Abstract
Background/aim: The current study aimed to identify biomarkers for differentiating between patients with oral cancer (OC) and healthy controls (HCs) on the basis of the comprehensive proteomic analyses of saliva samples by using liquid chromatography-mass spectrometry (LC-MS/MS). Patients and Methods: Unstimulated saliva samples were collected from 39 patients with OC and from 31 HCs. Proteins in the saliva were comprehensively analyzed using LC-MS/MS. To differentiate between patients with OC and HCs, a multiple logistic regression model was developed for evaluating the discriminatory ability of a combination of multiple markers. Results: A total of 23 proteins were significantly differentially expressed between the patients with OC and the HCs. Six out of the 23 proteins, namely α-2-macroglobulin-like protein 1, cornulin, hemoglobin subunit β, Ig ĸ chain V-II region Vĸ167, kininogen-1 and transmembrane protease serine 11D, were selected using the forward-selection method and applied to the multiple logistic regression model. The area under the curve for discriminating between patients with OC and HCs was 0.957 when the combination of the six metabolites was used (95% confidence interval=0.915-0.998; p<0.001). Furthermore, these candidate proteins did not show a stage-specific difference. Conclusion: The results of the current study showed that six salivary proteins are potential non-invasive biomarkers for OC screening.
Keywords: Proteomics, oral cancer, screening, saliva
Oral cancer (OC) is among the 15 most common types of cancer worldwide, with an incidence of 500,550 in 2018 (1); in addition, the incidence of OC has been steadily increasing during recent decades (2). The curability of OC is high if it is detected at an early stage; the 5-year survival rate of patients with OC detected at an early stage generally exceeds 80% (3). However, the curability of advanced OC is not high, and the 5-year survival rate of patients with OC at the late stage is generally below 50% (2-5). In addition to the lower survival rate, a reduced quality of life after surgery for OC is problematic. The oral cavity is an important organ for the functions of speech, swallowing, and chewing (6). Moreover, the oral cavity is a part of the face. Highly invasive surgery that is usually performed for advanced-stage OC can lead to oral dysfunction and cosmetic disfiguration (6). Therefore, it is important to detect OC at an earlier stage to the maximum extent possible. As the oral cavity is a commonly examined area, many physicians assume that OC is easy to detect. However, several lesions mimic OC, such as intractable stomatitis, bite wounds, and periodontitis, because of which accurate detection of OC is still difficult (7-9), leading to a delay in the detection of OC in early stages (10)
A conventional visual and tactile examination (CVTE) is the most common method for the screening of OC (11,12); however, physicians need sufficient experience to ensure that the results of the CVTE are highly accurate (13). Therefore, the CVTE is not a suitable high-precision method for OC screening for clinicians who are not specialists in oral surgery. Consequently, several light-based detection systems based on the optical properties of biological tissues, which have enhanced oral mucosal examinations and facilitated the detection of OC, have become commonly used as adjunctive screening aids for OC screening in recent years (4); however, there is limited evidence regarding the effectiveness of OC screening with such light-based detections systems (4,11). Therefore, a CVTE followed by open biopsy of suspicious lesions remains the gold standard for OC detection (11). Thus, the development of novel screening methods for OC is urgently needed.
Saliva has recently been evaluated as a notable biofluid for the detection of diseases (14,15). Saliva is a biological fluid comprising >99% water and <1% proteins, electrolytes, and other low-molecular-weight components (15,16), and saliva reflects systemic physiological conditions (17). Furthermore, several diseases can be detected using saliva specimens, such as cancer, cardiovascular disease, neurological disease, and metabolic disease (7,9,18-21). Saliva is an excellent source of biomarkers, as obtaining saliva is non-invasive, convenient, and safe. Compared to blood, saliva can be collected without any pain and risk of needle-stick injury. Therefore, the number of studies regarding the use of saliva for disease detection has been increasing.
Since the first report of using saliva as a diagnostic tool for OC detection was published in 2000 by Liao et al. (17,22), salivary genomics, transcriptomics, proteomics, metabolomics, and microbiomics have been investigated for biomarkers for OC detection (7,9,17,23). Salivary proteomics for biomarkers of OC detection is among the most popular fields, and various candidate proteomic biomarkers, such as interleukin (IL)-1, IL6, IL8, tumor necrosis factor α, and catalase have been reported (17,24-26). Furthermore, owing to recent innovative advancements in quantitative mass spectrometry (MS) technologies, comprehensive proteomic approaches can be performed (18,27). Therefore, we performed a comprehensive proteomic approach using liquid chromatography (LC)-MS/MS for salivary biomarkers for OC screening. To the best of our knowledge, only a few studies have performed comprehensive proteomic approaches for salivary biomarkers for OC screening. The current study aimed to identify salivary proteomic biomarkers for differentiating between patients with OC and healthy controls (HCs) on the basis of the comprehensive proteomic analyses of saliva samples using LC-MS/MS.
Patients and Methods
All procedures performed in the studies involving human participants were conducted in accordance with the ethical standards of the institutional and/or national research committee and 1964 Helsinki declaration and its later amendments or comparable ethical standards. The study protocol was approved by the Ethics Committee of Yamagata University (2015-288, 2017-18, 2017-180, 2017-182, 2018-106, and 2018-455). Written informed consent was obtained from all participants prior to sample collection. Patients with OC were recruited from the Department of Dentistry, Oral and Maxillofacial surgery, Yamagata University Hospital between April 2016 and March 2019. Only patients who had not received prior adjuvant therapy, such as chemotherapy or radiotherapy, were enrolled. The pathological diagnosis was obtained for all patients with OC via incisional open biopsy. HCs included individuals who underwent comprehensive medical examinations at Yamagata University Hospital between December 2017 and February 2019 and who had no health issues considering the results of these examinations. We collected saliva from patients with OC and from HCs after obtaining informed consent.
Saliva collection and sample preparation. A dentist or dental hygienist confirmed the oral hygiene of all the subjects before saliva collection. Dental plaque and calculus deposits, if remarkable, were removed by using a toothbrush without dentifrice and ultrasonic scaling at least 3 hours before saliva collection. All the participants were asked to refrain from eating and drinking for at least 1.5 h prior to saliva collection. Participants rinsed their mouths with water immediately before sample collection and were instructed to spit their saliva into 50-cc Falcon tubes (Corning, Inc., Corning, NY, USA) in a paper cup filled with crushed ice. Approximately 3 ml unstimulated whole saliva was collected over 5-10 min. The collected samples were centrifuged at 2,600 × g for 15 min at 4˚C. The supernatants were immediately treated with a protease inhibitor mixture. The samples were aliquoted into smaller volumes and stored at −80˚C.
Proteomic analysis of saliva. The preparation of clean peptide mixtures from saliva for MS analysis was performed following the protocol of the preparation kit (Pierce™ Mass Spec Sample Prep Kit for Cultured Cells; ThermoFisher Scientific, Waltham, MA, USA) of LC-MS/MS analysis. For sample lysis, lysis buffer was added to 20 μl of saliva, and the lysate was incubated for 5 min at 95˚C. After incubation, the lysate was centrifuged at 16,000 × g for 10 min at 4˚C. A total of 0.5 μg (0.5% w/w) of the internal control indicator (Pierce™ Digestion Indicator for Mass Spectrometry; ThermoFisher Scientific) was added to the supernatant. Samples were incubated with dithiothreitol for 45 min at 50˚C. After the incubation, samples were further incubated with iodoacetamide for 20 minutes at room temperature, followed by acetone washing. After washing the samples with acetone, the acetone-precipitated protein pellet was resuspended with a digestion buffer containing the Lys-C protease and then the samples were incubated for 2 h at 37˚C. After further overnight incubation with trypsin protease at 37˚C, the samples were stored at −80˚C. On the day of MS analysis, samples were first cleaned-up using a C18-spin column (Pierce™ C18 Spin Columns; ThermoFisher Scientific) and resolved with 0.1% (v/v) formic acid/water. A nanoLC system (EASY-nLC 1000; ThermoFisher Scientific) with C18 analytical column (NTCC-360/75-3-125; Nikkyo Technos, Tokyo, Japan) were used to separate peptides with a gradient of 100% acetonitrile/0.1% formic acid for 75 min (linear gradient at a flow rate 300 nl/min: 0 to 32.0% acetonitrile by 60 min, 95.0% by 65 min). The samples were analyzed using a hybrid quadrupole-Orbitrap MS system (Q-Exactive; ThermoFisher Scientific). For data analysis, Proteome Discoverer software (ThermoFisher Scientific) was used to search for MS/MS spectra against the UniProt human database using the SEQUEST search engine with a 1% false-discovery rate. To assess the digestion efficiency, a Digestion Indicator protein sequence was included in the protein database. The raw data were processed using the Mascot database search engines (Matrix Science, London, UK). Furthermore, the estimated absolute protein amount in proteomics was calculated as the Exponentially Modified Protein Abundance Index (emPAI) from the protein identification results obtained via the database search engines of Mascot (28). The emPAI is highly correlated with the actual protein amount in a complex mixture and can be routinely used for reporting the approximate absolute protein abundance in a large-scale analysis (28-30).
Statistical analyses. To determine the discriminatory ability of salivary proteins between patients with OC and HCs, a multiple logistic regression (MLR) model was developed. Firstly, proteins detected in >50% of individuals in at least one group were selected. Secondly, proteins found at levels with differences of p<0.05 (Mann–Whitney U-test) between patients with OC and HCs were selected. Thirdly, an MLR model was developed using the forward feature-selection method. The predictive performance of the multivariate model was evaluated using the value of the area under the receiver operating characteristic curve (AUC). Statistical analyses were performed with SPSS software, version 20 (IBM, Armonk, NY, USA).
Results
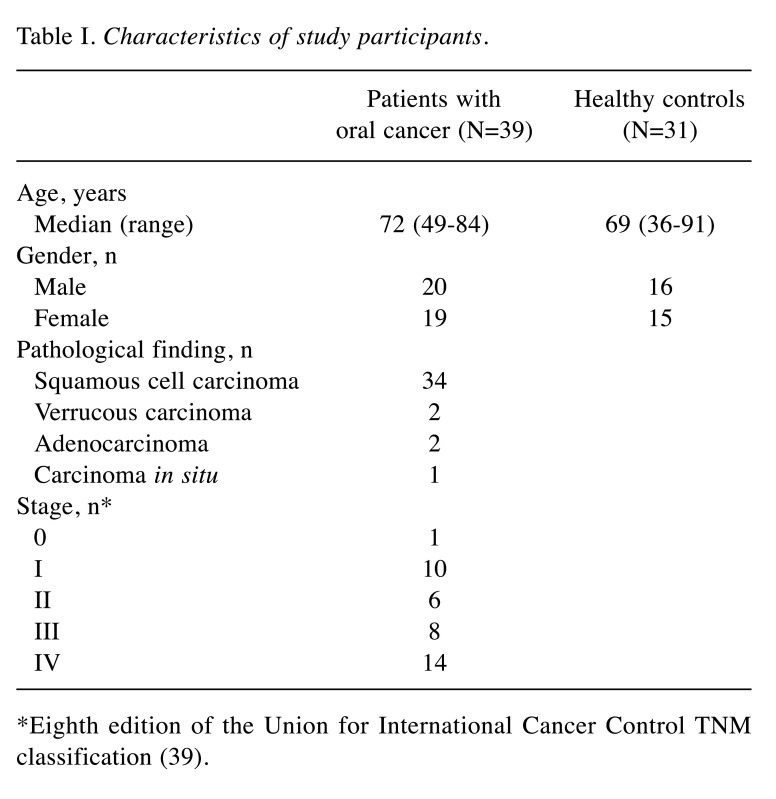
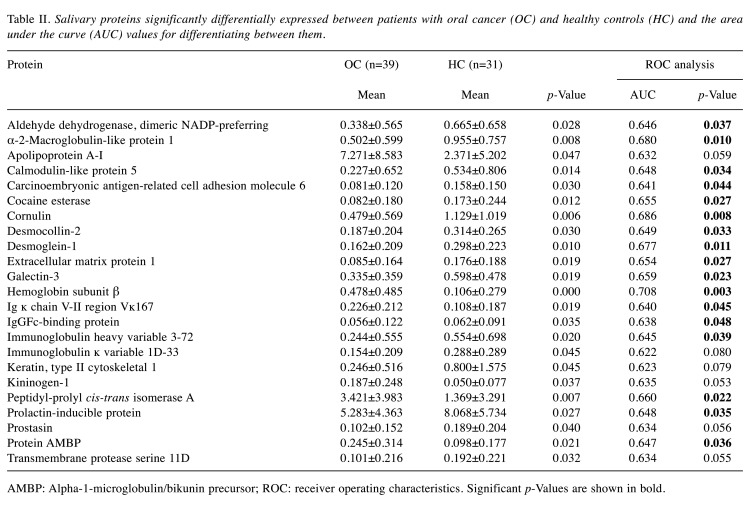
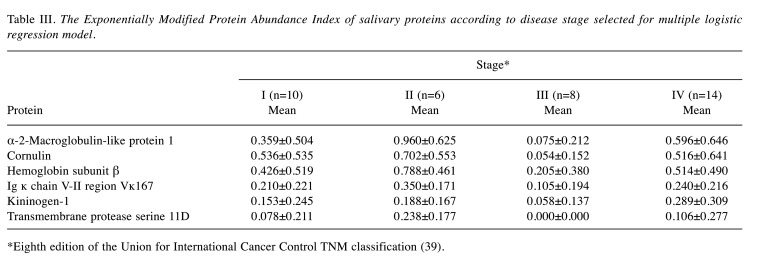
Table I shows the characteristics of the participants with respect to age and sex for both groups, as well as the pathological findings and staging for the OC group. Table II shows the expression of salivary proteins significantly differentially expressed between the OC and HC groups, and the AUC value for differentiating between patients with OC and HCs. Twenty-three proteins were significantly differentially expressed between the two groups. Six proteins, namely α-2-macroglobulin-like protein 1, cornulin, hemoglobin subunit β, Ig ĸ chain V-II region Vĸ167, kininogen-1 and transmembrane protease serine 11D, were selected using the forward selection method from among the significantly differentially expressed metabolites and were used for the MLR model owing to their discriminatory ability. Figure 1 shows the receiver operating characteristics curves and AUC value of the MLR model, respectively, for differentiating between patients with OC and HCs. The AUC for differentiating between patients with OC and HCs was 0.957 when this combination of six metabolites was used (95% confidence interval=0.915-0.998; p<0.001). Table III shows the comparisons of the emPAI of the proteins that were selected for the MLR model according to the OC stage, except stage 0. The candidate proteins did not show any stage-specific difference for differentiating between patients with OC and HCs.
Table I. Characteristics of study participants.
*Eighth edition of the Union for International Cancer Control TNM classification (39)
Table II. Salivary proteins significantly differentially expressed between patients with oral cancer (OC) and healthy controls (HC) and the area under the curve (AUC) values for differentiating between them.
AMBP: Alpha-1-microglobulin/bikunin precursor; ROC: receiver operating characteristics. Significant p-Values are shown in bold
Figure 1. Receiver operating characteristic curve of multiple logistic regression model using α-2-macroglobulin-like protein 1, cornulin, hemoglobin subunit β, Ig ĸ chain V-II region Vĸ167, kininogen-1 and transmembrane protease serine 11D for differentiating between patients with oral cancer and healthy controls. AUC: Area under the curve; CI: confidence interval.
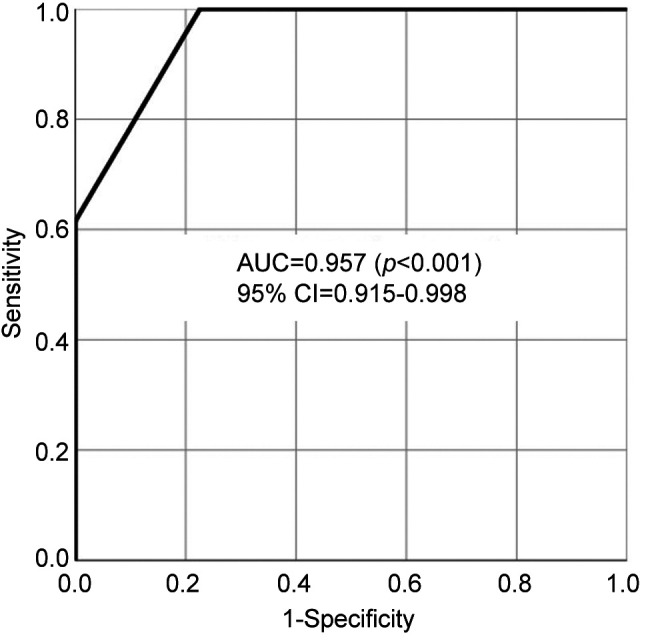
Table III. The Exponentially Modified Protein Abundance Index of salivary proteins according to disease stage selected for multiple logistic regression model.
*Eighth edition of the Union for International Cancer Control TNM classification (39)
Discussion
In the present study, we revealed that 23 salivary proteins were significantly differentially expressed between patients with OC and HCs using LC-MS/MS. Furthermore, we revealed that the combination of α-2-macroglobulin-like protein 1, cornulin, hemoglobin subunit β, Ig ĸ chain V-II region Vĸ167, kininogen-1 and transmembrane protease serine 11D has high accuracy for differentiating between patients with OC and HCs.
Most cases of OC are found by general dental practitioners, who are not specialists like general oral surgeons. Furthermore, most cases of OC are definitively diagnosed at a late stage (31). Moreover, it is not easy to differentiate between patients with OC and HCs by using a CVTE, especially for physicians who are not specialists in oral surgery. Accordingly, the salivary biomarkers identified in the current study appear to be good candidates for differentiating between patients with OC and HCs non-invasively and easily. These results have the potential to help clinicians to detect OC, especially those who are not specialists in oral surgery.
In the present study, six proteins, namely α-2-macroglobulin-like protein 1, cornulin, hemoglobin subunit β, Ig ĸ chain V-II region Vĸ167, kininogen-1 and transmembrane protease serine 11D, were selected for the MLR model to differentiate between patients with OC and HCs. Of these, cornulin is already a potential salivary proteomic biomarker for differentiating between patients with OC and HCs (32). Cornulin is expressed during epidermal differentiation and engaged in different biological processes, such as cell communication, cell cycle, cellular processes, immune system processes, and response to stimulus (32,33). On proteomic analysis of OC tissue, dysplastic epithelial tissue, and normal epithelial tissue, cornulin was the most down-regulated protein in epithelial dysplasia and OC tissue samples (32,34). Furthermore, salivary cornulin was lower in their OC group than in the HC group, and had a high AUC (of 0.91) for differentiating between patients with OC and the HCs, although the sample size was small (32). In the present study, salivary cornulin was significantly lower in patients with OC than in HCs, similar to the results of the previous studies.
We compared the emPAI of the proteins that were selected for the MLR model according to the OC stage, except stage 0, and we observed that our candidate biomarkers did not show a stage-specific difference. It is desirable for OC screening biomarkers to detect OC at an early stage, such as stage I. OC at an early stage is often asymptomatic, in contrast, at late stages OC causes evident symptoms such as pain, bleeding, and malodor. Therefore, it is not difficult to detect OC at late stages on a CVTE. Furthermore, the prognosis of OC at late stages is evidently poor (2-4). Therefore, screening biomarkers for detecting late-stage OC is not of great significance. Serum biomarkers such as squamous cell carcinoma antigen (SCC antigen) for oral squamous cell carcinoma, the most common pathology of OC, have already been used clinically. However, the level of SCC antigen is elevated, especially in later-stage OC (7,35,36). Therefore, although used practically for the diagnosis of OC, SCC antigen is not useful for the screening for OC (7). The candidate salivary biomarkers in the current study did not show any clear differences among the various stages. These findings indicate that these proteomic biomarkers should be clinically useful for screening patients with OC.
This study had several limitations. The first limitation was the small sample size. The second limitation pertains to the conditions of saliva collection. The collection time after meals affects the levels of salivary metabolites for OC screening (37). In that previous study, a longer fasting time after a meal was better for using salivary metabolites for differentiating between patients with OC and HCs (37). The levels of proteins in saliva are also affected by conditions such as fasting, smoking, the circadian and circannual cycles, use of medications, emotional state, size of the salivary glands, body weight, level of exercise, alcohol consumption, presence of systemic disease, nutritional status, presence of nausea, age, and sex (38). In the present study, all the participants were asked to refrain from eating and drinking for at least 1.5 hours prior to saliva collection; however, there was no strict fasting time such as fasting more than 1.5 hours. Consequently, in future studies, strict conditions might be necessary regarding the saliva collection time. The third limitation is the heterogeneity of the samples, especially owing to the pathological diagnosis. The OC samples included not only oral squamous cell carcinoma but also verrucous carcinoma, adenocarcinoma, and carcinoma in situ. To the best of our knowledge, no previous studies compared the differences in salivary proteins between oral squamous cell carcinoma and other pathological types of OC. Thus, future studies might need to include patients with only one type of OC.
In conclusion, we performed a comprehensive proteomic approach of saliva for identifying biomarkers for the screening of OC. The results of the study revealed that the proteomic profile in saliva was evidently different between patients with OC and HCs. Furthermore, we revealed that a combination of 6 proteins has a high accuracy for differentiating between patients with OC and HCs. These salivary proteins have the potential for non-invasive screening to differentiate between patients with OC and HCs.
Conflicts of Interest
No potential conflicts of interest were disclosed.
Authors’ Contributions
S.I., K.K., A.S., S.U. and K.E. collected the saliva samples. S.I. and A.T. conducted proteomic analysis. S.I., K.I. and M.I. designed the study. S.I. and H.K. conducted the statistical analysis. S.I. wrote the main article and prepared all tables and the figure. All Authors reviewed and edited the manuscript.
Acknowledgements
The Authors thank all the patients who provided the samples as well as Editage for editing our article. This work was supported by grants from YU-COE(C) from Yamagata University and the Ministry of Education, Culture, Sports, Science and Technology (MEXT) KAKENHI (16K11742, 17K11897, and 19K10304).
References
- 1.Peres MA, Macpherson LMD, Weyant RJ, Daly B, Venturelli R, Mathur MR, Listl S, Celeste RK, Guarnizo-Herreno CC, Kearns C, Benzian H, Allison P, Watt RG. Oral diseases: A global public health challenge. Lancet. 2019;394(10194):249–260. doi: 10.1016/S0140-6736(19)31146-8. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70(1):7–30. doi: 10.3322/caac.21590. [DOI] [PubMed] [Google Scholar]
- 3.Epstein JB, Zhang L, Rosin M. Advances in the diagnosis of oral premalignant and malignant lesions. J Can Dent Assoc. 2002;68(10):617–621. [PubMed] [Google Scholar]
- 4.Mascitti M, Orsini G, Tosco V, Monterubbianesi R, Balercia A, Putignano A, Procaccini M, Santarelli A. An overview on current non-invasive diagnostic devices in oral oncology. Front Physiol. 2018;9:1510. doi: 10.3389/fphys.2018.01510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li Q, Ouyang X, Chen J, Zhang P, Feng Y. A review on salivary proteomics for oral cancer screening. Curr Issues Mol Biol. 2020;37:47–56. doi: 10.21775/cimb.037.047. [DOI] [PubMed] [Google Scholar]
- 6.Chandu A, Sun KCV, DeSilva RN, Smith ACH. The assessment of quality of life in patients who have undergone surgery for oral cancer: A preliminary report. J Oral Maxillofac Surg. 2005;63(11):1606–1612. doi: 10.1016/j.joms.2005.07.012. [DOI] [PubMed] [Google Scholar]
- 7.Ishikawa S, Sugimoto M, Kitabatake K, Sugano A, Nakamura M, Kaneko M, Ota S, Hiwatari K, Enomoto A, Soga T, Tomita M, Iino M. Identification of salivary metabolomic biomarkers for oral cancer screening. Scientific Reports. 2016;6 doi: 10.1038/srep31520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ishikawa S, Wong DTW, Sugimoto M, Gleber-Netto FO, Li F, Tu M, Zhang Y, Akin D, Iino M. Identification of salivary metabolites for oral squamous cell carcinoma and oral epithelial dysplasia screening from persistent suspicious oral mucosal lesions. Clin Oral Investig. 2019;23(9):3557–3563. doi: 10.1007/s00784-018-2777-3. [DOI] [PubMed] [Google Scholar]
- 9.Ishikawa S, Sugimoto M, Edamatsu K, Sugano A, Kitabatake K, Iino M. Discrimination of oral squamous cell carcinoma from oral lichen planus by salivary metabolomics. Oral Dis. 2020;26(1):35–42. doi: 10.1111/odi.13209. [DOI] [PubMed] [Google Scholar]
- 10.Carvalho AL, Pintos J, Schlecht NF, Oliveira BV, Fava AS, Curado MP, Kowalski LP, Franco EL. Predictive factors for diagnosis of advanced-stage squamous cell carcinoma of the head and neck. Arch Otolaryngol Head Neck Surg. 2002;128(3):313–318. doi: 10.1001/archotol.128.3.313. [DOI] [PubMed] [Google Scholar]
- 11.Rashid A, Warnakulasuriya S. The use of light-based (optical) detection systems as adjuncts in the detection of oral cancer and oral potentially malignant disorders: A systematic review. J Oral Pathol Medi. 2015;44(5):307–328. doi: 10.1111/jop.12218. [DOI] [PubMed] [Google Scholar]
- 12.Liu D, Zhao X, Zeng X, Dan H, Chen Q. Non-invasive techniques for detection and diagnosis of oral potentially malignant disorders. Tohoku J Exp Med. 2016;238(2):165–177. doi: 10.1620/tjem.238.165. [DOI] [PubMed] [Google Scholar]
- 13.Nagi R, Reddy-Kantharaj YB, Rakesh N, Janardhan-Reddy S, Sahu S. Efficacy of light based detection systems for early detection of oral cancer and oral potentially malignant disorders: Systematic review. Med Oral Patol Oral Cir Bucal. 2016;21(4):e447–e455. doi: 10.4317/medoral.21104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schafer CA, Schafer JJ, Yakob M, Lima P, Camargo P, Wong DT. Saliva diagnostics: Utilizing oral fluids to determine health status. Monogr Oral Sci. 2014;24:88–98. doi: 10.1159/000358791. [DOI] [PubMed] [Google Scholar]
- 15.Wang X, Kaczor-Urbanowicz KE, Wong DT. Salivary biomarkers in cancer detection. Med Oncol. 2017;34(1):7. doi: 10.1007/s12032-016-0863-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Soini HA, Klouckova I, Wiesler D, Oberzaucher E, Grammer K, Dixon SJ, Xu Y, Brereton RG, Penn DJ, Novotny MV. Analysis of volatile organic compounds in human saliva by a static sorptive extraction method and gas chromatography-mass spectrometry. J Chem Ecol. 2010;36(9):1035–1042. doi: 10.1007/s10886-010-9846-7. [DOI] [PubMed] [Google Scholar]
- 17.Shah FD, Begum R, Vajaria BN, Patel KR, Patel JB, Shukla SN, Patel PS. A review on salivary genomics and proteomics biomarkers in oral cancer. Indian J Clin Biochem. 2011;26(4):326–334. doi: 10.1007/s12291-011-0149-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chu HW, Chang KP, Hsu CW, Chang IY, Liu HP, Chen YT, Wu CC. Identification of salivary biomarkers for oral cancer detection with untargeted and targeted quantitative proteomics approaches. Mol Cell Proteomics. 2019;18(9):1796–1806. doi: 10.1074/mcp.RA119.001530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang Y, Sun J, Li F, Grogan TR, Vergara JL, Luan Q, Park MS, Chia D, Elashoff D, Joshipura KJ, Wong DTW. Salivary extracellular RNA biomarkers for insulin resistance detection in Hispanics. Diabetes Res Clin Pract. 2017;132:85–94. doi: 10.1016/j.diabres.2017.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang X, Broszczak D, Kostner K, Guppy-Coles KB, Atherton JJ, Punyadeera C. Salivary protein panel to diagnose systolic heart failure. Biomolecules. 2019;9(12) doi: 10.3390/biom9120766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liang D, Lu H. Salivary biological biomarkers for Alzheimer’s disease. Arch Oral Biol. 2019;105:5–12. doi: 10.1016/j.archoralbio.2019.06.004. [DOI] [PubMed] [Google Scholar]
- 22.Liao PH, Chang YC, Huang MF, Tai KW, Chou MY. Mutation of p53 gene codon 63 in saliva as a molecular marker for oral squamous cell carcinomas. Oral Oncology. 2000;36(3):272–276. doi: 10.1016/S1368-8375(00)00005-1. [DOI] [PubMed] [Google Scholar]
- 23.Gleber-Netto FO, Yakob M, Li F, Feng Z, Dai J, Kao HK, Chang YL, Chang KP, Wong DT. Salivary biomarkers for detection of oral squamous cell carcinoma in a taiwanese population. Clin Cancer Res. 2016;22(13):3340–3347. doi: 10.1158/1078-0432.CCR-15-1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rhodus NL, Ho V, Miller CS, Myers S, Ondrey F. Nf-kappab dependent cytokine levels in saliva of patients with oral preneoplastic lesions and oral squamous cell carcinoma. Cancer Detect Prev. 2005;29(1):42–45. doi: 10.1016/j.cdp.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 25.Brailo V, Vucicevic-Boras V, Cekic-Arambasin A, Alajbeg IZ, Milenovic A, Lukac J. The significance of salivary interleukin 6 and tumor necrosis factor alpha in patients with oral leukoplakia. Oral Oncol. 2006;42(4):370–373. doi: 10.1016/j.oraloncology.2005.09.001. [DOI] [PubMed] [Google Scholar]
- 26.Hu S, Arellano M, Boontheung P, Wang J, Zhou H, Jiang J, Elashoff D, Wei R, Loo JA, Wong DT. Salivary proteomics for oral cancer biomarker discovery. Clin Cancer Res. 2008;14(19):6246–6252. doi: 10.1158/1078-0432.CCR-07-5037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang A, Sun H, Wang P, Wang X. Salivary proteomics in biomedical research. Clin Chim Acta. 2013;415:261–265. doi: 10.1016/j.cca.2012.11.001. [DOI] [PubMed] [Google Scholar]
- 28.Shinoda K, Tomita M, Ishihama Y. Empai calc—for the estimation of protein abundance from large-scale identification data by liquid chromatography-tandem mass spectrometry. Bioinformatics. 2010;26(4):576–577. doi: 10.1093/bioinformatics/btp700. [DOI] [PubMed] [Google Scholar]
- 29.Zhu W, Smith JW, Huang CM. Mass spectrometry-based label-free quantitative proteomics. J Biomed Biotechnol. 2010;2010:840518. doi: 10.1155/2010/840518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ishihama Y, Oda Y, Tabata T, Sato T, Nagasu T, Rappsilber J, Mann M. Exponentially modified protein abundance index (emPAI) for estimation of absolute protein amount in proteomics by the number of sequenced peptides per protein. Mol Cell Proteomics. 2005;4(9):1265–1272. doi: 10.1074/mcp.M500061-MCP200. [DOI] [PubMed] [Google Scholar]
- 31.Cristaldi M, Mauceri R, Di Fede O, Giuliana G, Campisi G, Panzarella V. Salivary biomarkers for oral squamous cell carcinoma diagnosis and follow-up: Current status and perspectives. Front Physiol. 2019;10 doi: 10.3389/fphys.2019.01476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xiao H, Langerman A, Zhang Y, Khalid O, Hu S, Cao CX, Lingen MW, Wong DTW. Quantitative proteomic analysis of microdissected oral epithelium for cancer biomarker discovery. Oral Oncol. 2015;51(11):1011–1019. doi: 10.1016/j.oraloncology.2015.08.008. [DOI] [PubMed] [Google Scholar]
- 33.Schaaij-Visser TBM, Graveland AP, Gauci S, Braakhuis BJM, Buijze M, Heck AJR, Kuik DJ, Bloemena E, Leemans CR, Slijper M, Brakenhoff RH. Differential proteomics identifies protein biomarkers that predict local relapse of head and neck squamous cell carcinomas. Clin Cancer Res. 2009;15(24):7666–7675. doi: 10.1158/1078-0432.Ccr-09-2134. [DOI] [PubMed] [Google Scholar]
- 34.Santosh N, McNamara KK, Beck FM, Kalmar JR. Expression of cornulin in oral premalignant lesions. Oral Surg Oral Med Oral Pathol Oral Radiol. 2019;127(6):526–534. doi: 10.1016/j.oooo.2019.02.003. [DOI] [PubMed] [Google Scholar]
- 35.Torre GC. SCC antigen in malignant and nonmalignant squamous lesions. Tumour Biol. 1998;19(6):517–526. doi: 10.1159/000030045. [DOI] [PubMed] [Google Scholar]
- 36.Feng XY, Li JH, Li JZ, Han ZX, Xing RD. Serum scca, cyfra 21-1, egfr and cyclin d1 levels in patients with oral squamous cell carcinoma. Int J Biol Markers. 2010;25(2):93–98. doi: 10.1177/172460081002500206. [DOI] [PubMed] [Google Scholar]
- 37.Ishikawa S, Sugimoto M, Kitabatake K, Tu M, Sugano A, Yamamori I, Iba A, Yusa K, Kaneko M, Ota S, Hiwatari K, Enomoto A, Masaru T, Iino M. Effect of timing of collection of salivary metabolomic biomarkers on oral cancer detection. Amino Acids. 2017;49(4):761–770. doi: 10.1007/s00726-017-2378-5. [DOI] [PubMed] [Google Scholar]
- 38.de Almeida Pdel V, Gregio AM, Machado MA, de Lima AA, Azevedo LR. Saliva composition and functions: A comprehensive review. J Contemp Dent Pract. 2008;9(3):72–80. [PubMed] [Google Scholar]
- 39.Brierly JD, Gospodarowicz MK, Wittekind C (eds.) UICC TNM Classification of Malignant Tumours. Wiley Blackwell. 2016;Eighth Edition [Google Scholar]