Newly identified mitovesicles are a type of extracellular vesicles whose levels and cargo are altered in Down syndrome.
Abstract
Mitochondrial dysfunction is an established hallmark of aging and neurodegenerative disorders such as Down syndrome (DS) and Alzheimer’s disease (AD). Using a high-resolution density gradient separation of extracellular vesicles (EVs) isolated from murine and human DS and diploid control brains, we identify and characterize a previously unknown population of double-membraned EVs containing multiple mitochondrial proteins distinct from previously described EV subtypes, including microvesicles and exosomes. We term these newly identified mitochondria-derived EVs “mitovesicles.” We demonstrate that brain-derived mitovesicles contain a specific subset of mitochondrial constituents and that their levels and cargo are altered during pathophysiological processes where mitochondrial dysfunction occurs, including in DS. The development of a method for the selective isolation of mitovesicles paves the way for the characterization in vivo of biological processes connecting EV biology and mitochondria dynamics and for innovative therapeutic and diagnostic strategies.
INTRODUCTION
Extracellular vesicles (EVs) are heterogeneous nanoscale vesicles secreted into the extracellular space by all cell types studied, including brain cells (1). Microvesicles and exosomes are the main EV subtypes identified to date. Microvesicles are 100- to 1000-nm EVs that bud from the plasma membrane and contain specific proteins, such as annexin A1 and annexin A2 (2). Exosomes are 50- to 150-nm EVs that originate from the endocytic pathway upon fusion of the late endosomes/multivesicular bodies (MVBs) with the plasma membrane and are enriched in endosomal sorting complex required for transport (ESCRT) proteins, such as the ALG-2-interacting protein X (Alix) and the tumor susceptibility gene 101 protein (TSG101), and in tetraspanins, such as CD63 and CD81 (2).
Previous in vitro studies demonstrated that multiple cell types secrete mitochondrial components into the culture media (3–7). It was suggested that entire mitochondria are transferred between cells and have a neuroprotective function (8). It was also demonstrated that mitochondrial stress stimulates the release of molecules, including mitochondrial DNA (mtDNA), and damage-associated molecular patterns (DAMPs) that have strong proinflammatory activity (9). Contradictory reports were published on the nature of secreted mitochondrial material, ranging from a proposal that whole mitochondria, mitochondria-encapsulated microvesicles, or exosomes packed with mitochondrial cargo can be secreted from cells in vitro (3–7). Notably, these studies relied on differential centrifugation-based methods to isolate EVs that failed to differentiate among EV subpopulations and between vesicular and nonvesicular components (2). Thus, mechanisms underlying how mitochondrial constituents are secreted and their biological functions remain unknown. Furthermore, in vivo release of mitochondrial components into brain extracellular space has not been described.
We previously developed a protocol to isolate small EVs from the brain, using a sucrose density gradient (10, 11). With this approach, we demonstrated that exosomes are key players in both Alzheimer’s disease (AD) and Down syndrome (DS) pathogenesis, exerting complex pleiotropic roles as neuroprotective and neuropathogenic agents (1, 12–16). Here, we apply a novel high-resolution density gradient on isolated brain EVs. We demonstrate separation of various brain EV subtypes and the existence of a previously unknown population of mitochondria-derived EVs. These EVs, which we term mitovesicles, differ morphologically, by size and content, from intracellular mitochondria and from canonical EV subtypes. We also show that the levels and cargo of mitovesicles are altered in DS brains, which have been characterized by the concomitant presence of mitochondrial and endosomal abnormalities (17). These data demonstrate that brain cells constitutively secrete mitochondrial vesicles and that this pathway is altered under conditions in which mitochondrial dysfunction occurs, providing new insights into heterogeneous EV biology and its relationship with mitochondrial dynamics.
RESULTS
Isolation of distinct subpopulations of brain EVs by a high-resolution density gradient
To separate brain EV subpopulations and determine which EV subtypes contain mitochondrial proteins, we modified our original method of isolation and fractionated brain EVs with an OptiPrep-based step gradient density column. Briefly, murine right hemibrains were dissected, digested in papain to loosen the extracellular matrix, filtrated, and differentially centrifuged to eliminate cells, undigested materials, and cellular debris. A crude EV pellet was subsequently washed and further purified through a high-resolution step gradient (table S1). Eight distinct fractions were obtained using this approach (Fig. 1A). Brain EV fractions were initially characterized in wild-type mice. Nanoparticle tracking analysis (NTA) demonstrated that the number of vesicles is higher in fractions 1, 2, 3, 4, and 5 (Fr1, Fr2, Fr3, F4, and Fr5) when compared with Fr6 and Fr7 and reached a minimum in Fr8 (Fig. 1B). EV protein content did not parallel the number of vesicles per fraction, suggesting differences in the amount of total proteins per vesicle (Fig. 1C). Normalization of the quantity of proteins to the number of EVs in each fraction revealed three patterns, with low-density fractions (LDFs; corresponding to Fr1, Fr2, and Fr3) having low protein amount per vesicle, intermediate-density fractions (IDFs; corresponding to Fr4, Fr5, and Fr6) showing a higher protein amount per vesicle, and Fr8 containing the highest protein amount per vesicle (Fig. 1D).
Fig. 1. Subpopulations of brain EVs with different morphometric features are separated through a high-resolution density gradient.
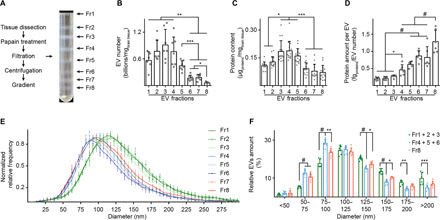
(A) Schematic protocol of the brain EV isolation and a representative picture of the OptiPrep density gradient fractionation. (B) NTA, (C) protein content, and (D) quantification of protein content per vesicle in EV fractions isolated from the brain of wild-type mice, normalized to the brain weight. (E) Diameter analysis of brain EVs by NTA. The bell curves are normalized to the mode of the distribution. (F) Percentage of vesicles with a diameter that falls within a 25-nm bin, as estimated in (E). The numbers of LDFs (Fr1 to Fr3, green) and of IDFs (Fr4 to Fr6, blue) were combined as they showed similar results. Fr8 is in red. Statistical test: (B) to (D), one-way ANOVA with Tukey’s multiple comparisons test; (F), two-way ANOVA with Tukey’s multiple comparisons test. Number of independent isolations: (B), 7; (C), 15; (D) to (F), 6. Data shown in (B) to (F): Means ± SD. *P < 0.05, **P < 0.01, ***P < 10−3, #P < 10−4.
EVs found in LDFs were larger as characterized by NTA than other EV subtypes (Fig. 1, E and F). A small size difference between IDFs and Fr8 was also observed, with IDF EVs being slightly smaller than EVs found in Fr8 (Fig. 1, E and F). These data indicate that the number, protein amount, and size of brain EVs differ substantially between OptiPrep fractions.
Morphological analysis of brain EVs by cryogenic electron microscopy
Imaging of unfixed, nonstained, hydrated brain EVs using cryogenic electron microscopy (cryoEM; Fig. 2A) preserves native EV morphology better than transmission electron microscopy (TEM; Fig. 2A, insets with red borders). The eight EV fractions contained three main types of EVs, each enriched in LDFs, IDFs, and Fr8 (Fig. 2A, magnifications a, b, and c), as well as several evenly distributed minor subtypes of EVs (Fig. 2, A to E). None of the samples visualized by either cryoEM or TEM contained debris, broken membranes, or protein aggregates, an indication of their purity. CryoEM revealed extensive heterogeneity of the brain EVs, including single-, double-, and triple-membrane–bound vesicles, with different luminal contents. LDFs were particularly homogeneous and contained mainly large (Fig. 2, B and C), single-membrane, electron-lucent EVs (about 75% of the total; Fig. 2, D and E) and a lesser number of large, single-membrane vesicles with an intraluminal vesicle-like body (11 to 15%). Some of these EVs bore spiked membrane microdomains (Fig. 2A, magnification a), usually at two opposite poles of the vesicle, which were occasionally found in other EV types (Fig. 2D, IV and VIII), although never on electron-dense EVs (Fig. 2A, magnifications b and c). IDFs were more varied, including a higher presence of smaller, granular, electron-dense EVs, whose frequency peaked in Fr5 (46% of the total) with decreased frequency in Fr6 and Fr7. The luminal granularity was, in most vesicles, confined to the center of the vesicles and absent from the area adjacent to the membrane (Fig. 2A, magnification b). Fr7 was particularly heterogeneous, while Fr8 was uniquely characterized by a high percentage of double membrane, highly electron-dense EVs [45% of the total; Fig. 2, A (magnification c), D, and E]. Infrequently (about 6%), these vesicles were surrounded by a single membrane.
Fig. 2. Cryogenic electron microscopy reveals enrichment of specific EV subtypes in each fraction.
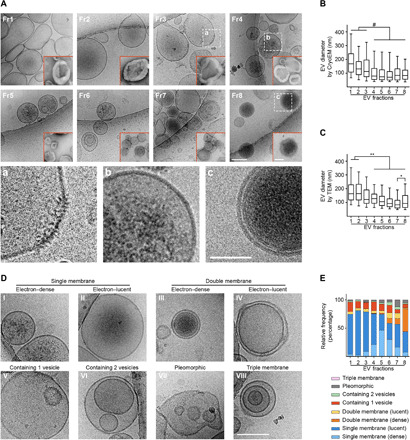
(A) Representative photomicrographs of brain EVs imaged by either cryoEM or TEM (insets with red borders). Unlike TEM, under cryoEM morphology was preserved. Dashed boxes: area magnified in a, b, and c, respectively. (B and C) Quantification of vesicle diameter in each EV fraction imaged under cryoEM or TEM. (D and E) Representative photomicrographs and relative percentage per fraction of morphologically different EV subtypes imaged under cryoEM. Statistical test in (B) and (C): Kruskal-Wallis with Dunn’s multiple comparisons test. Number of replicates in (B) and (C): 30 random photomicrographs per fraction from three independent isolations; (E): 30 random cryoEM photomicrographs per fraction from two independent isolations. Data shown in (B) and (C): box and whiskers plot (whiskers: 1st and 99th percentiles). Scale bars, 100 nm (A and D; main figure), 50 nm [(A) insets (a, b, and c magnifications and TEM photomicrographs)]. *P < 0.05, **P < 0.01, #P < 10−4.
EVs found in LDFs were larger by cryoEM and TEM than other EV subtypes (Fig. 2, B and C), consistent with NTA results. A small size difference between IDFs and Fr8 was also observed by NTA, consistent with a similar trend observed by cryoEM and TEM (Fig. 2, B and C). In summary, our data show that the OptiPrep density gradient identifies three brain EV subtypes based on total protein content, morphology, electron density, and size.
Protein analysis of EV components in LDFs, IDFs, and Fr8
Western blot analyses were performed to identify the type of vesicles contained in each EV fraction. Antibodies to annexin A1, annexin A2, and annexin V, markers of microvesicles (2), identified these plasma membrane–derived vesicles mainly in LDFs (Fig. 3, A and B). Caveolin-1, a protein found mainly in microvesicles (18), showed a similar distribution (Fig. 3E). Thus, brain microvesicles are concentrated in LDFs and likely correspond to the electron-lucent, single-membrane EVs observed by cryoEM.
Fig. 3. LDFs, IDFs, and Fr8 are enriched in microvesicles, exosomes, and mitovesicles, respectively.
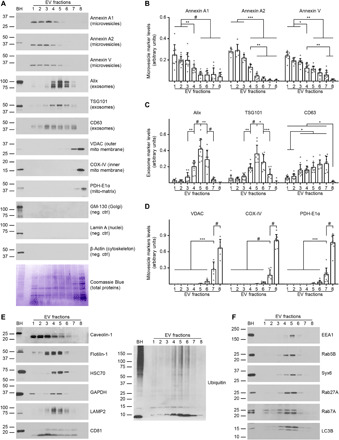
(A) Representative Western blot analysis and Coomassie Blue staining (loading control) of EVs isolated from the brain of wild-type mice and fractionated by an OptiPrep density gradient. The same volume of each fraction was loaded. (B) Densitometric quantifications of the microvesicle, (C) exosome, and (D) mitovesicle markers shown are normalized to the brain weight. (E) Representative Western blot analyses of EVs isolated from the brain of wild-type mice and fractionated by an OptiPrep density gradient. The same volume of each fraction was loaded. (F) Representative Western blot analyses of the endocytic proteins found in brain EVs. Brain homogenate (BH) was loaded as a reference. Statistical test in (B) to (D): one-way ANOVA with Tukey’s multiple comparisons test. Number of independent isolations: 10. Data shown in (B) to (D): Means ± SD. *P < 0.05, **P < 0.01, ***P < 10−3, #P < 10−4.
The ESCRT-associated proteins TSG101 and Alix, markers of ESCRT-dependent exosomes, were mostly found in IDFs with a peak in Fr5, paralleling the distribution of granular, single- membrane EVs (Fig. 3, A and C). Enriched within IDFs were also other exosomal markers such as the lysosome-associated membrane protein 2 (LAMP2), the heat shock cognate 71 kDa protein (HSC70), Flotilin-1, the glyceraldehyde-3-phosphate dehydrogenase (GAPDH), and poly-ubiquitylated proteins (2), as well as the endosomal proteins early endosome antigen 1 (EEA1), Rab5B, Rab7A, Rab27A, Syntaxin-6 (Syx6) and the autophagy protein Microtubule-associated proteins 1A/1B light chain 3B (LC3B) (Fig. 3, E and F), confirming the endosomal origin of these vesicles. The exosomal tetraspanins CD63 and CD81 (2), which are found on both ESCRT-dependent and ESCRT-independent exosomes (19), were detected in IDFs but showed a broader distribution across the fractions, enriched within Fr3 to Fr7 (Fig. 3, A, C, and E).
Microvesicle, exosome, and endocytic markers were undetectable in Fr8. This fraction, the only one that contains a high proportion of double membrane, electron-dense EVs (Fig. 2), was uniquely enriched in mitochondrial proteins found in the mitochondrial outer membrane (MOM), mitochondrial inner membrane (MIM), and mitochondrial matrix (mito-matrix), as assessed with antibodies directed against the voltage-dependent anion channel (VDAC), the cytochrome c oxidase subunit 4 (COX-IV), and the pyruvate dehydrogenase E1 component subunit alpha (PDH-E1α), respectively (Fig. 3, A and D).
Proteins found abundantly in the cytosol (β-actin) and the nucleus (Lamin A) and markers of unrelated intracellular compartments (the 130 kDa cis-Golgi matrix protein, GM-130), were not observed in any of the fractions, confirming a lack of intracellular contaminants.
These data demonstrate that microvesicles are enriched in LDFs, while exosomes are distributed from F3 to Fr7, with ESCRT-dependent exosomes concentrated in IDFs with a peak in Fr5. Fr8 is distinct from the other fractions and contains nonmicrovesicular, nonexosomal double-membrane EVs containing abundant mitochondrial markers. We have named these newly identified EVs mitovesicles.
Lipid analysis of LDF, IDF, and Fr8 brain EVs
Cellular membranes, including plasma membrane, endosomes, and mitochondria, differ in their lipid composition, and lipid content differences are reflected in EVs derived from these respective membranes, with some unique characteristics of each EV type. We characterized the EV content in each fraction of lipids that account for >95% of lipids found in EVs (20), including phosphatidylserine (PS), phosphatidylcholine (PC), phosphatidylethanolamine (PE), phosphatidic acid (PA), cardiolipin (CL), sterols [cholesterol and cholesteryl esters (CE)], acylglycerols [mono- (MAG), di- (DAG), and triacylglycerols (TAG)], lysobisphosphatidic acid (LBPA), and the sphingolipids ceramide, sphingomyelin (SM), galactocerebroside (GalCer), GD1a, and GT1b. The amount of lipids per microgam of protein was higher in LDFs compared with Fr8, with IDFs having an intermediate quantity, consistent with their differences in density (Fig. 4A). When analyzed by molar percentage, the amount of different classes of lipids in Fr1 to Fr5 (sterols: 39 to 43%, PC: 29 to 32%, sphingolipids: 8 to 11%, PS: 9 to 10%, PE: 4%, acylglycerols: 1%, and CL <1%) was similar to previous reports on small EVs isolated from cell culture media of several cell lines, with PA being the only exception, as previously it was found only in trace amounts (20). Fr8 displayed a different lipid composition (Fig. 4B). CL, a unique tetra-acyl lipid almost exclusively found in the MIM, peaked in Fr8 (Fig. 4C), which also displayed a higher amount of PC (48%) and PE (9%) and a lower content of cholesterol (18%) and SM (1.7%) compared with the other fractions (Fig. 4, D to F).
Fig. 4. Lipid analysis of brain EV fractions confirms the nature of LDF, IDF, and Fr8 EVs.
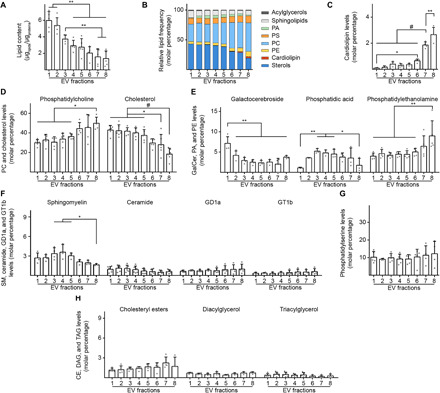
(A) Quantification of the total lipid content per fraction. The lipids analyzed were phosphatidylserine (PS), phosphatidylcholine (PC), phosphatidylethanolamine (PE), phosphatidic acid (PA), cardiolipin (CL), sterols [cholesterol and cholesteryl esters (CE)], acylglycerols [mono- (MAG), di- (DAG), and triacylglycerols (TAG)], and the sphingolipids ceramide, sphingomyelin (SM), galactocerebroside (GalCer), GD1a, and GT1b. The graph shows the total amount of these lipids normalized to the amount of proteins in each fraction as estimated by BCA. (B) Schematic lipid composition of the EVs found in each fraction expressed as a molar percentage. The lipids analyzed were described in (A) and grouped according to their families. (C) Cardiolipin molar percentage of brain EVs. (D) PC and cholesterol molar percentage of brain EVs. (E) GalCer, PA, and PE molar percentage of brain EVs. (F) Sphingomyelin, ceramide, GD1a, and GT1b content (expressed as a molar percentage over the total) of brain EVs. (G) Phosphatidylserine content (expressed as a molar percentage over the total) of brain EVs. No statistical differences were found. (H) Cholesteryl esters, DAG, and TAG content (expressed as a molar percentage over the total) of brain EVs. No statistical differences were found. Number of independent isolations: (A), 5; (B), 5; (C), 4; (D), 5; (E), 4 to 6; (F), 4; (G), 5; and (H), 4. Statistical test in (A) to (H): one-way ANOVA with Tukey’s multiple comparisons test. Data shown in (A) to (H): Means ± SD. *P < 0.05, **P < 0.01, #P < 10−4.
Fr8 was also poor in PA, which, in turn, was found at higher levels in IDFs, in agreement with the endosomal origin of IDF vesicles. PA was reported to be enriched in MVBs, closely associated with the ESCRT complex, where it facilitates membrane budding and fusion in cooperation with the Cbl-interacting protein of 85 kDa (CIN85) (21). Another lipid component showing a different pattern between fractions was GalCer, which peaked in Fr1. GalCer is found almost exclusively in the central nervous system, particularly in the plasma membrane of oligodendrocytes but also at low levels in neurons, where it colocalizes with Caveolin-1 (22). This is consistent with the presence of GalCer mainly in Fr1. Levels of the other lipids analyzed did not differ between fractions or were only marginally represented (Fig. 4, F to H). LBPA and MAG were below the level of detection. Exclusion of LBPA from the list of the brain EV lipids is consistent with the previous data on EVs isolated from cell media (20).
Based on a comprehensive analysis of morphology, protein markers, and lipid composition, we show that brain microvesicles and exosomes are enriched in distinct density fractions. Within these two groups, some heterogeneity of vesicle subtypes is apparent, for example, in the differences seen in ESCRT-dependent and overall exosome distribution. In addition, we have identified a type of brain EV isolated in the most-dense fraction: mitovesicles. Mitovesicles have morphological characteristics consistent with a mitochondrial origin (e.g., double membrane and electron density) and are uniquely enriched in mitochondrial proteins while lacking microvesicle and exosomal protein markers and containing a lipid profile that is consistent with a mitochondrial origin.
Mitovesicle proteomic and metabolic analyses
We undertook a qualitative liquid chromatography–mass spectrometry (LC-MS) analysis of Fr8 isolated from wild-type mouse brains to determine the full range of detectable mitochondrial proteins. We found 279 mitochondria-specific proteins of 673 proteins identified in Fr8, which were classified according to known function (Fig. 5A).
Fig. 5. Mitovesicles are metabolically active EVs with a specific subset of mitochondrial proteins.
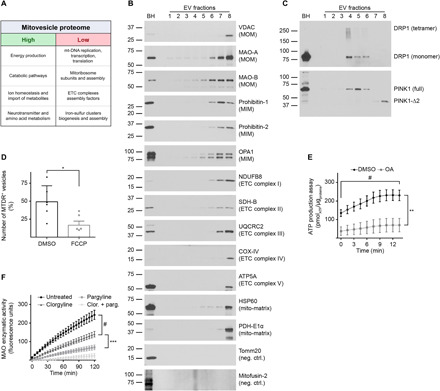
(A) EVs were isolated from brains of 12-month-old wild-type mice and fractionated through an OptiPrep step gradient. Fr8 EV lysates were analyzed by LC-MS, and the mitochondrial proteins found were clustered according to their known functions. Mitochondrial pathways that were highly underrepresented are indicated as “high” and “low,” respectively. (B) Representative Western blot analysis of brain EVs with antibodies to mitochondrial proteins. (C) Representative Western blot analyses of brain EVs using antibodies to DRP1 and PINK1. (D) Relative number of MTDR-positive (MTDR+) vesicles found in Fr8 EVs as quantified by NTA and expressed as percentage over the total number of EVs. As a control, samples were treated with the uncoupler FCCP that dissipates mitochondrial potential. (E) Analysis of ATP levels in brain Fr8 using a luciferase-based assay. As a control, samples were treated with oligomycin + antimycin-A (OA). Each experiment was run in triplicate. Chemiluminescence was quantified every 1.5 min. (F) MAO activity was measured in Fr8 brain EVs using a commercial MAO enzymatic activity assay. As controls, samples were treated with clorgyline (MAO-A inhibitor), pargyline (MAO-B inhibitor), or both. Fluorescence was read every 5 min. For additional data see table S2. Brain homogenate (BH) was loaded as a reference. Statistical test in (D): Student’s t test; (E) and (F): two-way ANOVA with Tukey’s multiple comparisons test. Number of independent isolations: (A), 3; (B) and (C), 10; (D), 6; (E) and (F), 4. Data shown in (D) to (F): Means ± SD. *P < 0.05, **P < 0.01, ***P < 10−3, and #P < 10−4.
The mitovesicle proteome was enriched in catabolic pathways (table S2). Electron transport chain (ETC) and the pyruvate dehydrogenase complex subunits were almost fully represented, as well as other energy production pathways, such as Krebs cycle, ketone body catabolism, fatty acid β-oxidation, neurotransmitter metabolism, and membrane transporters of metabolites directly linked to energy production. The mitochondrial intermembrane bridging (MIB) complex, which tethers the MIM to the MOM and promotes MIM curvature, was also detected.
In contrast, mitovesicles lacked several biosynthetic and import proteins. Proteins involved in the cytosol-to-mitochondria peptide import machinery (TOM/TIM23/PAM supercomplex), ETC assembly factors, iron-sulfur cluster formation and loading, ubiquinone biosynthesis, mitoribosome components, mitochondrial transfer RNA (mt-tRNA) metabolism, and mtDNA replication, transcription and translation were only scarcely found in mitovesicles, although relatively abundant in intracellular mitochondria. We also identified the presence of pro-fission proteins and a complete lack of pro-fusion factors.
Recent evidence indicates neuronal and astrocytic mitochondria contain different molecular profiles and have differential enrichment of specific proteins (23). Specifically, Sideroflexin-5 (Sfxn5) and the monoamine oxidase type B (MAO–B) are astrocyte specific, whereas the protein NipSnap homolog 1 (Nipsnap1), the OCIA domain-containing protein 2 (Ociad2), the mitochondial calcium uniporter (Mcu), and the monoamine oxidase type A (MAO-A) are predominantly expressed by neurons (23). Each of these proteins was identified in our Fr8 LC-MS analysis (table S2), suggesting that both astrocytes and neurons have the capability of secreting mitovesicles into the extracellular milieu.
LC-MS findings were validated by Western blot analysis of Fr8. The presence of the anion channel VDAC, neuroamine degrading enzymes MAO-A, MAO-B, PROHIBITIN 1 and 2, the optic atrophy protein 1 homolog (OPA1), ETC complex subunits NADH dehydrogenase beta subcomplex subunit 8 (NDUFB8), Succinate dehydrogenase subunit B (SDH-B), ubiquinol-cytochrome-c reductase complex core protein 2 (UQCRC2), COX-IV, and ATP synthase subunit alpha (ATP5A), chaperone heat shock protein 60 KDa (HSP60), and PDH-E1α were confirmed. Moreover, Tomm20, subunit of the TOM complex, and mitofusin-2, the main pro-fusion protein in the murine brain, were undetectable by both LC-MS and Western blot analysis (Fig. 5B). The shift toward pro-fission pathways in mitovesicles as indicated by LC-MS was also confirmed by the analysis of OPA1 isoforms. The indication of a fused network, revealed in brain homogenates by a stronger signal for OPA1-L (long isoforms) than for OPA1-S (short isoforms), was not observed in Fr8.
The dynamin-related protein 1 (DRP1), master regulator of mitochondrial dynamics, was not found in mitovesicles, confirming LC-MS data (Fig. 5C). However, both monomeric and tetrameric DRP1 were present in exosome-enriched fractions, as reported for the full-length isoform of the PTEN-induced kinase 1 PINK1 (fl-PINK1), a protein that can be found both in the cytosol and on the MOM (24). When imported into the mito-matrix, PINK1 is cleaved into a ~53-kDa fragment (PINK1-Δ1) or a ~44-kDa peptide (PINK1-Δ2), the latter being a mitophagy-independent isoform (25). Mitovesicles lacked fl-PINK1 and PINK1-Δ1 but retained PINK1-Δ2.
LC-MS analysis of Fr8 showed the presence of the core machinery to produce adenosine 5′-triphosphate (ATP) in mitovesicles (table S2). Accordingly, we posited that mitovesicles could retain a functional intermembrane H+ potential and synthetize ATP. To test this hypothesis, we isolated EVs from wild-type mouse hemibrains and incubated Fr8 with pyruvate, malate, and adenosine 5′-diphosphate (ADP) to boost oxidative phosphorylation and with MitoTracker Deep Red (MTDR), a dye that is incorporated more efficiently into polarized and actively respiring mitochondria (26). We measured by NTA the number of vesicles positive (i.e., above the detection limit) for MTDR. As a negative control, samples were treated with carbonyl cyanide 4-(trifluoromethoxy)phenylhydrazone (FCCP), an uncoupler that dissipates mitochondrial potential. Approximately 50% of identified vesicles in Fr8 were stained with MTDR, and FCCP treatment decreased the number of positive vesicles to ~17% (Fig. 5D), providing evidence for the presence of an active potential. To determine whether mitovesicles can synthetize ATP, we incubated EVs at 37°C and measured ATP using a commercially available ATP assay kit, resulting in a twofold increase in ATP. As negative controls, treatment of the mitovesicles with oligomycin + antimycin-A (OA), specific blockers of the ETC complexes V and III, respectively, did not affect ATP production (Fig. 5E), demonstrating that mitovesicles are metabolically active and able to autonomously produce ATP.
MAO enzymatic activity of mitovesicles was also analyzed because MAO-A and MAO-B were detected in Fr8. As negative controls, irreversible inhibitors of MAO-A (clorgyline), MAO-B (pargyline), or a combination of both were used, demonstrating that MAO activity is maintained in mitovesicles, with MAO-B inhibition having a stronger effect than MAO-A inhibition (Fig. 5F). Concomitant presence of both proteins and activities suggests that both neurons and astrocytes secrete enzymatically active mitovesicles into the murine brain extracellular space.
Proteomic analysis of EVs isolated from brains of a DS mouse model
DS brains are characterized by extensive mitochondrial abnormalities originating in fetal stages and persisting throughout life span of individuals with DS (27). Accordingly, we hypothesized that mitovesicles are aberrant in DS and DS models. To test our hypothesis, we isolated EVs from brains of the trisomic mouse model Ts[Rb(12.1716)]2Cje (hereafter Ts2) (28) and diploid (2N) littermates, combining all EV fractions for each genotype. Ten micrograms of total EVs per mouse was analyzed by label-free LC-MS–based proteomics (tables S3 to S7). Approximately 1600 proteins were identified in total, as detected in at least one mouse per genotype. A total of 16 and 81 proteins were exclusively found in 2N and Ts2 samples, respectively (tables S3 and S5). The quantitative analysis of the proteome revealed that 11 proteins were less abundant in Ts2 EVs compared with 2N (P < 0.05; fig. S1A and table S4). Among them was cystatin C (P = 0.023), consistent with our previous observations that a moderate higher cystatin C expression can mitigate Ts2 phenotypes in vivo (29). By contrast, 149 proteins were up-regulated in Ts2 EVs compared with 2N (P < 0.05; fig. S1A and table S6). As expected, several of these are due to higher expression of genes linked to the segmental trisomy of Ts2 mice. For example, Sod1, which is triplicated in Ts2 mice (28), was higher in Ts2 EVs compared with 2N levels (P = 0.001). The majority of identified proteins not from triplicated regions in Ts2 EVs are mitochondrial (fig. S1B). A total of 187 mitochondria-specific proteins were identified in the EVs (fig. S1C). Among them, a large cohort was greater in Ts2 compared with 2N EVs (73 proteins; P < 0.05), and 81 were exclusively found in Ts2 EVs (tables S5 and S6). Among the 50 most differentially present proteins by P value, 38 (76%) were mitochondrial. In addition, the four proteins with the highest fold change were mitochondrial (fig. S1D). Gene set enrichment analysis revealed that several mitochondrial pathways were up-regulated in Ts2 EVs (table S7). We conclude that the mitochondrial proteome found in brain EVs is enriched in Ts2 mice compared with 2N littermates and represents a prominent pathway altered in vivo within DS brain EVs.
Brain mitovesicle levels in DS
To determine whether the changes in EVs isolated from the brains of Ts2 mice as compared with 2N littermates are due to a modulation of mitovesicle secretion or cargo loading, we further characterized Ts2 and 2N mitovesicles. Brain-derived Fr8 showed a higher protein content (Fig. 6A) and higher mitovesicle numbers in Ts2 mice compared with 2N littermates as estimated by NTA (Fig. 6B). In agreement with analysis of total protein content, NTA, and LC-MS data, levels of mitochondrial proteins were also higher in Ts2 EVs compared with 2N littermates (Fig. 6, C and D) but did not change in the respective brain homogenates (Fig. 6, E to G), suggesting an increase in mitovesicle levels in the brain parenchyma. As we have previously reported (1, 12), the number of exosomes is higher in Ts2 brain compared with 2N littermates, consistent with the elevation of exosome markers and the increase in protein content and vesicle numbers in IDFs. Although annexin A2 levels were higher in the Ts2 LDFs (Fig. 6, C and D), microvesicle numbers were unaffected (Fig. 6, A and B). Consequently, annexin A2 alterations were likely related to an altered cargo loading of this protein in the Ts2 mice rather than an alteration of microvesicle secretion, consistent with the up-regulation of annexin A2 in Ts2 brain homogenates (Fig. 6, E and F).Analysis of EVs obtained from human postmortem brains of individuals with DS and age-matched cognitively normal controls devoid of neuropathology (table S8) confirmed our findings in the murine model (Fig. 6, H and I). These data demonstrate a higher number of mitovesicles in DS brain parenchyma, both in murine and human postmortem brains.
Fig. 6. Brain mitovesicle levels are altered by neuropathology.
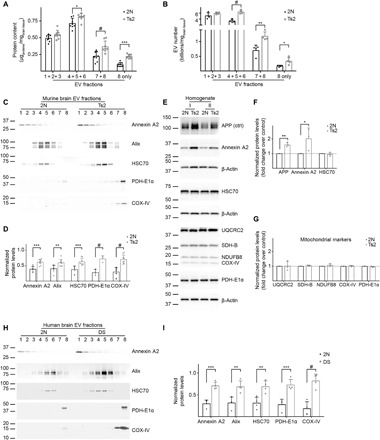
(A) Protein levels and (B) number of EVs normalized to the brain weight from 12-month-old 2N and Ts2 mice fractionated by an OptiPrep gradient. LDFs, IDFs, and Fr7/Fr8 were combined. Fr8 alone is also shown. (C) Representative Western blot analyses and (D) densitometric quantification of microvesicle, exosome, and mitovesicle markers in murine brain EVs. Quantifications were performed combining the signal from all the fractions normalized to the brain weight. (E to G) Representative Western blots and densitometric analyses of microvesicle, exosome, and mitovesicle markers in brain homogenates of 12-month-old Ts2 and 2N mice. β-actin was used as a loading control. APP, triplicated in Ts2 mice, was used as a positive control. Two experiments shown in the blot (I and II). Graphs are plotted as a fold change over the 2N controls. (H) Representative Western blot analyses and (I) densitometric quantification of microvesicle, exosome, and mitovesicle markers in human postmortem brain EVs. Quantifications were performed combining the signal from all the fractions normalized to the brain weight. For additional specifications. see table S8. Statistical test in (A) and (B): two-way ANOVA with Tukey’s multiple comparisons test; (D), (F), (G), and (I): Student’s t test. Number of independent isolations: (A) and (D), 8; (B) and (I), 4; (F) and (G), 3. Data shown in (A), (B), (D), (F), (G), and (I): Means ± SD. *P < 0.05, **P < 0.01, ***P < 10−3, #P < 10−4.
Brain mitovesicle cargo in DS
We sought to determine whether mitochondrial proteins, mtDNA, and mtRNA are altered in their abundance per mitovesicle by Ts2 trisomy. Equal numbers of mitovesicles as determined by NTA were analyzed, revealing that UQCRC2, VDAC, and SDH-B levels were lower in Ts2 mitovesicles compared with 2N littermates, while COX-IV, NDUFB8, and HSP60 were unchanged (Fig. 7, A and B). Quantification of mtDNA in Fr8 by quantitative polymerase chain reaction (qPCR) demonstrated that Ts2 mitovesicles had more mtDNA per vesicle than 2N littermate mitovesicles (Fig. 7C). No difference in the bulk amount of RNA between the genotypes was observed (Fig. 7D). By qPCR, the most abundant mitochondria-encoded RNA species in mitovesicles was ribosomal RNA, which did not differ between genotypes (Fig. 7E). Thus, the mitochondria-encoded 12S RNA (mt-RNR1) was used as a housekeeping gene in further analysis of mitochondria-encoded mRNA levels. Investigation of 5 of the 13 genes found in mtDNA (ND1, ND4L, Cyb, COX-II, and ATP8) revealed they were all significantly down-regulated in Ts2 mitovesicles compared with 2N littermates (Fig. 7F).
Fig. 7. Brain mitovesicle cargo is altered in DS.
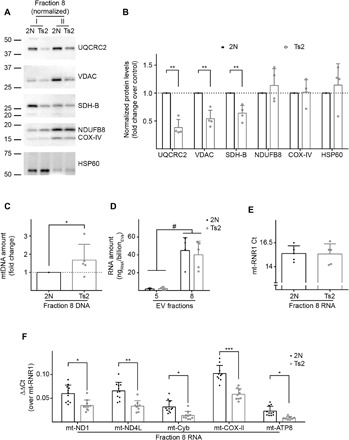
(A and B) Western blot analyses of EVs isolated from the brain of Ts2 and 2N control mice and fractionated through an OptiPrep gradient. Equal numbers of Fr8 EVs as estimated by NTA were loaded for each isolation and tested for mitochondrial markers. Two isolations are shown in the blot (I and II), the densitometric analysis is plotted as a fold change over the 2N controls. (C) Quantification of mtDNA (normalized to the number of vesicles and plotted as a fold change of trisomic versus control mice) by qPCR in murine brain Fr8 EVs. (D) Quantification of the total RNA (normalized to the number of vesicles) extracted from murine brain Fr5 and Fr8 EVs. (E) qPCR cycles of mt-RNR1 for equal amounts of Fr8 cDNA in Ts2 and 2N brain-derived EVs. (F) qPCR analysis of five mitochondria-encoded genes in Fr8 EV complementary DNAs, normalized to mt-RNR1. Statistical test in (D): two-way ANOVA with Tukey’s multiple comparisons test; (B), (C), and (E): Student’s t test; and (F): mixed effects models. Number of independent isolations: (B), 4; (C) to (F), 5. Data shown in (B) to (F): Means ± SD. *P < 0.05, **P < 0.01, ***P < 10−3, #P < 10−4.
These findings show that the protein and nucleic acid composition of mitovesicles is affected by DS genotype, with more mtDNA and lower mRNA levels per vesicle.
To directly link mitovesicle biology to mitochondrial integrity, we used an in vitro model of mitochondrial damage and analyzed the EVs released into the cell media. Human primary fibroblasts were treated with antimycin-A (anti-A), a specific inhibitor of the ETC complex III that causes mitochondrial reactive oxygen species (ROS) accumulation without inducing mitophagy (30). Mitochondrial stress following anti-A treatment was confirmed in our experimental conditions by Western blot analysis of cell lysates. OPA1 cleavage, a marker of mitochondrial fragmentation, was increased, and the levels of COX-IV decreased relative to controls, indicating oxidative phosphorylation impairment (fig. S2, A to C). Mito-matrix proteins PDH-E1α and HSP60, preferentially degraded through mitophagy (31), were unchanged (fig. S2, A to C). Western blot analysis of EVs isolated from the cell media show that the mitovesicle markers COX-IV, PDH-E1α, and HSP60 were up-regulated in EVs secreted by anti-A–treated cells, while exosome and microvesicle proteins were unaffected (fig. S2, D and E). These data indicate that mitochondrial damage directly perturbs mitovesicle biology, either by stimulating the release of these vesicles or by up-regulating mitovesicle cargo loading in single EV. However, COX-IV and HSP60 are among the proteins that were unchanged in their content per EV in Ts2 brain mitovesicles (Fig. 7, A and B), suggesting cargo loading for these proteins is not affected by mitochondrial dysfunction in vivo. Moreover, PDH-E1α and COX-IV are among the mitochondrial proteins that successfully predicted a higher release of mitovesicles in Ts2 brains (Fig. 6C, fig. S1D, and table S6). The preponderance of evidence supports the use of COX-IV, PDH-E1α, and HSP60 as bona fide markers for mitovesicle levels in total isolated EVs and reinforces the idea that mitochondrial damage directly stimulates mitovesicle secretion in a mitophagy-independent fashion, although we cannot exclude the less likely hypothesis of a coordinated perturbation in cargo loading for all these markers at the same time.
Overall, our data show that mitovesicle levels and composition reflect mitochondrial alterations within the cell of origin and may serve as a biomarker to assess brain mitochondrial dysfunctions in neurological disorders.
DISCUSSION
We have established and validated a new experimental approach to isolate and separate EV subpopulations in vivo from the brain extracellular matrix. We demonstrate that brain EV fractions diverge in morphometrical properties, morphology, protein, and lipid composition, as well as in in microvesicle and exosome markers. Other density gradients, mainly sucrose based, have been applied to purify EVs from the brain but have not been able to generate an unequivocal separation between EV subtypes (10). This report is novel in that it implements a high-resolution gradient and shows that EV subpopulations can be fractionated from solid tissues in vivo. Our data confirm that EV subtypes are not efficiently separated by a differential centrifugation approach alone, as they largely overlap in terms of dimension. However, EV subtypes can be reproducibly and rigorously separated by density, as previously demonstrated for EVs isolated from cell media, within density ranges that are highly consistent with what was used in vitro to achieve this goal (2).
Our high-resolution approach led to the identification of a previously unknown third pool of dense, metabolically competent EVs of mitochondrial origin, which we term mitovesicles, and reinforces the importance of high-resolution density step gradients in EV preparations. A large body of evidence demonstrates the presence of mitochondrial material in 100,000g EV pellets (3–7). These vesicles were interpreted as exosomes bearing mitochondrial proteins, lipids, and mtDNA. Although we cannot rule out that some mitochondrial constituents can be found in exosomes (as the case for full-length PINK1; Fig. 5C), this interpretation may be due to the fact that mitovesicles and exosomes copellet at 100,000g and previous studies that used low-resolution gradients failed to separate these two populations. Other independent studies have reported the existence of particularly large (>1 μm) vesicles containing intact mitochondria (3, 8). A limitation of our study is that we apply a 0.2-μm filtration step to further purify EVs from intracellular contaminants, and with this step, we lose large EVs that may contain entire mitochondria (or even mitochondrial clusters). In our experience, the 0.2-μm filtration step is unavoidable for clean, debris-free preparations from solid tissues, and therefore, the question remains open on whether these structures exist in vivo and what their specific contribution is to brain homeostasis and neuronal pathological processes when compared with mitovesicles.
Mitovesicles differ from intracellular mitochondria. They are up to 10-fold smaller, have a narrower intermembrane space [~6 nm, compared to 20 nm of native mitochondria (32)], and lack several mitochondrial structures, including cristae, mitochondrial ribosomes, and proteins constitutively found in mitochondria such as Tomm20 (23). On the other hand, these same features are shared with a specific subset of intracellular vesicles known as mitochondria-derived vesicles (MDVs). MDVs bud from the mitochondrial surface, may include the MOM, MIM, and mito-matrix, and are targeted to MVBs (33). This suggests MDVs might be released into the extracellular space as mitovesicles upon fusion of MVBs with the plasma membrane (34), although the mechanisms responsible for mitovesicle formation from the mitochondria and their secretion into the extracellular space need to be established. Nevertheless, MDVs are heterogeneous, and some characteristics of these intracellular vesicles are not shared with mitovesicles. Several subtypes of MDVs have been described in previous studies, including mitochondria-anchored protein ligase (MAPL)–containing MDVs, as well as Tomm20 or vacuolar protein sorting-associated protein 35 (Vps35)–positive MDVs (33). These proteins are not found in mitovesicle preparations by LC-MS, and Tomm20 negativity was validated by Western blot analysis. This would point to a model by which several subsets of MDVs with specific functions exist. One of these subsets would be secreted into the extracellular space (PDH-E1α+ Tomm20− MDVs), possibly exerting specific extracellular functions other than mere carriers of mitochondrial debris outside the cell. Supportive of this contention, our data demonstrate that mitovesicles are not encapsulated by an external membrane, enabling extracellularly the enzymatic activity of MOM-anchored proteins such as MAO-A and MAO-B and the ability to synthetize ATP at direct contact with the relative substrates (Fig. 5). This evidence would highlight a differential function for mitovesicles relative to an entire mitochondria encapsulated into microvesicles that are likely shielded from the extracellular space by the presence of the microvesicular membrane and that have been described to serve mainly as energy supply for receiving cells (8).
An unanswered question remains on the role that mitovesicles have during the neuropathological process, such as evidenced in DS. We show that the number of brain mitovesicles is higher (Fig. 6) and that their composition is altered (Fig. 7) in the brains of Ts2 mice and individuals with DS as compared with age-matched diploid controls. DS is characterized by mitochondrial dysfunction and ROS accumulation, as was previously demonstrated either in cell lines (17) or in DS brains (27), as well as in the brains of related murine models, independently of the amyloid β precursor protein (APP) and the superoxide dismutase isoform 1 (SOD1) triplication (35). In addition, it was shown that DS cells are mitophagy deficient (17), suggesting that brain cells potentially accumulate toxic mitochondria over time in the absence of a prosurvival mitochondria quality control mechanism, such as mitophagy, which is able to digest damaged mitochondria (36, 37). Mitovesicle secretion into the brain extracellular space of individuals with mitochondrial dysfunction may contribute to the disease or act to eliminate detrimental mitochondrial material from the cell and mitigate oxidative stress in a homeostatic feedback loop, although additional support is necessary to validate this notion.
In addition, mitovesicle release may be a mechanism to transfer bioenergetically competent packages from one cell to the other. A similar cooperative model has been proposed to explain the transfer of mitochondria encapsulated into microvesicles from astrocytes to neurons in vitro, a phenomenon up-regulated in the presence of oxidative stress and that may be considered a neuroprotective mechanism (8). In DS, the up-regulation of mitovesicle secretion follows this pattern; however, the alteration in the composition (and particularly, the down-regulation in EVs of proteins involved in ATP production such as UQCRC2 and SDH-B; Fig. 7, A and B) would be responsible of the failure in restoring normal ATP levels.
Mitovesicle-related MAO activity is preserved. Accordingly, it is reasonable to conclude that mitovesicles modulate levels of neurotransmitters in the extracellular space, potentially linking mitochondrial damage and neurotransmission in a different system. The increase in mitovesicle levels in DS brain may be related to an elevation of extracellular MAO activity, which likely leads to a decline in the levels of monoamines. Notably, DS brains are characterized by a specific decrease in monoamines such as serotonin and dopamine, but not glutamate or glycine (38). Further studies are needed to elucidate whether mitovesicles are a driver of this decline.
Last, we hypothesize a role for mitovesicles in the neuroinflammation process. Mitovesicles may activate microglia as proinflammatory agents given the presence of mitochondrial material and DAMPs in single vesicles. Support for this comes from Parkin–knockout (KO) mice, in which mitochondrial stress causes an increase in mtDNA levels in plasma. This drives a sustained up-regulation of neuroinflammation (9). How non-necrotic cells extrude DAMPs and mtDNA into the extracellular space has yet to be demonstrated, although mitovesicles would appear to be appropriate for this signaling activity. Although not demonstrated in the present report, the presence of mitovesicles in the blood, similarly to other EVs, is likely. A strong support for this assumption comes from public repositories of whole proteomes detected in human plasma by LC-MS, such as the Human Protein Atlas (39–41), available at https://proteinatlas.org/humanproteome/blood/proteins+detected+in+ms. These high-throughput data not only show that several mitochondrial proteins are present in human plasma but also that these mitochondrial datasets are highly selective and largely overlap with the mitovesicle proteome (table S2). For instance, mitochondrial proteins reported in human plasma comprise ETC complex subunits, Krebs cycle enzyme, HSP60, and MAO-B. However, no mitochondrial ribosomal proteins, mitofusins, TOM subunits, Coenzyme Q10 biosynthesis protein (COQ) members, or mtDNA replication/transcription regulators [e.g., polymerase gamma (POLG) or the mitochondrial transcription factor A (TFAM)] are reported in the list. Of 14 members of the Bcl2 family, the only reported protein found in our LC-MS data was Bax, and this overlaps with the human plasma dataset. Thus, the analysis of mitovesicles isolated from biofluids can represent an innovative, yet unexplored biomarker approach to test mitochondrial status in vivo in a noninvasive way, a procedure that we are currently exploring in neurological diseases such as AD.
In summary, similar to more extensively defined EV populations such as exosomes (16), mitovesicles may reflect and modulate pathological changes in the intracellular organelle from which they are derived as well as play important roles in cell-to-cell communication or in the remodeling of the extracellular environment. A rich appreciation has developed in the last decade of the multiple functions and clinical relevance of other EV subtypes. The mitovesicles defined and described herein are likely to contribute in similar ways to our understanding of the biology of mitochondrial regulation and dysfunction.
MATERIALS AND METHODS
Murine models
A breeding colony of B6EiC3Sn-Rb(12.Ts171665Dn)2Cje/CjeDnJ (abbreviated in Ts2, RRID:IMSR_JAX:004850) mice (28) was maintained on a B6EiC3SnF1/J (abbreviated in 2N, RRID:IMSR_JAX:001875) background. Only Rd.−/− mice (not carrying the retinal degeneration mutation found in some Ts2 colonies) have been used for our studies. Trisomic and 2N mice were examined at 12 months of age in all our experiments, because this is the age at which Ts2 mice start to show an EV-related phenotype (1, 12). C57BL/6J mice (indicated as wild-type mice in the text, RRID:IMSR_JAX:000664) were assessed at 12 months of age for the characterization of the protocol of isolation of brain EVs. Both females and males were used in all experiments. All animal procedures were performed following the National Institutes of Health guidelines with approval from the Institutional Animal Care and Use Committee at the Nathan S. Kline Institute for Psychiatric Research.
Cell lines
Foreskin male untransformed primary fibroblasts from a 2-year-old apparently healthy subject (abbreviated in fibroblasts, Coriell, catalog no. AG07095, RRID:CVCL_0N66) were grown at 37°C in Dulbecco’s modified Eagle’s medium (DMEM), supplemented with 10% (v/v) fetal bovine serum, penicillin (100 U/ml) + streptomycin (100 μg/ml), and 2 mM GlutaMAX (all reagents from Thermo Fisher Scientific, Waltham, MA, USA) at 5% CO2 in a humidified incubator. The EV-depleted cell medium (conditioned cell medium) was obtained by overnight ultracentrifugation of the supplemented media at 100,000g (36,000 rpm) at 4°C in a fixed-angled rotor (k-factor: 207.5, 45Ti rotor type, Beckman Coulter, Brea, CA, USA). To avoid cellular senescence, experiments were performed using cells at passage 14 or lower (starting from frozen stocks at passage 8). Mycoplasma contamination was routinely checked under a microscope through 4′,6-diamidino-2-phenylindole (DAPI) staining of an aliquot of the cells in use by ensuring the absence of intracellular, extranuclear DAPI signal.
Human subject details
Postmortem frozen brain tissues (Brodmann area 9) from human DS and control subjects (table S8) were provided by J. Wegiel, head of the Brain and Tissue Bank Department, Department of Developmental Neurobiology, New York State Institute for Basic Research in Developmental Disabilities, Staten Island, NY.
Isolation of EVs from brain parenchyma
Murine EVs were isolated from the right hemiforebrains (hereafter “hemibrain”) as previously described (10, 11). Briefly, hemibrains were minced and incubated with papain (20 U/ml) (Worthington, Lakewood, NJ, USA) in Hibernate A (BrainBits, Springfield, IL, USA) for 15 min at 37°C. The enzymatic digestion was stopped by the addition of ice-cold leupeptin (5 μg/ml), antipain dihydrochloride (5 μg/ml), pepstatin A (5 μg/ml), 1 mM phenylmethanesulfonyl fluoride, 1 μM E64 (all reagents from Sigma-Aldrich, St. Louis, MO, USA) in Hibernate A. The solution was gently disassociated by pipetting and centrifuged at 300g for 10 min at 4°C to eliminate undigested tissue and intact cells. The supernatant was subsequently filtered twice, first through a 40-μm cell strainer (Fisher Scientific, Pittsburgh, PA, USA) and then through a 0.2-μm surfactant-free cellulose acetate syringe filter (Corning Incorporated, Corning, NY, USA). The cleared mixture was centrifuged at 2000g for 10 min at 4°C to discard large debris and apoptotic bodies, and the supernatant was centrifuged at 10,000g for 30 min at 4°C to discard smaller debris. The supernatant was then ultracentrifuged at 100,000g for 70 min at 4°C. The pellet contained the crude EV preparation. The pellet was washed once in phosphate-buffered saline (PBS) (pH 7.4) (Corning Incorporated), recentrifuged at 100,000g for 70 min at 4°C, and resuspended in a 40% (v/v) OptiPrep (iodixanol) solution, containing 10 mM tris-HCl (pH7.4), 0.25 M sucrose, and 40% iodixanol (all reagents from Sigma-Aldrich). An OptiPrep density step gradient was set up by carefully layering on the top of the 40% OptiPrep-equilibrated EVs a decreasing scale of OptiPrep solutions (20, 15, 13, 11, 9, 7, and 5%). The column was centrifuged overnight at 4°C in a swinging bucket rotor at 200,000g (k-factor: 137.3, SW40Ti rotor, Beckman Coulter). Afterward, 1.5-ml fractions (eight fractions in total), corresponding to the different interphases, were collected, washed in PBS, and centrifuged at 100,000g for 70 min at 4°C in a fixed-angle rotor (k-factor: 61.2, MLA80 rotor, Beckman Coulter). Pellets were resuspended in PBS, buffer B (225 mM sucrose, 44 mM KH2PO4, 12.5 mM magnesium acetate, and 6 mM EDTA, all reagents from Sigma-Aldrich) or lysed in radioimmunoprecipitation assay (RIPA) buffer with protease inhibitors for further analysis, as described below.
EV isolation from cell media
Fibroblasts at 80% confluence in 150-mm plates (Corning Incorporated) were treated for 24 hours with 4 μM anti-A (Sigma-Aldrich) in DMSO (Sigma-Aldrich) or with DMSO only in EV-depleted cell medium. The medium was collected and centrifuged at 300g for 10 min at 4°C. The supernatant was serially centrifuged at 2000g for 10 min at 4°C to discard large debris and at 10,000g for 30 min at 4°C for large debris. The supernatant was then ultracentrifuged at 100,000g (36,000 rpm) for 70 min at 4°C in a fixed-angled rotor (45Ti rotor type, Beckman Coulter), and the pellet was washed in 1 ml of PBS and centrifuged again at 100,000g (50,000 rpm) for 70 min at 4°C in a fixed-angled rotor (k-factor: 79.9, TLA-55 rotor type, Beckman Coulter). The washed EV pellet was then resuspended in RIPA buffer and prepared for Western blot analysis.
Nanotrack analysis
Fresh, unfrozen brain EV OptiPrep fractions from wild-type, 2N, and Ts2 mice were resuspended in PBS and analyzed by NTA (ZetaView model PMX-120 basic or PMX-220 twin for fluorescence analysis, Particle Metrix, Meerbusch, Germany) to estimate the amount and the dimension of the vesicles. Before the acquisition, EV fractions were further diluted in degassed, particle-free, 0.2-μm-filtered PBS to reach the appropriate concentrations of particles to fall into the ideal reading range of the machine (around 100 particles per field of view). Empirically, we found that the most adequate concentrations were 1:50,000 to 1:25,000 for Fr1 to Fr4, 1:20,000 to 1:10,000 for Fr5, 1:10,000 for Fr6 and Fr7, and 1:3,000 to 1:1,000 for Fr8. Readings were performed as per the manufacturer’s instruction. The parameters set were as follows: sensitivity, 85; minimum size, 10; maximum size, 1000; shutter, 100; frame rate, 30; trace length, 15; bin size, 5 nm; and positions per single, reading 11. The temperature of the machine never exceeded room temperature during acquisitions.
For the MTDR analysis, Fr8 EVs were resuspended in buffer B with 1 mM ADP, 1 mM pyruvate, 1 mM malate (Sigma-Aldrich), and 500 nM MTDR (Thermo Fisher Scientific) and incubated at 37°C for 15 min with or without FCCP 20 μM (Sigma-Aldrich). Samples were eventually read using the ZetaView PMX-220 twin at a concentration of 1:10,000 to ensure proper dilution of the dye and minimization of the background.
BCA protein assay
Pierce BCA (bicinchoninic acid) Protein Assay Kit (Thermo Fisher Scientific) was used as per the manufacturer’s instructions with minimal adjustments. Two microliters of EVs diluted in PBS (in triplicate) were incubated at 65°C for 30 min in a polypropylene microplate (Corning Incorporated) with the BCA Reagent Solution (50:1 reagent A to reagent B, provided in the kit). A bovine serum albumin (BSA; provided in the kit) standard curve was loaded in parallel for each plate analyzed. The colorimetric reaction was quantified as the absorbance at 562 nm using the VersaMax microplate reader (Molecular Devices, San Jose, CA, USA).
ATP assay
Fr8 brain EVs were resuspended in buffer B, and protein content was quantified by the BCA assay as described above. An ATP standard curve and 5 μg of these preparations were plated in triplicate in white, opaque, MicroFluor1 plates (Thermo Fisher Scientific) and incubated in the presence or absence of 2 μM oligomycin + 4 μM antimycin-A (OA) in an ATP assay MasterMix [1 mM ADP, 1 mM pyruvate, 1 mM malate (reagents from Sigma-Aldrich), 1 mM DTT, 100 μM EDTA, 5 mM MgSO4, 25 mM tricine buffer (pH 7.8), 0.5 mM d-luciferin, and recombinant firefly luciferase (1.25 μg/ml) (reagents provided in the ATP Determination Kit, catalog no. A22066, from Invitrogen, Thermo Fisher Scientific)]. Chemiluminescence was read in a SpectraMax microplate reader (Molecular Devices) at 30°C and after reaching 37°C every 1.5 min.
Monoamine oxidase assay
The MAO enzymatic assay was performed using the MAO Assay Kit (catalog no. MAK136) from Sigma-Aldrich following the manufacturer’s instructions. Briefly, 2 μg of Fr8 brain EVs were incubated in the presence or absence of 1 μM clorgyline (irreversible MAO-A inhibitor), 1 μM pargyline (irreversible MAO-B inhibitor), or both together (provided with the kit) in triplicate in a fluorescence-optimized, dark microplate (Corning Incorporated) for 10 min at room temperature before the start of the assay. A MAO assay mix (provided with the kit) was then added to the samples, and the plate was read in a SpectraMax microplate reader (Molecular Device) at room temperature with excitation/emission wavelengths of 530/585 nm, respectively. Fluorescence was read every 5 min for 2 hours.
Negative stain of EVs and TEM
Brain EVs were fixed in a paraformaldehyde (PFA) solution [containing 2% (w/v) PFA in a 100 mM sodium cacodylate buffer, reagents from Electron Microscopy Sciences, Hatfield, PA, USA], while Formvar-covered carbon-coated copper, 200 mesh grids (Ted Pella Inc., Redding, CA, USA) were glow discharged in a PELCO easiGlow (Ted Pella Inc.). Three microliters of fixed EVs were added to the grid, held by self-clamping tweezers, and left for 20 s. The solution was then wicked off with a Whatman filter paper and negatively stained in 1% (w/v) uranyl acetate (Electron Microscopy Sciences) before being immediately wicked off using filter paper. This process was repeated three times before a final incubation of 5 min. The excess solution was wicked off, and the grid was allowed to air dry. Imaging took place on a Talos L120C transmission electron microscope operating at 120 kV (Thermo Fisher Scientific).
Cryo–electron microscopy
R 2/2 copper 200 mesh Quantifoil grids (Ted Pella Inc.) were glow discharged in a PELCO easiGlow (Ted Pella Inc.). Three microliters of a 1:10 (v/v) dilution of EVs in PBS were applied to the grid before being blotted with filter paper and then rapidly frozen in liquid ethane using a Vitrobot Mark IV (Thermo Fisher Scientific). The grids were then transferred to a 914 high tilt model cryo-tomography holder (Gatan, AMETEK, Berwyn, PA, USA) and imaged using low-dose conditions on a Talos L120C transmission electron microscope operating at 120 kV (Thermo Fisher Scientific).
Western blot analysis
Samples examined by Western blot were supplemented with a 6× Laemmli sample buffer [375 mM tris-HCl (pH 7.4), 9% SDS, 50% glycerol, 9% β-mercaptoethanol (β-ME), 0.03% bromophenol blue, all reagents from Sigma-Aldrich], boiled for 5 min at 95°C, and loaded on a 4 to 20% gradient precast polyacrylamide gel (Bio-Rad, Hercules, CA, USA) to run at 120 V for 2 hours. With the exception of the blots in Fig. 7 (A and B), equal volumes of brain EVs were loaded. For DRP1 immunoblots, gels were run in nonreducing conditions (no β-ME was added to the Laemmli buffer) to detect DRP1 tetramers. Gels were then either stained for 30 min at room temperature in a Coomassie solution [0.25% (w/v) Coomassie Brilliant Blue G-250 (Sigma-Aldrich) in 40% (v/v) methanol and 10% acetic acid (v/v) (Fisher Scientific)] or transferred overnight at 100 mA onto polyvinylidene difluoride membranes (Immobilon, EMD Millipore, Billerica, MA). When needed, as indicated in the figures, membranes were stained using SYPRO Ruby Protein Blot Stain (Lonza, Basilea, Switzerland) following the manufacturer’s instructions before the blocking step. Membranes were subsequently blocked for 1 hour in 5% (w/v) blotting-grade nonfat dry milk (Bio-Rad) or in 5% (w/v) BSA (Sigma-Aldrich), depending on the conditions suggested by the manufacturers for the primary antibody applied. A comprehensive list of the primary antibodies used is provided in table S9. The monoclonal antibody against APP (clone C1/6.1) (42) was a gift from P. M. Mathews. Horseradish peroxidase–conjugated secondary antibodies were purchased from Jackson ImmunoResearch (West Grove, PA, USA) and used as per the manufacturer’s instructions. Protein bands were visualized either on reflection autoradiography films or with the iBright FL1500 imaging system (Thermo Fisher Scientific) after incubation of the blots for 5 min with enhanced chemiluminesence (ECL) (Pierce, Rockford, IL, USA) or femto ECL (Pierce) when the signal was too faint.
Liquid chromatography–tandem mass spectrometry
2N and Ts2 fractions of brain EVs were combined for the analysis of the levels of mitochondrial proteins in Ts2 EVs (fig. S1). Ten micrograms of EV lysates were analyzed from three mice for each genotype. For the qualitative analysis of the mitovesicle proteome (Fig. 5A), Fr8 EVs were isolated from three wild-type mice and fractionated through an OptiPrep-based step gradient. EVs were run briefly into a 15% SDS–polyacrylamide gel electrophoresis gel until all proteins focused into a single 1-cm band. Gels were washed three times in ddH2O for 15 min each and visualized by overnight staining with EZ-Run Protein Gel Staining Solution (Thermo Fisher Scientific). Stacked and stained protein gel regions were excised and destained, using standard, published protocols (43). In-gel digestion was performed overnight, with MS-grade trypsin (Trypsin Gold, Promega, Madison, WI, USA) at a concentration of 5 ng/μl in a 50 mM NH4HCO3 digest buffer. After acidification with 10% (v/v) formic acid (final concentration of 0.5 to 1% formic acid), resulting peptides were desalted using hand-packed, reversed-phase Empore C18 Extraction Disks (3M, St. Paul, MN, USA).
Desalted peptides were concentrated to a very small droplet by vacuum centrifugation and reconstituted in 10 μl of 0.1% (v/v) formic acid in water. Around 90% of the peptide material was used for LC followed by data-dependent-acquisition (DDA) mode tandem MS (LC-MS/MS). A Q Exactive HF mass spectrometer was coupled directly to an EASY-nLC 1000 (Thermo Fisher Scientific) equipped with a self-packed 75 μm × 20 cm reverse-phase column (ReproSil-Pur C18, 3 μM, Dr. Maisch GmbH, Germany) for peptide separation. Analytical column temperature was maintained at 50°C by a column oven (Sonation GmBH, Germany). Peptides were eluted with a 3 to 40% acetonitrile gradient over 110 min at a flow rate of 250 nl/min. The mass spectrometer was operated in DDA mode with survey scans acquired at a resolution of 120,000 [at mass/charge ratio (m/z) 200] over a scan range of 300 to 1750 m/z. Up to 15 most abundant precursors from the survey scan were selected with an isolation window of 1.6 Th for fragmentation by higher-energy collisional dissociation with normalized collision energy of 27. The maximum injection time for the survey and MS/MS scans was 60 ms, and the ion target value (automatic gain control, AGC) for both scan modes was set to 3 × 106.
The mass spectra files were processed using the MaxQuant proteomics data analysis workflow (version 1.6.0.1) with the Andromeda search engine (44). Raw MS files were used to extract peak lists that were searched with the Andromeda search engine against human or mouse proteomes and a file containing contaminants such as human keratins. Trypsin was specified allowing up to two missed cleavages with the minimum required peptide length set to be seven amino acids. N-acetylation of protein N termini, oxidation of methionines, and deamidation of asparagines and glutamines were set as variable modifications. For the initial main search, parent peptide masses were allowed mass deviation of up to 20 parts per million. Peptide spectral matches and protein identifications were filtered using a target-decoy approach at a false discovery rate of 1%. We used the raw MS1 intensity for protein quantitation.
Lipid analysis of brain EVs
Each fraction obtained from the OptiPrep density gradient and suspended in 20 μl of PBS was lyophilized, and the lyophilized powder was sonicated in 200 μl of the mixture of methyl-tert-butyl ether and methanol (1:1, v/v). The resultant fine powder suspension was centrifuged at 10,000g for 5 min, and the precipitate was reextracted with 200 μl of the above solvent mixture. The supernatants were combined to obtain the total lipid extracts. The total lipid extracts were partitioned according to the method of Matyash et al. (45). The lower aqueous-methanol phase containing gangliosides was evaporated to dryness, applied to high-performance thin-layer chromatography (HPTLC) plates, and developed with chloroform/methanol/0.25% KCl (5:4:1). The plates including various concentrations of GM2 standards were stained with an orcinol reagent, scanned with the Gel Logic molecular imaging system (Carestream Health, Rochester, NY, USA) and analyzed by Multi Gauge version 2.0 (Fujifilm USA Medical Systems, Stamford, CT, USA). The concentration of each ganglioside was calculated using GM2 standards. The upper organic methyl-tert-butyl ether phase obtained by the partition described above was evaporated to dryness and separated into neutral and acidic lipids using DEAE-Sephadex columns. The neutral lipid fraction (including sterols, acylglycerols, ceramide, GalCer, PE, PC, and SM) and various concentrations of each lipid standard were applied on HPTLC plates and developed first with hexane/ethyl acetate/water/acetic acid (3:4:5:2) until the solvent front ascended to 4 cm from origin, second with hexane/methyl-tert-butyl ether/acetic acid (65:35:2) until 6.5 cm, and third with hexane/methyl-tert-butyl ether (98/2/1) until 9 cm above origin. The acidic lipid fraction (including PA, CL, and PS) and various concentrations of each lipid were developed on HPTLC with acetone/benzene/acetic acid/water (20:30:4:1). After development plates were first dipped in 20% methanol and then stained in 0.0001% primuline. Fluorescent lipid bands were scanned and analyzed as described above for ganglioside analyses.
EV RNA purification and qPCR
Fr5 and Fr8 brain EVs were resuspended in PBS, and the RNA was purified from 16 μl of the suspension (46) from each fraction using the miRNeasy Mini Kit per the manufacturer’s specifications (217004, Qiagen, Germantown, MD, USA). The deoxyribonuclease (DNase) digestion step was performed twice sequentially before proceeding with final washes and purification. This was performed to ensure no mtDNA contamination in the following qPCRs as mtDNA does not contain introns and could amplify with the TaqMan primers used for qPCR. RNA quantity and purification quality were measured using an Agilent 2100 Bioanalyzer (Pico 6000 kit, Agilent, Santa Clara, CA, USA). Reverse transcription of 40 ng of input RNA per sample was performed using random hexamers as primers (2.5 μM final concentration) and Superscript IV per the manufacturer’s specifications in a 50-μl reaction volume (18090050, Thermo Fisher Scientific). qPCR was performed using TaqMan qPCR primers (Life Technologies, Grand Island, NY, USA) for the following genes: mitochondrially encoded 12S RNA (mt-RNR1, Mm04260177_s1), ATP synthase F0 subunit 8 (Atp8; Mm04225236_g1), cytochrome b (Cytb; Mm04225271_g1), cytochrome c oxidase subunit II (COX-II; Mm03294838_g1), NADH (reduced form of nicotinamide adenine dinucleotide) dehydrogenase subunit 4 (ND4; Mm04225294_s1), and NADH dehydrogenase subunit 1 ND1 (ND1; Mm04225274_s1). Samples were assayed in triplicate on a real-time qPCR cycler (ABI 7900HT, Thermo Fisher Scientific) in 96-well optical plates with coverfilm as described previously (47–50). Standard curves and cycle threshold (Ct) were generated using standards obtained from total mouse brain RNA. The ΔΔCt method was used to determine relative gene-level differences between study groups (47, 51–53). Negative controls consisted of the reaction mixture without input RNA.
mtDNA extraction from Fr8 EVs and qPCR
To extract the DNA found in Fr8 brain EVs without RNA contamination, equal amounts of Fr8 EVs were incubated with DNase and exonuclease III (Thermo Fisher Scientific) for 30 min at 37°C and then inactivated at 65°C for 10 min. Protein kinase (Thermo Fisher Scientific) digestion was performed for 1 hour at 50°C and heat inactivated at 95°C for 5 min. DNA was then purified using AMPure XP magnetic beads (Beckman Coulter) and resuspended in nuclease-free water. DNA was measured by NanoDrop 2000c (Thermo Fisher Scientific) and directly used for qPCR for the mitochondrially encoded 12S RNA (mt-RNR1, Mm04260177_s1) gene.
Quantification and statistical analysis
Statistical analyses and graphs were performed using the software GraphPad Prism (version 6.01, GraphPad Holdings, San Diego, CA, USA). All data are shown as means ± SD. For clarity and transparency, each replicate is shown as a superimposed dot plot and represents a different isolation. None of the replicates were excluded from the analysis. When two groups were compared, a two-tailed, unpaired, Student’s t test was performed. When more than two groups were taken into consideration and one variable was analyzed (e.g., differences between the fractions), one-way analysis of variance (ANOVA) with Tukey’s multiple comparisons test was used. When two variables were analyzed simultaneously (e.g., changing of the enzymatic activity over time compared with a control condition), a two-way ANOVA with Tukey’s multiple comparisons was performed. Significance was set to a P value of <0.05. Variables considered, normalization procedures, exact value of n, and other specific statistical information are provided for each experiment in the respective figure legend or in the graph shown in the figure, when possible.
Protein bands were quantified using ImageJ (54) and analyzed and graphed through GraphPad Prism. EV proteins were routinely normalized to the net weight of brain tissue from which they were isolated. Densitometric analyses of EVs isolated from conditioned cell media were normalized to the SyproRuby total protein staining, as indicated in the respective figure legends. The level of the proteins found in the brain and cell homogenate were normalized over a housekeeping loading control, as indicated in the respective figure legends.
The concentration of each lipid is provided as lipid weight per microgram of proteins in each fraction. To estimate the lipid composition of each EV, we transformed the mass in moles and calculated the molar percentage of a specific lipid population over the total. The approximated molecular masses considered in our analysis were 649 g/mol for cholesteryl esters, 387 g/mol for cholesterol, 786 g/mol for PC, 743 g/mol for PE, 787 g/mol for PS, 682 g/mol for PA, 587 g/mol for DAG, 843 g/mol for TAG, 1893 g/mol for GD1a, 1989 g/mol for GT1b, 1466 g/mol for CL, 538 g/mol for ceramide, 737 g/mol for SM, and 718 for GalCer.
For calculating the relative frequency of different subtypes of EVs in the OptiPrep fractions (Fig. 2E), 30 random cryoEM photomicrographs per fraction from two independent isolations were acquired. Given the paucity of vesicles found in some of the fractions, especially for Fr8, data were pooled together, resulting in 192 vesicles counted in total for Fr1, 220 for Fr2, 269 for Fr3, 237 for Fr4, 193 for Fr5, 170 for Fr6, 169 for Fr7, and 93 for Fr8.
For the EV diameter estimation, we used two different approaches: electron microscopy (EM) and NTA. Thirty random cryoEM or TEM photomicrographs per fraction from three independent isolations were acquired. All the EVs in the images were analyzed except for the vesicles found at the border that were not fully included in the field of view (and so the diameter could not be estimated accurately) and pooled for each fraction. Given that many larger EVs, especially by TEM, were not exactly circular but tended to have an ovoid or an irregular shape (see Fig. 2A), we did not measure the diameter directly, as it would have been slightly different in each position. Accordingly, we quantified the area of each vesicle through the software ImageJ and calculated post hoc the diameter assuming a circular shape, by the formula d = 2√(A/π), where d is the diameter and A is the area. The resulting box and whiskers plot clearly showed that the distribution of this parameter was not normal but positively skewed for all the fractions, as the median did not divide the distribution in two symmetrical halves (Fig. 2, B and C). For this reason, we used the Kruskal-Wallis H test by ranks (with Dunn’s multiple comparisons test), which is the nonparametric equivalent of the one-way ANOVA (calculated through the software GraphPad Prism). The positive skewness of the curve was largely expected because of a well-established systematic error, called “fishnet artifact,” by which very small EVs tend to be more easily lost during EM sample preparation. Ultimately, this causes an overestimation of the proportion of the large EVs over the small ones (55).
A similar slight positive skewness could be observed by NTA (Fig. 1E). This was also predictable. In the density range of biological samples, NTA cannot detect vesicles that are smaller than 50 nm without catching noise signal, while no limitations are provided in the upper limit (at least for the dimension range of the EVs), as per the manufacturer’s guidelines. To circumvent the non-normality of the distribution, we performed the analysis using a binning system, as shown in the figure, and as successfully applied elsewhere (12).
With respect to the analysis of LC-MS data, identified proteins within EVs were quantified using 2N mice as the reference to calculate log-fold differences. Proteins with at least two reported intensities per group (Ts2 and 2N) were included in the analysis and compared using a Student’s t test with a P value cutoff (P < 0.05) deemed significant. Differentially regulated proteins were depicted using ggplot and pheatmap packages in R. Proteins were identified as “mitochondrial” if verified in mitochondria in at least two of these three databases: MitoCarta 2.0 (56), UniProt (available at https://uniprot.org) (57), and the Human Protein Atlas (available at http://proteinatlas.org) (39, 41). Gene set enrichment analysis (GSEA) of gene sets was performed using fgsea package (58) in R (table S5). Gene sets were determined to be significant if Padj < 0.05 with 100,000 permutations and a minimum gene set size of 10. Fgsea was used on Go gene sets from the Msigdb database available from the Broad Institute, available at https://cran.r-project.org/web/packages/msigdbr/index.html. Murine brain tissue expression and gene function indicated in table S2 were derived from UniProt and Human Protein Atlas (40) annotations.
Last, for qPCR analysis of mitochondrial mRNA, the signal intensity ratio of each gene was modeled as a function of mouse study group, using mixed effects models with random mouse effect to account for the correlation between repeated assays on the same mouse (59). Significance was judged at the level α = 0.01, two sided; false discovery rate based on an empirical null distribution due to strong correlation between genes was controlled at level α = 0.1.
Acknowledgments
We dedicate this work to the memory of Mitsuo Saito, our colleague and friend, whose contribution to understanding the role of lipids in neurotransmission was unsurpassed. His insight, knowledge, and passion for science will be missed by many. Mitsuo Saito passed away on November 20, 2020. We thank P. M. Mathews and L. Rachmany for discussions; J. Wegiel for providing human samples; B. Barreto, M. Kurz, and S. Bhamdeo for technical support; S. Das, S. DeRosa, and N. Patel for animal husbandry: S. H. Lee for statistical analysis; F. Huang for producing MS data shown in fig. S1; and G. Ferrari for coordinating and managing our laboratory. Funding: This work was supported by the National Institute on Aging (AG017617, AG056732, and AG057517) and the National Institute on Drug Abuse (DA044489) to E.L., and NIH Shared Instrumentation Grant (S10RR027990) to T.A.N. Author contributions: Conceptualization: P.D. and E.L.; methodology: P.D. and R.P.-G.; formal analysis: P.D. and S.C.P.; investigation: P.D., R.P.-G., T.H., H.E-B., Y.K., C.M., M.J.A., and C.N.G.; resources: M.P.; writing original and revised drafts: P.D. and E.L.; visualization: P.D. and E.L.; supervision: E.L., Mariko Saito, Mitsuo Saito, S.D.G., and T.A.N.; project administration: E.L. and P.D.; funding acquisition: E.L. and T.A.N. Competing interests: The authors declare that they have no competing interests. Data and materials availability: The raw MS data generated during this study are available at MassIVE (UCSD, https://massive.ucsd.edu/ProteoSAFe/static/massive.jsp) with accession number MSV000085447. All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/7/eabe5085/DC1
REFERENCES AND NOTES
- 1.Gauthier S. A., Pérez-González R., Sharma A., Huang F.-K., Alldred M. J., Pawlik M., Kaur G., Ginsberg S. D., Neubert T. A., Levy E., Enhanced exosome secretion in Down syndrome brain - A protective mechanism to alleviate neuronal endosomal abnormalities. Acta Neuropathol. Commun. 5, 65 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jeppesen D. K., Fenix A. M., Franklin J. L., Higginbotham J. N., Zhang Q., Zimmerman L. J., Liebler D. C., Ping J., Liu Q., Evans R., Fissell W. H., Patton J. G., Rome L. H., Burnette D. T., Coffey R. J., Reassessment of Exosome Composition. Cell 177, 428–445.e18 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Puhm F., Afonyushkin T., Resch U., Obermayer G., Rohde M., Penz T., Schuster M., Wagner G., Rendeiro A. F., Melki I., Kaun C., Wojta J., Bock C., Jilma B., Mackman N., Boilard E., Binder C. J., Mitochondria are a subset of extracellular vesicles released by activated monocytes and induce type I IFN and TNF responses in endothelial cells. Circ. Res. 125, 43–52 (2019). [DOI] [PubMed] [Google Scholar]
- 4.Jang S. C., Crescitelli R., Cvjetkovic A., Belgrano V., Olofsson Bagge R., Sundfeldt K., Ochiya T., Kalluri R., Lötvall J., Mitochondrial protein enriched extracellular vesicles discovered in human melanoma tissues can be detected in patient plasma. J. Extracell. Vesicles 8, 1635420 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guescini M., Genedani S., Stocchi V., Agnati L. F., Astrocytes and Glioblastoma cells release exosomes carrying mtDNA. J. Neural Transm. (Vienna) 117, 1 (2010). [DOI] [PubMed] [Google Scholar]
- 6.Phinney D. G., Giuseppe M. D., Njah J., Sala E., Shiva S., St Croix C. M., Stolz D. B., Watkins S. C., Di Y. P., Leikauf G. D., Kolls J., Riches D. W. H., Deiuliis G., Kaminski N., Boregowda S. V., McKenna D. H., Ortiz L. A., Mesenchymal stem cells use extracellular vesicles to outsource mitophagy and shuttle microRNAs. Nat. Commun. 6, 8472 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haraszti R. A., Haraszti R. A., Didiot M.-C., Sapp E., Leszyk J., Shaffer S. A., Rockwell H. E., Gao F., Narain N. R., Figlia M. D., Kiebish M. A., Aronin N., Khvorova A., High-resolution proteomic and lipidomic analysis of exosomes and microvesicles from different cell sources. J. Extracell. Vesicles 5, 32570 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hayakawa K., Esposito E., Wang X., Terasaki Y., Liu Y., Xing C., Ji X., Lo E. H., Transfer of mitochondria from astrocytes to neurons after stroke. Nature 535, 551–555 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sliter D. A., Martinez J., Hao L., Chen X., Sun N., Fischer T. D., Burman J. L., Li Y., Zhang Z., Narendra D. P., Cai H., Borsche M., Klein C., Youle R. J., Parkin and PINK1 mitigate STING-induced inflammation. Nature 561, 258–262 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Perez-Gonzalez R., Gauthier S. A., Kumar A., Levy E., The exosome secretory pathway transports amyloid precursor protein carboxyl-terminal fragments from the cell into the brain extracellular space. J. Biol. Chem. 287, 43108–43115 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pérez-González R., Gauthier S. A., Kumar A., Saito M., Saito M., Levy E., A method for isolation of extracellular vesicles and characterization of exosomes from brain extracellular space. Methods Mol. Biol. 1545, 139–151 (2017). [DOI] [PubMed] [Google Scholar]
- 12.D’Acunzo P., Hargash T., Pawlik M., Goulbourne C. N., Pérez-González R., Levy E., Enhanced generation of intraluminal vesicles in neuronal late endosomes in the brain of a Down syndrome mouse model with endosomal dysfunction. Dev. Neurobiol. 79, 656–663 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pérez-González R., Sahoo S., Gauthier S. A., Kim Y., Li M., Kumar A., Pawlik M., Benussi L., Ghidoni R., Levy E., Neuroprotection mediated by cystatin C-loaded extracellular vesicles. Sci. Rep. 9, 11104 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peng K. Y., Pérez-González R., Alldred M. J., Goulbourne C. N., Morales-Corraliza J., Saito M., Saito M., Ginsberg S. D., Mathews P. M., Levy E., Apolipoprotein E4 genotype compromises brain exosome production. Brain 142, 163–175 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pérez-González R., Gauthier S. A., Sharma A., Miller C., Pawlik M., Kaur G., Kim Y., Levy E., A pleiotropic role for exosomes loaded with the amyloid β precursor protein carboxyl-terminal fragments in the brain of Down syndrome patients. Neurobiol. Aging 84, 26–32 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mathews P. M., Levy E., Exosome production is key to neuronal endosomal pathway integrity in neurodegenerative diseases. Front. Neurosci. 13, 1347 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bordi M., Darji S., Sato Y., Mellén M., Berg M. J., Kumar A., Jiang Y., Nixon R. A., mTOR hyperactivation in Down syndrome underlies deficits in autophagy induction, autophagosome formation, and mitophagy. Cell Death Dis. 10, 563 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee H., Li C., Zhang Y., Zhang D., Otterbein L. E., Jin Y., Caveolin-1 selectively regulates microRNA sorting into microvesicles after noxious stimuli. J. Exp. Med. 216, 2202–2220 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van Niel G., Charrin S., Simoes S., Romao M., Rochin L., Saftig P., Marks M. S., Rubinstein E., Raposo G., The tetraspanin CD63 regulates ESCRT-independent and -dependent endosomal sorting during melanogenesis. Dev. Cell 21, 708–721 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Skotland T., Hessvik N. P., Sandvig K., Llorente A., Exosomal lipid composition and the role of ether lipids and phosphoinositides in exosome biology. J. Lipid Res. 60, 9–18 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang J., Zheng X., Yang X., Liao K., CIN85 associates with endosomal membrane and binds phosphatidic acid. Cell Res. 19, 733–746 (2009). [DOI] [PubMed] [Google Scholar]
- 22.Singh R. D., Puri V., Valiyaveettil J. T., Marks D. L., Bittman R., Pagano R. E., Selective caveolin-1-dependent endocytosis of glycosphingolipids. Mol. Biol. Cell 14, 3254–3265 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fecher C., Trovò L., Müller S. A., Snaidero N., Wettmarshausen J., Heink S., Ortiz O., Wagner I., Kühn R., Hartmann J., Karl R. M., Konnerth A., Korn T., Wurst W., Merkler D., Lichtenthaler S. F., Perocchi F., Misgeld T., Cell-type-specific profiling of brain mitochondria reveals functional and molecular diversity. Nat. Neurosci. 22, 1731–1742 (2019). [DOI] [PubMed] [Google Scholar]
- 24.Steer E. K., Dail M. K., Chu C. T., Beyond mitophagy: Cytosolic PINK1 as a messenger of mitochondrial health. Antioxid. Redox Signal. 22, 1047–1059 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lin W., Kang U. J., Characterization of PINK1 processing, stability, and subcellular localization. J. Neurochem. 106, 464–474 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou R., Yazdi A. S., Menu P., Tschopp J., A role for mitochondria in NLRP3 inflammasome activation. Nature 469, 221–225 (2011). [DOI] [PubMed] [Google Scholar]
- 27.Izzo A., Mollo N., Nitti M., Paladino S., Calì G., Genesio R., Bonfiglio F., Cicatiello R., Barbato M., Sarnataro V., Conti A., Nitsch L., Mitochondrial dysfunction in down syndrome: Molecular mechanisms and therapeutic targets. Mol. Med. 24, 2 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Villar A. J., Belichenko P. V., Gillespie A. M., Kozy H. M., Mobley W. C., Epstein C. J., Identification and characterization of a new Down syndrome model, Ts[Rb(12.1716)]2Cje, resulting from a spontaneous Robertsonian fusion between T(1716)65Dn and mouse chromosome 12. Mamm. Genome 16, 79–90 (2005). [DOI] [PubMed] [Google Scholar]
- 29.Kaur G., Gauthier S. A., Perez-Gonzalez R., Pawlik M., Singh A. B., Cosby B., Mohan P. S., Smiley J. F., Levy E., Cystatin C prevents neuronal loss and behavioral deficits via the endosomal pathway in a mouse model of down syndrome. Neurobiol. Dis. 120, 165–173 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vives-Bauza C., Zhou C., Huang Y., Cui M., de Vries R. L. A., Kim J., May J., Tocilescu M. A., Liu W., Ko H. S., Magrané J., Moore D. J., Dawson V. L., Grailhe R., Dawson T. M., Li C., Tieu K., Przedborski S., PINK1-dependent recruitment of Parkin to mitochondria in mitophagy. Proc. Natl. Acad. Sci. U.S.A. 107, 378–383 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.D’Acunzo P., Strappazzon F., Caruana I., Meneghetti G., Rita A. D., Simula L., Weber G., Bufalo F. D., Valle L. D., Campello S., Locatelli F., Cecconi F., Reversible induction of mitophagy by an optogenetic bimodular system. Nat. Commun. 10, 1533 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kühlbrandt W., Structure and function of mitochondrial membrane protein complexes. BMC Biol. 13, 89 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sugiura A., McLelland G. L., Fon E. A., McBride H. M., A new pathway for mitochondrial quality control: Mitochondrial-derived vesicles. EMBO J. 33, 2142–2156 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Picca A., Guerra F., Calvani R., Bucci C., Monaco M. R. L., Bentivoglio A. R., Landi F., Bernabei R., Marzetti E., Mitochondrial-derived vesicles as candidate biomarkers in parkinson’s disease: Rationale, design and methods of the exosomes in PArkinson disease (EXPAND) study. Int. J. Mol. Sci. 20, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shukkur E. A., Shimohata A., Akagi T., Yu W., Yamaguchi M., Murayama M., Chui D., Takeuchi T., Amano K., Subramhanya K. H., Hashikawa T., Sago H., Epstein C. J., Takashima A., Yamakawa K., Mitochondrial dysfunction and tau hyperphosphorylation in Ts1Cje, a mouse model for Down syndrome. Hum. Mol. Genet. 15, 2752–2762 (2006). [DOI] [PubMed] [Google Scholar]
- 36.Di Rita A., D’Acunzo P., Simula L., Campello S., Strappazzon F., Cecconi F., AMBRA1-mediated mitophagy counteracts oxidative stress and apoptosis induced by neurotoxicity in human neuroblastoma SH-SY5Y cells. Front. Cell. Neurosci. 12, 92 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Di Rita A., Peschiaroli A., D’Acunzo P., Strobbe D., Hu Z., Gruber J., Nygaard M., Lambrughi M., Melino G., Papaleo E., Dengjel J., Alaoui S. E., Campanella M., Dötsch V., Rogov V. V., Strappazzon F., Cecconi F., HUWE1 E3 ligase promotes PINK1/PARKIN-independent mitophagy by regulating AMBRA1 activation via IKKα. Nat. Commun. 9, 3755 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Whittle N., Sartori S. B., Dierssen M., Lubec G., Singewald N., Fetal Down syndrome brains exhibit aberrant levels of neurotransmitters critical for normal brain development. Pediatrics 120, e1465–e1471 (2007). [DOI] [PubMed] [Google Scholar]
- 39.Thul P. J., Åkesson L., Wiking M., Mahdessian D., Geladaki A., Blal H. A., Alm T., Asplund A., Björk L., Breckels L. M., Bäckström A., Danielsson F., Fagerberg L., Fall J., Gatto L., Gnann C., Hober S., Hjelmare M., Johansson F., Lee S., Lindskog C., Mulder J., Mulvey C. M., Nilsson P., Oksvold P., Rockberg J., Schutten R., Schwenk J. M., Sivertsson Å., Sjöstedt E., Skogs M., Stadler C., Sullivan D. P., Tegel H., Winsnes C., Zhang C., Zwahlen M., Mardinoglu A., Pontén F., von Feilitzen K., Lilley K. S., Uhlén M., Lundberg E., A subcellular map of the human proteome. Science 356, eaal3321 (2017). [DOI] [PubMed] [Google Scholar]
- 40.Uhlén M., Fagerberg L., Hallström B. M., Lindskog C., Oksvold P., Mardinoglu A., Sivertsson Å., Kampf C., Sjöstedt E., Asplund A., Olsson I. M., Edlund K., Lundberg E., Navani S., Szigyarto C. A.-K., Odeberg J., Djureinovic D., Takanen J. O., Hober S., Alm T., Edqvist P.-H., Berling H., Tegel H., Mulder J., Rockberg J., Nilsson P., Schwenk J. M., Hamsten M., von Feilitzen K., Forsberg M., Persson L., Johansson F., Zwahlen M., von Heijne G., Nielsen J., Pontén F., Proteomics. Tissue-based map of the human proteome. Science 347, 1260419 (2015). [DOI] [PubMed] [Google Scholar]
- 41.Uhlen M., Oksvold P., Fagerberg L., Lundberg E., Jonasson K., Forsberg M., Zwahlen M., Kampf C., Wester K., Hober S., Wernerus H., Björling L., Ponten F., Towards a knowledge-based human protein atlas. Nat. Biotechnol. 28, 1248–1250 (2010). [DOI] [PubMed] [Google Scholar]
- 42.Mathews P. M., Jiang Y., Schmidt S. D., Grbovic O. M., Mercken M., Nixon R. A., Calpain activity regulates the cell surface distribution of amyloid precursor protein. Inhibition of calpains enhances endosomal generation of β-cleaved C-terminal APP fragments. J. Biol. Chem. 277, 36415–36424 (2002). [DOI] [PubMed] [Google Scholar]
- 43.Kishinevsky S., Wang T., Rodina A., Chung S. Y., Xu C., Philip J., Taldone T., Joshi S., Alpaugh M. L., Bolaender A., Gutbier S., Sandhu D., Fattahi F., Zimmer B., Shah S. K., Chang E., Inda C., Koren J. III, Saurat N. G., Leist M., Gross S. S., Seshan V. E., Klein C., Tomishima M. J., Erdjument-Bromage H., Neubert T. A., Henrickson R. C., Chiosis G., Studer L., HSP90-incorporating chaperome networks as biosensor for disease-related pathways in patient-specific midbrain dopamine neurons. Nat. Commun. 9, 4345 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cox J., Neuhauser N., Michalski A., Scheltema R. A., Olsen J. V., Mann M., Andromeda: A peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 10, 1794–1805 (2011). [DOI] [PubMed] [Google Scholar]
- 45.Matyash V., Liebisch G., Kurzchalia T. V., Shevchenko A., Schwudke D., Lipid extraction by methyl-tert-butyl ether for high-throughput lipidomics. J. Lipid Res. 49, 1137–1146 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Eldh M., Lötvall J., Malmhäll C., Ekström K., Importance of RNA isolation methods for analysis of exosomal RNA: Evaluation of different methods. Mol. Immunol. 50, 278–286 (2012). [DOI] [PubMed] [Google Scholar]
- 47.Alldred M. J., Chao H. M., Lee S. H., Beilin J., Powers B. E., Petkova E., Strupp B. J., Ginsberg S. D., CA1 pyramidal neuron gene expression mosaics in the Ts65Dn murine model of Down syndrome and Alzheimer’s disease following maternal choline supplementation. Hippocampus 28, 251–268 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Alldred M. J., Lee S. H., Petkova E., Ginsberg S. D., Expression profile analysis of vulnerable CA1 pyramidal neurons in young-middle-aged Ts65Dn mice. J. Comp. Neurol. 523, 61–74 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Alldred M. J., Lee S. H., Petkova E., Ginsberg S. D., Expression profile analysis of hippocampal CA1 pyramidal neurons in aged Ts65Dn mice, a model of Down syndrome (DS) and Alzheimer’s disease (AD). Brain Struct. Funct. 220, 2983–2996 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Alldred M. J., Duff K. E., Ginsberg S. D., Microarray analysis of CA1 pyramidal neurons in a mouse model of tauopathy reveals progressive synaptic dysfunction. Neurobiol. Dis. 45, 751–762 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ginsberg S. D., Alldred M. J., Counts S. E., Cataldo A. M., Neve R. L., Jiang Y., Wuu J., Chao M. V., Mufson E. J., Nixon R. A., Che S., Microarray analysis of hippocampal CA1 neurons implicates early endosomal dysfunction during Alzheimer’s disease progression. Biol. Psychiatry 68, 885–893 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jiang Y., Mullaney K. A., Peterhoff C. M., Che S., Schmidt S. D., Boyer-Boiteau A., Ginsberg S. D., Cataldo A. M., Mathews P. M., Nixon R. A., Alzheimer’s-related endosome dysfunction in Down syndrome is Aβ-independent but requires APP and is reversed by BACE-1 inhibition. Proc. Natl. Acad. Sci. U.S.A. 107, 1630–1635 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Alldred M. J., Chao H. M., Lee S. H., Beilin J., Powers B. E., Petkova E., Strupp B. J., Ginsberg S. D., Long-term effects of maternal choline supplementation on CA1 pyramidal neuron gene expression in the Ts65Dn mouse model of Down syndrome and Alzheimer’s disease. FASEB J. 33, 9871–9884 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schneider C. A., Rasband W. S., Eliceiri K. W., NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Linares R., Tan S., Gounou C., Brisson A. R., Imaging and quantification of extracellular vesicles by transmission electron microscopy. Methods Mol. Biol. 1545, 43–54 (2017). [DOI] [PubMed] [Google Scholar]
- 56.Calvo S. E., Clauser K. R., Mootha V. K., MitoCarta2.0: An updated inventory of mammalian mitochondrial proteins. Nucleic Acids Res. 44, D1251–D1257 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.The UniProt Consortium , UniProt: A worldwide hub of protein knowledge. Nucleic Acids Res. 47, D506–D515 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Korotkevich G., Sukhov V., Sergushichev A., Fast gene set enrichment analysis. bioRxiv, 060012 (2019). [Google Scholar]
- 59.C. McCulloch, S. Searle, J. Neuhaus, Generalized, Linear, and Mixed Models (John Wiley & Sons, 2011). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/7/eabe5085/DC1


