To the editor:
Zinc-finger nucleases (ZFNs) consist of an engineered zinc-finger array fused to a nuclease domain. Dimers of ZFNs can create targeted double-strand DNA breaks, which can stimulate highly efficient gene targeting in many cell types1 (Supplementary Fig. 1 online). We found that the modular assembly method of engineering zinc-finger arrays has an unexpectedly higher failure rate than previously reported.
Modular assembly advocates linking individual zinc fingers, each of which typically binds to a 3-bp ‘subsite’ (Supplementary Fig. 2 online). Two large-scale surveys have suggested that modular assembly is highly effective (100% (ref. 2) and 60% (ref. 3) success rates) for making three-finger arrays designed to bind 9-bp target sites. The Zinc Finger Consortium recently assembled an archive of 141 previously published finger modules3–5 encoded on a standardized platform6. Our initial experiences using these reagents suggested that modular assembly was inefficient (Supplementary Discussion and Supplementary Table 1 online).
To perform a larger-scale test, we assembled 168 zinc-finger arrays designed for 104 diverse target DNA sites (Supplementary Table 2 and Supplementary Methods online). We tested these domains for DNA binding using a bacterial two-hybrid (B2H) assay6, which accurately identifies arrays that lack activity as ZFNs in human cells (Supplementary Discussion and Supplementary Figs. 3 and 4 online). For 79 of the 104 target sites, we did not obtain a single three-finger array that scored positively in the B2H assay (overall failure rate of ~76%; Fig. 1a and Supplementary Table 2). Notably, modular assembly was far less effective for target sites composed of two, one, or no GXX subsites (where X is any base) compared with those composed of three GXX subsites (Fig. 1a). Additionally, because ZFNs function as dimers, we would expect failure rates for making a functional ZFN pair to be even higher (Supplementary Discussion). Notably, these values are all likely underestimates of actual failure rates because not all zinc-finger arrays that are positive in the B2H assay will be active as ZFNs in human cells (Supplementary Fig. 4).
Figure 1 |.
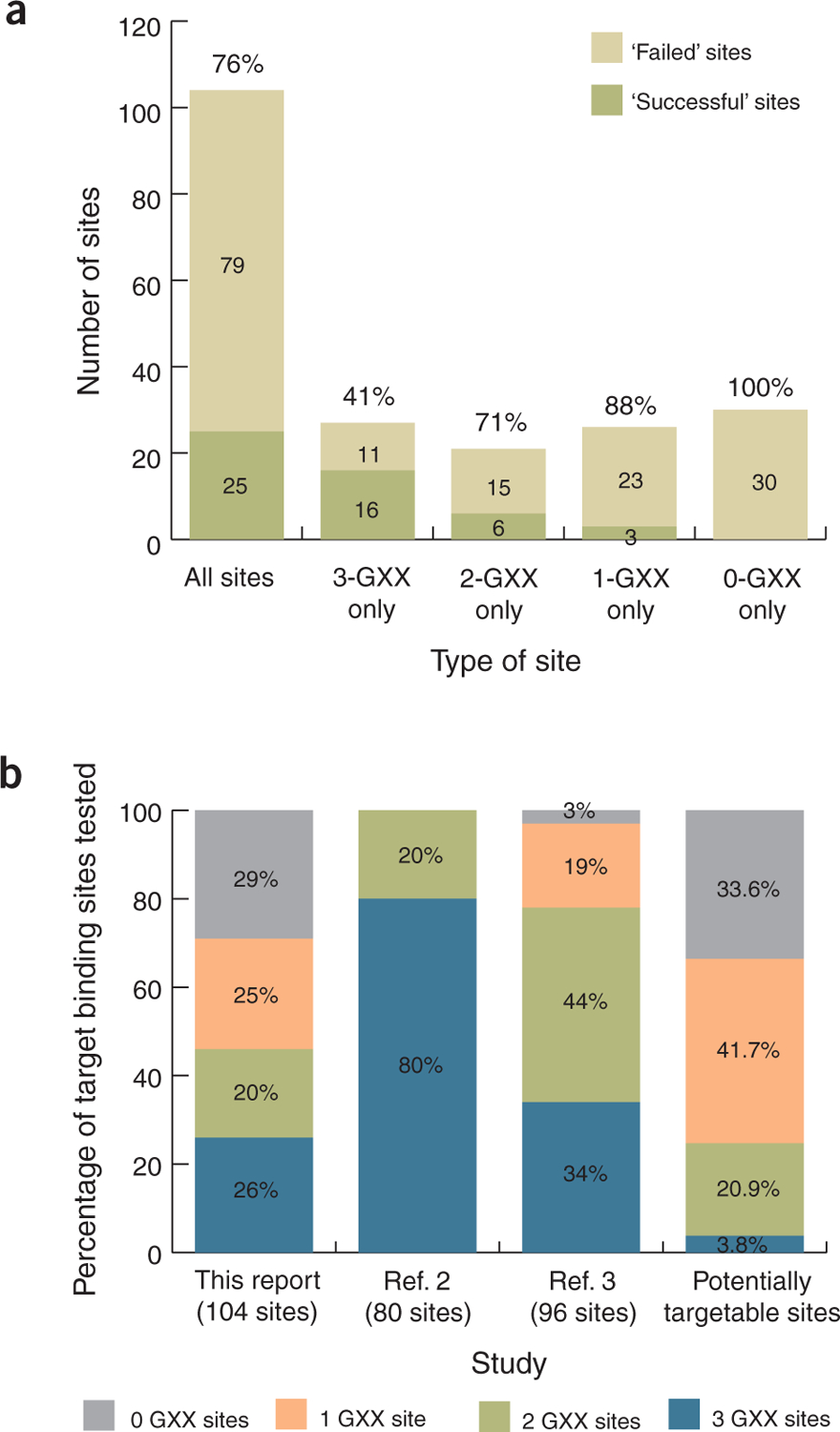
Large-scale evaluation of the modular assembly method for engineering zinc-finger arrays. (a) Data for success and failure of modular assembly as judged by the B2H assay for all 104 target DNA sites (see Supplementary Discussion for definition of success threshold) and for subsets of target sites containing three, two, one or no GXX subsites. ‘Successful’ sites are those for which at least one functional zinc-finger array was identified; ‘failed’ sites are those for which we failed to obtain a single successful array (see Supplementary Table 2 for details). Predicted failure rates for each set of target sites are indicated above the bars. (b) Distributions of target DNA sites used in different modular assembly evaluation studies according to the number of GXX subsites present. The distribution of all 107,011 potential 9-bp sites that can be targeted using 141 zinc-finger modules from the Zinc Finger Consortium Modular Assembly Kit 1.0 (ref. 6) is also shown (far right bar).
One reason for the apparent discrepancy between previously published studies2,3 and our results is that the former primarily used 9-bp sites composed of two or three GXX subsites, whereas we used sites with a more varied number of GXX subsites (Fig. 1b); this difference will critically affect the observed failure rate (Fig. 1a). Our study represents a more meaningful evaluation because target sites containing one or no GXX subsites (underrepresented in previous studies) encompass the majority (>75%) of the 107,011 potential 9-bp sites that can be targeted with existing modules (Fig. 1b).
Our results strongly suggest that potential users of modular assembly should expect that this method will fail to yield a functional three–zinc finger array for the majority of potentially targetable sites. To emphasize this we have modified content on the Zinc Finger Consortium website (http://www.zincfingers.org/; Supplementary Methods). Highly effective but more labor-intensive selection-based methods for engineering zinc-finger arrays have been previously described (Supplementary Discussion), and at present these are the only publicly available alternatives for academic researchers interested in using ZFN technology.
Supplementary Material
ACKNOWLEDGMENTS
We thank D. Dobbs and P. Zaback for helpful discussions and E. Unger-Wallace (Iowa State University) for contributions to zinc-finger array construction. J.K.J. is supported by the US National Institutes of Health (R21 RR024189), the Cystic Fibrosis Foundation (MCCRAY07G0) and the Massachusetts General Hospital Department of Pathology; D.F.V. is supported by a US National Science Foundation grant DBI 0501678; T.C. is supported by the German Research Foundation (SPP1230 CA311/2); C.L.R. is supported by a Ford Foundation Pre-Doctoral Diversity Fellowship and a National Science Foundation Graduate Research Fellowship; T.I.C. is supported by a fellowship from the Swiss Foundation for Grants in Biology and Medicine.
Footnotes
Note: Supplementary information is available on the Nature Methods website.
References
- 1.Porteus MH & Carroll D Nat. Biotechnol 23, 967–973 (2005). [DOI] [PubMed] [Google Scholar]
- 2.Segal DJ et al. Biochemistry 42, 2137–2148 (2003). [DOI] [PubMed] [Google Scholar]
- 3.Bae KH et al. Nat. Biotechnol 21, 275–280 (2003). [DOI] [PubMed] [Google Scholar]
- 4.Mandell JG & Barbas CF Nucleic Acids Res 34, W516–W523 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu Q, Xia Z, Zhong X & Case CC J. Biol. Chem 277, 3850–3856 (2002). [DOI] [PubMed] [Google Scholar]
- 6.Wright DA et al. Nat. Protoc 1, 1637–1652 (2006). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


