Abstract
Background
There is growing concern about individuals reported to suffer repeat COVID-19 disease episodes, these in a small number of cases characterised as de novo infections with distinct sequences, indicative of insufficient protective immunity even in the short term.
Methods
Observational case series and case-control studies reporting 33 cases of recurrent, symptomatic, qRT-PCR positive COVID-19. Recurrent disease was defined as symptomatic recurrence after symptom-free clinical recovery, with release from isolation >14 days from the beginning of symptoms confirmed by qRT-PCR. The case control study-design compared this group of patients with a control group of 62 patients randomly selected from the same COVID-19 database.
Results
Of 33 recurrent COVID-19 patients, 26 were female and 30 were HCW. Mean time to recurrence was 50.5 days which was associated with being a HCW (OR 36.4 (p <0.0001)), and blood type A (OR 4.8 (p = 0.002)). SARS-CoV-2 antibodies were signifcantly lower in recurrent patients after initial COVID-19 (2.4 ± 0.610; p<0.0001) and after recurrence (6.4 ± 11.34; p = 0.007). Virus genome sequencing identified reinfection by a different isolate in one patient.
Conclusions
This is the first detailed case series showing COVID-19 recurrence with qRT-PCR positivity. For one individual detection of phylogenetically distinct genomic sequences in the first and second episodes confirmed bona fide renfection, but in most cases the data do not formally distinguish between reinfection and re-emergence of a chronic infection reservoir. These episodes were significantly associated with reduced Ab response during initial disease and argue the need for ongoing vigilance without an assumption of protection after a first episode.
Keywords: COVID-19, Recurrence, Reinfection, SARS-CoV-2, Antibodies
Graphical abstract
Research in context.
Evidence before this study. Considerable attention has been paid to concerns over the potential for repeat episodes of COVID-19 in individuals who have recovered. We conducted a PubMed search on 26 October 2020 under the terms “recurrence” or “reinfection” and “COVID”, identifying 11 primary research studies. These were largely case reports and in the case of 5 individuals included definitive evidence of distinct reinfection through comparison of viral sequence between the first and second episode. Because it is often not possible during routine healthcare provision in the pandemic response to archive swabs for genomic sequencing in the case of reinfection, this data is generally unachievable outside of an academic setting. In the absence of definitive widespread access to sequence comparisons, the data can best be captured by adopting a conservative terminology that records “recurrence” after recovery from PCR-verified disease.
Added value of this study. It has been unclear until now whether or not repeat episodes of COVID-19 are rare exceptions. We describe 33 cases of recurrence at a single healthcare center in Brazil, 30 of these among HCW. Recurrence was associated with being a female HCW, with blood group A and with significantly reduced antibodies after the first episode and tended to be of enhanced severity, in one case, fatal.
Implications of all the available evidence. It is clear from this study and others that infected individuals mount a variable immune response and, in a significant minority, this may be insufficient to offer subsequent protection. Such individuals remain at risk, especially in highly exposed healthcare settings. These findings alert against complacency in those who have recovered from infection and offers a caution to pursuit of protection through natural herd immunity.
Alt-text: Unlabelled box
Introduction
The COVID-19 global pandemic caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) started in Wuhan, China, in December 2019, and has spread worldwide.1 The virus can infect the respiratory, gastrointestinal, hepatic and central nervous systems, with a high rate of severe cases requiring hospitalization.2 The average incubation period from SARS-CoV-2 exposure to symptom onset is approximately 4–5 days, with 97.5% of symptomatic patients experiencing symptoms within 11.5 days.3 , 4 While most infections are asymptomatic or mild, risk factors for severe COVID-19 include advanced age, cardiovascular diseases, chronic lung disease, systemic arterial hypertension, diabetes, obesity, black and minority ethnicity. At a time when millions of first-wave infections have occurred and many populations are experiencing a second wave, there is concern about whether disease can be suffered recurrently, whether by re-emergence of a poorly-cleared viral reservoir or by reinfection.5 Some patients re-experience COVID-19 symptoms after a considerable period of time again becoming qRT-PCR positive, not to be conflated with the different phenomenon of chronic post-viral symptoms or ‘long Covid’6 a spectrum of long-term, ongoing symptoms in people who are generally qRT-PCR negative. It is not always practicable to validate by sequencing whether specific cases constitute persistence or reinfection, so we have here used the conservative term ‘recurrence’, except where there are data on infection with a de novo viral sequence.7, 8, 9, 10 We report a case series of 33 patients with recurrence of COVID-19 symptoms and qRT-PCR positivity and compared them with a control group of 62 patients with a single episode of COVID 19, in a case-control design. Additionally, we were able to compare viral genomes recovered in the first and second episodes for one of these cases.
Methodology
This study is a mix of observational case series and case-control studies; samples were obtained by agreement through the “Monitora Corona”, an emergency project adopted by the Federal University of Sergipe for daily phone monitoring of confirmed COVID-19 cases diagnosed and treated at the Centro de Doenças Respiratórias (CDR) of the Instituto de Promoção e Assistência à Saúde dos Servidores do Estado de Sergipe (IPESAÚDE-SE). Medical students follow up patient symptoms by phone every 24 h or 48 h, registering details online on the Monitora Corona Platform and through the “Teleatendimento ambulatorial: novo cenário de Ensino Médico e cuidado ao paciente” Project by the University Hospital of Federal University of Sergipe with confirmed or suspected patients for COVID-19 admitted to the outpatient clinic of the University Hospital of the Federal University of Sergipe. Ethical Committee of the Federal University of Sergipe approval was received for the studies (CAAE 31079720.5.0000.5546), and informed consent of all participating subjects or their legal guardians was obtained.
Inclusion criteria: patients of any gender or age group >18 years who had symptoms and at least two positive results by qRT-PCR for SARS-CoV-2 (KIT BIOMOL OneStep/COVID-19; Instituto de Biologia Molecular do Paraná; Paraná, Brazil). Recurrent disease was defined as recurrence of symptoms after full clinical recovery and release from isolation for 14 days from the beginning of symptoms, and at least 7 days without symptoms, and confirmed by qRT-PCR in the first and second episode of COVID-19. Exclusion criteria: patients without a qRT-PCR result or with negative qRT-PCR from the first infection or in the second phase of symptoms of COVID-19. Questionnaire data collected included demographic features, comorbidities, clinical symptoms and treatment during the first and the recurrent episodes of COVID-19. Reported clinical symptoms included sore throat, anosmia, malaise, myalgia, arthralgia, cough, shortness of breath. Reported treatment data included medicines, methylprednisolone pulse therapy, emergency room admissions, hospitalization, oxygen therapy, ICU admission, respiratory support and ventilation.
To ascertain associations of the demographic and clinical data with the occurrence of recurrent COVID-19, a case control study was designed by comparing this group of patients with a control group (n = 62), randomly selected from the same follow-up database. Inclusion criteria for this group were patients of any gender or age group >18 years who had a single episode of COVID-19 symptoms with a positive result for qRT-PCR for SARS-CoV-2. Exclusion criteria for this group were patients without a qRT-PCR result or with negative qRT-PCR or with a history of a second episode of symptoms of COVID-19. All patients signed an informed consent form (ICF) confirming their participation in the study.
SARS-CoV-2 antibody testing was performed for 17 (51.5%) recurrent disease patients in the first and second episode of symptoms of COVID-19, and 31 (50%) of the control group with a single episode. The test used the AFIAS COVID-19 Ab fluorescence immunoassay for IgM/IgG which is against SARS-CoV-2 nucleocapsid (NP) (Boditech Med Incorporated 43, Geodudanji 1-gil, Dongnae-myeon, Chuncheon-si, Gang-won-do, 24,398 Republic of Korea). This kit has a 100% sensitivity, and 96,7% specificity in clinical tests. The laboratory considered values higher than 1.1 positive, 0.9 to 1.1 indeterminate, and lower than 0.90 negative, for both IgG and IgM.
Statistical analyses were performed using Jamovi Software (version 1.2). For comparison of continuous variables between the groups, the data were tested for normality, using D'Agostino and Pearson tests. Ages and titres of IgM and IgG between the groups were compared by Mann-Whitney U test. For the categorical data, the association of each variable with the groups were tested by calculating the Odds Ratio (OR) and 95% confidence interval (CI) using the Fisher exact test.
Complete viral genome sequences from specific cases were obtained with the Ion AmpliSeq™ SARS-CoV-2 Research Panel, using the Ion S5 NGS platform (Thermo Fisher). Details on the virus amplification and sequencing protocols and all bioinformatics analyses are presented as Supplementary Methods. SARS-CoV-2 lineage assignments were performed with Pangolin11 and phylogenetic analysis of sequenced viral genomes was conducted using Nextstrain, described in supplementary methods.12
Results
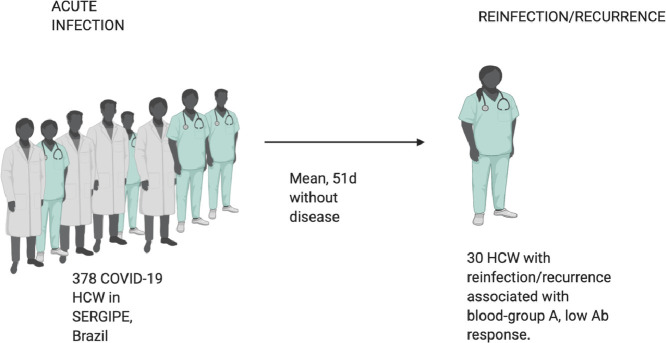
This study documents 33 patients who showed clear evidence of COVID-19 recurrence following an interval with recovery. Demographic and clinical data for each indivudal, including time interval between first and second episode and co-morbidities are supplied in Supplementary Table 1 and summarised medication data in Supplementary Table 2. Recurrence patients were mainly female, n = 26 (78.8%) and largely composed of healthcare workers (HCW) n = 30/33 (91%), aged from 22–58 years (mean 39.2 years, ±8.5SD). The denominator of all HCW with confirmed qRT-PCR from the database is 378, meaning that 7.9% suffered recurrence/reinfection. The interval between recurrent symptoms ranged from 8–130 days, with an average of 41 days ±24.0 SD. The most prevalent blood type was A+, 14 patients (42%), and O+, n = 10 (30%), but there were also 4 patients B+, 1 patient A-, 1 patient O- and 1 patient AB+ (and 2 for whom blood type was unavailable). Fifteen individuals (45.5%) had comorbidities of which the most prevalent was obesity (BMI > 30 Kg/m2), n = 10 (30.3%); we also observed overweight (BMI from >25 to <30 Kg/m2) in other 13 patients (39.4%), that was not considered for the analysis of comorbidities.
The time between the first and second qRT-PCR ranged from 18–134 days, with an average of 50.5 days ±19.37SD. The patient that had only 18 days of interval between the positive qRT-PCR, had 20 days of symptoms in the first episode, had 8 days without symptoms and returned to work with full recovery of COVID-19 symptoms, symptoms returning 8 days later. The CT from the qRT-PCR from the first episode ranged from 15–37, mean±SD = 30±4.0; and from the second episode, from 15–36, mean±SD = 29±9.0. No differences were detected between these two values. In this standard healthcare setting, the patients did not access a repeat qRT-PCR test after recovery from the symptoms of the first infection to confirm negativity, but 2 of these patients had a second qRT-PCR which tested negative after recovery from the first COVID-19 episode and 3 had negative results after the recurrence. The average total duration in days of first episode symptoms from all 33 patients was 16.2 days ± 9.65 SD, while in the recurrence it was 19.4 days ±10.13 SD.
Table 1 shows the frequency of clinical manifestations and outcomes in the first and subsequent COVID-19 episode. Patients tended to show a higher frequency of symptoms associated with disease severity in the recurrence, as compared to the first episode, but the differences did not reach statistical significance. For example, myalgia was seen in 72.7% of patients in the recurrence versus 48.5% in the first episode and dyspnea in 57.6% in the recurrence versus 33.3% in their first episode. Sixty percent of the patients attended at an emergency room during recurrence versus 57.6% in the first episode. Tomography was requested in 75.8% of the patients during recurrence, and 52% were compatible with a COVID-19 pattern (12% presenting moderate to severe lesions), while tomography was requested in 33.3% in the first episode, and 18.2% was compatible with a mild COVID-19 pattern. Hospitalization was required for 12.1% in the recurrence but none of the patients in the first episode of COVID-19, 2 requiring ICU admission, one of whom died (patient 32 from Supplementary Table 1). This patient was a 44-year-old male pharmacist, blood type O+, with obesity and systemic arterial hypertension. He had 9 days of mild symptoms during the first episode and a positive qRT-PCR in May. He returned to work and presented with recurrence of symptoms 38 days after the first episode, with a second positive PCR on June 13th. During the recurrent episode he had severe respiratory symptoms and was admitted to ICU, dying after 20 days of symptoms. It was not possible to recover the virus sample from his first episode, but the virus was isolated from the recurrence and analysed in this paper (K2).
Table 1.
Clinical manifestations observed in thirty-three patients during first and second COVID-19 episodes.
| Clinical manifestations | Frequency (n) |
p valuea | |
|---|---|---|---|
| First COVID-19 | Recurrent COVID-19 | ||
| Headache | 87.9 (29) | 84.8 (28) | >0.999 |
| Asthenia | 81.8 (27) | 87.9 (29) | 0.733 |
| Myalgia | 48.5 (16) | 72.7 (24) | 0.077 |
| Arthralgia | 30.3 (10) | 42.4 (14) | 0.443 |
| Sneeze/runny nose | 45.5 (15) | 66.7 (22) | 0.163 |
| Odynophagia | 57.6 (19) | 60.6 (20) | >0.999 |
| Dysgeusia | 30.3 (10) | 51.5 (17) | 0.130 |
| Anosmia | 24.2 (8) | 48.5 (16) | 0.072 |
| Dry cough | 45.5 (15) | 63.6 (21) | 0.216 |
| Productive cough | 6.1 (2) | 18.2 (6) | 0.258 |
| Dyspnea | 33.3 (11) | 57.6 (19) | 0.083 |
| Diarrhea | 48.5 (16) | 48.5 (16) | >0.999 |
| Hyporexia | 36.4 (12) | 45.5 (15) | 0.617 |
| Abdominal pain | 30.3 (10) | 36.4 (12) | 0.794 |
| Nausea/vomiting | 30.3 (10) | 36.4 (12) | 0.794 |
| Fever | 21.2 (7) | 36.4 (12) | 0.277 |
| Skin lesions | 24.2 (8) | 15.2 (5) | 0.537 |
| Dizziness | 27.3 (9) | 36.4 (12) | 0.598 |
| Mental confusion | 6.1 (2) | 15.2 (5) | 0.427 |
| Tomography data | |||
| Tomography requested | 33.3 (11) | 75.8 (25) | 0.001 |
| Pattern compatible with COVID-19 | 18.2 (2) | 52.0 (13) | 0.077 |
| Pattern compatible with mild COVID-19 | 18.2 (2) | 40 (10) | 0.268 |
| Pattern compatible with moderate COVID-19 | 0 | 4 (1) | >0.999 |
| Pattern compatible with severe COVID-19 | 0 | 8 (2) | >0.999 |
| Clinical evolution and outcome | |||
| Emergency room attendance | 57.6 (19) | 60.6 (20) | >0.999 |
| Hospitalization | 0 | 12.1 (4) | 0.114 |
| Oxygen therapy | 0 | 9.1 (3) | 0.239 |
| Intensive care unit admission | 0 | 6.1 (2) | 0.492 |
| Mechanical ventilation | 0 | 3.0 (1) | >0.999 |
| Death | 0 | 3.0 (1) | >0.999 |
Fisher exact test.
Treatments used in the first and second episodes are described in Supplementary Table 2. In the first episode, the most frequently used treatment was azithromycin in 20 patients (61%), followed by systemic corticosteroids 12 (36%), and other antibiotics 6 (18.2%). In the recurrence, the most used treatment was systemic corticosteroids in 26 patients (79%), followed by Ivermectin, 21 patients (63%), Azithromycin, 20 patients (61%), other antibiotics, 20 patients (61%), Heparin, 12 patients (36%) and Hydroxychloroquine, 3 patients (9%).
We compared within the recurrent COVID-19 group, classified by severity of recurrent episode symptoms. Mean age was similar 39.1 ± 10.05 versus 38.9 ± 5.93 (p = 0.93) comparing the moderate/severe (first group, n = 21) and the mild (second group, n = 12). However, associated comorbidities (systemic arterial hypertension, diabetes, obesity, asthma) were present in 15/21) of the moderate/severe group compared to 2/12) from the mild group; OR 12.5 [95% CI 12.09 – 74.84]; p = 0.004. Treatment with corticosteroids was also analysed to investigate any association of immunosuppression with severity of recurrence, but no differences were observed between therapy in the moderate/severe 10 (n = 21), versus 2 (n = 12) patients from the mild group; OR 4.6 [95% IC 0.80 – 25.99]; p value = 0.13.
Data from the case-control study design are summarized in the Table 2 . No differences were observed between age and sex frequencies between the recurrent cases (n = 33) and controls who experienced a single episode (n = 62). Associations were observed between recurrence and being a HCW (OR 36.4; 95%CI 9.67 – 137.20; p<0.0001), and being blood type A (OR 4.8; 95%CI 1.76 – 13.07; p = 0.002). Interestingly, there was a protective effect from recurrence of having had anosmia (OR 0.22; 95%CI 0.08 - 0.56; p = 0.001) and dry cough (OR 0.20; 95%CI 0.08 - 0.51; p = 0.0009) during the first episode. In the recurrent group, 97% worked in an environment with possible exposure to COVID-19, with a range of 4–90 h/week of exposure, mean ± SD 42.2 ± 18.94 h/week. Additionally, 48.5% worked in specific COVID-19 Units, with a range of 10–48 h/week of exposure, mean ± SD 32.7 ± 11.81.
Table 2.
Data from a case-control study design, comparing the recurrent COVID-19 patients with controls with a single episode of COVID-19.
| Variables | Recurrent COVID-19 | Controls with single episode | aOR | b95% CI | cp value |
|---|---|---|---|---|---|
| n = 33 | n = 62 | ||||
| Age (Mean ± SD) | 39.2 ± 8.53 | 39.2 ± 13.10 | – | – | d0.647 |
| Number of patients (Total) | |||||
| Females | 26 (33) | 39 (62) | 1.7 | 0.02 - 0.53 | 0.350 |
| Healthcare workers | 30 (33) | 14 (62) | 36.4 | 9.67 – 137.20 | < 0.0001 |
| Blood type A | 15 (31) | 9 (55) | 4.8 | 1.76 – 13.07 | 0.002 |
| Associated diseases (Systemic Arterial Hypertension, Diabetes, Obesity and Asthma) | 15 (33) | 18 (62) | 2.2 | 0.91 – 5.21 | 0.112 |
| Anosmia | 8 (33) | 37 (62) | 0.22 | 0.08 - 0.56 | 0.001 |
| Dry cough | 13 (33) | 46 (62) | 0.20 | 0.08 - 0.51 | 0.0009 |
| Corticoid treatment in the 1st COVID-19 episode | 12 (33) | 35 (62) | 1.96 | 0.78 - 4.95 | 0.226 |
| Recurrent COVID-19 | Controls with single episode |
dp value |
|||
| n = 17 | n = 31 | ||||
| Age (Mean ± SD) | 39 ± 8.27 | 46.45 ± 11.35 | 0.015 | ||
| Titre of IgG (Mean ± SD) | 0.82 ± 2.31 | 16.62 ± 15.86 | – | – | <0.0001 |
| Titre of IgG (Mean ± SD) after the recurrence of COVID 19 symptoms | 6.38 ± 11.34 | – | e0.007 | ||
OR = Odds ratio.
CI = Confidence interval.
Fisher exact test.
Mann-Whitney U test.
Comparing with the only measurement of IgG title from the controls (16.62 ± 15.86).
SARS-CoV-2 anti-NP antibody titres were available for the first episode of 17 from the 33 recurrence patients and for 31 from the 62 control group with a single COVID 19 episode. The mean titres for IgM (0.57 ± 0.82) and IgG (0.82 ± 2.31) were lower in the recurrence group after their first episode compared to titres in controls (IgM 1.73 ± 4.815 and IgG 16.62 ± 15.86); only IgG titres were significantly different from the controls (p <0.0001 by Mann-Whitney U test). Antibody was tested in the recurrence group at 22±14 days after symptoms, and, although we do not have the exact time for the control group, it was annotated as >14 days in the database. IgG titres in the recurrence group even after the second episode were still lower (6.38 ± 11.34) than the control group (16.62 ± 15.86); p = 0.007; Mann-Whitney U test.
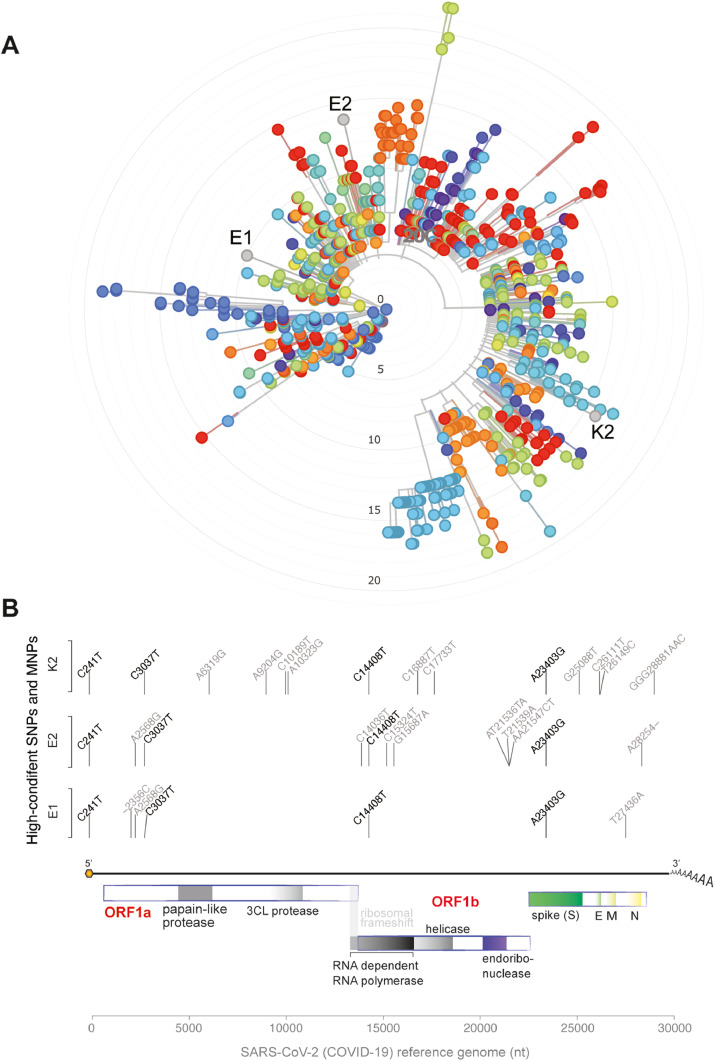
Next-generation sequencing of viral genomes recovered from two recurrent cases presenting with symptoms rendered full-length SARS-CoV-2 sequences with over 99.8% genome coverage in the second episodes (Supplementary Table 3). Viral sequences from the two cases were from phylogenetically distinct clades (Fig. 1 A). Of note, the two sequences from the first and second symptomatic episodes of the same patient (samples E1 and E2) were also from different clades (lineages B.1 and B.1.80) arguing for reinfection (Fig. 1A). All three genomic sequences presented mutations highly distributed among SARS-CoV-2 global lineages (C241T, C3037T, C14408T, A23403G) (Fig. 1B; Supplementary Tables 4 and 5). Some rare SARS-CoV-2 mutations (A2568G and C15324T) were shared only by sequences E1 and E2 (Supplementary Table 6). Additionally, exclusive mutations of sample K2 included a multi-nucleotide polymorphism (MNP) that is globally distributed (GGG28881AAC) and a SNP (G25088T) found with raised frequency in SARS-CoV-2 genomic sequences from fatal cases (Supplementary Table 7). Details on the mutations exclusive to each viral isolate are indicated in Supplementary Tables 7 and 8. Altogether, these results confirm that qRT-PCR re-positivity was not due to delayed nucleic acid conversion from positive to negative in these recurrent symptomatic cases, but rather, to active infection. The detection of phylogenetically distinct genomic sequences in the first and second episodes of one patient argues for de-novo reinfection.
Fig. 1.
Characterization of SARS-CoV-2 viral genomes in patients with recurrent COVID-19. Phylogenetic (A) and genomic variant (B) analysis of viral genomes recovered from individuals with recurrent COVID-19. In the phylogenetic tree, strains derived from Brazilian individuals are shown in orange. E1 and E2 represent samples of the patient E at first and second event of COVID-19, respectively. K2 represent the sample of patient K during the second episode of COVID-19. Vertical lines indicate the genomic position of the high-confidence mutation in SARS-CoV-2 genomic sequence. Only variants with frequency higher than 90% were considered in the analysis. Text above the vertical lines indicates the base transition from reference genomes to newly assembled genomes in this work. Genomic structure of SARS-CoV-2 is also plotted.
Discussion
This study reports 33 cases with recurrent COVID-19 episodes following recovery and confirmed by positive qRT-PCR in each of the first and second episodes. Recurrences tend to be more severe than the first episode, one individual dying. In a case-control design we identified risk factors associated with recurrence: being a HCW and being blood type A showed high odds ratio for recurrence. Symptoms of upper respiratory tract infections, anosmia, and dry cough during the primary episode were negatively associated with recurrence. Comorbidities such as arterial hypertension, obesity, diabetes, and asthma were not associated with recurrence, but were associated with moderate/severe clinical presentation of recurrent COVID-19. Genomic sequencing rendered full-length SARS-CoV-2 sequences with over 99.8% coverage in 2 of these recurrent symptomatic cases, confirming that qRT-PCR positivity is due to active infection. Additionally, the detection of phylogenetically distinct genomic sequences in one patient confirmed reinfection by a different strain.
There have thus far been rare instances where the clinical and laboratory context allowed for comparative genomic sequencing of virus from each of the two episodes to confirm reinfection, and we here add another such example.13 , 14 In general, though it can prove challenging to access stored, isolated, virus from each episode in patients during the heat of a pandemic, unless the individuals are enrolled within a study cohort. Where such data are absent, one cannot formally distinguish reinfection from the more general term, ‘recurrence’. The rationale for our case series and case-control studies is that in either case, this large HCW series constitute a concerning addition to our understanding of the disease spectrum.
The Korean Center for Disease Control and Prevention declared in the first week of April that there were 91 documented cases of patients in Daegu, South Korea, who had recovered, left quarantine, and then tested positive again. Although they could not rule out reactivation as a possibility, they stated that these 91 cases were more likely to be due to levels of the virus that fall below a detectable level, allowing symptoms to improve, then turned positive again, although the findings are compatible either with intermediate false negative tests under conditions of low viral load, or with reinfection. Based on data from China, the most used test type showed a false-negative rate of 30%.15 After that recurrence of qRT-PCR positivity was reported in several studies.16, 17, 18, 19, 20 These studies suggest virus persistence. However, in none of these studies are there reports of associated clinical symptoms. A study of 98 individuals recovering from COVID-19 who had been discharged following two negative qRT-PCR tests showed that during follow-up, 17 individuals (17.3%) returned to positivity on testing, though only one patient returned to clinically symptomatic disease.21
In the case series reported here, all 33 patients presented with a recurrence of COVID-19 symptoms, 30 of them HCW. All had recovered from first episode symptoms, returned to work and later suffered recurrent symptoms. This high frequency of recurrence in HCW may be due to the higher chance of SARS-CoV-2 exposure. While formal proof of reinfection was possible only in one case, we highlight the potential for reinfection. We cannot formally rule out an observation bias, since HCW may be better placed to note the recurrence of symptoms and may have more access to qRT-PCR testing, but this seems unlikely to explain the observed difference between case and controls (OR 36.4; p < 0.0001); during this pandemic all patients have become highly vigilant and have similar access to follow-up within the program to report clinical symptoms and access a new qRT-PCR test.
Recent reports of one case with recurrence of COVID-19, confirmed reinfection with a second virus sequence.13 , 22 Unfortunately, we do not have a negative qRT-PCR for all the recurrence cases after their initial recovery, but 2 of these patients had a second qRT-PCR test negative after recovery and 3 had negative results after the recurrence. This is because discharge before returning to work under Brazilian Ministry of Health criteria, during the time this study was performed, indicated a requirement of 14 days from first symptoms plus 3 days without symptoms, but does not advocate or allocate resource for repeat testing. Now the rule is a requirement of 10 days from first symptoms plus 24 h without symptoms.23 It has been proposed that tomographic patterns should be used for isolation, discharge, and transferring for hospitalized patients after recovery from clinically diagnosed COVID-19.18 , 19 , 24 , 25
Some studies have made a case for virus persistence.5 , 24 , 26, 27, 28 In a prospective cohort study including 131 patients who had been discharged for 4 weeks, some patients had new symptoms in this period, and 7 needed to be readmitted. Of the 94 individuals who underwent qRT-PCR, 8 (6.1%) showed positive reversion, of which 6 remained asymptomatic, 2 had a recurrence of fever and one exhibited worsening tomography.27 In a case control study of 414 COVID-194 patients, all who were discharged and underwent strict quarantine at a designated location for 14 days, it was observed that 69 patients (16.9%) showed recurrence of qRT-PCR positivity. Comparing these retested positive cases with a control group that did show recurrence, younger patients with less severe index illness were more likely to retest positive.24
There have been cases reporting COVID-19 symptom recurrence.29, 30, 31 It has been argued that virus may be present in a latent state, in the lysogenic part (viral reproduction), inactive or hidden in cells, not causing disease for some time and then reactivating.15 , 32 The main risk factors include: (1) host status, (2) viral factors and (3) environmental factors.17 Host factors may include sex, age, comorbidities, immunosuppression and genetic background. Environmental factors may include coinfections or immunosuppressive therapies.33 Immune dysfunction associated with SARS-CoV2 infection would be compatible with the notion of facilitating either reinfection or a potential persistence/reactivation of infection.34, 35, 36
In the present study the term “recurrence” was conservatively adopted because these case series might represent a group of patients, ones with reinfections and others with a bimodal disease and prolonged qRT-PCR positivity. However, there is evidence to indicate reinfection: 1) The association of recurrence with being a HCW; 2) Negative qRT-PCR after the first COVID-19 recovery in 2 patients; 3) Sequencing data from the recurrent episode of other 2 patients, with over 99.8% genome coverage in the second episodes confirms active infection, and not late conversion of PCR from positive to negative; 4) Viral genome sequencing data with the detection of phylogenetically distinct genomic sequences in the first and second episodes of one patient argues for de-novo reinfection, and 5) the association with low antiviral Ab titre.
We compared antibody levels in recurrence patients and controls. Only 17 recurrence patients and 31 controls had serological data available, but interestingly, the levels of IgG for SARS-CoV-2 were significantly lower in recurrence patients after the first episode, as compared to controls and even after recurrence. Some of us have recently reported a UK HCW cohort in which about 10% of previously infected HCW carried very low or absent neutralizing antibody levels at 4-months and would presumably be vulnerable to reinfection.37 Sekine et al. described a potent memory T cell response, similar to those desirable in the context of successful vaccines and that confer long-term immunity, independent of the absence or presence of circulating antibodies. Approximately twice as many people showed T-cell immunity compared to those in which they detected antibodies. Thus, even in the face of a negative serology, patients may potentially be protected from COVID-19 recurrence.38
In the present study, recurrent patients presented with somewhat more severe symptoms, though generally making a good recovery, except for one individual with a fatal outcome. Severity of the second COVID-19 episode was associated with presence of comorbid obesity, arterial hypertension, diabetes, and asthma.
We also detected an association of being blood type A with recurrence of COVID-19. A genetic association study, using a genome wide screen approach, reported an association of blood type A with a higher risk of infection.39 This offers further support for the hypotheses of persistence or reinfection.
Guidelines have suggested two, consecutive, negative qRT-PCR test results as a criterion for discharge. However, due to the high false negative rate of viral test or prolonged viral clearance, rather than qRT-PCR testing, additional clinical indicators should be used not only for diagnosis and treatment but also for isolation and discharge. The present study brings into question how much longer social isolation beyond the 14 days of symptoms should be adopted and for how long precautionary measures should be taken. A longer observation period should be considered for certain groups with COVID-19, especially HCW, blood type A group and with low Ab titre for SARS-CoV-2.
Next-generation sequencing of viral genomes recovered from two recurrent cases presenting with symptoms rendered full-length SARS-CoV-2 sequences with over 99.8% genome coverage in the recurrent episodes. Importantly, whole genome sequencing helps in excluding prolonged nucleic acid conversion from positive RT-qPCR to negative, as several studies have described cases with such delayed negative results, even without active infection.40 , 41 The genomic sequences, with higher than 99.8% genome coverage, recovered for both sequenced recurrent cases, also support that qRT-PCR positivity was not due to degraded virus genetic material.40, 41, 42
The sequencing results confirm that qRT-PCR re-positivity in these recurrent symptomatic cases is due to active infection. The detection of phylogenetically distinct genomic sequences in the first and second episodes of one particular patient is also suggestive of re-infection by a different strain. Unfortunately, the other patient, even with a positive PCR, had virus sequenced and analyzed only in the recurrent COVID-19 episode. Similar results were recently described in studies with smaller number of patients.13 , 22 , 43 The large number of cases of recurrence or reinfection may be related to the virus strains circulating locally, or more likely, may be occurring globally but follow-up of patients performed by the “Monitora Corona” and the Project from our University Hospital helped to document these cases.
Viral sequencing revealed the presence of the mutation A23403G in the spike region. This SNP promotes the change from adenine to guanine in the spike-encoding region of the virus genome (A23403G), causing a coding change from aspartate to glycine at position 614 of the protein spike (D614G) and associated with an increase in SARS-CoV-2 infectivity. However this variation is outside the binding domain between the spike and the ACE2 receptor.44 It is a common variant in SARS-CoV-2 isolates worldwide.45 , 46
Another important result from the SARS-CoV-2 sequencing data was the finding of the mutation G25088T, present only in the virus from the patient who had a fatal outcome. This mutation is characterized by the substitution of an amino acid in the spike protein (Val1176Phe). It is widely present in Brazil,47 and it is present more frequently in patients who died in a region of India.48 Another mutation present only in the virus of this patient, was GGG28881AAC, a MNP characterized by the changes of 3 nucleotides, which occurs in the genomic region that encodes the viral nucleocapsid protein, modifying two amino acids in the generated protein. The virus from this patient also has several other rare mutations that are distinctive from the 2 viruses of the other patient, that include one SNP in orf1ab (A6319G) and one in orf3a (T26149C) that were only reported in other 22 viral isolates from Brazil (Supplementary Table S7).
Conclusion
This study reports 33 individuals with recurrent COVID-19, these being largely HCW who had shown low SARS-CoV-2 antibody titres after the initial episode. Recurrence of SARS-CoV-2 cautions us against complacency about protection through natural immunity and poses an additional health problem over and above recovery from initial infection. While the data are compatible with de novo reinfection or virus persistence, the detection of phylogenetically distinct genomic sequences in the first and second episodes of one patient is evidence of reinfection. This relatively large case series in a single HCW cohort argues that recurrence may not be uncommon.
Declaration of Competing Interests
All authors report there are no conflicts of interest.
Funding
This work was supported by grant from CAPES [88887.506662/2020-00 to JSS]. ARJ, JSS, RPA are Scientists from the Brazilian Research Council (CNPq); CNOS, JNA and ILS have fellowships from CAPES - Finance Code 001, and LSM has a postdoctoral fellowship from CAPES, process 88887.506662/2020-00. ARJ, RPA, CNOS and JNA are from Health Science Graduate Program. DMA/RJB received support from UK Research and Innovation and Newton Fund.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.jinf.2021.01.020.
Appendix. Supplementary materials
References
- 1.Na Zhu, Dingyu Zhang, Wenling Wang, Xingwang Li, Bo Yang, Jingdong Song. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020 doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yu Chen, Qianyun Liu, Deyin Guo. Emerging coronaviruses: genome structure, replication, and pathogenesis. J Med Virol. 2020;92(4):418–423. doi: 10.1002/jmv.25681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gandhi Rajesh T., Lynch John B., del Rio Carlos. Mild or moderate covid-19. N Engl J Med. 2020:1–9. doi: 10.1056/nejmcp2009249. [DOI] [PubMed] [Google Scholar]
- 4.Lauer Stephen A., Grantz Kyra H., Qifang Bi, Jones Forrest K., Qulu Zheng, Meredith Hannah R. The incubation period of coronavirus disease 2019 (CoVID-19) from publicly reported confirmed cases: estimation and application. Ann Intern Med. 2020;172(9):577–582. doi: 10.7326/M20-0504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guangming Ye, Zhenyu Pan, Yunbao Pan, Qiaoling Deng, Liangjun Chen, Jin Li. Clinical characteristics of severe acute respiratory syndrome coronavirus 2 reactivation. J Infect. 2020;80(5):e14–e17. doi: 10.1016/j.jinf.2020.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mannan Baig Abdul. Chronic COVID syndrome: need for an appropriate medical terminology for Long-COVID and COVID long-Haulers. J Med Virol. 2020 doi: 10.1002/jmv.26624. [DOI] [PubMed] [Google Scholar]
- 7.Ren-zi Zhang, Wang Deng, Jing He, Yu-yan Song, Chun-fang Qian, Qian Yu. Case report: recurrence of positive SARS-CoV-2 results in patients recovered from COVID-19. Front Med. 2020;7 doi: 10.3389/fmed.2020.585485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marco Bongiovanni, Marco Vignati, Giuseppe Giuliani, Giampiero Manes, Stefania Arienti, Loris Pelucchi. The dilemma of COVID-19 recurrence after clinical recovery. J Infect. 2020;81(6):979–997. doi: 10.1016/j.jinf.2020.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mahalul Azam, Rina Sulistiana, Martha Ratnawati, Ika Fibriana Arulita, Udin Bahrudin, Dian Widyaningrum. Recurrent SARS-CoV-2 RNA positivity after COVID-19: a systematic review and meta-analysis. Sci Rep. 2020;10(1):20692. doi: 10.1038/s41598-020-77739-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.David Harrington, Beatrix Kele, Spiro Pereira, Xose Couto-Parada, Anna Riddell, Suzanne Forbes. Confirmed Reinfection with SARS-CoV-2 Variant VOC-202012/01. Clin Infect Dis. 2021 doi: 10.1093/cid/ciab014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Andrew Rambaut, C. Holmes Edward, Áine O'Toole, Verity Hill, McCrone John T., Christopher Ruis. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat Microbiol. 2020;5(11):1403–1407. doi: 10.1038/s41564-020-0770-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.James Hadfield, Colin Megill, Bell Sidney M., John Huddleston, Barney Potter, Charlton Callender. NextStrain: real-time tracking of pathogen evolution. Bioinformatics. 2018;34(23):4121–4123. doi: 10.1093/bioinformatics/bty407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tillett Richard L., Sevinsky Joel R., Hartley Paul D., Heather Kerwin, Natalie Crawford, Andrew Gorzalski. Genomic evidence for reinfection with SARS-CoV-2: a case study. Lancet Infect Dis. 2020;3099(20):1–7. doi: 10.1016/S1473-3099(20)30764-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kai-Wang To Kelvin, Fan-Ngai Hung Ivan, Daniel Ip Jonathan, Wing-Ho Chu Allen, Wan-Mui Chan, Raymond Tam Anthony. COVID-19 re-infection by a phylogenetically distinct SARS-coronavirus-2 strain confirmed by whole genome sequencing. Clin Infect Dis. 2020;2019(Xx):1–6. doi: 10.1093/cid/ciaa1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kayat S. 2021. Doctor's Note: Can the Coronavirus Reactivate?https://www.aljazeera.com/indepth/features/doctor-note-coronavirus-reactivate200412062905537.html Available from: n.d. [Google Scholar]
- 16.Javad Alizargar. Risk of reactivation or reinfection of novel coronavirus (COVID-19) J Formos Med Assoc. 2020;119(Juny):06. doi: 10.1016/j.jfma.2020.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smith David C., Berger J. South Korea reports more recovered coronavirus patients testing positive again. Reuters. Available at https://www.reuters.com/article/us-health-coronavirus-southkorea-idUSKCN21V0JQ 2020.
- 18.feng Zhang Jing, Kun Yan, hua Ye Hong, Jie Lin, jun Zheng Jian, Ting Cai. SARS-CoV-2 turned positive in a discharged patient with COVID-19 arouses concern regarding the present standards for discharge. Int J Infect Dis. 2020;97:212–214. doi: 10.1016/j.ijid.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yan Chen, Weizhi Bai, Bin Liu, Jian Huang, Irakoze Laurent, Feng Chen. Re-evaluation of retested nucleic acid-positive cases in recovered COVID-19 patients: report from a designated transfer hospital in Chongqing, China. J Infect Public Health. 2020;13(7):932–934. doi: 10.1016/j.jiph.2020.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Minlin Jiang, Ya Li, Mingli Han, Zhenhua Wang, Yuhang Zhang, Xinwei Du. Recurrent PCR positivity after hospital discharge of people with coronavirus disease 2019 (COVID-19) J Infect. 2020;81(1):147–178. doi: 10.1016/j.jinf.2020.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hui Zhu, Liyun Fu, Yinhua Jin, Jiale Shao, Shun Zhang, Nanhong Zheng. Clinical features of COVID-19 convalescent patients with re-positive nucleic acid detection. J Clin Lab Anal. 2020;(April):1–10. doi: 10.1002/jcla.23392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kai-Wang To Kelvin, Fan-Ngai Hung Ivan, Daniel Ip Jonathan, Wing-Ho Chu Allen, Wan-Mui Chan, Raymond Tam Anthony. COVID-19 re-infection by a phylogenetically distinct SARS-coronavirus-2 strain confirmed by whole genome sequencing. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Center of Disease Controle (CDC) 2020. Options to Reduce Quarantine for Contacts of Persons with SARS-CoV-2 Infection Using Symptom Monitoring and Diagnostic Testing. COVID-19.https://www.cdc.gov/coronavirus/2019-ncov/more/scientific-brief-options-to-reduce-quarantine.html Available at. Accessed January 7, 2021. [PubMed] [Google Scholar]
- 24.Jia Huang, Le Zheng, Zhen Li, Shiying Hao, Fangfan Ye, Jun Chen. Recurrence of SARS-CoV-2 PCR positivity in COVID-19 patients: a single center experience and potential implications. MedRxiv. 2020 [Google Scholar]
- 25.Yafang Li, Lin Yao, Jiawei Li, Lei Chen, Yiyan Song, Zhifang Cai. Stability issues of RT-PCR testing of SARS-CoV-2 for hospitalized patients clinically diagnosed with COVID-19. J Med Virol. 2020;92(7):903–908. doi: 10.1002/jmv.25786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.You Zou, Bin-Ru Wang, Liu Sun, Shan Xu, Yong-Gang Kong, Li-Jun Shen. The issue of recurrently positive patients who recovered from COVID-19 according to the current discharge criteria: investigation of patients from multiple medical institutions in Wuhan, China. J Infect Dis [Internet] 2020;1996 doi: 10.1093/infdis/jiaa301. Jun 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xingyu Wang, Hao Xu, Haini Jiang, Liuming Wang, Chao Lu, Xiang Wei. The Clinical Features and Outcomes of Discharged Coronavirus Disease 2019 Patients:A Prospective Cohort Study. QJM An Int J Med. 2020;2(April):1–9. doi: 10.1093/qjmed/hcaa178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bo Yuan, Yong-xin Chen, Kai Zhang, Cheng Wang. Recurrence of Positive SARS-CoV-2 viral RNA in recovered COVID-19 patients during medical isolation observation. Nat Res. 2021:1–12. doi: 10.21203/rs.3.rs-22829/v1. n.d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yoon Yoo Si, Youngseok Lee, Hee Lee Ga, Hyun Kim Dong. Reactivation of SARS-CoV-2 after recovery. Pediatr Int. 2020:10–13. doi: 10.1111/ped.14312. Accepted Author Manuscript. doi:10.1111/ped. Doi: 10.1111/ped.14312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Daniela Loconsole, Francesca Passerini, Vincenzo Ostilio Palmieri, Francesca Centrone, Anna Sallustio, Stefania Pugliese. Recurrence of COVID-19 after recovery: a case report from Italy. Infection. 2020;(0123456789):3–5. doi: 10.1007/s15010-020-01444-1. [DOI] [PubMed] [Google Scholar]
- 31.Araujo Torres Danielle de, Bueno Ribeiro Luciana do Carmo, Linhares Riello Anna Patrícia de Freitas, Gandelman Horovitz Dafne Dain, Ribeiro Pinto Luis Felipe, Julio Croda. Reinfection of COVID-19 after 3 months with a distinct and more aggressive clinical presentation: case report. J Med Virol. 2020 doi: 10.1002/jmv.26637. jmv.26637. [DOI] [PubMed] [Google Scholar]
- 32.Pankaj Gugnani. Coronavirus: it's reactivation in cured patients and role of artificial intelligence for infected patients. SSRN Electron J. 2020 doi: 10.2139/ssrn.3589222. (April 30, 2020). Available at SSRN: https://ssrn.com/abstract=3589222 or http://dx.doi.org/10.2139/ssrn.3589222) [DOI] [Google Scholar]
- 33.Mark Löwenberg, P Verhaar Auke, van den Brink Gijs R., Hommes Daniel W. Glucocorticoid signaling: a nongenomic mechanism for T-cell immunosuppression. Trends Mol Med. 2007;13(4):158–163. doi: 10.1016/j.molmed.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 34.Zhe Xu, Lei Shi, Yijin Wang, Jiyuan Zhang, Lei Huang, Chao Zhang. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir Med. 2020;8(4):420–422. doi: 10.1016/S2213-2600(20)30076-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wong Raymond S.M., Alan Wu, To K.F., Nelson Lee, Lam Christopher W.K., Wong C.K. Haematological manifestations in patients with severe acute respiratory syndrome: retrospective analysis. Br Med J. 2003;326(7403):1358–1362. doi: 10.1136/bmj.326.7403.1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tay Matthew Zirui Poh, Chek Meng Rénia, Laurent MacAry, Paul A., Ng Lisa F.P. The trinity of COVID-19: immunity, inflammation and intervention. Nat Rev Immunol. 2020;20(6):363–374. doi: 10.1038/s41577-020-0311-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reynolds C.J., Swadling L., Gibbons J.M., Pade C., Jensen M.P. Discordant neutralizing antibody and T cell responses in asymptomatic and mild SARS-CoV-2 infection. Sci. Immunol. 2020;5(54) doi: 10.1126/sciimmunol.abf3698. Dec 23eabf3698PMID: 33361161(2020) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sekine Takuya, Perez-potti André, Rivera-ballesteros Olga, Strålin Kristoffer. Robust T cell immunity in convalescent individuals with asymptomatic or mild COVID-19 2020. [DOI] [PMC free article] [PubMed]
- 39.Yuqin Wu, Zhicai Feng, Peng Li, Qizhi Yu. Relationship between ABO blood group distribution and clinical characteristics in patients with COVID-19. Clin Chim Acta. 2020;509(March):220–223. doi: 10.1016/j.cca.2020.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jing Lu, Jinju Peng, Qianling Xiong, Zhe Liu, Huifang Lin, Xiaohua Tan. Clinical, immunological and virological characterization of COVID-19 patients that test re-positive for SARS-CoV-2 by RT-PCR. EBioMedicine. 2020;59 doi: 10.1016/j.ebiom.2020.102960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tang Xiao Ai, Yi Xin Tong, Sheng Zhang. False negative of RT-PCR and prolonged nucleic acid conversion in COVID-19: rather than recurrence. J Med Virol. 2020;92(10):1755–1756. doi: 10.1002/jmv.25855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Viviana Simon, Harm van Bakel, Mia Sordillo Emilia. Positive, again! What to make of “re-positive” SARS-CoV-2 molecular test results. EBioMedicine. 2020;60 doi: 10.1016/j.ebiom.2020.103011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vivek Gupta, Bhoyar Rahul C., Abhinav Jain, Saurabh Srivastava, Rashmi Upadhayay, Mohamed Imran. Asymptomatic reinfection in two healthcare workers from India with genetically distinct SARS-CoV-2. Clin Infect Dis. 2020:1–53. doi: 10.1093/cid/ciaa1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Qihui Wang, Yanfang Zhang, Lili Wu, Sheng Niu, Chunli Song, Zengyuan Zhang. Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell. 2020;181(4):894–904. doi: 10.1016/j.cell.2020.03.045. e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Daniele Mercatelli, M Giorgi Federico. Geographic and genomic distribution of SARS-CoV-2 mutations. Front Microbiol. 2020;11(July):1–13. doi: 10.3389/fmicb.2020.01800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mutluhan Ugurel Osman, Oguz Ata. Turgut-Balik Dilek. An updated analysis of variations in SARS-CoV-2 genome. Turkish J Biol. 2020;44(Special issue 1):157–167. doi: 10.3906/biy-2005-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Candido Darlan S., Claro Ingra M., de Jesus Jaqueline G., Souza William M., Moreira Filipe R.R., Simon Dellicour. Evolution and epidemic spread of SARS-CoV-2 in Brazil. Science. 2020;369(80–):1255–1260. doi: 10.1126/SCIENCE.ABD2161. (6508) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Madhvi Joshi, Apurvasinh Puvar, Dinesh Kumar, Afzal Ansari, Maharshi Pandya, Janvi Raval. Genomic variations in SARS-CoV-2 genomes from Gujarat: underlying role of variants in disease epidemiology. BioRxiv. 2020 doi: 10.1101/2020.07.10.197095. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.