Abstract
Prostate cancer (PCa) is the second leading cause of cancer death in American men. Androgen receptor (AR) signaling is essential for PCa cell growth/survival and remains a key therapeutic target for lethal castration-resistant PCa (CRPC). GATA2 is a pioneer transcription factor crucial for inducing AR expression/activation. We recently reported that MAPK4, an atypical MAPK, promotes tumor progression via noncanonical activation of AKT. Here, we demonstrated that MAPK4 activated AR by enhancing GATA2 transcriptional expression and stabilizing GATA2 protein through repression of GATA2 ubiquitination/degradation. MAPK4 expression correlated with AR activation in human CRPC. Concerted activation of both GATA2/AR and AKT by MAPK4 promoted PCa cell proliferation, anchorage-independent growth, xenograft growth, and castration resistance. Conversely, knockdown of MAPK4 decreased activation of both AR and AKT and inhibited PCa cell and xenograft growth, including castration-resistant growth. Both GATA2/AR and AKT activation were necessary for MAPK4 tumor-promoting activity. Interestingly, combined overexpression of GATA2 plus a constitutively activated AKT was sufficient to drive PCa growth and castration resistance, shedding light on an alternative, MAPK4-independent tumor-promoting pathway in human PCa. We concluded that MAPK4 promotes PCa growth and castration resistance by cooperating parallel pathways of activating GATA2/AR and AKT and that MAPK4 is a novel therapeutic target in PCa, especially CRPC.
Keywords: Oncology
Keywords: Prostate cancer
Introduction
Prostate cancer (PCa) is the most common cancer after skin cancer and the second leading cause of cancer death in American men (1). Androgen receptor (AR) signaling is essential for PCa cell growth and survival. Therefore, androgen deprivation therapy (ADT) is the standard therapy for advanced/metastatic PCa (2). However, patients inevitably develop lethal castration-resistant PCa (CRPC), including resistance to the most advanced therapies: enzalutamide, apalutamide, and abiraterone (3–5). Most CRPCs express activated AR, and attacking AR signaling activation remains a major therapeutic avenue for CRPC. GATA2 is a transcription factor and pioneer factor crucial for inducing AR expression and activation in PCa (6–8). GATA2 protein expression in human PCa tissues correlates with advanced stages and worse prognosis; it is further elevated in metastatic therapy-resistant PCa (8–11). GATA2 protein is unstable (12, 13) and the molecular mechanisms that regulate GATA2 stability in PCa remain unknown.
AKT plays crucial roles in regulating cell survival, proliferation, and metabolism. In the canonical PI3K/AKT/mTOR pathway, PI3K catalyzes PIP3 production to recruit AKT to the cell membrane for activation (14–16). The 2 mTOR complexes, mTORC1 and mTORC2, act coordinately with AKT. mTORC2 is a major AKT S473 kinase (17), which works together with PDK1, an AKT T308 kinase (18), to fully activate AKT. In contrast, mTORC1 is activated by AKT and is a vital hub that integrates extracellular and nutrient signals to modulate cell growth, autophagy, and metabolism (19). PTEN, along with PHLPP phosphatases and INPP4B, offer the major brake to restrain the PI3K/AKT pathway (20–23). Aberrant activation of the PI3K/AKT/mTOR signaling leads to many pathological outcomes, including cancers (24, 25).
The PI3K/AKT/mTOR signaling pathway is frequently altered in PCa (26) and the key components of this pathway have emerged as attractive therapeutic targets in PCa and CRPC (27). Complex reciprocal crosstalk between the PI3K/AKT/mTOR and AR pathways at multiple levels has been reported. The central theme is mutually antagonistic activities of PI3K/AKT/mTOR and AR (28, 29), although other interactions have also been reported (30–32).
MAPK4 is an atypical MAPK that lacks the canonical Thr-X-Tyr activation motif for phosphorylation and activation by MAPK kinase (MAPKK) (33, 34). Little was known about MAPK4 physiological function and its involvement in diseases, including cancers. We recently reported that MAPK4 promotes tumor progression via noncanonical activation of AKT that is independent of PI3K/PDK1 (35). Here we report that MAPK4 also activates AR signaling by both enhancing GATA2 mRNA expression and inhibiting GATA2 protein ubiquitination and degradation. Concerted activation of both the GATA2/AR and AKT pathways is required to promote PCa cell growth.
Results
MAPK4 promotes PCa cell growth.
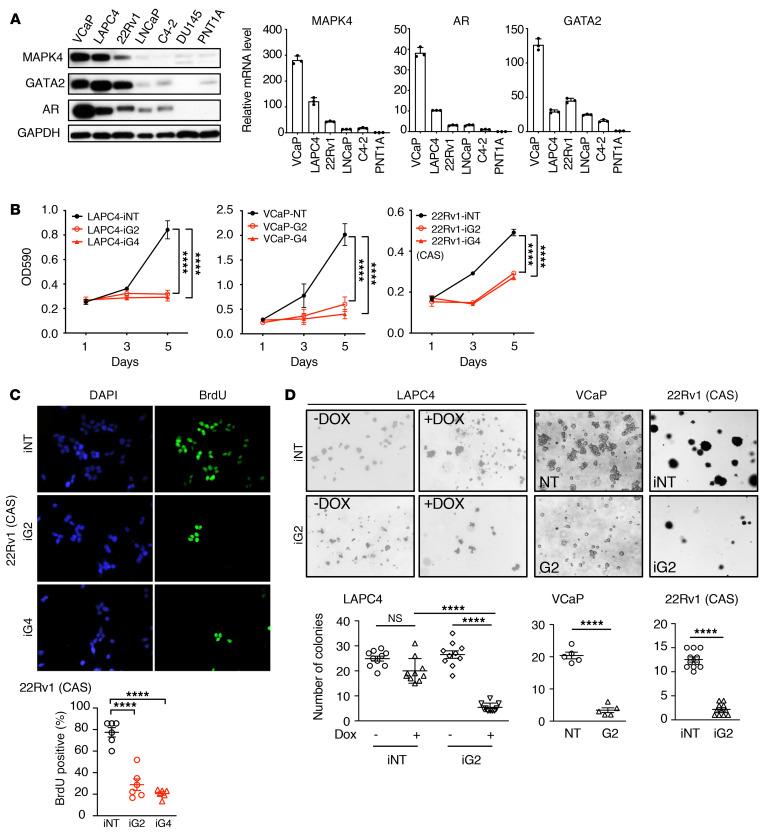
We recently discovered that MAPK4 promotes diverse types of human cancers (35). To characterize MAPK4 activity in androgen-dependent PCa, we first compared MAPK4 expression in various human PCa cell lines, including VCaP, LAPC4, 22Rv1, LNCaP, C4-2, and DU145 cells, as well as PNT1A cells, an immortalized human prostate epithelial cell line. MAPK4 is expressed at high levels in VCaP, LAPC4, and 22Rv1 cells, with low expression in LNCaP, C4-2, DU145, and PNT1A cells (Figure 1A). To assess MAPK4 function in PCa, we performed knockdown of MAPK4 in VCaP, LAPC4, and 22Rv1 cells (see below). As expected, MAPK4 knockdown greatly inhibited LAPC4, VCaP, and 22Rv1 cell proliferation (Figure 1, B and C) as well as anchorage-independent growth (Figure 1D). Thus, MAPK4 plays a critical role in promoting the growth of MAPK4-high PCa cells.
Figure 1. Knockdown of MAPK4 inhibits PCa cell growth.
(A) MAPK4, GATA2, and AR expression in human PCa cell lines as well as in PNT1A, an immortalized normal prostate epithelial cell line that does not express AR. Left panel: Western blots. Right panel: qPCR. (B) MTT assays comparing the growth of the LAPC4 cells with Dox-inducible knockdown of MAPK4 (iG2, iG4) or control (iNT) and VCaP cells with knockdown of MAPK4 (G2, G4) or control (NT). Also shown are 22Rv1 cells with Dox-inducible knockdown of MAPK4 (iG2, iG4) or control (iNT) cultured under complete castration (CAS) condition in media containing 5% charcoal-stripped serum (CSS) plus 10 μM MDV3100 for maximal androgen blockade. Data represent mean ± SD. (C) BrdU incorporation assays comparing the proliferation of 22Rv1 cells with Dox-inducible knockdown of MAPK4 (iG2, iG4) or control (iNT) cultured under CAS condition as described above. Original magnification: ×400. The percentage of BrdU-labeled cells was quantified and data shown as mean ± SEM. (D) Soft-agar assays comparing the anchorage-independent growth of the LAPC4 cells with Dox-inducible knockdown of MAPK4 (iG2) or control (iNT) and VCaP cells with knockdown of MAPK4 (G2) or control (NT). Also shown are 22Rv1 cells with Dox-inducible knockdown of MAPK4 (iG2) or control (iNT) cultured under CAS condition as described above. Original magnification: ×50. The colony numbers were quantified and data shown as mean ± SEM. P values determined by unpaired 2-tailed Student’s t test and adjusted P values determined by 1-way ANOVA followed by Dunnett’s multiple comparisons (B, C) or 2-way ANOVA followed by Sidak’s multiple comparisons (LAPC4 data in D). ****P ≤ 0.0001. Data are representative of at least 3 independent experiments.
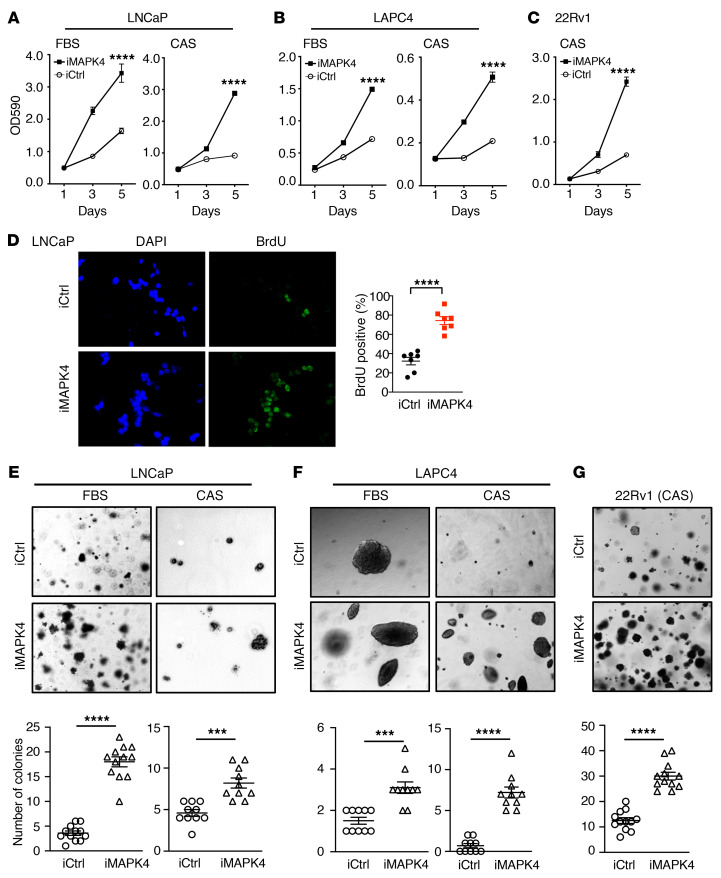
We next engineered ectopic MAPK4 overexpression in LNCaP, LAPC4, and 22Rv1 cells (see below). In general, MAPK4 overexpression promoted LNCaP, LAPC4, and 22Rv1 cell proliferation as well as anchorage-independent growth (Figure 2, A–G). More specifically, proliferation and anchorage-independent growth of the androgen-dependent control LNCaP and LAPC4 cells were minimal when cultured in complete androgen blockade conditions consisting of 5% charcoal-striped serum (CSS) deprived of androgen plus cotreatment of 10 μM MDV3100. However, MAPK4 overexpression potently stimulated LNCaP and LAPC4 cell growth, including anchorage-independent growth under such conditions (Figure 2, A, B, E, and F). MAPK4 overexpression also robustly promoted PNT1A cell growth and transformed these cells into anchorage-independent growth, as we previously reported (35). Altogether, these results demonstrate that MAPK4 promotes growth, including androgen-independent growth of PCa cells in vitro.
Figure 2. MAPK4 overexpression promotes PCa cell growth.
(A–C) MTT assays comparing the growth of LNCaP (A), LAPC4 (B), and 22Rv1 (C) cells with Dox-inducible overexpression of MAPK4 (iMAPK4) or control (iCtrl). FBS: cells cultured in media containing 10% FBS. CAS: cells cultured under CAS condition in media containing 5% CSS plus 10 μM MDV3100. Data represent mean ± SD. (D) BrdU incorporation assays comparing the proliferation of LNCaP cells with Dox-inducible expression of MAPK4 (iMAPK4) or control (iCtrl). Original magnification: ×400. The percentage of BrdU-labeled cells were quantified and data shown as mean ± SEM. (E–G) Soft-agar assays comparing the anchorage-independent growth of LNCaP (E), LAPC4 (F), and 22Rv1 (G) cells with Dox-inducible overexpression of MAPK4 (iMAPK4) or control (iCtrl). FBS and CAS: cell culture conditions as described above. Original magnification: ×50. The colony numbers were quantified and data shown as mean ± SEM. P values determined by unpaired 2-tailed Student’s t test. ***P ≤ 0.001. ****P ≤ 0.0001. Data are representative of at least 3 independent experiments.
MAPK4 induces GATA2 and AR expression in PCa.
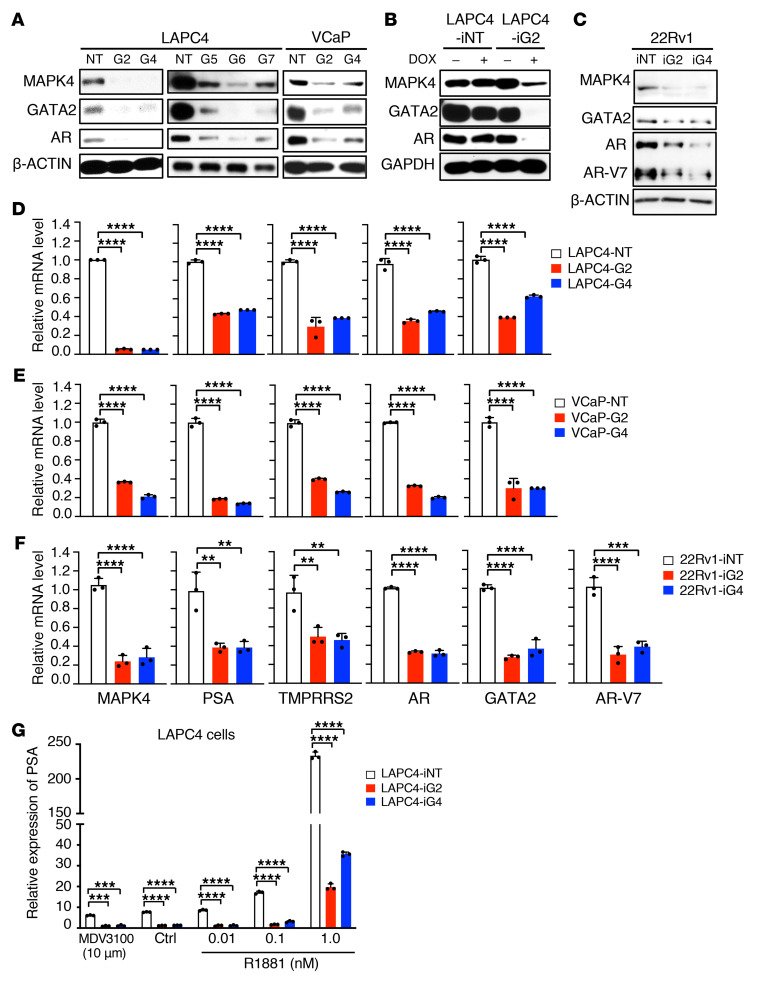
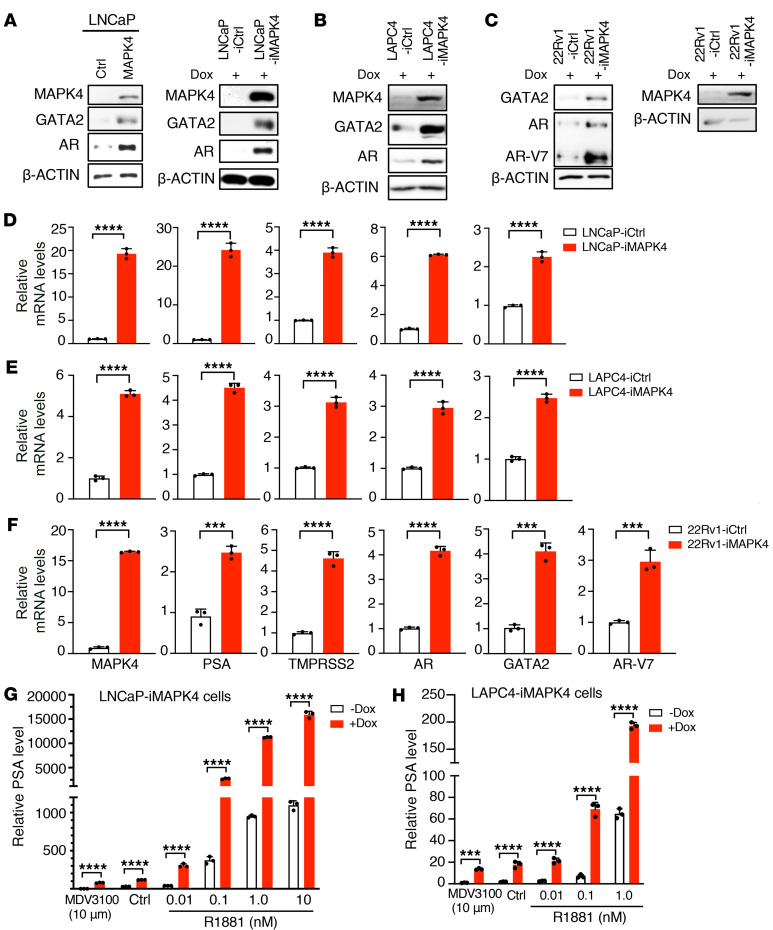
Since AR plays an essential role in driving most PCa and GATA2 is a key transcription factor regulating AR expression and activation, we examined the expression of MAPK4, AR, and GATA2 in human PCa cells (Figure 1A). Expression of AR and GATA2 generally followed that of MAPK4 in these cells, suggesting that MAPK4 may regulate GATA2 and AR expression. Indeed, knockdown of MAPK4 in the LAPC4 cells using 5 independent shRNAs greatly repressed GATA2 and AR protein expression (Figure 3A). Knockdown of MAPK4 in VCaP cells and Dox-induced knockdown of MAPK4 in LAPC4 cells similarly suppressed GATA2 and AR expression (Figure 3, A and B). In the opposite direction, constitutive overexpression of MAPK4 in LNCaP cells as well as Dox-induced overexpression of MAPK4 in LNCaP cells and LAPC4 cells all greatly increased GATA2 and AR protein levels (Figure 4, A and B). AR-V7 is an AR splicing variant with androgen-independent constitutive activity, and gain of AR-V7 expression is an established mechanism for castration-resistant growth in CRPC (36–41). 22Rv-1 cells highly express AR-V7 as well as full-length AR at lower level (38, 39). Interestingly, Dox-inducible knockdown of MAPK4 expression in 22Rv1 cells repressed expression of GATA2 and AR-V7, as well as full-length AR (Figure 3C). In accord with this, Dox-inducible ectopic overexpression of MAPK4 enhanced expression of GATA2, AR-V7, and full-length AR in 22Rv1 cells (Figure 4C). Altogether, these data strongly support that MAPK4 promotes GATA2 and AR/AR-V7 expression in PCa.
Figure 3. Knockdown of MAPK4 represses GATA2/AR expression and AR activation in PCa cells.
Western blots on (A) LAPC4 and VCaP cells with knockdown of MAPK4 (G2, G4, G5, G6, G7) or control (NT), (B) LAPC4 cells with Dox-inducible knockdown of MAPK4 (iG2) or control (iNT), and (C) 22Rv1 cells with Dox-inducible knockdown of MAPK4 (iG2 and iG4) or control (iNT). qPCR on (D) LAPC4, (E) VCaP, and (F) 22Rv1 cells with knockdown of MAPK4 (G2, G4) or control (NT), or Dox-inducible knockdown of MAPK4 (iG2, iG4) or control (iNT). (G) LAPC4 cells with Dox-inducible knockdown of MAPK4 (iG2, iG4) or control (iNT) were treated with 10 μM MDV3100, Ctrl, or increasing doses (0.01, 0.1, and 1 nM) of R1881 in 10% CSS in the presence of 2 μg/mL Dox. qPCR was used for assessing PSA expression. Data represent mean ± SD. Adjusted P values determined by 1-way (D–F) or 2-way ANOVA (G) followed by Dunnett’s multiple comparisons. **P ≤ 0.01. ***P ≤ 0.001. ****P ≤ 0.0001. Data are representative of at least 3 independent experiments.
Figure 4. MAPK4 induces GATA2/AR expression and AR activation in PCa cells.
Western blots on (A) LNCaP cells with overexpression of MAPK4, control (Ctrl), Dox-inducible overexpression of MAPK4 (iMAPK4) or control (iCtrl), (B) LAPC4 cells, and (C) 22Rv1 cells with Dox-inducible overexpression of MAPK4 (iMAPK4) or control (iCtrl). Additional loading control (β-ACTIN) was used for the ectopic MAPK4 expression in 22Rv1-iMAPK4 cells. The left and right panels of Western blots were on the same samples run at different times. qPCR on (D) LNCaP, (E) LAPC4, and (F) 22Rv1 cells with Dox-inducible overexpression of MAPK4 (iMAPK4) or control (iCtrl). (G) LNCaP-iMAPK4 cells and (H) LAPC4-iMAPK4 cells were treated with 10 μM MDV3100, vehicle control (Ctrl), or increasing doses (up to 10 nM) of R1881 in 10% of CSS in the presence (+) or absence (–) of 0.5 μg/mL Dox. qPCR was used for assessing PSA expression. Data represent mean ± SD. P values determined by unpaired 2-tailed Student’s t test. ***P ≤ 0.001. ****P ≤ 0.0001. Data are representative of at least 3 independent experiments.
MAPK4 induces androgen-independent activation of AR and greatly enhances the ligand-induced AR activation.
To further assess MAPK4 regulation of AR expression and activation in PCa, we measured expression of GATA2, AR/AR-V7, and the AR target genes PSA and TMPRSS2 in MAPK4-knockdown LAPC4, VCaP, and 22Rv1 cells and MAPK4-overexpressing LNCaP, LAPC4, and 22Rv1 cells. In line with MAPK4 regulation of GATA2 and AR/AR-V7 protein expression, MAPK4 promoted expression of the AR targets in all of the cell lines (Figure 3, D–F and Figure 4, D–F).
MAPK4 induction also both promoted basal, androgen-independent AR activation and supported androgen-dependent AR activation. Dox-induced MAPK4 overexpression significantly activated both basal AR activity in LNCaP and LAPC4 cells cultured in 10% charcoal-stripped serum (CSS) plus 10 μM MDV3100 for maximum androgen ligand blockade, as well as AR agonist R1881-induced AR activation of PSA mRNA expression (Figure 4, G and H). In accord with this, knockdown of MAPK4 greatly inhibited both baseline and the ligand-stimulated AR activation in LAPC4 cells (Figure 3G).
MAPK4-induced AR activation crucially supports the growth promoting activity of MAPK4 in PCa.
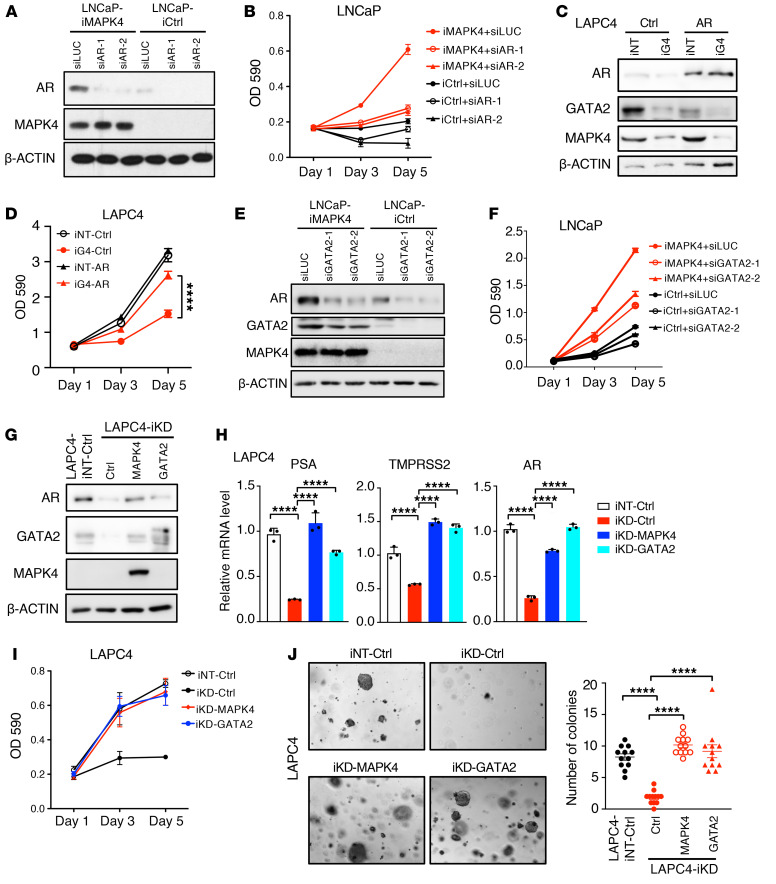
To assess the functional significance of AR signaling in mediating MAPK4 tumor-promoting activity in PCa, we used AR knockdown in the MAPK4-overexpressing LNCaP cells. As expected from the critical roles of AR in PCa, AR knockdown significantly repressed MAPK4-induced LNCaP cell growth (Figure 5, A and B; Supplemental Figure 1A; supplemental material available online with this article; https://doi.org/10.1172/JCI135465DS1). In accord with this, AR overexpression in control LAPC4 cells minimally affected cell growth, but largely rescued MAPK4-knockdown LAPC4 cell growth (Figure 5, C and D) and AR target gene expression (Supplemental Figure 1B). Altogether, these data suggest that MAPK4-induced AR signaling crucially supports MAPK4 growth promoting activity in PCa.
Figure 5. GATA2/AR signaling is essential for mediating MAPK4 tumor-promoting activity in PCa.
(A) Western blots and (B) Proliferation assays on the LNCaP cells with Dox-induced MAPK4 overexpression (iMAPK4) or control (iCtrl) transfected with siRNA against AR (siAR-1, siAR-2) or luciferase (siLUC, negative control). (C) Western blots and (D) Proliferation assays on the LAPC4 cells with Dox-induced MAPK4 knockdown (iG4) that also ectopically express AR or Control (Ctrl), or control (iNT). (E) Western blots and (F) Proliferation assays on the Dox-induced LNCaP-iMAPK4 or LNCaP-iCtrl cells transfected with siRNA against GATA2 (siGATA2-1, siGATA2-2) or luciferase (siLUC). (G) Western blots, (H) qPCR analysis, (I) proliferation assays, and (J) soft-agar assays (original magnification: ×50) on the Dox-induced MAPK4-knockdown LAPC4 (iKD) cells that also overexpress MAPK4, GATA2, or control (Ctrl). LAPC4-iNT: Dox-induced nontargeting control LAPC4 cells. In H, data represent mean ± SD. All other data represent mean ± SEM. P values determined by unpaired 2-tailed Student’s t test and adjusted P values determined by 1-way ANOVA followed by Sidak’s multiple comparisons. ****P ≤ 0.0001. Data are representative of at least 3 independent experiments.
MAPK4 enhances GATA2 to induce AR expression and activation and to promote PCa cell growth.
Since GATA2 is essential for AR expression and activation in PCa (7), we next investigated GATA2 function in mediating MAPK4-induced AR signaling and PCa cell growth. GATA2 knockdown in LNCaP-iMAPK4 cells greatly inhibited MAPK4-induced AR expression and PSA expression, and cell growth, supporting the critical roles of GATA2 in mediating MAPK4 biology in these cells (Figure 5, E and F; Supplemental Figure 1A). Furthermore, GATA2 knockdown in control LAPC4 cells reduced AR expression and cell growth to levels similar to those of MAPK4-knockdown cells, and GATA2 knockdown in MAPK4-knockdown LAPC4 cells further reduced cell growth (Supplemental Figure 1C). We made similar observations in VCaP cells (Supplemental Figure 1D). Therefore, knockdown of GATA2 in MAPK4-high PCa cells at least partially recapitulates MAPK4-knockdown phenotype.
To further define the functional significance of GATA2 in mediating MAPK4 activity in PCa, we also performed gain-of-function studies. Ectopic expression of GATA2, like MAPK4, largely rescued AR expression and activation as well as the growth, including anchorage-independent growth of MAPK4-knockdown LAPC4 cells (Figure 5, G–J). Altogether, these data suggest that MAPK4-induced GATA2/AR signaling crucially supports MAPK4 growth promoting activity in PCa.
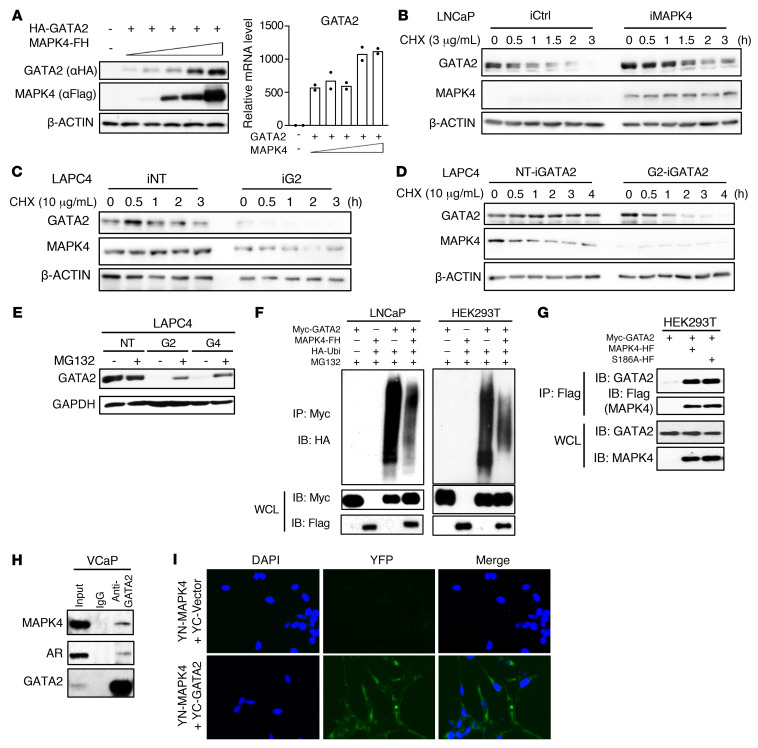
MAPK4 promotes GATA2 expression by both inducing GATA2 gene transcription and enhancing GATA2 protein stability.
MAPK4 promoted GATA2 gene transcription (Figure 3, D–F and Figure 4, D–F). However, GATA2 protein is unstable and subject to ubiquitin-proteasome mediated degradation (12, 13). To assess whether MAPK4 also regulates GATA2 protein stability, we first investigated MAPK4 regulation of GATA2 protein levels when GATA2 mRNA expression levels were comparable. Accordingly, we transfected HA-tagged GATA2 into HEK293T cells together with increasing doses of a Flag/His-tagged MAPK4. As expected, increasing doses of MAPK4 exhibited limited effect on mRNA levels of the ectopically expressed GATA2. In sharp contrast, MAPK4 substantially enhanced the protein levels of this ectopically overexpressed GATA2 in a dose-dependent manner (Figure 6A). This suggests that MAPK4 is capable of potently regulating GATA2 protein levels.
Figure 6. MAPK4 enhances GATA2 protein levels in PCa.
(A) HEK293T cells were cotransfected with vectors carrying HA-tagged GATA2 (HA-GATA2) and increasing doses of Flag/His-tagged MAPK4 (MAPK4-FH). Left panel: Western blots. Right panel: qPCR. Western blots were performed using cell lysates from (B) LNCaP-iMAPK4 (Dox-inducible MAPK4 expression) cells induced with 0.5 μg/mL Dox, (C) LAPC4-iNT (control) and LAPC4-iG2 (Dox-inducible knockdown of MAPK4) cells induced with 2 μg/mL Dox, (D) LAPC4-NT (control) and LAPC4-G2 (stable knockdown of MAPK4) cells with 0.5 μg/mL Dox-induced ectopic expression of GATA2 (iGATA2). All cells were also treated with (B) 3 μg/mL or (C, D) 10 μg/mL CHX for up to 4 hours as indicated. (E) Western blots on LAPC4 cells with stable knockdown of MAPK4 (G2, G4) or control (NT). All cells were also treated with 10 μM MG132 for 4 hours. (F) Ubiquitination assay. LNCaP cells and HEK293T cells were transfected with plasmids expressing Myc-tagged GATA2 (Myc-GATA2), Flag/His-tagged MAPK4 (MAPK4-FH), HA-tagged Ubiquitin (HA-Ubi), and/or vector controls. Twenty-four hours later, cells were treated with 10 μM MG132 for 6 hours before collection. Immunoprecipitation were performed using anti-Myc tag antibody, and Western blots were run using anti-HA antibody. WCL: whole cell lysate. Coimmunoprecipitation (co-IP) assays reveal (G) ectopically overexpressed Myc-GATA2 binding to Flag/His-tagged MAPK4 and MAPK4S186A mutant (MAPK4-HF and S186-HF) in the transfected HEK293T cells, and (H) endogenous GATA2 binding to endogenous MAPK4 in VCaP cells. (I) BiFC assay for GATA2 association with MAPK4 in cytoplasm. HeLa cells were cotransfected with YN-MAPK4 and YC-GATA2 or YC control vectors. Twenty-four hours later, the cells were fixed, counterstained with DAPI, and imaged for YFP fluorescence to indicate MAPK4-GATA2 interaction. Original magnification: ×400. Data are representative of at least 3 independent experiments.
We next examined how MAPK4 regulates endogenous GATA2 protein stability in PCa cells. While Dox-induced MAPK4 expression in the LNCaP-iMAPK4 cells induced GATA2 gene transcription (Figure 4D), blocking protein synthesis using cycloheximide (CHX) revealed that MAPK4 also increased GATA2 protein stability (Figure 6B). While endogenous and ectopically overexpressed GATA2 protein were relatively stable in CHX-treated LAPC4 cells, MAPK4 knockdown greatly reduced their stability (Figure 6, C and D), and proteasome inhibitor MG132 treatment partially blocked this reduction (Figure 6E). This partial rescue of GATA2 levels in the MG132-treated MAPK4-knockdown LAPC4 cells is consistent with the notion that MAPK4 promotes GATA2 expression through both enhancing GATA2 gene transcription (Figure 3D and Figure 4E) and inhibiting GATA2 protein degradation. Ubiquitination assays revealed that MAPK4 greatly reduced GATA2 ubiquitination in both LNCaP cells and HEK293T cells, further confirming that MAPK4 enhances GATA2 protein stability by repressing GATA2 protein ubiquitination and degradation (Figure 6F).
To assess whether MAPK4 directly regulates GATA2 protein ubiquitination and degradation, we further investigated MAPK4-GATA2 interaction. When ectopically overexpressed in HEK293T cells, a Myc-tagged GATA2 coimmunoprecipitated (co-IP) with a Flag-tagged MAPK4 (Figure 6G). We also detected the co-IP of endogenous MAPK4 and endogenous GATA2 in VCaP cells (Figure 6H). Bimolecular fluorescence complementation (BiFC) enables visualization of protein-protein interactions and subcellular locations in live cells (42). Accordingly, we co-transfected HeLa cells with a YN-MAPK4 construct encoding MAPK4 fused to the N-terminal fragment of YFP, along with a YC-GATA2 construct encoding GATA2 fused to the C-terminal fragment of YFP. Observation of a positive cytoplasmic signal in cells coexpressing YN-MAPK4 and YC-GATA2, but not in the YN-MAPK4 or YC vector control cotransfected cells, further confirmed the MAPK4-GATA2 interaction and revealed that this interaction occurs mainly in the cytoplasm (Figure 6I).
Altogether, these data suggest that besides promoting GATA2 gene transcription, MAPK4 binds GATA2 and further enhances GATA2 protein levels by stabilizing GATA2 protein through inhibiting its proteasome-mediated degradation.
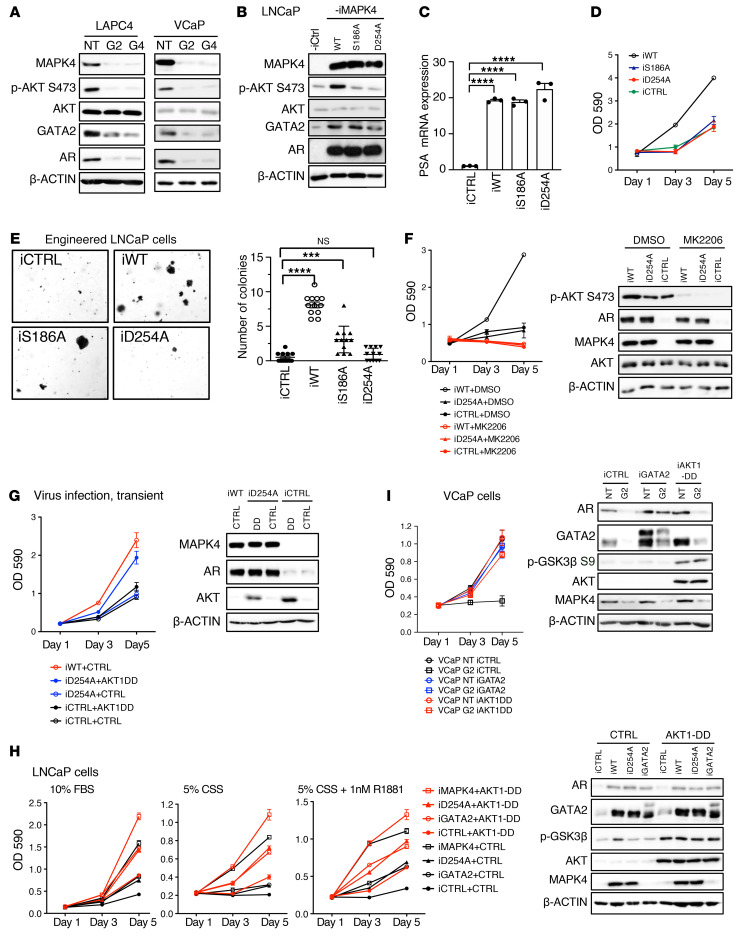
AKT activation is essential for mediating the growth-promoting activity of MAPK4 in PCa.
We previously reported that MAPK4 promotes tumor growth via noncanonically phosphorylating and activating AKT (35). As expected, MAPK4 knockdown also repressed AKT phosphorylation in PCa cells (Figure 7A). The mutation of MAPK4 residues D254A or S186A largely abolished MAPK4 activation of AKT via different mechanisms (35). Interestingly, when overexpressed in LNCaP cells to levels comparable to the endogenous MAPK4 levels in VCaP and LAPC4 cells (Supplemental Figure 2A), both MAPK4D254A and MAPK4S186A mutants retained their ability to induce GATA2 and AR expression and AR activation, indicating that MAPK4 regulates GATA2/AR signaling and AKT signaling through distinct mechanisms (Figure 7, B and C).
Figure 7. Concerted Activation of AKT and GATA2/AR signaling is essential for mediating MAPK4 tumor-promoting activity in PCa.
(A) Western blots on LAPC4 cells and VCaP cells with knockdown of MAPK4 (G2, G4) or control (NT). (B) Western blots, (C) qPCR, (D) Proliferation assays, and (E) Soft-agar assays (original magnification: × 50) on the engineered LNCaP cells with 0.5 μg/mL Dox-induced overexpression of MAPK4 (iWT), MAPK4S186A (iS186A), MAPK4D254A (iD254A), or control (iCtrl). Data represent mean ± SEM. Adjusted P values determined by 1-way ANOVA followed by Dunnett’s multiple comparisons. ***P ≤ 0.001. ****P ≤ 0.0001. (F) Proliferation assays and Western blots on the LNCaP cells with 0.5 μg/mL Dox-induced overexpression of MAPK4 (iWT), MAPK4D254A (iD254A), or control (iCtrl), treated with 1 μM of AKT inhibitor MK2206 or DMSO control. (G) Proliferation assays and Western blots on the LNCaP cells with 0.5 μg/mL Dox-induced expression of MAPK4, MAPK4D254A, or control, also infected with lentivirus expressing AKT1-DD mutant or control. (H) Proliferation assays on the LNCaP cells with 0.5 μg/mL Dox-induced overexpression of MAPK4, MAPK4D254A, GATA2, or control, also overexpressing AKT1-DD or control. The growth of these cells in 10% FBS, 5% CSS, and 5% CSS plus 1 nM R1881 were compared. Also shown are Western blots on these cells cultured in 10% FBS. (I) Proliferation assays and Western blots on the control (NT) or MAPK4-knockdown (G2) VCaP cells with 0.5 μg/mL Dox-induced expression of GATA2 (iGATA2), AKT1-DD (iAKT1DD), or control (iCtrl). Data are representative of at least 3 independent experiments.
Since AKT activation is essential for mediating MAPK4 oncogenic activity (35), we next compared the biological activities of WT MAPK4 and the MAPK4D254A and MAPK4S186A mutants in promoting LNCaP cell growth. Interestingly, WT MAPK4, but not MAPK4D254A or MAPK4S186A, robustly promoted LNCaP cell proliferation and anchorage-independent growth (Figure 7, D and E). In accord with this, the AKT inhibitor MK2206 greatly inhibited MAPK4-induced LNCaP cell growth (Figure 7F), as well as that of MAPK4-high LAPC4 cells and 22Rv1 cells (Supplemental Figure 2B). We also confirmed that MK2206 treatment did not repress MAPK4-induced GATA2 and AR expression and AR activation in LNCaP, LAPC4, and 22Rv1 cells (Figure 7F; Supplemental Figure 2, C-E). Altogether, these data are in accord with previous results showing that AKT activation is essential for mediating MAPK4 tumor-promoting activity (35).
Since MAPK4D254A and MAPK4S186A mutants were able to enhance GATA2/AR signaling but failed to robustly promote LNCaP cell growth, MAPK4-induced GATA2/AR signaling alone appeared not sufficient to mediate the growth-promoting effects of MAPK4. Activation of AKT alone by overexpression of a constitutively active AKT1T308D/S473D (DD) mutant was also not sufficient to substantially promote LNCaP cell growth. However, overexpression of this AKT1(DD) mutant largely rescued MAPK4D254A growth-promoting activity in LNCaP-iMAPK4D254A cells (Figure 7G) and in the stably engineered LNCaP cell lines (Figure 7H). These results further support the essential role of AKT activation in mediating MAPK4-dependent PCa cell growth.
Concerted activation of AKT and GATA2/AR signaling is sufficient to promote PCa growth and castration resistance.
Since AKT and GATA2/AR signaling pathways appeared to work concertedly downstream of MAPK4, we next investigated whether simultaneous activation of AKT (expression of AKT1[DD]) and GATA2 overexpression in the absence of ectopic MAPK4 overexpression promotes PCa growth and castration resistance. While expression of AKT1(DD) or GATA2 alone exhibited some activities, the coexpression of both substantially enhanced LNCaP cell growth (Figure 7H). Furthermore, while GATA2 growth-promoting activity was markedly reduced in engineered LNCaP cells cultured in 5% CSS deprived of androgen, coexpression of AKT1(DD) and GATA2 was sufficient to induce robust proliferation under the same condition (Figure 7H). These data support that the concerted activation of AKT and GATA2/AR signaling is sufficient to promote PCa growth and castration resistance. Finally, ectopic overexpression of GATA2 or AKT1(DD) alone was sufficient to mostly rescue the growth of the MAPK4-knockdown VCaP cells (Figure 7I). These data suggest that the remaining MAPK4 activity in the MAPK4-knockdown VCaP cells may be sufficient to support VCaP cell growth once supplemented with either GATA2 or AKT1 activation.
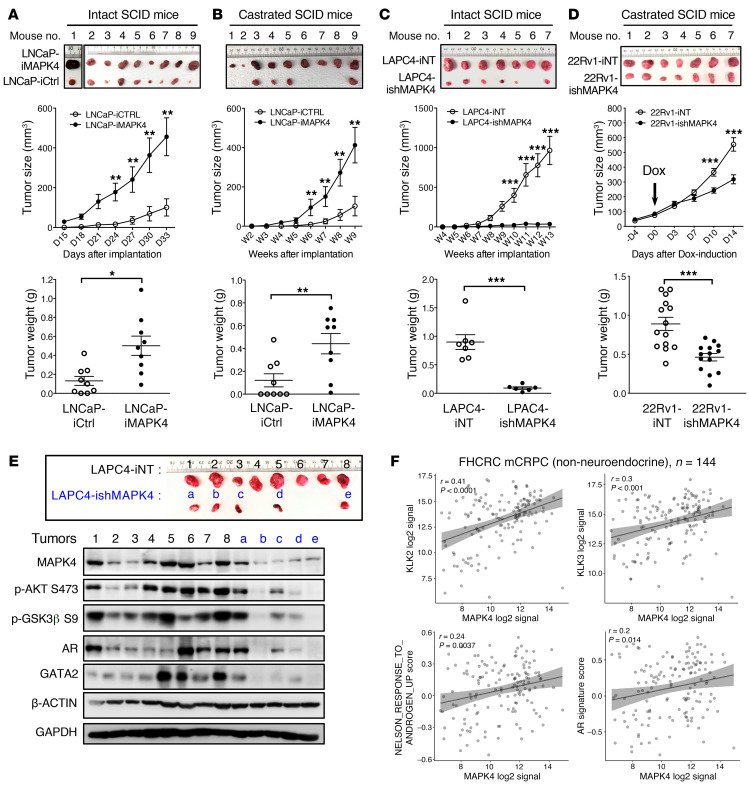
MAPK4 promotes PCa xenograft growth and castration resistance, and its expression correlates with AR activation in human CRPC tissues.
We further investigated MAPK4 regulation of prostate xenograft tumor growth and castration resistance in vivo. We first performed xenograft studies using the LNCaP-iMAPK4 cells in both intact and castrated SCID mice. Dox-induced MAPK4 overexpression greatly enhanced LNCaP xenograft growth in the intact mice, confirming the potent tumor-promoting activity of MAPK4 in PCa in vivo (Figure 8A). Furthermore, while only 4 of 9 control LNCaP xenografts formed detectable tumors in the castrated SCID mice 9 weeks after inoculation, MAPK4 overexpression both increased the tumor incidence to 100% and promoted the growth of these tumors (Figure 8B). Conversely, Dox-induced knockdown of MAPK4 greatly inhibited LAPC4 xenograft growth in vivo (Figure 8C). Western blot analysis confirmed the association of MAPK4 expression with GATA2, AR, and AKT activation in these tumors (Figure 8E).
Figure 8. MAPK4 promotes prostate tumor growth and its expression correlates with AR activation in human CRPC.
Dox-induced overexpression of MAPK4 promotes LNCaP xenograft growth in (A) intact SCID mice and (B) castrated SCID mice. (C) Dox-induced knockdown of MAPK4 inhibits LAPC4 xenograft growth in SCID mice. 2 × 106 of LNCaP-iMAPK4, LNCaP-iCtrl, LAPC4-iNT, or LAPC4-ishMAPK4 cells were injected into the lateral flanks (SubQ) of intact or castrated SCID mice (iCtrl or iNT on the left side; iMAPK4 or ishMAPK4 on the right side). Mice began receiving Dox (0.2 mg/mL for LNCaP and 4 mg/mL for LAPC4 xenografts) in 10% sucrose in drinking water on the day of xenograft implantation. Tumors were harvested as indicated. (D) Dox-induced knockdown of MAPK4 inhibits the growth of previously established 22Rv1 xenograft tumors in castrated SCID mice. A quantity of 2 × 106 22Rv1-iNT or 22Rv1-ishMAPK4 cells were injected into the lateral flanks (SubQ) of castrated SCID mice (iNT on the left side; ishMAPK4on the right side). Mice began receiving Dox (4 mg/ml) in 10% sucrose in drinking water when tumors grow into about 100 mm3. Tumors were harvested as indicated. iMAPK4 and iCtrl: Dox-inducible expression of MAPK4 or control. ishMAPK4 and iNT: Dox-inducible knockdown of MAPK4 or nontargeting control. Data represent mean ± SEM. P values determined by unpaired 2-tailed Student’s t test. *P ≤ 0.05. **P ≤ 0.01. ***P ≤ 0.001. Data are representative of at least 3 independent experiments. (E) Western blots on an independent set of LAPC4-iNT or LAPC4-ishMAPK4 tumors (a repeated study of panel C). Data are representative of at least 3 independent experiments. (F) MAPK4 mRNA expression correlates with AR activation status in human CRPC tissues.
Finally, to probe how knockdown of MAPK4 affects the growth of previously established CRPC tumors, we performed xenograft studies using the 22Rv1-ishMAPK4 cells and the control 22Rv1-iNT cells on castrated SCID mice. After xenografts grew into about 100 mm3, the mice began to receive 4 mg/mL Dox in drinking water to induce knockdown of MAPK4 in the 22Rv1-ishMAPK4 tumors but not in the control 22Rv1-iNT tumors. Knockdown of MAPK4 again greatly inhibited the growth of these previously established CRPC xenograft tumors (Figure 8D). Altogether, these data support a critical role of MAPK4 in driving PCa growth and castration resistance in vivo.
AR activation in human PCa tissues is subjected to complex regulation, including circulating/intratumor steroid levels, steroid synthesis/metabolism (43–46), AR amplification/overexpression/mutation (47–49), AR splice variants (36–41), AR coactivators/corepressors (50), protein kinases (31, 51–56), and tumor microenvironment (57). To enhance our ability to detect the impact of MAPK4 on AR activation in human PCa, we specifically analyzed CRPC from patients in whom circulating steroid levels and ligand-induced AR activation were reduced. Analysis of the gene expression profile of laser-captured PCa cells from nonneuroendocrine CRPC tissues of deceased patients (58) confirmed a positive correlation between MAPK4 expression and the expression of AR targets KLK2, PSA/KLK3, the Nelson AR signature (59), and the TCGA AR signature (http://software.broadinstitute.org/gsea/msigdb/cards/NELSON_RESPONSE_TO_ANDROGEN_UP.html) (Figure 8F). Altogether, these data support a positive impact of MAPK4 activation on AR in human PCa.
Discussion
Both AKT and AR signaling pathways play important roles in promoting PCa. Two landmark studies have revealed that AKT and AR antagonize each other in PCa (28, 29); repressing one signaling pathway led to the activation of the other. In contrast, our study demonstrated that MAPK4 concertedly activates both AR and AKT and promotes PCa growth and castration resistance. Accordingly, inhibiting MAPK4 simultaneously represses AR and AKT activation and inhibits PCa growth, including castration-resistant growth. We believe that these findings support MAPK4 as a novel therapeutic target for PCa, including CRPC.
Since the pioneering work from the Brown lab identified GATA2 as a key factor regulating AR signaling and PCa biology more than a decade ago (6), little progress has been made on understanding molecular mechanisms that regulate GATA2 expression and protein stability in PCa. Here we demonstrated that MAPK4 enhances GATA2 gene transcription through mechanisms that remain to be delineated. In addition, MAPK4 promotes GATA2 protein stability through inhibiting its ubiquitination and degradation. By greatly enhancing GATA2 expression/activation, MAPK4 promotes AR expression and activation, PCa growth, and castration resistance. In accord with this, GATA2 knockdown repressed MAPK4-induced AR expression/activation, and knockdown of either GATA2 or AR inhibited MAPK4-mediated PCa cell growth.
We previously identified 2 MAPK4 mutants (D254A and S186A) with greatly decreased ability to activate AKT (35). Interestingly, these 2 mutants exhibited few growth-promoting effects when overexpressed in LNCaP cells. Coexpression of a constitutively activated AKT (AKT1T308D/S473D) largely rescued the growth-promoting activities of these 2 mutants, supporting that AKT activation is essential for mediating MAPK4 activities in PCa. This is consistent with our previous results on the essential role of AKT in mediating MAPK4 tumor-promoting activities in other types of cancers (35). In accord with this, AKT inhibitor treatment blocked the growth promoting activities of MAPK4 in PCa. These data shed light on the notion that MAPK4-high PCa, especially the MAPK4-high therapy-resistant CRPC, may benefit from the AKT blockade therapy.
Overexpression of GATA2 alone or activation of AKT alone (overexpression of AKT1T308D/S473D) exhibited a moderate effect on LNCaP cell growth (Figure 7). In contrast, coexpression of both GATA2 and AKT1-DD mutant robustly promoted LNCaP cell growth, which largely recapitulated MAPK4 activity in these cells (Figure 7). These data support the concerted activation of both GATA2/AR and AKT as dual mediators of MAPK4 biology in PCa. The detailed molecular mechanisms underlying the synergistic activities of GATA2 and AKT in driving PCa remain to be defined.
MAPK4 activation promotes both hormone-independent and hormone-dependent AR transactivation (Figure 4). The augmentation of agonist response sensitizes PCa cells to subnanomolar androgen levels. Although the mechanisms remain to be determined, AR overexpression has been suggested to enhance AR activation in PCa and the strong induction of AR expression in response to MAPK4 presumably contributes to this potentiation. Importantly, the effect of MAPK4 extends to the clinically significant AR splicing variant AR-V7, which can drive castration resistance of PCa (36–41).
AKT activation has been indicated in the development of CRPC, and AR remains activated in most CRPC. MAPK4 can activate both AKT and AR, suggesting its potent activity in driving castration resistance of PCa. Our data from LNCaP, LAPC4, and 22Rv1-based models collectively support this notion. Analysis further reveals that MAPK4 expression correlates with AR activation in CRPC tissues (Figure 8F), supporting the human disease relevance of our discovery. Therefore, targeting MAPK4 emerges as a promising therapeutic avenue for a subset of CRPC with significant expression of MAPK4.
Methods
Reagents and antibodies.
Antibodies, including those against HA (catalog sc-805), Myc (catalog sc-40), GAPDH (catalog sc-32233), AR (catalog sc-816), Protein A-Agarose (catalog sc-2001), and Protein G PLUS-Agarose (catalog sc-2002) were purchased from Santa Cruz Biotechnology. Anti-MAPK4 antibodies used include AP7298b from Abgent, HPA007461 from MilliporeSigma, and TA505872 from Origene. Anti-GATA2 antibodies used include sc-9008 from Santa Cruz Biotechnology and A0677 from ABclonal. Antibodies against p-AKT T308 (catalog 4056), p-AKT S473 (catalog 9271), AKT (catalog 2920), and p-GSK3β S9 (catalog 9558) were from Cell Signaling Technology. EZview Red Anti-FLAG M2 Affinity Gel (catalog F2426), MG132 (catalog M7449), MTT (3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide), R1881 (catalog R0908), BrdU (catalog B9285), and antibodies against β-ACTIN (catalog A1978), BrdU (catalog B8434), and FLAG (catalog F3165) were purchased from MilliporeSigma. Enzalutamide (MDV3100, catalog S1250) and MK2206 (catalog S1078) were purchased from Selleckchem.
Plasmids.
The pGIPZ lentiviral shRNA constructs were purchased from Open Biosystem (Thermo Fisher Scientific). The pInducer10 and pInducer20 vectors were provided by Thomas Westbrook, Baylor College of Medicine, Houston, Texas, USA (60). pInducer20-YF vector was previously generated to allow cDNA subcloning independent of the Gateway system (35). For Dox-inducible knockdown, shRNA oligos were synthesized and cloned into the pLKO-Tet-On vector between AgeI and EcoRI sites. We also used the pInducier10 lentiviral system for the Dox-inducible knockdown of MAPK4, as previously described (35). GATA2 cDNA was PCR amplified from the cDNA prepared from the HEK293T cells and cloned into the pRK5 vector (35) between EcoRI and SalI sites. GATA2 cDNA was also subcloned into the pInducer20-YF vector (35) and the pCDH-CMV-MCS-EF1α-RFP+Puro vector (CD516B-2, System Biosciences) to generate the lentiviral vectors for Dox-inducible and constitutive overexpression of GATA2. The YN and YC vectors for BiFC assay were provided by Xin-Hua Feng, Baylor College of Medicine, Houston, Texas, USA. GATA2 was cloned into the YC vector, and the obtained YC-GATA2 vector was used in the BiFC assay along with the YN-MAPK4 vector, as previously described (35).
Cell culture, transfection, and lentivirus infection.
LNCaP, LAPC4, 22Rv1, and VCaP cells were acquired from ATCC. PNT1A cells were acquired from the European Collection of Authenticated Cell Cultures (ECACC). LNCaP and 22Rv1 cells were cultured in RPMI-1640 supplemented with 10% FBS (Hyclone or Invitrogen), and VCaP and PNT1A cells in DMEM supplemented with 10% FBS. LAPC4 cells were cultured in IMDM supplemented with 10% FBS and 1nM R1881. Charcoal-stripped FBS (catalog SH3007103) was from Hyclone.
GenMute siRNA transfection reagent (catalog SL100568) and LipoD293 DNA transfection reagent (catalog SL100668, SignaGen Laboratories) were used for siRNA and DNA transfection. For lentiviral-mediated gene delivery, lentiviral-based expression constructs were transfected into the HEK293T cells together with the packaging mix in the Trans-lentiviral packaging kit (catalog TLP5914, Thermo Fisher Scientific). Viruses in the conditioned media were harvested at 48 hours to 96 hours after the transfection and filtered (0.45 μm). The cells were infected with the viral medium for 3 days before drug selection. The established stabled cell lines were then expanded for further assays.
The pCDH and pInducer20-YF based lentiviral gene delivery system were used for stable and Dox-inducible overexpression of MAPK4, AR, or GATA2 in LNCaP, LAPC4, or 22Rv1 cells. The pGIPZ, pLKO-Tet-On, or pInducer10 vectors were used for lentiviral-mediated stable or Dox-inducible knockdown of MAPK4, AR, or GATA2 in VCaP, LAPC4, LNCaP, or 22Rv1 cells. The inducible cells were treated with 2 μg/mL Dox for at least 3 days to obtain significant MAPK4 knockdown or 0.2–0.5 μg/mL Dox for at least 3 days for MAPK4 overexpression.
Western blot.
Protein samples for Western blot assays were prepared in RIPA buffer. Cell lysate was applied to a brief sonication before protein quantification using the BCA protein assay kit (catalog 23225, Pierce). An equal amount (5–20 μg) of protein was used in Western blot analysis.
Bimolecular fluorescence complementation (BiFC) assay.
To directly visualize MAPK4 and GATA2 interaction in live cells, we performed bimolecular fluorescence complementation (BiFC) as previously described (35). In brief, we transfected the YN-MAPK4 and YC-GATA2 into HeLa cells and used transfection of YN-MAPK4 and YC-vector as negative control. At 24 hours after transfection, cells were fixed, and the fluorescence signal was detected under microscope as previously described (35).
BrdU assay.
Before being seeded on coverslips in 24-well plates, LNCaP-iCtrl/iMAPK4 cells were treated with 0.5 μg/mL Dox in 10% FBS RPMI-1640 and 22Rv1-iNT/iG2/iG4 cells were treated with 2 μg/ml Dox in 5% CSS RPMI-1640 plus 10 μM MDV3100 for 3 days. The cells were then cultured in presence of 20 μM BrdU for 16 hours and fixed with cold methanol at –20°C for 20 minutes. After treatment with 2 M HCl for 1 hour, cells were washed with 0.1 M boric acid 3 times and blocked with 1% BSA for 1 hour. Cells were then incubated with anti-BrdU antibody overnight at 4°C, followed by incubation with Dylight 488 goat anti-mouse secondary antibody (catalog 35502, Invitrogen) for 1 hour. After DAPI counterstaining, images were acquired using a fluorescence microscope. The percentage of BrdU-positive cells were analyzed and calculated using ImageJ as previously described (35).
Quantitative real-time PCR.
Total RNA was extracted using the Quick-RNA MiniPrep kit (R1054, Zymo Research). Reverse transcription reactions were performed using the amfiRivert cDNA Synthesis Master Mix (R5600, GenDEPOT). Quantitative real-time PCR was performed using the PerfeCTa SYBR Green FastMix reagent (95072, Quanta Biosciences) on the LightCycler 480 Real-Time PCR System (Roche Applied Science). PCR primers used are AR: 5′-gccttgctctctagcctcaa-3′ and 5′-gtcgtccacgtgtaagttgc-3′, PSA: 5′-ACCAGAGGAGTTCTTGACCCCAAA-3′ and 5′-CCCCAGAATCACCCGAGCAG-3′, TMPRSS2: 5′-cgctggcctactctggaa-3′ and 5′-ctgaggagtcgcactctatcc-3′, MAPK4: 5′-tacggggagaatgctctttg-3′ and 5′-cagggatggtctccaggat-3′, GATA2: 5′-ATCAAGCCCAAGCGAAGACT-3′ and 5′-CATGGTCAGTGGCCTGTTAAC-3′, GAPDH: 5′-AGCCACATCGCTCAGACAC-3′ and 5′-GCCCAATACGACCAAATCC-3′. Relative expression was determined using the ΔΔCt method and normalized to GAPDH.
Ubiquitination assay.
The FLAG-tagged GATA2 cDNA and HA-tagged ubiquitin in pRK5 vector were cotransfected into HEK293T cells. Forty-eight hours later, cells were treated with 10 μM MG132 for 4–6 hours. The cell lysate was prepared and immunoprecipitation was performed using EZview Red Anti-FLAG M2 Affinity Gel (MilliporeSigma). The IP products were then applied in Western blot assay using an anti-HA antibody to reveal GATA2 ubiquitination.
Proliferation assay.
For MTT assay, 5 mg/mL of MTT substrate dissolved in 1× PBS was added to the cell cultures and incubated at 37°C for 4 hours. The purple crystals were then dissolved in isopropanol with 0.1% Triton X-100. Absorbance was read at 590 nm. When applicable, cells were pretreated with 2 μg/mL Dox for 3 days for inducible gene knockdown or 0.5 μg/mL Dox for inducible overexpression. Dox was used throughout the assay.
For crystal violet staining–based cell proliferation assay, cells were fixed in 4% paraformaldehyde in PBS for 15 minutes and stained with 0.1% crystal violet in 10% ethanol for 20 minutes. After washing and air dry, crystal violet was extracted using 10% acetic acid solution (600 μL for 24-well plate). The absorbance was similarly at 590 nm. The first measurement was done 24 hours after cell seeding and cell proliferation was assessed every other day as described.
Soft agar colony formation assay.
Cells were mixed in full medium containing 0.35% low melting agarose at 37°C and then plated onto supporting gel (1% agarose). When applicable, 2 μg/mL Dox was used for inducible gene knockdown and 0.5 μg/mL Dox for inducible overexpression. Colony numbers were counted after inoculation for 3–4 weeks.
Xenograft tumor models.
Male SCID/beige mice, 8 to 10 weeks old, from Envigo were used for xenograft studies. Animals were housed in a pathogen-free facility, and all animal studies were performed under a Baylor College of Medicine IACUC–approved protocol. For LNCaP xenograft studies, 2 × 106 LNCaP-iMAPK4 or LNCaP-iCtrl cells (pretreated with 0.2 μg/mL Dox for 3 days) in 1:1 Matrigel (354234, Corning) were injected into the lateral flanks (subQ) of intact or castrated SCID mice (iCtrl on the left side and iMAPK4 on the right side). Mice began receiving 0.2 mg/mL Dox in 10% sucrose in drinking water on the day of tumor inoculation for Dox-induced MAPK4 overexpression in the LNCaP-iMAPK4 xenografts. For LAPC4 xenograft studies, 2 × 106 LAPC4-ishMAPK4 or LAPC4-iNT cells in 1:1 Matrigel were injected into the lateral flanks of SCID mice (ishMAPK4 on the left side and iNT on the right side). Mice began receiving 4 mg/mL Dox in 10% sucrose in drinking water on the day of tumor inoculation for induced MAPK4 knockdown in the LAPC4-ishMAPK4 xenografts. For 22Rv1 xenograft studies, 2 × 106 22Rv1-ishMAPK4 or 22Rv1-iNT cells in 1:1 Matrigel were injected into the lateral flanks of castrated SCID mice (ishMAPK4 on the left side and iNT on the right side). When tumors grew to about 100 mm3, mice began receiving 4 mg/mL Dox in 10% sucrose in drinking water for inducible knockdown of MAPK4 in the established 22Rv1-ishMAPK4 xenografts. In all studies, the tumors were monitored every 2 days and harvested as indicated and weighed. Tumor size was calculated as volume = 0.52 × abc (where a, b, and c are the maximum length of each dimension of the tumor). Average tumor weight was compared between groups for statistical relevance using the unpaired 2-tailed Student’s t test. P less than 0.05 was considered statistically significant.
siRNAs and shRNAs for knockdown.
Synthetic siRNAs targeting GATA2 and AR were obtained from Invitrogen as follows.
siRNA AR-1: CCGGAAGCUGAAGAAACUUGGUAAU (sense); AUUACCAAGUUUCUUCAGCUUCCGG (anti-sense). siRNA AR-2: GAUGAAGCUUCUGGGUGUCACUAUG (sense); CAUAGUGACACCCAGAAGCUUCAUC (anti-sense). siRNA GATA2-1: CCAGACGAGGUGGACGUCUUCUUCA (sense); UGAAGAAGACGUCCACCUCGUCUGG (anti-sense). siRNA GATA2-2: GGAAGAUGUCCAACAAGUCCAAGAA (sense); UUCUUGGACUUGUUGGACAUCUUCC (anti-sense). siRNA against luciferase (siLuc) was used as negative control: GCACUCUGAUUGACAAAUACGAUUU (sense); AAAUCGUAUUUGUCAAUCAGAGUGC (anti-sense).
Lentiviral shRNAs expression constructs were obtained from Open Biosystem (Thermo Fisher Scientific) with the following targeting sequences: pGIPZ-shMAPK4-G2: GGGTTGGTAACAAAGTGGT; pGIPZ-shMAPK4-G4: CGGGAGGAAGACAAGGACG; pGIPZ-shMAPK4-G5: CGGAAGGTCGCTGTGAAGA; pGIPZ-shMAPK4-G6: AGGATCGTTGATCAGCATT; pGIPZ-shMAPK4-G7: ACAAGGGTTATCTGTCAGA. The following synthesized oligos containing G2 and G4 targeting sequences were cloned into the pLKO-Tet-On vector between AgeI and EcoRI to generate pLKO-G2 (also named as iG2) and pLKO-G4 (also as iG4). pLKO-G2 (iG2): CCGGGGGTTGGTAACAAAGTGGTCTCGAGACCACTTTGTTACCAACCCTTTTT (top); AATTAAAAAGGGTTGGTAACAAAGTGGTCTCGAGACCACTTTGTTACCAACCC (bottom). pLKO-G4 (iG4): CCGGCGGGAGGAAGACAAGGACGCTCGAGCGTCCTTGTCTTCCTCCCGTTTTT (top); AATTAAAAACGGGAGGAAGACAAGGACGCTCGAGCGTCCTTGTCTTCCTCCCG (bottom).
Analysis of human tumor molecular data sets.
Published microarray data (GEO GSE77930) of 171 mCRPC tumors were laser-capture micro-dissected, hybridized to Agilent whole-genome microarrays, and analyzed as previously described (58). Sample phenotypic groups were assigned by clustering using classical multidimensional scaling (MDS) calculated with the cmdscale function in R using the expression profiles of genes from androgen and neuroendocrine signatures (58). The profiles of those samples not characterized as neuroendocrine by transcriptomic signature (n = 144) were further used to analyze gene expression associations. The signature-normalized enrichment scores were calculated in R using the GSVA Bioconductor package with default parameters (61). Pearson correlations of gene expression were calculated by the cor function in R.
Statistics.
The statistical relevance in the cell culture studies and xenograft tumor studies was analyzed using the unpaired 2-tailed Student’s t test. When there were multiple groups in the analysis, 1-way or 2-way ANOVA followed by Dunnett’s multiple comparisons test or Sidak’s multiple comparisons test was performed using GraphPad Prism 9. P less than 0.05 was considered significant.
Study approval.
All animal studies were approved by the Institutional Animal Care and Use Committee of Baylor College of Medicine.
Author contributions
TS, WW, BD, and FY designed the experiments. TS, IC, BD, MMI, DRR, PSN, DDM, and FY wrote and revised the manuscript. TS, WW, WZ, QC, YB, and YM performed the experiments. TS, WW, IC, BD, CJC, and FY analyzed the data.
Supplementary Material
Acknowledgments
This research was supported by grants from the Department of Defense Congressionally Directed Medical Research Programs (W81XWH-13-1-0162, W81XWH-13-1-0163, W81XWH-17-1-0043 to FY) and the Cancer Prevention and Research Institute of Texas (RP130651 to FY). We acknowledge the joint participation by Adrienne Helis Malvin Medical Research Foundation through its direct engagement in the continuous active conduct of medical research in conjunction with Baylor College of Medicine and the MAPK4 as a Novel Therapeutic Target for Human Cancers Cancer Program.
Version 1. 02/15/2021
Electronic publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2021, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2021;131(4):e135465.https://doi.org/10.1172/JCI135465.
Contributor Information
Tao Shen, Email: tshen@bcm.edu.
Wei Wang, Email: dingowang@gmail.com.
Wolong Zhou, Email: wollongzhou@gmail.com.
Ilsa Coleman, Email: icoleman@fredhutch.org.
Qinbo Cai, Email: qinboc@bcm.edu.
Bingning Dong, Email: bd148049@bcm.edu.
Chad J. Creighton, Email: creighto@bcm.edu.
Yingnan Bian, Email: bianyingnan@yeah.net.
Yanling Meng, Email: mengyanl@hotmail.com.
David D. Moore, Email: moore@bcm.edu.
Feng Yang, Email: fyang@bcm.edu.
References
- 1.Siegel RL, et al. Cancer statistics, 2019. CA Cancer J Clin. 2019;69(1):7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 2.Huggins C, Hodges CV. Studies on prostatic cancer. I. The effect of castration, of estrogen and androgen injection on serum phosphatases in metastatic carcinoma of the prostate. CA Cancer J Clin. 1972;22(4):232–240. doi: 10.3322/canjclin.22.4.232. [DOI] [PubMed] [Google Scholar]
- 3.de Bono JS, et al. Abiraterone and increased survival in metastatic prostate cancer. N Engl J Med. 2011;364(21):1995–2005. doi: 10.1056/NEJMoa1014618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Scher HI, et al. Increased survival with enzalutamide in prostate cancer after chemotherapy. N Engl J Med. 2012;367(13):1187–1197. doi: 10.1056/NEJMoa1207506. [DOI] [PubMed] [Google Scholar]
- 5.Smith MR, et al. Apalutamide treatment and metastasis-free survival in prostate cancer. N Engl J Med. 2018;378(15):1408–1418. doi: 10.1056/NEJMoa1715546. [DOI] [PubMed] [Google Scholar]
- 6.Wang Q, et al. A hierarchical network of transcription factors governs androgen receptor-dependent prostate cancer growth. Mol Cell. 2007;27(3):380–392. doi: 10.1016/j.molcel.2007.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu D, et al. Three-tiered role of the pioneer factor GATA2 in promoting androgen-dependent gene expression in prostate cancer. Nucleic Acids Res. 2014;42(6):3607–3622. doi: 10.1093/nar/gkt1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.He B, et al. GATA2 facilitates steroid receptor coactivator recruitment to the androgen receptor complex. Proc Natl Acad Sci U S A. 2014;111(51):18261–18266. doi: 10.1073/pnas.1421415111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bohm M, et al. A role for GATA-2 in transition to an aggressive phenotype in prostate cancer through modulation of key androgen-regulated genes. Oncogene. 2009;28(43):3847–3856. doi: 10.1038/onc.2009.243. [DOI] [PubMed] [Google Scholar]
- 10.Chiang YT, et al. GATA2 as a potential metastasis-driving gene in prostate cancer. Oncotarget. 2014;5(2):451–461. doi: 10.18632/oncotarget.1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vidal SJ, et al. A targetable GATA2-IGF2 axis confers aggressiveness in lethal prostate cancer. Cancer Cell. 2015;27(2):223–239. doi: 10.1016/j.ccell.2014.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Minegishi N, et al. Rapid turnover of GATA-2 via ubiquitin-proteasome protein degradation pathway. Genes Cells. 2005;10(7):693–704. doi: 10.1111/j.1365-2443.2005.00864.x. [DOI] [PubMed] [Google Scholar]
- 13.Lurie LJ, et al. Differential GATA factor stabilities: implications for chromatin occupancy by structurally similar transcription factors. Biochemistry. 2008;47(3):859–869. doi: 10.1021/bi701692p. [DOI] [PubMed] [Google Scholar]
- 14.Whitman M, et al. Association of phosphatidylinositol kinase activity with polyoma middle-T competent for transformation. Nature. 1985;315(6016):239–242. doi: 10.1038/315239a0. [DOI] [PubMed] [Google Scholar]
- 15.Auger KR, et al. PDGF-dependent tyrosine phosphorylation stimulates production of novel polyphosphoinositides in intact cells. Cell. 1989;57(1):167–175. doi: 10.1016/0092-8674(89)90182-7. [DOI] [PubMed] [Google Scholar]
- 16.Franke TF, et al. The protein kinase encoded by the Akt proto-oncogene is a target of the PDGF-activated phosphatidylinositol 3-kinase. Cell. 1995;81(5):727–736. doi: 10.1016/0092-8674(95)90534-0. [DOI] [PubMed] [Google Scholar]
- 17.Sarbassov DD, et al. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science. 2005;307(5712):1098–1101. doi: 10.1126/science.1106148. [DOI] [PubMed] [Google Scholar]
- 18.Alessi DR, et al. Characterization of a 3-phosphoinositide-dependent protein kinase which phosphorylates and activates protein kinase Balpha. Curr Biol. 1997;7(4):261–269. doi: 10.1016/S0960-9822(06)00122-9. [DOI] [PubMed] [Google Scholar]
- 19.Zoncu R, et al. mTOR: from growth signal integration to cancer, diabetes and ageing. Nat Rev Mol Cell Biol. 2011;12(1):21–35. doi: 10.1038/nrm3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Steck PA, et al. Identification of a candidate tumour suppressor gene, MMAC1, at chromosome 10q23.3 that is mutated in multiple advanced cancers. Nat Genet. 1997;15(4):356–362. doi: 10.1038/ng0497-356. [DOI] [PubMed] [Google Scholar]
- 21.Maehama T, Dixon JE. The tumor suppressor, PTEN/MMAC1, dephosphorylates the lipid second messenger, phosphatidylinositol 3,4,5-trisphosphate. J Biol Chem. 1998;273(22):13375–13378. doi: 10.1074/jbc.273.22.13375. [DOI] [PubMed] [Google Scholar]
- 22.Gao T, et al. PHLPP: a phosphatase that directly dephosphorylates Akt, promotes apoptosis, and suppresses tumor growth. Mol Cell. 2005;18(1):13–24. doi: 10.1016/j.molcel.2005.03.008. [DOI] [PubMed] [Google Scholar]
- 23.Gewinner C, et al. Evidence that inositol polyphosphate 4-phosphatase type II is a tumor suppressor that inhibits PI3K signaling. Cancer Cell. 2009;16(2):115–125. doi: 10.1016/j.ccr.2009.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Klempner SJ, et al. What a tangled web we weave: emerging resistance mechanisms to inhibition of the phosphoinositide 3-kinase pathway. Cancer Discov. 2013;3(12):1345–1354. doi: 10.1158/2159-8290.CD-13-0063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fruman DA, Rommel C. PI3K and cancer: lessons, challenges and opportunities. Nat Rev Drug Discov. 2014;13(2):140–156. doi: 10.1038/nrd4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Taylor BS, et al. Integrative genomic profiling of human prostate cancer. Cancer Cell. 2010;18(1):11–22. doi: 10.1016/j.ccr.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bitting RL, Armstrong AJ. Targeting the PI3K/Akt/mTOR pathway in castration-resistant prostate cancer. Endocr Relat Cancer. 2013;20(3):R83–R99. doi: 10.1530/ERC-12-0394. [DOI] [PubMed] [Google Scholar]
- 28.Carver BS, et al. Reciprocal feedback regulation of PI3K and androgen receptor signaling in PTEN-deficient prostate cancer. Cancer Cell. 2011;19(5):575–586. doi: 10.1016/j.ccr.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mulholland DJ, et al. Cell autonomous role of PTEN in regulating castration-resistant prostate cancer growth. Cancer Cell. 2011;19(6):792–804. doi: 10.1016/j.ccr.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wen Y, et al. HER-2/neu promotes androgen-independent survival and growth of prostate cancer cells through the Akt pathway. Cancer Res. 2000;60(24):6841–6845. [PubMed] [Google Scholar]
- 31.Mellinghoff IK, et al. HER2/neu kinase-dependent modulation of androgen receptor function through effects on DNA binding and stability. Cancer Cell. 2004;6(5):517–527. doi: 10.1016/j.ccr.2004.09.031. [DOI] [PubMed] [Google Scholar]
- 32.Fang Z, et al. Androgen receptor enhances p27 degradation in prostate cancer cells through rapid and selective TORC2 activation. J Biol Chem. 2012;287(3):2090–2098. doi: 10.1074/jbc.M111.323303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aberg E, et al. Regulation of MAPK-activated protein kinase 5 activity and subcellular localization by the atypical MAPK ERK4/MAPK4. J Biol Chem. 2006;281(46):35499–35510. doi: 10.1074/jbc.M606225200. [DOI] [PubMed] [Google Scholar]
- 34.Kant S, et al. Characterization of the atypical MAPK ERK4 and its activation of the MAPK-activated protein kinase MK5. J Biol Chem. 2006;281(46):35511–35519. doi: 10.1074/jbc.M606693200. [DOI] [PubMed] [Google Scholar]
- 35.Wang W, et al. MAPK4 overexpression promotes tumor progression via noncanonical activation of AKT/mTOR signaling. J Clin Invest. 2019;129(3):1015–1029. doi: 10.1172/JCI97712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dehm SM, et al. Splicing of a novel androgen receptor exon generates a constitutively active androgen receptor that mediates prostate cancer therapy resistance. Cancer Res. 2008;68(13):5469–5477. doi: 10.1158/0008-5472.CAN-08-0594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dehm SM, Tindall DJ. Alternatively spliced androgen receptor variants. Endocr Relat Cancer. 2011;18(5):R183–R196. doi: 10.1530/ERC-11-0141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guo Z, et al. A novel androgen receptor splice variant is up-regulated during prostate cancer progression and promotes androgen depletion-resistant growth. Cancer Res. 2009;69(6):2305–2313. doi: 10.1158/0008-5472.CAN-08-3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hu R, et al. Ligand-independent androgen receptor variants derived from splicing of cryptic exons signify hormone-refractory prostate cancer. Cancer Res. 2009;69(1):16–22. doi: 10.1158/0008-5472.CAN-08-2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sun S, et al. Castration resistance in human prostate cancer is conferred by a frequently occurring androgen receptor splice variant. J Clin Invest. 2010;120(8):2715–2730. doi: 10.1172/JCI41824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Antonarakis ES, et al. AR-V7 and resistance to enzalutamide and abiraterone in prostate cancer. N Engl J Med. 2014;371(11):1028–1038. doi: 10.1056/NEJMoa1315815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hu CD, et al. Visualization of interactions among bZIP and Rel family proteins in living cells using bimolecular fluorescence complementation. Mol Cell. 2002;9(4):789–798. doi: 10.1016/S1097-2765(02)00496-3. [DOI] [PubMed] [Google Scholar]
- 43.Geller J, et al. Steroid levels in cancer of the prostate—markers of tumor differentiation and adequacy of anti-androgen therapy. Prog Clin Biol Res. 1979;33:103–111. [PubMed] [Google Scholar]
- 44.Mohler JL, et al. The androgen axis in recurrent prostate cancer. Clin Cancer Res. 2004;10(2):440–448. doi: 10.1158/1078-0432.CCR-1146-03. [DOI] [PubMed] [Google Scholar]
- 45.Montgomery RB, et al. Maintenance of intratumoral androgens in metastatic prostate cancer: a mechanism for castration-resistant tumor growth. Cancer Res. 2008;68(11):4447–4454. doi: 10.1158/0008-5472.CAN-08-0249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Massie CE, et al. The androgen receptor fuels prostate cancer by regulating central metabolism and biosynthesis. EMBO J. 2011;30(13):2719–2733. doi: 10.1038/emboj.2011.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Visakorpi T, et al. In vivo amplification of the androgen receptor gene and progression of human prostate cancer. Nat Genet. 1995;9(4):401–406. doi: 10.1038/ng0495-401. [DOI] [PubMed] [Google Scholar]
- 48.Chen CD, et al. Molecular determinants of resistance to antiandrogen therapy. Nat Med. 2004;10(1):33–39. doi: 10.1038/nm972. [DOI] [PubMed] [Google Scholar]
- 49.Taplin ME, et al. Androgen receptor mutations in androgen-independent prostate cancer: cancer and leukemia Group B study 9663. J Clin Oncol. 2003;21(14):2673–2678. doi: 10.1200/JCO.2003.11.102. [DOI] [PubMed] [Google Scholar]
- 50.Heemers HV, Tindall DJ. Androgen receptor (AR) coregulators: a diversity of functions converging on and regulating the AR transcriptional complex. Endocr Rev. 2007;28(7):778–808. doi: 10.1210/er.2007-0019. [DOI] [PubMed] [Google Scholar]
- 51.Culig Z, et al. Androgen receptor activation in prostatic tumor cell lines by insulin-like growth factor-I, keratinocyte growth factor, and epidermal growth factor. Cancer Res. 1994;54(20):5474–5478. [PubMed] [Google Scholar]
- 52.Hobisch A, et al. Interleukin-6 regulates prostate-specific protein expression in prostate carcinoma cells by activation of the androgen receptor. Cancer Res. 1998;58(20):4640–4645. [PubMed] [Google Scholar]
- 53.Craft N, et al. A mechanism for hormone-independent prostate cancer through modulation of androgen receptor signaling by the HER-2/neu tyrosine kinase. Nat Med. 1999;5(3):280–285. doi: 10.1038/6495. [DOI] [PubMed] [Google Scholar]
- 54.Signoretti S, et al. Her-2-neu expression and progression toward androgen independence in human prostate cancer. J Natl Cancer Inst. 2000;92(23):1918–1925. doi: 10.1093/jnci/92.23.1918. [DOI] [PubMed] [Google Scholar]
- 55.Guo Z, et al. Regulation of androgen receptor activity by tyrosine phosphorylation. Cancer Cell. 2006;10(4):309–319. doi: 10.1016/j.ccr.2006.08.021. [DOI] [PubMed] [Google Scholar]
- 56.Mahajan NP, et al. Activated Cdc42-associated kinase Ack1 promotes prostate cancer progression via androgen receptor tyrosine phosphorylation. Proc Natl Acad Sci U S A. 2007;104(20):8438–8443. doi: 10.1073/pnas.0700420104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yang F, et al. Stromal TGF-β signaling induces AR activation in prostate cancer. Oncotarget. 2014;5(21):10854–10869. doi: 10.18632/oncotarget.2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kumar A, et al. Substantial interindividual and limited intraindividual genomic diversity among tumors from men with metastatic prostate cancer. Nat Med. 2016;22(4):369–378. doi: 10.1038/nm.4053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bluemn EG, et al. Androgen receptor pathway-independent prostate cancer is sustained through FGF signaling. Cancer Cell. 2017;32(4):474–489.e6. doi: 10.1016/j.ccell.2017.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Meerbrey KL, et al. The pINDUCER lentiviral toolkit for inducible RNA interference in vitro and in vivo. Proc Natl Acad Sci U S A. 2011;108(9):3665–3670. doi: 10.1073/pnas.1019736108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hanzelmann S, et al. GSVA: gene set variation analysis for microarray and RNA-seqdata. BMC Bioinformatics. 2013;14:7. doi: 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.