Abstract
Purpose:
Ras pathway mutations are one of the most common type of alterations in pediatric hematologic malignancies and are frequently associated with adverse outcomes. Despite ongoing efforts to use targeted treatments, there remain no food and drug administration (FDA) approved medications specifically for children with Ras pathway mutated leukemia. This review will summarize the role of Ras pathway mutations in pediatric leukemia, discuss the current state of RAS pathway inhibitors and highlight the most promising agents currently being evaluated in clinical trials.
Recent findings:
Efficacy of RAF and MEK inhibitors has been demonstrated across multiple solid and brain tumors and these are now considered the standard of care for treatment of certain tumor types in adults and children. Clinical trials are now testing these medications for the first time in pediatric hematologic disorders such as acute lymphoblastic leukemia, juvenile myelomonocytic leukemia and histiocytic disorders. Novel inhibitors of the Ras pathway, including direct RAS inhibitors, are now being tested in clinical trials across a spectrum of pediatric and adult malignancies.
Summary:
Activation of the Ras pathway is a common finding in pediatric hematologic neoplasms. Implementation of precision medicine with a goal of improving outcomes for these patients will require testing of Ras pathway inhibitors in combination with other drugs in the context of current and future clinical trials.
Keywords: Ras, targeted therapy, pediatric leukemia
Introduction
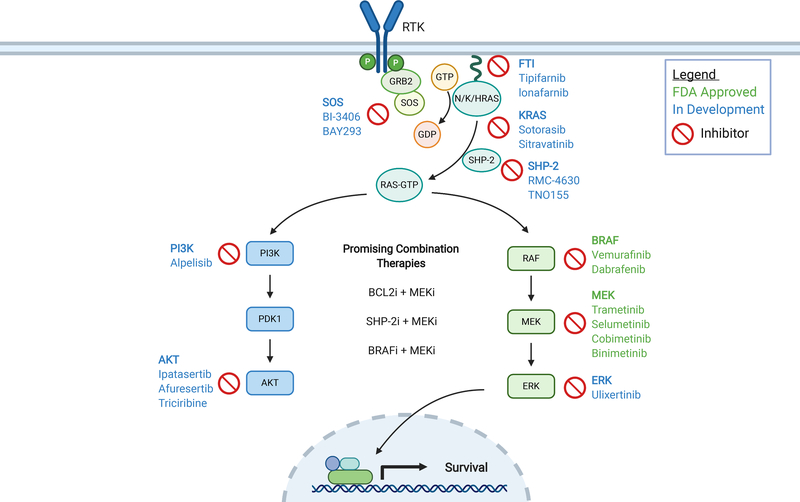
Activation of the Ras signaling pathway is one of the most common findings in cancer. RAS proteins act as molecular switches that cycle between the active, GTP-bound, state, and the inactive, GDP-bound, state. RAS is activated by guanine nucleotide exchange factors (GEFs), and the GTP bound RAS interacts with a number of effectors. GTPase-activating proteins (GAPs) downregulate RAS by accelerating its intrinsic GTPase activity. Alterations including point mutations, insertion/deletions, rearrangements, amplifications and deletions have been reported in nearly every gene in this pathway. NRAS, KRAS, HRAS, NF1, PTPN11, BRAF, and CBL are among the most commonly altered genes and lead to hyperactive signaling in effector pathways including PI3K/mTOR/AKT and RAF/MEK/ERK which are mitogen-activated protein kinases (MAPK). The final result is upregulation of pro-survival transcription factors resulting in increased cellular proliferation and enhanced survival (Figure 1).
Figure 1. Clinically relevant inhibitors of the RAS pathway.
Approved inhibitors are listed in green and inhibitors currently in development are listed in blue. Figure was created in Biorender.com.
Abbreviations: FTI, farnesyltransferase inhibitor; i, inhibitor; RTK, receptor tyrosine kinase; GTP, guanosine triphosphate; GDP, guanosine diphosphate.
In pediatrics, Ras pathway mutations are among the most common genomic alterations in both solid and hematologic malignancies. This review will focus on pediatric hematological malignancies with an emphasis on recent advances in pharmacological targeting of the Ras signaling pathway.
Incidence of Ras mutations in pediatric leukemia
Ras pathway activation is common in pediatric hematological malignancies (Table 1). Ras pathway mutations are found in over 30% of infant B-cell acute lymphoblastic leukemia (B-ALL)(1–3), non-infant B-ALL(4, 5) and high hyperdiploid B-ALL(6–8). Ras pathway and receptor tyrosine kinase (RTK) mutations are present in 70% of near haploid B-ALL(9). Ras (NRAS, KRAS, BRAF, NF1, and PTPN11) mutations are present in approximately 14% of T-ALL(10) and up to 30% of early T-cell precursor ALL(11). Ras mutations are also frequent in myeloid malignancies with nearly half of pediatric myelodysplastic syndromes (MDS)(12) and one-third of acute myeloid leukemia (AML)(13, 14) patients harboring Ras mutations. In ALL and AML, the mutations are often subclonal and can be gained or lost at diagnosis and relapse(1, 15). Despite the frequent sub-clonal nature at diagnosis, Ras mutations appear to retain prognostic relevance. Ras pathway mutations have been associated with early relapse, chemotherapy resistance and poor survival in both patients with infant KMT2A-rearranged ALL(3), non-infant ALL(16) and AML(17). While not all Ras mutations from diagnosis persist at relapse, de novo mutation in Ras pathway genes can also appear at the time of disease progression(18). In distinction to ALL and AML, the Ras pathway is universally activated in juvenile myelomonocytic leukemia (JMML), with 95% of patients harboring founding Ras pathway mutations that always persist in the event of relapse(19).
Table 1:
Frequency of Ras pathway mutations in pediatric hematological malignancies.
| Disease | Genes | Ras pathway mutation frequencies | References |
|---|---|---|---|
| B-ALL | FLT3, KRAS, NRAS, PTPN11 | 35% diagnosis, 25% relapse | (4) |
| KRAS and NRAS, codons 12 and 13 | 20% | (6) | |
| KRAS, NRAS, PTPN11 | 28% (35/125) | (15) | |
| KRAS, NRAS | 30% (18/60) | (15) | |
| Infant | KRAS, NRAS, NF1, PIK3CA, PIK3R1, PTPN11 | 42% | (1) |
| KRAS, NRAS | 22% in KMT2Ar ALL | (2) | |
| BRAF, KRAS, NRAS | 14% overall; 24% in KMT2A-AF4 | (3) | |
| iAMP21 | BRAF, CBL, KRAS, MAPK1, NRAS, NF1, PTPN11 | 57% (24/42) | (5) |
| High hyperdiploid | KRAS and NRAS, codons 12 and 13 | 30% | (6) |
| FLT3, KRAS, NRAS, PTPN11 | 53% | (7) | |
| KRAS, NRAS, PTPN11 | 52% (26/50) | (8) | |
| Near haploid | FLT3, KRAS, NRAS, MAPK1, NF1, PTPN11 | 71% | (9) |
| T-ALL | BRAF, KRAS, NF1, NRAS, PTPN11 | 14% | (10) |
| ETP | BRAF, KRAS, NF1, NRAS, PTPN11 | 28% (18/64) | (11) |
| AML | KRAS, NRAS, PTPN11 | >30% | (14) |
| KRAS, NRAS, PTPN11 | 43% (diagnosis) 49% (relapse) | (13) | |
| MDS | Ras/MAPK pathway | 55% | (12) |
| JMML | CBL, KRAS, NRAS, NF1, PTPN11, RRAS, RRAS2 | 95% | (19) |
While mutations in the Ras pathway are the most common mechanism of upregulated RAS signaling, other mechanisms have been reported as well. Methylation, leading to silencing of RASSF1A and RASD1 gene expression have also been implicated in RAS activated hematologic diseases, most notably multiple myeloma(20, 21). Point mutations and fusions involving RTKs, such as CSF1R, ALK, PDGFRB and FLT3, have been reported in a variety of myeloid and lymphoid neoplasms leading to constitutive activation of the Ras pathway with elevated levels of phosphorylated ERK(22–26).
Associations with specific subtypes of leukemia
Ras pathway mutations are often found in association with other genetic features in acute leukemia. For KMT2A rearranged acute leukemia, Ras mutations are found in over 30% of ALL (1, 27) and AML(14, 28). These mutations appear to cooperate in leukemogenesis as the presence of NRAS p.G12D and FLT3 mutations have been shown to accelerate leukemia onset in a KMT2A-MLLT3 driven AML and lead to a more aggressive disease(29).
Patients with hyperdiploid ALL have excellent outcomes with overall survival greater than 90% at 5 years(30). Despite the favorable prognosis, hyperdiploid ALL is responsible for a disproportionate number of relapses because it is one of the most common subtypes of ALL in children. Studies have implicated the presence of Ras mutations, in particular KRAS and NRAS, as poor predictors of outcome when they co-exist with CREBBP even if they are all subclonal at diagnosis. This “malicious liaison” of CREBBP with Ras mutations has been associated with early relapse due to a possible combinatorial effect that leads to resistance to chemotherapy(8).
Pediatric patients with Ph-like, or BCR-ABL1-like, disease have inferior outcomes compared to those without these gene expression signatures(26, 31). While most patients with this subtype of ALL have CRLF2 over-expressing or ABL class kinase fusion lesions, approximately 5% of patients have been found to harbor Ras mutations leading to a Ph-like designation (26). One recent study among pediatric Korean patients demonstrated that 68% of patients with a BCR-ABL1-like signature harbored mutations in the Ras pathway, higher than any other subtype in that study(32).
FDA approved compounds targeting the MAPK signaling pathway
Given the frequent prevalence of Ras mutations in cancer, efforts to pharmacologically target this pathway have been a longstanding goal in cancer therapy. Despite early setbacks in targeting RAS itself, novel insights into molecular biology and advances in structural chemistry have led to the development of clinically relevant medications for the treatment of Ras pathway mutant cancers (Table 2). First, we will review two classes of FDA approved inhibitors of the Ras pathway, RAF and MEK inhibitors. Despite FDA approval for their use in subsets of solid tumors, RAF and MEK inhibitors do not have FDA approval for any hematological malignancy.
Table 2.
Clinical trials in pediatric hematologic malignancies testing RAS pathway inhibitors.
| Trial identifier | Study name | Disease | Targeted agent | Class |
|---|---|---|---|---|
| Hematologic malignancy | ||||
| NCT03190915 | Trametinib in Treating Patients With Relapsed or Refractory Juvenile Myelomonocytic Leukemia | JMML | Trametinib | MEK inhibitor |
| NCT03705507 | International Trial of Selumetinib in Combination With Dexamethasone for the Treatment of Acute Lymphoblastic Leukaemia (SeluDex) | ALL | Selumetinib | MEK inhibitor |
| NCT03585686 | A Combination of Vemurafenib, Cytarabine and 2-chlorodeoxyadenosine in Children With LCH and BRAF V600E Mutation | LCH | Vemurafenib | BRAF inhibitor |
| Pediatric MATCH | ||||
| NCT04284774 | Tipifarnib for the Treatment of Advanced Solid Tumors, Lymphoma, or Histiocytic Disorders With HRAS Gene Alterations, a Pediatric MATCH Treatment Trial | Solid Tumors, non-Hodgkin lymphoma, or histiocytic Disorders with HRAS mutations | Tipifarnib | Farnesyltransferase inhibitor |
| NCT03220035 | Vemurafenib in Treating Patients With Relapsed or Refractory Advanced Solid Tumors, Non-Hodgkin Lymphoma, or Histiocytic Disorders With BRAF V600 Mutations (A Pediatric MATCH Treatment Trial) | Solid Tumors, non-Hodgkin lymphoma, or histiocytic Disorders with BRAF V600E mutations | Vemurafenib | BRAF inhibitor |
| NCT03213691 | Selumetinib Sulfate in Treating Patients With Relapsed or Refractory Advanced Solid Tumors, Non-Hodgkin Lymphoma, or Histiocytic Disorders With Activating MAPK Pathway Mutations (A Pediatric MATCH Treatment Trial) | Solid Tumors, non-Hodgkin lymphoma, or histiocytic Disorders with MAPK pathway mutations | Selumetinib | MEK inhibitor |
| NCT03698994 | Ulixertinib in Treating Patients With Advanced Solid Tumors, Non-Hodgkin Lymphoma, or Histiocytic Disorders With MAPK Pathway Mutations (A Pediatric MATCH Treatment Trial) | Solid Tumors, non-Hodgkin lymphoma, or histiocytic Disorders with MAPK pathway mutations | Ulixertinib | ERK inhibitor |
RAF inhibitors
BRAF is the most frequently mutated gene in the MAPK pathway with nearly 10% of all human cancers harboring an alteration. Mutations in ARAF and CRAF are infrequent compared to BRAF, which can be categorized into 3 distinct classes (I, II and III). Under physiologic conditions, RAS proteins interact with BRAF, undergo dimerization and subsequent activation. Mutations can alter this balanced process and the effects are dependent on the type of mutation. Class I mutations affect codon V600 and represent ~90% of all BRAF alterations. These mutations allow BRAF to signal as a monomer, resulting in constitutive activation and upregulation of MAPK signaling leading to elevated levels of phosphorylated ERK, independent of RAS. Class II mutations include activating point mutations, fusions and in-frame deletions which all lead to activated BRAF dimers that also function independent of RAS activation. Class III alterations are unique in that they are dependent on RAS activation, frequently co-occur with N/K/HRAS mutations and lead to high receptor tyrosine kinase activity.
There are reports of BRAF alterations in chronic lymphocytic leukemia, chronic myeloid leukemia and hairy cell leukemia, all diseases that predominantly affect adults. BRAF mutations in pediatric AML, ALL, MDS are also rare but are frequent in histiocytic disorders(33). Histiocytic disorders can be broadly grouped into Langerhans cell histiocytosis (LCH) and non-Langerhans cell histiocytoses (non-LCH) of which several varieties have been named including Rosai Dorfman disease, Erdheim-Chester disease (ECD), juvenile xanthogranulomatous disease and histiocytic sarcoma. In contrast to acute leukemias, histiocytic disorders harbor ARAF alterations alone or in combination with other MAPK mutations (most commonly NRAS) in approximately 20% of cases (34, 35). BRAF p.V600E mutations occur in approximately 65% of LCH patients(36). NRAS and KRAS mutations are also seen in both LCH and non-LCH patients. More recent reports have identified mutations in the gene MAP2K1 (which encodes for MEK1) in ~ 20% of LCH and ECD patients without BRAF or NRAS/KRAS mutations. Elevated MAPK signaling is the hallmark of LCH and non-LCH and therefore attempts to treat these disorders with both RAF inhibitors and MEK inhibitors have been tested. LCH patients with BRAF p.V600E mutations have been treated with single agent RAF inhibitor, vemurafenib, with one basket study demonstrating an overall response rate (ORR) of 43% in LCH and ECD(37). Another ongoing study in pediatrics combines vemurafenib with cytarabine and cladribine in the treatment of newly-diagnosed BRAF p.V600E mutated LCH (NCT03585686). Vemurafenib and dabrafenib are FDA approved for the treatment of BRAF mutant melanoma.
MEK inhibitors
MEK is downstream of RAS in the MAPK pathway and has been shown to be upregulated in a variety of leukemias. While directly inhibiting RAS has been vexing, pre-clinical data provided the justification to test inhibiting the downstream effector, MEK, as a strategy in pediatric leukemia. Trametinib, cobimetinib, selumetinib and binimetinib inhibited the growth of primary Ras pathway mutant leukemia cells in vitro and in vivo in different subtypes of ALL(5, 16, 38). Interestingly, KMT2A-rearranged samples without Ras mutations have also been noted to respond to MEK inhibition, suggesting activation of the pathway via other mechanisms besides genetic mutations(38, 39).
An adult phase 1 study tested the MEK inhibitor trametinib in adult patients with a variety of relapsed or refractory hematologic malignancies, including MDS, chronic myelomonocytic leukemia and AML. Responses were seen in nearly 30% of Ras mutated leukemias but only 3% of non-mutated cases(40). Cobimetinib has been tested in adults with advanced histiocytic disorders with an ORR of nearly 90%, irrespective of genotype(41). Due to the unique dependency on the Ras pathway in JMML, a phase II trial of trametinib in patients with relapsed or refractory disease is under way (NCT03190915). While Ras pathway mutant ALL has been associated with steroid resistance (42), glucocorticoids and the MEK inhibitor selumetinib, have been shown to be synergistic in pre-clinical models(43) and led to the ongoing Seludex clinical trial (NCT03705507). There are four FDA approved MEK inhibitors including trametinib, selumetinib, binimetinib and cobimetinib in a variety of solid tumors. Selumetinib is FDA approved for patients greater than 2 years old with NF1 and plexiform neurofibromas.
Combination therapy with MEK inhibitors
Signaling pathway inhibition along two nodes has the potential for more durable inhibition and prevention of upregulation of pathway enzymes as a mechanism of resistance to a single drug. BRAF inhibition causes upregulation of ERK signaling pathway, supporting targeting of both proteins. This approach may be efficacious in targeting the signaling pathway even in the setting of wildtype BRAF(44). The combination of a BRAF inhibitor with a MEK inhibitor has proven effective in melanoma(45, 46) and the combination of dabrafenib and trametinib was recently tested in adult patients with ECD (NCT02281760).
Given the prevalence of Ras pathway mutations in hematological malignancies, combining inhibition of this pathway with other novel compounds may offer additional therapeutic benefit. For example, the oral BCL2 inhibitor, venetoclax, has transformed treatment of both newly diagnosed and relapsed adult patients with AML. Venetoclax has now been tested in combination with the hypomethylating agents, decitabine and azacitidine (47) as well as with conventional chemotherapy including cytarabine(48). The first published prospective study in relapsed pediatric AML using venetoclax was in combination with cytarabine with or without idarubicin and demonstrated an overall response rate of 69% in the 35 patients treated on the phase I/II study(49). However, resistance to venetoclax is common and combinatorial approaches beyond hypomethylating agents and cytarabine are therefore being explored. Preclinical testing demonstrated synergy between venetoclax and the MEK1/2 inhibitor, cobimetinib, in 7 of 11 AML cell lines tested including in lines resistant to the individual medications(50). One mechanism for this synergy was the downregulation of MCL-1 protein levels and disruption of BCL2:BIM complexes, leading to the release of BIM and eventual cell death. A clinical trial is now underway in adults testing the combination of venetoclax, azacitidine and trametinib for patients with relapsed or refractory AML or MDS (NCT04487106). Similar work in ALL had previously demonstrated synergy of BCL-2/BCL-XL inhibitors, venetoclax or ABT-263, and the MEK1/2 inhibitor, trametinib, across multiple B-ALL cell lines with or without MAPK mutations(51). That preclinical study also identified BIM as the potential mediator of synergy. BIM is dephosphorylated as a result of MEK inhibition which then neutralizes MCL-1, and eventually leads to BCL2 mediated apoptosis. A phase 1B trial combined cobimetinib with venetoclax in elderly patients with relapsed or refractory AML but data from the trial are not yet published (NCT02670044). Combinations of MAPK targeted agents with venetoclax and/or other novel drugs will need to be explored further in clinical trials.
Novel Ras pathway inhibitor development
While Ras isoforms have historically been considered directly “undruggable”, recent efforts have focused on alternate strategies. Here we will discuss drugs targeting the Ras pathway that are still in clinical development and therefore not FDA approved for use outside of clinical trials.
Farnesyl transferase inhibitors
Another approach to target RAS focuses on inhibiting its necessary interaction with the inner plasma membrane. This interaction is mediated by the addition of a farnesyl lipid to its carboxy-terminal CAAX motif. Efforts to directly interfere with RAS GTPase have centered on farnesyltransferase, a member of the prenyltransferase family, involved in catalyzing the chemical reaction between farnesyl diphosphate and protein-cysteine during the protein post-translational modification, with RAS being one of its targets. This is one of the first modification steps leading to the active RAS protein. Farnesyltransferarse inhibitors (FTIs) were trialed in several phase 3 studies across different cancers but few if any responses were seen in KRAS or NRAS mutated cancers(52). Specifically in pediatric hematologic malignancies, tipifarnib, an FTI, was tested in newly diagnosed JMML patients in a window setting followed by stem cell transplant. However, outcomes were not significantly different than in prior studies and the addition of an FTI was not determined to be beneficial(53). Tipifarnib was also tested in older adults with AML with acceptable toxicities but limited efficacy(54). A mechanism of resistance to FTIs was identified which involved an alternative post-translational modification by an enzyme geranylgeranyltransferase I (GGTase I) via a process called prenylation. Recently however, there has been interest in resurrecting FTIs for HRAS mutated cancers because they lack this alternative mechanism in response to FTI. Tipifarnib and a more potent FTI, lonafarnib, are now being tested in more rationally designed clinical trials focusing on HRAS mutant cancers including within the MATCH study being conducted through the Children’s Oncology Group (NCT04284774) (55, 56).
KRAS G12C inhibitors
RAS proteins act as a binary switch in either the active or inactive state. In the active state, Switch I (residues 32–38) and Switch II (residues 60–75) regions undergo conformational changes leading to activation of downstream signaling of the MAPK pathway(57). Oncogenic mutations in KRAS at codon G12, G13 and Q61 result in decreased stimulation of GAPs and therefore higher levels of GTP-bound KRAS. This active form of the protein leads to downstream activation of signaling pathways via interactions at the GTPase-binding-domain (GBD) of effector proteins including RAF, MEK and ERK among others. GTPases can be recharged by GEFs, which weaken the binding of GDP and catalyze its replacement with GTP.
Previous attempts at inhibiting RAS included strategies to identify GTP-competitive inhibitors of RAS(58). However, GTP binds to RAS proteins with picomolar activity, effectively precluding that as a feasible approach. Several groups have recently demonstrated that allele specific inhibition of KRAS may be a tractable approach(59). Small molecules were discovered that covalently bind to G12C mutant form of KRAS during a screen utilizing GDP-bound KRAS-G12C tethering approach. The compounds bind to a region of the Switch 2 region and block nucleotide exchange and therefore decrease the binding of RAS to both BRAF and CRAF. These compounds were found to have selectivity for KRAS p.G12C mutated cancers. Drugs in this class have been rapidly shepherded into the clinic and are now the focus of several ongoing studies(60). One phase I/II clinical trial testing sotorasib, enrolled 129 patients who harbored KRAS G12C mutations across a variety of solid tumors. No dose limiting toxicity was observed. Activity of sotorasib was most pronounced in patients with non-small cell lung cancer where 88% of patients had disease control which included objective responses and stable disease. Responses were also observed in a smaller proportion of patients with colorectal, pancreatic, endometrial, appendiceal cancers and melanoma. However, median progression-free survival was 4 months(61). KRAS p.G12C is a rare variant in leukemia in general and even more so in pediatric patients. As such, there have yet to be any clinical trials specifically designed using an allele specific approach for patients with hematologic malignancies but a proof of principle regarding direct inhibition of RAS has now been demonstrated.
SHP-2 inhibitors
The protein tyrosine phosphatase SHP-2, encoded by PTPN11, is a critical regulator of the Ras signaling pathway. Activating, somatic mutations in PTPN11 are the most common cause of JMML and germline mutations are the most common cause of Noonan syndrome. SHP099 is a selective small-molecule SHP-2 inhibitor, which stabilizes SHP-2 in an auto-inhibited conformation(62). SHP099 suppresses RAS-ERK signaling and has been shown to have activity in vitro and in vivo(63). Although PTPN11 mutations are common in AML and JMML, SHP099, an allosteric inhibitor does not have activity for the most common alterations including D61Y, A72V and E76K due to conformational selection to the closed state that reduces drug affinity(64).
Previous studies have demonstrated that compensatory upregulation of upstream pathways including FGFR1 are possible mechanisms of resistance to long-term MEK inhibition. In a preclinical study using KRAS mutant colorectal cancer cell lines, SHP099 blocked activation of RAS signaling through several RTKs. In addition, synergy was noted between combined MEK and SHP2 inhibition, revealing a potentially strategy to prevent MEK inhibitor mediated resistance(65). A clinical trial involving the allosteric SHP2 inhibitor furthest along in clinical development, RMC-4630, is now being tested in combination with cobimetinib in adults relapsed and refractory solid tumors (NCT03989115). Early studies of TNO155, an allosteric inhibitor of SHP-2, are currently in clinical trials for adults with solid tumors (NCT03114319, NCT04000529, NCT04330664, NCT04294160).
SOS1 inhibitors
SOS1 is a KRAS activator and a major control point for Ras pathway regulation. SOS1 can be activated in response to MEK inhibition, a mechanism of resistance to RAS pathway inhibition. SOS1 inhibitors, BI-3406 and BAY293, interfere with SOS1 function. BI-3406 binds to the catalytic domain of SOS1, preventing its interaction with KRAS(66). KRAS mutant cells lines with a variety of G12 and G13 mutations, are sensitive to BI-3406. Cell lines with KRAS/NRAS Q61 mutations are less sensitive to BI-3406, possibly due to their lowest intrinsic GTPase activity. Interestingly, 7 out of 14 tested cell lines carrying NF1 aberrations were also sensitive to BI-3406(66). Sensitive cell lines show sustained inhibition of ERK1/2 phosphorylation. BI-3406 attenuates feedback activation of MEK signaling in response to MEK inhibitors, enhancing sensitivity to MEK inhibitors and promising for potential combination use(66). BAY293 is another inhibitor of the KRAS::SOS1 interaction, interfering with the KRASG12C-SOS1 complex and showing synergy when combined with a covalent KRAS G12C inhibitor, ARS-853(67). BAY293 did not show selectivity for KRAS mutant cells when tested head to head with BI-3406(66).
PI3K/AKT inhibitors
The PI3K/mTOR/AKT signaling pathway is upregulated in many Ras pathway mutant leukemias. In addition, there are several types of genomic alterations in the pathway itself, including loss of function alterations in the tumor suppressor PTEN, activating mutations in PI3K, and amplifications that can also result in increased signaling. One effector protein in this pathway is AKT and there have been several attempts to target this protein. MK-2206, an oral, allosteric inhibitor of AKT, was evaluated in a phase II clinical trial for adult patients with relapsed or refractory AML. Among the 18 patients evaluated, only 1 had an objective response leading to study termination(68). In addition, correlative biology assays performed on the study using a reverse phase protein array indicated that even at the maximum tolerated dose there was only modest decrease in phosphorylated levels of AKT. Upregulation of upstream signaling pathways including PI3K and mTOR were also noted as a possible mechanism of resistance. There have more recently been attempts to trial traditional ATP-competitive AKT inhibitors such as ipatasertib which is now being tested in adults with a variety of solid tumors. Afuresertib, another oral, reversible, ATP-competitive, pan-AKT kinase inhibitor was first tested in a phase 1 trial for adults with a variety of hematologic malignancies with the most promising results seen in patients with multiple myeloma (69). A phase IIa study using the same compound was then tested in adult LCH patients with an ORR of 33% and 28% in treatment naive and relapsed/refractory patients, respectively(70). PTX-200 is a synthetic tricyclic nucleoside inhibitor that inhibits the phosphorylation of AKT1/2/3 but does not have activity on the kinase itself. There is an ongoing trial of PTX-200 (triciribine) in combination with cytarabine for adult patients with relapsed or refractory AML (NCT02930109). In general, development of clinically active AKT inhibitors has lagged behind inhibitors of RAF, MEK and more recently ERK owing to structural limitations in targeting AKT.
ERK inhibitors
ERK1 and ERK2 are encoded by the same gene and are splice variants. When GTP-bound RAS recruits and activates RAF, it phosphorylates and activates MEK which then phosphorylates and activates ERK which eventually translocates to the nucleus. There are multiple substrates of ERK including transcription factors and kinases that regulate key cellular functions including differentiation, proliferation and death. There are multiple feedback mechanisms in this pathway; ERK1/2 can phosphorylate BRAF/CRAF which then inhibit phosphorylation of MEK. Additionally, Sprouty proteins and dual-specificity phosphatases provide negative feedback by dephosphorylating ERK1/2. ERK is the last effector in the MAPK pathway and is therefore an attractive treatment strategy in Ras mutated cancers. Targeting ERK may also overcome resistance mechanisms that arise in the setting of inhibiting MEK which include amplification of RAF or downregulation/mutations in MEK. Ulixertinib is an oral ERK1/2 inhibitor that is furthest along in clinical trial development. In vitro, ulixertinib resulted in reduced proliferation and inhibited phosphorylation of target substrates despite increased phosphorylation of ERK1/2. In vivo studies demonstrated efficacy even in models with acquired resistance to MEK or combined BRAF and MEK therapy(71). A phase 1 study was completed in patients with advanced solid tumors that resulted in an overall response rate of 15%(72). A phase 1/2 study of ulixertinib in adults with AML and MDS has been completed but results are not yet published (NCT02296242). An ongoing study in pediatrics using ulixertinib is being conducted in patients with relapsed or refractory solid tumors and histiocytic disorders with MAPK alterations (NCT03698994).
Conclusion
Despite initial obstacles in targeting the MAPK pathway for the treatment of cancer, there has been a recent surge in the development of compounds with potential to inhibit the Ras signaling pathway including KRAS itself. There is newfound optimism that the success of Ras pathway targeting agents in solid tumors will be translated to the treatment of pediatric patients with hematological malignancies, where Ras pathway mutations are among the most common genomic alterations. Translation to pediatric leukemia patients has the greatest likelihood of being effective by using a combination of the agents described above, thereby preventing upregulation of parallel or upstream pathways.
Key points:
Ras mutations are among the most common alterations in pediatric hematologic malignancies and are frequently associated with adverse outcomes.
RAF and MEK inhibitors are the Ras pathway inhibitors furthest along in clinical development and are now being tested in pediatric leukemia patients.
Novel Ras pathway inhibitors including directly inhibiting KRAS p.G12C are now being tested in adults with solid tumors.
A combination of agents is the most likely approach to have clinical activity in Ras mutated pediatric leukemia.
Acknowledgements:
We would like to thank our mentors Drs. Kimberly Stegmaier and Mignon Loh.
Financial support and sponsorship:
This work was supported by the St. Baldrick’s Foundation Consortium grant and Hannah’s Heroes (Y.P.); the Joshua Rappaport Pediatric Oncology Fellowship (Y.P.); the National Institutes of Health, National Cancer Institute grants K08CA222684 (Y.P.) and 1U54CA196519 (E.S.), National Heart, Lung, and Blood Institute grant K08HL135434 (E.S.).
Footnotes
Conflicts of interest:
Yana Pikman has no conflicts of interest. Elliot Stieglitz has received honoraria from Onconova.
References:
- 1.Andersson AK, Ma J, Wang J, Chen X, Gedman AL, Dang J, et al. The landscape of somatic mutations in infant MLL-rearranged acute lymphoblastic leukemias. Nat Genet. 2015;47(4):330–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Emerenciano M, Barbosa TdC, de Almeida Lopes B, Meyer C, Marschalek R, Pombo-de-Oliveira MS. Subclonality and prenatal origin of RAS mutations in KMT2A (MLL)-rearranged infant acute lymphoblastic leukaemia. Br J Haematol. 2015;170(2):268–71. [DOI] [PubMed] [Google Scholar]
- 3.Driessen EMC, van Roon EHJ, Spijkers-Hagelstein JAP, Schneider P, de Lorenzo P, Valsecchi MG, et al. Frequencies and prognostic impact of RAS mutations in MLL-rearranged acute lymphoblastic leukemia in infants. Haematologica. 2013;98(6):937–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Case M, Matheson E, Minto L, Hassan R, Harrison CJ, Bown N, et al. Mutation of genes affecting the RAS pathway is common in childhood acute lymphoblastic leukemia. Cancer research. 2008;68(16):6803–9. [DOI] [PubMed] [Google Scholar]
- 5.Ryan SL, Matheson E, Grossmann V, Sinclair P, Bashton M, Schwab C, et al. The role of the RAS pathway in iAMP21-ALL. Leukemia. 2016;30(9):1824–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wiemels JL, Zhang Y, Chang J, Zheng S, Metayer C, Zhang L, et al. RAS mutation is associated with hyperdiploidy and parental characteristics in pediatric acute lymphoblastic leukemia. Leukemia. 2005;19(3):415–9. [DOI] [PubMed] [Google Scholar]
- 7.Paulsson K, Lilljebjörn H, Biloglav A, Olsson L, Rissler M, Castor A, et al. The genomic landscape of high hyperdiploid childhood acute lymphoblastic leukemia. Nat Genet. 2015;47(6):672–6. [DOI] [PubMed] [Google Scholar]
- 8.Malinowska-Ozdowy K, Frech C, Schönegger A, Eckert C, Cazzaniga G, Stanulla M, et al. KRAS and CREBBP mutations: a relapse-linked malicious liaison in childhood high hyperdiploid acute lymphoblastic leukemia. Leukemia. 2015;29:1656–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Holmfeldt L, Wei L, Diaz-Flores E, Walsh M, Zhang J, Ding L, et al. The genomic landscape of hypodiploid acute lymphoblastic leukemia. Nat Genet. 2013;45:242–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu Y, Easton J, Shao Y, Maciaszek J, Wang Z, Wilkinson MR, et al. The genomic landscape of pediatric and young adult T-lineage acute lymphoblastic leukemia. Nat Genet. 2017;49(8):1211–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang J, Ding L, Holmfeldt L, Wu G, Heatley SL, Payne-Turner D, et al. The genetic basis of early T-cell precursor acute lymphoblastic leukaemia. Nature. 2012;481:157–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schwartz JR, Ma J, Lamprecht T, Walsh M, Wang S, Bryant V, et al. The genomic landscape of pediatric myelodysplastic syndromes. Nat Commun. 2017;8(1):1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pikman Y, Tasian SK, Sulis ML, Cooper TM, Pauly M, Maloney KW, et al. Matched Targeted Therapy for Pediatric Patients with Relapsed, Refractory or High-Risk Leukemias: A Report from the LEAP Consortium. Blood. 2018;132:261–. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bolouri H, Farrar JE, Triche T Jr., Ries RE, Lim EL, Alonzo TA, et al. The molecular landscape of pediatric acute myeloid leukemia reveals recurrent structural alterations and age-specific mutational interactions. Nature medicine. 2018;24(1):103–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mar BG, Bullinger LB, McLean KM, Grauman PV, Harris MH, Stevenson K, et al. Mutations in epigenetic regulators including SETD2 are gained during relapse in paediatric acute lymphoblastic leukaemia. Nat Commun. 2014;5:3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Irving J, Matheson E, Minto L, Blair H, Case M, Halsey C, et al. Ras pathway mutations are prevalent in relapsed childhood acute lymphoblastic leukemia and confer sensitivity to MEK inhibition. Blood. 2014;124(23):3420–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ney GM, Anderson B, Bender J, Kumar-Sinha C, Wu Y-M, Vats P, et al. Mutations predictive of hyperactive Ras signaling correlate with inferior survival across high-risk pediatric acute leukemia. Transl Pediatr. 2020;9(1):43–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Farrar JE, Schuback HL, Ries RE, Wai D, Hampton OA, Trevino LR, et al. Genomic profiling of pediatric acute myeloid leukemia reveals a changing mutational landscape from disease diagnosis to relapse. Cancer research. 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stieglitz E, Taylor-Weiner AN, Chang TY, Gelston LC, Wang YD, Mazor T, et al. The genomic landscape of juvenile myelomonocytic leukemia. Nature genetics. 2015;47(11):1326–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ng MH, Lau KM, Wong WS, To KW, Cheng SH, Tsang KS, et al. Alterations of RAS signalling in Chinese multiple myeloma patients: absent BRAF and rare RAS mutations, but frequent inactivation of RASSF1A by transcriptional silencing or expression of a non-functional variant transcript. British journal of haematology. 2003;123(4):637–45. [DOI] [PubMed] [Google Scholar]
- 21.Nojima M, Maruyama R, Yasui H, Suzuki H, Maruyama Y, Tarasawa I, et al. Genomic screening for genes silenced by DNA methylation revealed an association between RASD1 inactivation and dexamethasone resistance in multiple myeloma. Clinical cancer research : an official journal of the American Association for Cancer Research. 2009;15(13):4356–64. [DOI] [PubMed] [Google Scholar]
- 22.Ridge SA, Worwood M, Oscier D, Jacobs A, Padua RA. FMS mutations in myelodysplastic, leukemic, and normal subjects. Proceedings of the National Academy of Sciences of the United States of America. 1990;87(4):1377–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Arefi M, Garcia JL, Penarrubia MJ, Queizan JA, Hermosin L, Lopez-Corral L, et al. Incidence and clinical characteristics of myeloproliferative neoplasms displaying a PDGFRB rearrangement. European journal of haematology. 2012;89(1):37–41. [DOI] [PubMed] [Google Scholar]
- 24.Murakami N, Okuno Y, Yoshida K, Shiraishi Y, Nagae G, Suzuki K, et al. Integrated molecular profiling of juvenile myelomonocytic leukemia. Blood. 2018. [DOI] [PubMed] [Google Scholar]
- 25.Chao AK, Meyer JA, Lee AG, Hecht A, Tarver T, Van Ziffle J, et al. Fusion driven JMML: a novel CCDC88C-FLT3 fusion responsive to sorafenib identified by RNA sequencing. Leukemia. 2020;34(2):662–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roberts KG, Li Y, Payne-Turner D, Harvey RC, Yang YL, Pei D, et al. Targetable kinase-activating lesions in Ph-like acute lymphoblastic leukemia. The New England journal of medicine. 2014;371(11):1005–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jerchel IS, Hoogkamer AQ, Ariës IM, Steeghs EMP, Boer JM, Besselink NJM, et al. RAS pathway mutations as a predictive biomarker for treatment adaptation in pediatric B-cell precursor acute lymphoblastic leukemia. Leukemia. 2018;32(4):931–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bill M, Mrózek K, Kohlschmidt J, Eisfeld A-K, Walker CJ, Nicolet D, et al. Mutational landscape and clinical outcome of patients with de novo acute myeloid leukemia and rearrangements involving 11q23/KMT2A. Proc Natl Acad Sci U S A. 2020.* This study demonstrated a high incidence of mutually exclusive Ras pathway mutations in KMT2A-rearranged AML.
- 29.Hyrenius-Wittsten A, Pilheden M, Sturesson H, Hansson J, Walsh MP, Song G, et al. De novo activating mutations drive clonal evolution and enhance clonal fitness in KMT2A-rearranged leukemia. Nat Commun. 2018;9(1):1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Paulsson K, Forestier E, Lilljebjorn H, Heldrup J, Behrendtz M, Young BD, et al. Genetic landscape of high hyperdiploid childhood acute lymphoblastic leukemia. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(50):21719–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boer JM, Koenders JE, van der Holt B, Exalto C, Sanders MA, Cornelissen JJ, et al. Expression profiling of adult acute lymphoblastic leukemia identifies a BCR-ABL1-like subgroup characterized by high non-response and relapse rates. Haematologica. 2015;100(7):e261–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee JW, Kim Y, Cho B, Kim S, Jang PS, Lee J, et al. High incidence of RAS pathway mutations among sentinel genetic lesions of Korean pediatric BCR-ABL1-like acute lymphoblastic leukemia. Cancer Med. 2020;9(13):4632–9.* This study highlighted a surprisingly high incidence of Ras pathway mutation amongst a cohort of pediatric BCR-ABL-like patients.
- 33.Pakneshan S, Salajegheh A, Smith RA, Lam AK. Clinicopathological relevance of BRAF mutations in human cancer. Pathology. 2013;45(4):346–56. [DOI] [PubMed] [Google Scholar]
- 34.Diamond EL, Durham BH, Haroche J, Yao Z, Ma J, Parikh SA, et al. Diverse and Targetable Kinase Alterations Drive Histiocytic Neoplasms. Cancer discovery. 2016;6(2):154–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nelson DS, van Halteren A, Quispel WT, van den Bos C, Bovee JV, Patel B, et al. MAP2K1 and MAP3K1 mutations in Langerhans cell histiocytosis. Genes, chromosomes & cancer. 2015;54(6):361–8. [DOI] [PubMed] [Google Scholar]
- 36.Berres ML, Lim KP, Peters T, Price J, Takizawa H, Salmon H, et al. BRAF-V600E expression in precursor versus differentiated dendritic cells defines clinically distinct LCH risk groups. The Journal of experimental medicine. 2014;211(4):669–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hyman DM, Puzanov I, Subbiah V, Faris JE, Chau I, Blay JY, et al. Vemurafenib in Multiple Nonmelanoma Cancers with BRAF V600 Mutations. The New England journal of medicine. 2015;373(8):726–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kerstjens M, Driessen EMC, Willekes M, Pinhanços SS, Schneider P, Pieters R, et al. MEK inhibition is a promising therapeutic strategy for MLL-rearranged infant acute lymphoblastic leukemia patients carrying RAS mutations. Oncotarget. 2017;8(9):14835–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Loftus JP, Yahiaoui A, Brown PA, Niswander LM, Bagashev A, Wang M, et al. Combinatorial efficacy of entospletinib and chemotherapy in patient-derived xenograft models of infant acute lymphoblastic leukemia. Haematologica. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Borthakur GPL, Boyiadzis M, et al. Phase I/II trial of the MEK1/2 inhibitor trametinib (GSK1120212) in relapsed/refractory myeloid malignancies: evidence of activity in patients with RAS mutation-positive disease.. Blood (ASH Annual Meeting Abstracts). 2012;120(21):677. [Google Scholar]
- 41.Diamond EL, Durham BH, Ulaner GA, Drill E, Buthorn J, Ki M, et al. Efficacy of MEK inhibition in patients with histiocytic neoplasms. Nature. 2019;567(7749):521–4.** This study demonstrated a high overall response rate in histiocytic disorders using the MEK inhibitor, cobimetinib, irrespective of genotype.
- 42.Ariës IM, van den Dungen RE, Koudijs MJ, Cuppen E, Voest E, Molenaar JJ, et al. Towards personalized therapy in pediatric acute lymphoblastic leukemia: RAS mutations and prednisolone resistance. Haematologica. 2015;100(4):e132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Matheson EC, Thomas H, Case M, Blair H, Jackson RK, Masic D, et al. Glucocorticoids and selumetinib are highly synergistic in RAS pathway-mutated childhood acute lymphoblastic leukemia through upregulation of BIM. Haematologica. 2019;104(9):1804–11.** This study highlighted synergy between BCL2 and MEK inhibition in preclinical models of B-ALL and elucidated a possible mechanism of action involving BIM.
- 44.Del Curatolo A, Conciatori F, Cesta Incani U, Bazzichetto C, Falcone I, Corbo V, et al. Therapeutic potential of combined BRAF/MEK blockade in BRAF-wild type preclinical tumor models. J Exp Clin Cancer Res. 2018;37(1):140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Long GV, Hauschild A, Santinami M, Atkinson V, Mandalà M, Chiarion-Sileni V, et al. Adjuvant Dabrafenib plus Trametinib in Stage III BRAF-Mutated Melanoma. N Engl J Med. 2017;377(19):1813–23. [DOI] [PubMed] [Google Scholar]
- 46.Dummer R, Hauschild A, Santinami M, Atkinson V, Mandalà M, Kirkwood JM, et al. Five-Year Analysis of Adjuvant Dabrafenib plus Trametinib in Stage III Melanoma. N Engl J Med. 2020;383(12):1139–48. [DOI] [PubMed] [Google Scholar]
- 47.DiNardo CD, Jonas BA, Pullarkat V, Thirman MJ, Garcia JS, Wei AH, et al. Azacitidine and Venetoclax in Previously Untreated Acute Myeloid Leukemia. The New England journal of medicine. 2020;383(7):617–29.** This study has transformed the landscape of AML treatment in adults by demonstrating high ORRs in untreated patients using the BCL2 inhibitor venetoclax, eventually leading to FDA approval.
- 48.Wei AH, Strickland SA Jr., Hou JZ, Fiedler W, Lin TL, Walter RB, et al. Venetoclax Combined With Low-Dose Cytarabine for Previously Untreated Patients With Acute Myeloid Leukemia: Results From a Phase Ib/II Study. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2019;37(15):1277–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Karol SE, Alexander TB, Budhraja A, Pounds SB, Canavera K, Wang L, et al. Venetoclax in combination with cytarabine with or without idarubicin in children with relapsed or refractory acute myeloid leukaemia: a phase 1, dose-escalation study. The Lancet Oncology. 2020;21(4):551–60.** This is the first prospective study in pediatric AML using venetoclax in combination with chemotherapy. Responses were seen in nearly 70% of patients and a possible biomarker using BH3 profiling was identified.
- 50.Han L, Zhang Q, Dail M, Shi C, Cavazos A, Ruvolo VR, et al. Concomitant targeting of BCL2 with venetoclax and MAPK signaling with cobimetinib in acute myeloid leukemia models. Haematologica. 2020;105(3):697–707.** This study highlights possible synergy between BCL2 and MEK inhibition in preclinical models of AML. This approach is now being tested in a phase I/II in adults.
- 51.Korfi K, Smith M, Swan J, Somervaille TC, Dhomen N, Marais R. BIM mediates synergistic killing of B-cell acute lymphoblastic leukemia cells by BCL-2 and MEK inhibitors. Cell Death Dis. 2016;7:e2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Berndt N, Hamilton AD, Sebti SM. Targeting protein prenylation for cancer therapy. Nature reviews Cancer. 2011;11(11):775–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stieglitz E, Ward AF, Gerbing RB, Alonzo TA, Arceci RJ, Liu YL, et al. Phase II/III trial of a pre-transplant farnesyl transferase inhibitor in juvenile myelomonocytic leukemia: a report from the Children’s Oncology Group. Pediatric blood & cancer. 2015;62(4):629–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Erba HP, Othus M, Walter RB, Kirschbaum MH, Tallman MS, Larson RA, et al. Four different regimens of farnesyltransferase inhibitor tipifarnib in older, untreated acute myeloid leukemia patients: North American Intergroup Phase II study SWOG S0432. Leuk Res. 2014;38(3):329–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hanna GJ, Guenette JP, Chau NG, Sayehli CM, Wilhelm C, Metcalf R, et al. Tipifarnib in recurrent, metastatic HRAS-mutant salivary gland cancer. Cancer. 2020;126(17):3972–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lee HW, Sa JK, Gualberto A, Scholz C, Sung HH, Jeong BC, et al. A Phase II Trial of Tipifarnib for Patients with Previously Treated, Metastatic Urothelial Carcinoma Harboring HRAS Mutations. Clin Cancer Res. 2020;26(19):5113–9. [DOI] [PubMed] [Google Scholar]
- 57.Cherfils J, Zeghouf M. Chronicles of the GTPase switch. Nat Chem Biol. 2011;7(8):493–5. [DOI] [PubMed] [Google Scholar]
- 58.Taveras AG, Remiszewski SW, Doll RJ, Cesarz D, Huang EC, Kirschmeier P, et al. Ras oncoprotein inhibitors: The discovery of potent, ras nucleotide exchange inhibitors and the structural determination of a drug-protein complex. Bioorganic & Medicinal Chemistry. 1997;5(1):125–33. [DOI] [PubMed] [Google Scholar]
- 59.Ostrem JM, Peters U, Sos ML, Wells JA, Shokat KM. K-Ras(G12C) inhibitors allosterically control GTP affinity and effector interactions. Nature. 2013;503(7477):548–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nagasaka M, Li Y, Sukari A, Ou SI, Al-Hallak MN, Azmi AS. KRAS G12C Game of Thrones, which direct KRAS inhibitor will claim the iron throne? Cancer Treat Rev. 2020;84:101974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hong DS, Fakih MG, Strickler JH, Desai J, Durm GA, Shapiro GI, et al. KRAS(G12C) Inhibition with Sotorasib in Advanced Solid Tumors. The New England journal of medicine. 2020;383(13):1207–17.**This study provided results from one of the first direct KRAS G12C inhibitors to reach the clinic. Responses although short lived were seen across tumor types.
- 62.Chen YN, LaMarche MJ, Chan HM, Fekkes P, Garcia-Fortanet J, Acker MG, et al. Allosteric inhibition of SHP2 phosphatase inhibits cancers driven by receptor tyrosine kinases. Nature. 2016;535(7610):148–52. [DOI] [PubMed] [Google Scholar]
- 63.Hao H-X, Wang H, Liu C, Kovats S, Velazquez R, Lu H, et al. Tumor Intrinsic Efficacy by SHP2 and RTK Inhibitors in KRAS-Mutant Cancers. Mol Cancer Ther. 2019;18(12):2368–80. [DOI] [PubMed] [Google Scholar]
- 64.Sun X, Ren Y, Gunawan S, Teng P, Chen Z, Lawrence HR, et al. Selective inhibition of leukemia-associated SHP2E69K mutant by the allosteric SHP2 inhibitor SHP099. Leukemia. 2018;32(5):1246–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lu H, Liu C, Velazquez R, Wang H, Dunkl LM, Kazic-Legueux M, et al. SHP2 Inhibition Overcomes RTK-Mediated Pathway Reactivation in KRAS-Mutant Tumors Treated with MEK Inhibitors. Mol Cancer Ther. 2019;18(7):1323–34.** This study demonstrated efficacy by combining SHP2 and MEK inhibitors to prevent upstream pathway activation in KRAS-mutant tumors.
- 66.Hofmann MH, Gmachl M, Ramharter J, Savarese F, Gerlach D, Marszalek JR, et al. BI-3406, a potent and selective SOS1::KRAS interaction inhibitor, is effective in KRAS-driven cancers through combined MEK inhibition. Cancer Discov. 2020.* This study demonstrates a proof of principle using a SOS1::KRAS inhibitor. Combining this compound with MEK inhibitors demonstrated activity across KRAS-mutated tumor types.
- 67.Hillig RC, Sautier B, Schroeder J, Moosmayer D, Hilpmann A, Stegmann CM, et al. Discovery of potent SOS1 inhibitors that block RAS activation via disruption of the RAS-SOS1 interaction. Proc Natl Acad Sci U S A. 2019;116(7):2551–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Konopleva MY, Walter RB, Faderl SH, Jabbour EJ, Zeng Z, Borthakur G, et al. Preclinical and early clinical evaluation of the oral AKT inhibitor, MK-2206, for the treatment of acute myelogenous leukemia. Clinical cancer research : an official journal of the American Association for Cancer Research. 2014;20(8):2226–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Spencer A, Yoon SS, Harrison SJ, Morris SR, Smith DA, Brigandi RA, et al. The novel AKT inhibitor afuresertib shows favorable safety, pharmacokinetics, and clinical activity in multiple myeloma. Blood. 2014;124(14):2190–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Arceci RJ, Allen CE, Dunkel IJ, Jacobsen E, Whitlock J, Vassallo R, et al. A phase IIa study of afuresertib, an oral pan-AKT inhibitor, in patients with Langerhans cell histiocytosis. Pediatric blood & cancer. 2017;64(5). [DOI] [PubMed] [Google Scholar]
- 71.Germann UA, Furey BF, Markland W, Hoover RR, Aronov AM, Roix JJ, et al. Targeting the MAPK Signaling Pathway in Cancer: Promising Preclinical Activity with the Novel Selective ERK1/2 Inhibitor BVD-523 (Ulixertinib). Molecular cancer therapeutics. 2017;16(11):2351–63. [DOI] [PubMed] [Google Scholar]
- 72.Sullivan RJ, Infante JR, Janku F, Wong DJL, Sosman JA, Keedy V, et al. First-in-Class ERK1/2 Inhibitor Ulixertinib (BVD-523) in Patients with MAPK Mutant Advanced Solid Tumors: Results of a Phase I Dose-Escalation and Expansion Study. Cancer Discov. 2018;8(2):184–95. [DOI] [PubMed] [Google Scholar]