Abstract
A novel synthesis of C(2)-modified peptide nucleic acids (PNAs) is proposed, using a submonomeric strategy with minimally protected building blocks, which allowed a reduction in the required synthetic steps. N(3)-unprotected, d-Lys- and d-Arg-based backbones were used to obtain positively charged PNAs with high optical purity, as inferred from chiral GC measurements. “Chiral-box” PNAs targeting the G12D point mutation of the KRAS gene were produced using this method, showing improved sequence selectivity for the mutated- vs wild-type DNA strand with respect to unmodified PNAs.
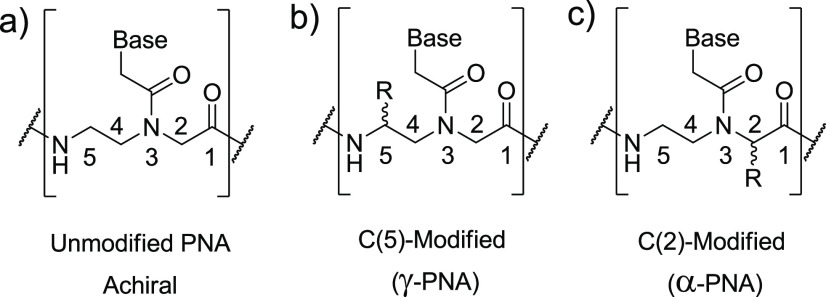
Peptide nucleic acids (PNAs, Figure 1a)1 are synthetic analogs of DNA with a poly-N-(2-aminoethyl)glycine backbone, which are largely used in biological applications due to their high affinity and very high sequence selectivity for complementary nucleic acids.2 Thanks to these properties, PNAs can be used as gene modulators using antisense,3 antigene,4 and anti-miR approaches;5 they have also been shown to promote gene-editing with high precision6 and to be suitable materials in a plethora of other applications.7 In diagnostics, PNAs have been used as probes for the detection of DNA and RNA, resulting in being particularly suited for the discrimination of single-point mutations8 and for the development of ultrasensitive devices exploiting the so-called “liquid biopsy” approach.9
Figure 1.
Structure of (a) unmodified PNA, (b) C5-modified- (γ-PNA), and (c) C2-modified (α-PNA) chiral PNA structure. Base: nucleobase (A, T, G, C).
Modified PNAs bearing positively charged amino or guanidino side chains on their backbone can display improved performances,10 allowing for the production of multifunctional derivatives11 and facilitating their cellular uptake.12 These modifications also affect their ability in interacting with complementary DNA or RNA strands, depending on the configuration of the chiral center introduced in the backbone: l-amino acid synthons in the C(5)-position (γ-PNAs) are ideal for increasing the binding affinity for complementary oligonucleotides, while d-side chains in the C(2)-position (α-PNAs) are known to increase the selectivity for single-mismatched sequences (Figure 1b and c, respectively).13 The latter issue is crucial for the diagnosis of genetic diseases or tumors, and stretches of three consecutive C(2)-modified monomers (“Chiral boxes”) derived from either d-Lys or d-Arg have been found to be effective in inducing the best single-base selectivity for target mutated-DNAs,14 with complete control in the orientation (antiparallel) of the resulting PNA:DNA duplex.15
On the other hand, some drawbacks prevent C2-modified PNAs to be used on large scales: (i) the relatively long and challenging synthetic routes to produce the corresponding monomers; (ii) the occurrence of epimerization reactions during the PNA synthesis, which generate mixtures of stereoisomers with different properties.16
The main strategies proposed to solve the first problem exploit reductive amination,17 Mitsunobu reaction,18 and alkylation of Nosyl- protected amino acids19 for the synthesis of the chiral backbone, followed by introduction of the carboxymethyl nucleobase. However, the use of complete monomers (i.e., bearing the nucleobase) require careful control of the reaction conditions for the synthesis of C(2)-modified PNAs, as, being α-acilated amino acid derivatives, they are prone to racemization on their chiral center.16 The epimerization process can be minimized by following a “submonomeric strategy”, in which the modified monomers are built directly on the solid support by the sequential attachment of the backbone (the submonomer) and of the nucleobase on a growing PNA chain.17b,17c Although optically pure PNAs could be produced in this way,14,20,21 this protocol remains challenging from a synthetic point of view since it requires the introduction of an additional protecting group at the N(3)-position of a fully protected submonomer (Scheme 1, top left), thus restricting the variety of reaction conditions that can be used subsequently and creating issues of unwanted deprotection. These points are particularly problematic in the case of Fmoc/Bhoc conditions, which are the most suited for automatic synthesizers.17c As an alternative, a submonomeric approach based on an Ugi three-component reaction was also proposed.22 In this paper, we describe the development of a shorter strategy for the production of optically pure “Chiral box” PNAs according to a submonomeric Fmoc/Bhoc or Fmoc/Boc protocol (Scheme 1, bottom route), based on the use of minimally protected building blocks (i.e., bearing protecting groups only at the N(6)-moiety and on the side chain attached at the C(2)-position) which were obtained with a simplified synthetic route.
Scheme 1. Comparison of Solid Phase Synthesis of “Chiral Box” PNAs (Top, Right Panel) with Fully and Minimally Protected Submonomers (Top Left and Bottom Routes, Respectively).
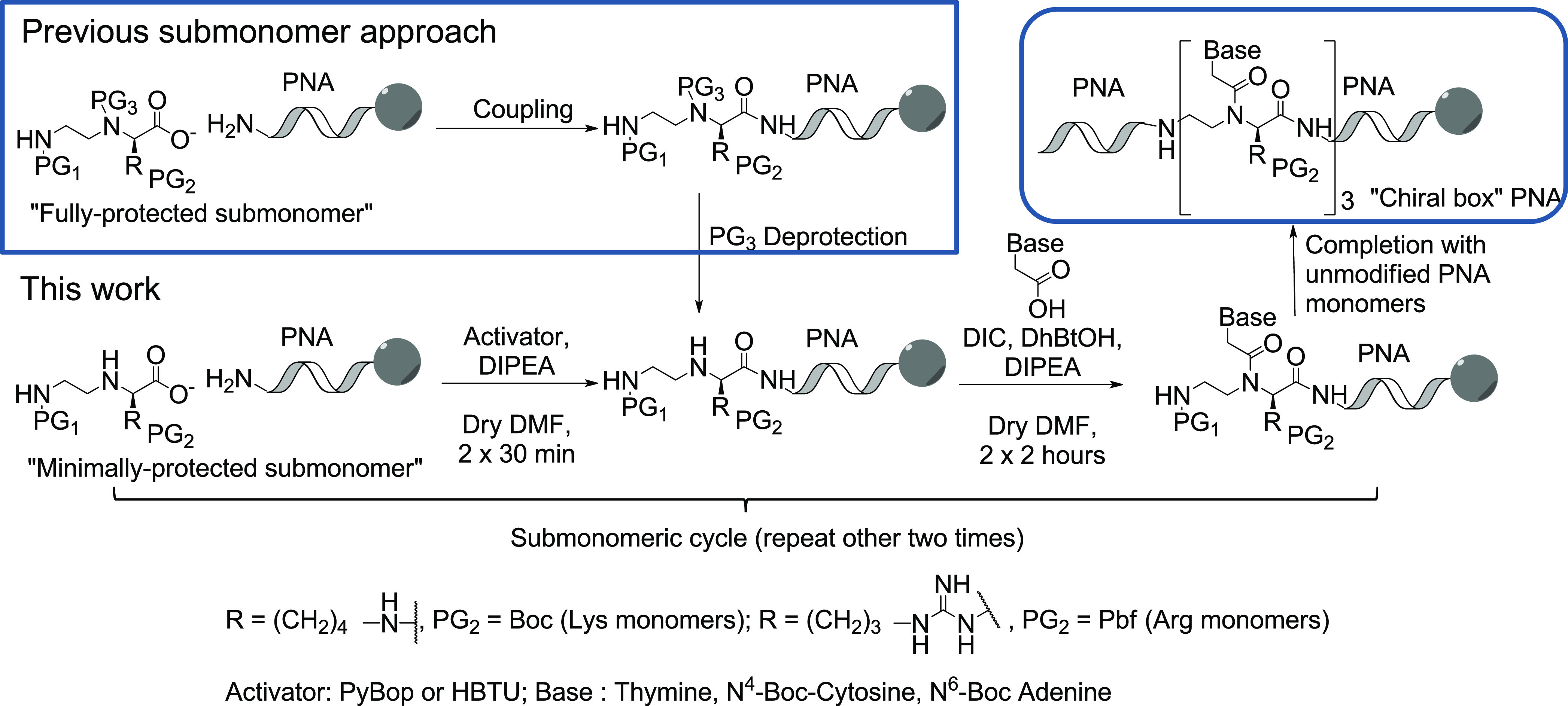
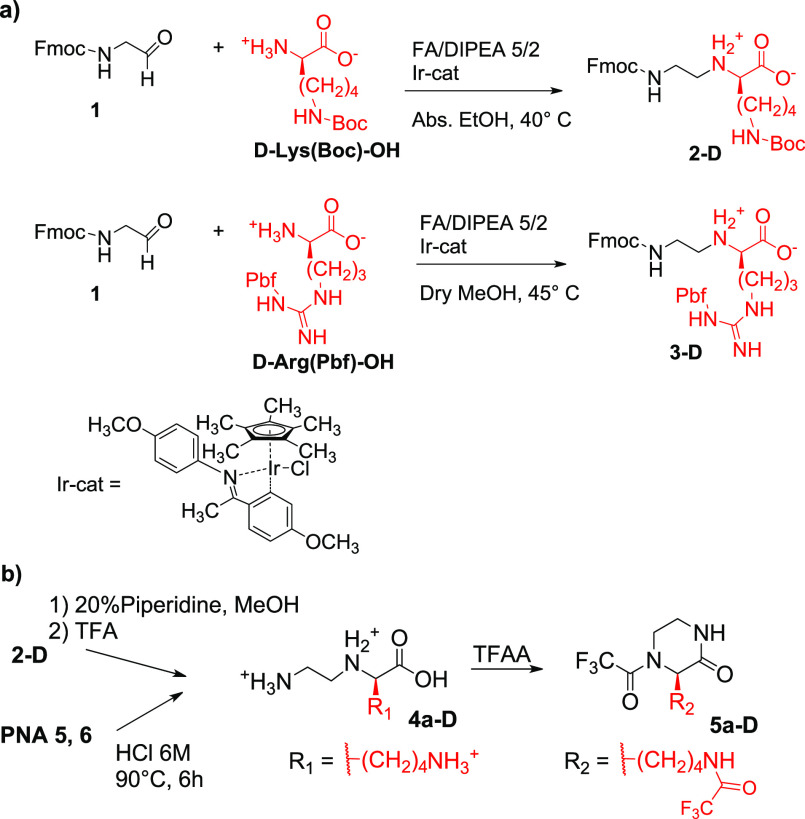
PG1, PG2, and PG3 represent orthogonal protecting groups.
During the synthesis of PNAs by the submonomeric approach, the coupling of the primary amine at the N-term with the incoming submonomer is much faster than that involving the secondary amino group of the backbone and the nucleobase in the next step; thus, we reasoned that building blocks lacking protecting groups on their N(3)-position could be suitable to perform the former reaction. Accordingly, also the protection of the C-term carboxylate could be unnecessary during the synthesis of the “minimally protected” submonomers, which could be directly performed in a single reductive amination step. This reaction can be performed with standard reducing agents (i.e., NaBH3CN), but we also explored an alternative procedure reported by Wang et al.,23 which has never been tested for the synthesis of PNA backbones. In this case, the reductive amination takes place by transfer hydrogenation, promoted by an Ir(III) catalyst in the presence of a 5/2 mixture of formic acid (FA) and DIPEA. Compounds 2-D and 3-D—the most commonly used submonomers for the synthesis of C(2)-modified PNAs—were successfully obtained by adding this mixture of reagents to Fmoc-aminoacetaldehyde 1 and d-Lys(Boc)-OH or d-Arg(Pbf)-OH in dry alcohol (EtOH or MeOH, respectively, Scheme 2a). Remarkably, the Lys-based synthon was isolated from the reaction medium by simple filtration, while the Arg-modified backbone required a reversed-phase chromatography purification step. This procedure gave 2-D and 3-D in 61% and 49% yield, respectively, which are similar or slightly higher values than those reported for standard reductive amination protocols on the same substrates (see ref (17c) or Supporting Information (SI), section 2.2, respectively). Being performed in a single step and in the presence of less toxic reagents, we suggest this strategy as a very convenient method for the synthesis of C(2)-modified PNA submonomers.
Scheme 2. (a) Synthesis of the Minimally Protected d-Submonomers by Adapting the Procedure Reported in ref (23) and (b) Derivatization of d-Lys-Based Submonomers (top) and “Chiral Box” PNAs (bottom) for Chiral GC Analysis.
Both the Lys- or Arg-based backbones were then used to produce different “Chiral Box” PNAs presenting a fully complementary sequence for the G12D point mutation of the KRAS gene (Figure 2, top), which is highly relevant for monitoring the efficacy of antibody-based therapies in colorectal cancer.24 The “Chiral box” moiety was constituted by an ATC stretch of nucleobases attached on three consecutive modified backbones, where the central T was expected to face the single point mutation of the target DNA.
Figure 2.
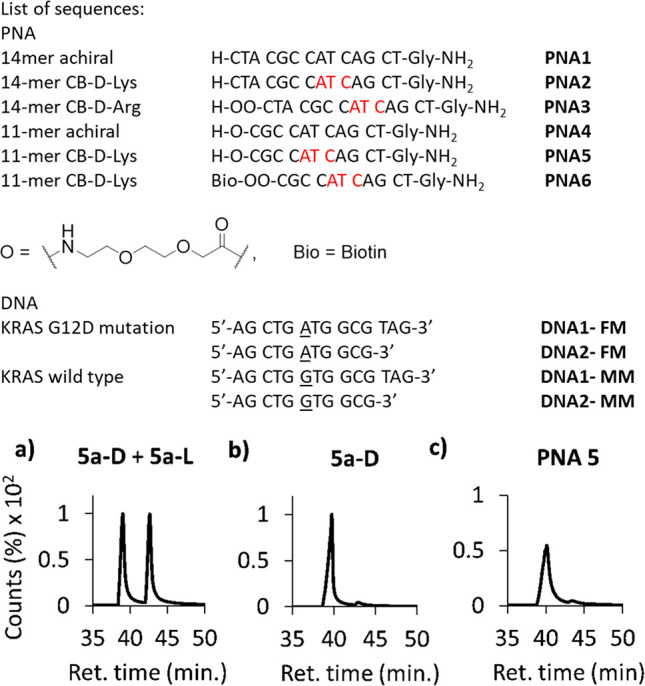
(Top) “Chiral Box” (CB) PNA, unmodified PNA and DNA sequences used in this work. C(2)-modified monomers are highlighted in red. (Bottom) Chiral GC analysis of (a) 1:1 mixture of compounds 5a-D and 5a-L, (b) 5a-D, and (c) PNA 5.
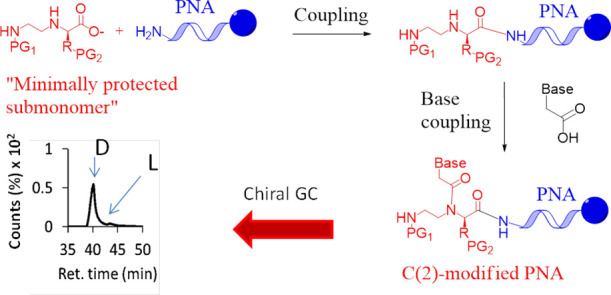
The PNAs were synthesized by adapting known submonomeric protocols for solid phase synthesis,17c in which the minimally protected building block 2-D or 3-D is attached on the N-term of a growing oligomer (Scheme 1, bottom). UPLC-MS analyses did not reveal any traces of double backbone attachment in this step (Figures S21 and S22). PyBop and HBTU were used as activating agents with similar overall efficiency, although the occurrence of uronium-based adducts on the unprotected N(3)-position was in principle suggested in the latter case.25
Subsequently, the appropriated (Boc-protected) carboxymethylnucleobase26 (in our case A, C, and T, but the same can be extended to G, as shown in other “sumbonomeric” syntheses21) was directly attached on the backbone. As expected, this step was much slower and required a combination of strong coupling agents (DIC/DhBtOH) and a long reaction time (2 × 2 h) to be finalized. The procedure was repeated for each C(2)-modified monomer and then the PNAs were completed according to standard Fmoc/Bhoc synthetic routes. In this way, Lys- and Arg-based oligomers (Figure 2, top) were obtained as probes for a fully complementary KRAS G12D-mutated DNA strand, and their selectivity for the recognition of the complementary oligonucleotide over its wild-type version will be briefly discussed below. The “Chiral box” PNAs were obtained with 5–8% yield (after purification), in line with what was reported for analogous derivatives with previous methodologies.14c The couplings of both the chiral submonomers and the nucleobases were found to be the harder steps, as evaluated by UPLC-MS after the completion of the “Chiral box” part for the d-Lys-based PNAs (Figures S23 and S24).
At this point, it was crucial to verify that the protocols presented here do not induce significant racemization in both the C(2)-modified synthons and the final “Chiral box” oligomers. For this purpose, we took advantage of a gaschromatographic method developed by some of us for the direct chiral analysis of PNAs and of their submonomers, after conversion in the corresponding trifluoroacetylated piperazine-2-ones.27 The Lys-based PNA 5 and 6 were digested in concentrated HCl to give a mixture of N-(2-aminoethyl)amino acids 4-D,27 which were in turn derivatized with trifluoroacetic anhydride (TFAA) for GC analysis on a Chirasil-Val column (Scheme 2b).
An aliquot of the submonomer 2-D and of its l-analog 2-L were instead converted to piperazine-2-ones 5a-D and 5a-L(27) (Scheme 2b and Scheme S5) after deprotection of their N-term amino group (Scheme S3, compounds 6a-D, 6a-L) and then submitted to analogous investigations.
For the d- and l-submonomers the amount of the undesired enantiomer was 3.3 ± 0.5% and 2.8 ± 1.5%, respectively (Figure 2b and S37), as reported for the same compounds obtained by regular reductive amination.27 For PNA 5 and 6 the racemization to the l-form was found to be dependent on the activator used for the introduction of the C(2)-modified backbones, being estimated to be 2.7 ± 0.3% in the former case, where PyBop was used, and 5.0 ± 2.3% for the latter PNA, which was obtained with HBTU (Figure 2c and S39, respectively). This was probably due to the different electron-withdrawing effects in the first steps of activation, which correspond to the formation of acylphosphonium vs acyluronium adducts, both of them leading to the same N-hydroxybenzotriazolyl activated ester.
These data indicate that “Chiral box” PNAs can be effectively obtained from minimally protected synthons with minimal epimerization during solid-phase synthesis, especially when proper coupling agents (i.e., PyBop) are used.
Attempts to perform the same analyses on the modified-backbones 3-D and 3-L and the Arg-based PNA 3 failed because, as for most of the arginine derivatives injected in fused-silica columns,28 their derivatization yielded piperazine-2-ones which were not suitable for chiral GC analysis (SI, sections 6 and 7).
However, we suggest that for these compounds the electronic effects on the chiral center should be similar to those affecting their Lys-based analogs, thus limiting the racemization process during the synthesis of both the submonomers and the corresponding “Chiral box” PNAs.
Finally, the recognition properties of these optically pure PNAs were tested by evaluation of the thermal stabilities for the complexes formed with complementary DNA strands presenting full-matched (G12D-mutated) and mismatched (wild type) sequences (Figure 2 top).
The change in melting temperature between the two types of duplexes (ΔTm) increased, respectively, by 1.6 and 0.9 °C when the 14-mer PNA2 and 3 were used in place of the corresponding unmodified PNA 1 (Table 1), indicating a higher performance of the C(2)-modified oligomers for the discrimination of single point mutations. A further increase of selectivity was recorded by shortening the sequences of the tested probes, thus increasing the influence of the “Chiral box” stretch. In fact, for the Lys-based, 11-mer PNA 5 the best discrimination ability was obtained, giving a remarkable ΔTm value of 19.2 °C between the full-matched and the mismatched complexes, which was 3.3 °C higher than that afforded by its unmodified analog PNA 4.
Table 1. Comparison of Melting Temperatures for Unmodified and “Chiral Box” PNAs with Full-Matched and Mismatched Target DNA. ΔTm = Tm(Full Match) – Tm(Mismatch).
| Hybrid type | DNA | Tm [°C] | ΔTm [°C] |
|---|---|---|---|
| PNA 1 | DNA1-FM | 76.7 ± 0.9 | 14.1 |
| PNA 1 | DNA1-MM | 62.6 ± 0.6 | |
| PNA 2 | DNA1-FM | 72.4 ± 0.8 | 15.7 |
| PNA 2 | DNA1-MM | 56.7 ± 0.8 | |
| PNA 3 | DNA1-FM | 73.0 ± 2.0 | 15.0 |
| PNA 3 | DNA1-MM | 58.0 ± 0.9 | |
| PNA 4 | DNA2-FM | 65.9 ± 0.5 | 15.9 |
| PNA 4 | DNA2-MM | 50.0 ± 0.5 | |
| PNA 5 | DNA2-FM | 59.9 ± 0.2 | 19.2 |
| PNA 5 | DNA2-MM | 40.7 ± 0.5 |
It is worth noting that “Chiral-box” PNAs form less stable adducts with complementary DNAs in comparison to their unmodified version (i.e., Tm = 59.9 °C vs 65.9 °C for PNA 5 and 4, respectively), due to the sum of destabilizing steric effects generated by the three adjacent modified monomers. This apparent disadvantage is balanced by the higher selectivity of the PNA:DNA interaction, resulting in a higher sensitivity for a single mismatch in cognate DNA strands.
In conclusion, the described submonomeric strategy for obtaining “Chiral Box” PNAs has significant advantages in terms of simplicity and time consumption for the synthesis of both the monomers and the corresponding oligomers. In particular, a careful evaluation of the reactivity for the substrates possibly undergoing acylation during the insertion of the modified-backbones (primary vs hindered secondary amino groups) has allowed the elimination of undue protection/deprotection steps. The given PNAs show high optical purity and increased performance in terms of mismatch discrimination for cognate DNA strands, resulting in suitability for sensing devices relying on advanced optical or electronic techniques such as Surface Plasmon Fluorescence Spectroscopy (SPFS)29—whose development is currently underway—or Field Effect Transistors (FETs).30
Acknowledgments
This work was partially funded by the H2020 ULTRAPLACAD project (Project No. 633937) and has benefited from the equipment and framework of the COMP-HUB Initiative, funded by the “Departments of Excellence” program of the Italian Ministry for Education, University and Research (MIUR, 2018-2022) for the Department of Chemistry, Life Sciences and Environmental Sustainability of the University of Parma. The TeachInParma Program is also acknowledged for sponsoring the supervision and collaborative work of W.K. at the University of Parma premises.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.orglett.0c04116.
Experimental procedures, characterizations, NMR spectra for all compounds, UPLC-MS traces for pure PNAs, GC chromatograms, and hybridization studies (PDF)
Author Contributions
§ S.V. and A.R. contributed equally.
The authors declare no competing financial interest.
Supplementary Material
References
- a Nielsen P. E.; Egholm M.; Berg R. H.; Buchardt O. Sequence-Selective Recognition of DNA by Strand Displacement with a Thymine-Substituted Polyamide. Science 1991, 254 (5037), 1497–1500. 10.1126/science.1962210. [DOI] [PubMed] [Google Scholar]; b Nielsen P. E. Peptide Nucleic Acid. A Molecule with Two Identities. Acc. Chem. Res. 1999, 32 (7), 624–630. 10.1021/ar980010t. [DOI] [Google Scholar]
- Egholm M.; Buchardt O.; Christensen L.; Behrens C.; Freier S. M.; Driver D. A.; Berg R. H.; Kim S. K.; Norden B.; Nielsen P. E. PNA Hybridizes to Complementary Oligonucleotides Obeying the Watson-Crick Hydrogen-Bonding Rules. Nature 1993, 365, 566–568. 10.1038/365566a0. [DOI] [PubMed] [Google Scholar]
- a Gait M. J.; Arzumanov A. A.; McClorey G.; Godfrey C.; Betts C.; Hammond S.; Wood M. J. A. Cell-Penetrating Peptide Conjugates of Steric Blocking Oligonucleotides as Therapeutics for Neuromuscular Diseases from a Historical Perspective to Current Prospects of Treatment. Nucleic Acid Ther. 2019, 29 (1), 1–12. 10.1089/nat.2018.0747. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Delgado E.; Bahal R.; Yang J.; Lee J. M.; Ly D. H.; Monga S. P. S. β-Catenin Knockdown in Liver Tumor Cells by a Cell Permeable Gamma Guanidine-Based Peptide Nucleic Acid. Curr. Cancer Drug Targets 2013, 13 (8), 867–878. 10.2174/15680096113139990081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Cutrona G.; Carpaneto E. M.; Ulivi M.; Roncella S.; Landt O.; Ferrarini M.; Boffa L. C. Effects in Live Cells of a C-Myc Anti-Gene PNA Linked to a Nuclear Localization Signal. Nat. Biotechnol. 2000, 18 (3), 300–303. 10.1038/73745. [DOI] [PubMed] [Google Scholar]; b Janowski B. A.; Kaihatsu K.; Huffman K. E.; Schwartz J. C.; Ram R.; Hardy D.; Mendelson C. R.; Corey D. R. Inhibiting Transcription of Chromosomal Dna With Antigene Peptide Nucleic Acids. Nat. Chem. Biol. 2005, 1 (4), 210–215. 10.1038/nchembio724. [DOI] [PubMed] [Google Scholar]
- a Fabani M. M.; Abreu-Goodger C.; Williams D.; Lyons P. A.; Torres A. G.; Smith K. G. C.; Enright A. J.; Gait M. J.; Vigorito E. Efficient Inhibition of MiR-155 Function in Vivo by Peptide Nucleic Acids. Nucleic Acids Res. 2010, 38 (13), 4466–4475. 10.1093/nar/gkq160. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Babar I. A.; Cheng C. J.; Booth C. J.; Liang X.; Weidhaas J. B.; Saltzman W. M.; Slack F. J. Nanoparticle-Based Therapy in an in Vivo MicroRNA-155 (MiR-155)-Dependent Mouse Model of Lymphoma. Proc. Natl. Acad. Sci. U S.A. 2012, 109 (26), E1695. 10.1073/pnas.1201516109. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Torres A. G.; Fabani M. M.; Vigorito E.; Williams D.; Al-Obaidi N.; Wojciechowski F.; Hudson R. H. E.; Seitz O.; Gait M. J. Chemical Structure Requirements and Cellular Targeting of MicroRNA-122 by Peptide Nucleic Acids Anti-MiRs. Nucleic Acids Res. 2012, 40 (5), 2152–2167. 10.1093/nar/gkr885. [DOI] [PMC free article] [PubMed] [Google Scholar]; d Oh S. Y.; Ju Y.; Kim S.; Park H. PNA-Based Antisense Oligonucleotides for MicroRNAs Inhibition in the Absence of a Transfection Reagent. Oligonucleotides 2010, 20 (5), 225–230. 10.1089/oli.2010.0238. [DOI] [PubMed] [Google Scholar]; e Gambari R.; Fabbri E.; Borgatti M.; Lampronti I.; Finotti A.; Brognara E.; Bianchi N.; Manicardi A.; Marchelli R.; Corradini R. Targeting MicroRNAs Involved in Human Diseases: A Novel Approach for Modification of Gene Expression and Drug Development. Biochem. Pharmacol. 2011, 82 (10), 1416–1429. 10.1016/j.bcp.2011.08.007. [DOI] [PubMed] [Google Scholar]
- a Bahal R.; Ali McNeer N.; Quijano E.; Liu Y.; Sulkowski P.; Turchick A.; Lu Y. C.; Bhunia D. C.; Manna A.; Greiner D. L.; Brehm M. A.; Cheng C. J.; López-Giráldez F.; Ricciardi A.; Beloor J.; Krause D. S.; Kumar P.; Gallagher P. G.; Braddock D. T.; Mark Saltzman W.; Ly D. H.; Glazer P. M. In Vivo Correction of Anaemia in β-Thalassemic Mice by Γ3PNA-Mediated Gene Editing with Nanoparticle Delivery. Nat. Commun. 2016, 7. 10.1038/ncomms13304. [DOI] [PMC free article] [PubMed] [Google Scholar]; b McNeer N. A.; Anandalingam K.; Fields R. J.; Caputo C.; Kopic S.; Gupta A.; Quijano E.; Polikoff L.; Kong Y.; Bahal R.; Geibel J. P.; Glazer P. M.; Mark Saltzman W.; Egan M. E.. Nanoparticles That Deliver Triplex-Forming Peptide Nucleic Acid Molecules Correct F508del CFTR in Airway Epithelium. Nat. Commun. 2015, 6. 10.1038/ncomms7952. [DOI] [PMC free article] [PubMed] [Google Scholar]; c McNeer N. A.; Schleifman E. B.; Cuthbert A.; Brehm M.; Jackson A.; Cheng C.; Anandalingam K.; Kumar P.; Shultz L. D.; Greiner D. L.; Mark Saltzman W.; Glazer P. M. Systemic Delivery of Triplex-Forming PNA and Donor DNA by Nanoparticles Mediates Site-Specific Genome Editing of Human Hematopoietic Cells in Vivo. Gene Ther. 2013, 20 (6), 658–669. 10.1038/gt.2012.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Sharma C.; Awasthi S. K. Versatility of Peptide Nucleic Acids (PNAs): Role in Chemical Biology, Drug Discovery, and Origins of Life. Chem. Biol. Drug Des. 2017, 89 (1), 16–37. 10.1111/cbdd.12833. [DOI] [PubMed] [Google Scholar]; b Gupta A.; Mishra A.; Puri N. Peptide Nucleic Acids: Advanced Tools for Biomedical Applications. J. Biotechnol. 2017, 259 (March), 148–159. 10.1016/j.jbiotec.2017.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Saarbach J.; Sabale P. M.; Winssinger N. Peptide Nucleic Acid (PNA) and Its Applications in Chemical Biology, Diagnostics, and Therapeutics. Curr. Opin. Chem. Biol. 2019, 52 (September), 112–124. 10.1016/j.cbpa.2019.06.006. [DOI] [PubMed] [Google Scholar]
- a Dong B.; Nie K.; Shi H.; Chao L.; Ma M.; Gao F.; Liang B.; Chen W.; Long M.; Liu Z. Film-Spotting Chiral MiniPEG-ΓPNA Array for BRCA1 Gene Mutation Detection. Biosens. Bioelectron. 2019, 136 (February), 1–7. 10.1016/j.bios.2019.04.027. [DOI] [PubMed] [Google Scholar]; b Zhang N.; Appella D. H. Colorimetric Detection of Anthrax DNA with a Peptide Nucleic Acid Sandwich-Hybridization Assay. J. Am. Chem. Soc. 2007, 129 (27), 8424–8425. 10.1021/ja072744j. [DOI] [PubMed] [Google Scholar]
- D’Agata R.; Bellassai N.; Allegretti M.; Rozzi A.; Korom S.; Manicardi A.; Melucci E.; Pescarmona E.; Corradini R.; Giacomini P.; Spoto G. Direct Plasmonic Detection of Circulating RAS Mutated DNA in Colorectal Cancer Patients. Biosens. Bioelectron. 2020, 170 (September), 112648. 10.1016/j.bios.2020.112648. [DOI] [PubMed] [Google Scholar]
- a Kumar V. A.; Ganesh K. N. Conformationally Constrained PNA Analogues: Structural Evolution toward DNA/RNA Binding Selectivity. Acc. Chem. Res. 2005, 38 (5), 404–412. 10.1021/ar030277e. [DOI] [PubMed] [Google Scholar]; b Corradini R.; Sforza S.; Tedeschi T.; Totsingan F.; Manicardi A.; Marchelli R. Peptide Nucleic Acids with a Structurally Biased Backbone. Updated Review and Emerging Challenges. Curr. Top. Med. Chem. 2011, 11 (12), 1535–1554. 10.2174/156802611795860979. [DOI] [PubMed] [Google Scholar]; c Sugiyama T.; Kittaka A. Chiral Peptide Nucleic Acids with a Substituent in the N-(2-Aminoethy) Glycine Backbone. Molecules 2013, 18 (1), 287–310. 10.3390/molecules18010287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Englund E. A.; Appella D. H. γ-Substituted Peptide Nucleic Acids Constructed from L-Lysine Are a Versatile Scaffold for Multifunctional Display. Angew. Chem., Int. Ed. 2007, 46 (9), 1414–1418. 10.1002/anie.200603483. [DOI] [PubMed] [Google Scholar]; b Englund E. A.; Appella D. H. Synthesis of γ-Substituted Peptide Nucleic Acids: A New Place to Attach Fluorophores without Affecting DNA Binding. Org. Lett. 2005, 7 (16), 3465–3467. 10.1021/ol051143z. [DOI] [PubMed] [Google Scholar]; c Ishizuka T.; Yoshida J.; Yamamoto Y.; Sumaoka J.; Tedeschi T.; Corradini R.; Sforza S.; Komiyama M. Chiral Introduction of Positive Charges to PNA for Double-Duplex Invasion to Versatile Sequences. Nucleic Acids Res. 2008, 36 (5), 1464–1471. 10.1093/nar/gkm1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Zhou P.; Wang M.; Du L.; Fisher G. W.; Waggoner A.; Ly D. H. Novel Binding and Efficient Cellular Uptake of Guanidine-Based Peptide Nucleic Acids (GPNA). J. Am. Chem. Soc. 2003, 125 (23), 6878–6879. 10.1021/ja029665m. [DOI] [PubMed] [Google Scholar]; b Dragulescu-Andrasi A.; Zhou P.; He G.; Ly D. H. Cell-Permeable GPNA with Appropriate Backbone Stereochemistry and Spacing Binds Sequence-Specifically to RNA. Chem. Commun. 2005, (2), 244–246. 10.1039/b412522c. [DOI] [PubMed] [Google Scholar]; c Dragulescu-Andrasi A.; Rapireddy S.; He G.; Bhattacharya B.; Hyldig-Nielsen J. J.; Zon G.; Ly D. H. Cell-Permeable Peptide Nucleic Acid Designed to Bind to the 5′-Untranslated Region of E-Cadherin Transcript Induces Potent and Sequence-Specific Antisense Effects. J. Am. Chem. Soc. 2006, 128 (50), 16104–16112. 10.1021/ja063383v. [DOI] [PubMed] [Google Scholar]; d Sahu B.; Chenna V.; Lathrop K. L.; Thomas S. M.; Zon G.; Livak K. J.; Ly D. H. Synthesis of Conformationaly Preorganized and Cell-Permeable Guanidine-Based γ-Peptide Nucleic Acids (γGPNAs). J. Org. Chem. 2009, 74 (4), 1509–1516. 10.1021/jo802211n. [DOI] [PMC free article] [PubMed] [Google Scholar]; e Thomas S. M.; Sahu B.; Rapireddy S.; Bahal R.; Wheeler S. E.; Procopio E. M.; Kim J.; Joyce S. C.; Contrucci S.; Wang Y.; Chiosea S. I.; Lathrop K. L.; Watkins S.; Grandis J. R.; Armitage B. A.; Ly D. H. Antitumor Effects of EGFR Antisense Guanidine-Based Peptide Nucleic Acids in Cancer Models. ACS Chem. Biol. 2013, 8 (2), 345–352. 10.1021/cb3003946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Sforza S.; Tedeschi T.; Corradini R.; Marchelli R. Induction of Helical Handedness and DNA Binding Properties of Peptide Nucleic Acids (PNAs) with Two Stereogenic Centres. Eur. J. Org. Chem. 2007, 2007 (35), 5879–5885. 10.1002/ejoc.200700644. [DOI] [Google Scholar]; b Mitra R.; Ganesh K. N. Aminomethylene Peptide Nucleic Acid (Am -PNA): Synthesis, Regio-/Stereospecific DNA Binding, and Differential Cell Uptake of (α/γ, R/S) Am- PNA Analogues. J. Org. Chem. 2012, 77 (13), 5696–5704. 10.1021/jo300860f. [DOI] [PubMed] [Google Scholar]
- a Feriotto G.; Corradini R.; Sforza S.; Bianchi N.; Mischiati C.; Marchelli R.; Gambari R. Peptide Nucleic Acids and Biosensor Technology for Real-Time Detection of the Cystic Fibrosis W1282X Mutation by Surface Plasmon Resonance. Lab. Invest. 2001, 81 (10), 1415–1427. 10.1038/labinvest.3780355. [DOI] [PubMed] [Google Scholar]; b Tedeschi T.; Chiari M.; Galaverna G.; Sforza S.; Cretich M.; Corradini R.; Marchelli R. Detection of the R553X DNA Single Point Mutation Related to Cystic Fibrosis by a “Chiral Box” D-Lysine-Peptide Nucleic Acid Probe by Capillary Electrophoresis. Electrophoresis 2005, 26 (22), 4310–4316. 10.1002/elps.200410390. [DOI] [PubMed] [Google Scholar]; c Manicardi A.; Calabretta A.; Bencivenni M.; Tedeschi T.; Sforza S.; Corradini R.; Marchelli R. Affinity and Selectivity of C2- and C5-Substituted ‘“ Chiral-Box ”’ PNA in Solution and on Microarrays. Chirality 2010, 22 (May), E161–E172. 10.1002/chir.20865. [DOI] [PubMed] [Google Scholar]
- Sforza S.; Corradini R.; Ghirardi S.; Dossena A.; Marchelli R. DNA Binding of a D-Lysine-Based Chiral PNA: Direction Control and Mismatch Recognition. Eur. J. Org. Chem. 2000, 2000 (16), 2905–2913. . [DOI] [Google Scholar]
- a Corradini R.; Sforza S.; Dossena A.; Palla G.; Rocchi R.; Filira F.; Nastri F.; Marchelli R. Epimerization of Peptide Nucleic Acids Analogs during Solid-Phase Synthesis: Optimization of the Coupling Conditions for Increasing the Optical Purity. J. Chem. Soc. Perkin 1 2001, 1 (20), 2690–2696. 10.1039/b104146k. [DOI] [Google Scholar]; b Tedeschi T.; Corradini R.; Marchelli R.; Nielsen P. E. Racemization of Chiral PNAs during Solid-Phase Synthesis : Effect of the Coupling Conditions on Enantiomeric Purity. Tetrahedron: Asymmetry 2002, 13, 1629–1636. 10.1016/S0957-4166(02)00413-5. [DOI] [Google Scholar]
- a Püschl A.; Sforza S.; Haaima G.; Dahl O.; Nielsen P. E. Peptide Nucleic Acids (PNAs) with a Functional Backbone. Tetrahedron Lett. 1998, 39 (26), 4707–4710. 10.1016/S0040-4039(98)00862-4. [DOI] [Google Scholar]; b Sforza S.; Tedeschi T.; Corradini R.; Ciavardelli D.; Dossena A.; Marchelli R. Fast, Solid-Phase Synthesis of Chiral Peptide Nucleic Acids with a High Optical Purity by a Submonomeric Strategy. Eur. J. Org. Chem. 2003, 2003 (6), 1056–1063. 10.1002/ejoc.200390148. [DOI] [Google Scholar]; c Tedeschi T.; Sforza S.; Maffei F.; Corradini R.; Marchelli R. A Fmoc-Based Submonomeric Strategy for the Solid Phase Synthesis of Optically Pure Chiral PNAs. Tetrahedron Lett. 2008, 49 (33), 4958–4961. 10.1016/j.tetlet.2008.05.114. [DOI] [Google Scholar]
- a Falkiewicz B.; Wiśniowski W.; Kolodziejczyk A. S.; Wiśniewski K. Synthesis of New Chiral Peptide Nucleic Acid (PNA) Monomers. Nucleosides, Nucleotides Nucleic Acids 2001, 20 (4–7), 1393–1397. 10.1081/NCN-100002563. [DOI] [PubMed] [Google Scholar]; b Abdelbaky A. S.; Prokhorov I. A.; Smirnov I. P.; Koroleva K. M.; Shvets V. I.; Kirillova Y. G. Synthesis of α-(R)-/γ-(S)-Dimethyl Substituted Peptide Nucleic Acid Submonomer Using Mitsunobu Reaction. Lett. Org. Chem. 2019, 16 (5), 437–446. 10.2174/1570178616666190118155031. [DOI] [Google Scholar]
- Malamgari S. R.; Manikandan P.; Ramani P.; Katta V. R. Synthesis of Peptide Nucleic Acid Monomers via N-Alkylation of Nosyl-Protected Amino Acids with N-Boc Bromoethyl Amine. ChemistrySelect 2018, 3 (14), 3948–3951. 10.1002/slct.201800202. [DOI] [Google Scholar]
- Menchise V.; De Simone G.; Tedeschi T.; Corradini R.; Sforza S.; Marchelli R.; Capasso D.; Saviano M.; Pedone C. Insights into Peptide Nucleic Acid (PNA) Structural Features: The Crystal Structure of a D-Lysine-Based Chiral PNA-DNA Duplex. Proc. Natl. Acad. Sci. U. S. A. 2003, 100 (21), 12021–12026. 10.1073/pnas.2034746100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manicardi A.; Fabbri E.; Tedeschi T.; Sforza S.; Bianchi N.; Brognara E.; Gambari R.; Marchelli R.; Corradini R. Cellular Uptakes, Biostabilities and Anti-MiR-210 Activities of Chiral Arginine-PNAs in Leukaemic K562 Cells. ChemBioChem 2012, 13 (9), 1327–1337. 10.1002/cbic.201100745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saarbach J.; Masi D.; Zambaldo C.; Winssinger N. Facile Access to Modified and Functionalized PNAs through Ugi-Based Solid Phase Oligomerization. Bioorg. Med. Chem. 2017, 25 (19), 5171–5177. 10.1016/j.bmc.2017.05.064. [DOI] [PubMed] [Google Scholar]
- Wang C.; Pettman A.; Basca J.; Xiao J. A Versatile Catalyst for Reductive Amination by Transfer Hydrogenation. Angew. Chem. 2010, 122 (41), 7710–7714. 10.1002/ange.201002944. [DOI] [PubMed] [Google Scholar]
- Mullard A. Cracking KRAS. Nat. Rev. Drug Discovery 2019, 18 (12), 887–891. 10.1038/d41573-019-00195-5. [DOI] [PubMed] [Google Scholar]
- El-Faham A.; Albericio F. Peptide Coupling Reagents, More than a Letter Soup. Chem. Rev. 2011, 111 (11), 6557–6602. 10.1021/cr100048w. [DOI] [PubMed] [Google Scholar]
- Pothukanuri S.; Pianowski Z.; Winssinger N. Expanding the Scope and Orthogonality of PNA Synthesis. Eur. J. Org. Chem. 2008, 2008 (18), 3141–3148. 10.1002/ejoc.200800141. [DOI] [Google Scholar]
- Corradini R.; Di Silvestro G.; Sforza S.; Palla G.; Dossena A.; Nielsen P. E.; Marchelli R. Direct Enantiomeric Separation of N-Aminoethylamino Acids: Determination of the Enantiomeric Excess of Chiral Peptide Nucleic Acids (PNAs) by GC. Tetrahedron: Asymmetry 1999, 10 (11), 2063–2066. 10.1016/S0957-4166(99)00210-4. [DOI] [Google Scholar]
- a Pätzold R.; Brückner H. Chiral Separation of Amino Acids by Gas Chromatography. J. Chromatogr. Libr. 2005, 70 (C), 98–118. 10.1016/S0301-4770(05)80005-3. [DOI] [Google Scholar]; b Schurig V. Gas Chromatographic Enantioseparation of Derivatized α-Amino Acids on Chiral Stationary Phases-Past and Present. J. Chromatogr. B: Anal. Technol. Biomed. Life Sci. 2011, 879 (29), 3122–3140. 10.1016/j.jchromb.2011.04.005. [DOI] [PubMed] [Google Scholar]
- Park H.; Germini A.; Sforza S.; Corradini R.; Marchelli R.; Knoll W. Kinetic and Affinity Analyses of Hybridization Reactions between Peptide Nucleic Acid Probes and DNA Targets Using Surface Plasmon Field-Enhanced Fluorescence Spectroscopy. Biointerphases 2006, 1 (4), 113–122. 10.1116/1.2365386. [DOI] [PubMed] [Google Scholar]
- Khan H. U.; Roberts M. E.; Johnson O.; Förch R.; Knoll W.; Bao Z. In Situ, Label-Free DNA Detection Using Organic Transistor Sensors. Adv. Mater. 2010, 22 (40), 4452–4456. 10.1002/adma.201000790. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.