Abstract
Background
Recent studies have focused on the correlation between N6-methyladenosine (m6A) modification and specific tumor-infiltrating immune cells. However, the potential roles of m6A modification in the tumor immune landscape remain elusive.
Methods
We comprehensively evaluated the m6A modification patterns and tumor immune landscape of 513 clear cell renal cell carcinoma (ccRCC) patients, and correlated the m6A modification patterns with the immune landscape. The m6Ascore was established using principal component analysis. Multivariate Cox regression analysis was performed to evaluate the prognostic value of the m6Ascore.
Results
We identified three m6Aclusters—characterized by differences in Th17 signature, extent of intratumor heterogeneity, overall cell proliferation, aneuploidy, expression of immunomodulatory genes, overall somatic copy number alterations, and prognosis. The m6Ascore was established to quantify the m6A modification pattern of individual ccRCC patients. Further analyses revealed that the m6Ascore was an independent prognostic factor of ccRCC. Finally, we verified the prognostic value of the m6Ascore in the programmed cell death protein 1 (PD-1) blockade therapy of patients with advanced ccRCC.
Conclusions
This study demonstrated the correlation between m6A modification and the tumor immune landscape in ccRCC. The comprehensive evaluation of m6A modification patterns in individual ccRCC patients enhances our understanding of the tumor immune landscape and provides a new approach toward new and improved immunotherapeutic strategies for ccRCC patients.
Keywords: immunotherapy, kidney neoplasms
Background
Methylation of N6 adenosine to produce N6-methyladenosine (m6A) is the most common type of RNA modification1; it is thought to regulate multiple RNA-related processes, such as RNA stability,2 translation,3 alternative splicing4 5 and nuclear export.6 m6A modification is a dynamic and reversible process which is regulated by m6A methyltransferases (‘writers’), m6A demethylases (‘erasers’) and m6A-binding proteins (‘readers’).7 The m6A methyltransferases—consisting of METTL3, METTL14, WTAP, RBM15, RBM15B, ZC3H13, CBLL1 and VIRMA—catalyze m6A modification as m6A writers, while a set of m6A demethylases— including ALKBH5 and FTO—mediate the reversal of m6A modification of RNA as m6A erasers.8 9 Moreover, m6A-binding proteins—such as IGF2BP1/2/3, YTHDF1/2/3 and YTHDC1/2—recognize and bind to the m6A methylation sites in RNA as m6A readers.8 9 m6A is an essential RNA modification that regulates multiple key cellular processes including cellular differentiation, stem cell renewal and response to DNA damage.10 Evidently, aberrant expression of m6A regulators is associated with tumorigenesis, malignant tumor progression and immunomodulatory abnormality.10 11
Immune checkpoint therapy (ICT)—such as programmed cell death protein 1 (PD-1)/PD ligand 1 (PD-L1) blockade therapy—is transformative in the treatment of advanced clear cell renal cell carcinoma (ccRCC).12 13 However, there remains a considerable proportion of patients with no response or resistance to ICT.14 In solid malignant tumors, the PD-1 blocking response is associated with numerous tumor-intrinsic15 16 and tumor immune microenvironment (TIME) characteristics.17 18 A common paradigm in the immunology of solid tumors is that effective responses to anti-PD-1 therapy occur when the TIME is characterized by high infiltration of CD8+ T cells and that resistance to this therapy occurs when the TIME is characterized by the lack of such an infiltration.19 20 Understanding the biology of the TIME that drives the ICT response is crucial to the design of immunotherapeutic strategies.21 22
Several studies have recently focused on the special relationship between m6A regulators and immune cells. Wang et al23 reported that METTL3-mediated m6A modification increased the translation of certain immune transcripts and physiologically promoted the activation of dendritic cells (DCs) and DC-based T-cell responses. Li et al24 showed that deletion of METTL3 in T cells disrupted the homeostasis and differentiation of T cells. Han et al25 found that deletion of YTHDF1 elevated the antitumor response of antigen-specific CD8+ T cells and enhanced the efficacy of anti-PD-L1 therapy. However, limited by existing experimental technology, the above research is confined to a limited number of m6A regulators and cell types, while the development and progression of cancers depend on cross-talk among multiple m6A regulators of RNA methylation.9 Therefore, a comprehensive evaluation of the immune landscape mediated by a variety of m6A regulators will enhance our overall understanding of the immunomodulatory effect of m6A regulators on the TIME. Recently, the m6A modification patterns of gastric cancer were comprehensively evaluated based on multiple m6A regulators and systematically correlated with the tumor immune landscape, indicating the important role of m6A modification in TIME diversity in gastric cancer.26
In the present study, we integrated the molecular and clinical data of 513 ccRCC patients to comprehensively evaluate the m6A modification patterns and tumor immune landscape and correlated the m6A modification patterns with the immune landscape. We identified three distinct m6A modification patterns and were surprised to find that they had distinct immune landscapes and prognoses, indicating the crucial roles of m6A modification in the formation of individual tumor immune landscapes in ccRCC patients. We went on to quantify the m6A modification patterns of individual ccRCC patients by establishing the gene signature of m6A regulators.
Methods
Molecular and clinical data
The workflow of our study is shown in online supplemental figure S1. RNA sequencing data (count values) for gene expression analysis, genetic mutations (VarScan), and clinical data were downloaded from the Genomic Data Commons (https://portal.gdc.cancer.gov/).27 The count values were transformed into transcripts per kilobase million (TPM) values (the gene lengths used for the above transformation were measured as total non-overlapping exon length) and the Ensembl gene IDs of the RNA-seq data were converted to gene symbols by referring to the annotation file (https://www.gencodegenes.org/human/release_22.html). The copy number variation (CNV) data were downloaded from the Broad GDAC Firehose (https://gdac.broadinstitute.org/). The normalized data from another ccRCC cohort (91 cases) were downloaded from the International Cancer Genome Consortium (ICGC, https://dcc.icgc.org/).
jitc-2020-001646supp002.pdf (369.7KB, pdf)
Model-based clustering analysis for m6A regulators
Gene expression levels were quantified using the metric log2 (TPM +1), then used to identify m6A modification patterns based on the expression of 24 m6A regulators by model-based clustering analysis implemented in the R package ‘mclust’.28 The optimal number of clusters was determined based on the Bayesian information criterion.
Immune cellular fraction estimates
CIBERSORT—a deconvolution algorithm reported by Newman et al29 and verified by fluorescence-activated cell sorting—was used to quantify the 22 infiltrated immune cells according to normalized gene expression profiles. The 22 immune cells included memory B cells, naïve B cells, plasma cells, resting/activated DCs, resting/activated natural killer (NK) cells, resting/activated mast cells, eosinophils, neutrophils, monocytes, M0–M2 macrophages, and seven T-cell types (CD8+ T cells, regulatory T cells (Tregs), resting/activated memory CD4+ T cells, follicular helper T cells, naïve CD4+ T cells and gammadelta T cells (γδ T cells)) For each sample, the sum of all estimated values for the proportion of immune cells was equal to 1. CIBERSORT results were obtained from the following website: https://gdc.cancer.gov/about-data/publications/panimmune (online supplemental table S1).30 The relative abundance of Th1/Th2/Th17 cell infiltration in the ccRCC TIME was quantified by single-sample gene-set enrichment analysis. The gene sets for marking the Th1/Th2/Th17 cell types were obtained from a study published by Thorsson et al.30 The prognostic value of infiltrated immune cells was assessed by univariate Cox regression analysis.
jitc-2020-001646supp001.xlsx (345.5KB, xlsx)
Evaluation of values of key immune characteristics and measures of DNA damage among m6Aclusters
Values of key immune characteristics (including leukocyte fraction, Th1/Th2/Th17 cells, single nucleotide variant neoantigens, indel neoantigens, proliferation, aneuploidy score, intratumor heterogeneity (ITH), B-cell receptor (BCR) evenness, T-cell receptor (TCR) evenness and cancer testis antigens (CTA) score) and measures of DNA damage (including CNV burden (number of segments and fraction of genome alterations, respectively), loss of heterozygosity (LOH; number of segments with LOH events, and fraction of bases with LOH events, respectively), homologous recombination deficiency, and mutation load (non-silent mutation)) were obtained from the following website: https://gdc.cancer.gov/about-data/publications/panimmune (online supplemental table S1).
Correlations between the expression characteristics of m6A regulators and immunomodulators
A list of 78 immunomodulators (IMs) was obtained (online supplemental table S2),30 three of which (HLA-DRB3, HLA-DRB4 and KIR2DL2) had no corresponding mRNA expression and were excluded from subsequent analysis. The median expression levels of the samples were used to represent the expression of each ccRCC subtype. In order to examine the differences in IM expression among different subtypes, we carried out the Kruskal-Wallis test on the gene expression levels for each ccRCC subtype. CNVs for each IM gene were obtained from the following website: https://gdc.cancer.gov/about-data/publications/panimmune.30 We calculated the difference between the observed and expected amplification frequencies (deletions) for each IM gene in each ccRCC subtype, where the expected frequency is the overall amplification frequency (deletions) of all ccRCC cases.
Gene set variation analysis
Gene set variation analysis (GSVA)—a non-parametric and unsupervised method commonly used for estimating pathway variations in the samples of expression datasets—was performed to explore the differences in biological processes among m6A modification patterns.31 The ‘c2.cp.kegg.v6.2.symbols’ gene sets for GSVA were downloaded from the Molecular Signatures Database (MSigDB). A p<0.05 was considered statistically significant.
Identification of differentially expressed genes among m6Aclusters
To identify genes related to m6A modification patterns, we classified patients into m6Aclusters based on the expression of 24 m6A regulators. Differentially expressed genes (DEGs) among these clusters were determined using the R package ‘DESeq2’, which was applied using the raw count values of RNA sequencing data. Genes with adjusted p<0.01 and at least two-fold changes in expression were identified as DEGs.
Construction of the m6A gene signature
We applied a methodology to quantify the m6A modification pattern (m6Ascore) of individual ccRCC patients. The m6Ascore was established as follows. First, we extracted the overlapping DEGs among m6Aclusters and classified the ccRCC patients into several groups using model-based clustering to analyze overlapping DEGs. Univariate Cox regression analysis was performed to evaluate the prognosis of each overlapping DEG. Genes with a significant prognosis (p<0.05) were extracted for further analysis. Next, principal component analysis (PCA) was performed to establish the m6A gene signature. We selected both principal components 1 and 2 as signature scores. Finally, the m6Ascore was defined using a method similar to Genomic Grade Index26 32 33:
where i is the expression of overlapping genes with a significant prognosis of DEGs among m6Aclusters.
Correlation between m6Ascore and other relevant biological processes
Spearman’s correlation analysis was performed to investigate the correlation between m6Ascore and other relevant biological processes using the gene sets reported by Mariathasan et al (online supplemental table S3),18 including (1) antigen processing machinery (APM), (2) effector CD8 T-cell signature, (3) immune checkpoint, (4) nucleotide excision repair, (5) mismatch repair, (6) DNA replication, (7) DNA damage repair, (8) epithelial-mesenchymal transition markers, (9) Wnt targets, (10) pan-fibroblast transforming growth factor-β response signature, and (11) angiogenesis signature.
Genomic and clinical data with anti-PD-1 therapy for ccRCC
A systematic search for the genomic and transcriptomic datasets of ccRCC patients treated with anti-PD-1 therapy was performed. We ultimately included one immunotherapeutic cohort—advanced ccRCC with treatment of PD-1 blockade and mammalian target of rapamycin (mTOR) inhibition—obtaining the genomic, transcriptomic and clinical data (online supplemental table S4) from the online supplemental data appended to the published paper.34
jitc-2020-001646supp008.pdf (86.5KB, pdf)
Statistical analysis
Statistical significance for three or more groups was estimated using the Kruskal-Wallis test and association between categorical variables was explored using the χ2 test. The correlation coefficient was calculated via Spearman’s correlation analysis. Continuous variables were dichotomized for patient survival using optimal cut-off values determined by ‘survminer’ R package. The Kaplan-Meier method was used to generate survival curves and the log-rank test was used to determine the statistical significance of differences. The independent prognostic factors, determined by multivariate Cox regression analysis, were visualized by ‘forestplot’ R package. The ‘oncoplot’ function of R package ‘maftools’ was used to depict the mutation landscape of The Cancer Genome Atlas Kidney Renal Clear Cell Carcinoma (TCGA-KIRC) cohort and immunotherapeutic cohort. The protein–protein interaction (PPI) networks among m6A regulators were identified based on the STRING interaction database35 and visualized by Cytoscape.36 All tests were two sided, and p<0.05 was regarded as significant. All analyses were performed with R software V.3.62 (http://www.R-project.org).
Results
Molecular characteristics and clinical relevance of m6A regulators in ccRCC
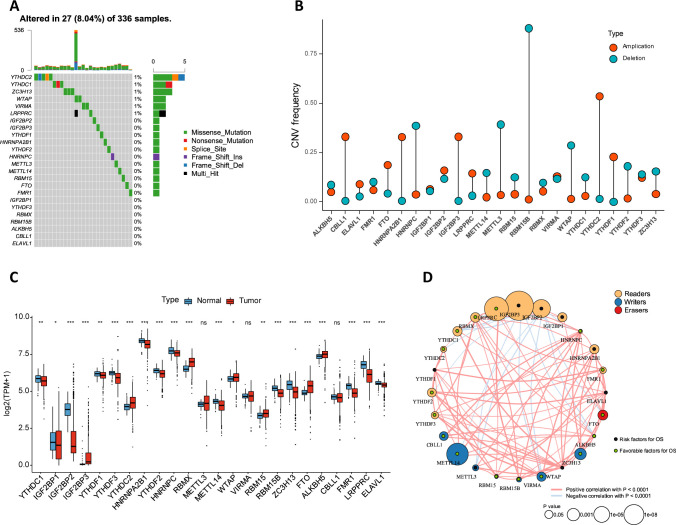
On reviewing the literature, we identified 24 genes that mainly regulate RNA methylation including 8 writers (RBM15/RBM15B, METTL14, METTL3, WTAP, CBLL1, VIRMA and ZC3H13), 2 erasers (FTO and ALKBH5) and 14 readers (FMR1, ELAVL1, HNRNPC, HNRNPA2B1, YTHDF1/2/3, YTHDC1/2, RBMX, IGF2BP1/2/3 and LRPPPRC). Somatic mutations and CNVs were integrated to explore the prevalence of m6A regulator variations in ccRCC. The overall average mutation frequency of m6A regulators was low, with only 27 of 336 samples having m6A regulator mutations (figure 1A). We then studied the CNV alteration frequency of the m6A regulators and demonstrated that CNV alterations were prevalent (figure 1B). The mRNA expression levels of m6A regulators in ccRCC and adjacent tissues were also explored, revealing that 21 out of 24 m6A regulators were differentially expressed (figure 1C). The above analyses showed that the genetic and expressional variations in m6A regulators were highly heterogeneous between ccRCC and adjacent tissues, suggesting a crucial role for the imbalance of m6A regulator expression in the development and progression of ccRCC. Moreover, the function of genes is not isolated, in that it has been shown that collaboration among m6A regulators exists in the context of cancer.37 38 Thus, the correlation of mRNA expression among m6A regulators was explored. We identified that writers, erasers, and readers had a high expression correlation (figure 1D, (online supplemental table S5) and interacted with each other frequently in PPI networks (online supplemental figure S2). Taken together, these results indicate crucial cross-talk roles among m6A regulators of RNA methylation in the formation of distinct m6A modification patterns.
Figure 1.
Molecular characteristics and clinical relevance of m6A regulators in ccRCC. (A) The mutation frequency of m6A regulators in ccRCC. (B) Dumbbell plot depicted the CNV alteration frequency of m6A regulators in ccRCC. The deletion (amplification) frequency was marked with blue (red) dot. (C) The gene expression alterations among m6A regulators. (D) Interaction of m6A regulators in ccRCC. Readers, yellow; Writers, blue; Erasers, red. The size of each circle represented survival impact of each m6A regulator, calculation used the formula log10 (unicox p values indicated). Green (black) dots represented favorable (risk) factors of overall survival. The lines connecting m6A regulators presented their interactions, and thickness of the lines represented the correlation strength among regulators. Positive (negative) correlation was indicated in red (blue). ccRCC, clear cell renal cell carcinoma; CNV, copy number variation; m6A, N6-methyladenosine; OS, overall survival.
jitc-2020-001646supp003.pdf (1.7MB, pdf)
Next, the clinical relevance of m6A regulators in ccRCC patients was explored. We found that many m6A regulators were related to prognosis in patients with ccRCC (figure 1D). Several m6A regulators (eg, IGF2BP1 and IGF2BP3) presented oncogenic characteristics, with higher expression levels of these genes related to poor prognosis in ccRCC patients. In contrast, we found that several m6A regulators (eg, LRPPRC and METTL14) presented characteristics of tumor suppressors, with higher expression levels of these genes correlated with favorable prognosis in ccRCC patients.
m6A modification patterns mediated by 24 m6A regulators
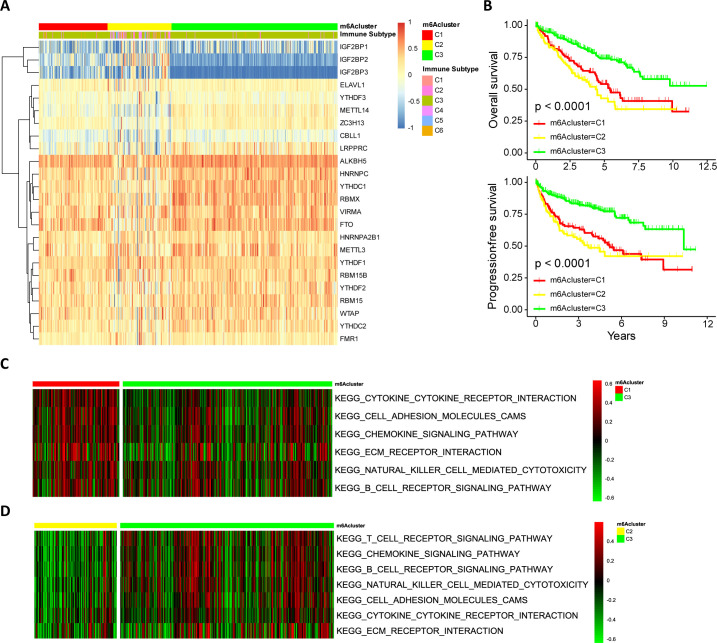
Model-based clustering was performed to classify ccRCC patients based on the expression of 24 m6A regulators. We ultimately uncovered three distinct methylation modification patterns (identified as m6Aclusters C1–C3), including 118 cases in m6Acluster-C1, 110 cases in m6Acluster-C2, and 285 cases in m6Acluster-C3 (figure 2A). The expression of m6A regulators with the greatest differences among subtypes (p<10−15) included two risk factors for overall survival (OS) (IGF2BP2 and IGF2BP3) and four favorable factors for OS (YTHDC1, RBMX, METTL14 and FTO) (online supplemental figure S3). m6Acluster-C3 was characterized by low expression levels of IGF2BP2 and IGF2BP3 and high expression levels of YTHDC1, RBMX, METTL14 and FTO (online supplemental figure S3). Therefore, it was not surprizing that m6Acluster-C3 had the most favorable prognosis (figure 2B).
Figure 2.
m6A modification patterns in ccRCC and biological characteristics of m6A subtypes. (A) Model-based clustering of ccRCC yields three subtypes in the TCGA-KIRC dataset. C1, cluster1; C2, cluster2; C3, cluster3. (B) Comparison of prognosis among ccRCC subtypes (Kaplan-Meier analysis). (C, D) The heatmap depicted the activation states of biological processes (evaluated by GSVA) among m6Aclusters, and activated and inhibited biological processes were marked with red and green, respectively. (C) m6Acluster-C1 vs m6Acluster-C3; (D) m6Acluster-C2 vs m6Acluster-C3. ccRCC, clear cell renal cell carcinoma; GSVA, gene set variation analysis; m6A, N6-methyladenosine.
jitc-2020-001646supp004.pdf (1,019.8KB, pdf)
GSVA was performed to investigate the activity of biological processes among these distinct m6A modification patterns. As shown in figure 2C–D and online supplemental table S6, m6Aclusters-C1 and -C3 were markedly enriched in pathways related to immune activation, including the activation of cytokine–cytokine receptor interaction and the chemokine signaling pathway, TCR signaling pathway and BCR signaling pathway. Meanwhile, m6Acluster-C1 showed enrichment in stromal activation pathways such as cell adhesion and ECM receptor interaction. In contrast, m6Acluster-C2 was predominantly associated with the biological process of immunosuppression.
Immune characteristics in distinct m6A modification patterns
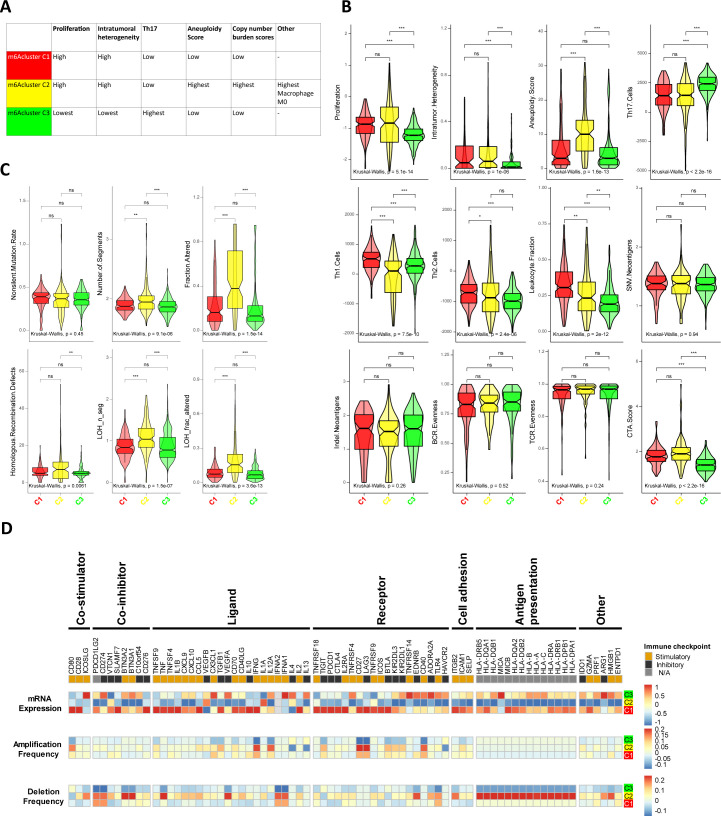
Thorsson et al30 explored the pan-cancer immune landscape and finally identified six immune subtypes (C1–C6) covering 30 cancer types that were assumed to define immune response patterns with implications for further exploration of immunotherapy. Immune subtype C3—characterized by elevated Th17, low to moderate tumor cell proliferation, and lower levels of overall CNVs and aneuploidy than the other immune subtypes—was enriched in most ccRCC patients. Strikingly, the three distinct methylation modification patterns had distinct proportions of the C3 immune subtype, with m6Acluster-C3 having the highest (96.14%), followed by m6Acluster-C1 (90.68%) and C2 (57.27%) (p<0.001) (figure 2A). We then explored the detailed immune characteristics in distinct m6A modification patterns. As shown in figure 3A–C, m6Acluster-C1 had a high proliferation rate, ITH, and lower levels of aneuploidy and overall CNVs. m6Acluster-C2 had the highest aneuploidy score and overall CNVs, as well as a high proliferation rate and ITH, and presented a more prominent macrophage signature dominated by M0 macrophages (online supplemental figure S4). m6Acluster-C3 was defined by elevated Th17, low tumor cell proliferation, ITH and lower levels of aneuploidy and overall CNVs.
Figure 3.
The immune landscape in distinct m6A modification patterns. (A) Key characteristics of m6Aclusters. (B) Values of key immune characteristics in m6Aclusters. (C) DNA damage measures of m6Aclusters, including non-silent mutation rate, copy number burden scores (number of segments, and fraction of genome alterations. respectively), homologous recombination deficiency and loss of heterozygosity (LOH; fraction of bases with LOH events, and number of segments with LOH events, respectively). (D) Regulation of Immunomodulators in distinct m6Aclusters. From top to bottom: mRNA expression (median normalized expression levels); amplification frequency (the difference between the fraction of samples in which an IM is amplified in a particular subtype and the amplification fraction in all samples); and the deletion frequency (as amplifications) for 75 IM genes by m6Aclusters. IM, immunomodulator; m6A, N6-methyladenosine; ns, not significant. The asterisks represented the statistical p value (*P < 0.05; **P < 0.01; ***P < 0.001).
jitc-2020-001646supp005.pdf (566.5KB, pdf)
IMs are essential for cancer immunotherapy with multiple IM agonists and antagonists being investigated in clinical oncology.39 To advance this research, an understanding of their expression in different m6A modification patterns is needed. We explored IM gene expression and CNVs among the m6A subtypes (figure 3D). IM gene expression varied across m6A subtypes and genes with the greatest differences between subtypes (p<10−15) including ADORA2A, CX3CL1, EDNRB, ENTPD1, HMGB1, TNFRSF4, VEGFA and C10orf54 were most highly expressed in m6Acluster-C3 (online supplemental figure S5). CNVs affected numerous IMs and varied across m6A subtypes. m6Acluster-C1 and C2 exhibited frequent amplification and deletion of most IM genes.
jitc-2020-001646supp006.pdf (567.3KB, pdf)
Immune landscape was significantly associated with the expression of m6A regulators
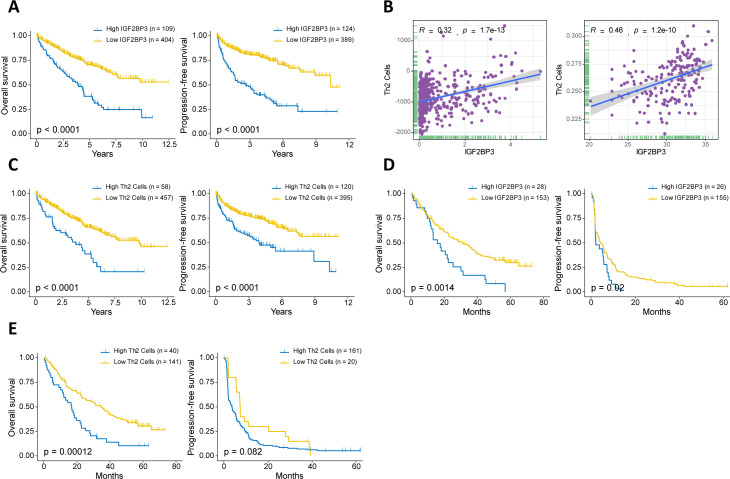
Spearman’s correlation analysis was performed to explore the specific correlation between each m6A regulator and immune cell infiltration. As shown in online supplemental figure S6, there was a widespread correlation between the expression of m6A regulators and immune cell infiltration. We focused on the regulator IGF2BP3—an m6A reader—demonstrating its association with poor survival in ccRCC patients (figure 4A). We revealed that ccRCC samples with high expression levels of IGF2BP3 demonstrated greater Th2-cell infiltration enrichment in both the TCGA-KIRC dataset and the immunotherapeutic cohort (figure 4B). Consistent with a previous study,40 Th2 cells were associated with negative outcomes in ccRCC patients (figure 4C). Furthermore, we explored whether the expression of IGF2BP3 and Th2-cell infiltration affected the efficacy of anti-PD-1 therapy. In the anti-PD-1 cohort, a trend in impaired survival was observed in patients with high expression levels of IGF2BP3 (figure 4D). As expected, high Th2-cell infiltration was also associated with poor survival with PD-1 blockade (figure 4E).
Figure 4.
Relationship between the expression of m6A regulators and immune cell infiltration. (A) Kaplan-Meier curves for patients with high or low IGF2BP3 expression in the TCGA-KIRC cohort. (B) The correlation between IGF2BP3 and the infiltration of Th2 cell in the TCGA-KIRC cohort (Left) and anti-PD-1 therapy cohort (Right). (C) Kaplan-Meier curves for patients with high or low Th2 cell infiltration in the TCGA-KIRC cohort. (D, E) Kaplan-Meier curves depicted the survival differences between patients with high and low IGF2BP3 expression (D) and Th2 cell infiltration (E) in the anti-PD-1 therapy cohort. m6A, N6-methyladenosine.
jitc-2020-001646supp007.pdf (641.3KB, pdf)
Construction of the m6A gene signature
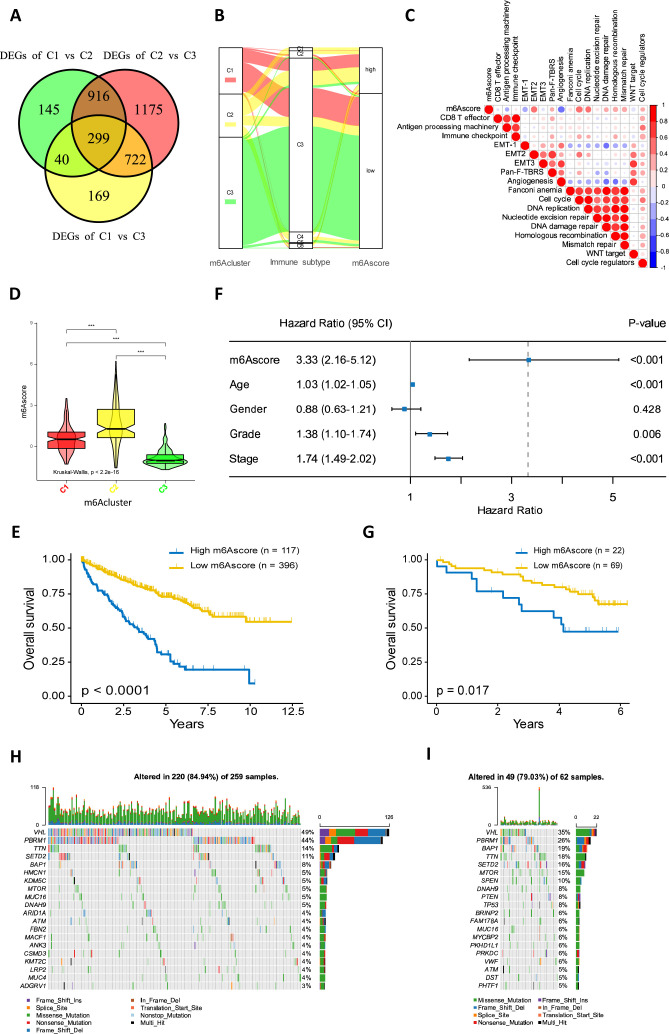
We applied a methodology (known as m6Ascore) to accurately evaluate the m6A modification pattern in individual ccRCC patients. 299 m6A subtype-related DEGs (figure 5A, (online supplemental table S6) were identified using the DESeq2 package of R software. Univariate Cox regression analysis was performed to evaluate the prognosis of each gene in the m6A subtype-related DEGs; 190 genes conferring significant prognoses were extracted for further PCA to establish the m6A gene signature. Changes in the attributes of individual ccRCC patients were visualized with an alluvial diagram which showed that m6Acluster-C2 had the lowest proportion of the C3 immune subtype and was linked to a high m6Ascore (figure 5B).
Figure 5.
Construction of the m6A gene signature. (A) 299 m6A subtype-related genes shown in venn diagram. (B) The changes of m6Aclusters, immune subtypes and m6Ascore depicted by alluvial diagram. (C) Correlations between m6Ascore and the known biological processes in the TCGA-KIRC cohort. (D) Differences in m6Ascore among m6Aclusters. The statistical difference among the clusters was tested by Kruskal-Wallis test. (E) Kaplan-Meier curves depicted the survival difference between low and high m6Ascore patient groups. (F) Multivariate Cox regression analysis for m6Ascore in the TCGA-KIRC cohort shown by the forest plot. (G) Kaplan-Meier curves depicted the survival difference between low and high m6Ascore patient groups in the ICGC dataset. (H, I) The waterfall plot depicted tumor somatic mutation of those with low m6Ascore (H) and high m6Ascore (I). Individual patients represented in each column. The numbers and bar plot on the right showed the mutation frequency of each gene and the proportion of each variant type, respectively. The top bar plot showed tumor mutation burden. DEGs, differentially expressed genes; EMT, epithelial-mesenchymal transition; ICGC, International Cancer Genome Consortium; m6A, N6-methyladenosine.
The correlation between m6Ascore and the known biological processes was analyzed to better demonstrate the features of the m6A gene signature (figure 5C, online supplemental table S8). It was shown that m6Ascore was negatively correlated with APM (r = –0.22, p<0.001), but positively correlated with mismatch repair-relevant signatures, including mismatch repair, DNA damage repair and DNA replication. Furthermore, the Kruskal-Wallis test showed a significant difference in m6Ascore among m6Aclusters (figure 5D). Next, the prognostic value of the m6Ascore in patients with ccRCC was explored. The patients were divided into high or low m6Ascore groups, with optimal cut-off values determined by the ‘survminer’ R package. Patients with high m6Ascores demonstrated significant survival impairment (figure 5E). We further performed multivariate Cox regression analysis (with factors related to patient sex, age, grade and TNM status included) to investigate the independent prognostic value of m6Ascore, revealing that m6Ascore serves as an independent prognostic biomarker for ccRCC patients (figure 5F). The prognostic value of m6Ascore in ccRCC was validated in another cohort (91 cases) from the ICGC database (figure 5G). The differences in the distribution of somatic mutations between the high and low m6Ascores in the TCGA-KIRC cohort were explored using the maftools package (figure 5H, I). We also found that high m6Ascore was relatively depleted for PBRM1 mutations (26% vs 44%) (p=0.009).
m6A modification patterns in the role of anti-PD-1 therapy
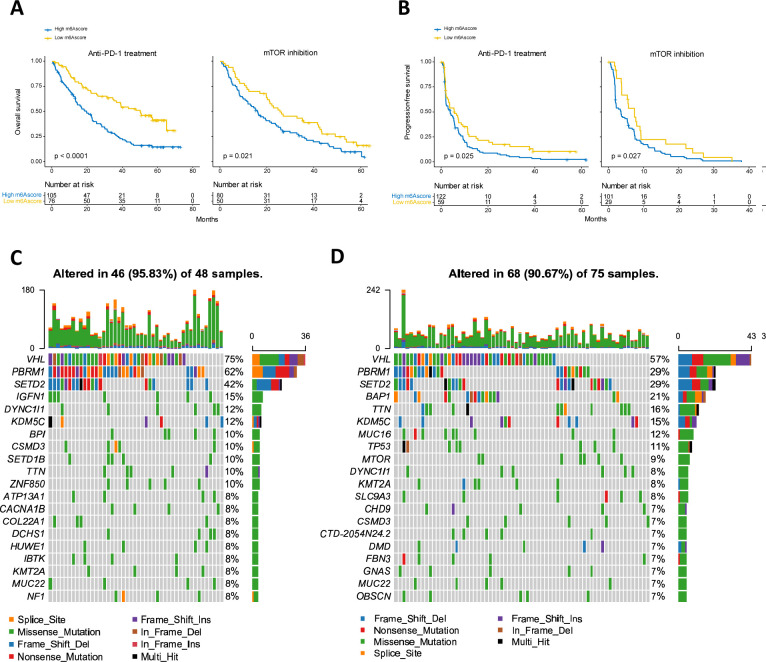
ICTs (eg, anti-PD-1/PD-L1 therapies) have emerged as a critical breakthrough in the field of tumor therapy. We explored the prognostic value of the m6A modification signature in the anti-PD-1 therapy of patients with ccRCC based on one immunotherapeutic cohort. In the anti-PD-1 cohort, the low m6Ascore group presented a markedly prolonged survival (figure 6A, B). In addition, we found that high m6Ascore was relatively depleted for PBRM1 mutations (29% vs 62%) (p<0.001) (figure 6C, D).
Figure 6.
m6A modification patterns in the role of anti-PD-1 therapy. m6Ascore was related to improved progression-free survival (A) and overall survival (B) following anti-PD-1 and mTOR inhibition therapies. (C, D) The waterfall plot depicted tumor somatic mutation of those with low m6Ascore (C) and high m6Ascore (D) in the anti-PD-1 cohort. Individual patients represented in each column. The numbers and bar plot on the right showed the mutation frequency of each gene and the proportion of each variant type, respectively. The top bar plot showed tumor mutation burden. m6A, N6-methyladenosine.
Discussion
Increasing evidence reveals that m6A modification plays critical roles in tumorigenesis, therapeutic resistance and immune response via collaboration among m6A regulators.41 Recently, the role of m6A modification in the tumor immune landscape was comprehensively explored in gastric cancer.26 In this study, we focused on the role of m6A modification in the immune landscape in ccRCC to enhance our understanding of the TIME antitumor immune response and provide more effective immunotherapeutic strategies for patients with ccRCC.
Many previous studies have identified ccRCC subtypes based on genomic profiling,42–44 improving the future application of precision-focused, personalized treatments for ccRCC. A 4-mRNA pattern with significant differences in patient survival was identified by unsupervised analyses based on mRNA expression data.42 In the present study—based on 24 m6A regulators—we identified three m6A modification patterns with significantly distinct immune landscapes, characterized by differences in Th17 signature, extent of ITH, overall cell proliferation, aneuploidy, overall somatic copy number alterations, expression of immunomodulatory genes and prognosis. m6Acluster-C3 was the lowest in both proliferation and ITH, suggesting low tumor growth rates in C3. In addition, C3 presented enrichment pathways related to full immune activation and demonstrated the most pronounced Th17 signature, consistent with a previous study demonstrating that Th17 expression is generally related to improved prognosis.45 m6Acluster-C1 also presented enrichment pathways related to full immune activation and relatively high infiltration of CD8+ T cells, while exhibiting high proliferation and ITH, suggesting high tumor growth rates in C1. Consequently, it was not surprizing that C1 exhibited activated immunity but poor survival. m6Acluster-C2 was predominantly associated with immune suppression of biological processes and relatively low infiltration of CD8+ T cells, exhibiting high proliferation and ITH.
In addition, the correlation between each m6A regulator and each TIME infiltration cell type was explored. Our results revealed that high expression levels of IGF2BP3 demonstrated significantly greater enrichment of Th2-cell infiltration. Strikingly, the high infiltration of Th2 cells and expression of IGF2BP3 were both associated with poor survival with PD-1 blockade. It has been reported that Th2-mediated immunosuppression reduced protective cellular immunity and was found to be related to tumor progression.46 Based on the results described above, we speculated that IGF2BP3-mediated m6A modification may promote the infiltration of Th2 cells, thus decreasing the intratumoral antitumor immune response.
Considering the individual heterogeneity of m6A modification, we applied a methodology to accurately evaluate the m6A methylation pattern of individual ccRCC patients known as m6Ascore. Integrated analyses revealed that m6Ascore is a robust and independent prognostic factor for ccRCC. Our study also found that m6Ascore was negatively correlated with APM and high m6Ascore was relatively depleted for PBRM1 mutations. It has been shown that APM was elevated in patients with a partial or complete response to anti-PD-1 therapy but decreased in those with progressive disease on anti-PD-1 therapy.40 PBRM1 mutations were found to be related to improved response, progression-free survival (PFS) and OS with anti-PD-1 therapy in patients with advanced ccRCC.34 Therefore, the above results indirectly revealed that m6A modification may be a critical factor mediating the clinical response to immunotherapy and indirectly confirmed the value of m6Ascore in predicting immunotherapeutic outcomes.
ICTs (such as anti-PD-1/PD-L1 therapies) have revolutionized the treatment of multiple advanced cancers, including ccRCC.12 13 Although significant clinical benefits are achievable in ccRCC patients receiving ICTs, the immunotherapeutic outcomes exhibited individual heterogeneity.14 Therefore, it is of clinical significance to search for markers to predict the outcomes of immunotherapy. The common paradigm in solid tumor immunology is that pre-existing CD8+ T cell infiltration, coupled with a high number of non-synonymous mutations, drives the response to anti-PD-1 therapy.15 16 19 47 48 However, in contrast to other cancer types, neoantigen load, tumor mutation burden and HLA zygosity were not related to anti-PD-1 therapy response in advanced ccRCC.34 Importantly, it was found that there were no statistical differences in response to or survival following anti-PD-1 therapy between immune infiltrated tumors and immune deserts/excluded tumors in advanced ccRCC.34 In this study, we verified the prognostic value of the m6Ascore in the anti-PD-1 therapy of patients with advanced ccRCC. Thus, the m6Ascore may serve as a predictive strategy for anti-PD-1 therapy.
Consequently, we herein provided a new perspective of immuno-oncology and individualized immunotherapy in ccRCC. However, several limitations should be addressed in our study. First, the infiltration of tumor immune cells was obtained based on algorithms owing to technical limitations. Our analyses were also limited by the lack of clinical cohorts to verify the correlation between m6A modification and tumor immune landscape and the prognostic value of m6Ascore in ccRCC. Therefore, further validation based on large-cohort prospective clinical trials are warranted in the future.
In conclusion, this study revealed the correlation between m6A modification and the tumor immune landscape in ccRCC. Our comprehensive evaluation of m6A modification patterns in individual ccRCC patients enhances our understanding of the tumor immune landscape and provides a new approach toward new and improved immunotherapeutic strategies for ccRCC patients.
Footnotes
JZ and ZL contributed equally.
Contributors: GZ and JZ contributed to the study conception; JZ and ZL conducted the data analysis and were responsible for writing the first draft of the paper. CC, XD and TD revised the paper; and all authors read and approved the final version of the manuscript.
Funding: This work was financed by grants from the National Natural Science Foundation of China (No.81670643, No.81802821, No.81872437 and No.81870483), the Collaborative Innovation Project of Guangzhou Education Bureau (No.1201620011) the Guangzhou Science Technology and Innovation Commission (No.201704020193) and the Science and Technology Planning Project of Guangdong Province (No.2017B030314108).
Competing interests: None declared.
Patient consent for publication: Not required.
Provenance and peer review: Not commissioned; externally peer reviewed.
Data availability statement: All data relevant to the study are included in the article or uploaded as online supplemental information. All data used in this work can be acquired from the GDC portal (https://portal.gdc.cancer.gov/), Broad GDAC Firehose (https://gdac.broadinstitute.org/) and the website (https://gdc.cancer.gov/about-data/publications/panimmune).
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
References
- 1.Roundtree IA, Evans ME, Pan T, et al. Dynamic RNA modifications in gene expression regulation. Cell 2017;169:1187–200. 10.1016/j.cell.2017.05.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang X, Lu Z, Gomez A, et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 2014;505:117–20. 10.1038/nature12730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang X, Zhao BS, Roundtree IA, et al. N(6)-methyladenosine Modulates Messenger RNA Translation Efficiency. Cell 2015;161:1388–99. 10.1016/j.cell.2015.05.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bartosovic M, Molares HC, Gregorova P, et al. N6-Methyladenosine demethylase FTO targets pre-mRNAs and regulates alternative splicing and 3'-end processing. Nucleic Acids Res 2017;45:11356–70. 10.1093/nar/gkx778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xiao W, Adhikari S, Dahal U, et al. Nuclear m(6)A Reader YTHDC1 Regulates mRNA Splicing. Mol Cell 2016;61:507–19. 10.1016/j.molcel.2016.01.012 [DOI] [PubMed] [Google Scholar]
- 6.Roundtree IA, Luo G-Z, Zhang Z, et al. YTHDC1 mediates nuclear export of N6-methyladenosine methylated mRNAs. Elife 2017;6. 10.7554/eLife.31311. [Epub ahead of print: 06 Oct 2017]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang Y, Hsu PJ, Chen Y-S, et al. Dynamic transcriptomic m6A decoration: writers, erasers, readers and functions in RNA metabolism. Cell Res 2018;28:616–24. 10.1038/s41422-018-0040-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen X-Y, Zhang J, Zhu J-S. The role of m6A RNA methylation in human cancer. Mol Cancer 2019;18:103. 10.1186/s12943-019-1033-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li Y, Xiao J, Bai J, et al. Molecular characterization and clinical relevance of m6A regulators across 33 cancer types. Mol Cancer 2019;18:137. 10.1186/s12943-019-1066-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pinello N, Sun S, Wong JJ-L. Aberrant expression of enzymes regulating m6A mRNA methylation: implication in cancer. Cancer Biol Med 2018;15:323–34. 10.20892/j.issn.2095-3941.2018.0365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tong J, Cao G, Zhang T, et al. m6A mRNA methylation sustains Treg suppressive functions. Cell Res 2018;28:253–6. 10.1038/cr.2018.7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rini BI, Plimack ER, Stus V, et al. Pembrolizumab plus axitinib versus sunitinib for advanced renal-cell carcinoma. N Engl J Med 2019;380:1116–27. 10.1056/NEJMoa1816714 [DOI] [PubMed] [Google Scholar]
- 13.Rini BI, Powles T, Atkins MB, et al. Atezolizumab plus bevacizumab versus sunitinib in patients with previously untreated metastatic renal cell carcinoma (IMmotion151): a multicentre, open-label, phase 3, randomised controlled trial. Lancet 2019;393:2404–15. 10.1016/S0140-6736(19)30723-8 [DOI] [PubMed] [Google Scholar]
- 14.Roviello G, Corona SP, Nesi G, et al. Results from a meta-analysis of immune checkpoint inhibitors in first-line renal cancer patients: does PD-L1 matter? Ther Adv Med Oncol 2019;11:1758835919861905. 10.1177/1758835919861905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rizvi NA, Hellmann MD, Snyder A, et al. Cancer immunology. mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science 2015;348:124–8. 10.1126/science.aaa1348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yarchoan M, Hopkins A, Jaffee EM. Tumor mutational burden and response rate to PD-1 inhibition. N Engl J Med 2017;377:2500–1. 10.1056/NEJMc1713444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cristescu R, Mogg R, Ayers M, et al. Pan-tumor genomic biomarkers for PD-1 checkpoint blockade-based immunotherapy. Science 2018;362. 10.1126/science.aar3593. [Epub ahead of print: 12 Oct 2018]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mariathasan S, Turley SJ, Nickles D, et al. Tgfβ attenuates tumour response to PD-L1 blockade by contributing to exclusion of T cells. Nature 2018;554:544–8. 10.1038/nature25501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Binnewies M, Roberts EW, Kersten K, et al. Understanding the tumor immune microenvironment (time) for effective therapy. Nat Med 2018;24:541–50. 10.1038/s41591-018-0014-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen DS, Mellman I. Elements of cancer immunity and the cancer-immune set point. Nature 2017;541:321–30. 10.1038/nature21349 [DOI] [PubMed] [Google Scholar]
- 21.Hugo W, Zaretsky JM, Sun L, et al. Genomic and transcriptomic features of response to anti-PD-1 therapy in metastatic melanoma. Cell 2016;165:35–44. 10.1016/j.cell.2016.02.065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McGranahan N, Furness AJS, Rosenthal R, et al. Clonal neoantigens elicit T cell immunoreactivity and sensitivity to immune checkpoint blockade. Science 2016;351:1463–9. 10.1126/science.aaf1490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang H, Hu X, Huang M, et al. Mettl3-mediated mRNA M6a methylation promotes dendritic cell activation. Nat Commun 2019;10:10. 10.1038/s41467-019-09903-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li H-B, Tong J, Zhu S, et al. m6A mRNA methylation controls T cell homeostasis by targeting the IL-7/STAT5/SOCS pathways. Nature 2017;548:338–42. 10.1038/nature23450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Han D, Liu J, Chen C, et al. Anti-tumour immunity controlled through mRNA m6A methylation and YTHDF1 in dendritic cells. Nature 2019;566:270–4. 10.1038/s41586-019-0916-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang B, Wu Q, Li B, et al. m6A regulator-mediated methylation modification patterns and tumor microenvironment infiltration characterization in gastric cancer. Mol Cancer 2020;19:53. 10.1186/s12943-020-01170-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Colaprico A, Silva TC, Olsen C, et al. TCGAbiolinks: an R/Bioconductor package for integrative analysis of TCGA data. Nucleic Acids Res 2016;44:e71. 10.1093/nar/gkv1507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Scrucca L, Fop M, Murphy TB, et al. mclust 5: clustering, classification and density estimation using Gaussian finite mixture models. R J 2016;8:289–317. 10.32614/RJ-2016-021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Newman AM, Liu CL, Green MR, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods 2015;12:453–7. 10.1038/nmeth.3337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thorsson V, Gibbs DL, Brown SD, et al. The immune landscape of cancer. Immunity 2018;48:812–30. 10.1016/j.immuni.2018.03.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hänzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-Seq data. BMC Bioinformatics 2013;14:7. 10.1186/1471-2105-14-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sotiriou C, Wirapati P, Loi S, et al. Gene expression profiling in breast cancer: understanding the molecular basis of histologic grade to improve prognosis. J Natl Cancer Inst 2006;98:262–72. 10.1093/jnci/djj052 [DOI] [PubMed] [Google Scholar]
- 33.Zeng D, Li M, Zhou R, et al. Tumor microenvironment characterization in gastric cancer identifies prognostic and Immunotherapeutically relevant gene signatures. Cancer Immunol Res 2019;7:737–50. 10.1158/2326-6066.CIR-18-0436 [DOI] [PubMed] [Google Scholar]
- 34.Braun DA, Hou Y, Bakouny Z, et al. Interplay of somatic alterations and immune infiltration modulates response to PD-1 blockade in advanced clear cell renal cell carcinoma. Nat Med 2020;26:909–18. 10.1038/s41591-020-0839-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Szklarczyk D, Franceschini A, Wyder S, et al. String v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res 2015;43:D447–52. 10.1093/nar/gku1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 2003;13:2498–504. 10.1101/gr.1239303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Barabási A-L, Gulbahce N, Loscalzo J. Network medicine: a network-based approach to human disease. Nat Rev Genet 2011;12:56–68. 10.1038/nrg2918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Panneerdoss S, Eedunuri VK, Yadav P, et al. Cross-talk among writers, readers, and erasers of m6A regulates cancer growth and progression. Sci Adv 2018;4:eaar8263. 10.1126/sciadv.aar8263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tang J, Shalabi A, Hubbard-Lucey VM. Comprehensive analysis of the clinical immuno-oncology landscape. Ann Oncol 2018;29:84–91. 10.1093/annonc/mdx755 [DOI] [PubMed] [Google Scholar]
- 40.Şenbabaoğlu Y, Gejman RS, Winer AG, et al. Tumor immune microenvironment characterization in clear cell renal cell carcinoma identifies prognostic and immunotherapeutically relevant messenger RNA signatures. Genome Biol 2016;17:231. 10.1186/s13059-016-1092-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chang G, Leu J-S, Ma L, et al. Methylation of RNA N6-methyladenosine in modulation of cytokine responses and tumorigenesis. Cytokine 2019;118:35–41. 10.1016/j.cyto.2018.06.018 [DOI] [PubMed] [Google Scholar]
- 42.Cancer Genome Atlas Research Network Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature 2013;499:43–9. 10.1038/nature12222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brooks SA, Brannon AR, Parker JS, et al. ClearCode34: a prognostic risk predictor for localized clear cell renal cell carcinoma. Eur Urol 2014;66:77–84. 10.1016/j.eururo.2014.02.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wu P, Liu J-L, Pei S-M, et al. Integrated genomic analysis identifies clinically relevant subtypes of renal clear cell carcinoma. BMC Cancer 2018;18:287. 10.1186/s12885-018-4176-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Punt S, Langenhoff JM, Putter H, et al. The correlations between IL-17 vs. Th17 cells and cancer patient survival: a systematic review. Oncoimmunology 2015;4:e984547. 10.4161/2162402X.2014.984547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chtanova T, Mackay CR. T cell effector subsets: extending the Th1/Th2 paradigm. Adv Immunol 2001;78:233–66. 10.1016/s0065-2776(01)78005-4 [DOI] [PubMed] [Google Scholar]
- 47.Snyder A, Makarov V, Merghoub T, et al. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N Engl J Med 2014;371:2189–99. 10.1056/NEJMoa1406498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Van Allen EM, Miao D, Schilling B, et al. Genomic correlates of response to CTLA-4 blockade in metastatic melanoma. Science 2015;350:207–11. 10.1126/science.aad0095 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
jitc-2020-001646supp002.pdf (369.7KB, pdf)
jitc-2020-001646supp001.xlsx (345.5KB, xlsx)
jitc-2020-001646supp008.pdf (86.5KB, pdf)
jitc-2020-001646supp003.pdf (1.7MB, pdf)
jitc-2020-001646supp004.pdf (1,019.8KB, pdf)
jitc-2020-001646supp005.pdf (566.5KB, pdf)
jitc-2020-001646supp006.pdf (567.3KB, pdf)
jitc-2020-001646supp007.pdf (641.3KB, pdf)