yang L., Nilsson-Payant B.E., Wang P., Duan F., Tang X., Yaron T.M., Zhang T., Uhl S. Identification of SARS-CoV-2 inhibitors using lung and colonic organoids. Nature. 2020;589:270–275. doi: 10.1038/s41586-020-2901-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helms J., Kremer S., Merdji H., Clere-Jehl R., Schenck M., Kummerlen C., Collange O., Boulay C., Fafi-Kremer S., Ohana M. Neurologic features in severe SARS-CoV-2 infection. N. Engl. J. Med. 2020;382:2268–2270. doi: 10.1056/NEJMc2008597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Kruger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob F., Pather S.R., Huang W.K., Zhang F., Wong S.Z.H., Zhou H., Cubitt B., Fan W., Chen C.Z., Xu M. Human pluripotent stem cell-derived neural cells and brain organoids reveal SARS-CoV-2 neurotropism predominates in choroid plexus epithelium. Cell Stem Cell. 2020;27:937–950.e9. doi: 10.1016/j.stem.2020.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsura H., Sontake V., Tata A., Kobayashi Y., Edwards C.E., Heaton B.E., Konkimalla A., Asakura T., Mikami Y., Fritch E.J. Human lung stem cell-based alveolospheres provide insights into SARS-CoV-2-mediated interferon responses and pneumocyte dysfunction. Cell Stem Cell. 2020;27:890–904.e8. doi: 10.1016/j.stem.2020.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremer S., Lersy F., Anheim M., Merdji H., Schenck M., Oesterle H., Bolognini F., Messie J., Henri-Feugeas M.C., Khalil A. Neurologic and neuroimaging findings in COVID-19 patients: a retrospective multicenter study. Neurology. 2020;95:e1868–e1882. doi: 10.1212/WNL.0000000000010112. [DOI] [PubMed] [Google Scholar]
- Lamers M.M., van der Vaart J., Knoops K., Riesebosch S., Breugem T.I., Mykytyn A.Z., Beumer J., Schipper D., Bezstarosti K., Koopman C.D. An organoid-derived bronchioalveolar model for SARS-CoV-2 infection of human alveolar-type II-like cells. EMBO J. 2020:e105912. doi: 10.15252/embj.2020105912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leibel S.L., Winquist A., Tseu I., Wang J., Luo D., Shojaie S., Nathan N., Snyder E., Post M. Reversal of surfactant protein B deficiency in patient specific human induced pluripotent stem cell derived lung organoids by gene therapy. Sci. Rep. 2019;9:13450. doi: 10.1038/s41598-019-49696-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., Wang W., Song H., Huang B., Zhu N. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao L., Jin H., Wang M., Hu Y., Chen S., He Q., Chang J., Hong C., Zhou Y., Wang D. Neurologic manifestations of hospitalized patients with coronavirus disease 2019 in Wuhan, China. JAMA Neurol. 2020;77:683–690. doi: 10.1001/jamaneurol.2020.1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller A.J., Dye B.R., Ferrer-Torres D., Hill D.R., Overeem A.W., Shea L.D., Spence J.R. Generation of lung organoids from human pluripotent stem cells in vitro. Nat. Protoc. 2019;14:518–540. doi: 10.1038/s41596-018-0104-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muffat J., Li Y., Omer A., Durbin A., Bosch I., Bakiasi G., Richards E., Meyer A., Gehrke L., Jaenisch R. Human induced pluripotent stem cell-derived glial cells and neural progenitors display divergent responses to Zika and dengue infections. Proc. Natl. Acad. Sci. U S A. 2018;115:7117–7122. doi: 10.1073/pnas.1719266115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mykytyn A.Z., Breugem T.I., Riesebosch S., Schipper D., van den Doel P.B., Rottier R.J., Lamers M.M., Haagmans B.L. SARS-CoV-2 entry into human airway organoids is serine protease-mediated and facilitated by the multibasic cleavage site. eLife. 2021;10:e64508. doi: 10.7554/eLife.64508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrin E.J., Little D.R., Gerner-Mauro K.N., Sumner E.A., Rios-Corzo R., Ambrosio E., Holt S.E., Forcioli-Conti N., Akiyama H., Hanash S.M. beta-Catenin maintains lung epithelial progenitors after lung specification. Development. 2018;145:dev160788. doi: 10.1242/dev.160788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrini L., Albecka A., Mallery D.L., Kellner M.J., Paul D., Carter A.P., James L.C., Lancaster M.A. SARS-CoV-2 infects the brain choroid plexus and disrupts the blood-CSF barrier in human brain organoids. Cell Stem Cell. 2020;27:951–961 e955. doi: 10.1016/j.stem.2020.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puelles V.G., Lutgehetmann M., Lindenmeyer M.T., Sperhake J.P., Wong M.N., Allweiss L., Chilla S., Heinemann A., Wanner N., Liu S. Multiorgan and renal tropism of SARS-CoV-2. N. Engl. J. Med. 2020;383:590–592. doi: 10.1056/NEJMc2011400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramani A., Muller L., Ostermann P.N., Gabriel E., Abida-Islam P., Muller-Schiffmann A., Mariappan A., Goureau O., Gruell H., Walker A. SARS-CoV-2 targets neurons of 3D human brain organoids. Embo J. 2020;39:e106230. doi: 10.15252/embj.2020106230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Li W., Hui H., Tiwari S.K., Zhang Q., Croker B.A., Rawlings S., Smith D., Carlin A.F., Rana T.M. Cholesterol 25-hydroxylase inhibits SARS-CoV-2 and coronaviruses by depleting membrane cholesterol. EMBO J. 2020;39:e106057. doi: 10.15252/embj.2020106057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia S., Liu M., Wang C., Xu W., Lan Q., Feng S., Qi F., Bao L., Du L., Liu S. Inhibition of SARS-CoV-2 (previously 2019-nCoV) infection by a highly potent pan-coronavirus fusion inhibitor targeting its spike protein that harbors a high capacity to mediate membrane fusion. Cell Res. 2020;30:343–355. doi: 10.1038/s41422-020-0305-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M., Lee E.M., Wen Z., Cheng Y., Huang W.K., Qian X., Tcw J., Kouznetsova J., Ogden S.C., Hammack C. Identification of small-molecule inhibitors of Zika virus infection and induced neural cell death via a drug repurposing screen. Nat. Med. 2016;22:1101–1107. doi: 10.1038/nm.4184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L., Han Y., Nilsson-Payant B.E., Gupta V., Wang P., Duan X., Tang X., Zhu J., Zhao Z., Jaffre F. A human pluripotent stem cell-based platform to study SARS-CoV-2 tropism and model virus infection in human cells and organoids. Cell Stem Cell. 2020;27:125–136.e7. doi: 10.1016/j.stem.2020.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.