Abstract
The rapid worldwide spread of the COVID-19 pandemic, caused by the severe acute respiratory SARS-CoV-2, has created an urgent need for its diagnosis and treatment. As a result, many researchers have sought to find the most efficient and appropriate methods to detect and treat the SARS-CoV-2 virus over the past few months. Real-time reverse-transcriptase polymerase chain reaction (RT-PCR) testing is currently used as one of the most reliable methods to detect the new virus; however, this method is time-consuming, labor-intensive, and requires trained laboratory workers. Moreover, despite its high sensitivity and specificity, false negatives are reported, especially in non-nasopharyngeal swab samples that yield lower viral loads. Therefore, designing and employing faster and more reliable methods seems necessary. In recent years, many attempts have been made to fabricate various nanomaterial-based biosensors to detect viruses and bacteria in clinical samples. The use of nanomaterials plays a significant role in improving the performance of biosensors. Plasmonic biosensors, field-effect transistor (FET)-based biosensors, electrochemical biosensors, and reverse transcription loop-mediated isothermal amplification (RT-LAMP) methods are only some of the effective ways to detect viruses. However, to use these biosensors to detect the SARS-CoV-2 virus, modifications must be performed to increase sensitivity and speed of testing due to the rapidly spreading nature of SARS-CoV-2, which requires an early point of care detection and treatment for pandemic control. Several studies have been carried out to show the nanomaterial-based biosensors’ performance and success in detecting the novel virus. The limit of detection, accuracy, selectivity, and detection speed are some vital features that should be considered during the design of the SARS-CoV-2 biosensors. This review summarizes various nanomaterials-based sensor platforms to detect the SARS-CoV-2, and their design, advantages, and limitations.
Keywords: COVID-19, SARS-CoV-2, Nanomaterials, Biosensors, Diagnostic test
Introduction
In December 2019, a new respiratory disease emerged in Wuhan, China, and spread rapidly throughout the world [1, 2]. It was later discovered that this was a new type of coronavirus, existed in the past; therefore, it was named 2019 novel coronavirus (2019-nCoV) by the world health organization (WHO). Due to the genetic similarity of the new coronavirus to the SARS (severe acute respiratory syndrome), it was later renamed as Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) by the coronavirus study group of the international committee on taxonomy of viruses [3, 4]. In addition to causing health problems, this virus has brought a socioeconomic crunch worldwide [5]. Even though at this point, a few vaccines (e.g., mRNA vaccines from Pfizer or Moderna) are approved and administered in public, it does not mean the importance of diagnostic testing being diminished by any means [6]. As a result, devising accurate methods to diagnose and detect the SARS-CoV-2 virus rapidly is vital.
Chest computed tomography (CT) is used to examine whether lungs are infected or not [7–10], and real-time reverse-transcriptase polymerase chain reaction (RT-PCR) is the standard method for detecting of SARS-CoV-2 virus [11–15]. However, the RT-PCR method has some drawbacks, such as being time-consuming, labor-intensive, and a slow detection process [16]. Biosensors can be an ideal alternative to the RT-PCR method because of their real-time detection and continuous monitoring [17, 18]. Biosensors are devices with a biological element (i.e., tissue, microorganisms, organelles, cell receptors, enzymes, antibodies, nucleic acids) that can recognize an analyte and generate a signal proportional to the concentration of the analyte [19]. Biosensors can be classified into three general categories, including electrochemical biosensors [20–22], optic biosensors [23–25], and piezoelectric biosensors [26, 27]. Electrochemical biosensors, such as FET-based biosensors, and optical biosensors, such as plasmonic biosensors, are the proposed platforms so far for detecting the SARS-CoV-2 virus. Electrochemical biosensors have been used for the detection of viruses and bacteria for a long time. These biosensors consist of semi-conductors and screen-printed electrodes. After forming the antibody-antigen conjugates on the electrode surface, electrochemical biosensors can monitor the change in dielectric properties and charge distribution [28]. Surface plasmon resonance (SPR) biosensing is another method for detecting the SARS-CoV-2 virus. This method is a strong photon-driven coherent oscillation of the surface conduction electrons, which can be modulated when coupling occurs at the surface of the plasmonic materials. Plasmonic biosensors can be successfully employed for real-time and label-free detection of microscale and nanoscale analytes. It has been shown that the localized surface plasmon resonance (LSPR) method used for nucleic acid detection can be employed as an exciting alternative for the detection of SARS-CoV-2 [29]. Field effect transistor (FET)-based biosensors can also be used for clinical diagnosis, point-of-care testing, and on-site detection. It has been reported that graphene-based FET biosensors can be employed for simple, rapid, and highly responsive detection of the SARS-CoV-2 virus in clinical samples [30]. The nanomaterial-enabled biosensors have several advantages due to materials' extraordinary physicochemical properties such as high surface-to-volume ratio, quantum size effects, high adsorption, and reaction capacity compared to their bulk form. Consequently, a small amount of analyte requirement, fast and sensitive signal delivery, low-cost, and ease of operations are granted [31].
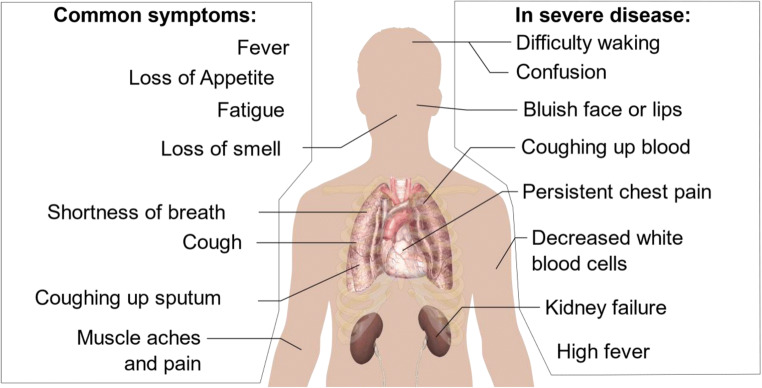
Nanomaterials-based biosensing has been successfully used for the detection of various viruses such as HIV/AIDS [33], hepatitis B virus [34], influenza virus [35, 36], and herpes virus [37]. Gold nanostructures-enabled biosensing, magnetic nanoparticles-enabled biosensing, and lanthanide-doped polystyrene nanoparticles-enabled biosensing systems have been recently published for sensitive detection of the SARS-CoV-2 virus. It has been reported that different nanomaterials-enabled biosensors can detect SARS-CoV-2 RNA, antigen, or antibody within 10 to 100 min [31]. Figure 1 shows SARS-CoV-2 symptoms [32]. This review focuses on the design, applications, and performance of the various nanomaterial-based biosensors for detecting the SARS-CoV-2 virus. For this purpose, different SARS-CoV-2 biomarkers, electrochemical biosensors, plasmonic biosensors, FET-based biosensors, and nanomaterials’ vital role in detecting the SARS-CoV-2 virus are reviewed. Therefore, this article aims to review and compare different biosensors and materials with the potential to replace the RT-PCR test.
Fig. 1.
SARS-CoV-2 symptoms, Ref: [32]
Biomarkers and indicators
Some different biomarkers and indicators can be used for the detection of SARS-CoV-2 by employing biosensors and nanomaterials. SARS-CoV-2 is a single-stranded positive-sense RNA virus having a large genome (29.8 kb) that encodes four structural proteins, which are the membrane (M), small envelope (E), spike (S), and nucleocapsid phosphoprotein (N) [38]. Therefore, single-stranded RNA, which is the most common biomarker used in the RT-PCR method, is one of the most important biomarkers for detecting SARS-CoV-2 [11, 12]. Moreover, each of those structural proteins can be used as antigens to detect SARS-CoV-2 [38]. It has been reported that M and E proteins are the most important ones in the formation of the SARS-CoV-2 virus structure [39]. Because the S protein combines with the host cells and its receptor-binding site interacts with ACE2 receptors, this protein is vital for detecting SARS-CoV-2. Generally, S and N proteins are usually used as biomarkers to detect both SARS-CoV and SARS-CoV-2 viruses [40, 41]. Furthermore, it has also been reported that by detecting some antibodies in the serological samples, the detection of COVID-19 becomes possible. It has been shown that the concentration of IgM and IgG in serological samples of patients is very low on day 0, increasing a detectable level on day 5 of the symptom [42]. Antibody detection is not only useful for patients during recovery but also afterward for epidemiological analysis. It can help make a vaccine because its exact level correlates with virus neutralization titer [43].
Plasmonic biosensors
Plasmonic biosensors play an essential role in detecting viruses, bacteria, and proteins. These highly sensitive biosensors are label-free and can be used for a wide range of analytes. The surface plasmon resonance (SPR)-based biosensors, which are highly sensitive, have been widely used to detect different viruses and bacteria. Moreover, recent advances in nanobiotechnology have made it possible to develop the SPR-based biosensor to detect biological targets using nanomaterials on the substrate. Localized surface plasmon resonance (LSPR) biosensors have been recently used instead of SPR biosensors because of their highly localized electromagnetic fields forming at nanoparticle surfaces make the detection of biological analytes possible [44–46]. Many different studies have been done on using these biosensors to detect SARS-CoV-2 [43]. Qiu et al. investigated a dual-functional plasmonic biosensor, including plasmonic photothermal (PPT) effect and LSPR for the clinical detection of SARS-CoV-2. The reason for choosing LSPR for detecting SARS-CoV-2 is that due to the enhanced plasmonic field around the nanostructures, the LSPR method shows great sensitivity to local variation, including the refractive index change molecular binding. Because of the exceptional properties of plasmonic nanoparticles, heating energy is localized in the vicinity of the nanoparticles. Therefore, these nanoparticles can be used as a heat source for thermal processing. On the other hand, the PPT effect can increase the sensitivity of the biosensor. For this purpose, two-dimensional gold nanoisland (AuNI) chips were fabricated by employing the self-assembly process of thermal dewetted Au nanofilm. The thickness of magnetron-sputtered Au nanofilms on the BK7 glass surface was within the range of 5 to 5.2 nm. The AuNIs surface functionalization was carried out by using the step-by-step injection of 0.1 nmol thiol-cDNA. Subsequently, 200 μL of the target DNA was entered into the AuNI microfluidic chamber for 800 s, and the hybridization reaction occurred under the PPT heat. The limit of detection of this system for RNA-dependent RNA polymerase-COVID (RdRp-COVID) was about 0.22 ± 0.08 pM [29].
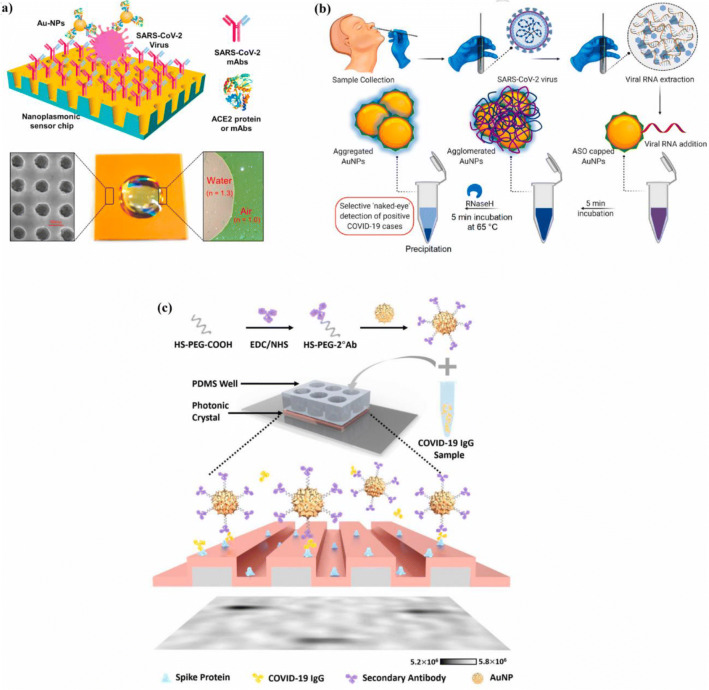
Huang et al. [47] investigated a nanoplasmonic biosensor to detect the SARS-CoV-2 virus without sample preparation (Fig. 2a). An Au-TiO2-Au nano-cup array chip, fabricated by the replica molding process, with a drop of water on top of it, was used as the sensor chip on a substrate made of silicon oxide wafer. Due to the use of this chip, the periodic nanostructure design of the biosensor, the plasmon resonance wavelength, and intensity change on the virus-capturing sensor could be detected by transmission light spectroscopy without any need for external coupling optics. The thicknesses of Au and Ti on the nano-cap array were 70 nm and 10 nm, respectively. For surface functionalization, immobilizing SARS-CoV-2 mAbs of 12.0 μg/mL to the activated chips was carried out. The nanoplasmonic biosensor was integrated with a standard 96-well plate to detect the SARS-CoV-2 virus in one step. The biomarker in this study was the spike protein. The limit of the biosensor was 30 SARS-CoV-2 virus particles in one step, within 15 min. The authors also fabricated a gold nanoparticle-enhanced nanoplasmonic biosensor that enabled rapid and single-step coronavirus detection. The binding of SARS-CoV-2 mAbs to the SARS-CoV-2 spike protein and Au NP-labeled ACE2 protein to the receptor-binding domain (RBD) was used for the detection of the virus.
Fig. 2.
a Nanoplasmonic biosensor with Au-TiO2-Au nanocup array chip with a drop of water [47], b naked-eye detection of SARS-CoV-2 by ASO capped gold nanoparticles [49], c PRAM-based AC + DC immunoassay [54]
Peng et al. [48] investigated a near-infrared plasmonic biosensor to detect SARS-CoV-2 and its spike (S) glycoprotein. Two-dimensional (2D) Van der Waals heterostructures (carboxyl-functionalized molybdenum disulfide (MoS2) layers and tellurene) with transparent indium tin oxide film were used to fabricate this plasmonic biosensor. They used theoretical predictions to determine the thickness of indium tin oxide (ITO) film, tellurene nanosheets, and MoS2–COOH to maximize the biosensor's sensitivity. Therefore, it was shown that the 121 nm ITO film/three-layer tellurene/ten-layer MoS2–COOH was the best configuration to obtain the maximum detection sensitivity, 8.4069 × 104 deg/RIU. It was reported that the carboxyl-functionalized MoS2 increases the detection sensitivity; moreover, it was also able to capture target protein amide bonds (-NH2).
Moitra et al. reported a naked-eye detection of SARS-CoV-2 by using antisense oligonucleotide capped plasmonic nanoparticles (Fig. 2b). Due to the lower sensitivity of the biosensors for the detection of the N gene (nucleocapsid phosphoprotein gene) compared to the RdRP gene (RNA-dependent RNA polymerase gene) and E gene (envelope protein gene), they decided to fabricate this biosensor and improve its sensitivity for the detection of N gene. Four antisense oligonucleotides (ASOs) sequences used to cap AuNPs were selected according to their closely target following position, binding disruption energies, and binding energies. Two of these ASOs were functionalized from 5′, and the other two ones were functionalized from the 3′ with thiol moieties. All AuNPs capped with these four ASOs were dispersed very well without forming a large entity. Mixing all ASO-capped AuNPs (Au-ASO1M, Au-ASO2L, Au-ASO3H, and Au-ASO4M), resulting in the formation of Au-ASOmix, increased the sensitivity of the AuNPs for the detection of SARS-CoV-2 RNA. Surface plasmon bands verified the formation of ASO-conjugated thiol-stabilized AuNPs. These Au-ASOmix nanoparticles tended to disperse individually in the samples before adding viral load; however, they were agglomerated and formed large clusters in the presence of SARS-CoV-2 RNA. For the naked-eye detection of the SARS-CoV-2 RNA, RNaseH was added to the solution. In the presence of RNaseH, the RNA strand was cleaved from the RNA−DNA hybrid leading to detectable precipitation in the solution, mediated by the additional agglomeration of the AuNPs. This biosensor also showed good selectivity for the detection of SARS-CoV-2 in the presence of the MERS-CoV virus. The detection limit was 0.18 ng/μL for the SARS-CoV-2 RNA in the viral load [49].
Ahmadivand et al. fabricated another type of plasmonic biosensor, called toroidal plasmonic metasensor, to detect SARS-CoV-2 spike protein. One of the most significant features of the plasmonic metasensors is that “it can squeeze electromagnetic fields in frequency, time, and space” simultaneously. However, these biosensors usually cannot detect small biomolecules at low amounts. Ahmadivand et al. fabricated a miniaturized plasmonic immunosensor based on toroidal electrodynamics to overcome this limitation. A mixture of 0.1 M of reactant buffer with 50 μL of purified spike S1 antibody was utilized to conjugate SARS-CoV-2 Spike S1 antibody with the NHS activated AuNPs. The average diameter of AuNPs was around 45 nm. For dissolving the immunoreagents, both bovine serum albumin (BSA) and a phosphate buffer solution (PBS) were used. To enhance the binding of biomarkers to the sensor surface, functionalize AuNPs were dispersed on the biosensor's surface. The ultratight field confinement of toroidal metasensor makes it possible to achieve high sensitivity like the LSPR biosensors. To evaluate the toroidal dipole position variations, both a solution of functionalized AuNPs conjugated with the SARS-CoV-2 antibody and PBS (without spike protein) and a solution containing spike proteins were tested. The results showed that gold nanoparticles conjugated with antibodies play an important role in capturing spike proteins and detecting SARS-CoV-2. Due to the exceptional properties of the plasmonic metasensor, including high Q-factor, “ultratight field confinement, and high sensitivity of toroidal resonances,” the detection limit of this plasmonic metasensor was 4.2 fmol [50].
Colorimetric biosensors, which are based on LSPR, can also be used for detecting the SARS-CoV-2 virus. Metal nanoparticles are the most critical part of these biosensors because their unique optical properties make the rapid detection of the targets possible. AuNPs are usually used in these biosensors because of their surface chemistry and biocompatibility. Using AuNPs in the colorimetric method, the color changes from red to blue in a colloidal suspension due to LSPR coupling among the gold nanoparticles. Ventura et al. reported a colorimetric biosensor based on AuNPs to detect SARS-CoV-2 in nasal and throat swabs. They used 94 clinical samples, tested before by standard RT-PCR, 45 of which were positive, and 49 were negative, to test this biosensor. AuNPs were functionalized with antibodies to detect SARS-CoV-2 spike, envelope, and membrane proteins in clinical samples; therefore, there were three types of functionalized AuNPs whose ratio was 1: 1: 1. It was reported that one of the most significant advantages of this biosensor was its sensitivity to the virion instead of its content, which is RNA. The test results showed that the colorimetric biosensor was able to detect a very low amount of target proteins, and its limit of detection was very close to that of the real-time PCR method. Therefore, this colorimetric detection platform’s limit was reported based on the real-time PCR cycle threshold (Ct) and was Ct = 36.5 [51, 52].
Funari et al. fabricated an opto-microfluidic sensing platform with gold nano spikes based on LSPR to detect SARS-CoV-2 spike protein in 30 min. 1000 Å of gold was electrodeposited on a 50 Å chromium layer on the glass substrates to produce the gold nano spikes. These gold nano spikes were functionalized by immersing them in the thiol mixture. 1:1 solution of 10 mM NHS (N-Hydroxysuccinimide) and 40 mM of EDC (1-Ethyl-3-(3-dimethyl aminopropyl) carbodiimide) was used to activate the surface gold nano spikes. To produce an opto-microfluidic platform, the functionalized Au nano spikes were bonded to a polydimethylsiloxane (PDMS) slab using an 85 μm thick adhesive polyester layer. The limit of detection for the LSRP-based biosensor was around 0.08 ng/mL (∼ 0.5 pM) [53].
Zhao et al. investigated a single-step, wash-free digital immunoassay for the qualitative detection of IgG against SARS-CoV-2 (Fig. 2c). For this purpose, the linear grating period of the photonic crystal (PC)-based biosensor was 380 nm. Its grating depth was 97 nm etched into a glass substrate using the reactive ion etching method. This granting was next coated with a 98.5-nm-thick TiO2 layer. By employing the AC + DC assay, SARS-CoV-2 IgG proteins activated the functionalized AuNPs in solution and the binding of activated AuNPs to the PC. To activate the PC surface, oxygen plasma treatment was used. Subsequently, a coating of the recombinant COVID-19 spike protein was applied on the PC’s surface to capture COVID-19 IgG. Finally, COVID-19 IgG was incubated with secondary antibody functionalized AuNPs (2°Ab-AuNPs). After mixing the sample and 2°Ab-AuNPs, there was no incubation step, and the mixture was exposed to the spike protein-coated PC biosensor. Finally, a photonic resonator absorption microscopy (PRAM) method was used for imaging. The limit of quantification and detection of this method was 32.0 ± 8.9 pg/mL and 26.7 ± 7.7 pg/mL, respectively [54].
As mentioned, plasmonic biosensors have high sensitivity and a low limit of detection. According to reviewed articles, their detection limit for the SARS-CoV-2 varies approximately from 0.08 to 180 ng/mL. However, it should be noted that plasmonic biosensors have some drawbacks, such as specialized instrumentation, challenging portability, challenging point of care applications, and hands-on experience [55, 56]. Different plasmonic-based biosensors used for the detection of SARS-CoV-2 are reported in Table 1.
Table 1.
Plasmonic biosensors for the detection of SARS-CoV-2
| Biosensing technique | Material selection and design | Biomarker | Limit of detection |
|---|---|---|---|
|
Dual-functional plasmonic photothermal Biosensor [29] |
Two-dimensional AuNI chips were fabricated using the self-assembly process of thermal dewetted Au nanofilm. The thickness of magnetron-sputtered Au nanofilms on the BK7 glass surface was within the range of 5 to 5.2 nm | RdRp-COVID (SARS-CoV-2 RNA) | 0.22 ± 0.08 pM |
| Nanoplasmonic biosensor [47] | An Au-TiO2-Au nano-cup array chip, fabricated by the replica molding process, with a drop of water on top of it, was used as the sensor chip on the silicon oxide wafer. the thicknesses of Au and Ti on the nano-cap array were 70 nm and 10 nm, respectively | Spike protein of SARS-CoV-2 | 30 virus particles in one step |
| Near-infrared plasmonic biosensor [48] | integrating two-dimensional (2D) Van der Waals heterostructures, including tellurene and carboxyl-functionalized molybdenum disulfide layers, with transparent indium tin oxide film | SARS-CoV-2 spike (S) glycoprotein | Sensitivity = 8.4069 × 104 deg/RIU |
| N gene-targeted antisense oligonucleotide capped plasmonic nanoparticles (naked-eye detection) [49] | Four ASOs sequences used to cap AuNPs were selected according to their closely target following position, binding disruption energies, and binding energies. Mixing all ASO-capped AuNPs, which resulted in the formation of Au-ASOmix, increased the sensitivity of the gold nanoparticles for the detection of SARS-CoV-2 RNA | SARS-CoV-2 N gene (nucleocapsid phosphoprotein gene) | 0.18 ng/μL |
| Toroidal plasmonic metasensor [50] | A mixture of 0.1 M of reactant buffer with 50 μL of purified spike S1 antibody was utilized to conjugate SARS-CoV-2 Spike S1 antibody with the NHS activated gold nanoparticles. The average diameter of AuNPs was around 45 nm. For dissolving the immunoreagents, both bovine serum albumin and a phosphate buffer solution were used. The functionalized AuNPs were dispersed on the sensor surface to enhance the binding events. | SARS-CoV-2 spike protein | 4.2 fmol |
| Colorimetric biosensor based on localized surface plasmon resonance (LSPR) [51] | AuNPs were used in these biosensors because of their surface chemistry and biocompatibility. By using gold nanoparticles in the colorimetric method, the color changes from red to blue in a colloidal suspension because of LSPR coupling among the AuNPs, which were functionalized with antibodies | SARS-CoV-2 spike, envelope, and membrane proteins | Ct = 36.5 (the limit of detection of this colorimetric biosensor was reported based on the real-time PCR cycle threshold (Ct)) |
| Gold nano spikes in an opto-microfluidic chip (based on localized surface plasmon resonance) [53] | 1000 Å of Gold was electrodeposited on a 50 Å of chromium layer on the glass substrates. These gold nano spikes were functionalized by immersing them in the thiol mixture. 1:1 solution of 10 mM NHS (N-Hydroxysuccinimide) and 40 mM of EDC (1-Ethyl-3-(3-dimethyl aminopropyl) carbodiimide) was used to activate the surface gold nano spikes | SARS-CoV-2 spike protein | ∼ 0.08 ng/mL (∼ 0.5 pM) |
| Single-step and washing-free immunoassay for the detection of SARS-CoV-2 by photonic resonator absorption microscopy (PRAM) [54] | the linear grating period of the PC-based biosensor was 380 nm. Its grating depth was 97 nm, etched into a glass substrate using the reactive ion etching method. This granting was next coated with a 98.5 nm thick TiO2 layer. By employing the AC + DC assay, SARS-CoV-2 IgG proteins activated the functionalized AuNPs in solution, which triggered the binding of activated AuNPs to the PC. To activate the PC surface, oxygen plasma treatment was used. Subsequently, a coating of the recombinant COVID-19 spike protein was applied on the PC's surface to capture COVID-19 IgG. Finally, COVID-19 IgG was incubated with secondary antibody functionalized AuNPs | SARS-CoV-2 IgG | limit of detecting = 26.7 ± 7.7 pg/mL limit of quantification = 32.0 ± 8.9 pg/mL |
Electrochemical biosensing
Electrochemical biosensors are another class of biosensors that have widely been investigated by researchers to detect the SARS-CoV-2 virus. An electrochemical transducer that transforms biochemical information is used in this type of biosensors. The high sensitivity, simple instrumentation, cost-effectiveness, and miniaturization possibility can be considered the most critical advantages of electrochemical biosensors [57]. The basis of the electrochemical biosensors is the reaction between the immobilized biomolecules and the analyte. As a result of this reaction, the solution's electrical properties will be affected, leading to analyte detection. Potentiometric, amperometric, conductometric, voltametric, polarographic, impedimetric, capacitive, and piezoelectric biosensors are different electrochemical biosensors [58].
Rashed et al. investigated a rapid label-free impedance sensing platform to detect the SARS-CoV-2 virus using its spike protein. For this purpose, 2.5 μg/mL of the receptor-binding domain (RBD) of SARS-CoV-2 spike protein was coated on a 16-well container, and sensing electrodes were located inside the container. To incubate wells, they were filled with a blocking solution containing 3% milk. First, this apparatus was tested with anti-SARS-CoV-2 monoclonal antibody CR3022 with different concentrations, including 0.1 μg/ml, 1.0 μg/ml, and 10 μg/ml to validate the designed system. This apparatus was then used to test six serological samples taken from six different people, some of whom had COVID-19, and others did not. However, the research team had not been informed about whether they were positive or negative. The well plate was connected to a laptop to measure the impedance changes with time. The results were shown as changes in impedance magnitude between the real-time measured impedance (푍) with respect to a measured background impedance, shown by 푍표 (훥푍 = 푍 – 푍표). Finally, the results obtained from impedance measurements were compared to ELISA results of the same serum samples, and it was reported that there was a strong correlation between them. The limit of this biosensing method's detection was not reported because of two reasons, hardware noise and sample handling variations [59].
Vadlamani et al. studied the fabrication of low-cost, rapid, and highly sensitive cobalt-functionalized TiO2 nanotubes (Co-TNTs)-based electrochemical sensors to detect the SARS-CoV-2 virus. The RBD of the SARS-CoV-2 spike protein on the viral surface was chosen as the analyte for detecting the virus. TNTs were synthesized by employing a low cost, simple, one-step electrochemical anodization of G1 grade titanium sheets of size 1.5 × 1.5 cm. The anodization of the Ti sheets was carried out by employing a two-electrode method in which Ti sheet was a working electrode and platinum acted as a counter electrode. Cobalt functionalization of TNTs was also done by an incipient wetting method called the wet ion exchange process. The system was connected to a potentiostat to record the data obtained from the experiments. An amperometry electrochemical method at a bias voltage of −0.8 V was used to evaluate the fabricated Co-TNT biosensor's ability to detect the SARS-CoV-2 spike protein. It was reported that when the electrode was exposed to the SARS-CoV-2 spike protein, its response current increased suddenly and sharply. The decrease in the protein concentration from 1400 to 14 nM decreased the peak sensor current from 0.74 to 0.23 μA. Within this concentration range of the protein, the detection time of the sensor was around 30 s. However, it only took around 2 s for this sensor to reach the current peak; in other words, the average sensor response time was 2 s. The rapid increase of the current after exposing the sensor to the SARS-CoV-2 spike protein was related to the unfolding of the protein structure. As a result of this phenomenon, the unfolded protein and Co formed a complex (the Co cation in the Co-TNT reacts with the O anion in the protein), leading to the rapid increase of the current. The limit of detection of this sensor for the detection of the SARS-CoV-2 spike protein was 0.7 nM. This electrochemical-based biosensor can be used to detect SARS-CoV-2 in nasal, nasopharyngeal swabs, and saliva samples [60].
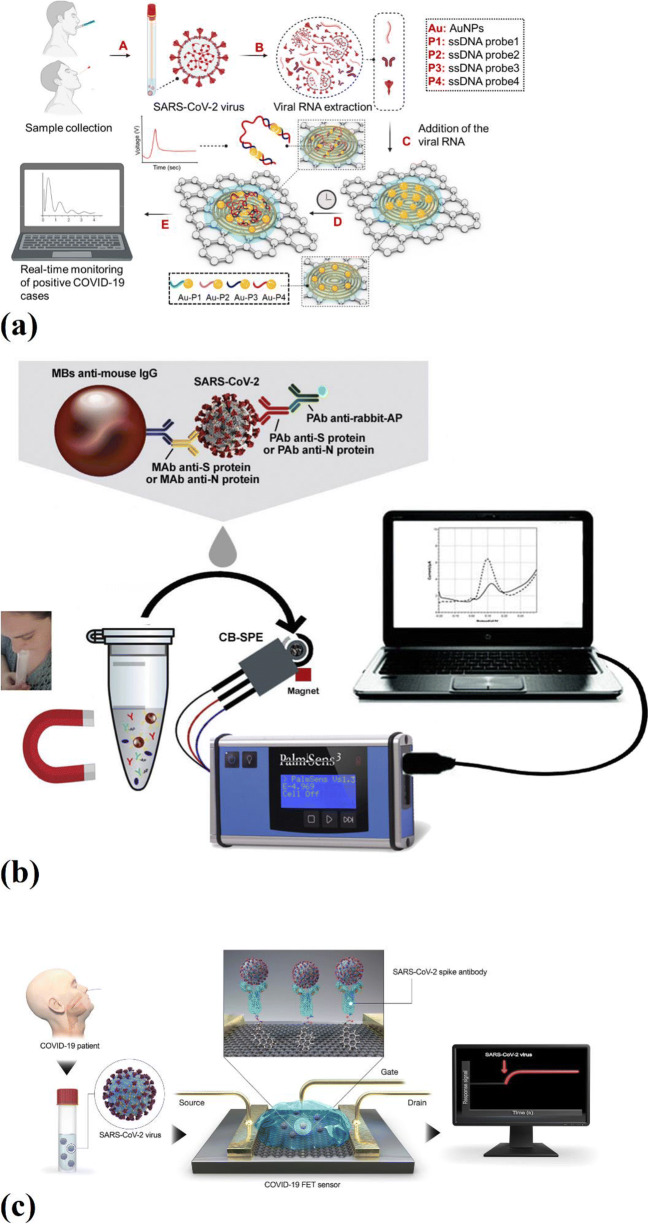
A rapid and quantitative electrochemical biosensor chip was investigated by Alafeef et al. (Fig. 3a). This biosensor was easy to implement and a quantitative paper-based electrochemical biosensor chip that could detect the SARS-CoV-2 viral RNA in less than 5 min. AuNPs that were capped with specific antisense oligonucleotides (ssDNA) were employed to detect viral N-gene. A graphene film was coated to form a conductive layer on the surface of the filter paper. The high carrier mobility of graphene, which is higher than 2000 cm2 V s−1, provides a highly sensitive layer for interaction and adsorption of the charged target. Three different graphene suspension concentrations, 5, 10, and 20 mg/mL, were used to find the optimum graphene film. The suspension with the graphene concentration of 20 mg/mL did not form a uniform film due to its high concentration. On the other hand, the suspensions with graphene concentrations of 5 and 10 mg/mL resulted in a uniform layer of graphene on the filter paper surface. AuNPs are usually used to increase the sensitivity of the biosensors because of their excellent unmatched properties. Therefore, ssDNA-capped AuNPs were deposited on the surface of the biosensor platform to increase the electrochemical sensitivity. The antisense oligonucleotides were incorporated on the sensor chip in two different ways. First, antisense oligonucleotides were conjugated directly to the gold surface. Second, the surface of the nanoparticles was capped with antisense oligonucleotides. However, the test results showed that capped Au-NPs had a higher response to the SARS-CoV-2 RNA related to antisense oligonucleotides' higher reactivity when they were capped on the AuNPs and then deposited on the surface of the electrode. This configuration encourages the electron transfer kinetics; moreover, due to the high surface area provided by AuNPs in this configuration for the interaction of antisense oligonucleotides, signal amplification occurs. The limit of detection of this biosensor without nucleic acid amplification was 6.9 copies/μL [61].
Fig. 3.
a Real-time electrochemical biosensing of SARS-CoV-2 RNA [61], b magnetic beads-based assay for the detection of SARS-CoV-2 in untreated saliva [63], c graphene-based FET biosensor for the detection of SARS-CoV-2 [30]
Torrente-Rodriguez studied a multiplexed, portable, wireless graphene-based electrochemical biosensor, which was named the SARS-CoV-2 RapidPlex, for the rapid detection and monitoring of the COVID-19. It was reported that this electrochemical biosensor was able to detect viral antigen nucleocapsid protein, IgM and IgG antibodies, and the inflammatory biomarker C-reactive protein (CRP). This SARS-CoV-2 RapidPlex consisted of five graphene electrodes, four of which were working electrodes, and one of them was a counter electrode and an Ag/AgCl reference electrode. These four working electrodes made it possible to detect nucleocapsid protein (NP), S1-IgG, S1-IgM, and CRP in only one step. By employing CO2 laser engraving, all these electrodes were patterned on a Polyimide substrate. High charge mobility, surface area, sensitivity, and selectivity of graphene made this biosensor an accurate device for rapid detection of SARS-CoV-2. Functionalization and modification for each of the receptors’ covalent attachments were carried out on the laser engraved graphene (LEG) surfaces. In this study, pyrene derivatives were used to introduce the functional groups on the sensing layer. Finally, target proteins, which were NP and CRP, were detected by sandwich-based immunosensing onto the LEG electrodes because this method was highly sensitive due to the involvement of two different antibodies as the capture and the detector molecules. On the other hand, immunoglobulins S1-IgG and S1-IgM were detected by indirect-based immunosensing onto the LEG electrodes because this method is suitable for detecting circulating macromolecules in antisera and other biofluids. An amperometric technique was employed to evaluate the selectivity and crosstalk of the SARS-CoV-2 RapidPlex. The results showed no cross-reaction for NP, S1-IgG, S1-IgM, and CRP assays in the presence of the interferents. One of the significant advantages of this electrochemical biosensor over the other is that only 1-min incubation time was enough to ensure the highest possible sensitivity [62].
Fabiani et al. [63] investigated a miniaturized electrochemical immunosensor to detect the SARS-CoV-2 virus in saliva (Fig. 3b). This work’s novelty was the combination of electrochemical detection with miniaturization, resulting in a highly sensitive method for detecting SARS-CoV-2. In this study, NS and NP's detection was carried out using magnetic beads to support immunological recognition. The secondary antibody with a linked alkaline phosphatase was used as the immunological label. A three-electrode cell was used for performing electrochemical tests. For this purpose, an electrode made of graphite was chosen as working and counter electrodes, and the reference electrode was silver-based. Screen-printed electrodes (SPEs) were fabricated onto transparent and flexible polyester support. Six microliters of carbon black (CB) N220 dispersion (1 mg/mL in N, N-dimethylformamide: distilled H2O 1:1 v/v) was used for the modification of the SPEs to increase their sensitivity. First, the ELISA test was performed to verify the possibility of binding target proteins with antibodies. After this step, the electrochemical magnetic beads-based technique was employed to detect NS and NP. This process included three sequential steps. First, a blocking coating of IgG was produced to store them at a temperature of 4°C for several months. Second, sequential incubations for the immuno-recognition events were combined to make a single incubation of 30 min. Finally, the electrochemical detection was carried out using the SPEs. After the immunoassay procedure, 20 μL of the beads’ suspension in 100 μL of DEA buffer was drop cast on the working electrode made of graphite and magnetically concentrated on the surface of the working electrode. The differential pulse voltammetry (DPV) method was employed to measure the electroactive enzymatic product (1- naphthol) after 2 min of the enzymatic reaction. In this experiment, Ebegin was - 0.2 V, Eend was 0.4 V, Estep was 0.016 V, Epulse was 0.05 V, tpulse was 0.06 s, and the scan rate was 0.016 V/s. It was shown that by employing antibodies directed against spike protein, the assay’s sensitivity became excellent, and it was able to detect 6.5 PFU/mL. However, the NP-based assay’s sensitivity was lower because of the lower NP amount compared to spike protein in the SARS-CoV-2 virus. This electrochemical biosensor limit was reported as 19 ng/mL for spike protein and 8 ng/mL for NP. This MBs-based assay was also used to test clinical samples, fresh and frozen saliva to evaluate its effectiveness for clinical samples. The results showed that the frozen samples of both negative and positive patients resulted in a reduced signal. Therefore, the threshold was increased to 1.8 μA for frozen samples, while its value for fresh samples was 1 μA [63].
A super sandwich-type electrochemical biosensor for detecting the SARS-CoV-2 was reported by Zhao et al. This biosensor was based on calixarene functionalized graphene oxide for targeting RNA of SARS-CoV-2. It was reported that by employing the super sandwich-type detection method, this electrochemical biosensor was able to detect the RNA of SARS-CoV-2 without nucleic acid amplification and reverse-transcription by using a portable electrochemical smartphone. In this study, both artificial targets and clinical RNA samples were tested. Clinical samples were collected from patients whose COVID-19 were confirmed by CT scans and nucleic acid testing. To prepare premix A, Fe3O4 nanoparticles were dissolved, and then PEG400, trisodium citrate, HAuCl4, and ascorbic acid were added for producing the Au@Fe3O4 nanocomposite. Then, 100 μL of 1 mg/m of this nanocomposite was dissolved in Buffer I, containing 10 mM Tris-HCl, 1 mM EDTA, 300 mM NaCl, and 10 mM TCEP. Finally, the produced precipitation was dissolved with 100 μL of Buffer II containing 10 mM Tris-HCl containing 1 mM EDTA, 300 mM NaCl, and 1 mM MgCl2. 50 μL of premix A was mixed with 10 μL of detection samples, and the mixture was incubated for one hour. Subsequently, 50 μL of premix B was added to the mixture and incubated for two hours. The obtained nanocomposite was dissolved in 50 μL of PBS and dropped on screen printing carbon electrode (SPCE) for electrochemical measurement. The capture probes (CPs) were immobilized on the surfaces of the Au@Fe3O4 nanoparticles to produce CP/Au@Fe3O4 nanocomposites. Then p-sulfocalix[8]arene (SCX8) was immobilized on the functionalized graphene (RGO), and Au@SCX8-TB-RGO-LP bioconjugate was produced. Finally, a CP-target-LP sandwich structure was fabricated. The limit of detection was found 3 aM for SARS-CoV-2 in the artificial sample. On the other hand, the limit of detection from clinical samples was 200 copies/mL [64].
Mahari et al. [65], investigated a biosensor to detect SARS-Cov-2 spike antigen in spiked saliva samples and compared it to a potentiostat. For this purpose, they fabricated a potentiostat based sensor and an eCovSens (the fabricated biosensor’s name). The potentiostat-based sensor was fabricated using a fluorine-doped tin oxide electrode (FTO) with AuNPs. It was also immobilized by a nCovid-19 monoclonal antibody (nCovid-19Ab). In the case of eCovSens was immobilized with a nCovid-19 Ab on the SPCE. The heat-reflux citrate reduction method was employed to synthesize AuNPs. For this purpose, 0.01 ml of 10% gold chloride was mixed with milli-Q water and heated until the solution started boiling, and then 1 mL of 1 % sodium citrate tribasic was mixed with the boiling mixture. 90 μg of nCovid-19 Ab was added to 1 mL of AuNPs solution in phosphate buffer (PB) to label nCovid-19 Ab AuNPs. The fluorine-doped tin oxide (FTO) electrode was fabricated by using glass coated with FTO. For the fabrication of the FTO electrode with AuNPs/nCovid-19 Ab, 200 μl of AuNPs were drop casted on the FTO electrode surface and dried. Subsequently, 40 μl of 1 μg/mL nCovid-19 Ab was immobilized on the FTO/AuNPs electrode. Due to the good electrical conductivity of AuNPs, they could amplify the electrochemical signal. On the other hand, nCovid-19 Ab was attached to the AuNPs by electrostatic interactions. The electrochemical device was composed of a central processing unit (CPU) and a memory chip. By adding nCovid-19 Ag, the electrical current was changed because of the proteins' polarity that affected the charge transfer from the electrode surface. Various concentrations of nCovid-19 Ag were tested on the prepared electrode (from 1 fM to 1 μM) using differential pulse voltammetry. However, the maximum signal was observed at the concentration of 100 nM, and a further increase in the concentration did not affect the obtained signal. Finally, it was shown that this electrochemical biosensor was as sensitive as a potentiostat, and its limit of detection was 10 fM for nCovid-19 Ag.
Since COVID-19 infection demands diagnostics at the point-of-care (POC) Kaushik et al. [66] investigated the importance of developing a nano-enabled electrochemical biosensor for COVID-19 diagnostics at point-of-care (POC) application. In order to manage the coronavirus epidemic, they proposed the design and development of a smart biosensor. It was reported that artificial intelligence (AI)-supported nano-enabled electrochemical biosensors were required to better manage the COVID-19 pandemic.
The field-effect transistor (FET)-based biosensors are another type of electrochemical biosensors widely studied by researchers. A FET-based biosensor for the detection of SARS-CoV-2 in clinical samples was reported by Seo et al. (Fig. 3c) [30]. In the study, graphene sheets of FET were coated with a specific antibody to detect SARS-CoV-2 spike protein. This protein was chosen because of being a major transmembrane protein of the virus and highly immunogenic. The nasopharyngeal swab samples of COVID-19 patients, antigens, and the cultured virus were used to assess this FET-based biosensor’s performance. By using a conventional wet-transfer method, graphene was coated on the SiO2/Si substrate. Next, a coating of poly(methyl methacrylate) (PMMA) C4 was applied to the graphene layer. After transferring the PMMA/graphene layer to the SiO2/Si substrate, the PMMA layer was removed using acetone. A gold-chromium electrode layer was later fabricated on the etched graphene layer. For the immobilization of the SARS-CoV-2 antibody on the graphene layer, first, it was immersed in 2 mM PBASE and methanol, then the functionalized surface was exposed to 250 μg/mL SARS-CoV-2 spike antibody. Since this biosensor did not show any responses to MERS-CoV spike proteins, it was verified that the fabricated COVID-19 FET-based biosensor was highly sensitive and specific for the SARS-CoV-2 spike antigen protein. Moreover, this biosensor was able to detect SARS-CoV-2 spike antigen proteins in clinical samples without any preparation. The limit of detection of this FET-based biosensor for the detection of SARS-CoV-2 spike protein was 1 fg/mL and lower than that of the ELISA method.
Gaurav et al. also reported a graphene bio-FET-based biosensor to detect the SARS-CoV-2 virus. The graphene was soaked with a PBASE solution on the surface of the graphene FET biosensor. PBASE has two ends: a pyrene group that non-covalently attaches to graphene through π-π (pi-pi) stacking, and the other one is s an activated ester that reacts with the means. The sensitivity of this biosensor was tested after the graphene was attached to the antibody. The binding of spike protein to the antibody altered the distribution in the vicinity of the graphene layer, and therefore, changed its electrical conductivity. Consequently, the current flowing between the source and drain electrodes was changed, and the biosensor was able to detect the spike protein in clinical samples with the limit of detection of 1 fg/mL. One of these biosensor features was distinguishing between the noninfected and infected people with the SARS-CoV-2 virus. Moreover, this sensor did not respond to the spike proteins of other viruses, such as SARS and MERS [66].
A graphene FET (Gr-FET) biosensor for detecting SARS-CoV-2 spike protein was also investigated by Zhang et al. this biosensor was able to detect SARS-CoV-2 spike protein in 2 min. In this study, extremely sensitive Gr-FET was combined with selective SARS-COV spike protein antibody (CSAb) - COVID-19 spike protein antigen for the real-time detection of SARS-CoV-2. The chemical vapor deposition (CVD) method was employed to synthesize a single-crystal graphene layer on the single crystal copper. Functionalization of the graphene surface to bind the COVID-19 spike protein was carried out by immobilizing CSAb and ACE2 receptors on graphene’s surface. During the incubation of the positively charged CSAb and negatively charged ACE2 in PBS buffer solution, negative and positive potentials were applied, respectively. As a result, their immobilization on the surface of the graphene was enhanced. The test results showed that the CSAb-based FET biosensor had a higher affinity than the ACE2-based FET biosensor to the spike protein RBD. The limit of detection was reported as 0.2 pM [67].
The electrochemical biosensors have a low detection limit for detecting the SARS-CoV-2 virus, varying approximately from 10-6 to 19 ng/mL, and they have been successfully used to detect the virus despite the limitations. Electrochemical biosensors could be sensitive to the sample matrix effects. It has been reported that they were not as sensitive as the RT-PCR tests. Compared to RT-PCR, their lower shelf life is another critical disadvantage of this type of biosensor [68]. Different electrochemical biosensors used for the detection of SARS-CoV-2 are reported in Table 2.
Table 2.
Electrochemical biosensors for the detection of SARS-CoV-2
| Biosensing technique | Material selection and design | Biomarker | Limit of detection |
|---|---|---|---|
| Electrochemical impedance-based detector [59] | A 16-well container sensing electrodes and coated with 2.5 μg/mL of the RBD of SARS-CoV-2 spike protein | SARS-CoV-2 spike protein | It was not reported because of the hardware noise and variations in sample handling |
| Functionalized TiO2 Nanotube-Based Electrochemical Biosensor [60] | TiO2 nanotubes were synthesized by employing a low cost, simple, and one step electrochemical anodization of G1 grade titanium. Cobalt functionalization of the TiO2 nanotubes was also done by an incipient wetting method | SARS-CoV-2 spike protein | 0.7 nM |
| Antisense oligonucleotides directed electrochemical biosensor [61] | AuNPs-capped with specific antisense oligonucleotides (ssDNA) on a filter paper coated with graphene | SARS-CoV-2 viral RNA | 6.9 copies/μL |
| Graphene-based multiplexed telemedicine electrochemical biosensor [62] | Four graphene working electrodes, one graphene counter electrode, and one Ag/AgCl reference electrode patterned on a polyimide substrate | Nucleocapsid protein, IgM and IgG antibodies, and the inflammatory biomarker C-reactive protein (CRP) | - |
| Magnetic beads combined with carbon black-based screen-printed electrodes (a miniaturized electrochemical immunosensor) [63] | magnetic beads as support of immunological chain and secondary antibody with alkaline phosphatase as the immunological label; a three-electrode electrochemical cell including a graphite working electrode and counter electrode and a silver-based reference electrode | Spike (S) protein and nucleocapsid (N) protein | 19 ng/mL for spike protein and 8 ng/mL for nucleocapsid protein |
| Super sandwich-type electrochemical biosensor [64] | The CPs were immobilized on the surfaces of the Au@Fe3O4 nanoparticles to produce CP/Au@Fe3O4 nanocomposites. Then SCX8 was immobilized on the functionalized graphene (RGO), and Au@SCX8-TB-RGO-LP bioconjugate was produced. Finally, a CP-target-LP sandwich structure was fabricated. |
SARS-CoV-2 RNA (artificial and clinical samples) |
3 aM for artificial target and 200 copies/mL for clinical specimens |
| Novel printed circuit board-based electrochemical device [65] | FTO with gold nanoparticle (AuNPs). It was also immobilized by a nCovid-19 monoclonal antibody (nCovid-19Ab) | SARS-Cov-2 spike antigen | 10 fM |
| Field-effect transistor-based biosensor [30] | Graphene was coated on the SiO2/Si substrate. A coating of PMMA C4 was applied to the graphene layer. After transferring the PMMA/graphene layer to the SiO2/Si substrate, the PMMA layer was removed using acetone. A gold-chromium electrode layer was later fabricated on the etched graphene layer. | SARS-CoV-2 spike protein | 1 fg/mL (lower than the limit of detection of ELISA) |
| A field-effect transistor (Bio-FET)-based biosensor [66] | The graphene was soaked with a PBASE solution on the surface of the graphene FET biosensor. | SARS-CoV-2 spike protein | 1 fg/mL |
| Label-free graphene field-effect transistor (Gr-FET) biosensor [67] | The CVD method was employed to synthesize a single-crystal graphene layer on the single crystal copper. Functionalization of the graphene surface to bind the COVID-19 spike protein was carried out by immobilizing CSAb and ACE2 receptors on graphene's surface. During the incubation of the positively charged CSAB and negatively charged ACE2 in PBS buffer solution, negative and positive potentials were applied, respectively, to the graphene to enhance their immobilization on the surface of the graphene. | SARS-CoV-2 spike protein | 0.2 pM |
Other methods and materials
Some researchers have reported the advantages of using different nanoparticles for the detection of the SARS-CoV-2 virus. Aminul Islam et al. studied magnetic nanoparticle-based biosensors’ application to detect the SARS-CoV-2 virus rapidly [69]. There are different methods for the synthesis of magnetic nanoparticles including wet chemical [70], template-directed [71], microemulsion [72], thermal decomposition [73], solvothermal method [74], solid-state [75], deposition method [76], spray pyrolysis [77], and self-assembly [78]. However, it has been reported that the thermal decomposition and hydrothermal methods were the best ones for the synthesis of magnetic nanoparticles because of the drawbacks of the other methods, such as irregular shape and contamination of the nanoparticles during the synthesis process. In the thermal decomposition method, no stabilizer is required. The surface functionalization of the synthesized magnetic nanoparticles is required to increase their bio-detection and bio-sensing ability. Both organic and inorganic coatings can be used for this purpose. Viruses are attached to the magnetic nanoparticles and form supramolecular architectural design with unique building blocks. Therefore, the optical and magnetic properties of the magnetic nanoparticles are changed, and the viruses can be detected. Aminul Islam et al. reported that these magnetic nanoparticle-based biosensors could successfully detect the SARS-CoV-2 spike protein and +ssRNA. Superparamagnetic nanoparticles are one of the widely used magnetic nanoparticles for the detection of viruses. These nanoparticles are magnetized by applying the magnetic field and redisperse in the solution to remove the magnetic field. These magnetic nanosensors with superparamagnetic nanoparticles can detect complex targets. On the other hand, giant magneto resistive (GMR) sensors and magnetic nanoparticles can be employed for the rapid real-time detection of the SARS-CoV-2 virus. The spin collision between magnetic nanoparticles and non-magnetic biomolecules changes the electrical resistance—the alternation of magnetization with changing the electrical resistance results in detecting the viruses using giant magnetoresistance-based biosensors. Because SARS-CoV-2 spike protein and +ssRNA do not have ferromagnetism property, magnetic signals can be detected with a minimal amount of background noise [69].
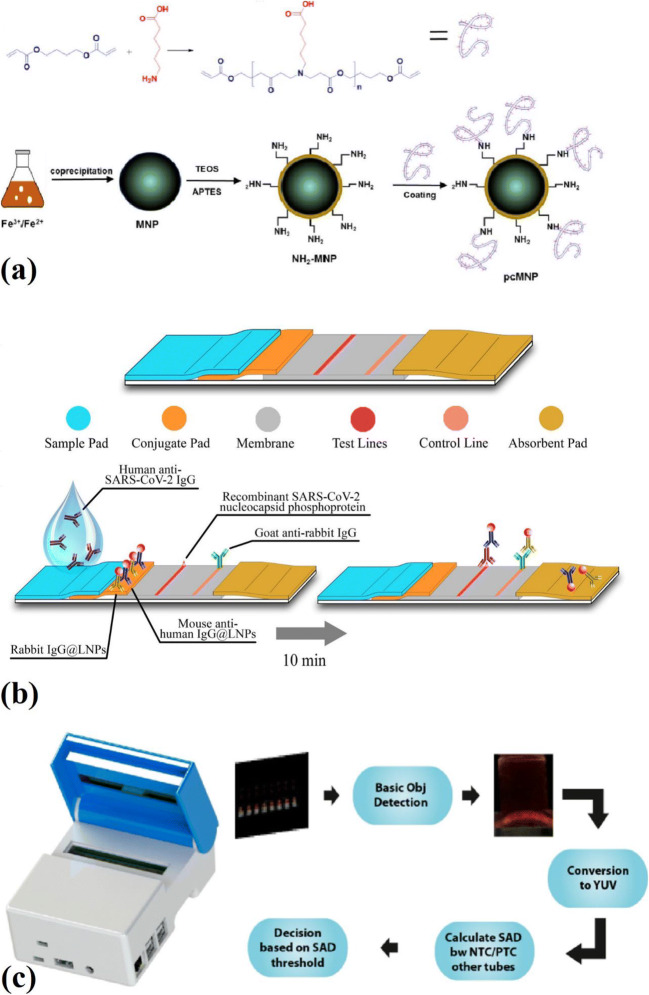
Zhao et al. also investigated a magnetic nanoparticles-based viral RNA extraction method to detect the SARS-CoV-2 virus (Fig. 4a). They synthesized poly (amino ester) with carboxyl groups (PC) coated magnetic nanoparticles (pcMNPs) for the detection of this virus. The magnetic nanoparticles were synthesized using Iron (III) chloride and a co-precipitation method. Then a silica layer was coated on the surface of the magnetic nanoparticles. These silica-coated magnetic nanoparticles were separated from the solution by using a magnet. Then silica-coated magnetic nanoparticles were dispersed into a mixture of 50 mL isopropanol and 0.2 mL of APTES to prepare amino-modified magnetic nanoparticles (NH2-MNPs). Finally, a solution containing poly (amino ester) with PC was used to coat amino-modified magnetic nanoparticles with PC. By employing this method, lysis and binding steps were combined into a single step, and these pc-magnetic nanoparticles were directly introduced into the RT-PCR. This method could purify viral RNA from different samples in only 20 min. Since PC magnetic nanoparticles have excellent viral RNA binding performance, a 10-copy sensitivity was achieved by employing this method. Moreover, the obtained pcMNPs-RNA complexes using this method were also compatible with other isothermal amplification methods, including recombinase polymerase amplification (RPA) and loop-mediated isothermal amplification (LAMP) [79]. One of the most important limitations of using metallic MNPs is their chemical instability and toxicity; therefore, they require an external coating, usually a silica coating, to overcome this limitation. However, this silica coating is also unstable under harsh conditions, where Si-O-Si bonds are readily hydrolyzed under basic conditions [80].
Fig. 4.
a Synthesis of PC polymer and the preparation of pcMNP for the detection of SARS-CoV-2 [80], b fabrication of the developed assay for LNPs-based lateral flow immunoassay [82], c AI-assisted algorithm and image processing for the detection of SARS-CoV-2 using AI-LAMP method [86]
Chen et al. reported a lateral flow immunoassay (LFIA) that employs lanthanide-doped polystyrene nanoparticles (LNPs), which were synthesized using the mini emulsion polymerization process to detect anti-SARS-CoV-2 IgG (Fig. 4b). Mouse anti-human IgG antibody (MHIgG) and rabbit IgG (RIgG) were used to functionalize LNPs. After diluting human serum samples with assay buffer, it was added to the sample well of an LFIA strip. The specific IgG of SARS-CoV-2 was captured by dispensing the virus’s nucleocapsid phosphoprotein onto a nitrocellulose membrane. This biosensor was also used for some clinical samples tested by RT-PCR before. Among 12 clinical samples that were suspicious for the presence of anti-SARS-CoV-2 IgG, one of them was determined to be SARS-CoV-2 IgG positive by using this biosensor [81].
G-quadruplex-based biosensors can also be used to detect the SARS-CoV-2 virus. G-quadruplex, guanine (G) rich sequence, can arrange four guanine bases in a square plane. Strands of DNA or RNA with G-rich sequence can make G-quadruplex. Based on the numbers of strands of DNA or RNA, there can be unimolecular, bimolecular, and tetramolecular G-quadruplexes. The sequence of G-quadruplex is usually G(≥2)NxG(≥2)NyG(≥2)NzG(≥2), where Nx, Ny, and Nz can be any combination of nucleotides. G-quadruplexes with three or more G-tetrads is usually more stable than the G-quadruplexes with two G-tetrads. Metal cations are used to neutralize the negative electrostatic potential caused by oxygen atoms in eight guanines. These metal cations, temperature, and pH affect the stability of the G-quadruplex. G-quadruplex can be used to fabricate biosensors because of its ability to inhibit transcription, DNA replication, and translation. Moreover, it can convert binding events into signals and carry nanoparticles, vital in fabricating biosensors to detect the SARS-CoV-2 virus. In G-quadruplex based biosensors, the G-quadruplex nucleic acid (DNA or RNA) is the recognition part that works with a converter to produce detectable signals. G-quadruplex can be used with electrochemical biosensors as a label-free probe molecule. There are both DNA and RNA-based G-quadruplex electrochemical biosensors. Moreover, some optical G-quadruplex biosensors such as fluorescent G-quadruplex biosensors and colorimetry G-quadruplex biosensors can also be used to detect viruses. The mixture of G-quadruplex and metal nanoparticles in colorimetry G-quadruplex biosensors can detect different targets based on the color changes. However, the G-quadruplex biosensors had some limitations, including preparing and screening suitable G-quadruplex molecules as the sensing probes, the biological complexity of the samples, and the need to improve the final sensitivity and the selectivity [82].
Some other biosensors and materials have been fabricated and used to detect SARS-CoV-2 in artificial and clinical samples. One of these methods is loop-mediated isothermal amplification (LAMP). Thi et al. reported a colorimetric reverse transcription LAMP (RT-LAMP) assay to detect SARS-CoV-2 RNA in clinical samples. It was reported that the RT-LAMP is a more straightforward method than RT-qPCR to detect viruses. In this study, a two-color RT-LAMP assay protocol was utilized to detect the SARS-CoV-2 N gene in clinical samples. The color of the phenol-red dye was changed within 30 min of the reaction in the presence of positive clinical samples with a cycle threshold (CT) less than 30. Because after 35 min of the reaction, some negative samples also changed the color of the phenol-red dye. The value of the difference in absorbance (∆OD) was used at 30 min to distinguish the positive samples from the negative samples. . In this way, the RT-LAMP method’s sensitivity for detecting SARS-CoV-2 RNA in clinical samples with CT < 30 was confirmed [83]. Zhang et al. also investigated a colorimetric LAMP biosensor. This biosensor was used to detect SARS-CoV-2 RNA from purified RNA or cell lysis. Moreover, some RNA samples purified, using RNA clean up columns, from respiratory swabs of some positive COVID-19 patients were used to verify the test results. Five LAMP primers sets were designed to detect the SARS-CoV-2 RNA (ORF1a gene and Gene N), and they all showed very close detection sensitivity (around 4.8 copies/μL). It was reported that the results obtained from colorimetric tests were in total agreement with real-time PCR method results [84]. Rohaim et al. fabricated an artificial intelligence-assisted LAMP (AI-LAMP) biosensor to detect the SARS-CoV-2 RNA-dependent RNA polymerase gene (Fig. 4c). They made a novel hand-held smart diagnostic device to eliminate any subjectivity related to the operator interpretation of results. The AI pipelines were used to observe and detect color changes through the image processing to decrease the assay run time of the colorimetric LAMP detection. The detection of SARS-CoV-2 using a LAMP assay-based biosensor (based on color changes) took around 30 min. However, by employing AI, color changes detection, and as a result, total test time was reduced to less than 30 min. This RT-LAMP assay-based biosensor detected as low as 100 copies of SARS-CoV-2 RNA [85]. Despite all the features mentioned, LAMP biosensors have some drawbacks, such as their low versatility and the possibility of primer-primer interactions. Moreover, the presence of some constituents within the samples can inhibit the detection process [86]. Some other methods and materials used for the detection of the SARS-CoV-2 virus are reported in Table 3. Table 4 also compares the advantages and disadvantages of different biosensors used to detect the SARS-CoV-2 virus.
Table 3.
Other methods and materials used for the detection of SARS-CoV-2
| Biosensing technique | Material selection and design | Biomarker | Limit of detection |
|---|---|---|---|
| Poly (amino ester) with carboxyl groups (PC)-coated magnetic nanoparticles (pcMNPs) – based viral RNA extraction [79] | The magnetic nanoparticles were synthesized using Iron (III) chloride and a co-precipitation method. Then a silica layer was coated on the surface of the magnetic nanoparticles. Then silica-coated magnetic nanoparticles were dispersed into a mixture of 50 mL isopropanol and 0.2 mL of APTES to prepare NH2-MNPs. Finally, a solution containing poly (amino ester) with PC was used to coat amino-modified magnetic nanoparticles with PC | SARS-CoV-2 RNA (N gene) | A 10-copy sensitivity |
| Lanthanide-doped nanoparticles-based lateral flow immunoassay [81] | LNPs were synthesized using the mini emulsion polymerization process to detect anti-SARS-CoV-2 IgG. Mouse anti-human IgG antibody (MHIgG) and rabbit IgG (RIgG) were used to functionalize LNPs. By dispensing a recombinant nucleocapsid phosphoprotein of SARS-CoV-2 onto a nitrocellulose membrane, the specific IgG of SARS-CoV-2 was captured | Anti-SARV-CoV-2 IgG | - |
| Reverse transcription loop-mediated isothermal amplification (RT-LAMP) [83] | A two-color RT-LAMP assay protocol was utilized to detect the SARS-CoV-2 N gene in clinical samples. The color of the phenol-red dye was changed within 30 min of the reaction in the presence of positive clinical samples with a CT of less than 30 | SARS-CoV-2 RNA (N gene) | Cycle threshold (CT) less than 30 |
| Colorimetric Loop-Mediated Isothermal Amplification (LAMP) biosensor [84] | Five LAMP primers sets were designed to detect the SARS-CoV-2 RNA (ORF1a gene and Gene N). DNAs containing these regions were synthesized as gBlocks | SARS-CoV-2 RNA (ORF1a gene and Gene N) | 4.8 copies/μL |
| Artificial intelligence-assisted loop-mediated isothermal amplification (AI-LAMP)-based biosensor [85] | A novel hand-held smart diagnostic device was designed to eliminate any subjectivity related to the operator interpretation of results. To decrease the assay run time of the colorimetric LAMP detection, AI pipelines were used to observe and detect color changes through AI image processing | SARS-CoV-2 RNA | 100 copies of SARS-CoV-2 RNA |
Table 4.
Advantages and disadvantages of different biosensors used for the detection of SARS-CoV-2 virus
| Biosensing method | Advantages | Disadvantages |
|---|---|---|
| Plasmonic |
- High sensitivity - High selectivity - Quick response time - Can be performed in an automated fashion - Label-free detection [56] |
- Specialized instrumentation - Challenging portability - Challenging point of care applications |
| Electrochemical |
- Low detection limits - A wide linear response range - Good stability - Good reproducibility [87] |
- Sensitive to sample matrix effects - Not as sensitive as the RT-PCR test - Lower shelf life compared to RT-PCR [68] |
| LAMP |
- Low-cost equipment - No need for thermal alternations - Can be reported with naked eyes [86] |
- Low versatility - Possibility of primer-primer interactions - Some constituents within the samples can inhibit the detection process [86] |
| G-quadruplex |
- High affinity - High stability - Easy regeneration [82] |
- The preparation and screening of suitable G-quadruplex molecules as probes - The biological complexity of samples - The need for further improvement of sensitivity and selectivity [82] |
Conclusion and future trends
Due to the pandemic of COVID-19, researchers have made efforts to develop efficient and accurate methods to make the early diagnosis possible. In this review, different methods of detecting the SARS-CoV-2, including plasmonic biosensors, electrochemical biosensors, and some other methods and materials that can replace the RT-PCR method due to high stress on it and its limitations, were discussed. For this purpose, the design and capabilities of these biosensors and methods were reviewed. These biosensors have several advantages, including high detection capability, stability, simple design, reliability, and affordability. Moreover, they can detect different biomarkers and indicators. Employing nanomaterials such as nanotubes, gold nanoparticles, lanthanide-doped polystyrene nanoparticles, and graphene nanoparticles significantly improve biosensors’ performance for detecting the SARS-CoV-2 virus, as they have shown their potential for detecting other viruses. However, some issues need further research and attention. First, most of these methods and materials have been investigated at a lab-scale, while using these methods in real situations may not be as accurate as in laboratory conditions. Moreover, none of these biosensors has been commercialized for the detection of the SARS-CoV-2 virus yet. Therefore, the commercialization of various efficient biosensors should be accellerated. Apart from the presented techniques and biosensors, novel methods such as AI-based technologies, wearable biosensors for continuous monitoring of the public, and single-use disposable sensors for individual testing need to be investigated for the mass-screening of SARS-CoV-2.
Declarations
Conflict of interest
The authors declare that they have no conflict of interest.
References
- 1.Chan JFW, Yuan S, Kok KH, To KKW, Chu H, Yang J, Xing F, Liu J, Yip CCY, Poon RWS, Tsoi HW, Lo SKF, Chan KH, Poon VKM, Chan WM, Ip JD, Cai JP, Cheng VCC, Chen H, Hui CKM, Yuen KY. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet. 2020;395:514–523. doi: 10.1016/S0140-6736(20)30154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen L, Liu W, Zhang Q, Xu K, Ye G, Wu W, Sun Z, Liu F, Wu K, Zhong B, Mei Y, Zhang W, Chen Y, Li Y, Shi M, Lan K, Liu Y. RNA based mNGS approach identifies a novel human coronavirus from two individual pneumonia cases in 2019 Wuhan outbreak. Emerg. Microbes. Infect. 2020;9:313–319. doi: 10.1080/22221751.2020.1725399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gorbalenya A, Baker S, Baric R, de Groot R, Drosten C, Gulyaeva A, Haagmans B, Lauber C, Leontovich A, Neuman B, Penzar D, Perlman S, Poon L, Samborskiy D, Sidorov I, Sola I, Ziebuhr J. The species Severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020;5:536–544. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lu R, Zhao X, Li J, Niu P, Yang B, Wu H, Wang W, Song H, Huang B, Zhu N, Bi Y, Ma X, Zhan F, Wang L, Hu T, Zhou H, Hu Z, Zhou W, Zhao L, Chen J, Meng Y, Wang J, Lin Y, Yuan J, Xie Z, Ma J, Liu WJ, Wang D, Xu W, Holmes EC, Gao GF, Wu G, Chen W, Shi W, Tan W. Lancet. 2020;395:565. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nicola M, Alsafi Z, Sohrabi C, Kerwan A, Al-Jabir A, Iosifidis C, Agha M, Agha R. The socio-economic implications of the coronavirus pandemic (COVID-19): A review. Int. J. Surg. 2020;78:185–193. doi: 10.1016/j.ijsu.2020.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tumban E. Lead SARS-CoV-2 Candidate Vaccines: Expectations from Phase III Trials and Recommendations Post-Vaccine Approval. Viruses. 2021;13(1):54. doi: 10.3390/v13010054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bai HX, Hsieh B, Xiong Z, Halsey K, Choi W, My T, Tran L, Pan I, Wang DC, Mei J, Jiang XL, Zeng QH, Egglin TK, Hu PF, Agarwal S, Xie F, Li S, Healey T, Atalay MK, Liao WH. Performance of Radiologists in Differentiating COVID-19 from Non-COVID-19 Viral Pneumonia at Chest CT. Radiology. 2020;1:1–E54. doi: 10.1148/radiol.2020200823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bernheim A, Mei X, Huang M, Yang Y, Fayad ZA, Zhang N, Diao K, Lin B, Zhu X, Li K, Li S, Shan H, Jacobi A, Chung M. J. Coll. Physicians Surg. Pak. 2020;295:685. doi: 10.1148/radiol.2020200463. [DOI] [Google Scholar]
- 9.Pan F, Ye T, Sun P, Gui S, Liang B, Li L, Zheng D, Wang J, Hesketh RL, Yang L, Zheng C. Time Course of Lung Changes at Chest CT during Recovery from Coronavirus Disease 2019 (COVID-19) Radiology. 2020;295:715–721. doi: 10.1148/radiol.2020200370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li Y, Xia L. Am. J. Roentgenol. 2020;214:1280. doi: 10.2214/AJR.20.22954. [DOI] [PubMed] [Google Scholar]
- 11.Chu DKW, Pan Y, Cheng SMS, Hui KPY, Krishnan P, Liu Y, Ng DYM, Wan CKC, Yang P, Wang Q, Peiris M, Poon LLM. Molecular Diagnosis of a Novel Coronavirus (2019-nCoV) Causing an Outbreak of Pneumonia. Clin. Chem. 2020;555:549–555. doi: 10.1093/clinchem/hvaa029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Corman VM, Landt O, Kaiser M, Molenkamp R, Meijer A, Chu DK, Bleicker T, Brünink S, Schneider J, Luisa Schmidt M, Mulders DGJC, Haagmans BL, van der Veer B, van den Brink S, Wijsman L, Goderski G, Romette JL, Ellis J, Zambon M, Peiris M, Goossens H, Reusken C, Koopmans MP, Drosten C. Euro Surveill. 2020;25:1. doi: 10.2807/2F1560-7917.ES.2020.25.3.2000045. [DOI] [Google Scholar]
- 13.Loeffelholz MJ, Tang YW. Laboratory diagnosis of emerging human coronavirus infections – the state of the art. Emerg. Microbes. Infect. 2020;9:747–756. doi: 10.1080/22221751.2020.1745095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lan L, Xu D, Ye G, Xia C, Wang S, Li Y, Xu H. JAMA - J. Am. Med. Assoc. 2020;323:1502–1503. doi: 10.1001/jama.2020.2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yüce M, Filiztekin E, Özkaya KG. COVID-19 diagnosis —A review of current methods. Biosens. Bioelectron. 2021;172:112752. doi: 10.1016/j.bios.2020.112752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.An J, Liao X, Xiao T, Qian S, Yuan J, Ye H, Qi F, Shen C, Wang L, Liu Y, Cheng X, Li N, Cai Q, Wang F, Chen J, Li G, Cai Q, Liu Y, Wang Y, Zhang F, Fu Y, He Q, Tan X, Liu L, Zhang Z. Clinical characteristics of recovered COVID-19 patients with re-detectable positive RNA test. Ann. Transl. Med. 2020;8:1084. doi: 10.1101/2020.03.26.20044222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Soler M, Huertas CS, Lechuga LM. Label-free plasmonic biosensors for point-of-care diagnostics: a review. Expert. Rev. Mol. Diagn. 2019;19:71–81. doi: 10.1080/14737159.2019.1554435. [DOI] [PubMed] [Google Scholar]
- 18.Masson JF. Surface Plasmon Resonance Clinical Biosensors for Medical Diagnostics. ACS Sens. 2017;2:16–30. doi: 10.1021/acssensors.6b00763. [DOI] [PubMed] [Google Scholar]
- 19.Bahl S, Javaid M, Bagha A, Singh R, Haleem A, Vaishya R, Suman R. Biosensors applications in fighting COVID-19 pandemic. Apollo Med. 2020;17(3):221. doi: 10.4103/am.am_56_20. [DOI] [Google Scholar]
- 20.Eissa S, Siaj M, Zourob M. Aptamer-based competitive electrochemical biosensor for brevetoxin-2. Biosens. 2015;69:148–154. doi: 10.1016/j.bios.2015.01.055. [DOI] [PubMed] [Google Scholar]
- 21.Hu T, Zhang L, Wen W, Zhang X, Wang S. Enzyme catalytic amplification of miRNA-155 detection with graphene quantum dot-based electrochemical biosensor. Biosens. Bioelectron. 2016;77:451–456. doi: 10.1016/j.bios.2015.09.068. [DOI] [PubMed] [Google Scholar]
- 22.Shin SR, Zhang YS, Kim DJ, Manbohi A, Avci H, Silvestri A, Aleman J, Hu N, Kilic T, Keung W, Righi M, Assawes P, Alhadrami HA, Li RA, Dokmeci MR, Khademhosseini A. Aptamer-Based Microfluidic Electrochemical Biosensor for Monitoring Cell-Secreted Trace Cardiac Biomarkers. Anal. Chem. 2016;88:10019–10027. doi: 10.1021/acs.analchem.6b02028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang X, Wolfbeis OS. Fiber-Optic Chemical Sensors and Biosensors (2013–2015) Anal. Chem. 2015;88:203–227. doi: 10.1021/acs.analchem.5b04298. [DOI] [PubMed] [Google Scholar]
- 24.Bharadwaj R, Mukherji S, Mukherji S. Probing the Localized Surface Plasmon Field of a Gold Nanoparticle-Based Fibre Optic Biosensor. Plasmonics. 2016;11:753–761. doi: 10.1007/s11468-015-0106-0. [DOI] [Google Scholar]
- 25.Saylan Y, Yılmaz F, Derazshamshir A, Yılmaz E, Denizli A. J. Mol. Recognit. 2017;30(9):1. doi: 10.1002/jmr.2631. [DOI] [PubMed] [Google Scholar]
- 26.do Nascimento NM, Juste-Dolz A, Grau-García E, Román-Ivorra JA, Puchades R, Maquieira A, Morais S, Gimenez-Romero D. Biosens. Bioelectron. 2017;90:166. doi: 10.1016/j.bios.2016.11.004. [DOI] [PubMed] [Google Scholar]
- 27.Pohanka M. Piezoelectric biosensor for the determination of Tumor Necrosis Factor Alpha. Talanta. 2018;178:970–973. doi: 10.1016/j.talanta.2017.10.031. [DOI] [PubMed] [Google Scholar]
- 28.Saylan Y, Erdem Ö, Ünal S, Denizli A. An Alternative Medical Diagnosis Method: Biosensors for Virus Detection. Biosensors. 2019;9(2):65. doi: 10.3390/bios9020065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qiu G, Gai Z, Tao Y, Schmitt J, Kullak-Ublick GA, Wang J. Dual-Functional Plasmonic Photothermal Biosensors for Highly Accurate Severe Acute Respiratory Syndrome Coronavirus 2 Detection. ACS Nano. 2020;14:5268–5277. doi: 10.1021/acsnano.0c02439. [DOI] [PubMed] [Google Scholar]
- 30.Seo G, Lee G, Kim MJ, Baek SH, Choi M, Ku KB, Lee CS, Jun S, Park D, Kim HG, Kim SJ, Lee JO, Kim BT, Park EC, Kim SI. Rapid Detection of COVID-19 Causative Virus (SARS-CoV-2) in Human Nasopharyngeal Swab Specimens Using Field-Effect Transistor-Based Biosensor. ACS Nano. 2020;14:5135–5142. doi: 10.1021/acsnano.0c02823. [DOI] [PubMed] [Google Scholar]
- 31.Srivastava M, Srivastava N, Mishra PK, Malhotra BD. Prospects of nanomaterials-enabled biosensors for COVID-19 detection. Sci. Total Environ. 2021;754:142363. doi: 10.1016/j.scitotenv.2020.142363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.M. Häggström, Symptoms of coronavirus disease 2019. (Reusing image, 2020), https://en.wikipedia.org/wiki/Symptoms_of_COVID19#/media/File:Symptoms_of_coronavirus_disease_2019_4.0.svg
- 33.Farzin L, Shamsipur M, Samandari L, Sheibani S. HIV biosensors for early diagnosis of infection: The intertwine of nanotechnology with sensing strategies. Talanta. 2020;206:120201. doi: 10.1016/j.talanta.2019.120201. [DOI] [PubMed] [Google Scholar]
- 34.Negahdari B, Darvishi M, Saeedi AA. Artif. Cells, Nanomed. Biotechnol. 2019;47:469. doi: 10.1080/21691401.2018.1546185. [DOI] [PubMed] [Google Scholar]
- 35.Siuzdak K, Niedziałkowski P, Sobaszek M, Łęga T, Sawczak M, Czaczyk E, Dziąbowska K, Ossowski T, Nidzworski D, Bogdanowicz R. Sensors Actuators. B Chem. 2019;280:263–271. doi: 10.1016/j.snb.2018.10.005. [DOI] [Google Scholar]
- 36.Lee T, Park SY, Jang H, Kim GH, Lee Y, Park C, Mohammadniaei M, Lee MH, Min J. Fabrication of electrochemical biosensor consisted of multi-functional DNA structure/porous au nanoparticle for avian influenza virus (H5N1) in chicken serum. Mater. Sci. Eng. C. 2019;99:511–519. doi: 10.1016/j.msec.2019.02.001. [DOI] [PubMed] [Google Scholar]
- 37.Narang J, Singhal C, Mathur A, Sharma S, Singla V, Pundir CS. Portable bioactive paper based genosensor incorporated with Zn-Ag nanoblooms for herpes detection at the point-of-care. Int. J. Biol. Macromol. 2018;107:2559–2565. doi: 10.1016/j.ijbiomac.2017.10.146. [DOI] [PubMed] [Google Scholar]
- 38.Dinesh DC, Chalupska D, Silhan J, Koutna E, Nencka R, Veverka V, Boura E. Structural basis of RNA recognition by the SARS-CoV-2 nucleocapsid phosphoprotein. PLoS Pathog. 2020;16(12):e1009100. doi: 10.1371/journal.ppat.1009100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nieto-Torres JL, DeDiego ML, Verdiá-Báguena C, Jimenez-Guardeño JM, Regla-Nava JA, Fernandez-Delgado R, Castaño-Rodriguez C, Alcaraz A, Torres J, Aguilella VM, Enjuanes L. Severe Acute Respiratory Syndrome Coronavirus Envelope Protein Ion Channel Activity Promotes Virus Fitness and Pathogenesis. PLoS Pathog. 2014;10(5):e1004077. doi: 10.1371/journal.ppat.1004077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou P, Lou Yang X, Wang XG, Hu B, Zhang L, Zhang W, Si HR, Zhu Y, Li B, Huang CL, Chen HD, Chen J, Luo Y, Guo H, Di Jiang R, Liu MQ, Chen Y, Shen XR, Wang X, Zheng XS, Zhao K, Chen QJ, Deng F, Liu LL, Yan B, Zhan FX, Wang YY, Xiao GF, Shi ZL. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Woo PCY, Lau SKP, Wong BHL, Tsoi HW, Fung AMY, Kao RYT, Chan KH, Peiris JSM, Yuen KY. Differential Sensitivities of Severe Acute Respiratory Syndrome (SARS) Coronavirus Spike Polypeptide Enzyme-Linked Immunosorbent Assay (ELISA) and SARS Coronavirus Nucleocapsid Protein ELISA for Serodiagnosis of SARS Coronavirus Pneumonia. J. Clin. Microbiol. 2005;43:3054–3058. doi: 10.1128/jcm.43.7.3054-3058.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang W, Du RH, Li B, Zheng XS, Lou Yang X, Hu B, Wang YY, Xiao GF, Yan B, Shi ZL, Zhou P. Molecular and serological investigation of 2019-nCoV infected patients: implication of multiple shedding routes. Emerg. Microbes. Infect. 2020;9:386–389. doi: 10.1080/22221751.2020.1729071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Asif M, Ajmal M, Ashraf G, Muhammad N, Aziz A, Iftikhar T, Wang J, Liu H. The role of biosensors in coronavirus disease-2019 outbreak. Curr. Opin. Electrochem. 2020;23:174–184. doi: 10.1016/j.coelec.2020.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee T, Ahn J, Park SY, Kim G, Kim J, Kim T, Nam I, Park C, Lee M. Recent Advances in AIV Biosensors Composed of Nanobio Hybrid Material. Micromachines. 2018;9(12):651. doi: 10.3390/mi9120651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Anker JN, Hall WP, Lyandres O, Shah NC, Zhao J, Van Duyne RP. Biosensing with plasmonic nanosensors. Nat. Mater. 2008;7:442–453. doi: 10.1038/nmat2162. [DOI] [PubMed] [Google Scholar]
- 46.Hall WP, Ngatia SN, Van Duyne RP. LSPR Biosensor Signal Enhancement Using Nanoparticle−Antibody Conjugates. J. Phys. Chem. 2011;115(5):1410–1414. doi: 10.1021/jp106912p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huang L, Ding L, Zhou J, Chen S, Chen F, Zhao C, Zhang Y, Xu J, Hu W, Ji J, Xu H, Liu GL. Biosens. 2021;171:112685. doi: 10.1016/j.bios.2020.112685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Peng X, Zhou Y, Nie K, Zhou F, Yuan Y, Song J, Qu J. New J. Phys. 2020;22:103046. doi: 10.1088/1367-2630/abbe53. [DOI] [Google Scholar]
- 49.Moitra P, Alafeef M, Dighe K, Frieman MB, Pan D. Selective Naked-Eye Detection of SARS-CoV-2 Mediated by N Gene Targeted Antisense Oligonucleotide Capped Plasmonic Nanoparticles. ACS Nano. 2020;14:7617–7627. doi: 10.1021/acsnano.0c03822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.A. Ahmadivand, B. Gerislioglu, Z. Ramezani, A. Kaushik, P. Manickam, S.A. Ghoreishi, ArXiv 1 (2020) arXiv:2006.08536 [DOI] [PMC free article] [PubMed]
- 51.Ventura BD, Cennamo M, Minopoli A, Campanile R, Censi SB, Terracciano D, Portella G, Velotta R. Colorimetric Test for Fast Detection of SARS-CoV-2 in Nasal and Throat Swabs. ACS Sens. 2020;5:3043–3048. doi: 10.1021/acssensors.0c01742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li M, Cushing SK, Wu N. Plasmon-enhanced optical sensors: a review. Analyst. 2015;140:386–406. doi: 10.1039/C4AN01079E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Funari R, Chu K, Shen AQ. Detection of antibodies against SARS-CoV-2 spike protein by gold nanospikes in an opto-microfluidic chip. Biosens. Bioelectron. 2020;169:112578. doi: 10.1016/j.bios.2020.112578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhao B, Che C, Wang W, Li N, Cunningham BT. Single-step, wash-free digital immunoassay for rapid quantitative analysis of serological antibody against SARS-CoV-2 by photonic resonator absorption microscopy. Talanta. 2021;225:122004. doi: 10.1016/j.talanta.2020.122004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tymm C, Zhou J, Tadimety A, Burklund A, Zhang JXJ. Scalable COVID-19 Detection Enabled by Lab-on-Chip Biosensors. Cell. Mol. Bioeng. 2020;13:313–329. doi: 10.1007/s12195-020-00642-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bhalla N, Pan Y, Yang Z, Payam AF. Opportunities and Challenges for Biosensors and Nanoscale Analytical Tools for Pandemics: COVID-19. ACS Nano. 2020;14:7783–7807. doi: 10.1021/acsnano.0c04421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Faridbod F, Gupta VK, Zamani HA. Electrochemical Sensors and Biosensors. Int. J. Electrochem. 2011;2011:352546–352542. doi: 10.4061/2011/352546. [DOI] [Google Scholar]
- 58.Khan MZH, Hasan MR, Hossain SI, Ahommed MS, Daizy M. Ultrasensitive detection of pathogenic viruses with electrochemical biosensor: State of the art. Biosens. Bioelectron. 2020;166:112431. doi: 10.1016/j.bios.2020.112431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rashed MZ, Kopechek JA, Priddy MC, Hamorsky KT, Palmer KE, Mittal N, Valdez J, Flynn J, Williams SJ. Rapid detection of SARS-CoV-2 antibodies using electrochemical impedance-based detector. Biosens. Bioelectron. 2021;171:112709. doi: 10.1016/j.bios.2020.112709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vadlamani BS, Uppal T, Verma SC, Misra M. Functionalized TiO2 Nanotube-Based Electrochemical Biosensor for Rapid Detection of SARS-CoV-2. Sensors. 2020;20(20):5871. doi: 10.3390/s20205871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Alafeef M, Dighe K, Moitra P, Pan D. Rapid, Ultrasensitive, and Quantitative Detection of SARS-CoV-2 Using Antisense Oligonucleotides Directed Electrochemical Biosensor Chip. ACS Nano. 2020;14(12):17028–17045. doi: 10.1021/acsnano.0c06392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Torrente-Rodríguez RM, Lukas H, Tu J, Min J, Yang Y, Xu C, Rossiter HB, Gao W. SARS-CoV-2 RapidPlex: A Graphene-Based Multiplexed Telemedicine Platform for Rapid and Low-Cost COVID-19 Diagnosis and Monitoring. Matter. 2020;3:1981–1998. doi: 10.1016/j.matt.2020.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fabiani L, Saroglia M, Galatà G, De Santis R, Fillo S, Luca V, Faggioni G, D’Amore N, Regalbuto E, Salvatori P, Terova G, Moscone D, Lista F, Arduini F. Magnetic beads combined with carbon black-based screen-printed electrodes for COVID-19: A reliable and miniaturized electrochemical immunosensor for SARS-CoV-2 detection in saliva. Biosens. Bioelectron. 2021;171:112686. doi: 10.1016/j.bios.2020.112686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhao H, Liu F, Xie W, Zhou TC, OuYang J, Jin L, Li H, Zhao CY, Zhang L, Wei J, Zhang YP, Li CP. Sensors Actuators. B Chem. 2021;327:128899. doi: 10.1016/j.snb.2020.128899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.S. Mahari, A. Roberts, D. Shahdeo, S. Gandhi, ArXiv (2020). 10.1101/2020.04.24.059204
- 66.Kaushik AK, Dhau JS, Gohel H, Mishra YK, Kateb B, Kim N, Goswami DY. Electrochemical SARS-CoV-2 Sensing at Point-of-Care and Artificial Intelligence for Intelligent COVID-19 Management. ACS Appl. Bio. Mater. 2020;3(11):7306–7325. doi: 10.1021/acsabm.0c01004. [DOI] [PubMed] [Google Scholar]
- 67.Gaurav A, Shukla P. Int. Res. J. Mod. Eng. Technol. Sci. 2020;2(5):1207. [Google Scholar]
- 68.Zhang X, Qi Q, Jing Q, Ao S, Zhang Z, Ding M, Wu M, Liu K, Wang W, Ling Y, Zhang Z, Fu W. ArXiv 1. 2020. [Google Scholar]
- 69.Menon S, Mathew MR, Sam S, Keerthi K, Kumar KG. Recent advances and challenges in electrochemical biosensors for emerging and re-emerging infectious diseases. J. Electroanal. Chem. 2020;878:114596. doi: 10.1016/j.jelechem.2020.114596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Aminul Islam M, Ziaul Ahsan M. Am. J. Nanosci. 2020;6(2):6. doi: 10.11648/j.ajn.20200602.11. [DOI] [Google Scholar]
- 71.Sun J, Zhou S, Hou P, Yang Y, Weng J, Li X, Li M. Synthesis and characterization of biocompatible Fe3O4 nanoparticles. J. Biomed. Mater. Res. A. 2007;80(2):333–341. doi: 10.1002/jbm.a.30909. [DOI] [PubMed] [Google Scholar]
- 72.Jiao JC, Jumas M, Womes AV, Chadwick A, Harrison PG, Bruce J. Synthesis of Ordered Mesoporous Fe3O4and γ-Fe2O3with Crystalline Walls Using Post-Template Reduction/Oxidation. Am. Chem. Soc. 2006;128:12905–12909. doi: 10.1021/ja063662i. [DOI] [PubMed] [Google Scholar]
- 73.Wang G, Wang C, Dou W, Ma Q, Yuan P, Su X. The Synthesis of Magnetic and Fluorescent Bi-functional Silica Composite Nanoparticles via Reverse Microemulsion Method. J. Fluoresc. 2009;19:939–946. doi: 10.1007/s10895-009-0493-8. [DOI] [PubMed] [Google Scholar]
- 74.Han YC, Cha HG, Kim CW, Kim YH, Kang YS. Synthesis of Highly Magnetized Iron Nanoparticles by a Solventless Thermal Decomposition Method. J. Phys. Chem. C. 2007;111:6275–6280. doi: 10.1021/jp0686285. [DOI] [Google Scholar]
- 75.Kang M. Synthesis of Fe/TiO2 photocatalyst with nanometer size by solvothermal method and the effect of H2O addition on structural stability and photodecomposition of methanol. J. Mol. Catal. A Chem. 2003;197:173–183. doi: 10.1016/S1381-1169(02)00586-1. [DOI] [Google Scholar]
- 76.Ai L, Zhang C, Chen Z. Removal of methylene blue from aqueous solution by a solvothermal-synthesized graphene/magnetite composite. J. Hazard. Mater. 2011;192:1515–1524. doi: 10.1016/j.jhazmat.2011.06.068. [DOI] [PubMed] [Google Scholar]
- 77.Teja AS, Koh PY. Synthesis, properties, and applications of magnetic iron oxide nanoparticles. Prog. Cryst. Growth Charact. Mater. 2009;55:22–45. doi: 10.1016/j.pcrysgrow.2008.08.003. [DOI] [Google Scholar]
- 78.Taniguchi I. Powder properties of partially substituted LiMxMn2−xO4 (M=Al, Cr, Fe and Co) synthesized by ultrasonic spray pyrolysis. Mater. Chem. Phys. 2005;92:172–179. doi: 10.1016/j.matchemphys.2005.01.020. [DOI] [Google Scholar]
- 79.Polshettiwar V, Baruwati B, Varma RS. Self-Assembly of Metal Oxides into Three-Dimensional Nanostructures: Synthesis and Application in Catalysis. ACS Nano. 2009;3:728–736. doi: 10.1021/nn800903p. [DOI] [PubMed] [Google Scholar]
- 80.Zhao Z, Cui H, Song W, Ru X, Zhou W, Yu X. ArXiv. 2020;1:518055. doi: 10.1101/2020.02.22.961268. [DOI] [Google Scholar]
- 81.Chen Y, Kolhatkar AG, Zenasni O, Xu S, Lee TR. Sensors. 2017;17:2300. doi: 10.3390/s17102300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chen Z, Zhang Z, Zhai X, Li Y, Lin L, Zhao H, Bian L, Li P, Yu L, Wu Y, Lin G. Rapid and Sensitive Detection of anti-SARS-CoV-2 IgG, Using Lanthanide-Doped Nanoparticles-Based Lateral Flow Immunoassay. Anal. Chem. 2020;92:7226–7231. doi: 10.1021/acs.analchem.0c00784. [DOI] [PubMed] [Google Scholar]
- 83.Xi H, Juhas M, Zhang Y. G-quadruplex based biosensor: A potential tool for SARS-CoV-2 detection. Biosens. Bioelectron. 2020;167:112494. doi: 10.1016/j.bios.2020.112494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Dao Thi VL, Herbst K, Boerner K, Meurer M, Kremer LPM, Kirrmaier D, Freistaedter A, Papagiannidis D, Galmozzi C, Stanifer ML, Boulant S, Klein S, Chlanda P, Khalid D, Miranda IB, Schnitzler P, Kräusslich HG, Knop M, Anders S. A colorimetric RT-LAMP assay and LAMP-sequencing for detecting SARS-CoV-2 RNA in clinical samples. Sci. Transl. Med. 2020;12:7075. doi: 10.1126/scitranslmed.abc7075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhang Y, Odiwuor N, Xiong J, Sun L, Nyaruaba RO, Wei H, Tanner N. MedrXiv 2. 2020. [Google Scholar]
- 86.Rohaim MA, Clayton E, Sahin I, Vilela J, Khalifa ME, Al-natour MQ, Bayoumi M, Poirier AC, Branavan M, Tharmakulasingam M, Chaudhry NS, Sodi R, Brown A, Burkhart P, Hacking W, Botham J, Boyce J, Wilkinson H, Williams C, Whittingham-dowd J, Shaw E, Hodges M, Butler L, Bates MD, La Ragione R, Balachandran W, Fernando A, Munir M. Artificial Intelligence-Assisted Loop Mediated Isothermal Amplification (AI-LAMP) for Rapid Detection of SARS-CoV-2. Viruses. 2020;12:972. doi: 10.3390/v12090972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Orooji Y, Sohrabi H, Hemmat N, Oroojalian F, Baradaran B, Mokhtarzadeh A, Mohaghegh M, Karimi-Maleh H. An Overview on SARS-CoV-2 (COVID-19) and Other Human Coronaviruses and Their Detection Capability via Amplification Assay, Chemical Sensing, Biosensing, Immunosensing, and Clinical Assays. Nano-Micro. Lett. 2021;13:18. doi: 10.1007/s40820-020-00533-y. [DOI] [PMC free article] [PubMed] [Google Scholar]