Abstract
Epitranscriptomic analysis has recently led to the profiling of modified nucleosides in cancer cell biological matrices, helping to elucidate their functional roles in cancer and reigniting interest in exploring their use as potential markers of cancer development and progression. Pseudouridine, one of the most well-known and the most abundant of the RNA nucleotide modifications, is the C5-glycoside isomer of uridine and its distinctive physiochemical properties allows it to perform many essential functions. Pseudouridine functionally (a) confers rigidity to local RNA structure by enhancing RNA stacking, engaging in a cooperative effect on neighboring nucleosides that overall contributes to RNA stabilization (b) refines the structure of tRNAs, which influences their decoding activity (c) facilitates the accuracy of decoding and proofreading during translation and efficiency of peptide bond formation, thus collectively improving the fidelity of protein biosynthesis and (e) dynamically regulates mRNA coding and translation. Biochemical synthesis of pseudouridine is carried out by pseudouridine synthases. In this review we discuss the evidence supporting an association between elevated pseudouridine levels with the incidence and progression of human prostate cancer and the translational significance of the value of this modified nucleotide as a novel biomarker in prostate cancer progression to advanced disease.
Keywords: Biomarker, DKC1, H/ACA snoRNA, Modified nucleosides, Pseudouridine
1. Introduction
Prostate cancer (PCa) is the second leading cause of death in American men, with around 33,330 deaths predicted in 2020 [1]. Serum prostate-specific antigen (PSA) levels are frequently used for clinical diagnosis [2] however it lacks specificity for PCa [3] and its use results in overdiagnosis leading to unnecessary biopsies and overtreatment of indolent disease [4,5] as well as poor risk stratification [6]. In a report released by the US Preventive Services Task Force which systematically reviewed the effectiveness of PSA-based screening, between 20.7% to 50.4% of screen-detected cancers were estimated to be overdiagnosed [7] and overtreatment of low-grade non-lethal disease was found to cost over 1.3 billion dollars in the US annually [8]. Not only does PSA-based screening for PCa result in overdiagnosis and overtreatment, it can fail to detect cancer in almost 15% of men screened when levels are ≤ 4 ng/ml [9]. Although many PSA-derived or inclusive (e.g., free:total PSA ratio [10], PSA kinetic factors [11], the Prostate Health Index [12], Progensa Prostate Cancer Antigen 3 test, 4K Score test [13]), genomic (e.g., the Oncotype DX test, TMPRSS2-ERG and PTEN status [13] etc.) or other (e.g., circulating tumor cells, microRNAs, exosomal markers [13]) biomarkers and clinical tests have been shown to have improved specificity or performance over PSA alone in cancer detection or prediction of high-grade disease [13], PSA still remains an important and standard part of both diagnostic and prognostic procedures. In view of the challenges and limitations associated with the interpretation of PSA values as a biomarker of PCa progression, there is a huge clinically unmet need for novel biomarkers that can detect PCa at the onset of tumor development, distinguish between indolent and aggressive disease and predict therapeutic resistance [14]. In this review we discuss the current knowledge of RNA derived-modified molecules identified by epitranscriptomics that can be exploited for their potential biomarker role in PCa progression, with specific focus on the modified nucleoside pseudouridine.
Epitranscriptomics, the study of post-transcriptional modifications of RNA analogous to epigenetics is currently experiencing a revival of interest [15] as the techniques used to detect, sequence, and analyze them at the transcriptome-wide level have only been recently developed [16–22]. These modifications to RNA include methylation, hydroxylation, reduction, isomerization, sulfur or oxygen substitutions, or the addition of side chains [23,24]. Although modified RNA bases have been known to science for decades [25], modern next-generation sequencing (NGS) technologies [26] have enabled the discovery of over 170 of these post-transcriptional modifications [24,27] and several of them are now implicated in various human diseases, including cancer [28,29]. Interestingly in the years before the introduction of NGS many of these post-transcriptional modifications were evaluated for their use as biomarkers for malignant diseases [30–33], and are in fact still being exploited towards that scope [34–36]. Profiling of modified nucleosides (mNS) with an emphasis on biomarker discovery can be accomplished using a variety of biological matrices [23] (such as cancer cell lines [37], tissue [33], blood [38] and urine [39], although urine is most commonly used [23]) and is likely made possible by an impaired RNA metabolism, the increase in whole-body turnover of RNA in rapidly-proliferating tumors [36,40], and the inability of mNS to be recycled by cells [41,42], hence their excretion into bodily fluids [23]. Amongst the approximately 20–25 different modified RNA nucleosides that have been examined for their use as diagnostic biomarkers for cancer [43,44], pseudouridine (Ψ), an isomer of uridine (U) has repeatedly been seen to hold diagnostic potential for several kinds of cancers [44], implying that carcinogenesis may rely on or lead to increased rates of pseudouridylation. Excitingly, recent evidence may suggest its clinical utility as a novel biomarker for PCa as well.
2. The evolution of the modified nucleoside pseudouridine: Significance as a marker
Historically, RNA nucleosides with peculiar intramolecular arrangements can be traced back to the late 1930s [45], although definitive proof of the existence of RNA modifications occurred later on in the 1950s with the discovery of pseudouridine [46]. By examining degradation products of acid-catalyzed hydrolysis of RNA, the four ribonucleosides (i.e. adenosine, uridine, guanosine, and cytidine) were typically only observed with phosphoryl linkages in either the C-2 or C-3 positions, with the assumption that C-5 linkages were not possible due to the inability to isolate such products using this method [46]. In historical context, in 1938, Gulland and Jackson first reported the hydrolysis of yeast RNA using a phosphodiesterase and the enzyme 5’-nucleotidase contained in snake venom, producing inorganic phosphate in yields that could only suggest that the phosphoryl groups were attached at the C-5 position [45]. Indirect evidence for C-5 phosphoryl linkages later appeared in 1951 when Schmidt et al. performed periodate titrations with hydrolyzed yeast ribonucleic acid and confirmed that some nucleotide groups were linked to positions other than C-2 or C-3 of the ribose [46,47]. Moreover in 1951, Cohn and Volkin [46] isolated an unknown nucleoside 5’-phosphate from enzymatically hydrolyzed calf liver transfer RNA (tRNA) [48]. Originally designated “?”, this unknown compound would also be successfully isolated from yeast RNA in 1957 by Davis and Allen [49], with other researchers making similar reports, isolating the nucleoside from bacteria, dog pancreas and rat liver [48,49]. Remarkably, considering how novel and revolutionary the discovery of an entirely new “building block” of RNA might have seemed at the time, researchers were forward-thinking enough to pioneer medical research of the nucleoside, measuring its levels in the urine of patients with various diseases (e.g., gout and leukemia [48]), and even observed abnormally high levels of the substance in the urine of “mentally defective” patients in 1967 [50]. The structure of this novel nucleoside was solved by Cohn in 1959 [51], and was first known as 5-ribosyl uracil, but was later renamed and is now more commonly known as pseudouridine [51].
3. Pseudouridine: Physiochemical properties, biosynthesis and detection
Since its discovery, pseudouridine was commonly referred to as “the fifth nucleotide” and has been chemically characterized as the C5-glycoside isomer of uridine [52]. Pseudouridine, also known as β-pseudouridine and 5-(β-D-ribofuranosyl)uracil has a molar mass identical to uridine (244.2 g/mol) but differs in its mass spectrometric dissociation [53,54]. Pseudouridine is also among the most well-known of the RNA modifications, perhaps due to the relative abundance [55–57] and inertness of the isomer compared to other mNS within a cell, and is estimated to comprise ~5% of the total of all cellular RNA nucleotides [49,58]. The isomerization of uridine to Ψ confers to pseudouridine some additional and distinctive properties (Fig. 1). The C-C glycosyl bond, unique amongst modified nucleosides, is more freely able to rotate than the N-C bond in uridine and thus allows for greater conformational flexibility; when part of a polynucleotide chain however it has only been found to confer rigidity instead of flexibility on both single and double-stranded regions of RNA [59]. Pseudouridine also slightly prefers the syn rather than anti conformation that uridine and other nucleosides adopt and therefore can function as a conformational switch in RNA [59]. Moreover, Ψ has an increased capacity for hydrogen bonding and can donate an additional bond [59] (Fig. 1), allowing for enhanced local RNA stacking which is cooperatively increased through neighboring nucleosides [59]. Since this effect is propagated throughout adjacent helical regions, it is believed to be the most critical contribution of Ψ to the stabilization of RNA structure [59].
Fig. 1.
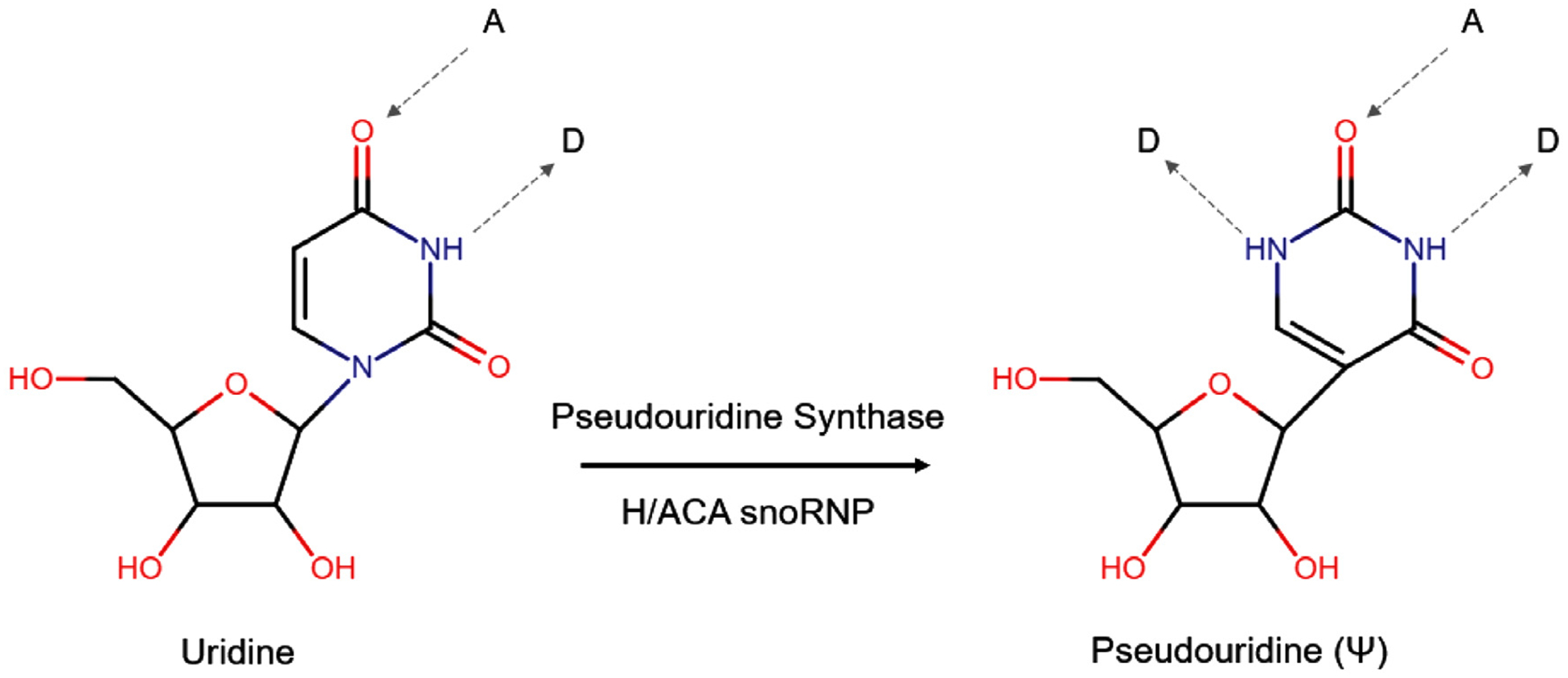
Biosynthesis and chemical properties of pseudouridine. The isomerization of uridine to pseudouridine is performed by either standalone pseudouridine synthases (PUSs) or H/ACA snoRNP complexes containing the catalytic component dyskerin (DKC1), additional core proteins (NOP10, GAR1 and NHP2) and an H/ACA snoRNA which serves as a guide. Uridine can accept (A) and donate (D) one hydrogen bond (gray dashed arrow) each whereas pseudouridine can donate an additional hydrogen bond.
Biosynthesis of Ψ is carried out by a class of enzymes known as pseudouridine synthases (PUSs), which are classified into 6 different families: (TruD, TruA, TruB, RsuA [which is not present in eukaryotes] RluA, and PUS10 [which is not present in E. coli] [60]). In eukaryotes, pseudouridine synthases can either function as standalone enzymes (i.e., PUS1 through PUS9 in yeast, although very similar enzymes exist in humans) or as part of RNA-guided ribonucleoprotein (RNP) complexes [60] (Fig. 1). These RNP complexes containing the TruB family member [61] dyskerin (DKC1), the enzymatic component that catalyzes the isomerization reaction and additional core proteins (NOP10, GAR1 and NHP2) are guided to the appropriate, specific uridines to be modified by non-coding RNAs (ncRNAs) called H/ACA box snoRNAs (H/ACA snoRNAs) [62]. Together, they comprise the H/ACA small nucleolar RNA-ribonucleoprotein complex (H/ACA snoRNP) [62]. To date, 12 pseudouridine synthase genes have been discovered in humans: PUS1, TRUB2, PUS3, PUS4, RPUSD1, RPUSD2, RPUSD3, RPUSD4, PUS7, PUS7L, PUS10, and DKC1 [60].
Pseudouridine can be detected using a number of different methods, including high performance liquid chromatography (HPLC) [63], liquid chromatography-mass spectrometry (LC-MS) [64], capillary electrophoresis (CE) [65] and immunological antibody-based methods [66] (including flow cytometry [60,67]). Deep sequencing methods include pseudo-seq [68], Ψ-seq [69], PSI-seq [70], and CeU-seq [71], and are all capable of mapping Ψ positions across the entire transcriptome with single nucleotide resolution [72]. These methods all work by using the chemical N-cyclohexyl-N′-(2-morpholinoethyl)-carbodiimide metho-p-toluenesulfonate (CMC) to selectively label Ψ [72]. These pseudouridine-CMC adducts efficiently block reverse transcription one base downstream of Ψ, thus making pseudouridylated residues detectable as a distinct stop [72]. NGS techniques have presently identified more than 9500 putative Ψ modification sites across most types of RNA [60].
4. Functions of pseudouridine
Functionally Ψ can provide enhanced rigidity to local RNA moieties but how that relates to RNA biology is not yet fully understood. Pseudouridine was first discovered in ncRNAs such as ribosomal RNA (rRNA), tRNA and small nuclear RNA (snRNA) [59,60] however recent sequencing studies have discovered modifications sites in many other types of RNA including messenger RNA (mRNA) [19,69] microRNAs [60] long non-coding RNAs (lncRNAs) [21] small Cajal Body-specific RNAs (scaRNAs), small nucleolar RNAs (snoRNAs), and other exotic kinds of RNA [60], indicating that proper pseudouridylation is likely essential to overall RNA integrity and that Ψ has many diverse RNA-specific functions. For example Ψ is present in both large and small subunits of rRNA, clustering in domains II, IV, and V and contributes to the proper functioning of ribosomes [62]. Domain V contains the peptidyltransferase center (PTC) [59], where peptide bonds are formed between amino acids during protein biosynthesis. As such, Ψ in rRNA has been implicated in ribosomal folding and assembly, and given its presence in functional domains (i.e. the PTC), it directly impacts the speed and accuracy of decoding and proofreading during translation and alters the efficiency of peptide bond formation [59]. In tRNA, while Ψ does not affect overall structure it does affect the structure of the local domains in which it resides, and these modifications may in fact be critical for proper binding of tRNA to ribosomes [59]. Overall Ψ can fine tune the structure of tRNAs, which influences their decoding activity and improves the fidelity of protein biosynthesis [59]. When incorporated into in some of the major spliceosomal small nuclear RNAs of eukaryotes, particularly in regions that are important for RNA-RNA or RNA-protein interactions within a spliceosome, Ψ contributes to proper spliceosome formation, rendering such modifications essential for premRNA processing [59].
Pseudouridinylation has recently been identified as a dynamic process with context-dependent effects. Thus 28S rRNA was found to be inducibly pseudouridylated by the mammalian target of rapamycin (mTOR) pathway in Chinese hamster ovary (CHO) cells [73]. This study found that the mTOR pathway controls cellular growth via inducible pseudouridylation in CHO cell cultures, and that treatment with rapamycin (an mTOR inhibitor) increases 28S rRNA pseudouridinylation without conferring enhanced ribosomal functionality; instead it’s more associated with the negative effects of rapamycin treatment on the cellular phenotype [73]. Pseudouridine profiling in yeast and human cells revealed that the majority of Ψs in mRNA are regulated in response to environmental signals, such as nutrient deprivation in yeast and serum starvation in HeLa cells [19]. Additional pseudouridylation sites have also been discovered in response to heat and cold shock and treatment with H2O2, cycloheximide, and hepatocyte growth factor, further suggesting that pseudouridylation plays a role in dynamically modulating RNA function [72]. Pseudouridylated mRNAs exhibit enhanced stability and translation efficiency [72,74]; targeting pseudouridylation to specific U residues within nonsense codons converts them to sense codons in budding yeast [72,75]. Overall this evidence suggests that dynamic mRNA pseudouridylation could be a mechanism for rapid rewiring of the genetic code [19].
5. Role of pseudouridine as a cancer biomarker
As mentioned previously, approximately 20–25 different mNS have been examined for their use as diagnostic biomarkers for cancer [43,44], and out of these modifications Ψ has been one of the most frequently studied (Table 1). Increased levels of Ψ have been observed in the blood, urine or tissue of patients with breast cancer [76], colorectal cancer [77], esophageal cancer [33], gallbladder cancer [78], hepatocellular carcinoma [79,80], leukemia [81], lymphoma [82,83], ovarian cancer [84,85], and small cell lung cancer [86,87]. A summary of 25 different studies evaluating the efficacy of Ψ as a biomarker are referenced in Table 1 and organized by sample type and method of detection used. Analysis of Ψ in urine is the most common as sample collection is easy and non-invasive. Urinary analysis is also ideal because the levels of mNS in urine reflect the rate of RNA (particularly tRNA) degradation within an organism, a process that is known to be hyperactivated in neoplastic tissue [43,88] hence the increased urinary excretion of Ψ seen in cancer patients. In terms of clinical feasibility some methods of detection (such as those utilizing mass spectrometry) may be prohibitively expensive or require a certain level of expertise [89] whereas other methods such as ELISA (enzyme-linked immunosorbent assay) are routinely used in clinical diagnostics. Improvements upon already-established methods such as those developed for analysis of mNS in urinary samples using CE [65] offer other advantages. Altogether there are multiple proven methodologies available for the detection, analysis and quantification of Ψ in human samples that are amendable for biomarker discovery in PCa.
Table 1.
Pseudouridine as a cancer biomarker - methods of detection. Pseudouridine can be detected in samples of human blood (using plasma or serum), urine and tissue using a variety of methods, and 25 different studies evaluating the potential biomarker value of pseudouridine for several different types of cancer are summarized in Table 1. The protocols contained within can be easily modified or are readily available to evaluate the diagnostic or prognostic potential of pseudouridine for PCa.
| Method | Blood (Plasma or Serum) | Urine | Tissue |
|---|---|---|---|
| 1H-NMR (1H-nuclear magnetic resonance) | Perez-Rambla et al., 2017 [95] | ||
| ELISA (enzyme-linked immunosorbent assay) | Itoh et al., 1989 [66] | ||
| Itoh et al., 1992 [105] | |||
| Masaki et al., 2006 [82] | |||
| GC-TOF-MS (gas chromatography-time of flight-mass spectrometry) | Thysell et al., 2010 [98] | ||
| HPLC (high performance liquid chromatography) | Tamura et al., 1987 [86] | Rasmuson et al., 1983 [83] | |
| Amuro et al., 1988 [79] | Tamura et al., 1986 [87] | ||
| Pane et al., 1993 [106] | Tamura et al., 1986 [80] | ||
| Li et al., 1992 [81] | |||
| Seidel et al., 2006 [43] | |||
| HPLC-ESI-MS/MS (high-performance liquid chromatography-electrospray ionization-tandem mass spectrometry) | Bond et al., 2006 [107] | ||
| IHC (immunohistochemistry) | Itoh et al., 1992 [108] | ||
| Masuda et al., 1993 [33] | |||
| Stockert et al., 2019 [96] | |||
| LC-MS (liquid chromatography-mass spectrometry) | Chen et al., 2012 [84] | ||
| LC-MS/MS (liquid chromatography-tandem mass spectrometry) | Zeleznik et al., 2020 [85] | ||
| MEKC (micellar electrokinetic chromatography) | Zheng et al., 2005 [76] | ||
| RP-HPLC (reverse phase-high performance liquid chromatography) | Jiao et al., 2014 [78] | Feng et al., 2005 [77] | Jiao et al., 2014 [78] |
| Jiao et al., 2014 [78] | |||
| UPLC-MS (ultra performance liquid chromatography-mass spectrometry) | Ke et al., 2015 [109] | ||
| UPLC-QTOF-MS (ultra performance liquid chromatography-quadrupole time of flight-mass spectrometry | Zhang et al., 2013 [110] | ||
| Jiang et al., 2015 [111] |
6. Pseudouridine in prostate cancer
Growing evidence suggests a potential association between elevated levels of Ψ with the incidence and/or progression of PCa. Recently it has been shown that there is an increased expression of H/ACA snoRNAs associated with PCa progression [62], including SNORA74A, SNORA42, and SNORA64 [62,90]. A study by Martens-Uzunova et al. that examined the small noncoding transcriptomes of organ-confined and metastatic lymph node PCa LN-PCa in an effort to identify novel diagnostic and prognostic ncRNA expression profiles revealed that the total amount of snoRNA fragments (including H/ACA snoRNAs) in metastatic tumors increased by >20% (20,569 counts in LN-PCa compared to 16,762 counts in organ-confined PCa) [90] and approximately 14 to 16 non-putative H/ACA snoRNAs were unique to and detected only in LN-PCa [90]. In a separate study published by Crea et al. [91] SNORA55 was found to be upregulated during PCa progression and predicts significantly shorter recurrence-free survival after prostatectomy [91]. Furthermore silencing of SNORA55 in PCa cell lines reduced their proliferation and metastatic potential [91,92]. Increased expression of pseudouridine synthases has also been associated with the progression of PCa. Sieron et al. had shown that prostate carcinomas were found to have a higher expression of DKC1, particularly in high-stage and recurring cases, and overexpression of the protein was associated with the progression of the disease [93]. Additionally, increased abundance of PUS1 was recently found to be associated with increased risk for biochemical relapse [94]. Taken together, this evidence suggests that increased pseudouridylation may result from the upregulation of these components that arise from or contribute to the progression of PCa. Indeed, 1H nuclear magnetic resonance metabolomic analysis of urine sampled from men with PCa revealed that these men, compared to men diagnosed with benign prostatic hyperplasia have different metabolite profiles including increased levels of urinary Ψ [95,96]. In a study using gas chromatography-time of flight-mass spectrometry (GC-TOF-MS) to discover new biomarkers for metastatic PCa, Ψ was found to be significantly increased in the plasma analyzed from PCa patients with bone metastases (M1 disease) compared to men with benign disease [97,98]. Work done by our group recently provided new evidence on establishing the relationship between pseudouridine expression and clinical progression of PCa [96]. Using antibody-based methods of detection [66] to examine the levels of pseudouridine in RNA extracted from PCa cell lines representative of different stages of disease progression we observed the highest levels of Ψ in androgen-independent PCa cells (PC3 and Du145), followed by castration-resistant PCa cells (22Rv1), compared to androgen-sensitive (LNCaP) PCa cells and normal prostate cells [96]. Immunohistochemical analysis of a prostate tissue micro array (TMA) containing normal adjacent tissues and tissue sampled from adenocarcinomas with Gleason sums ranging from 6 to 9 revealed that pseudouridine was highly expressed in and localized to glandular cells belonging to adenocarcinomas with negligible staining of the glandular cells of normal adjacent tissues [96].
7. Future directions
Evaluating pseudouridine levels in the blood, urine or tissue of patients in the context of relevant clinical (e.g., Gleason grade, biochemical recurrence, PSA levels, tumor volume, TNM stage, development of metastasis, presence of neuroendocrine disease, etc.), and genetic factors (e.g., PTEN loss, TMPRSS2-ERG fusions, genomic alterations in AR, PI3K/AKT, and/or DNA repair pathways [99] etc.) would greatly aid in parsing the potential value of Ψ as a diagnostic or prognostic tool for PCa. Transcriptomic analysis of Ψ in malignant vs. normal or benign patient tumors would be an innovative approach to study the differences between tumors of various grades or clinical parameters and could potentially yield novel biomarkers or therapeutic targets for PCa. The true relationship between pseudouridine and prostate cancer, and the potential functional contributions of elevated Ψ levels to PCa initiation, regardless of whether or not these levels result from increased expression of H/ACA snoRNAs, DKC1 or other pseudouridine synthases remains to be elucidated.
Pseudouridine synthases may one day become viable drug targets or biomarkers themselves. Besides catalyzing the isomerization of uridine to Ψ, DKC1 is also required for ribosome biogenesis and telomerase complex stabilization, processes that are hyper-active during neoplastic transformation, and as such have been and are still currently being explored as targets for the development of drugs to selectively or preferentially kill cancer cells [100,101]. Homology modelling and virtual ligand screening have yielded several novel inhibitors of DKC1 [100,101]. Rocchi et al. had found that the drug pyrazofurin was able to reduce the viability of breast cancer cells (MCF-7) as well as the in vitro pseudouridylation activity of DKC1 [100]. Armando et al. identified three novel compounds that successfully inhibit DKC1 and disrupt the formation of the telomerase complex, as they were observed to significantly decrease telomerase activity in vitro [101]. Furthermore, downregulation of DKC1 reduced the abundance of snoRNAs in neuroblastoma cells, inducing ribosomal stress which leads to G1 cell cycle arrest and cell death [61,102]. Evaluating the expression of DKC1 in PCa cell lines and testing their sensitivity to pyrazofurin or other DKC1 inhibitors would be useful in regards to evaluating the therapeutic potential of DKC1 inhibition to treat different stages of PCa. As mentioned previously, increased expression of PUS1 was found to be associated with increased risk for biochemical relapse [94] and interestingly, PUS1 modifications are required for the proper interaction of one of its targets, the ncRNA steroid receptor RNA activator 1 with several kinds of nuclear receptors, in particular the RARγ receptor in melanoma cells and the oestrogen receptor in breast cancer cells [103,104]; therefore PUS1 may be contributing to prostate carcinogenesis in yet unknown ways via interaction with SRA1. The upregulation of H/ACA snoRNAs in PCa progression [91], the differential expression of H/ACA snoRNAs in organ-confined vs. LN-PCa [90] and the reduction of proliferation and metastatic potential of PCa cells via silencing of SNORA55 suggests that H/ACA snoRNAs may be contributing to more advanced stages of PCa.
8. Conclusions
The detection and clinical management of prostate cancer was revolutionized by the introduction of PSA testing more than 30 years ago, an innovation which has reduced overall PCa mortality rates and improved the overall survival of men with PCa. Despite this progress, men are still being overdiagnosed and overtreated for prostate cancer, and significant cancer is missed in many men, thus there is a crucial need for novel biomarkers for PCa. Modified nucleosides have repeatedly been shown to hold diagnostic and prognostic potential for a number of malignancies, of which few have greater potential than pseudouridine. As a non-invasive marker that can be easily collected and detected from patient samples, coupled with the increasing evidence suggesting an association of Ψ with the incidence or progression of PCa as well as the introduction of novel Ψ-sequencing technologies, the time has never been better than now to explore the potential of pseudouridine as a novel biomarker for prostate cancer.
Acknowledgments
This work was supported by the Department of Urology at the Icahn School of Medicine at Mount Sinai, the Dean Prostate Health Fund and the Prostate Cancer Foundation. MarvinJS was used for drawing, displaying and characterizing chemical structures, substructures and reactions. MarvinJS 2020, ChemAxon (http://www.chemaxon.com).
Abbreviations:
- 1H-NMR
1H-nuclear magnetic resonance
- BPH
Benign prostatic hyperplasia
- CE
Capillary electrophoresis
- CHO
Chinese hamster ovary
- CMC
N-cyclohexyl-N′-(2-morpholinoethyl)-carbodiimide metho-p-toluenesulfonate
- CTCs
Circulating tumor cells
- DKC1
Dyskerin
- ELISA
Enzyme-linked immunosorbent assay
- GC-TOF-MS
Gas chromatography-time of flight-mass spectrometry
- H/ACA snoRNA
H/ACA box small nucleolar RNA
- H/ACA snoRNP
H/ACA small nucleolar RNA-ribonucleoprotein complex
- HPLC
High performance liquid chromatography
- HPLC-ESI-MS/MS
High-performance liquid chromatography-electrospray ionization-tandem mass spectrometry
- IHC
Immunohistochemistry
- LC-MS
Liquid chromatography-mass spectrometry
- LC-MS/MS
Liquid chromatography-tandem mass spectrometry
- LN-PCa
Metastatic lymph node prostate cancer
- lncRNA
Long non-coding RNA
- MEKC
Micellar electrokinetic chromatography
- miRNA
MicroRNA
- mNS
Modified nucleosides
- mRNA
Messenger RNA
- mTOR
Mammalian target of rapamycin
- NAT
Normal adjacent tissue
- ncRNA
Non-coding RNA
- NGS
Next-generation sequencing
- PCa
Prostate cancer
- PCA3
Prostate cancer antigen 3
- PHI
Prostate health index
- PSA
Prostate-specific antigen
- PTC
Peptidyltransferase center
- PUS
Pseudouridine synthase
- RNA
Ribonucleic acid
- RNP
Ribonucleoprotein
- RP-HPLC
Reverse phase-high performance liquid chromatography
- rRNA
Ribosomal RNA
- scaRNA
Small Cajal body-specific RNA
- snoRNA
Small nucleolar RNA
- snRNA
Small nuclear RNA
- SRA1
Steroid receptor RNA activator 1
- TMA
Tissue micro array
- TNM
Tumor, node, metastasis
- tRNA
Transfer RNA
- U
Uridine
- UPLC-MS
Ultra performance liquid chromatography-mass spectrometry
- UPLC-QTOFMS
Ultra performance liquid chromatography-quadrupole time of flight-mass spectrometry
- Ψ
Pseudouridine
References
- [1].Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin 2020;70:7–30. [DOI] [PubMed] [Google Scholar]
- [2].Merriel SWD, Funston G, Hamilton W. Prostate cancer in primary care. Adv Ther 2018;35:1285–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Tabayoyong W, Abouassaly R. Prostate cancer screening and the associated controversy. Surg Clin North Am 2015;95:1023–39. [DOI] [PubMed] [Google Scholar]
- [4].Sandhu GS, Andriole GL. Overdiagnosis of prostate cancer. J Natl Cancer Inst Monogr 2012;2012:146–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Grossman DC, Curry SJ, Owens DK, Bibbins-Domingo K, Caughey AB, Davidson KW, et al. Screening for prostate cancer: US preventive services task force recommendation statement. JAMA 2018;319:1901–13. [DOI] [PubMed] [Google Scholar]
- [6].Gnanapragasam VJ, Lophatananon A, Wright KA, Muir KR, Gavin A, Greenberg DC. Improving clinical risk stratification at diagnosis in primary prostate cancer: A prognostic modelling study. PLoS Med 2016;13:e1002063–e1002063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Fenton JJ, Weyrich MS, Durbin S, Liu Y, Bang H, Melnikow J. Prostate-specific antigen-based screening for prostate cancer: Evidence report and systematic review for the US preventive services task force. JAMA 2018;319:1914–31. [DOI] [PubMed] [Google Scholar]
- [8].Aizer AA, Gu X, Chen MH, Choueiri TK, Martin NE, Efstathiou JA, et al. Cost implications and complications of overtreatment of low-risk prostate cancer in the United States. J Natl Compr Canc Netw 2015;13:61–8. [DOI] [PubMed] [Google Scholar]
- [9].Thompson IM, Pauler DK, Goodman PJ, Tangen CM, Lucia MS, Parnes HL, et al. Prevalence of prostate cancer among men with a prostate-specific antigen level ≤4.0 ng per milliliter. N Engl J Med 2004;350:2239–46. [DOI] [PubMed] [Google Scholar]
- [10].Ayyildiz SN, Ayyildiz A. PSA, PSA derivatives, proPSA and prostate health index in the diagnosis of prostate cancer. Turk J Urol 2014;40:82–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Salman JW, Schoots IG, Carlsson SV, Jenster G, Roobol MJ. Prostate specific antigen as a tumor marker in prostate cancer: Biochemical and clinical aspects. Adv Exp Med Biol 2015;867:93–114. [DOI] [PubMed] [Google Scholar]
- [12].Loeb S, Catalona WJ. The Prostate Health Index: a new test for the detection of prostate cancer. Ther Adv Urol 2014;6:74–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Saini S PSA and beyond: alternative prostate cancer biomarkers. Cell Oncol 2016;39:97–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Lieberman HB, Rai AJ, Friedman RA, Hopkins KM, Broustas CG. Prostate cancer: unmet clinical needs and RAD9 as a candidate biomarker for patient management. Transl Cancer Res 2018;7:S651–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Xiong X, Yi C, Peng J. Epitranscriptomics: Toward a better understanding of RNA modifications. Genomics Proteomics Bioinformatics 2017;15:147–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Dominissini D, Moshitch-Moshkovitz S, Salmon-Divon M, Amariglio N, Rechavi G. Transcriptome-wide mapping of N(6)-methyladenosine by m(6)A-seq based on immunocapturing and massively parallel sequencing. Nat Protoc 2013;8:176–89. [DOI] [PubMed] [Google Scholar]
- [17].Dominissini D, Nachtergaele S, Moshitch-Moshkovitz S, Peer E, Kol N, Ben-Haim MS, et al. The dynamic N(1)-methyladenosine methylome in eukaryotic messenger RNA. Nature 2016; 530:441–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Li X, Xiong X, Wang K, Wang L, Shu X, Ma S, et al. Transcriptome-wide mapping reveals reversible and dynamic N(1)-methyladenosine methylome. Nat Chem Biol 2016;12:311–6. [DOI] [PubMed] [Google Scholar]
- [19].Carlile TM, Rojas-Duran MF, Zinshteyn B, Shin H, Bartoli KM, Gilbert WV. Pseudouridine profiling reveals regulated mRNA pseudouridylation in yeast and human cells. Nature 2014;515: 143–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Edelheit S, Schwartz S, Mumbach MR, Wurtzel O, Sorek R. Transcriptome-wide mapping of 5-methylcytidine RNA modifications in bacteria, archaea, and yeast reveals m5C within archaeal mRNAs. PLoS Genet 2013;9:e1003602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Shafik A, Schumann U, Evers M, Sibbritt T, Preiss T. The emerging epitranscriptomics of long noncoding RNAs. Biochim Biophys Acta 2016;1859:59–70. [DOI] [PubMed] [Google Scholar]
- [22].Chen W, Lin H. Recent Advances in Identification of RNA Modifications. Non-Coding RNA 2016;3:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Godoy AT, Eberlin MN, Simionato AVC. Targeted metabolomics: Liquid chromatography coupled to mass spectrometry method development and validation for the identification and quantitation of modified nucleosides as putative cancer biomarkers. Talanta 2020; 210:120640. [DOI] [PubMed] [Google Scholar]
- [24].Boccaletto P, Machnicka MA, Purta E, Piatkowski P, Baginski B, Wirecki TK, et al. MODOMICS: a database of RNA modification pathways. 2017 update. Nucleic Acids Res 2018;46:D303–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].COHN WE. Some results of the applications of ion-exchange chromatography to nucleic acid chemistry. J Cell Physiol Suppl 1951;38:21–40. [DOI] [PubMed] [Google Scholar]
- [26].Schwartz S, Motorin Y. Next-generation sequencing technologies for detection of modified nucleotides in RNAs. RNA Biol 2017; 14:1124–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Liang W, Lin Z, Du C, Qiu D, Zhang Q. mRNA modification orchestrates cancer stem cell fate decisions. Mol Cancer 2020;19:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Jonkhout N, Tran J, Smith MA, Schonrock N, Mattick JS, Novoa EM. The RNA modification landscape in human disease. RNA 2017;23:1754–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Delaunay S, Frye M. RNA modifications regulating cell fate in cancer. Nat Cell Biol 2019;21:552–9. [DOI] [PubMed] [Google Scholar]
- [30].Davis GE, Suits RD, Kuo KC, Gehrke CW, Waalkes TP, Borek E. High-performance liquid chromatographic separation and quantitation of nucleosides in urine and some other biological fluids. Clin Chem 1977;23:1427–35. [PubMed] [Google Scholar]
- [31].Waalkes TP, Abeloff MD, Ettinger DS, Woo KB, Gehrke CW, Kuo KC, et al. Modified ribonucleosides as biological markers for patients with small cell carcinoma of the lung. Eur J Cancer Clin Oncol 1982;18:1267–74. [DOI] [PubMed] [Google Scholar]
- [32].Langridge JI, McClure TD, el-Shakawi S, Fielding A, Schram KH, Newton RP. Gas chromatography/mass spectrometric analysis of urinary nucleosides in cancer patients; potential of modified nucleosides as tumour markers. Rapid Commun Mass Spectrom 1993; 7:427–34. [DOI] [PubMed] [Google Scholar]
- [33].Masuda M, Nishihira T, Itoh K, Mizugaki M, Ishida N, Mori S. An immunohistochemical analysis for cancer of the esophagus using monoclonal antibodies specific for modified nucleosides. Cancer 1993;72:3571–8. [DOI] [PubMed] [Google Scholar]
- [34].Guo C, Chen Q, Chen J, Yu J, Hu Y, Zhang S, et al. 8-Hydroxyguanosine as a possible RNA oxidative modification marker in urine from colorectal cancer patients: Evaluation by ultra performance liquid chromatography-tandem mass spectrometry. J Chromatogr B, Anal Technol Biomed Life Sci 2020;1136:121931. [DOI] [PubMed] [Google Scholar]
- [35].Opitz P, Herbarth O, Seidel A, Boehm A, Fischer M, Mozet C, et al. Modified nucleosides - molecular markers suitable for small-volume cancer? Anticancer Res 2018;38:6113–9. [DOI] [PubMed] [Google Scholar]
- [36].Seidel A, Seidel P, Manuwald O, Herbarth O. Modified nucleosides as biomarkers for early cancer diagnose in exposed populations. Env Toxicol 2015;30:956–67. [DOI] [PubMed] [Google Scholar]
- [37].Willmann L, Erbes T, Halbach S, Brummer T, Jäger M, Hirschfeld M, et al. Exometabolom analysis of breast cancer cell lines: Metabolic signature. Sci Rep 2015;5:13374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Buzatto AZ, de Oliveira Silva M, Poppi RJ, Simionato AVC. Assessment of nucleosides as putative tumor biomarkers in prostate cancer screening by CE-UV. Anal Bioanal Chem 2017;409:3289–97. [DOI] [PubMed] [Google Scholar]
- [39].Zhang Y-R, Shi L, Wu H, Tang D-D, Wang S-M, Liu H-M, et al. Urinary modified nucleosides as novel biomarkers for diagnosis and prognostic monitoring of urothelial bladder cancer. Tumori 2014; 100:660–6. [DOI] [PubMed] [Google Scholar]
- [40].Nakano K, Nakao T, Schram KH, Hammargren WM, McClure TD, Katz M, et al. Urinary excretion of modified nucleosides as biological marker of RNA turnover in patients with cancer and AIDS. Clin Chim Acta 1993;218:169–83. [DOI] [PubMed] [Google Scholar]
- [41].Schram KH. Urinary nucleosides. Mass Spectrom Rev 1998;17: 131–251. [DOI] [PubMed] [Google Scholar]
- [42].Henneges C, Bullinger D, Fux R, Friese N, Seeger H, Neubauer H, et al. Prediction of breast cancer by profiling of urinary RNA metabolites using Support Vector Machine-based feature selection. BMC Cancer 2009;9:104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Seidel A, Brunner S, Seidel P, Fritz GI, Herbarth O. Modified nucleosides: an accurate tumour marker for clinical diagnosis of cancer, early detection and therapy control. Br J Cancer 2006;94: 1726–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Patejko M, Struck-Lewicka W, Siluk D, Waszczuk-Jankowska M, Markuszewski MJ. In: Makowski GSBT-A in CC, ed. Chapter One - Urinary Nucleosides and Deoxynucleosides, 83, Elsevier; 2018:1–51. 10.1016/bs.acc.2017.10.001:. [DOI] [PubMed] [Google Scholar]
- [45].Gulland JM, Jackson EM. The constitution of yeast nucleic acid. J Chem Soc 1938;284:1492–8. [Google Scholar]
- [46].Cohn WE, Volkin E. Nucleoside-5′-phosphates from ribonucleic acid. Nature 1951;167:483–4. [Google Scholar]
- [47].Schmidt G, Cubiles R, Thannhauser SJ. On the nature of the products formed by the action of crystalline ribonuclease (Kunitz’s ribonuclease) on yeast ribonucleic acid. J Cell Physiol Suppl 1951; 38:61–70. 10.1002/jcp.1030380407. [DOI] [PubMed] [Google Scholar]
- [48].Shaban MAE, Nasr AZ. The chemistry of C-Nucleosides and their analogs I: C-Nucleosides of hetero monocyclic bases. Adv. Heterocycl. Chem 1997;68:223–432. [Google Scholar]
- [49].Davis FF, Allen FW. Ribonucleic acids from yeast which contain a fifth nucleotide. J Biol Chem 1957;227:907–15. [PubMed] [Google Scholar]
- [50].Kihara H Pseudouridinuria in mentally defective siblings. Am J Ment Defic 1967;71:593–6. [PubMed] [Google Scholar]
- [51].Cohn WE. 5-Ribosyl uracil, a carbon-carbon ribofuranosyl nucleoside in ribonucleic acids. Biochim Biophys Acta 1959;32:569–71. [DOI] [PubMed] [Google Scholar]
- [52].Mueller EG, F-DAR. Pseudouridine Formation, the Most Common Transglycosylation in RNA In: Grosjean H, ed. DNA RNA Modif Enzym Struct Mech Funct Evol n.d Austin, TX: Landes Bioscience; 2009:363–76. [Google Scholar]
- [53].Kim S, Chen J, Cheng T, Gindulyte A, He J, He S, et al. PubChem 2019 update: improved access to chemical data. Nucleic Acids Res 2018;47:D1102–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Spenkuch F, Motorin Y, Helm M. Pseudouridine: still mysterious, but never a fake (uridine)!. RNA Biol 2014;11:1540–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Reddy R, Busch H. Small Nuclear RNAs: RNA Sequences, Structure, and Modificationseditor In: Birnstiel ML, ed. Struct. Funct. Major Minor Small Nucl. Ribonucleoprotein Part, Berlin, Heidelberg: Springer Berlin Heidelberg; 1988:1–37. 10.1007/978-3-642-73020-7_1. [DOI] [Google Scholar]
- [56].Massenet S, Mougin A, Branlant C. Posttranscriptional Modifications in the U Small Nuclear RNAsModif. Ed. RNA, Modification and Editing of RNA; 1998201–27. 10.1128/9781555818296.ch11. [DOI] [Google Scholar]
- [57].Yu Y, Yu Y, Scharl EC, Smith CM, Steitz JA. The growing world of small nuclear ribonucleoproteins. RNA WORLD (Gestel1999. [Google Scholar]
- [58].Wu G, Huang C, Yu Y-T. Pseudouridine in mRNA: Incorporation, detection, and recoding. Methods Enzymol 2015;560:187–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Charette M, Gray MW. Pseudouridine in RNA: what, where, how, and why. IUBMB Life 2000;49:341–51. [DOI] [PubMed] [Google Scholar]
- [60].Penzo M, Guerrieri AN, Zacchini F, Trere D, Montanaro L. RNA pseudouridylation in physiology and medicine: For better and for worse. Genes (Basel) 2017:8 10.3390/genes8110301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Arndt GM, MacKenzie KL. New prospects for targeting telomerase beyond the telomere. Nat Rev Cancer 2016;16:508–24. [DOI] [PubMed] [Google Scholar]
- [62].McMahon M, Contreras A, Ruggero D. Small RNAs with big implications: new insights into H/ACA snoRNA function and their role in human disease. Wiley Interdiscip Rev RNA 2015; 6:173–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Montanaro L, Brigotti M, Clohessy J, Barbieri S, Ceccarelli C, Santini D, et al. Dyskerin expression influences the level of ribosomal RNA pseudo-uridylation and telomerase RNA component in human breast cancer. J Pathol 2006;210:10–8. [DOI] [PubMed] [Google Scholar]
- [64].Addepalli B, Limbach PA. Mass spectrometry-based quantification of pseudouridine in RNA. J Am Soc Mass Spectrom 2011;22:1363–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Jiang Y, Ma Y. A fast capillary electrophoresis method for separation and quantification of modified nucleosides in urinary samples. Anal Chem 2009;81:6474–80. [DOI] [PubMed] [Google Scholar]
- [66].Itoh K, Mizugaki M, Ishida N. Detection of elevated amounts of urinary pseudouridine in cancer patients by use of a monoclonal antibody. Clin Chim Acta 1989;181:305–15. [DOI] [PubMed] [Google Scholar]
- [67].Hoshino A, Honda I, Ishimori A, Itoh K, Mizugaki M, Nose M. [Molecular and immunological approach to hematological disease: detection and analysis of intracellular modified nucleosides by flow cytometry]. Rinsho Byori 1990;38:756–64. [PubMed] [Google Scholar]
- [68].Carlile TM, Rojas-Duran MF, Gilbert WV. Pseudo-Seq: Genome-Wide detection of pseudouridine modifications in RNA. Methods Enzymol 2015;560:219–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Schwartz S, Bernstein DA, Mumbach MR, Jovanovic M, Herbst RH, León-Ricardo BX, et al. Transcriptome-wide mapping reveals widespread dynamic-regulated pseudouridylation of ncRNA and mRNA. Cell 2014;159:148–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Lovejoy AF, Riordan DP, Brown PO. Transcriptome-wide mapping of pseudouridines: pseudouridine synthases modify specific mRNAs in S. cerevisiae. PLoS One 2014;9:e110799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Li X, Zhu P, Ma S, Song J, Bai J, Sun F, et al. Chemical pulldown reveals dynamic pseudouridylation of the mammalian transcriptome. Nat Chem Biol 2015;11:592–7. [DOI] [PubMed] [Google Scholar]
- [72].Zaringhalam M, Papavasiliou FN. Pseudouridylation meets next-generation sequencing. Methods 2016;107:63–72. [DOI] [PubMed] [Google Scholar]
- [73].Courtes FC, Gu C, Wong NSC, Dedon PC, Yap MGS, Lee D-Y. 28S rRNA is inducibly pseudouridylated by the mTOR pathway translational control in CHO cell cultures. J Biotechnol 2014; 174:16–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Karikó K, Muramatsu H, Welsh FA, Ludwig J, Kato H, Akira S, et al. Incorporation of Pseudouridine Into mRNA Yields Superior Nonimmunogenic Vector With Increased Translational Capacity and Biological Stability. Mol Ther 2008;16:1833–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Karijolich J, Yu Y-T. Converting nonsense codons into sense codons by targeted pseudouridylation. Nature 2011;474:395–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Zheng Y-F, Kong H-W, Xiong J-H, Lv S, Xu G-W. Clinical significance and prognostic value of urinary nucleosides in breast cancer patients. Clin Biochem 2005;38:24–30. [DOI] [PubMed] [Google Scholar]
- [77].Feng B, Zheng M-H, Zheng Y-F, Lu A-G, Li J-W, Wang M-L, et al. Normal and modified urinary nucleosides represent novel biomarkers for colorectal cancer diagnosis and surgery monitoring. J Gastroenterol Hepatol 2005;20:1913–9. [DOI] [PubMed] [Google Scholar]
- [78].Jiao X, Mo Y, Wu Y, He J, Zhang P, Hu R, et al. Upregulated plasma and urinary levels of nucleosides as biological markers in the diagnosis of primary gallbladder cancer. J Sep Sci 2014;37:3033–44. [DOI] [PubMed] [Google Scholar]
- [79].Amuro Y, Nakaoka H, Shimomura S, Fujikura M, Yamamoto T, Tamura S, et al. Serum pseudouridine as a biochemical marker in patients with hepatocellular carcinoma. Clin Chim Acta 1988;178: 151–8. [DOI] [PubMed] [Google Scholar]
- [80].Tamura S, Amuro Y, Nakano T, Fujii J, Moriwaki Y, Yamamoto T, et al. Urinary excretion of pseudouridine in patients with hepatocellular carcinoma. Cancer 1986;57:1571–5. [DOI] [PubMed] [Google Scholar]
- [81].Li Y, Wang S, Zhong N. Simultaneous determination of pseudouridine and creatinine in urine of normal children and patients with leukaemia by high performance liquid chromatography. Biomed Chromatogr 1992;6:191–3. [DOI] [PubMed] [Google Scholar]
- [82].Masaki Y, Itoh K, Sawaki T, Karasawa H, Kawanami T, Fukushima T, et al. Urinary pseudouridine in patients with lymphoma: comparison with other clinical parameters. Clin Chim Acta 2006;371:148–51. [DOI] [PubMed] [Google Scholar]
- [83].Rasmuson T, Bjork GR. Pseudouridine: a modified nucleoside as biological marker in malignant lymphomas. Cancer Detect Prev 1983;6:293–6. [PubMed] [Google Scholar]
- [84].Chen J, Zhou L, Zhang X, Lu X, Cao R, Xu C, et al. Urinary hydrophilic and hydrophobic metabolic profiling based on liquid chromatography-mass spectrometry methods: Differential metabolite discovery specific to ovarian cancer. Electrophoresis 2012;33:3361–9. [DOI] [PubMed] [Google Scholar]
- [85].Zeleznik OA, Eliassen AH, Kraft P, Poole EM, Rosner BA, Jeanfavre S, et al. A Prospective Analysis of Circulating Plasma Metabolites Associated with Ovarian Cancer Risk. Cancer Res 2020; 80:1357–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Tamura S, Fujioka H, Nakano T, Hada T, Higashino K. Serum pseudouridine as a biochemical marker in small cell lung cancer. Cancer Res 1987;47:6138–41. [PubMed] [Google Scholar]
- [87].Tamura S, Fujii J, Nakano T, Hada T, Higashino K. Urinary pseudouridine as a tumor marker in patients with small cell lung cancer. Clin Chim Acta 1986;154:125–32. [DOI] [PubMed] [Google Scholar]
- [88].Borek E, Baliga BS, Gehrke CW, Kuo CW, Belman S, Troll W, et al. High turnover rate of transfer RNA in tumor tissue. Cancer Res 1977;37:3362–6. [PubMed] [Google Scholar]
- [89].Heaney LM, Jones DJ, Suzuki T. Mass spectrometry in medicine: a technology for the future? Futur Sci OA 2017;3: FSO213–FSO213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Martens-Uzunova ES, Jalava SE, Dits NF, van Leenders GJ, Moller S, Trapman J, et al. Diagnostic and prognostic signatures from the small non-coding RNA transcriptome in prostate cancer. Oncogene 2012;31:978–91. [DOI] [PubMed] [Google Scholar]
- [91].Crea F, Quagliata L, Michael A, Liu HH, Frumento P, Azad AA, et al. Integrated analysis of the prostate cancer small-nucleolar transcriptome reveals SNORA55 as a driver of prostate cancer progression. Mol Oncol 2016;10:693–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Gong J, Li Y, Liu C-J, Xiang Y, Li C, Ye Y, et al. A pan-cancer analysis of the expression and clinical relevance of small nucleolar RNAs in human cancer. Cell Rep 2017;21:1968–81. [DOI] [PubMed] [Google Scholar]
- [93].Sieron P, Hader C, Hatina J, Engers R, Wlazlinski A, Muller M, et al. DKC1 overexpression associated with prostate cancer progression. Br J Cancer 2009;101:1410–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Sinha A, Huang V, Livingstone J, Wang J, Fox NS, Kurganovs N, et al. The proteogenomic landscape of curable prostate cancer. Cancer Cell 2019;35:414–427.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Perez-Rambla C, Puchades-Carrasco L, Garcia-Flores M, Rubio-Briones J, Lopez-Guerrero JA, Pineda-Lucena A. Non-invasive urinary metabolomic profiling discriminates prostate cancer from benign prostatic hyperplasia. Metabolomics 2017;13:52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Stockert JA, Gupta A, Herzog B, Yadav SS, Tewari AK, Yadav KK. Predictive value of pseudouridine in prostate cancer. Am J Clin Exp Urol 2019;7:262–72. [PMC free article] [PubMed] [Google Scholar]
- [97].Kdadra M, Höckner S, Leung H, Kremer W, Schiffer E. Metabolomics biomarkers of prostate cancer: A systematic review. Diagnostics 2019;9 10.3390/diagnostics9010021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Thysell E, Surowiec I, Hornberg E, Crnalic S, Widmark A, Johansson AI, et al. Metabolomic characterization of human prostate cancer bone metastases reveals increased levels of cholesterol. PLoS One 2010;5:e14175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Wang G, Zhao D, Spring DJ, DePinho RA. Genetics and biology of prostate cancer. Genes Dev 2018;32:1105–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Rocchi L, Barbosa AJ, Onofrillo C, Del Rio A, Montanaro L. Inhibition of human dyskerin as a new approach to target ribosome biogenesis. PLoS One 2014;9:e101971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Armando RG, Mengual Gomez DL, Juritz EI, Lorenzano Menna P, Gomez DE. Homology model and docking-based virtual screening for ligands of human dyskerin as new inhibitors of telomerase for cancer treatment. Int J Mol Sci 2018;19 10.3390/ijms19103216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].O’Brien R, Tran SL, Maritz MF, Liu B, Kong CF, Purgato S, et al. MYC-Driven neuroblastomas are addicted to a telomerase-independent function of dyskerin. Cancer Res 2016;76:3604:LP–3617. [DOI] [PubMed] [Google Scholar]
- [103].Zhao X, Patton JR, Davis SL, Florence B, Ames SJ, Spanjaard RA. Regulation of nuclear receptor activity by a pseudouridine synthase through posttranscriptional modification of steroid receptor RNA activator. Mol Cell 2004;15:549–58. [DOI] [PubMed] [Google Scholar]
- [104].Barbieri I, Kouzarides T. Role of RNA modifications in cancer. Nat Rev Cancer 2020. 10.1038/s41568-020-0253-2. [DOI] [PubMed] [Google Scholar]
- [105].Itoh K, Konno T, Sasaki T, Ishiwata S, Ishida N, Misugaki M. Relationship of urinary pseudouridine and 1-methyladenosine to activity of leukemia and lymphoma. Clin Chim Acta 1992;206:181–9. [DOI] [PubMed] [Google Scholar]
- [106].Pane F, Savoia M, Fortunato G, Camera A, Rotoli B, Salvatore F, et al. Serum pseudouridine in the diagnosis of acute leukaemias and as a novel prognostic indicator in acute lymphoblastic leukaemia. Clin Biochem 1993;26:513–20. [DOI] [PubMed] [Google Scholar]
- [107].Bond A, Dudley E, Lemiere F, Tuytten R, El-Sharkawi S, Brenton AG, et al. Analysis of urinary nucleosides. V. Identification of urinary pyrimidine nucleosides by liquid chromatography/electrospray mass spectrometry. Rapid Commun Mass Spectrom 2006;20:137–50. [DOI] [PubMed] [Google Scholar]
- [108].Itoh K, Ishiwata S, Ishida N, Mizugaki M. Diagnostic use of anti-modified nucleoside monoclonal antibody. Tohoku J Exp Med 1992;168:329–31. [DOI] [PubMed] [Google Scholar]
- [109].Ke C, Hou Y, Zhang H, Fan L, Ge T, Guo B, et al. Large-scale profiling of metabolic dysregulation in ovarian cancer. Int J Cancer 2015;136:516–26. [DOI] [PubMed] [Google Scholar]
- [110].Zhang T, Wu X, Ke C, Yin M, Li Z, Fan L, et al. Identification of potential biomarkers for ovarian cancer by urinary metabolomic profiling. J Proteome Res 2013;12:505–12. [DOI] [PubMed] [Google Scholar]
- [111].Jiang T, Lin Y, Yin H, Wang S, Sun Q, Zhang P, et al. Correlation analysis of urine metabolites and clinical staging in patients with ovarian cancer. Int J Clin Exp Med 2015;8:18165–71. [PMC free article] [PubMed] [Google Scholar]


