Abstract
The emergence of next-generation genomic sequencing (NGS) tests for use in clinical care has generated widespread interest around the globe, but little is known about the availability and funding of these tests worldwide. We examined NGS availability across world regions and countries, with a particular focus on availability of three key NGS tests—Whole-Exome Sequencing or Whole-Genome Sequencing for diagnosis of suspected genetic diseases such as intellectual disability disorders or rare diseases, non-invasive prenatal testing for common genetic abnormalities in fetuses and tumor sequencing for therapy selection and monitoring of cancer treatment. We found that these NGS tests are available or becoming available in every major region of the world. This includes both high-income countries with robust genomic programmes such as the USA and the UK, and growing availability in countries with upper-middle-income economies. We used exploratory case studies across three diverse health care systems (publicly funded/national (UK), publicly funded/provincial (Canada) and mixed private/public system (USA)) to illustrate the funding challenges and approaches used to address those challenges that might be adopted by other countries. We conclude by assessing what type of data and initiatives will be needed to better track and understand the use of NGS around the world as such testing continues to expand.
Keywords: health services research, health policy
Summary box.
The emergence of next-generation genomic sequencing (NGS) tests for use in clinical care has generated widespread interest around the globe, but little is known about the availability and funding of these tests worldwide.
Key NGS tests are available or becoming available in every major region of the world and across a broad range of countries.
There are wide gaps in available data on NGS implementation.
In order to better track and understand NGS use, data from a range of sources will be needed as well as coordinated testing infrastructures and stakeholder engagement.
Introduction
The human genome was mapped 11 years ago, and since that time there has been a rapid growth in the number and scope of genetic tests. Historically, genetic testing was used for a limited set of diseases such as Down syndrome and sickle cell anaemia. However, tools such as ‘next-generation genomic sequencing’ (NGS) have emerged that can measure multiple genes and even the whole genome quickly and at a much lower cost. NGS is a broad term for a technology that measures genetic variation that is either present at birth or that emerges later in life. The emergence of NGS has generated widespread interest around the globe, with growth being driven by the need for better tools to predict, diagnose, treat and monitor disease in conjunction with increasingly efficient sequencing technologies.
NGS is used for a range of clinical applications:
Risk assessment and disease screening, for example, germline cancer risk testing for Hereditary Breast and Ovarian Cancer or Lynch Syndrome
Reproductive health decision making, for example, non-invasive prenatal testing (NIPT) for fetal genetic abnormalities, carrier screening for recessive genetic disorders such as cystic fibrosis.
Diagnosis of an existing condition, for example, whole-exome sequencing (WES) or whole-genome sequencing (WGS) for diagnosis of suspected genetic diseases (SGD) such as intellectual disability disorders or rare diseases
Diagnosis of infectious diseases, for example, SARS-CoV-2.
Prognosis for a diagnosed disease, for example, FLT3 (fms-like tyrosine kinase 3) in acute myeloid leukaemia.
Prediction/monitoring of treatment response or adverse events, for example, tumour sequencing (TS) for therapy selection and monitoring of cancer treatment, pharmacogenomics panels to target current drug selection.
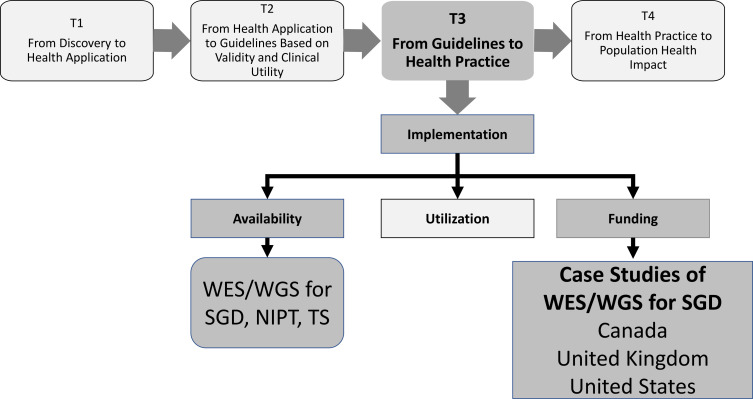
Understanding what tests are available is a key component in the ‘translational continuum’ for genomic medicine as this moves from discovery (T1) to evaluation of validity and clinical utility (T2) to health practice (T3) and then to population health impact (T4).1 However, there has not been an assessment of either the current availability of NGS or the process for developing a robust evidence base. Published studies have focused on specific testing applications such as BRCA1/2 (breast cancer type 1 and breast cancer type 2) testing for hereditary breast and ovarian cancer within specific healthcare systems,2 global governance,3 standardisation and data sharing,4–6 and projected market trends.7 Our study encourages the expansion of data collection with respect to NGS testing. Additional data collection on global availability and funding of NGS tests would benefit patients, providers, researchers and policy-makers, both in countries with established availability and those with emerging availability.
Our objective is to expand our prior work on this topic8 by describing the availability and funding of NGS globally (figure 1). We begin by describing the availability of three key uses of NGS (WES/WGS, NIPT and TS) across world regions and countries. We then use exploratory case studies across three diverse healthcare systems to examine funding for WES/WGS in the context of SGDs, and illustrate the challenges and potential solutions that may be applicable to other countries. WES and WGS are starting to be used to diagnose SGDs, but countries have different approaches to payer coverage and public funding (‘funding’) between countries and across areas (provinces/regions) within countries. This variation can lead to differences in an individual’s chance of disease diagnosis and potential treatments according to which country they live in and where in that country they reside. Understanding the differences in funding and the evolution of these differences may help inform future challenges and potential solutions for implementation of genomic technologies into practice. Our final section describes what type of data will be needed and where such data might be obtained, and we conclude with next steps.
Figure 1.
Study framework: focus on T3 translation, implementation, and case studies of funding. NIPT, non-invasive prenatal testing; SGD; suspected genetic diseases, TS, tumour sequencing, WES; whole-exome sequencing; WGS, whole-genome sequencing.
Clinical genomic sequencing is available across world regions and many countries
WES/WGS, NIPT and/or TS are available in many countries in all major regions of the world (table 1). ‘Availability’ measures where tests are available for clinical (vs research) use; although availability does not always translate into utilisation, it suggests where NGS is moving into clinical care.
Table 1.
Availability of clinical genomic sequencing tests globally: whole-exome sequencing, non-invasive prenatal testing and tumour sequencing
| Test | North America | Central/ South America | Europe | Middle east | Asia/Oceania | Other countries |
| Whole-exome sequencing | USA Canada Mexico |
Brazil Argentina Colombia Argentina Peru |
UK, Germany Belgium, Denmark, Netherlands, Sweden, Italy, Spain, Ireland, Estonia, Finland, France | Saudi Arabia Qatar Turkey Israel |
Australia China Japan South Korea Taiwan |
South Africa |
| Non-invasive prenatal testing | USA Canada Mexico |
Brazil Argentina Colombia Argentina Peru |
UK, France, Belgium, Denmark, Netherlands, Switzerland, Czech Republic, Sweden, Germany, Italy, Spain Ireland | Saudi Arabia Qatar Turkey Israel |
Australia China Japan South Korea Taiwan |
South Africa |
| Tumour sequencing | USA Canada Mexico |
Brazil Argentina Chile Colombia Venezuela Peru |
UK, Belgium, Czech Republic, Netherlands, Sweden, Norway, France, Germany, Spain, Italy, Estonia, Finland, Ireland | Saudi Arabia Qatar Turkey Israel |
Australia China Japan South Korea Taiwan |
South Africa Egypt |
Availability is based on the best available information obtained using a range of sources, including the grey literature and interviews with experts (personal communications, Veronique Forest, Marius Van Den Berg, 10/20/20).30 However, this table is illustrative and thus may not include all relevant countries. Also, we could not assess the availability of whole genome (vs exome) independently but many countries have both tests available.
Although it is widely known that NGS tests are available in high-income countries with robust genomic programmes such as the USA and the UK, tests are also becoming available in a range of other countries. Many European countries other than the UK have increasing availability of these tests and countries such as Australia have robust implementation. Availability is also increasing in countries with upper-middle-income economies. Although there are limited English language, peer-reviewed publications that describe implementation in Asia, the available studies illustrate the growth of genomic medicine (in general) and NGS (specifically). For example, Chong et al describe the landscape of genomic medicine adoption and implementation in four southeast Asian countries: Indonesia, Malaysia, Singapore and Thailand, noting that all four countries have made progress but that there is significant heterogeneity across countries in clinical implementation.9 Another upper-middle-income country that illustrates the slow but significant growth of NGS is South Africa, where genetic testing services have been established.10 However, African countries face numerous challenges in increasing their research and clinical NGS capabilities.11
Exploratory case studies of three countries with varying funding of whole-exome/genome-sequencing for SGD
Our case studies, based on published and unpublished literature as well as discussions with key experts in each country, are illustrative of three types of funding systems:
Publicly funded/national system: UK, which is one of a few countries with national, government-based funding for NGS (others include Belgium, Denmark, the Netherlands and Australia).
Publicly funded/provincial system: Canada, for which public funding of tests is provided at the provincial level after approval based on meeting specific criteria.
Mixed private/public system: USA, which has variable private and public insurer coverage.
Summaries of these case studies are in table 2.
Table 2.
Funding comparisons across countries for WES/WGS for suspected genetic diseases
| Themes | UK | Canada | USA |
| Current funding |
|
|
|
| Key factors influencing funding |
|
|
|
| Solutions for challenges |
|
|
|
WES, whole-exome sequencing; WGS, whole-fenome sequencing.
UK
In the UK, coverage and funding for most genomic tests, including WES/WGS for SGD, is generally provided at the national level within the National Health Service (NHS). Private ordering and funding for WES/WGS is very limited.
A key driver for increased coverage of SGD testing in recent years has been the Genomics England 100 000 Genomes Project. The project ran from 2013 to 2018 and sequenced 100 000 genomes from around 85 000 individuals, using WGS. All the participants were NHS patients affected by a SGD or cancer, recruited via one of 13 NHS Genomic Medicine Centres following referrals from physicians. The UK Department of Health considered the project to be a success due to its completion (demonstrating ability to test at scale) and because, especially for SGD, more diagnoses were made (greater diagnostic yield) than had been via non-WGS technologies. As WGS prices fell during the Project, WGS became a more affordable option if undertaken at large scale rather than in small testing laboratories. Increasing genomic sequencing also became a priority for the UK Life Sciences government agenda so there was significant government will and backing to see its use increased.
To build on this impetus, the Department of Health then set up a new national Genomic Medicine Service (GMS), which aims to take what had been achieved during the 100 000 Genomes Project and make WGS for certain conditions part of a ‘normal’ mainstreamed genomic testing offering. The GMS will include WGS, WES and some panels, which will be the first time that WGS has been used in routine NHS care rather than within research projects. A key aim of the national genomic testing service is to reduce testing inequality and ensure that patients with similar conditions receive the same tests, regardless of where they live. The GMS includes a national genomic laboratory network made up of seven genomic laboratory hubs, a National Genomic Test Directory that specifies which suspected rare diseases should have first-line genomic tests, and an integrated clinical service. This test directory also specifies when a panel, WES or WGS should be used.12
The GMS, thus, reflects one solution that other countries could consider—centralising and mainstreaming NGS test practices and policies. However, one of the main challenges of transitioning WGS into routine care in this way is the complexity of establishing the infrastructure to deliver a cutting-edge service with equitable provision nationally. One of the key issues to address is ensuring that there is capacity in the entire testing pipeline, from clinical referral for a test, through testing, bioinformatics analysis, interpreting results and then reporting the results back to patients. Delivering this at a national level requires significant organisation and coordination, which was achievable in the 100 000 Genomes Project, but has proven more challenging at very large scale.
Canada
Clinical diagnostic WGS is not reimbursed in any provincial jurisdiction, while WES funding is provided in some but not all provinces. WES funding emerged from the use of other panels; panels for cancer and cardiovascular testing specifically have been in place for at least a decade because of their use in obtaining a diagnosis.
The need to control expenditures has contributed to a centralised system and review process for approving genetic testing.13 14 Provinces are similar in that they all have an application and approval process for WES; approved tests are mostly sent out of province, and tests are predominantly requested by medical geneticists. In general, the criteria for WES approval are: (1) it is deemed medically necessary by a qualified health professional; (2) no other alternative exists and (3) the results of the genetic test have an impact on clinical decision making.15 Provinces differ in the specific requirements, the steps in the process and the organisational body that approves the request.16 17
One of the key challenges with expanding the use of WES/WGS further is the lack of infrastructure to deliver timely clinical exomes. Consequently, most WES testing is sent out of country. There are multiple layers of decision-making and a substantial amount of paperwork required to request testing. Since most requests are ultimately approved, the review process does not significantly reduce volume of testing but the time lag for approval (2–6 months in some provinces) can delay access to testing and appropriate follow-up. In some cases, patients are offered testing via private insurance or self-pay to reduce the time to testing and receipt of test results. Although this approach may reduce the current unmet need for WES/WGS, it remains controversial due to equity concerns (based on geographic location and the existence of financial for some patients).
Going forward, efforts are required to build appropriate infrastructure to undertake testing within provinces and for clinical applications in a timely way that is equitable to all patients. One approach to addressing infrastructure and equity concerns— which may be applicable to other countries—is the provincial Genomic Applications Partnership Programme (GAPP), which is aimed at translation and implementation of genomics into routine clinical practice. The platform being used—‘All for One’—engages a wide array of stakeholders including institutions, regulators, and members of the rare disease clinical research community to work collaboratively to determine policy, governance, areas of need and an ethical and legal framework. Although this platform is being developed within the context of rare diseases, it also has future applications as a model for other initiatives.18 The GAPP programmes are funded through Genome Canada and the respective provincial Genome agency in each province in partnership with a receptor organisation (such as the provincial laboratory and health services) and academic organisations.19 For example, in Alberta, the Translational Implementation of Genomics for Rare Diseases (TIGeR) GAPP programme was approved in 2020.20 21 TIGeR aims to address the lack of clinical sequencing infrastructure to deliver timely clinical exomes by optimising clinical workflows, building clinical genomics capacity within the province and driving the implementation of clinical genome wide sequencing and integrating genomic data into clinical practice in Alberta.
USA
There has been increased payer coverage of WES/WGS recently, but this varies by payer and is not universal. Currently, over half of insured individuals (private or public) have coverage for WES and/or WGS (63%).22 This compares to the situation in 2015, when our study did not find any positive coverage policies among the five largest US private payers and few payers even had policies.23 Among individuals with private insurance, 71% have coverage for WES/WGS while among individuals with Medicaid insurance, only 39% have coverage. There are 27 state Medicaid programmes (as well as the District of Columbia) that do not provide any WES/WGS coverage. However, fewer insured individuals have coverage for WGS in addition to WES; only 8% of individuals with private insurance have coverage for both WES and WGS.
We previously examined why private payers may be increasing their coverage of WES/WGS and found that a key factor was that many payers were adopting an expanded view of its clinical utility.24 Payers stated that they were willing to provide coverage because they saw merit in using available interventions or ending the diagnostic odyssey—factors that previously had been insufficient to justify coverage, for example, half agreed that clinical utility extended beyond its impact on clinical outcomes and management to include ending the diagnostic odyssey, informational utility (directing family to disease-specific support, education and research) and family utility for reproductive decision making.
Another factor that has led to increased funding is acceptance of coverage for rapid WES/WGS in neonatal or paediatric intensive care units. Over half of insured individuals with coverage for both WES and WGS only have coverage in these settings.22 Coverage for rapid testing in the neonatal intensive care unit (NICU) has emerged because several studies have found evidence of improved outcomes including time to diagnosis, diagnostic yield, and changes to patient management over non-rapid testing.25 Of note is a recent study examining the impact of expanding Medicaid coverage for rapid WGS in the NICU in California. ‘Project Baby Bear’ found that rapid WGS ‘improves clinical outcomes, improves the experience of care for families and clinicians, and reduces net healthcare expenditures’ and recommended that rapid WGS be covered by Medicaid.26 It is unknown whether the outcomes of this study have changed Medicaid policies in California or other states or had a spillover effect on private payer policies, but it does provide an example of how even ‘safety net’ programmes can be innovative in developing the evidence to support changes in implementation and coverage policies.
The USA does not have a government organisation tasked with developing consistent policies and funding, hence other organisations have emerged to fill this gap—a situation that other countries without a national health system may face. One example is the Patient-Centred Laboratory Utilisation Guidance Services programme (PLUGS), which is a non-profit laboratory stewardship collaboration within Seattle Children’s Hospital Department of Laboratories. Their mission is to improve laboratory test ordering, retrieval, interpretation and reimbursement.
PLUGS uses a two-pronged approach to develop consistent and medically appropriate policies and funding that may be applicable to other countries:
An inclusionary approach that obtains perspectives from providers, labs and payers to seek consensus (they also solicit patient/parent input and share their results with patients/parents).
A focus on creating initial policies with narrow inclusion criteria rather than attempting to broadly cover tests, which then provides coverage for many of the sickest patients and creates a pathway for eventual expansion of criteria over time as clinically appropriate.
In 2016, PLUGS developed a consensus WES policy that was subsequently adopted and implemented by a national private payer, a lab benefit management company (that develops draft coverage policies for payers), and a state Medicaid plan among others. More recently, PLUGS created a WGS policy with criteria limited to the rapid inpatient setting based on clinical utility evidence in this patient population and knowledge of payer readiness to adopt coverage for this novel test.
Data needed for implementation studies and potential sources
As noted previously, few studies have been published that empirically examine NGS implementation across countries. We, therefore, used grey literature and expert interviews to identify data gaps and possible data sources for three key implementation factors: availability, utilisation and funding. Table 3 shows that for each of these factors there are wide data gaps, particularly outside the USA and Western Europe. There is some limited data on availability and utilisation of specific tests in targeted locations and for government-funded testing programmes. We note that a diverse range of data sources will be needed to obtain needed data, including grey literature and data from administrative and/or clinical databases.
Table 3.
Data needed on implementation, data gaps and possible data sources
| Implementation factors |
Data gaps (based on publicly available, accessible data sources) |
Possible data sources with illustrative examples22 30 |
| Availability of NGS tests for clinical use |
|
Published journal articles For example, Article from global collaborative that focuses on enabling the implementation of genomic medicine Grey literature* For example., Online news source such as GenomeWeb that reports on genomic test availability and utilisation Administrative and clinical data† For example, Registries such as the US National Institutes of Health (NIH) Genetic Testing Registry that consists of voluntary submissions by laboratories of their available tests |
| Utilisation (# tests ordered) |
|
Published journal articles For example, Article that describes US genetic test availability and spending based on claims data Grey literature* For example, Market reports such as investor analyses of NGS Market by product type Administrative and clinical data† For example, white papers such as Personalised Medicine Coalition’s report that used data integrated from claims, census and proprietary databases |
| Funding |
|
Published journal articles For example, Article that reviewed coverage policies for ctDNA (liquid biopsy) tests Grey literature* For example, Advocacy group website such as Coalition for Access to Prenatal Screening Administrative and clinical data† For example, Proprietary and academic databases such as those developed by TRANSPERS and Canary Insights |
*Includes white papers, health system reports, market analyses, regulatory filings, company websites, news reports, national/international consortia websites.
†Includes electronic health records, claims data, fee schedules, industry databases, registries.
NGS, next-generation genomic sequencing.
CONCLUSION
We found that NGS tests are available in many countries across the globe and that many payers and health systems are funding NGS tests in at least some clinical scenarios. However, there are limited data, and thus, it is difficult to assess whether and how implementation is successful. Surprisingly few publications have addressed NGS global implementation especially in upper-middle-income countries, and thus, creative approaches are needed to identify and assess the required evidence. More data would be helpful to patients, providers, researchers and policy-makers. For example, one study found that the vast majority of studies are done on discovery (T1 in the translational continuum), demonstrating a need to increase research efforts in this area to implement promising genomic interventions into practice.27
We also found that some countries are implementing new approaches to improve implementation of NGS tests. By sharing experiences, lessons learnt can be leveraged and applied more broadly. Each country will have its own culture and constraints, but our case studies illustrated key leverage points. A key recommendation that emerges from the case studies is the need for coordinated, standardised, systematic testing infrastructure as well as stakeholder engagement in order to develop consistent, efficient and equitable practices and policies.
We examined whether and how tests are being used in clinical care, but we did not examine another topic—‘implementation science’—the application of methods that promote uptake of research findings into practice. It has been noted by influential organisations that more emphasis needs to be placed on using such methods to define and measure successful implementation, identify efficient approaches, and increase study rigour.28 29 We also did not examine the clinical utility of tests—how tests improve patient outcomes—although we must consider not only what tests are implemented but whether they are appropriately implemented and provide value to patients and populations.
We acknowledge that we may have missed relevant studies or misclassified some countries as having test availability, given the lack of data and inconsistencies in definitions and measurements. Also, we note that many other factors determine whether NGS will be implemented into routine clinical care including patient and provider preferences and decision making, regulatory actions, infrastructure development and dissemination of information.5
In conclusion, the emergence of genomic sequencing tests for use in clinical care has generated widespread interest around the globe, but more information is required on how these tests are being used in clinical care particularly in upper-middle-income countries that are implementing NGS but where there have been few published studies. Key recommendations for advancing NGS are the development of coordinated, standardised, systematic testing infrastructure and stakeholder engagement.
Acknowledgments
The authors gratefully acknowledge Dean Regier PhD, University of British Columbia for his contribution to the conceptualisation of the study in conjunction with the Global Economics and Evaluation of Clinical Genomics Sequencing (GEECS) Working Group. The authors also acknowledge the clinical experts and payers with whom we spoke to inform the case studies. Written permission has been obtained.
Footnotes
Handling editor: Soumitra S Bhuyan
Twitter: @KathrynP_phd
Contributors: Substantial contributions to the conception or design of the work: all. Acquisition, analysis or interpretation of data: KAP and MPD. Drafting the work: KAP. Revising the work critically for important intellectual content: all. Final approval of the version published: all. Agreement to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved: all.
Funding: This study was funded by grants from the National Cancer Institutes (R01 CA221870) and National Human Genome Research Institute (U01 HG009599) and consulting contracts to KAP and MPD. DAM is supported by the Arthur J.E. Child Chair in Rheumatology. SW and JB are partly funded by the Oxford National Institute for Health Research Biomedical Research Centre.
Competing interests: KAP receives consulting income from Illumina. Disclosures have been reviewed by the University of California San Francisco. MPD receives consulting income from Illumina. DAM, JB and SW have received travel expense reimbursement from Illumina to attend meetings.
Patient consent for publication: Not required.
Provenance and peer review: Not commissioned; externally peer reviewed.
Data availability statement: The data that support the findings of this study are available from the corresponding author, KAP, upon reasonable request.
References
- 1.Khoury MJ, Gwinn M, Yoon PW, et al. The continuum of translation research in genomic medicine: how can we accelerate the appropriate integration of human genome discoveries into health care and disease prevention? Genet Med 2007;9:665–74. 10.1097/GIM.0b013e31815699d0 [DOI] [PubMed] [Google Scholar]
- 2.Miller I Acceleration of adoption of high complexity precision diagnostics by global public healthcare systems: a case study of Europe and beyond. J Pers Med 2019;5:7–13. [Google Scholar]
- 3.Belsey J, Chaihorsky L, Chediak L, et al. World economic forum white paper: global data access for solving rare disease: a health economics value framework. Geneva, Switzerland, 2020. [Google Scholar]
- 4.Ginsburg GS A global collaborative to advance genomic medicine. Am J Hum Genet 2019;104:407–9. 10.1016/j.ajhg.2019.02.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Manolio TA, Rowley R, Williams MS, et al. Opportunities, resources, and techniques for implementing genomics in clinical care. Lancet 2019;394:511–20. 10.1016/S0140-6736(19)31140-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stark Z, Dolman L, Manolio TA, et al. Integrating genomics into healthcare: a global responsibility. Am J Hum Genet 2019;104:13–20. 10.1016/j.ajhg.2018.11.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Phillips KA, Douglas MP. The global market for next-generation sequencing tests continues its torrid PACE. J Precis Med 2018;4. [PMC free article] [PubMed] [Google Scholar]
- 8.Phillips KA, Douglas MP, Marshall DA. Expanding use of clinical genome sequencing and the need for more data on implementation. JAMA 2020;324:2029. 10.1001/jama.2020.19933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chong HY, Allotey PA, Chaiyakunapruk N. Current landscape of personalized medicine adoption and implementation in Southeast Asia. BMC Med Genomics 2018;11:94. 10.1186/s12920-018-0420-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kromberg JGR, Sizer EB, Christianson AL. Genetic services and testing in South Africa. J Community Genet 2013;4:413–23. 10.1007/s12687-012-0101-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Adebamowo SN, Francis V, Tambo E, et al. Implementation of genomics research in Africa: challenges and recommendations. Glob Health Action 2018;11:1419033. 10.1080/16549716.2017.1419033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.National Health Service National genomic test directory: National health service, 2020. Available: https://www.england.nhs.uk/publication/national-genomic-test-directories/
- 13.Lilley M, Christian S, Blumenschein P, et al. A centralized approach to out-of-province genetic testing leads to cost savings: the Alberta experience. Clin Genet 2013;84:373–7. 10.1111/cge.12077 [DOI] [PubMed] [Google Scholar]
- 14.Christian S, Blumenschein P, Lilley M. An assessment of Canadian systems for triaging referred out genetic testing. Clin Genet 2015;88:90–4. 10.1111/cge.12435 [DOI] [PubMed] [Google Scholar]
- 15.Waddell K, Wilson M. Rapid synthesis: examining the public provision and funding of clinical genetic tests. Hamilton, Canada: McMaster Health Forum, 2017. http://hdl.handle.net/11375/22549 [Google Scholar]
- 16.Genetic Testing Advisory Committee Use of Genome‐Wide sequencing for undiagnosed rare genetic diseases in Ontario, 2016. Available: http://www.health.gov.on.ca/en/pro/programs/gtac/docs/gtac_report_use_of_gws_for_undiagnosed_rare_genetic_diseases.pdf
- 17.Elliott AM, du Souich C, Adam S, et al. The genomic consultation service: a clinical service designed to improve patient selection for genome-wide sequencing in British Columbia. Mol Genet Genomic Med 2018 10.1002/mgg3.410. [Epub ahead of print: 30 May 2018]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.GenomeCanada All for one policy toolkit: GenomeCanada, 2020. Available: https://www.genomecanada.ca/en/all-one-policy-toolkit
- 19.GenomeCanada Genomic applications partnership program: GenomeCanada, 2020. Available: https://www.genomecanada.ca/en/programs/translation/funding-opportunities/genomic-applications-partnership-program
- 20.GenomeCanada Major investment in genomics research will improve the health and wellbeing of Canadians: GenomeCanada, 2020. Available: https://www.genomecanada.ca/en/news/major-investment-genomics-research-will-improve-health-and-wellbeing-canadians
- 21.GenomeCanada Seven genomic applications partnership program projects funded: GenomeCanada, 2020. Available: https://www.genomecanada.ca/sites/default/files/bk-gapp_15_16_english.pdf
- 22.UCSF TRANSPERS Illustrative data sources and citations for NGS test coverage, prices, and reimbursement in the US, 2020. Available: https://pharm.ucsf.edu/transpers/node/18041
- 23.Douglas MP, Parker SL, Trosman JR, et al. Private payer coverage policies for exome sequencing (ES) in pediatric patients: trends over time and analysis of evidence cited. Genet Med 2019;21:152–60. 10.1038/s41436-018-0043-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Trosman JR, Weldon CB, Slavotinek A, et al. Perspectives of US private payers on insurance coverage for pediatric and prenatal exome sequencing: results of a study from the program in prenatal and pediatric genomic sequencing (P3EGS). Genet Med 2020;22:283–91. 10.1038/s41436-019-0650-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.BlueCross Blue Shield Association Evidence street report: whole exome and whole genome sequencing for diagnosis of genetic disorders, 2020. [Google Scholar]
- 26.Norton ME Project Baby Bear - Final Report, 2020. Available: https://radygenomics.org/wp-content/uploads/2020/07/PBB-Final-Report_07.14.20.pdf
- 27.Clyne M, Schully SD, Dotson WD, et al. Horizon scanning for translational genomic research beyond bench to bedside. Genet Med 2014;16:535–8. 10.1038/gim.2013.184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roberts MC, Clyne M, Kennedy AE, et al. The current state of funded NIH grants in implementation science in genomic medicine: a portfolio analysis. Genet Med 2019;21:1218–23. 10.1038/gim.2017.180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chambers DA, Feero WG, Khoury MJ. Convergence of implementation science, precision medicine, and the learning health care system: a new model for biomedical research. JAMA 2016;315:1941–2. 10.1001/jama.2016.3867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.UCSF TRANSPERS Illustrative data sources and citations for NGS test availability, 2020. Available: https://pharm.ucsf.edu/transpers/node/18311