Abstract
Nuclear localization of the androgen receptor (AR) is necessary for its activation as a transcription factor. Defining the mechanisms regulating AR nuclear localization in androgen-sensitive cells and how these mechanisms are dysregulated in castration-resistant prostate cancer (CRPC) cells is fundamentally important and clinically relevant. According to the classical model of AR intracellular trafficking, androgens induce AR nuclear import and androgen withdrawal causes AR nuclear export. The present study has led to an updated model that AR could be imported in the absence of androgens, ubiquitinated, and degraded in the nucleus. Androgen withdrawal caused nuclear AR degradation, but not export. In comparison with their parental androgen-sensitive LNCaP prostate cancer cells, castration-resistant C4-2 cells exhibited reduced nuclear AR polyubiquitination and increased nuclear AR level. We previously identified 3-(4-chlorophenyl)-6,7-dihydro-5H-pyrrolo[1,2-a]imidazole (CPPI) in a high-throughput screen for its inhibition of androgen-independent AR nuclear localization in CRPC cells. The current study shows that CPPI is a competitive AR antagonist capable of enhancing AR interaction with its E3 ligase MDM2 and degradation of AR in the nuclei of CRPC cells. Also, CPPI blocked androgen-independent AR nuclear import. Overall, these findings suggest the feasibility of targeting androgen-independent AR nuclear import and stabilization, two necessary steps leading to AR nuclear localization and activation in CRPC cells, with small molecule inhibitors.
Keywords: Oncology, Therapeutics
Keywords: Drug therapy, Prostate cancer
Introduction
Understanding mechanisms regulating androgen receptor (AR) nuclear localization may lead to new approaches to inhibiting AR signaling, which is arguably the best therapeutic target for prostate cancer (PCa) — the most commonly diagnosed cancer and the second leading cause of cancer death in US male patients (1). Since PCa growth requires AR signaling, androgen-deprivation therapy (ADT) is a front-line treatment in addition to radical prostatectomy and radiotherapy for patients with high-risk localized disease or metastatic PCa. However, ADT is not curative and becomes ineffective when patients relapse with castration-resistant prostate cancer (CRPC), mainly due to reactivation of AR signaling. Reactivated AR can be inhibited by second-generation antiandrogens including abiraterone and enzalutamide, which were approved by the FDA for the treatment of CRPC (2–7). However, patients develop resistance to enzalutamide and/or abiraterone (8, 9), again mainly caused by AR reactivation (10, 11). Novel approaches to disabling AR may lead to new therapies for CRPC, including those resistant to second-generation antiandrogens.
AR is a ligand-dependent nuclear transcription factor (TF) and member of the steroid hormone receptor superfamily (12). Like other nuclear hormone receptors, the structure of AR comprises 4 distinct functional domains: an amino-terminal domain (NTD), a carboxy-terminal ligand-binding domain (LBD), a DNA-binding domain (DBD), and a flexible hinge region (H), which joins the LBD and DBD (13). Dihydrotestosterone (DHT) is the most active physiological agonist of AR. Without ligand, the AR is localized primarily in the cytoplasm and associated with heat shock protein chaperones (HSP90, HSP70, HSP56) (reviewed in ref. 14). Binding of DHT leads to AR homodimerization, translocation to nucleus, and recruitment to the androgen response elements (AREs) to initiate transcription (15–17). In CRPC, AR appears to localize predominantly to the nucleus even in the absence of androgens (18). Thus, defining the mechanisms leading to androgen-independent AR nuclear localization may lead to new approaches to targeting AR in CRPC.
Intracellular trafficking of AR is thought to play an important role in regulating AR levels in the nucleus. Our group previously identified a potential nuclear export signal (NES) in the LBD of AR, termed NESAR (19). NESAR consists of amino acids 743-817 of AR and appears to play an important role in driving AR cytoplasmic localization. A subsequent study showed that NESAR is a major signal for AR polyubiquitination and proteasomal degradation (20). However, the mechanism of NESAR-mediated export and how NESAR could function both as an export signal and a degradation signal are not clear.
Here, we reevaluated the role of intracellular trafficking and turnover in regulating AR nuclear localization in PCa cells, using fluorescent microscopy and nucleocytoplasmic fractionation coupled with 2 different pulse-chase methods. We also investigated the inhibition of androgen-independent AR nuclear localization in CRPC by a small molecule, 3-(4-chlorophenyl)-6,7-dihydro-5H-pyrrolo[1,2-a]imidazole (CPPI). Our findings provide insights into the mechanisms of regulation and potential targeting of androgen-independent AR nuclear localization in CRPC.
Results
NESAR modulates AR nuclear degradation.
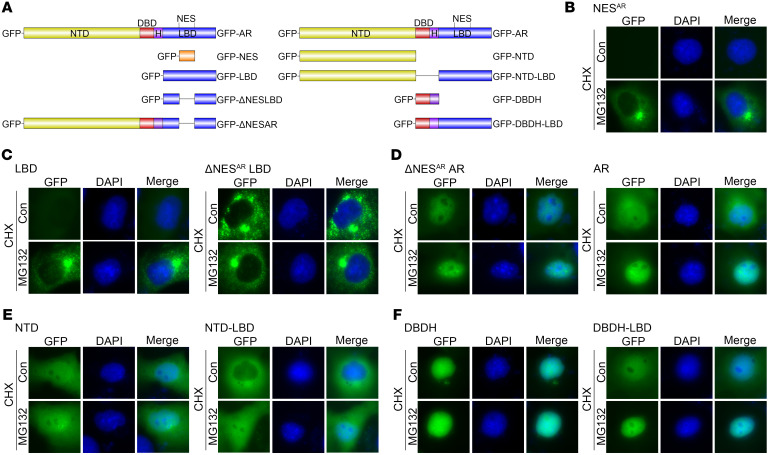
To study the role of NESAR as an export signal versus a degradation signal in regulating AR subcellular localization, we tested the impact of the proteasome inhibitor MG132 on a series of GFP-tagged AR deletion constructs (Figure 1A) expressed in transfected COS-7 cells (Supplemental Figure 1A; supplemental material available online with this article; https://doi.org/10.1172/JCI141335DS1). Cycloheximide (CHX) and MG132 were used to block de novo protein synthesis and proteasomal degradation of the expressed proteins, respectively, which allowed for tracking the subcellular localization of the expressed AR deletion constructs. Figure 1B and Supplemental Figure 1B show that the GFP-NESAR was almost undetectable in the CHX treatment group, indicating rapid degradation of the construct. In contrast, MG132 treatment prevented the decay of intracellular levels of GFP-NESAR, which is consistent with NESAR being a potent degron. In Figure 1C and Supplemental Figure 1C, it is shown that MG132 increased the level of GFP-LBD, but not GFP-ΔNESARLBD, indicating a critical role for NESAR in mediating LBD degradation. MG132 did not influence the subcellular localization of additional constructs lacking NESAR, including GFP-ΔNESARAR, GFP-NTD, and GFP-DBDH; these constructs were localized predominantly to the nucleus (Figure 1, D–F, and Supplemental Figure 1, D–F). In contrast, GFP-AR, GFP-NTD-LBD, and GFP-DBDH-LBD, which contain NESAR, exhibited a shift toward the nucleus in the presence of MG132 (Figure 1, D–F, and Supplemental Figure 1, D–F). These findings suggest that these NESAR-containing protein constructs can undergo nuclear import and that their degradation in the nucleus can be inhibited by the MG132. These observations also suggest that NESAR in these constructs could enhance cytoplasmic localization through promoting nuclear degradation. Thus, NESAR is potentially a potent nuclear degradation signal in AR.
Figure 1. Effect of MG132 on subcellular localization of GFP-tagged AR and AR deletion constructs.
(A) Diagrams of GFP-tagged AR and AR deletion constructs: GFP-AR, GFP-NESAR, GFP-LBD, GFP-ΔNESARLBD, GFP-ΔNESARAR, GFP-NTD, GFP-NTD-LBD, GFP-DBDH, and GFP-DBDH-LBD. (B–F) Representative fluorescent images of transfected GFP-NESAR (B), GFP-LBD, GFP-ΔNESARLBD (C), GFP-ΔNESARAR, GFP-AR (D), GFP-NTD, GFP-NTD-LBD (E), GFP-DBDH, and GFP-DBDH-LBD (F) in COS-7 cells 24 hours after treatment with DMSO, CHX, and/or MG132. Con, control. Data represent 1 of 2 independent experiments with consistent results. Original magnification, ×40.
Imported AR is degraded in the nucleus following DHT withdrawal.
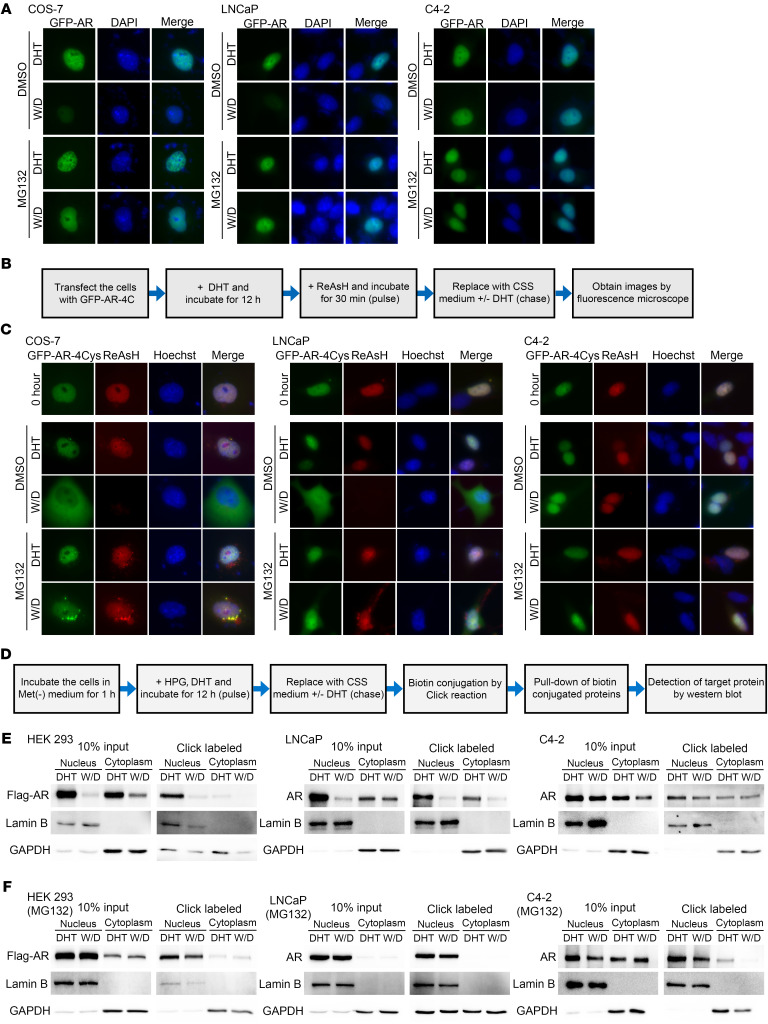
Identifying NESAR as a potential potent nuclear degradation signal in AR raised a question regarding the paradigm that AR is exported after androgen withdrawal (21, 22). To explore the fate of imported AR upon androgen withdrawal, we transfected GFP-AR into COS-7 (AR negative), LNCaP (AR positive, hormone sensitive), and C4-2 (AR positive, castration resistant) cells. AR nuclear import was induced with DHT, followed by DHT withdrawal. The GFP expression vector served as the negative control, showing that all treatments did not influence the overall intracellular distribution of GFP protein in COS-7, LNCaP, and C4-2 cells (Supplemental Figure 1G). Moreover, MG132 did not influence the nuclear export of NESPKI, a classic export signal (ref. 19 and Supplemental Figure 1H). As shown in Figure 2A and Supplemental Figure 2A, after DHT withdrawal, the nuclear GFP-AR signal decreased dramatically in COS-7 and LNCaP cells and, to a lesser extent, in C4-2 cells. MG132 effectively prevented the decrease of the nuclear GFP-AR signal after DHT withdrawal in all 3 cell lines. We did not observe an increase in cytoplasmic GFP-AR signal following DHT withdrawal, either in the presence or absence of MG132, suggesting that AR was not exported after DHT withdrawal. These observations were also confirmed by immunofluorescence staining and Western blot analysis for endogenous AR in LNCaP cells (Supplemental Figure 2, B and C).
Figure 2. Imported nuclear AR is degraded and not exported following DHT withdrawal.
(A) Representative fluorescent images of transfected GFP-AR in COS-7, LNCaP, or C4-2 cells treated with 10 nM DHT followed by DHT withdrawal (W/D) and CHX in presence or absence of MG132. (B) Schematic of ReAsH pulse-chase experiments. (C) Representative fluorescent images of ReAsH pulse-labeled GFP-AR-4Cys. Green signal represented total GFP-AR-4Cys protein, whereas red signal was derived from ReAsH-labeled GFP-AR-4Cys protein. Original magnification, ×40.(D) Schematic of Click pulse-chase analysis. (E and F) Western blot detection of the pulsed labeled protein in cells cultured in the absence (E) or presence (F) of MG132, with (DHT) or without (W/D) DHT. Data represent 1 of at least 2 independent experiments with consistent results.
To further explore this observation, we used 2 different pulse-chase methods, ReAsH and Click chemistry. COS-7, LNCaP, and C4-2 cells were transfected with GFP-AR containing a tetra-cysteine tag (GFP-AR-4Cys). ReAsH, a red fluorescent dye that forms a covalent link with the tetra-cysteine motif and can be used for live cell imaging (23), was then used to label GFP-AR-4Cys (Figure 2B). As shown in Supplemental Figure 2D, GFP-AR-4Cys, but not GFP-AR, exhibited a red fluorescent signal after wash, suggesting specific pulse labeling of GFP-AR-4Cys by ReAsH. After DHT withdrawal, the pulse-labeled GFP-AR-4Cys (ReAsH, red) in the nucleus was reduced dramatically in COS-7 and LNCaP cells and slightly in C4-2 cells (Figure 2C and Supplemental Figure 2, E and F). Although the ReAsH signal in the nucleus was significantly decreased (P < 0.01) in COS-7 and LNCaP cells, it was virtually undetectable in the cytoplasm following DHT withdrawal. As expected, MG132 maintained the ReAsH-labeled nuclear AR level without enhancing its level in the cytoplasm, further indicating that ReAsH-labeled nuclear AR was degraded instead of being exported following DHT withdrawal in COS-7 and LNCaP cells. In C4-2 CRPC cells, ReAsH-labeled nuclear AR appeared to be more stable, but also was not exported following DHT withdrawal.
A second approach was to pulse-chase exogenous or endogenous AR with the methionine analog homopropargylglycine (HPG) coupled with Click chemistry (24–26), nucleocytoplasmic fractionation, and Western blot analysis (Figure 2D). Figure 2E shows that the pulse-labeled nuclear AR was markedly reduced after DHT withdrawal in HEK293 and LNCaP cells and slightly decreased in C4-2 cells, while cytoplasmic pulse-labeled AR was not increased. Consistent with CHX and ReAsH assay results, MG132 stabilized the pulse-labeled AR level in the nuclei of both HEK293 and LNCaP (Figure 2F). MG132 did not enhance the level of the pulse-labeled AR in C4-2 nuclei following DHT withdrawal (Figure 2F). Also, MG132 did not enhance the pulse-labeled cytoplasmic AR level in all 3 cell lines (Figure 2F). Together, these results suggest that after DHT withdrawal, nuclear AR is degraded via the proteasome pathway and is not exported in COS-7 and LNCaP cells. In contrast, nuclear AR was less sensitive to DHT withdrawal and proteasome-mediated degradation in C4-2 cells, suggesting that AR is stabilized in the nuclei of CRPC cells in the absence of androgens.
AR can be imported into the nucleus in the absence of androgens.
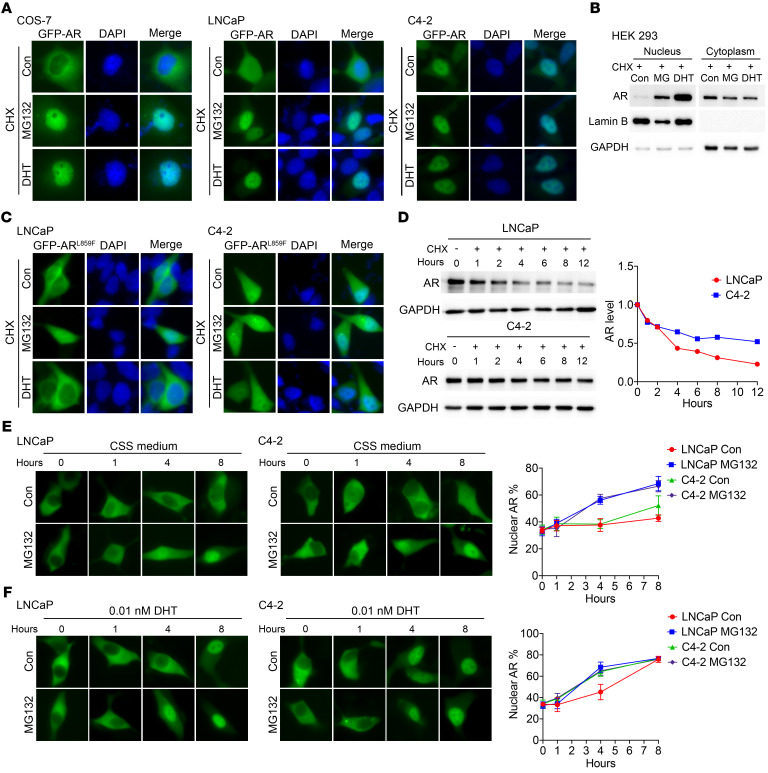
GFP-AR was localized to the nuclei in the presence of MG132 (Figure 1D), suggesting that AR can be imported to the nucleus in the absence of androgens. To determine whether AR could indeed be imported in the absence of androgens, COS-7, LNCaP, and C4-2 cells were transfected with GFP-AR in androgen-free medium. Figure 3A and Supplemental Figure 3A show differential GFP-AR localization in COS-7, LNCaP, and C4-2 cells in the absence of androgens, with GFP-AR being localized predominantly in the cytoplasm in COS-7, almost evenly distributed in the hormone-sensitive PCa (HSPC) LNCaP cells, and predominantly in the nucleus in castration-resistant C4-2 cells. As expected, in the presence of MG132, GFP-AR was imported into the nucleus, either in the presence or absence of DHT. The Western blot results of transfected AR in HEK293 cells confirmed that AR can be imported into the nucleus in the absence of androgens (Figure 3B). This result was also reproduced using the ReAsH pulse-chase assay (Supplemental Figure 3, B and C).
Figure 3. Unliganded AR can be imported into the nucleus in the presence of MG132.
(A) Representative fluorescent images of GFP-AR in COS-7, LNCaP, and C4-2 cells 24 hours after treatment with MG132 or DHT in CSS medium containing CHX. (B) Western blot analysis of nuclear and cytoplasmic extracts of transiently transfected Flag-AR in HEK293 cells after treatment with MG132 or DHT in CSS medium containing CHX. (C) Representative fluorescent images of GFP-ARL859F after treatment with MG132 or DHT for 24 hours in LNCaP or C4-2 cells in CSS medium containing CHX. (D) Effect of CHX on the protein levels of endogenous AR at indicated time points in LNCaP or C4-2 cells. (E and F) GFP-AR–transfected LNCaP and C4-2 cells were pretreated with CPPI for 24 hours and then replaced with fresh CSS medium without CPPI, but containing no DHT (E) (n = 6) or 0.01 nM DHT (F) (n = 6) with or without MG132. The GFP-AR images were detected at indicated time points after the medium replacement. Nuclear GFP-AR quantification data are shown at right. Quantitative data are presented as mean ± SD, and all data represent 1 of at least 2 independent experiments with consistent results. Original magnification, ×40.
Next, we constructed an AR mutant, GFP-ARL859F, which has been found in patients with androgen-insensitivity syndromes and does not respond to androgens (27). Transfected GFP-ARL859F was localized predominantly in the cytoplasm in LNCaP cells and evenly distributed in C4-2 cells cultured in androgen-free media. The ARL859F did not respond to DHT, but translocated to the nucleus in the presence of MG132 in LNCaP cells (Figure 3C and Supplemental Figure 3D). In CRPC C4-2 cells, MG132 only slightly increased the nuclear localization of ARL859F (Figure 3C and Supplemental Figure 3D), indicating that ARL859F was stabilized and less sensitive to proteasome degradation in C4-2 nuclei. These results suggest that AR can be imported into the nucleus independently of androgens and that increased nuclear stability of AR is responsible for androgen-independent AR nuclear localization in CRPC. The above finding was consistent with the increased stability of endogenous AR in C4-2 relative to LNCaP (Figure 3D and ref. 18).
We also determine whether the AR nuclear import rate is different between LNCaP and C4-2 cells. First, cells were transfected with GFP-AR and then treated with CPPI, a small molecule that can cause AR cytoplasmic localization in PCa cells (28). GFP-AR was localized predominantly in the cytoplasm in C4-2 cells in the presence of CPPI. After switching to CPPI-free medium, the cytoplasmic GFP-AR underwent nuclear import in response to DHT in a dose-dependent manner (Supplemental Figure 4A), suggesting that GFP-AR remains active after CPPI treatment. Again, we used MG132 to suppress protein degradation so that GFP-AR could be stabilized and visualized after nuclear import. As expected, in the presence MG132, the imported nuclear GFP-AR level increased in both LNCaP and C4-2 cells either in the absence (Figure 3E) or presence of 0.01 nM DHT (Figure 3F). In this experiment, GFP-AR nuclear import time courses were virtually identical between LNCaP and C4-2 cells after CPPI withdrawal in the presence of MG132. However, in the absence of MG132, the nuclear level of GFP-AR appeared to increase faster in C4-2 cells than in LNCaP cells, which is consistent with the increased stability of imported GFP-AR in C4-2 cells. This finding was reproduced using the ReAsH pulse-chase assay (Supplemental Figure 4, B and C). These results suggest that LNCaP and C4-2 cells have similar AR nuclear import rates but different AR degradation rates in the nucleus, with AR being more stable in C4-2 nuclei.
AR is polyubiquitinated in the nucleus.
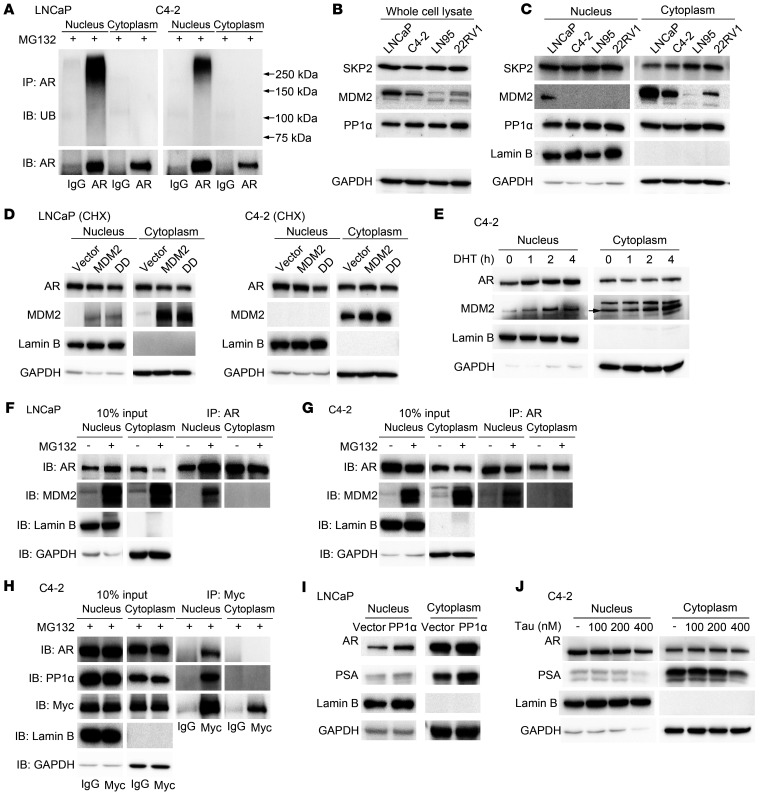
Since proteasomal AR nuclear degradation appeared to play an important role in regulating nuclear AR levels, we tested to determine whether AR polyubiquitination occurs efficiently in the nucleus. Figure 4A shows that AR polyubiquitination is readily detectable in the nuclear, but not cytoplasmic extracts, in both LNCaP and C4-2 cells. Data shown in Figure 4A also suggests that nuclear AR polyubiquitination was more efficient in LNCaP than in C4-2 cells, which is consistent with the observation that AR was less stable in LNCaP than in C4-2 cells (Figure 3D and ref. 18).
Figure 4. AR is polyubiquitinated and associated with E3 ligase MDM2 in the nuclei.
(A) Western blot analysis of ubiquitin (UB) and AR in LNCaP and C4-2 cells in the presence of MG132. (B and C) Western blot analysis of AR, SKP2, MDM2, and PP1α in whole-cell lysate (B) or nucleocytoplasmic fractions (C). (D) LNCaP and C4-2 cells were transfected with MDM2 or MDM2S166D, S186D (DD) expression vector. Expression levels of nuclear and cytoplasmic AR in presence of CHX were detected through Western blot. (E) C4-2 cells were cultured in CSS medium for 48 hours, and the time course of AR and MDM2 localization after DHT treatment was detected through Western blot. (F and G) Nuclear and cytoplasmic extracts were prepared from the MG132-treated LNCaP (F) and C4-2 (G) for IP with anti-AR antibody. Immunoblotting was performed using indicated antibodies. (H) IP was performed in myc-MDM2–transfected C4-2 cells with anti-myc antibody. IB was conducted with indicated antibodies. (I) Western blot analysis of protein levels of AR in LNCaP cells after PP1α transfection in CSS medium. (J) Effect of tautomycin (Tau) on expression levels of nuclear and cytoplasm AR was detected through Western blot in C4-2 cells. Data represent 1 of at least 2 independent experiments with consistent results.
AR polyubiquitination is catalyzed by E3 ligases, and several AR E3 ligases were identified previously, including SKP2 (29), MDM2 (30, 31), and CHIP (32). To explore the potential mechanism(s) regulating AR polyubiquitination in the nucleus, the expression levels and subcellular localization of 2 extensively studied E3 ligases, SKP2, MDM2, and one of their regulators, PP1α (33), were tested via Western blot in several PCa cell lines. The E3 ligase MDM2 was downregulated in CRPC cell lines C4-2, LNCaP95, and 22RV1 as compared with the HSPC cell line, LNCaP (Figure 4, B and C). To test the role of MDM2 in AR nuclear degradation, we transfected the myc-MDM2 and MDM2S166D,S186D constitutively active mutant (34) in LNCaP and C4-2 cells. The results showed that the nuclear AR level appeared to decrease in MDM2S166D,S186D transfected LNCaP and C4-2 cells, with more nuclear AR reduction in C4-2 cells (Figure 4D). However, MDM2 was mainly localized in the cytoplasm (Figure 4, C and D). This raised a question regarding how cytoplasmic MDM2 could induce AR degradation in the nucleus. Western blot analysis of nuclear and cytoplasmic extracts from DHT-treated C4-2 cells showed a time-dependent nuclear import of AR accompanied by an increased nuclear MDM2 level (Figure 4E), suggesting that cytoplasmic MDM2 could also be imported with AR into the nucleus. Furthermore, co-IP detected a physical interaction between endogenous MDM2 and endogenous AR in the nucleus, but not in the cytoplasm, in LNCaP (Figure 4F) and C4-2 (Figure 4G) cells in the presence of MG132, suggesting that MDM2 binds to AR and causes its degradation in the nucleus when the proteasome is not inhibited. Transfected myc-MDM2 also interacted with AR in the nucleus, but not in the cytoplasm in C4-2 cells in co-IP analysis (Figure 4H). The interaction between MDM2 and AR was also confirmed by MDM2-knockdown assay. The increased nuclear AR and decreased polyubiquitination were observed after MDM2 siRNA transfection in C4-2 cells (Supplemental Figure 5, A and B). These findings suggest that MDM2 is one of the E3 ligases that can regulate the nuclear degradation of AR in PCa cells.
We also tested the function of PP1α in regulating AR levels in the nucleus, because PP1α was increased in CRPC cell lines in both the nucleus and cytoplasm (Figure 4, B and C) and this protein phosphatase can suppress SKP2 and MDM2 activity by dephosphorylation (33). Co-IP showed PP1α interaction with myc-MDM2 in the nucleus, but not in the cytoplasm, in C4-2 cells (Figure 4H). Transfection of PP1α expression vector in LNCaP cells (Supplemental Figure 5C), which express PP1α at a low level (Figure 4, B and C), increased nuclear AR level and its target gene PSA expression in the absence of androgens (Figure 4I). Meanwhile, tautomycin, a PP1α inhibitor (35), reduced the nuclear AR level in a dose-dependent manner (Figure 4J). These findings suggest that PP1α could induce AR nuclear localization and activation in PCa cells, possibly via inhibiting MDM2 (35).
Since androgens can induce AR nuclear localization and stabilize AR via inhibiting polyubiquitination (20), it is likely that androgens suppress AR polyubiquitination in the nucleus. Supplemental Figure 5, D and E, showed that synthetic androgen R1881 suppressed polyubiquitination of transfected flag-AR in the nuclei of HEK293 cells or endogenous AR in LNCaP cells. This result suggests that androgens not only induce AR nuclear import, but also inhibited AR polyubiquitination and subsequent proteasome-mediated degradation.
CPPI induces AR nuclear degradation and inhibits AR nuclear import.
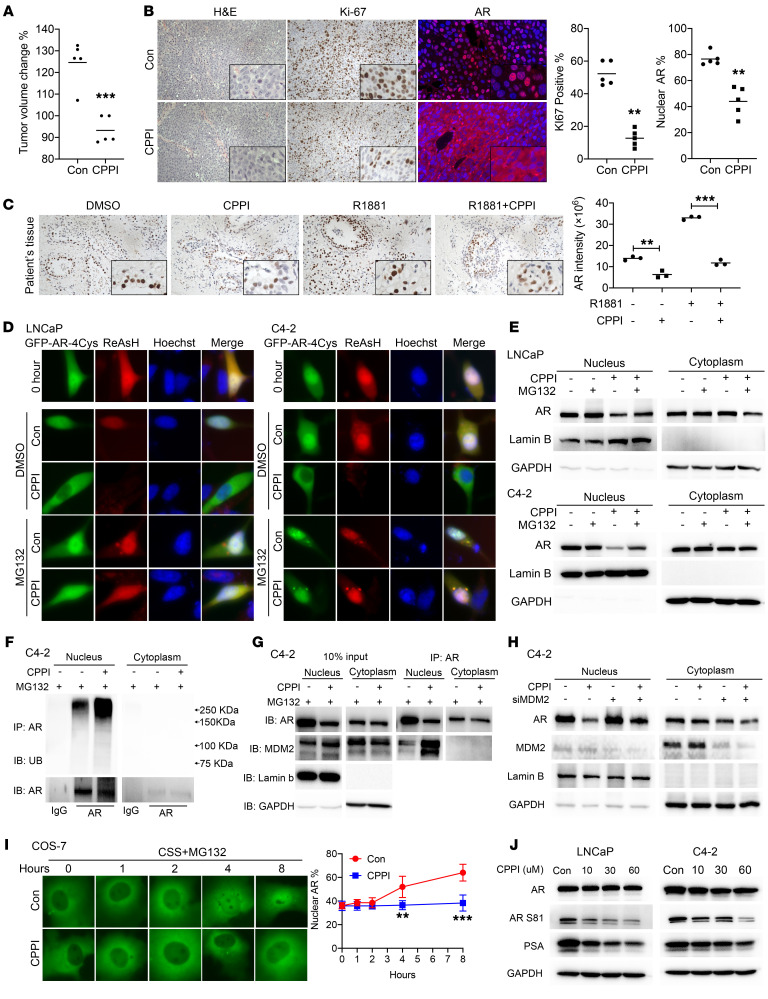
Since nuclear AR does not seem to undergo export, agents causing AR cytoplasmic localization in CRPC cells are likely acting through enhancing nuclear AR degradation. We previously showed that the small molecule CPPI can inhibit AR nuclear localization in cultured CRPC cells (28). We also reported that CPPI could significantly (P < 0.001) inhibit growth of relapsed LNCaP xenograft tumors over a 26-day treatment period (28). Here, we extended this finding by showing CPPI inhibition of endogenous AR nuclear localization and proliferation in C4-2 CRPC tumor xenografts (Figure 5, A and B), while not inducing apoptosis (Supplemental Figure 6A). Moreover, we tested CPPI inhibition of nuclear AR levels in patient-derived explants (PDEs) (36). Treatment of PDEs with CPPI significantly (P < 0.01) decreased the nuclear AR level (Figure 5C and Supplemental Figure 6B). Thus, CPPI may represent a class of small molecules that can specifically enhance AR degradation in the nucleus in PCa.
Figure 5. CPPI enhanced nuclear AR polyubiquitination and degradation, increased AR association with MDM2 in the nucleus, and inhibited AR nuclear import.
(A) Quantification of tumor volume changes after CPPI treatment (50 mg/kg/d) in C4-2 xenograft tumors (n = 5). (B) Representative images of H&E, Ki-67 immunostaining, and AR immunofluorescent staining in C4-2 xenograft tumors treated with CPPI or vehicle (n = 3). (C) Patient-derived explants were treated with CPPI. Effects of CPPI on AR expression with representative sections are shown. (D) Representative fluorescent images of total (green signal) and ReAsH pulse-labeled (red signal) GFP-AR-4Cys in response to CPPI (30 μM) in LNCaP or C4-2 cells with or without MG132. (E) Western blot analysis of endogenous AR in nuclear and cytoplasmic extracts of LNCaP and C4-2 cells after CPPI treatment with or without MG132 as in (C). (F) Western blot analysis of ubiquitin and AR in C4-2 cells treated with or without CPPI for 24 hours in the presence of MG132. (G) IP samples shown in E were also analyzed by Western blot with additional indicated antibodies. (H) Western blot analysis of AR expression in MDM2 knockdown or control C4-2 cells after CPPI treatment. (I) Time course of GFP-AR localization in transfected COS-7 cells treated with or without CPPI in the presence of MG132 in CSS medium. Nuclear GFP-AR quantification data are shown at right (n = 6). (J) AR, AR S81, and PSA were detected by Western blot in LNCaP and C4-2 cells treated with CPPI. Quantitative data are presented as mean ± SD, and all data represent 1 of at least 2 independent experiments with consistent results. Unpaired t test (I) was used to determine statistical significance. **P < 0.01; ***P < 0.001. Original magnification, ×40.
To explore the mechanism of CPPI inhibition of AR nuclear localization, ReAsH pulse chase was used to follow the effect of CPPI on GFP-AR-4Cys in LNCaP and C4-2 cells in the absence or presence of MG132. Figure 5D and Supplemental Figure 7A show that CPPI treatment caused cytoplasmic localization of total GFP-AR-4Cys and elimination of ReAsH-labeled GFP-AR-4Cys in both LNCaP and C4-2 cells. In CPPI-treated cells, the cytoplasmic GFP-AR-4Cys was not labeled by ReAsH and thus should be have been newly synthesized. As expected, MG132 suppressed CPPI induction of GFP-AR-4Cys cytoplasmic localization and degradation of ReAsH-labeled GFP-AR-4Cys in both LNCaP and C4-2 cells (Figure 5D). Western blot analysis of nuclear and cytoplasmic extracts from LNCaP and C4-2 cells showed that the endogenous nuclear AR level was reduced by CPPI and the reduction was partially blocked by MG132 (Figure 5E). Further analysis showed that CPPI enhanced AR polyubiquitination (Figure 5F) and AR co-IP with MDM2 in the nucleus in C4-2 cells (Figure 5G). MDM2 knockdown could partially reverse nuclear AR degradation induced by CPPI (Figure 5H). The increased AR polyubiquitination after CPPI treatment was also observed in LNCaP cells (Supplemental Figure 7B). Moreover, using Flag-AR–transfected HEK293 cells, we further confirmed CPPI enhancement of WT AR polyubiquitination and interaction with MDM2 in the nucleus (Supplemental Figure 7C).
Another potential mechanism contributing to CPPI inhibition of AR nuclear localization in CRPC cells may be through inhibiting AR nuclear import. Figure 5I shows that androgen-independent GFP-AR nuclear import was inhibited by CPPI in COS-7 cells in the presence of MG132, while enzalutamide did not block, but accelerated androgen-independent GFP-AR nuclear import in COS-7 cells in the presence of MG132 (Supplemental Figure 7D). CPPI also retarded androgen-induced GFP-AR nuclear import in the presence of MG132 (Supplemental Figure 7E).
Consistent with CPPI inhibition of AR nuclear localization, CPPI also inhibited AR phosphorylation at S81, which is required for AR nuclear translocation and association with chromatin (37). S81 phosphorylation plays an important role in androgen stimulation of endogenous AR transcription activation and subsequent cell growth (37). CPPI treatment markedly decreased S81-phosphorylated AR and AR-target gene PSA expression in LNCaP and C4-2 cells (Figure 5J). Cell-cycle analysis revealed that CPPI caused accumulation of cells in the G1 phase of the cell cycle (Supplemental Figure 7F). The luciferase assay also showed the antiandrogenic effect of CPPI on WT AR (Supplemental Figure 7G).
CPPI is a competitive AR antagonist.
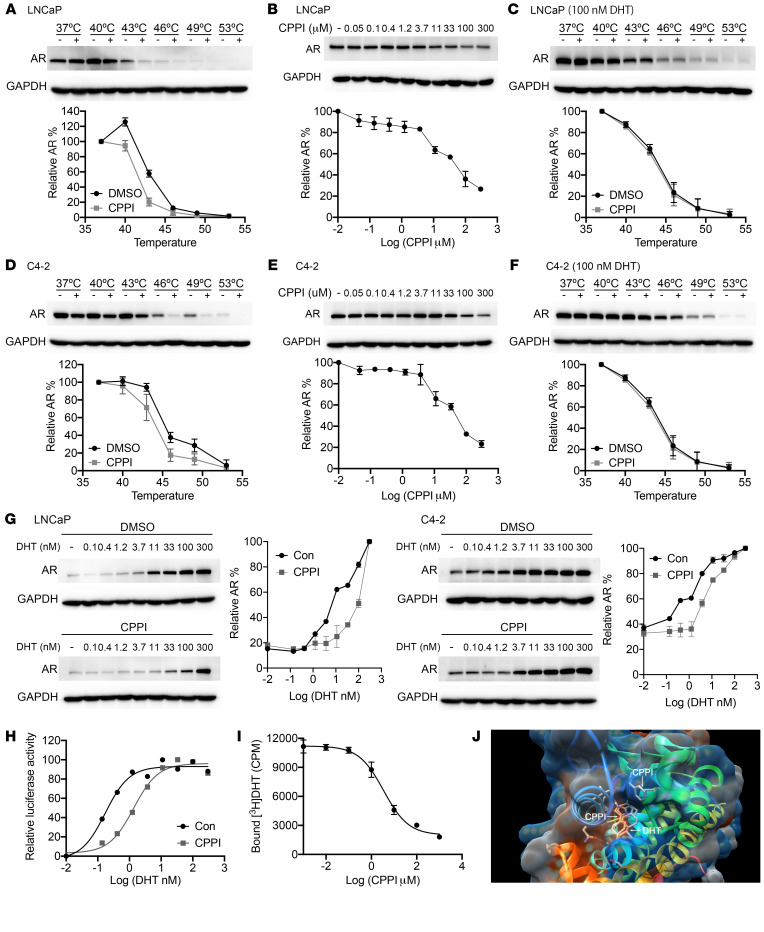
To explore the mechanism of CPPI inhibition of AR, we employed the cellular thermal shift assay (CETSA), which can detect drug-target engagement in live cells based on enhancement or reduction of protein thermostability (38–41). This method has been reported to determine whether a small molecule can directly bind to AR (42). As shown in Figure 6A, we observed destabilization of AR in CPPI-treated LNCaP cells. An isothermal drug-response fingerprinting (ITDRF) assay showed a fingerprint consistent with target engagement of CPPI (Figure 6B). Meanwhile, a high concentration of DHT (100 nM) prevented CPPI destabilization of AR (Figure 6C). Similar results were obtained in C4-2 cells (Figure 6, D–F), suggesting that DHT can compete with CPPI for AR binding. To determine whether CPPI is a competitive antagonist of AR, LNCaP and C4-2 cells were treated with increasing concentrations of DHT in the presence of CPPI or vehicle. As expected, DHT led to a concentration-dependent increase in the thermal stability of AR, while CPPI effectively antagonized DHT-mediated thermal stabilization in both cell lines (Figure 6G). As a negative control, MG132 did not affect DHT-mediated thermal stabilization (Supplemental Figure 8A). Furthermore, CPPI caused a shift toward higher dosages in the DHT-induced dose-response curve of AR transcriptional activity in a PSA luciferase assay, indicative of a competitive inhibition by CPPI (Figure 6H). To further determine whether CPPI is a competitive AR antagonist, we performed a [3H]-DHT–binding competition assay. CPPI treatment for 90 minutes inhibited [3H]-DHT binding to AR in the typical dose-dependent fashion of a competitive inhibitor, exhibiting an EC50 of 3.45 μM in C4-2 cells (Figure 6I). As a control, Western blot showed that CPPI treatment for 90 minutes had no marked effect on the endogenous AR level in C4-2 cells in a parallel experiment (Supplemental Figure 8B). Similarly, CPPI also inhibited [3H]-DHT binding to exogenous AR, with an EC50 of 5.70 μM in GFP-AR–transfected COS-7 cells (Supplemental Figure 8B). We also performed a molecular docking simulation with the x-ray crystal structure of the AR LBD (43). Two potential CPPI-binding sites were identified in the LBD, with 1 potential binding site in the DHT-binding pocket and a binding affinity of approximately –7.5 kcal/mol (Figure 6J and Supplemental Figure 8D). Taken together, the above findings argue that CPPI is a competitive antagonist of AR.
Figure 6. CPPI is an AR competitive inhibitor.
(A) Western blots showing thermostable AR following indicated heat shocks in the presence (+) or absence (−) of 50 μM CPPI in LNCaP cells (n = 2). (B) ITDRFCETSA experiments to determine the potency of CPPI target engagement in LNCaP cells (n = 2). (C) Western blots showing thermostable AR following indicated heat shocks in the presence (+) or absence (−) of 50 μM CPPI with 100 nM DHT in LNCaP cells (n = 2). (D–F) Western blots showing thermostable AR (D) (n = 2), the potency of CPPI target engagement (E) (n = 2),and the thermostable AR with 100 nM DHT (F) (n = 2) in C4-2 cells. (G) ITDRFCETSA experiments performed for DHT in the presence of 50 μM CPPI in LNCaP and C4-2 cells (n = 2). (H) C4-2-PSA-rl cells were treated with a gradient DHT dose with or without 50 μM CPPI. Firefly luciferase values were determined and normalized to Renilla (n = 3). (I) C4-2 cells were incubated with 1 nM [3H] DHT and the indicated amount of CPPI. The retained [3H] DHT in C4-2 cells was counted (n = 3). (J) Illustration of in silico predicted CPPI-binding sites in the LBD of AR. Quantitative data are presented as mean ± SD, and all data represent 1 of 2 independent experiments with consistent results.
CPPI attenuated the proliferation of enzalutamide-resistant LNCaP95 cells.
Our previous studies suggested that CPPI can inhibit the enzalutamide-resistant PCa cell line 22Rv1, which expresses both full-length AR (AR-FL) and AR splice variants (ARVs), particularly ARv7 (28). To further evaluate CPPI inhibition of enzalutamide-resistant PCa cells, we tested to determine whether CPPI can inhibit proliferation of another well-established enzalutamide-resistant PCa cell line, LNCaP95, that also expresses both AR-FL and ARv7 (44).
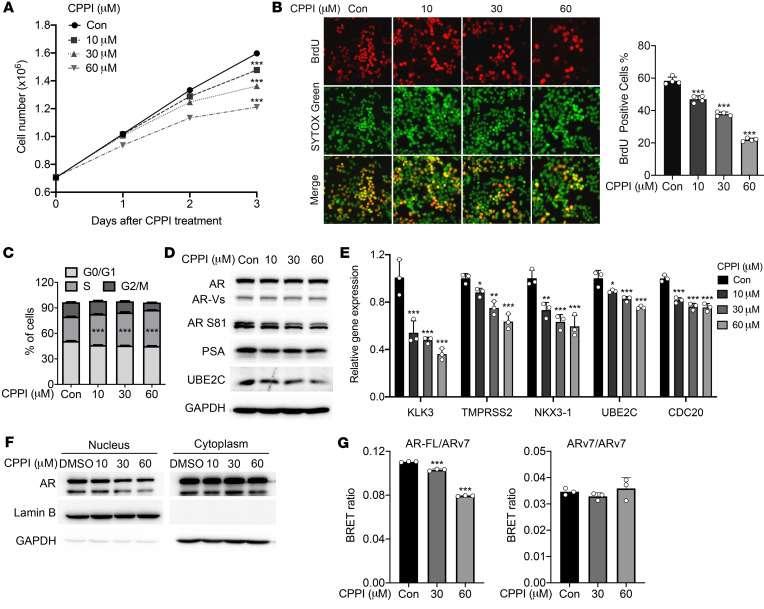
Consistent with the CPPI inhibition of 22RV1 cells, CPPI inhibited LNCaP95 cell proliferation in a cell-growth assay (Figure 7A) and a BrdU incorporation assay (Figure 7B). Cell-cycle analysis indicated that CPPI blocked the cell cycle at S phase (Figure 7C). Western blot showed that CPPI suppressed AR S81 phosphorylation, which is indicative of AR activation. The levels of the AR-FL target gene PSA and the ARv7 target gene UBE2C were also decreased by CPPI (Figure 7D). This result was supported by a quantitative reverse transcription PCR (qRT-PCR) assay showing CPPI inhibition of mRNAs of AR-FL target genes, KLK3 (PSA), TMPRSS2, and NKX3-1 as well as ARv7 target genes, UBE2C, and CDC20 (Figure 7E). Nucleocytoplasmic fractionation showed that CPPI reduced the levels of nuclear AR-FL and ARv7 (Figure 7F). However, in GFP-ARv7-transfected COS-7 cells, CPPI showed no effect on ARv7 nuclear localization (Supplemental Figure 9A). In addition, CPPI did not affect BrdU incorporation in GFP-ARv7:PC3, a PC3 subline stably transfected GFP-ARv7 expression vector (ref. 45 and Supplemental Figure 9B). As CPPI is a competitive AR antagonist targeting AR-FL through LBD, but not ARv7, which lacks LBD, the effect of CPPI on ARv7 in LNCaP95 cells might be mediated through AR-FL:ARv7 heterodimerization. Using the bioluminescence resonance energy transfer (BRET) technique described by Xu et al. (46), we tested to determine whether CPPI can inhibit AR-FL and ARv7 heterodimerization. The BRET plasmids were transfected into HEK293 cells, the expression of these fusion proteins was confirmed by Western blot, and the BRET saturation curve of AR-FL and ARv7 was reproduced (Supplemental Figure 9C). As shown in Figure 7G, CPPI inhibited heterodimerization of AR-FL and ARv7, but not ARv7 homodimerization. Collectively, these data show that CPPI attenuated proliferation of enzalutamide-resistant LNCaP95 cells and suppressed AR-FL as well as ARv7, possibly through blocking the AR-FL and ARv7 interaction.
Figure 7. CPPI inhibited LNCaP95 cell proliferation and full-length AR interaction with ARv7.
(A) LNCaP95 cell proliferation following treatment with indicated concentrations of CPPI for 1, 2, and 3 days (n = 3). (B) BrdU incorporation in LNCaP95 cells treated with CPPI. Right panel shows percentages of the LNCaP95 cells stained with BrdU (n = 4). Original magnification, ×40. (C) Cell-cycle analysis of LNCaP95 cells treated with CPPI (n = 3). (D) Western blot of AR, AR S81, PSA, and UBE2C in LNCaP95 cells treated with CPPI. GAPDH was probed as loading control. (E) qPCR analysis of AR target genes (KLK3, TMPRSS2, and NKX3-1) and ARV target genes (UBE2C and CDC20) in LNCaP95 cells treated with CPPI (n = 3). (F) Western blot analysis of AR and ARv7 in the nuclear and cytoplasmic extracts of LNCaP95 cells treated with CPPI. (G) BRET assay of AR-FL and ARv7 interaction following CPPI treatment (n = 3). Quantitative data are presented as mean ± SEM, and all data represent 1 of at least 2 independent experiments with consistent results. Unpaired t test (A) or 1-way ANOVA with Dunnett’s multiple-comparison post test (B, C, E, and G) was used to determine statistical significance. *P < 0.05; **P < 0.01; ***P < 0.001.
Discussion
This study suggests a paradigm shift regarding the mechanisms regulating AR levels in the nuclei of PCa cells. We present evidence that imported endogenous or exogenous nuclear AR is degraded, but not exported, upon androgen deprivation and that AR can be imported into the nucleus in the absence of androgens. AR polyubiquitination was detected in the nucleus, but not the cytoplasm, using nucleocytoplasmic fractionation coupled with Western blot analysis. We also observed reduced AR polyubiquitination in the nuclei of CRPC cells. These observations provide mechanistic insights into AR intracellular trafficking, AR turnover, and androgen-independent AR nuclear localization in CRPC. Furthermore, our studies identified CPPI as a small molecule nuclear AR degrader, which may lead to new therapeutics targeting nuclear AR in CRPC.
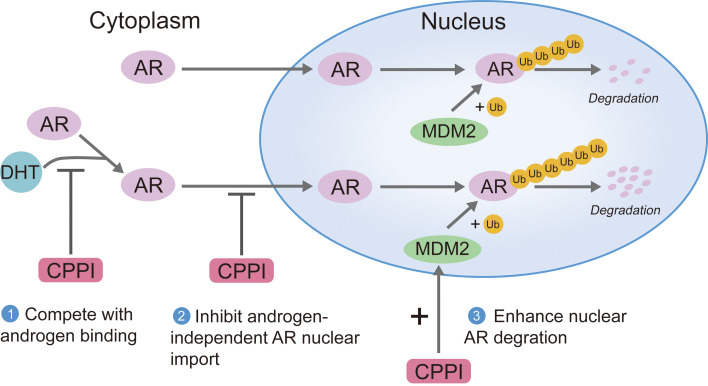
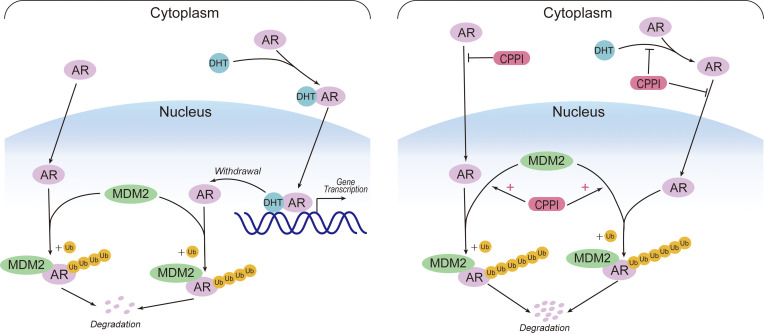
According to the classical model for mechanisms regulating AR intracellular trafficking, newly synthesized AR is localized to the cytoplasm and is imported into the nucleus upon androgen induction to transactive target genes. Upon androgen withdrawal, unliganded AR can be exported to the cytoplasm (21, 22). However, this model is not compatible with our observations of AR nuclear import in the absence of androgens and lack of AR export from the nucleus to the cytoplasm upon androgen withdrawal. Thus, we propose an updated model (Figure 8). In this new model, AR can be imported both in the absence and presence of androgens. Androgen binding can enhance AR nuclear import and suppress AR polyubiquitination and degradation in the nucleus. Once AR is in the nucleus, it will not be exported. Instead, unliganded AR will undergo rapid polyubiquitination and proteasomal degradation. Nuclear stabilization of AR can profoundly influence AR intracellular distribution. In CRPC cells, AR nuclear localization was reported as a common phenomenon (18, 47, 48). Increased AR stabilization appears to be the major mechanism leading to androgen-independent AR nuclear localization. Novel agents such as CPPI, capable of competing with ligand binding, inhibiting both androgen-induced or androgen-independent AR import, and enhancing nuclear AR degradation, should eliminate nuclear AR in CRPC cells (Figure 8).
Figure 8. An updated model for AR intracellular trafficking and the mechanisms of CPPI targeting nuclear AR.
AR can be imported in the absence of androgens, and the imported AR will be efficiently polyubiquitinated and degraded via proteasomes in the nucleus. DHT binding enhances AR nuclear import and inhibits AR degradation in the nucleus. Once AR is in the nucleus, it will not be exported. After DHT withdrawal, unliganded AR will undergo degradation in the nucleus immediately (left). In CRPC cells, the increased nuclear AR stability will lead to AR nuclear localization. CPPI, a pyrroloimidazole, can block AR ligand binding, inhibit both androgen-induced and androgen-independent AR import, and act as a nuclear AR degrader in CRPC cells (right).
NESAR was initially discovered based on its ability to drive AR cytoplasmic localization (19) and subsequently shown as the most potent signal for AR polyubiquitination (20). Previous efforts on elucidating the mechanism by which NESAR drives cytoplasmic localization did not identify any link between NESAR and known export machinery (19, 20, 49, 50). The present findings suggest that NESAR drives cytoplasmic localization via inducing robust polyubiquitination and proteasome-mediated degradation in the nucleus, rather than through the exportin pathway. The observation of cytoplasmic localization of AR constructs containing NESAR is due to the nuclear degradation role of NESAR. Thus, NESAR is not an export signal and should be renamed as a nuclear degradation signal of AR (NDSAR).
Our studies detected polyubiquitination of endogenous AR in the nuclei of PCa cells and transfected AR in the nuclei of nonprostatic HEK293 cells. This suggests that preferential AR nuclear degradation is not limited to prostatic cells. Detection of AR polyubiquitination in the nucleus, but not in the cytoplasm, also indicates that AR interacts with its E3 ligases preferentially in the nucleus. This hypothesis is supported by the AR co-IP with its E3 ligase MDM2 only in nuclear extracts. Moreover, decreased expression and/or activity of AR E3 ligases was identified in CRPC cells, while overexpression or knockdown of MDM2 expression was able to reduce or enhance nuclear AR levels, respectively. These findings further argue that the E3 ligases play important roles in AR nuclear turnover. In addition, overexpression of PP1α, a protein phosphatase that can suppress SKP2 and MDM2 activity by dephosphorylation (33), enhanced nuclear AR levels, while tautomycin, a PP1α inhibitor, inhibited nuclear AR levels. Thus, other molecules capable of regulating MDM2 and/or other AR E3 ligases may also influence AR nuclear localization and activity. Identification and characterization of signaling pathways that can regulate AR polyubiquitination in the nucleus may lead to new approaches to preventing and/or treating CRPC.
In addition to targeting the machinery regulating AR polyubiquitination, small molecules targeting AR could also modulate AR polyubiquitination. Androgen was previously reported to suppress AR polyubiquitination (24) and is shown here to suppress AR polyubiquitination in the nucleus. In contrast, the small molecule CPPI is an AR antagonist capable of enhancing AR polyubiquitination in the nucleus. We previously reported an IC50 of 4.3 μM for CPPI in its inhibition of GFP-AR nuclear localization in C4-2 cells (28), which is very similar to its IC50 in DHT competition assays. Also, in silico modeling predicted 2 potential CPPI-binding sites on the LBD of AR, with the stronger interaction at the ligand-binding site. These findings suggest that CPPI binding to AR is associated with and likely responsible for its inhibition of AR nuclear localization. CPPI represents what we believe to be a novel class of AR antagonists because none of the known AR antagonists can cause AR cytoplasmic localization in CRPC cells. CPPI is structurally different from other known AR antagonists (51–54), and CPPI binding is likely to induce a more labile structure for AR as compared with other AR antagonists. CPPI is also different from known AR degraders, such as ARCC-4 (55), UT69, and UT155 (56), because these degraders eliminate both nuclear and cytoplasmic AR. Considering the importance of AR as a therapeutic target for PCa, developing novel classes of AR antagonists is desirable because they may lead to more effective AR-targeting agents. CPPI can serve as a lead compound for developing novel AR antagonists that induce nuclear degradation with high potency and specificity.
In addition to inhibiting androgen-sensitive LNCaP and castration-resistant C4-2 cells, CPPI suppressed cell-cycle progression and proliferation of enzalutamide-resistant LNCaP95 PCa cells that express both AR-FL and ARv7. CPPI caused G0/G1 phase cell-cycle arrest in LNCaP and C4-2 cells, which is similar to the effect of other AR antagonists, such as enzalutamide (57, 58). However, CPPI caused a different type of cell-cycle perturbation in LNCaP95 cells, suggesting that AR regulation of cell-cycle progression in LNCaP95 cells may be different from that in LNCaP and C4-2 cells. Our study does not address whether CPPI suppression of LNCaP95 cell growth is mediated through its suppression of AR-FL and/or ARv7. The role of full-length AR vs ARv7 in driving androgen-independent PCa growth is still not clear (59). One study reported that the growth-promoting effects of ARvs are mediated through AR-FL (60). A recent paper provided evidence that AR-FL and ARv7 are both required for proliferation of CRPC under castration conditions (61). Another recent report suggested that the degradation of AR-FL is responsible for the growth suppression of a novel AR degrader in PCa cells expressing both full-length AR and ARv7 (62). Further studies will be needed to determine whether CPPI suppression of PCa cell growth is also mediated through AR-FL.
In summary, our studies suggest a mechanism for regulating AR subcellular localization that involves AR nuclear import and nuclear degradation via polyubiquitination, but not export. Reduced AR nuclear degradation appears to be responsible for androgen-independent AR nuclear localization in CRPC cells. We also showed that CPPI is an AR antagonist capable of promoting nuclear AR degradation via enhancing its polyubiquitination in the nucleus. Future studies will aim at understanding how AR is ubiquitinated in the nucleus and developing more potent analogs of CPPI.
Methods
Cell culture.
PCa cell lines C4-2 and LNCaP95 were provided by Leland W.K. Chung (Cedars-Sinai Medical Center, Los Angeles, California, USA) and Jun Luo (Johns Hopkins University, Baltimore, Maryland, USA), respectively. The ARv7:PC3 subline was generated by Michael A. Mancini (Baylor College of Medicine, Houston, Texas, USA; ref. 45). Other cell lines were purchased from ATCC. COS-7 and HEK293 cells were cultured in DMEM (Lonza), while PCa cells were cultured in RPMI 1640 (Corning). Media were supplemented with 10% FBS (Atlanta Biologicals) or charcoal-stripped FBS (for LNCaP95 cell line) and 1% penicillin/streptomycin. Mycoplasma contamination was tested by PCR.
Plasmid construction and transfection.
Constructs with full-length AR or various AR fragments fused to GFP at the N terminus were generated by cloning into the expression vector pEGFP-C1 (Clontech) as described previously (63). GFP-ARL859F was generated from GFP-AR vector using the Q5 site-directed mutagenesis kit (NEB). The GFP-AR-4Cys expression vector was cloned on the basis of the GFP-AR vector using a PCR-based approach and sequencing confirmed. The Myc-MDM2, MDM2S166D,S186D mutant, and PP1α plasmids were purchased from Addgene. PolyJet DNA In Vitro Transfection Reagent (SignaGen Laboratories) was used according to the manufacturer’s protocols. For knockdown study, MDM2 siRNA (AM51331, Thermo Fisher) and negative control siRNA (Horizon, D-001210-01-05) were chosen. Lipofectamine 2000 (Thermo Fisher) was used to transfect these siRNAs into cells.
Western blot.
Cells were lysed by RIPA buffer containing proteasome inhibitor cocktail (MilliporeSigma) or nucleocytoplasmic fractionation was performed according to the manufacturer’s instructions (G-Biosciences). The precipitates were resolved on SDS–polyacrylamide gel electrophoresis and subjected to Western blot analysis. As described previously (64), blots were probed with primary antibodies to the following: AR (Santa Cruz Biotechnology Inc., catalog sc-7305, 1:1000), lamin B (CST, catalog 13435, 1:1000), SKP2 (Santa Cruz Biotechnology Inc. catalog sc-74477, 1:1000), MDM2 (CST, catalog 86934, 1:1000), PP1α (Santa Cruz Biotechnology Inc., catalog sc-271762, 1:1000), Myc-tag (CST, catalog 2276, 1:1000), PSA (CST, catalog 5365, 1:1000), ubiquitin (EMD Millipore, catalog 07-375, 1:1000; abcam, catalog ab7780, 1:1000; CST, catalog 3936, 1:1000), AR S81 (EMD Millipore, catalog 07-1375, 1:1000), UBE2C (CST, catalog 14234, 1:200), GAPDH-HRP (Santa Cruz Biotechnology Inc., catalog sc-47724 HRP, 1:5000), and DYKDDDDK Tag HRP conjugate (CST, catalog 86861, 1:5000).
GFP-AR subcellular localization analysis.
COS-7, LNCaP, and C4-2 cells were transfected with GFP-AR or other GFP-AR deletion constructs and treated with CHX (50 μg/mL), MG132 (10 μM), or DHT (10 nM). Then, cells were fixed and permeabilized by ice-cold methanol for 15 minutes at –20 °C. Nucleus were labeled by DAPI (0.5 μg/mL, MilliporeSigma).
ReAsH pulse-chase assay.
Cells were transfected with GFP-AR-4Cys plasmids in phenol red–free media containing 10% CSS. For the AR nuclear export assay, DHT (10 nM) was added 12 hours prior to labeling. At 48 hours after transfection, cells were first incubated with 2 μM ReAsH-EDT2 (Cayman Chemical) in Opti-MDM for 30 minutes at 37°C. Cells were then washed with 2× BAL wash buffer in Opti-MDM for 15 minutes at 37°C. Then, BAL wash medium was replaced with cell culture medium and cells were treated as described in Results. Nuclei were labeled by NucBlue Live ReadyProbes Reagent (Hoechst 33342, Thermo Fisher). Images were obtained using a fluorescence microscope (Nikon T-2000).
Click chemistry pulse-chase assay.
Cells were washed in PBS and starved in cysteine- and methionine-free medium (MilliporeSigma) containing 5% dialyzed FBS (Gibco, Thermo Fisher Scientific) for 1 hour. Cells were then labeled with 50 μM HPG in the presence or absence of 10 nM DHT for 12 hours. The labeling medium was removed, and cells were washed twice in PBS before chasing in 10% CSS medium with indicated treatment. After 24 hours, cells were washed twice in PBS for nucleocytoplasmic fractionation. Lysates were quantified using bicinchoninic acid (BCA) assay (Thermo Fisher), and 250 μg protein was used for Click chemistry reaction according to the manufacturer’s protocols (Click-&-Go Click Chemistry Reaction Buffer Kit, Click Chemistry Tools). The suspension of Streptavidin Resin (GenScript) was dispensed into the samples for IP. The samples were incubated at room temperature for 1 hour, then washed 3 times with IP wash buffer.
Co-IP.
LNCaP-, C4-2–, and Myc-MDM2–transfected C4-2– and Flag-AR–transfected HEK293 cells were treated with MG132 (10 μM), DHT (10 nM) or CPPI (30 μM). After 24 hours, nucleocytoplasmic fractionation was performed. The cells lysed were incubated with 2 μg antibody to AR (Santa Cruz Biotechnology Inc., catalog sc-7305) or Myc-tag (CST, catalog 2276), then 35 μl of protein A/G (Thermo Fisher) was added and lysates were rocked for 4 hours at 4°C. The protein A/G beads were pelleted and washed 3 times with IP wash buffer. For Flag-AR– or GFP-AR–transfected HEK293 cells, Anti-DYKDDDDK G1 Affinity Resin (L00432, GenScript) or GFP Agarose Beads (D153-8, MBL International Corp.) were used to precipitate Flag-AR.
Xenograft tumor model.
Male Crl:SHO-PrkdcscidHrhr mice (6 to 8 weeks old) were purchased from Charles River Laboratory and maintained under pathogen-free conditions. C4-2 cells (3 × 106) suspended in 150 μl medium were gently mixed with 150 μl of Matrigel (Corning) and then inoculated subcutaneously into the right flank region of each mouse. Castration was performed after tumor volume reached 300 mm3, and treatment was initiated 4 days later. Tumor-bearing mice were randomly assigned into 2 groups and treated with CPPI (50 mg/kg/d) or vehicle (DMSO:EtOH:Kalliphor/PBS 1:1:8/10) for another 4 days.
Histology and immunohistochemistry/immunofluorescence.
Tumors were immediately fixed in 10% neutral buffered formalin for 24 hours, progressively dehydrated in solutions containing an increasing percentage of ethanol, and embedded into paraffin blocks. Consecutive 4 μm sections were obtained from paraffin blocks. Sections were counterstained with H&E or immunoassayed using antibody to Ki-67 (Dako, M7240, 1:100) using the immunoperoxidase technique or antibody to AR (Santa Cruz Biotechnology Inc., catalog sc-7305, 1:200) and labeled by secondary fluorescent antibody conjugated with Cy3. The Ki-67–positive cells were quantified as a percentage of total epithelial cells in 3 random ×40 fields. Nuclear AR was quantified as a percentage of nuclear AR intensity to the total the total intensity of AR in 3 random ×40 fields.
TUNEL assay.
Formalin-fixed, paraffin-embedded tumor tissue slides were used to detect apoptosis using the TACS 2 TdT-DAB In Situ Apoptosis Detection Kit (R & D Systems) according to the manufacturer’s protocol.
PDE model.
Excised prostatic tissue explants contained cancerous and/or nonmalignant tissues from PCa patients and were cultured ex vivo as described (65). Explants were treated with 30 μM CPPI in the absence or presence of 1 nM R1881 for 48 hours.
Cell-cycle analysis.
Cells were plated in 6-well plates and treated with CPPI at indicated concentrations for 48 hours. Cell-cycle distribution was analyzed with PI staining (BD Biosciences). The stained cells were acquired by flow cytometry (BD Biosciences) and analyzed by FlowJo software.
CETSA.
CETSA was performed in living LNCaP and C4-2 cells. Cells were divided into separate aliquots and exposed to vehicle control or compounds at the indicated concentrations for 1 hour at 37°C. Treated cells were heated at different temperatures for 3 minutes and then cooled at room temperature for 3 minutes. All samples were supplemented with protease inhibitor cocktail before lysis by ×3 freeze-thaw using liquid nitrogen and a heat block. Samples were then clarified by centrifugation at 20,000 g for 20 minutes. Supernatants were transferred to new tubes and detected by Western blot.
ITDRFCETSA.
LNCaP and C4-2 cells were divided into separate aliquots and exposed to compounds at a gradient concentration for 1 hour in the incubator. Heat shock was performed at 46°C for 3 minutes and lysed by 3× freeze-thaw using liquid nitrogen. Following centrifugation at 20,000 g for 20 minutes at 4°C, supernatants were transferred to new tubes and detected by Western blot.
Dual luciferase screening assay.
The DHT-induced PSA promoter luciferase assay was performed in C4-2-PSA-rl cells, a C4-2 subline containing a PSA-luciferase reporter vector (pPSA6.1-Luc) and a Renilla luciferase reporter vector (pRL-TK). The cells were treated with a gradient DHT dose with or without 50 μM CPPI for 24 hours prior to luciferase assays. PC3 and HEK293 cells were transiently transfected with GFP-ARWT, pPSA6.1-Luc, and pRL-TK. Cells were treated with a gradient R1881 with or without 30 μM CPPI. Renilla and firefly activities were then determined by luminometry using the Dual-Luciferase Reporter Assay System (Promega). Results were expressed as the ratio of firefly to Renilla luciferase activity. Curve fitting was performed using Prism software (GraphPad).
Radioligand competition binding assay.
C4-2– and GFP-AR–transfected COS-7 cells were seeded in 12-well plates in phenol red–free media containing 10% CSS. After 3 days, media were replaced with serum-free media containing 1 nM [3H] DHT in the presence or absence of CPPI at a gradient concentration for 90 minutes in an incubator at 37°C with 5% CO2. Cells were washed with phosphate buffer, and bound ligand was extracted in ethanol for 30 minutes at room temperature and detected using a scintillation counter. Curve fitting and IC50 calculations were performed using GraphPad Prism (version 8.2.1, for macOS, GraphPad Software).
CPPI-AR docking studies.
The PDB file was downloaded from the Protein Data Bank (PDB) (www.rcsb.org, ID: 4OEA), and the protein structure of LBD of AR was visualized in PyMOL (PyMOL Molecular Graphics System, version 2.3.1, Schrödinger LLC). Molecular docking studies were performed by Autodock Vina combined with PyRx (66). Illustrations of the AR-LBD and CPPI complex were generated using PyMOL.
In vitro cell growth assay.
Cells were plated in 6-well plates and treated with CPPI at indicated concentrations. Cell numbers were counted using a Coulter Counter (Beckman Coulter) at 1, 2, and 3 days after treatment. Each assay was performed in triplicate.
Brdu cell proliferation assays.
Cells were seeded in 24-well plates and treated with CPPI at indicated concentrations for 48 hours. BrdU labeling was performed 12 hours prior to staining. Cells were then fixed in Carnoy’s fixative and washed with 0.1 M boric acid. Cells were incubated in 2 M HCl for DNA hydrolysis and 3% BSA was used to block cells. Subsequently, cells were stained overnight with primary antibodies to BrdU (Santa Cruz Biotechnology Inc., catalog sc-51514, 1:500) and labeled by secondary fluorescent antibody conjugated with Cy3. Nuclei were stained with 1 μM SYTOX Green (Life Technologies). BrdU-positive cell density was determined according to the presence or absence of nuclear specific staining when compared with the negative controls.
qRT-PCR.
Cells were seeded in 6-well plates and treated with CPPI at indicated concentrations. After 48 hours, RNA from cells was isolated by TRIzol Reagent (Invitrogen). Reverse transcription was performed with 1 μg RNA using the PrimeScript RT Reagent Kit (Takara). cDNA was amplified with gene-specific primers (Supplemental Table 1) and the SYBR Premix Ex Taq II Kit (TaKaRa). Data were analyzed using a 2–ΔΔCt method.
BRET assay.
The plasmids used in BRET assay were provided by Yan Dong (Tulane University School of Medicine, New Orleans, Louisiana, USA). Cells were either transfected with an RLuc BRET fusion plasmid or cotransfected with an RLuc and a TFP BRET fusion plasmid (46). After 24 hours, the medium was changed and cells were treated with CPPI at indicated concentrations for another 48 hours. Then cells were detached with 5 mM EDTA in PBS and resuspended in PBS with 1% sucrose. Cells were reseeded in triplicate into a 96-well white-wall microplate at 105 cells per well. Freshly prepared coelenterazine (Nanolight Technology) in water was added to the cells at a final concentration of 25 mM. The BRET readings at 528 nm and 635 nm were obtained by LMax II Microplate Reader (Molecular Devices). The following equation was used: BRET ratio = (emission at 635 nm)/(emission at 528 nm) – (emission at 635 nm RLuc only)/(emission at 528 nm RLuc only).
Statistics.
Statistical analysis was performed using GraphPad Prism (version 8.2.1, for macOS, GraphPad Software). Data are presented as mean ± SEM. A parametric 2-tailed t test (2 groups) and 1-way ANOVA followed by Dunnett’s multiple-comparison post test (for more than 2 groups) were used when the data sets were found to be normally distributed, with F test comparison of variances or Bartlett’s test of equal variances, respectively. Otherwise, Mann-Whitney U test was used (2 groups). P < 0.05 was considered statistically significant.
Study approval.
Two prostatic tissues were obtained from the Pitt Biospecimen Core under an approved IRB protocol. The animal use protocol was approved by the University of Pittsburgh Institutional Animal Care and Use Committee.
Author contributions
SL designed and performed most of the experiments. QS generated preliminary data supporting the overall concept of this study. GC, EC, WC, RC, Z Wu, KW, and Z Wang performed some of the experiments. SL, WC, and LEP helped with mouse experiments and the PDE model. JBN and PW provided advice about experimental design. WH and QW are listed as coauthors since they are the thesis advisors of SL. Z Wang conceived and supervised the project as well as designed the experiments. SL, LEP, and Z Wang generated the figures and wrote the manuscript. All authors discussed the results and approved the manuscript.
Supplementary Material
Acknowledgments
We would like to thank Yan Dong for providing the plasmids to detect the AR-FL and ARv7 interaction based on the BRET6 system, Leland W.K. Chung for C4-2 cells, Jun Luo for LNCaP95 cells, and Ziyan Yin for statistical consultation. This work was funded in part by DOD Award W81XWH-16-1-0659 and by NIH grants R01 CA186780 (to Z Wang) and R50 CA211242 (to LEP) as well as by the Department of Urology, University of Pittsburgh. This project used the UPMC Hillman Cancer Center Animal Facility, and the Tissue and Research Pathology Services/Pitt Biospecimen Core, which were supported in part by NCI award P30 CA047904 and the Senior Vice Chancellor’s Office at the University of Pittsburgh.
Version 1. 12/17/2020
In-Press Preview
Version 2. 02/15/2021
Print issue publication
Funding Statement
R01 CA186780 (to Z. Wang), and R50 CA211242 (to L.E. Pascal)
the Tissue and Research Pathology Services/Pitt Biospecimen Core which were supported in part by NCI award P30 CA047904
Footnotes
Conflict of interest: JBN, PW, and Z Wang are listed as co-inventors on a US patent held by the University of Pittsburgh (US 10004730, “Small molecules targeting androgen receptor nuclear localization and/or level in prostate cancer”).
Copyright: © 2021, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2021;131(4):e141335.https://doi.org/10.1172/JCI141335.
Contributor Information
Shidong Lv, Email: lsd990@163.com.
Qiong Song, Email: songqem@163.com.
Guang Chen, Email: yeux92@163.com.
Erdong Cheng, Email: aaaced@gmail.com.
Wei Chen, Email: weichen@pitt.edu.
Ryan Cole, Email: cole.ryan.n@gmail.com.
Zeyu Wu, Email: 253957713@qq.com.
Laura E. Pascal, Email: pascalle@upmc.edu.
Ke Wang, Email: shishan_86@163.com.
Peter Wipf, Email: pwipf@pitt.edu.
Joel B. Nelson, Email: nelsonjb@upmc.edu.
Qiang Wei, Email: qwei@smu.edu.cn.
Wenhua Huang, Email: huangwenhua2009@139.com.
Zhou Wang, Email: wangz2@upmc.edu.
References
- 1.Siegel RL, et al. Cancer statistics, 2020. CA Cancer J Clin. 2020;70(1):7–30. doi: 10.3322/caac.21590. [DOI] [PubMed] [Google Scholar]
- 2.de Bono JS, et al. Abiraterone and increased survival in metastatic prostate cancer. N Engl J Med. 2011;364(21):1995–2005. doi: 10.1056/NEJMoa1014618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ryan CJ, et al. Abiraterone in metastatic prostate cancer without previous chemotherapy. N Engl J Med. 2013;368(2):138–148. doi: 10.1056/NEJMoa1209096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Salem M, Garcia JA. Abiraterone acetate, a novel adrenal inhibitor in metastatic castration-resistant prostate cancer. Curr Oncol Rep. 2011;13(2):92–96. doi: 10.1007/s11912-011-0153-4. [DOI] [PubMed] [Google Scholar]
- 5.Tran C, et al. Development of a second-generation antiandrogen for treatment of advanced prostate cancer. Science. 2009;324(5928):787–790. doi: 10.1126/science.1168175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Scher HI, et al. Increased survival with enzalutamide in prostate cancer after chemotherapy. N Engl J Med. 2012;367(13):1187–1197. doi: 10.1056/NEJMoa1207506. [DOI] [PubMed] [Google Scholar]
- 7.Beer TM, et al. Enzalutamide in metastatic prostate cancer before chemotherapy. N Engl J Med. 2014;371(5):424–433. doi: 10.1056/NEJMoa1405095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Claessens F, et al. Emerging mechanisms of enzalutamide resistance in prostate cancer. Nat Rev Urol. 2014;11(12):712–716. doi: 10.1038/nrurol.2014.243. [DOI] [PubMed] [Google Scholar]
- 9.Giacinti S, et al. Resistance to abiraterone in castration-resistant prostate cancer: a review of the literature. Anticancer Res. 2014;34(11):6265–6269. [PubMed] [Google Scholar]
- 10.Yuan X, et al. Androgen receptor functions in castration-resistant prostate cancer and mechanisms of resistance to new agents targeting the androgen axis. Oncogene. 2014;33(22):2815–2825. doi: 10.1038/onc.2013.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Watson PA, et al. Emerging mechanisms of resistance to androgen receptor inhibitors in prostate cancer. Nat Rev Cancer. 2015;15(12):701–711. doi: 10.1038/nrc4016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dai C, et al. Androgen signaling in prostate cancer. Cold Spring Harb Perspect Med. 2017;7(9):a030452. doi: 10.1101/cshperspect.a030452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Claessens F, et al. Diverse roles of androgen receptor (AR) domains in AR-mediated signaling. Nucl Recept Signal. 2008;6:e008. doi: 10.1621/nrs.06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smith DF, Toft DO. Minireview: the intersection of steroid receptors with molecular chaperones: observations and questions. Mol Endocrinol. 2008;22(10):2229–2240. doi: 10.1210/me.2008-0089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Claessens F, et al. Selective DNA binding by the androgen receptor as a mechanism for hormone-specific gene regulation. J Steroid Biochem Mol Biol. 2001;76(1–5):23–30. doi: 10.1016/s0960-0760(00)00154-0. [DOI] [PubMed] [Google Scholar]
- 16.Doesburg P, et al. Functional in vivo interaction between the amino-terminal, transactivation domain and the ligand binding domain of the androgen receptor. Biochemistry. 1997;36(5):1052–1064. doi: 10.1021/bi961775g. [DOI] [PubMed] [Google Scholar]
- 17.Heemers HV, et al. Androgen modulation of coregulator expression in prostate cancer cells. Mol Endocrinol. 2009;23(4):572–583. doi: 10.1210/me.2008-0363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gregory CW, et al. Androgen receptor stabilization in recurrent prostate cancer is associated with hypersensitivity to low androgen. Cancer Res. 2001;61(7):2892–2898. [PubMed] [Google Scholar]
- 19.Saporita AJ, et al. Identification and characterization of a ligand-regulated nuclear export signal in androgen receptor. J Biol Chem. 2003;278(43):41998–42005. doi: 10.1074/jbc.M302460200. [DOI] [PubMed] [Google Scholar]
- 20.Gong Y, et al. Nuclear export signal of androgen receptor (NESAR) regulation of androgen receptor level in human prostate cell lines via ubiquitination and proteasome-dependent degradation. Endocrinology. 2012;153(12):5716–5725. doi: 10.1210/en.2012-1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kumar S, et al. Intracellular localization and nucleocytoplasmic trafficking of steroid receptors: an overview. Mol Cell Endocrinol. 2006;246(1-2):147–156. doi: 10.1016/j.mce.2005.11.028. [DOI] [PubMed] [Google Scholar]
- 22.Tyagi RK, et al. Dynamics of intracellular movement and nucleocytoplasmic recycling of the ligand-activated androgen receptor in living cells. Mol Endocrinol. 2000;14(8):1162–1174. doi: 10.1210/mend.14.8.0497. [DOI] [PubMed] [Google Scholar]
- 23.Griffin BA, et al. Specific covalent labeling of recombinant protein molecules inside live cells. Science. 1998;281(5374):269–272. doi: 10.1126/science.281.5374.269. [DOI] [PubMed] [Google Scholar]
- 24.Wu P, et al. A high-throughput-compatible assay to measure the degradation of endogenous Huntingtin proteins. Acta Pharmacol Sin. 2016;37(10):1307–1314. doi: 10.1038/aps.2016.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Su Hui Teo C, et al. Spatial and temporal resolution of global protein synthesis during HSV infection using bioorthogonal precursors and click chemistry. PLoS Pathog. 2016;12(10):e1005927. doi: 10.1371/journal.ppat.1005927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hein CD, et al. Click chemistry, a powerful tool for pharmaceutical sciences. Pharm Res. 2008;25(10):2216–2230. doi: 10.1007/s11095-008-9616-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rajender S, et al. L859F mutation in androgen receptor gene results in complete loss of androgen binding to the receptor. J Androl. 2007;28(5):772–776. doi: 10.2164/jandrol.107.002691. [DOI] [PubMed] [Google Scholar]
- 28.Masoodi KZ, et al. Inhibition of androgen receptor nuclear localization and castration-resistant prostate tumor growth by pyrroloimidazole-based small molecules. Mol Cancer Ther. 2017;16(10):2120–2129. doi: 10.1158/1535-7163.MCT-17-0176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li B, et al. Skp2 regulates androgen receptor through ubiquitin-mediated degradation independent of Akt/mTOR pathways in prostate cancer. Prostate. 2014;74(4):421–432. doi: 10.1002/pros.22763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin H-K, et al. Phosphorylation-dependent ubiquitylation and degradation of androgen receptor by Akt require Mdm2 E3 ligase. EMBO J. 2002;21(15):4037–4048. doi: 10.1093/emboj/cdf406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vummidi Giridhar P, et al. Constant degradation of the androgen receptor by MDM2 conserves prostate cancer stem cell integrity. Cancer Res. 2019;79(6):1124–1137. doi: 10.1158/0008-5472.CAN-18-1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sarkar S, et al. Aurora kinase A Promotes AR degradation via the E3 ligase CHIP. Mol Cancer Res. 2017;15(8):1063–1072. doi: 10.1158/1541-7786.MCR-17-0062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu X, et al. Protein phosphatase 1 suppresses androgen receptor ubiquitylation and degradation. Oncotarget. 2016;7(2):1754–1764. doi: 10.18632/oncotarget.6434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou BP, et al. HER-2/neu induces p53 ubiquitination via Akt-mediated MDM2 phosphorylation. Nat Cell Biol. 2001;3(11):973–982. doi: 10.1038/ncb1101-973. [DOI] [PubMed] [Google Scholar]
- 35.Sydnes MO, Isobe M. Tautomycin’s interactions with protein phosphatase 1. Chem Asian J. 2010;5(3):410–420. doi: 10.1002/asia.200900394. [DOI] [PubMed] [Google Scholar]
- 36.Centenera MM, et al. A patient-derived explant (PDE) model of hormone-dependent cancer. Mol Oncol. 2018;12(9):1608–1622. doi: 10.1002/1878-0261.12354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen S, et al. Androgen receptor serine 81 phosphorylation mediates chromatin binding and transcriptional activation. J Biol Chem. 2012;287(11):8571–8583. doi: 10.1074/jbc.M111.325290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jafari R, et al. The cellular thermal shift assay for evaluating drug target interactions in cells. Nat Protoc. 2014;9(9):2100–2122. doi: 10.1038/nprot.2014.138. [DOI] [PubMed] [Google Scholar]
- 39.Molina DM, et al. Monitoring drug target engagement in cells and tissues using the cellular thermal shift assay. Science. 2013;341(6141):84–87. doi: 10.1126/science.1233606. [DOI] [PubMed] [Google Scholar]
- 40.Reinhard FB, et al. Thermal proteome profiling monitors ligand interactions with cellular membrane proteins. Nat Methods. 2015;12(12):1129–1131. doi: 10.1038/nmeth.3652. [DOI] [PubMed] [Google Scholar]
- 41.Wu J, et al. Momordin Ic, a new natural SENP1 inhibitor, inhibits prostate cancer cell proliferation. Oncotarget. 2016;7(37):58995–59005. doi: 10.18632/oncotarget.10636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shaw J, et al. Determining direct binders of the androgen receptor using a high-throughput cellular thermal shift assay. Sci Rep. 2018;8(1):163. doi: 10.1038/s41598-017-18650-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hsu CL, et al. Identification of a new androgen receptor (AR) co-regulator BUD31 and related peptides to suppress wild-type and mutated AR-mediated prostate cancer growth via peptide screening and X-ray structure analysis. Mol Oncol. 2014;8(8):1575–1587. doi: 10.1016/j.molonc.2014.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang YC, et al. Targeting androgen receptor activation function-1 with EPI to overcome resistance mechanisms in castration-resistant prostate cancer. Clin Cancer Res. 2016;22(17):4466–4477. doi: 10.1158/1078-0432.CCR-15-2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Szafran AT, et al. High-content screening identifies src family kinases as potential regulators of AR-V7 Expression and androgen-independent cell growth. Prostate. 2017;77(1):82–93. doi: 10.1002/pros.23251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xu D, et al. Androgen receptor splice variants dimerize to transactivate target genes. Cancer Res. 2015;75(17):3663–3671. doi: 10.1158/0008-5472.CAN-15-0381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mohler JL, et al. The androgen axis in recurrent prostate cancer. Clin Cancer Res. 2004;10(2):440–448. doi: 10.1158/1078-0432.CCR-1146-03. [DOI] [PubMed] [Google Scholar]
- 48.Gregory CW, et al. Androgen receptor expression in androgen-independent prostate cancer is associated with increased expression of androgen-regulated genes. Cancer Res. 1998;58(24):5718–5724. [PubMed] [Google Scholar]
- 49.Nguyen MM, et al. Cytoplasmic localization of the androgen receptor is independent of calreticulin. Mol Cell Endocrinol. 2009;302(1):65–72. doi: 10.1016/j.mce.2008.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nguyen MM, et al. Characterization of karyopherins in androgen receptor intracellular trafficking in the yeast model. Int J Clin Exp Pathol. 2014;7(6):2768–2779. [PMC free article] [PubMed] [Google Scholar]
- 51.Fujita K, Nonomura N. Role of androgen receptor in prostate cancer: a review. World J Mens Health. 2019;37(3):288–295. doi: 10.5534/wjmh.180040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Helsen C, et al. Androgen receptor antagonists for prostate cancer therapy. Endocr Relat Cancer. 2014;21(4):T105–T118. doi: 10.1530/ERC-13-0545. [DOI] [PubMed] [Google Scholar]
- 53.Masoodi KZ, et al. Inhibition of androgen receptor function and level in castration-resistant prostate cancer cells by 2-[(isoxazol-4-ylmethyl)thio]-1-(4-phenylpiperazin-1-yl)ethanone. Endocrinology. 2017;158(10):3152–3161. doi: 10.1210/en.2017-00408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Johnston PA, et al. Development and implementation of a high-throughput high-content screening assay to identify inhibitors of androgen receptor nuclear localization in castration-resistant prostate cancer cells. Assay Drug Dev Technol. 2016;14(4):226–239. doi: 10.1089/adt.2016.716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Salami J, et al. Androgen receptor degradation by the proteolysis-targeting chimera ARCC-4 outperforms enzalutamide in cellular models of prostate cancer drug resistance. Commun Biol. 2018;1(1):1–9. doi: 10.1038/s42003-017-0002-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Narayanan R, et al. Destroying the androgen receptor (AR)-potential strategy to treat advanced prostate cancer. Oncoscience. 2017;4(11–12):175–177. doi: 10.18632/oncoscience.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Qi W, et al. Reciprocal feedback inhibition of the androgen receptor and PI3K as a novel therapy for castrate-sensitive and -resistant prostate cancer. Oncotarget. 2015;6(39):41976–41987. doi: 10.18632/oncotarget.5659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sharp A, et al. Androgen receptor splice variant-7 expression emerges with castration resistance in prostate cancer. J Clin Invest. 2019;129(1):192–208. doi: 10.1172/JCI122819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Luo J, et al. Role of androgen receptor variants in prostate cancer: report from the 2017 Mission Androgen Receptor Variants meeting. Eur Urol. 2018;73(5):715–723. doi: 10.1016/j.eururo.2017.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Watson PA, et al. Constitutively active androgen receptor splice variants expressed in castration-resistant prostate cancer require full-length androgen receptor. Proc Natl Acad Sci U S A. 2010;107(39):16759–16765. doi: 10.1073/pnas.1012443107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cato L, et al. Arv7 represses tumor-suppressor genes in castration-resistant prostate cancer. Cancer Cell. 2019;35(3):401–413.e6. doi: 10.1016/j.ccell.2019.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kregel S, et al. Androgen receptor degraders overcome common resistance mechanisms developed during prostate cancer treatment. Neoplasia. 2020;22(2):111–119. doi: 10.1016/j.neo.2019.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dar JA, et al. N-terminal domain of the androgen receptor contains a region that can promote cytoplasmic localization. J Steroid Biochem Mol Biol. 2014;139:16–24. doi: 10.1016/j.jsbmb.2013.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lv S, et al. Loss of KMT2D induces prostate cancer ROS-mediated DNA damage by suppressing the enhancer activity and DNA binding of antioxidant transcription factor FOXO3. Epigenetics. 2019;14(12):1194–1208. doi: 10.1080/15592294.2019.1634985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schiewer MJ, et al. Dual roles of PARP-1 promote cancer growth and progression. Cancer Discov. 2012;2(12):1134–1149. doi: 10.1158/2159-8290.CD-12-0120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dallakyan S, Olson AJ. Small-molecule library screening by docking with PyRx. Methods Mol Biol. 2015;1263:243–250. doi: 10.1007/978-1-4939-2269-7_19. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.