Abstract
Background: MicroRNAs (miRNAs) are non-coding regulatory molecules 18-25 nucleotides in length that act as post-transcriptional regulators of gene expression. MiRNAs affect various biological processes including carcinogenesis. Deregulation of miRNAa expression has been described in a variety of tumors including papillary thyroid carcinoma (PTC). The aim of the present study was to investigate the role of selected miRNAs in PTC and find associations between miRNA expression and the BRAF (V600E) mutation. Materials and Methods: The study group comprised a total of 62 patients with surgically treated PTC. The control group consisted of 30 patients with nodular goitre that were surgically treated in the same time period. The expression status of miR-146b, miR-181a, miR-187, miR-221 and miR-222 was determined using quantitative real-time PCR. BRAF mutation analysis was performed by PCR with reverse hybridization. Results: MiR-146b, miR-181a, miR-187, miR-221 and miR-222 were up-regulated in PTC compared to normal thyroid gland tissue of the same patient. MiR-146b, miR-187, miR-221 and miR-222 were also up-regulated in PTC compared to nodular goitre. The recurrent tumors were statistically significantly associated with up-regulation of miR-221. The mutation V600E of BRAF gene was significantly associated with up-regulation of miR-146b and with down-regulation of miR-187. Conclusion: Over-expression of selected miRNAs in PTC compared to normal thyroid gland tissue and nodular goitre was found. Moreover, miR-221 may serve as a prognostic marker as its over-expression was significantly associated with recurrent tumors.
Keywords: MicroRNA, papillary thyroid cancer, BRAF mutation
Thyroid cancer is the most common malignant tumor of the endocrine system (1-2). The incidence of thyroid cancer has increased dramatically over last three decades (3). Papillary thyroid carcinoma (PTC) represents the majority of malignant thyroid tumors, accounting for approximately 80% of all thyroid cancers (4-5). Its overall prognosis is favourable (6).
In spite of this fact, some PTCs have aggressive biological behaviour characterized by local recurrence and dissemination (6). The most important clinical prognostic factors include tumor size, regional and distant metastases and extrathyroidal extension (4). Genetically, BRAF (V600E) mutation is the most common alteration in PTC (7). This mutation is significantly associated with extrathyroidal invasion, regional lymph node metastases, advanced TNM stages and recurrent disease (4).
MicroRNA (miRNA) deregulation is another important event in the pathogenesis of PTC (3,8). MiRNAs are non-coding regulatory molecules 18-25 nucleotides in length that act as post-transcriptional regulators of gene expression (9-10). MiRNAs affect various biological processes including carcinogenesis (11). Expression of miRNAs is tissue specific and varies between cancer and normal cells and also among different types of cancers (5,12-13). Deregulation of miRNAs expression has been described in a variety of tumors including pancreas, lung, breast and sinonasal tract (6,12-13). Alterations in miRNAs expression have been also described in thyroid tumors including PTC (14-18). The aim of the present study was to investigate the role of selected miRNAs in PTC and find some associations between miRNAs expression and the BRAF (V600E) mutation.
Materials and Methods
Patients. The study group comprised a total of 62 patients with PTC, treated from January 2009 to December 2014. All patients underwent surgery and the diagnosis of PTC was confirmed by microscopic examination. Patients were aged between 18-82 years (median=48 years), with a male to female ratio 1:3. A subgroup of 32 patients had recurrent disease (repeated surgery or repeated radioactive iodine ablation), the rest of the study group (30 patients) had non-recurrent disease. The control group consisted of 30 patients with nodular goitre, who were treated surgically in the same time period. The age range in the control group varied between 30 and 69 years (median=50 years). Male to female ratio in control group was 1:6. Studied miRNAs were miR-146b, miR-181a, miR-187, miR-221 and miR-222. These miRNAs were selected based on review of literature dealing with deregulation of miRNAs in PTC.
Total RNA isolation. Two to four 5-μm-thick sections were cut from FFPE tissue samples and deparaffinised using xylene and ethanol. Total RNA including miRNAs was isolated from FFPE tissue samples using RNeasy FFPE Kit (Qiagen, Hilden, Germany) according to the manufacturer’s protocol. The extracted RNA was ultimately eluted in 30 μl of RNase free water. Concentration and purity of the isolated RNA was determined by NanoDrop ND 1000 spectrophotometer (Thermo Fisher Scientific, Wilmington, DE, USA) by measuring the optical density at 260 nm and 280 nm (A260/280 ratio). After the isolation, the samples were immediately processed or stored at –80˚C.
Quantitative real-time PCR of microRNAs. The synthesis of cDNA was done using TaqMan® Advanced miRNA cDNA Synthesis Kit with universal reverse transcription primers (Applied Biosystems, Foster City, CA, USA) according to the manufacturer’s protocol with 8-10 ng of total RNA in the reaction. Real time PCR was done with TaqMan® Fast Advanced Master Mix (Applied Biosystems) and specific TaqMan® Advanced miRNA Assays (Applied Biosystems) on Rotor Gene Q (Qiagen). Assays for hsa-miR-146b-5p, hsa-miR-181a-5p, hsa-miR-187-3p, hsa-miR-221-3p, has-miR-222-3p and has-miR-361-5p (endogenous control) were used. All steps were performed following the manufacturer’s protocol. All reactions were performed in triplicates and the reaction volume was 10 μl with 2.5 μl of the sample. The reaction conditions were set according to the manufacturer’s protocol and it involved enzyme activation at 95˚C for 20 s followed by 40 cycles of denaturation at 95˚C for 3 s and annealing/extension at 60˚C for 30 s. Fluorescence data were analyzed in the Rotor Gene Q Series Software. Relative expression of each miRNA was determined using the 2ΔΔCt method (19) with expression levels of miR-361 for data normalization. This workflow was chosen based on literature review and manufacturer’s recommendation for endogenous controls listed in the user guide for TaqMan® Advanced miRNA Assays.
Determination of BRAF mutation. DNA from the paraffin-embedded tissue was extracted with the MagCore Genomic DNA FFPE One-Step Kit (RBC Bioscience, Taiwan) according to the manufacturer’s protocol. The amount of extracted DNA was measured with Qubit® dsDNA BR Assay Kit (Invitrogen). BRAF mutation analysis was performed by PCR with reverse hybridization with BRAF 600/601 StripAssay (ViennaLab). The test detects mutations V600A, V600D, V600E (c.1799T>A), V600E (c. 1799_1800 TG>AA), V600G, V600K, V600M, V600R, K601E. The analytical sensitivity given by manufacturer is the detection of 1% mutant alleles on wild-type background.
Statistical analysis. All statistical analyses were performed using data analysis software system STATISTICA version 13 (TIBCO Software Inc., Tulsa, OK, USA). MiRNA expression values were log transformed to a normal distribution of data for parametric tests. Student’s t-test was used to compare level of expression of miRNAs in tumor and control samples. The null hypothesis was based on theory that there was no difference between expression levels of studied mirRNAs between tumor samples and control samples. All tests were two tailed and p<0.05 was considered statistically significant. All data were reported as mean±standard error of the mean (SEM).
Ethical approval. All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and national research committee and with the 1964 Helsinki declaration. Experimental protocol was approved by a named institutional ethical committee (Ethical committee University Hospital Hradec Králové), reference number 201711S09P.
Results
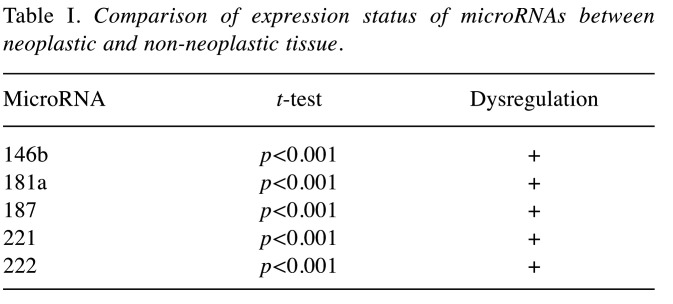
Comparison of expression status of microRNAs between neoplastic and non-neoplastic tissue. The comparison of the expression value of the studied miRNAs in PTC and in normal thyroid gland tissue of the same patient confirmed statistically significant differences: all five studied miRNAs (miR-146b, miR-181a, miR-187, miR-221, miR-222) were up-regulated in PTC tumor tissue (Table I).
Table I. Comparison of expression status of microRNAs between neoplastic and non-neoplastic tissue.
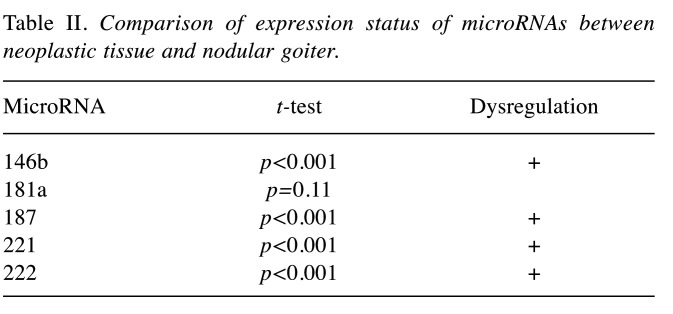
Comparison of expression status of microRNAs between neoplastic tissue and nodular goitre. The comparison of the expression value of the studied miRNAs in PTC and in nodular goitre (control group) proved to be statistically significant up-regulation of miR-146b, miR-187, miR-221 and miR-222 in PTC tumor tissue (Table II). The only exception was expression value of miR-181a without significant difference between the studied tissues.
Table II. Comparison of expression status of microRNAs between neoplastic tissue and nodular goiter.
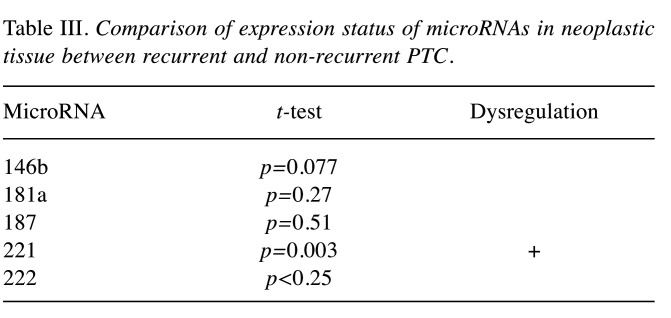
Comparison of expression status of microRNAs in neoplastic tissue between recurrent and non-recurrent PTC. The comparison of the expression value of the studied miRNAs in recurrent and non-recurrent PTC revealed the only statistically significant difference in miR-221 expression value, which was up-regulated in recurrent tumors (Table III).
Table III. Comparison of expression status of microRNAs in neoplastic tissue between recurrent and non-recurrent PTC.
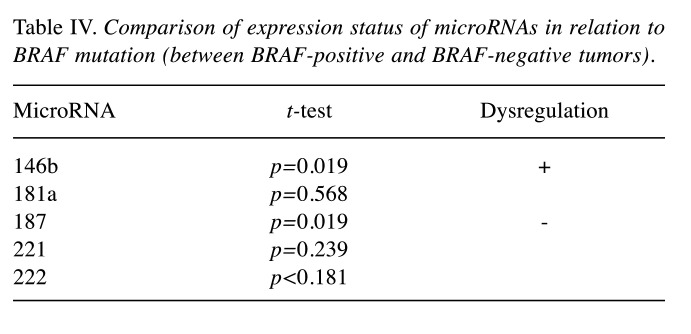
Comparison of expression status of microRNAs in relation to BRAF mutation. The mutation V600E of BRAF gene was significantly associated with up-regulation of miR-146b and down-regulation of miR-187 (Table IV).
Table IV. Comparison of expression status of microRNAs in relation to BRAF mutation (between BRAF-positive and BRAF-negative tumors).
Discussion
Micro-RNAs in papillary thyroid cancer. Nowadays, there is an increasing interest in the role of miRNAs in physiological and pathological cell processes which bring new insights into understanding cancer pathobiology (9). MiRNAs are 20 to 22 nucleotides long and bind to complementary messenger RNA. Their main function is to regulate gene expression at the post-transcriptional level. MiRNAs play an important role in proliferation, apoptosis, differentiation of cells and also in cancer formation. The development and progression of malignant tumors is influenced by deregulation of miRNA expression (20). Therefore, expression of micro-RNAs differs between PTC, benign thyroid lesions and normal thyroid tissue. Several studies have demonstrated over-expression of selected miRNAs in PTC, particularly miR-146b, mir-221, miR-222, miR-181a, miR-181b, miR-187, miR-155, miR-224 or miR-197 are most consistently over-expressed in well differentiated thyroid cancer (11,20). However, the miRNA profile varied significantly between individual types of tumor (20). Chen et al. (11) compared the expression of miR-146b, miR-221 and miR-222 in PTC with normal thyroid, hyperplastic nodules, follicular adenomas and follicular carcinomas. Mir-146b was significantly over-expressed and also MiR-221 and miR-222 showed higher expression in PTC. The authors suggested that miR-146b can be used as a marker in diagnostics of PTC in both histopathology and fine needle aspiration specimens. In our study, miR-146b, miR-181a, miR-187, miR-221 and miR-222 were up-regulated in PTC compared to normal thyroid gland tissue of the same patient. MiR-146b, miR-187, miR-221 and miR-222 were also up-regulated in PTC compared to benign nodular goitre.
MicroRNAs as a prognostic marker in PTC. Several studies have already reported that over-expression of selected miRNAs can play an important role in PTC progression and some miRNAs can also be used as a prognostic marker (4,8,21-22). Some authors also reported that deregulation of selected miRNAs can be associated with size and extension of primary tumor and presence of regional neck metastases. MiRNAs, most often described as a possible marker of tumor progression and prognosis are miR-146b, miR-222, miR-221 and miR-718. The deregulation of miR-222 and miR-146b is commonly associated with cancer recurrence (8). Chou et al. (4) demonstrated that over-expression of miR-146b may be associated with extrathyroidal extension and advanced stages of PTC. Also Yip et al. (6) concluded that miR-146b, miR-222, miR-34b, miR-130b are up-regulated in more aggressive cases of PTC. The author also described an association between over-expression of miR-146b and aggressive behaviour of BRAF-positive tumors. In our study, miR-221 was significantly up-regulated in recurrent tumors.
The relationship between microRNAs and BRAF mutation. RAF proteins are serine-threonine protein kinases. The family of RAF proteins has three isoforms: ARaf, BRaf and CRaf. These proteins have an important function in cellular proliferation, differentiation and gene expression. BRaf is the predominant isoform in thyroid cells. BRAF mutations occur in 15-30% of all cancers and are most frequent in carcinoma of thyroid gland, especially in PTC (7,20). BRAF mutation represents a single amino acid substitution of valine by glutamate at position 600. As previously reported, this mutation is often associated with advanced TNM stages of PTC, extrathyroidal extension of tumor, metastatic involvement of regional lymph nodes and also recurrent disease (4). Some authors also suggested that BRAF-positive PTCs are associated with a higher expression level of miR-146b than BRAF wild-type tumors (4). Moreover, over-expression of miR-146b among BRAF-positive tumors is associated with aggressive behaviour (6). Also, in our present study, the mutation V600E of BRAF gene was significantly associated with up-regulation of miR-146b. Another interesting result of our study was down-regulation of miR-187 in BRAF-positive tumors.
Conclusion
In summary, we demonstrated over-expression of miR-146b, miR-181a, miR-187, miR-221 and miR-222 in PTC compared to normal thyroid gland tissue and over-expression of miR-146b, miR-187, miR-221 and miR-222 compared PTC to nodular goitre. Moreover, miR-221 may serve as a prognostic marker as its over-expression was significantly associated with recurrent tumors. BRAF-positive tumors were significantly associated with up-regulation of miR-146b and with down-regulation of miR-187.
Conflicts of Interest
The Authors declared no potential conflicts of interest regarding this study.
Authors’ Contributions
P.C. and V.CH. wrote the main manuscript. H.K. and M.CH. performed Quantitative real-time PCR of microRNAs and statistical analysis. J.L., A.R. and H.V. were responsible for determination of BRAF mutation
Acknowledgements
This article was supported by the project (Ministry of Health, Czech Republic) for conceptual development of research organization 00179906 (research number 8158)
References
- 1.Braun J, Hoang-Vu C, Dralle H, Huttelmaier S. Downregulation of microRNAs directs the EMT and invasive potential of anaplastic thyroid carcinomas. Oncogene. 2010;29:4237–4244. doi: 10.1038/onc.2010.169. [DOI] [PubMed] [Google Scholar]
- 2.Keutgen XM, Filicori F, Crowley MJ, Wang Y, Scognamiglio T, Hoda R, Buitrago D, Cooper D, Zeiger MA, Zarnegar R, Elemento O, Fahey TJ. A Panel of four miRNAs accurately differentiates malignant from benign indeterminate thyroid lesions on fine needle aspiration. Clin Cancer Res. 2012;18:2032–2038. doi: 10.1158/1078-0432.CCR-11-2487. [DOI] [PubMed] [Google Scholar]
- 3.Lee JC, Gundara JS, Glover A, Serpell J, Sindhu SB. MicroRNA expression profiles in the management of papillary thyroid cancer. Oncologist. 2014;19:1141–1147. doi: 10.1634/theoncologist.2014-0135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chou CK, Liu RT, Kang HY. MicroRNA-146b: A novel biomarker and therapeutic target for human papillary thyroid cancer. Int J Mol Sci. 2017;18:636–645. doi: 10.3390/ijms18030636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.He H, Jazdzewski K, Li W, Liyanarachchi S, Nagy R, Volinia S, Calin GA, Liu C, Franssila K, Sester S, Kloos RT, Croce CM, de la Chapelle A. The role of microRNA genes in papillary thyroid carcinoma. PNAS. 2005;102:19075–19080. doi: 10.1073/pnas.0509603102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yip L, Kelly L, Shuai Y, Armstrong MJ, Nikiforov YE, Carty SE, Nikiforova MN. MicroRNA signature distinguishes the degree of aggressiveness of papillary thyroid carcinoma. Ann Surg Oncol. 2011;18:2035–2041. doi: 10.1245/s10434-011-1733-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang Y, Liao D, Pan L, Ye R, Li X, Wang S, Ye C, Chen L. Expressions of miRNAs in papillary thyroid carcinoma and their associations with BRAFV600E mutation. Eur J Endocrinol. 2013;168:675–681. doi: 10.1530/EJE-12-1029. [DOI] [PubMed] [Google Scholar]
- 8.Chruscik A, Lam AK. Clinical pathological impact of micro-RNAs in papillary thyroid carcinoma: A crucial review. Exp Mol Pathol. 2015;99:393–398. doi: 10.1016/j.yexmp.2015.08.013. [DOI] [PubMed] [Google Scholar]
- 9.Kalfert D, Pesta M, Kulda V, Topolcan O, Ryska A, Celakovky P, Ludvikova M. MicroRNA profile in site – specific head and neck squamous cell cancer. Anticancer Res. 2015;35:2455–2463. [PubMed] [Google Scholar]
- 10.Visone R, Russo L, Pallante P, De Martino I, Ferraro A, Leone V, Bonbone E, Petrocca F, Alder H, Croce CM, Fusco A. MicroRNAs (miR)-221 and miR-222, both overexpressed in human thyroid papillary carcinomas, regulate p27Kip1 protein levels and cell cycle. Endocrin Rel Cancer. 2007;14:791–798. doi: 10.1677/ERC-07-0129. [DOI] [PubMed] [Google Scholar]
- 11.Chen YT, Kitabayashi N, Zhou XK, Fandy TJ, Scognamiglio T. MicroRNA analysis as a potential diagnostic tool for papillary thyroid carcinoma. Modern Pathol. 2008;21:1139–1146. doi: 10.1038/modpathol.2008.105. [DOI] [PubMed] [Google Scholar]
- 12.Kovarikova H, Bubancova I, Laco J, Sieglova K, Vosmikova H, Dundr P, Nemejcova K, Michalek J, Vosmik M, Palicka V, Chmelarova M. Deregulation of selected microRNAs in sinonasal carcinoma: value of miR 21 as prognostic biomarker in sinonasal squamous cell carcinoma. Head Neck. 2017;39:2528–2536. doi: 10.1002/hed.24930. [DOI] [PubMed] [Google Scholar]
- 13.Laco J, Kovarikova H, Chmelarova M, Vosmikova H, Sieglova K, Bubancova I, Dundr P, Nemejcova K, Michalek J, Celakovsky P, Mottl R, Sirak I, Vosmik M, Marek I, Geryk T, Mejzlik J, Satankova J, Ryška A. Analysis of DNA methylation and microRNA expression in NUT (nuclear protein in testis) midline carcinoma of the sinonasal tract: a clinicopathological, immunohistochemical and molecular genetic study. Neoplasma. 2018;65:113–123. doi: 10.4149/neo_161122N581. [DOI] [PubMed] [Google Scholar]
- 14.Ludvikova M, Kalfert D, Kholova I. Pathobiology of microRNAs and their emerging role in thyroid fine-needle aspiration. Acta Cytol. 2015;59:435–444. doi: 10.1159/000442145. [DOI] [PubMed] [Google Scholar]
- 15.Ludvikova M, Kholova I, Kalfert D. Molecular aspects of thyroid tumors with emphasis on MicroRNA and their clinical implications. Clin Onkol. 2017;30:167–174. doi: 10.14735/amko2017167. [DOI] [PubMed] [Google Scholar]
- 16.Pallante P, Visone R, Ferracin M, Ferraro A, Berlingieri MT, Troncone G, Chiappetta G, Liu CG, Santoro M, Negrini M, Croce M, Fusco A. MicroRNA deregulation in human thyroid papillary carcinomas. Endocrin Rel Cancer. 2006;13:497–508. doi: 10.1677/erc.1.01209. [DOI] [PubMed] [Google Scholar]
- 17.Nikiforova MN, Tseng GC, Steward D, Diorit D, Nikiforov YE. MicroRNA expression profiling of thyroid tumors: Biological significance and diagnostic utility. J Clin Endocrinol Metab. 2008;93:1600–1608. doi: 10.1210/jc.2007-2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang Y, Yu S, Cao S, Yin Y, Hong A, Guan H, Li Y, Xiao A. MicroRNA-222 promotes invasion and metastasis of papillary thyroid cancer through targeting protein phosphatase 2 regulatory subunit B alpha expression. Thyroid. 2018;28:1162–1173. doi: 10.1089/thy.2017.0665. [DOI] [PubMed] [Google Scholar]
- 19.Schmittgen TD, Lee EJ, Jiang J, Sarkar A, Yang L, Elton TS, Chen C. Real-time PCR quantification of precursor and mature microRNA. Methods. 2008;44:31–38. doi: 10.1016/j.ymeth.2007.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.D’cruz AK, Vaish R, Vaidya A, Nixon IJ, Williams MD, Poorten VV, López F, Angelos P, Shaha AR, Khafif A, Skalova A, Rinaldo A, Hunt JL, Ferlito A. Molecular markers in well-differentiated thyroid cancer. Eur Arch Oto-Rhino-Laryngol. 2018;275:1375–1384. doi: 10.1007/s00405-018-4944-1. [DOI] [PubMed] [Google Scholar]
- 21.Zembska A, Jawiarczyk-Przybylowska A, Wojtczak B, Bolanowski M. MicroRNA expression in the progression and aggressiveness of papillary thyroid carcinoma. Anticancer Res. 2019;39:33–40. doi: 10.21873/anticanres.13077. [DOI] [PubMed] [Google Scholar]
- 22.Wang X, Qi M. miR-718 is involved in malignancy of papillary thyroid cancer through repression of PDPK1. Pathol Res Pract. 2018;214:1787–1793. doi: 10.1016/j.prp.2018.08.022. [DOI] [PubMed] [Google Scholar]