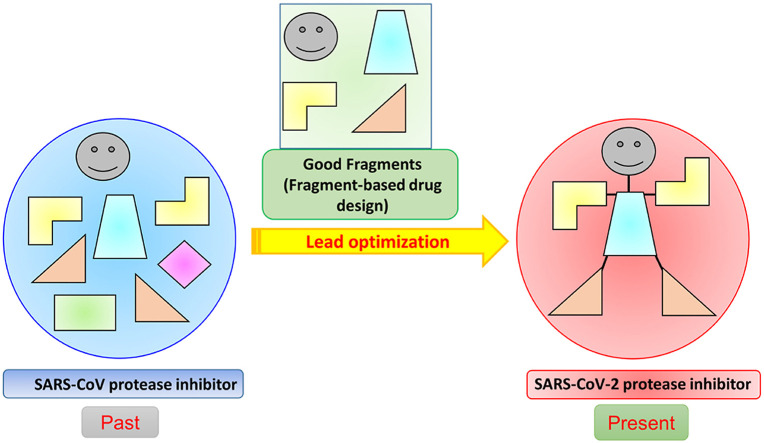
Abstract
The fascinating similarity between the SARS-CoV and SARS-CoV-2, inspires scientific community to investigate deeper into the SARS-CoV proteases such as main protease (Mpro) and papain-like protease (PLpro) and their inhibitors for the discovery of SARS-CoV-2 protease inhibitors. Because of the similarity in the proteases of these two corona viruses, there is a greater chance for the previous SARS-CoV Mpro and PLpro inhibitors to provide effective results against SARS-CoV-2. In this context, the molecular fragments from the SARS-CoV protease inhibitors through the fragment-based drug design and discovery technique can be useful guidance for COVID-19 drug discovery. Here, we have focused on the structure-activity relationship studies of previous SARS-CoV protease inhibitors and discussed about crucial fragments generated from previous SARS-CoV protease inhibitors important for the lead optimization of SARS-CoV-2 protease inhibitors. This study surely offers different strategic options of lead optimization to the medicinal chemists to discover effective anti-viral agent against the devastating disease, COVID-19.
Keywords: COVID-19, SARS-CoV-2, Fragment-based drug design, Mpro, PLpro, Structure-activity relationship (SAR)
Graphical abstract
1. Introduction
The new viral infection COVID-19 was first appeared in the Wuhan province of China at the very end of 2019 and dragged the entire human civilization into a global pandemic by affecting millions of people across the globe [[1], [2], [3], [4], [5]]. This COVID-19 is caused by the novel severe acute respiratory syndrome (SARS) coronavirus-2 (SARS-CoV-2) which belongs to the Beta coronavirus genus and is responsible for lower respiratory tract infection similar to SARS-CoV and MERS-CoV [1,6]. The SARS-CoV-2 is, a 30,000 base pair single-stranded positive-sense RNA virus with an envelope containing spike (S) proteins on the surface, providing a crown-like mien [1,7]. It bears 79% genetic similarity to SARS-CoV and is most similar to bat coronavirus RaTG13 [1]. The incubation period of the COVID-19 infection is 2–14 days and can be up to 24 days [6]. These longer incubation periods, for their transmissibility and asymptomatic nature, are responsible for a large number of infections [6]. To date, billions confirmed COVID-19 cases have been reported with million deaths worldwide [5]. The increasing numbers of COVID-19 cases depict the severity of the current situation and demand an effective solution.
In this situation, no effective drugs have been discovered to combat SARS-CoV-2 infections except a handful of repurposed drugs like chloroquine, hydroxychloroquine, remdesivir, etc. which are being used for the treatment of COVID-19 [2,6]. In case of vaccine development against COVID-19, the safety and efficacy are of major concerns. The majority of the vaccines developed against previous SARS-CoV and MERS-CoV are either inactivated or live-attenuated vaccine in nature [6]. Therefore, systematic rational drug discovery against different targets of COVID-19 is getting increasing attention of different researchers throughout the world.
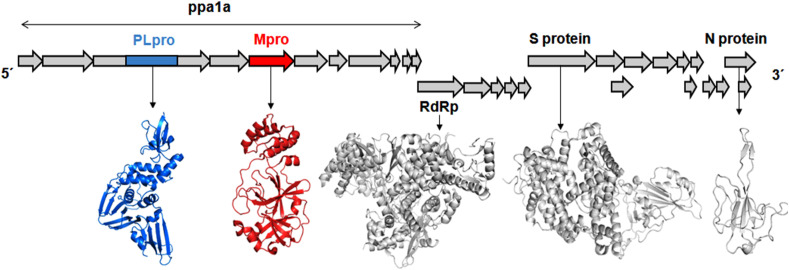
Researches related to COVID-19 were able to elucidate several druggable targets of SARS-CoV-2 including spike (S) protein, 3-chymotrypsin-like protease/main protease (3CLpro/Mpro), papain-like protease (PLpro), RNA dependent RNA polymerase, etc [3,7]. Out of these, viral proteases (PLpro and Mpro) are considered important targets for drug development (Fig. 1 ). In corona viruses, viral proteases are responsible for non-structural proteins (nsps) production by processing viral RNA translated polyproteins [3,4,7]. Hence, the Mpro and PLpro acknowledged great attention for their significant role in the enzymatic activity leading to their post-translational processing of replicase polyproteins those are crucial in the corona virus lifecycle [[8], [9], [10], [11], [12], [13], [14], [15], [16]].
Fig. 1.
Schematic plot of the SARS-CoV-2 genome and proteomes encoding the large replicase polyprotein 1a (pp1a) and pp1ab. Two proteases namely papain-like cysteine protease (PLpro) and 3-chymotrypsin-like protease/main protease (3CLpro/Mpro) are responsible for cleaving these polyproteins to produce important enzymes like RNA-dependent RNA polymerase (RdRp) and helicase, necessary in the transcription and replication of the virus.
The fascinating similarity between the SARS-CoV and SARS-CoV-2 [[2], [3], [4]], inspires us to investigate deeper into the SARS-CoV proteases and their inhibitors for the discovery of SARS-CoV-2 protease inhibitors [[17], [18], [19], [20]]. Because of the similarity in the proteases of these two corona viruses, there is a greater chance for the previous SARS-CoV inhibitors [[21], [22], [23], [24], [25], [26], [27], [28], [29], [30], [31], [32], [33], [34], [35], [36], [37], [38], [39], [40], [41], [42], [43], [44], [45], [46], [47], [48], [49], [50], [51], [52], [53], [54], [55], [56], [57], [58], [59], [60], [61], [62], [63], [64], [65], [66], [67], [68]] to provide effective results against SARS-CoV-2. In this context, the molecular fragments from the SARS-CoV protease inhibitors through the fragment-based drug design and discovery technique can be useful guidance for COVID-19 drug discovery. Thus, in this review, we have focused on the important molecular fragments of SARS-CoV inhibitors to catalyze the drug discovery process for COVID-19.
2. A short trip to human corona virus
SARS-CoV-2 is not the only human corona virus outbreak reported in the 21st century [69,70]. In November 2002, clusters of pneumonia of unknown cause were, disclosed in Guangdong province of China, which was later known as the SARS-CoV outbreak. It infected at least 8422 people globally in 32 countries and causing 916 deaths (fatality rate of ∼10%) [71]. Later in 2012, Middle East respiratory syndrome (MERS) coronavirus (MERS-CoV), epidemic surfaced in Middle Eastern countries, has infected more than 1700 people (fatality rate of ∼36%) [72,73].
It was more than sixty years past when the first identification of HCoV was proclaimed for respiratory tract infections [74,75]. As of seven species of HCoVs were reported for their association with respiratory tract infections, these strains are - (a) HCoV-229E, (b) HCoV-OC43, (c) HCoV- Hong Kong University 1 (HCoV-HKU1), (d) HCoV-NL63, (e) SARS-CoV, (f) MERS-CoV and (g) 2019-nCoV (now officially renamed as SARS-CoV-2). The seventh strain of HCoV, SARS-CoV-2, is taxonomically belongs to the Betacoronavirus genre [7].
HCoV contains a single-stranded positive sense RNA genome [[76], [77], [78], [79], [80]]. The spike glycoprotein of HCoV is the critical conciliator of entry into the host cells [81]. In consequence of virion entry into the host cells, two polyproteins namely pp1a and pp1ab are promptly translated and subsequently, split by two viral proteases such as 3C-like protease (3CLpro) and papain-like protease (PLpro) (Fig. 1). The proteolytic cleavage of these two viral polyproteins fabricated sixteen non-structural proteins (nsp1 to nsp16). The 3CLpro manages the proteolytic cleaving of all junctions downstream of nsp4 while PLpro cleaves nsp1, 2 and 3 [80,82]. Hence, both are climacteric for CoV replication, these two can be considered as druggable targets [[82], [83], [84], [85], [86], [87], [88]].
3. SARS-CoV proteases: a structural overview
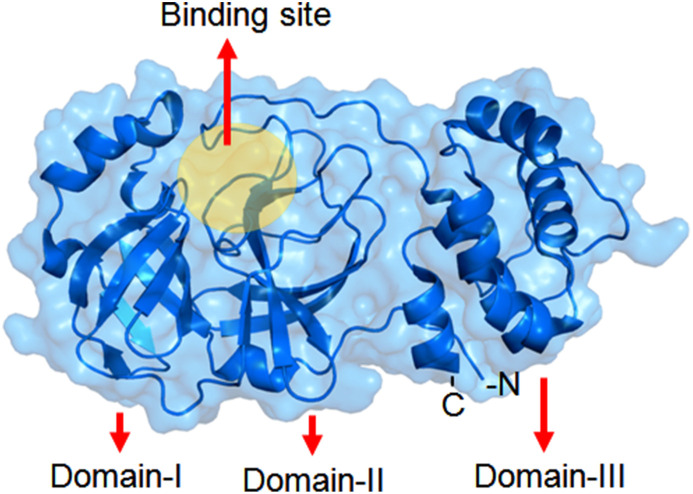
SARS-CoV M-pro is a homodimeric protein [7,89]. Its each subunit is also termed as protomer. A number of 306 amino acid residue is found in each protomer and is constructed by three domains [7]. The domain I is 8–100 residues long followed by domain II (101–184 residues) and domain III (from 199 to 306 residues) [83] (Fig. 2 ).
Fig. 2.
Structure of SARS-CoV Mpro.
Domains I and II allocated the same fold i.e., an antiparallel six stranded β-barrel structure. The substrate-binding site or catalytic site of SARS-CoV-2 Mpro is located at a cleft between domains I and II. Notably, the domain III helps in the regulation of Mpro dimerization. N-terminal amino acid residue of a protomer interacts with another to form the S1 subsite of the substrate-binding pocket [78]. Apart from S1 amino acid residue (from a protomer), F140, L141, N142, H163, E166 (from the other protomer) are involved in the formation of S1 site. Moreover, the S2 site is formed by M49, Y54, H164, D187 and R188. S4 site is formed by M165, L167, Q189, T190 and Q192. Notably, the S1′ is formed by H41, G143, S144 and C145. Meanwhile, H41 and C145 formed a catalytic dyad which modulates the catalysis [87].
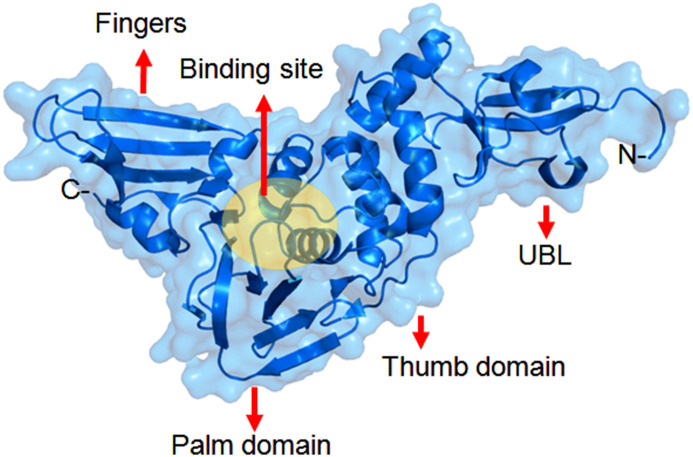
The X-ray structure of SARS-CoV PLpro of the nsp3 polyprotein domains was elucidated [33]. There are three domains in the structure: the thumb, the palm (contains catalytic triad) and the fingers (contains Zn finger motif). The substrate binding site is situated between the thumb and the palm domains. The highly conserved N-terminal region of the nsp3 contains an ubiquitin-like (UBL) domain also (Fig. 3 ). The PLpro enzyme can recognize and cleave ubiquitin, ISG15 (interferon-induced gene 15) proteins. The preference for cleavage of these substrates is after the LXGG motif. The fold of different PLpro enzymes in CoVs are very similar [20]. Therefore, it is considered as an important target for the development of broad spectrum inhibitors against different CoVs.
Fig. 3.
Structure of SARS-CoV PLpro.
Moreover, PLpro not only reveals proteolytic property but also it shows deubiquitinating (DUB) and deISGylating activities [90]. It deubiquitinate or deISGylate host cell proteins and inactivate the NF-κB pathway leading to host cells immune suppression [91,92]. Therefore, inhibiting the PLpro enzyme would potentially block viral replication as well as PLpro-derived host cell immune suppression [82].
The structures of these proteases are also important for the drug development against COVID-19. SARS-CoV-2 proteases and SARS-CoV proteases exhibit high sequence similarities. We have already done a detail structural biology of SARS-CoV-2 proteases in our early communication [69,76]. Thus, in order to minimize the time, cost and uncertainty in drug discovery knowledge from the past SARS-CoV proteases and its inhibitors should be efficiently used into the present drug development/lead optimization against SARS-CoV-2 proteases.
4. An insight into the SARS-CoV protease inhibitors
Since HIV and HCV proteases are fascinating targets for the successful development of antiviral drugs, it is proving the potential of HCoV protease inhibitors to hamper viral replication. In this connection, an enormous medicinal chemistry effort was dedicated to pinpoint effective peptidomimetics and/or small molecules as inhibitors of SARS-CoV Mpro [[21], [22], [23], [24], [25], [26], [27], [28], [29], [30]] and PLpro [[31], [32], [33], [34], [35], [36], [37], [38], [39], [40], [41], [42]]. However, most of the developed compounds displayed SARS-CoV protease inhibitory activities in micromolar (μM) level, only few of them exhibited activity in nanomolar (nM) ranges. Notably, the promising inhibitors with nM affinity towards the protease targets remained in the preclinical or early clinical stage. Nevertheless, these inhibitors are pretended to be an attentive starting point for lead optimization. Hence, the structure-activity relationships (SARs) studies may give rationale behind the effective CoV protease targeting drug design. Besides, we would consider SARs studies the first priority to include a detailed discussion and thus this study offers more insight than the reported studies [[17], [18], [19], [20]]. In addition, we also showed how the fragment-based drug design is preferable towards protease-based drug discovery.
4.1. Understanding chemico-biological interactions of SARS-CoV Mpro inhibitors
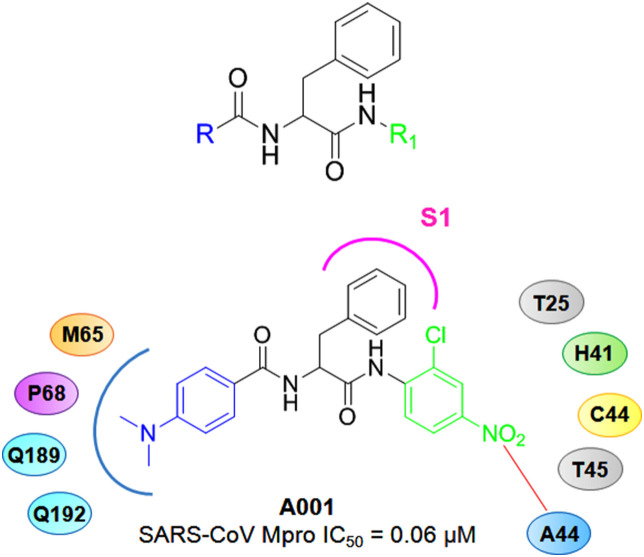
With a target to improve SARS-CoV Mpro inhibitory potency of anilide derivatives Shie and co-workers reported a group of anilides as potent Mpro inhibitors [23]. Although these 2-chloro-4-nitroanilides exhibited a decent Mpro inhibition, the ketomethylene isosters displayed comparatively lower potency in compare to the other derivatives. Compound A001 (Fig. 4 ) containing a N,N-dimethylaminophenyl and 2-chloro-4-nitrophenyl features at the R and R1 position, respectively exhibited promising SARS-CoV Mpro inhibition (IC50 = 0.06 μM) as well as excellent binding affinity (Ki = 0.03 μM).
Fig. 4.
Structures of A001 along with the parent molecule.
The docking study of A001 inside the SARS-CoV Mpro active site demonstrated the binding pattern of A001. The dimethylaminophenyl moiety fitted inside the P68, M165, Q189 and Q192 created cleft while the phenyl ring of the benzyl moiety entered the S1 pocket of the SARS-CoV Mpro. In addition, the 2-chloro-4-nitorphenyl function fitted inside the pocket formed by T25, H41, C44, A46 and T45 where the NO2 group interacted with amino acid residue A46 [23].
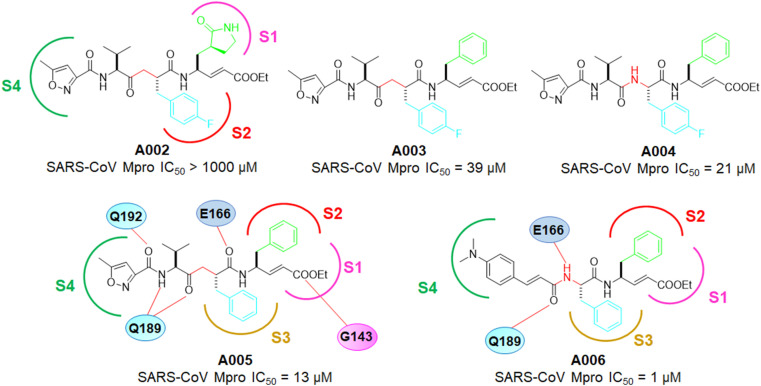
Inspired by the human rhinovirus (HRV) 3Cpro inhibitor, A002, further convening their study on SARS-CoV Mpro inhibitors. The same group developed a set of α,β-unsaturated peptidomimetic molecules as potent inhibitors of coronavirus main protease [22]. From the inhibition assay of these compounds, it has been observed that presence of lactam moiety at P1 (Fig. 5 ) was the detrimental for Mpro inhibition. Replacement of the lactam moiety with the phenyl ring improved the activity of these molecules (A002 vs A003, A004).
Fig. 5.
Structures of A002-A006.
Among the phenyl containing molecules both the ketomethylene isosteric compounds and tetrapeptidomimetic esters exhibited comparatively similar potency. Form the docking study of A002 with SARS-CoV Mpro, it would be observed the insertion of fluorophenyl moiety of A002 into the S2 site. However, the improper fitting of the lactam moiety inside the S1 pocket and disposition of the ester function into the S1ʹ site led to the prevention of suitable bond formation. In contrast, compound A005 (Fig. 5), an analog of A003, fitted well into the Mpro active site and interacted with H163, G143, E166, Q189 leading to its improved Mpro inhibitory activity than A002-A004. Notably, A005 was also unable to form a covalent bond with C145. As a consequence, the authors again design and reported a group of dipeptide inhibitors possessing a Michael acceptor at both ends of these molecules. Among these, A006 (Fig. 5) showed the highest SARS-CoV Mpro inhibitory potency (IC50 = 1 μM) as well as binding affinity (Ki = 0.52 μM). Furthermore, the molecular docking study A006 with SARS-CoV Mpro displayed that the phenyl rings of the molecules entered into the S2 and S3 pockets whilst the cinnamoyl moiety formed interactions with E166, Q189 and E192 [22].
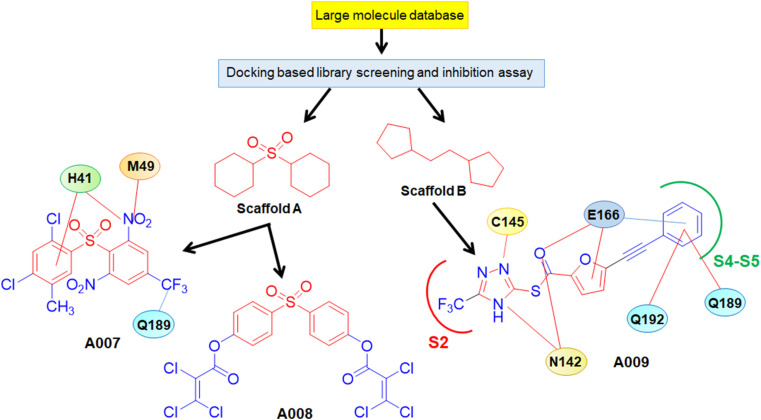
In a structure-based drug design approach to identify potent non-peptide mimetic novel SARS-CoV Mpro inhibitors, Lu et al. [21] screened a wide array of 58,855 compounds and identified two potential hits. Further, the core structures of these two hits were employed to analogue search which gave rise to 21 analogs having promising Mpro inhibitory activities. Among these, compounds of A007 and A008 containing scaffold A and A009 bearing scaffold B possessed potent inhibition against SARS-CoV Mpro (Fig. 6 ).
Fig. 6.
Development of A007-A009.
A007 occupied S3 and S5 pockets of Mpro as suggested by the structural analysis. The 2,4-dichloromethyl phenyl ring of A007 entered deep inside the hydrophobic pocket formed by P39, C145, H41, H163, H164, F182, F185 and Y182. Moreover, the phenyl ring of A007 and amino acid residue H41 formed a π-π interaction. Furthermore, 2,4-dichloro-5-methyl phenyl moiety of resides between the H41 and C145 allowed by shifting of H41 from C145 causing blocking of catalytic dyad, thus inhibiting SARS-CoV Mpro activity. Interestingly, the triazole functions of A009 (Fig. 6) entered into the S2 pocket. Moreover, the trifluoromethyl function of triazole moiety made close contact with the catalytic dyad. The furan ring interacted with E166 while the phenyl associated with the furan ring reached the S4 and S5 pockets to form hydrophobic interaction with Q192, P168, Q189, M165 and E166. Besides, carbothioate linker oxygen of compound A009 was found to interact with E166 and P140 [21].
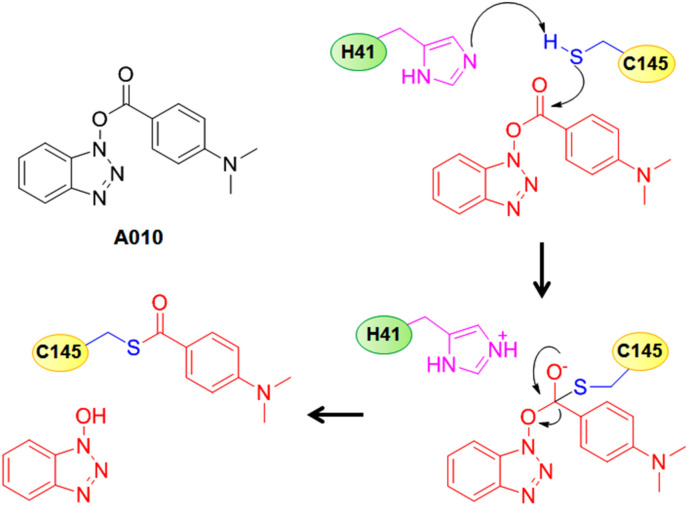
Wu et al. [24] reported a set of benzotriazole ester-based molecules as potent SARS-CoV Mpro inhibitors. These authors found that among those molecules, benzotriazole esters derived from electron donating group containing benzoic acids are more stable compared to the electron withdrawing group containing analogs which were more susceptible to hydrolysis. Upon analyzing the nature of inhibited enzyme, these authors proposed that the 4-dimethylamino benzoyl analogue, A010 interacted with the thiol group of C145 to acylate it and thus, irreversibly deactivating the enzyme (Fig. 7 ) [24].
Fig. 7.
Structure of A010 along with its SARS-CoV Mpro inhibitory mechanism.
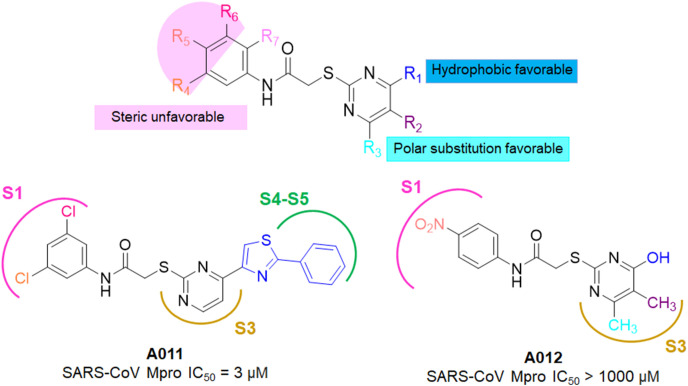
Tsai and collaborators utilized docking based virtual screening technique and screened 93 compounds on the basis of binding interaction patterns and affinity towards SARS-CoV Mpro (PDB: 1UK4) [25]. Further, twentyone compounds were selected with inhibition against SARS-CoV 3CLpro (IC50 ≤ 30 μM). A common N-phenyl-2-(2-pyrimidinylthio)acetamide core structure was pinpointed which was further utilities for analogue search and resulted twentyeight analogs having IC50 values in the range of 3–1000 μM. Meanwhile, the 3D-QSAR study on these twenty eight identified analogs and pharmacophore mapping suggested the important sites where steric and electrostatic interactions along with hydrogen-bond acceptor (HBA), hydrogen-bond donor (HBD) and hydrophobic (HY) features that could significantly contribute to SARS-CoV Mpro inhibition (Fig. 8 ) [25].
Fig. 8.
SARs of N-phenyl-2-(2-pyrimidinylthio)acetamides along with the structures of A011-A012.
In 2008, Mukherjee and colleagues identified two hits, A013 and A014 after extensive virtual screening (VS) study (Fig. 9 ) [26]. The binding poses of A013 and A014 revealed that both of these inhibitors interacted with binding site residues and form hydrophobic as well as hydrogen bond interactions to maintain their Mpro inhibition.
Fig. 9.
Structures of A013-A014.
In spite of occupancy of S1, S1ʹ, and S2 pockets by these inhibitors, the S4 pocket remains unoccupied (Fig. 9). Notably, the thiomethylene carboxamido linker of compound A013 (marked red in Fig. 9) was observed in the A011 as identified by Tsai et al. [25].
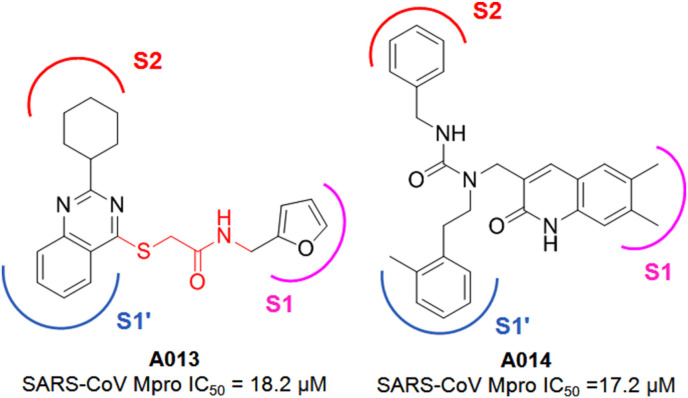
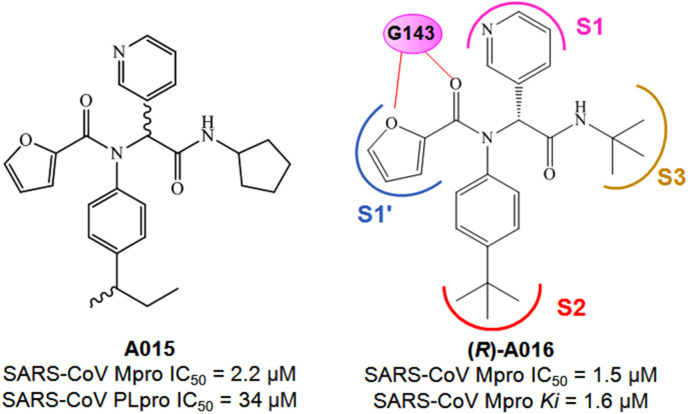
Jacobs and colleagues reported the discovery of several acetamide derivatives using high throughput screening (HTS) and structure-based optimization techniques [28]. In this HTS study, from a large library of molecules, 44 active molecules with Mpro IC50 < 10 μM were identified whereas subsequent counter-screening of these compounds against PLpro activity enabled the identification of hit compound A015 (Fig. 10 ).
Fig. 10.
Structures of A015-A016.
In order to optimize the A015 for more potent Mpro inhibition, these authors utilizing the HTS data and designed a library of 80 molecules bearing a similar scaffold. From this library of molecules, compound A016 (Fig. 10) was identified as a potent Mpro inhibitor. Interestingly, the R-enantiomer of A016 was highly active against Mpro whereas the S-enantiomer of the molecule exhibited its inactive nature. Also, the R-enantiomer displayed its Mpro selective nature while being inactive against PLpro.
From the compound A016 bound crystal structure of SARS-CoV Mpro (PDB: 3V3M), the molecule was found to occupy the S1–S3 pockets of the enzyme active site (Fig. 10). The 3-pyridyl group entered into the S1 pocket whereas the tert-butylamido group occupied the S3 pocket. Also, the tert-butyl anilido moiety (P2) of the molecule occupied the S2 pocket of the enzyme (Fig. 10). Moreover, the oxygen atom from the furan ring and its adjacent carbonyl function were found to interact with G143 amino acid via hydrogen bond interaction.
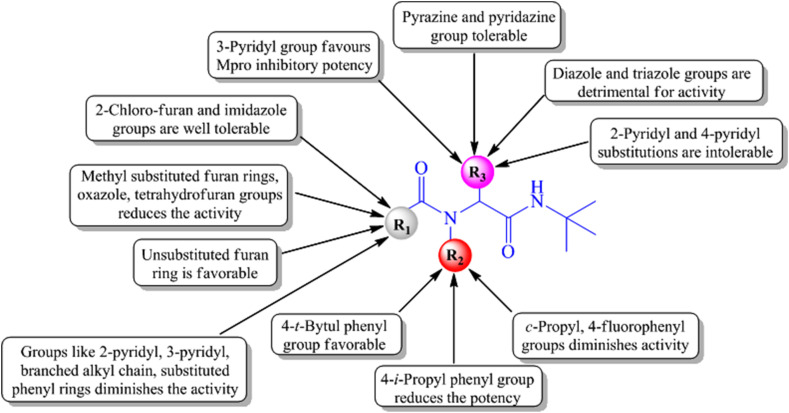
Although further optimization of A016 was also attempted to improve the Mpro inhibitory potency, the optimized compounds were unable to deliver higher Mpro inhibition than A016. The overall structure-activity relationship of these compounds suggested that the 4-tert-butyl phenyl group substitution at the R2 position (P2 substituent) of the scaffold (Fig. 11 ) is beneficial for Mpro inhibition whereas 4-iso-propyl phenyl group at that position reduces the activity. Also, the presence of cyclo-propyl or 4-fluoro phenyl groups at R2 diminished the activity. Regarding the R3 substitution, the 3-pyridyl group was found favorable for Mpro inhibition whereas 3-thienyl group as R3 substituent was deleterious for the activity.
Fig. 11.
Structure-activity relationship of acetamide derivatives.
Additionally, replacement of the R3 (P1 substituent) 3-pyridyl group of A016 with pyrazine or pyridazine groups exhibited well tolerability but were unable to improve the Mpro inhibition. On the other hand, substituting the 3-pyridyl group of A016 with diazole and triazole groups as well as 2-pyridyl and 4-pyridyl groups exhibited detrimental effects on the activity. Concerning the five-membered furan ring present at the R1 position (P1’ substituent) of A016, substituting that furan ring with 2-chloro furan or imidazole groups was well tolerated (Fig. 11). Moreover, substituents like branched alkyl group, 2-pyridyl and 3-pyridyl groups as well as substituted phenyl rings at R1 position showed detrimental effects on the activity [28].
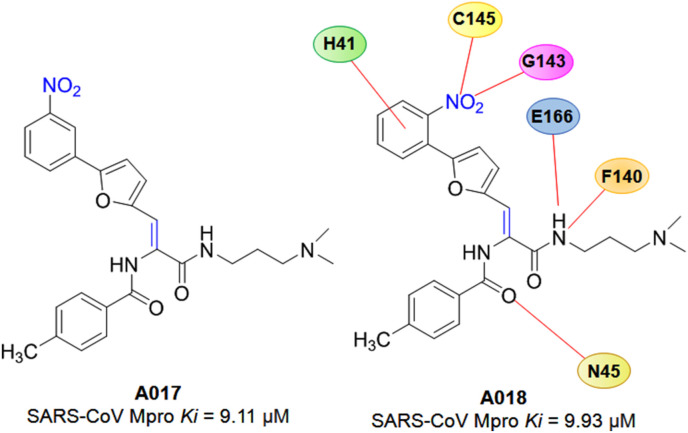
In 2017, a molecular docking-based VS study of 3,08,307 chemical compounds was adopted by Nguyen et al. [29] in an urge to identify SARS-CoV Mpro inhibitors. Finally, seven compounds were selected with their SARS-CoV Mpro IC50 ranged from 38.57 to 101.38 μM. The detailed molecular docking interaction of potent compounds A017 and A018 (Fig. 12 ) referred that the inhibitors formed several hydrogen bonds with catalytic residues and also established hydrophobic contacts with hotspot amino acid residues of SARS-CoV Mpro. Moreover, inhibitors, A017 and A018, competitively inhibit Mpro with Ki values of 9.11 and 9.93 μM, respectively [29].
Fig. 12.
Structures of A017-A018.
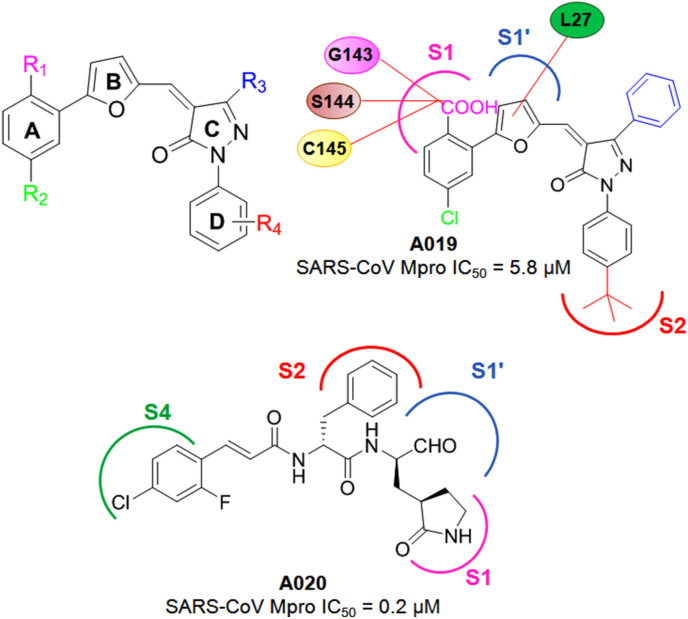
Kumar et al. [30] reported a group of neuramide inhibitors by adopting the screening and synthetic approaches on basis of their previous inhibitors. The phenyl function (ring A) connected to a furan ring (ring B) of these new neuramide inhibitors was found to pivotal while, the presence of carboxylic acid groups at both R1and R4 positions possessed detrimental effect as far as the SARS-CoV Mpro inhibitory activity was concerned (Fig. 13 ). The SAR study suggested that iso-butyl or tert-butyl methyl groups at the R4 position delivered higher SARS-CoV Mpo inhibition compared to their fluorine, carboxylic acid, cyano and methoxy substituted analogs. Additionally, bulky hydrophobic groups such as phenyl ring substitution at the pyrazolone (ring C) R3 position was found to be beneficial compared to the corresponding trifluoromethyl substituted analogs. Besides, pyrazolone carbonyl function was suggested as an important feature to interact with H41, thus inactivating the catalytic dyad. Moreover, the chlorine substitution at the R2 position was found to be important for the activity. The molecular docking study of the most active compound of this series, A019 (Fig. 13), showed that carboxylate function inserted at the S1 sub-site and forming interactions with S144, E143, C145 and H163 whereas, the furan ring was found to interact with L27 side chain via hydrophobic contact [30].
Fig. 13.
Structures of A019-A020.
In another study, the same group identified a set of potent SARS-CoV and MERS-CoV Mpro inhibitors by screening the inhibitors of enterovirus 71 (EV71) Mpro [31]. Among these, aldehyde derivatives in presence of fused heterocyclic ring were found to detrimental whilst the substituted phenyl analogs exhibited potent SARS-CoV Mpro inhibition. Also, the compound A020 containing a 2-fluoro-4-chloro-phenyl moiety showed a 0.2 μM IC50 value against SARS-CoV Mpro along with a MERS-CoV Mpro IC50 value of 4.7 μM (Fig. 13) [31].
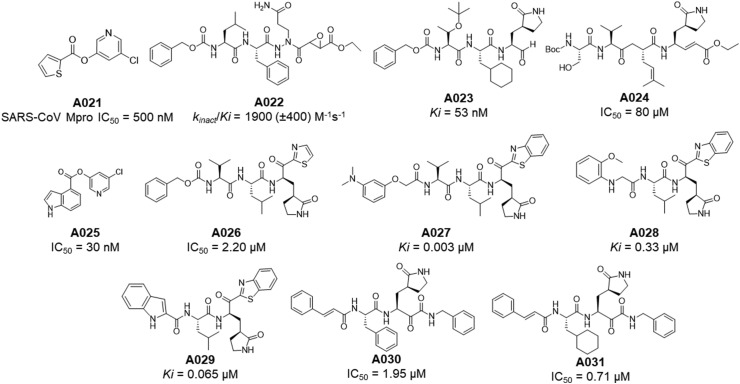
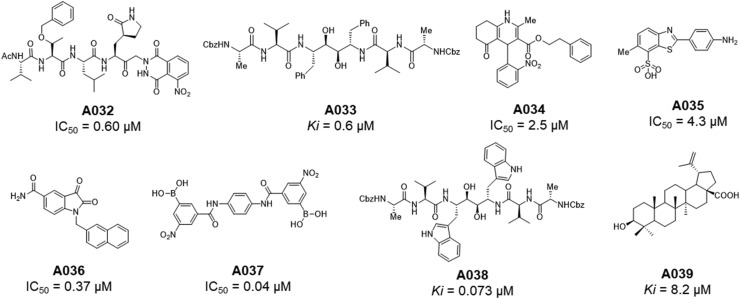
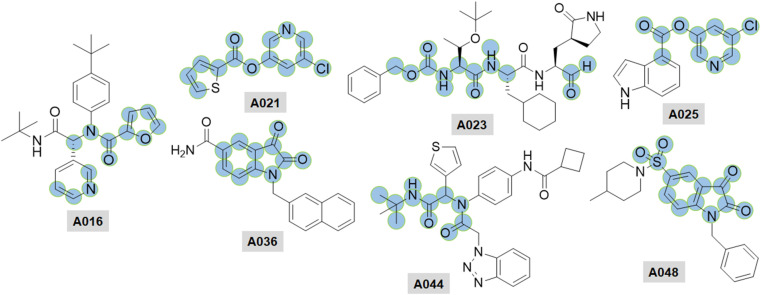
Apart from the above mentioned SARS-CoV Mpro inhibitors, some important peptidomimetic as well as small molecular covalent and non-covalent CoV Mpro inhibitors [27,[43], [44], [45], [46], [47], [48], [49], [50], [51], [52], [53], [54], [55], [56], [57], [58], [59], [60], [61], [62], [63], [64], [65], [66], [67], [68]] are summarised in Table 4, Table 5 , respectively (Fig. 14, Fig. 15, Fig. 16 ).
Table 4.
Peptidomimetic and small molecular covalent SARS-CoV Mpro inhibitors.
| Entry | Comp | PDB | Activity against Mpro | Year | Reference |
|---|---|---|---|---|---|
| 1 | A021 | -- | IC50 = 50 nM | 2004 | [27] |
| 2 | A022 | 2A5K | kinact/Ki = 1900 (±400) M−1s−1 | 2005 | [44] |
| 3 | A023 | 2GX4 | Ki = 53 nM | 2006 | [45] |
| 4 | A024 | 2QIQ | IC50 = 80 μM | 2007 | [46] |
| 5 | A025 | -- | IC50 = 50 nM; antiviral activity EC50 = 6.9 μM | 2008 | [47] |
| 6 | A026 | -- | IC50 = 2.20 μM | 2009 | [48] |
| 7 | A027 | -- | Ki = 0.003 μM | 2013 | [49] |
| 8 | A028 | -- | Ki = 0.33 μM | 2013 | [50] |
| 9 | A029 | -- | Ki = 0.065 μM | 2013 | [51] |
| 10 | A030 | 5N19 | IC50 = 1.95 μM | 2020 | [3] |
| 11 | A031 | -- | IC50 = 0.71 μM | 2020 | [3] |
Table 5.
Peptidomimetic and small molecular non-covalent SARS-CoV Mpro inhibitors.
| Entry | Comp | PDB | Activity against Mpro | Year | Reference |
|---|---|---|---|---|---|
| 1 | A032 | -- | IC50 = 0.60 μM | 2004 | [52] |
| 2 | A033 | -- | Ki = 0.6 μM | 2004 | [53] |
| 3 | A034 | -- | IC50 = 2.5 μM | 2004 | [54] |
| 4 | A035 | -- | IC50 = 4.3 μM | 2004 | [27] |
| 5 | A036 | -- | IC50 = 0.37 μM | 2006 | [56] |
| 6 | A037 | -- | IC50 = 0.04 μM | 2006 | [56] |
| 7 | A038 | -- | Ki = 0.073 μM | 2007 | [57] |
| 8 | A039 | -- | Ki = 8.2 μM | 2007 | [58] |
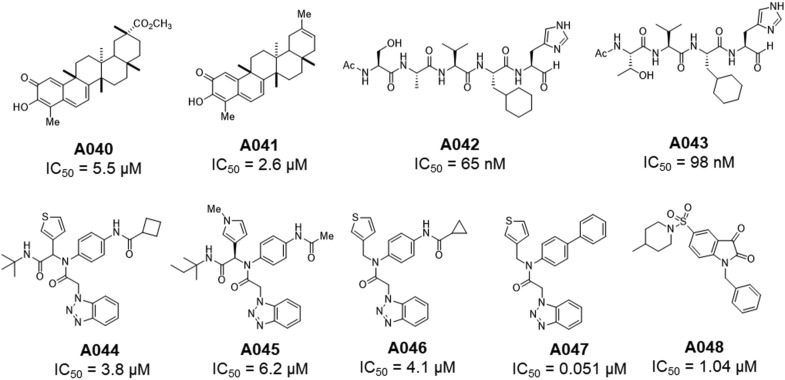
| 9 | A040 | -- | IC50 = 5.5 μM | 2010 | [62] |
| 10 | A041 | -- | IC50 = 2.6 μM | 2010 | [62] |
| 11 | A042 | -- | IC50 = 65 nM | 2011 | [63] |
| 12 | A043 | 3ATW | IC50 = 98 nM | 2011 | [63] |
| 13 | A044 | -- | IC50 = 3.8 μM | 2013 | [64] |
| 14 | A045 | 4MDS | IC50 = 8.2 μM | 2013 | [64] |
| 15 | A046 | -- | IC50 = 4.1 μM | 2013 | [64] |
| 16 | A047 | -- | IC50 = 0.051 μM | 2013 | [64] |
| 17 | A048 | -- | IC50 = 1.04 μM | 2014 | [65] |
Fig. 14.
Structures of A021-A031.
Fig. 15.
Structures of A032-A039.
Fig. 16.
Structures of A040-A048.
The close associateship of SARS-CoV-2 Mpro to homologues SARS-CoV Mpro exhibits a high sequence identity (∼96.1%). It confers the chance of efficiency of SARS-CoV Mpro inhibitors towards SARS-CoV-2.
4.2. Understanding chemico-biological interactions of SARS-CoV PLpro inhibitors
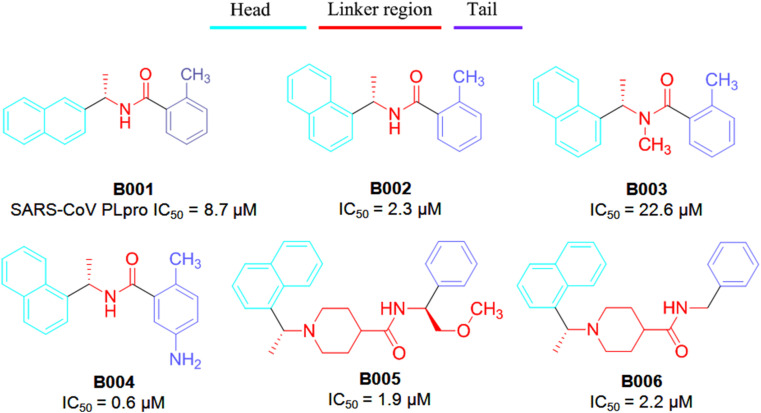
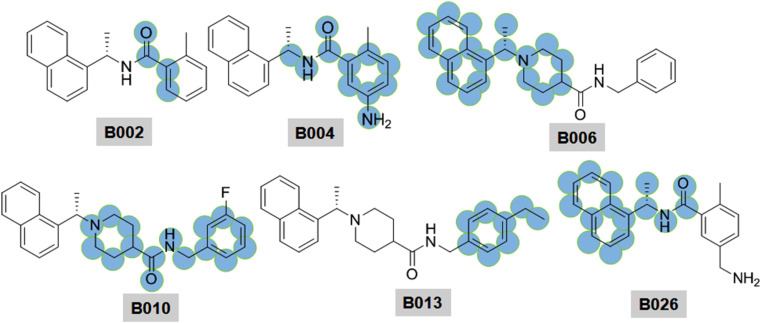
PLpro reveals proteolytic activity. Besides, it shows deubiquitinating (DUB) and deISGylating activities [82]. Ratia et al. [33] reported naphthyl derivatives were as non-covalent competitive inhibitors of SARS-CoV PLpro. Interestingly, these investigated naphthyl derivatives possessing acanonical pharmacophoric features such as head, linker and tail (Fig. 17 ).
Fig. 17.
Structure of SARS-CoV PLpro inhibitors (B001–B006).
The head region is mostly conserved in between 1 and 2-naphthyl features [[33], [34], [35], [36]]. SAR studies revealed that the 2-naphthyl head disfavors PLpro inhibition over 1-naphthyl analog (compound B001 vs B002–B006). The structure-based studies revealed that naphthyl derivatives bind within the S4–S3 sub-sites of PLpro which likely to induce a loop closure and subsequently, undergone conformational change and thereby, led to PLpro inactivation.
Besides, the stereochemical pattern of the methyl substituent served as a critical factor to modulate PLpro bining affinity. Báez-Santos et al. [34] reported that increasing steric properties at that position potentially hampered the PLpro inhibitory activity. The potential entropic gain may displace the water molecules resulted in a larger enthalpic penalty of breaking hydrogen bonds. Moreover, the stereo-chemistry of the methyl substituent at R3 influences the PLpro binding affinity. The (R)-methyl enantiomer extends into an interior of the PLpro enzyme between Y265 and T302 [34].
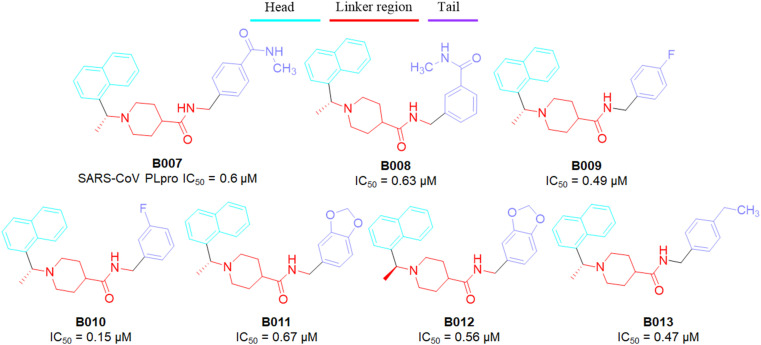
The amide NH (as linker) was found to be a key regulator of PLpro inhibition. Methylation at the aminde nitrogen atom led to loss in SARS-CoV PLpro inhibitory activity (the N-methyl derivative B003, IC50 = 22.6 μM vs compound B002, IC50 = 2.3 μM). Piperidine function has been pleasurable towards biological activity against PLpro enzyme. It exhibited positive effects on SARS-CoV PLpro inhibitory activity. In addition, the SAR study suggested that bulky R4 substituents had little influences towards PLpro inhibition (compound B005 vs B006 in Fig. 17) [34]. At the tail portion, substitutions at the 3rd and/or 4th position of the phenyl ring were suitable (compound B007–B010 vs B006) to achieve potency in nano-molar ranges (Fig. 18 ).
Fig. 18.
Structure of SARS-CoV PLpro inhibitors (B007–B013).
Notably, dioxolane derivatives B011–B012 and 4-ethyl prototype B013 exhibited excellent PLpro inhibitory activities. The interaction between the dioxolane group and catalytic site amino acid Q270 has contributed positively to the PLpro inhibition. Although there should be another influences which likely to contribute largely to the substituted phenyls. Unlike fluorine substitution at the phenyl ring induced conspicuous polarization effects in the π-system of associated phenyl which may elevate the binding affinity [34]. However, the methoxy group at the terminal phenyl ring displayed the negative influence towards SARS-CoV PLpro inhibition.
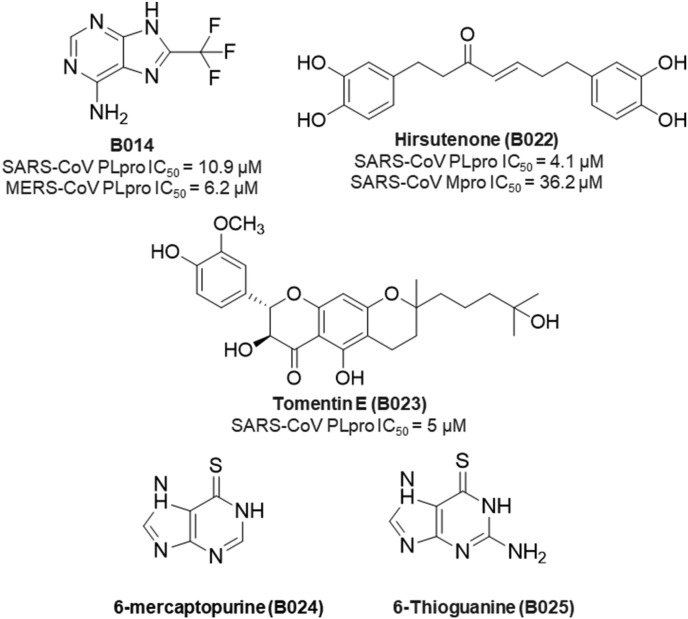
In spite of promising SARS-CoV PLpro inhibition, naphthyl derivatives exhibited poor MERS-CoV PLpro inhibitory activity. Lee and co-workers very nicely explain the reason behind this poor activity [42]. These authors also reported compound B014 (Fig. 19 ) as promising dual PLpro inhibitors of SARS-CoV (IC50 = 10.9 μM) and MERS-CoV (IC50 = 6.2 μM) after screening 25,000 compounds [42].
Fig. 19.
Structure of some SARS-CoV PLpro inhibitors.
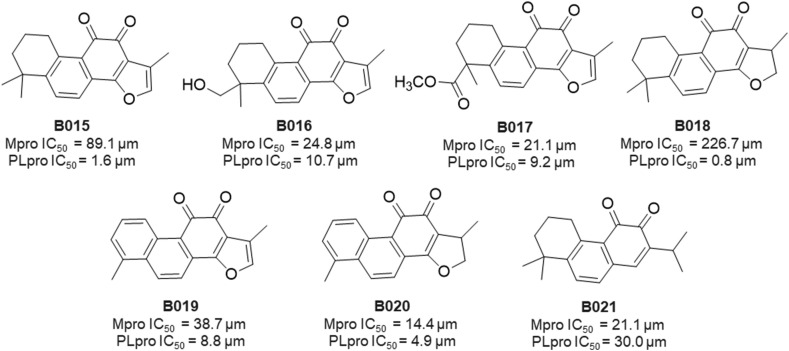
In 2012, a group of Scientists from Republic of Korea reported SARS-CoV Mpro and PLpro inhibitory properties of seven isolated tanshinones (B015–B021) from Salvia miltiorrhiza (Laminaceae) [37]. Significantly, all of these isolated compounds exhibited inhibitory activities against both cysteine proteases. In particular, these compounds (B015–B021) displayed significant SARS-CoV PLpro inhibition ranging from 0.8 to 30.0 μM (Fig. 20 ) [37].
Fig. 20.
Structure of SARS-CoV PLpro inhibitors (B015–B021).
Diarylheptanoids, isolated from Alnus japonica, exhibited promising SARS-CoV PLpro inhibitory activities. Among all these compounds, hirsutenone (B022) possessed the most active PLpro inhibitory activity (IC50 = 4.1 μM) comparatively better than curcumin (IC50 = 5.7 μM) [40].
Later, Cho and collaborators reported geranylated flavonoids from the fruits of Paulownia tomentosa to exert SARS-CoV PLpro inhibitory activities [41]. Of the isolated dihydro-2H-pyran moiety, tomentin E (B023, Fig. 19) showed the most potent activity against SARS-CoV PLpro (IC50 = 5 μM).
Purine analogs such as 6-mercaptopurine (6 MP, B024) and 6-thioguanine (6 TG, B025) as shown in Fig. 19 were found to inhibit SARS-CoV PLpro. These were categories as reversible and slow-binding inhibitors of SARS-CoV PLpro [39].
5. How the fragment-based drug design is preferable towards protease-based drug discovery?
In the field of drug development, the utilization of different computer-aided drug design techniques such as structure-based and ligand-based drug design approaches, virtual screening of molecule libraries have become efficient means for hit identification and lead optimization [7,[93], [94], [95], [96], [97], [98], [99], [100], [101]]. Fragment-based drug design (FBDD) approaches for lead optimization has also gained quite a reputation in both industrial and academic levels becoming a successful tool for modern drug discovery [99]. FBDD approaches have several advantages such as a) this method can provide lead molecules with better physicochemical properties, b) fragments have chances to produce good interactions, c) fragments with lesser complex structures can give higher rates of hit molecules, d) fragment-derived leads can be less hydrophobic and smaller and e) able to deliver high-quality lead molecules.
Choudhury from the PGIMER, Chandigarh, India screened a huge library of 191678 fragments against the SARS-CoV-2 Mpro binding cavity to pick fragments with high affinity [8]. Depending on the higher binding affinity, new molecules were designed. Interestingly, fifteen designed molecules formed stable complexes with SARS-CoV-2 Mpro as suggested by the molecular docking and Molecular Mechanics-Generalized Born Surface Area (MM/GBSA) based binding free energy.
In a recent study, Douangamath et al. [9] reported 74 high-value fragment hits after screening 1250 unique fragments against Mpro of SARS-CoV-2. Out of these 74 fragments, 23 non-covalent and 48 covalent hits in the active site and rest hits were distinguished at the vital dimerization interface.
Depending on our previous reported molecular modelling studies [69,76,102], we have identified fragments and marked in Fig. 21, Fig. 22 . A compound with furan and/or pyridine exerts effective SARS-CoV Mpro inhibitory activity (Fig. 21). Hence, it may be anticipated that furan and/or pyridine obviously put some more interaction benefit towards biological activity.
Fig. 21.
Important fingerprints are highlighted in the SARS-CoV Mpro inhibitors.
Fig. 22.
Important fingerprints are highlighted in the SARS-CoV PLpro inhibitors.
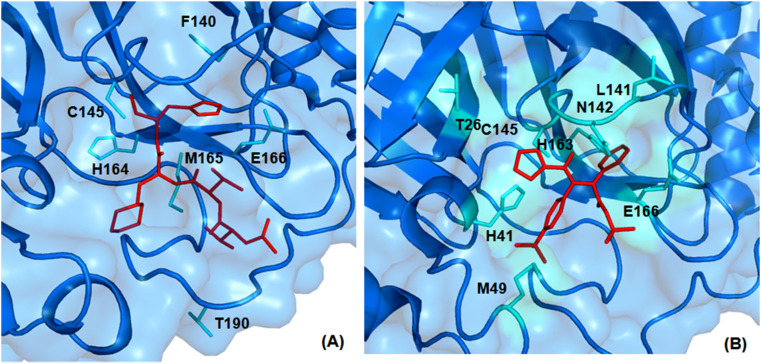
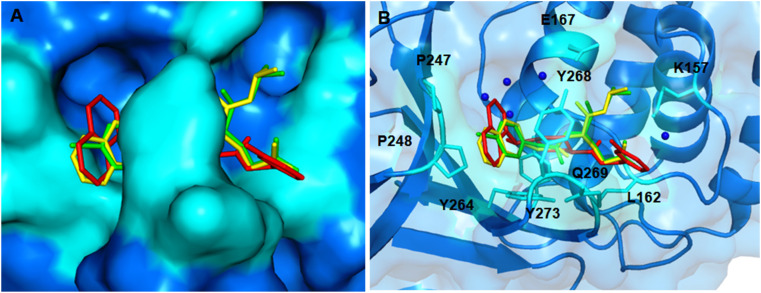
For instance, the furan oxygen atom of compound A016 interacts with the backbone NH of G143 by forming hydrogen bonding interaction (PDB: 3V3M). Likewise, the fingerprint CC(C)NC(=O)CNC(=O) in compounds found to interact with several active site amino acid residues of SARS-CoV Mpro (PDB: 3ATW) as illustrated in Fig. 23 .
Fig. 23.
3D interaction plot of prototype compounds with SARS-CoV 3CLpro active site amino acid residues (A) PDB: 3ATW and (B) PDB: 3V3M.
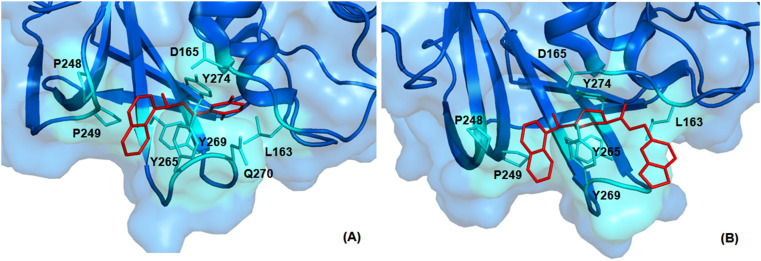
These distinguishing key molecular fragments revealed influences towards the SARS-CoV Mpro inhibitory activities. The studied fragments may leave considerable space to follow-up anti-SARS-CoV-2 drug design. Similarly, the 1-naphthalene (at the head), piperidine (at the linker) and benzamide (at the tail) moieties are the fragment of interest for the SARS-CoV PLpro inhibitory activities. The naphthalene ring forms interactions with amino acids P248, P249 and Y269 at the solvent exposed site of SARS-CoV PLpro (PDB: 3MJ5) as shown in Fig. 24 A and B. Very recently, 1-naphthalene based derivatives were reported to display SARS-CoV-2 PLpro inhibition [103].
Fig. 24.
3D interaction plots of naphthyl based PLpro inhibitors with SARS-CoV PLpro active site amino acid residues (A) PDB: 3E9S and (B) PDB: 3MJ5.
The piperidine ring is also found to be pivotal for SARS-CoV PLpro inhibition since it engages in π-sigma interaction with the Y265 of the enzyme (PDB: 3MJ5), as depicted in Fig. 24B. Furthermore, the amide group of benzamide moiety of a compound interacts with the side chain of D165 and the backbone nitrogen atom of Q270 (PDB: 3E9S) by forming hydrogen bonding interaction (Fig. 24A).
In an endeavour, our research team explored the crucial structural fingerprints modulating SARS-CoV PLpro inhibitory activities by the aid of 2D-QSAR, SPCI analysis as well as Monte Carlo optimization based QSAR [69]. Our research group also performed a molecular docking study of some in-house isoglutamine derivatives against putative target SARS-CoV-2 PLpro (Fig. 25 ). These derivatives may serve as a seed which sustain significant hope against corona virsus.
Fig. 25.
Binding interaction of a prototype in-house naphthyl derivative with SARS-CoV-2 PLpro.
Hence, these fragments identified by the previous studies may be an effective approach to accelerate drug design against SARS-CoV-2. Notably, the identified fragments could be implemented in the medicinal chemistry endeavors of COVID-19 and other CoV drug discovery also. Our modeling approach will give several strategic options to the medicinal chemists for the lead optimization of these protease inhibitors. With the incorporation of these fragments, potent inhibitors may be designed and synthesized. Some fragments are matching for SARS-CoV-2 PLpro, 3CLpro inhibitors and other fragments may give encouraging results in future. Since the protease based COVID-19 drug discovery is in its very early stage, the transferability of patterns may be judged with patience.
6. Conclusion
It is well known fact that many of these viruses encode one or more proteases employed in processes like maturation, viral poly protein processing, etc [104]. As a boon of such involvements, viral proteases have been lucrative targets to design potent viral protease inhibitors. Historically, in diseases like human immune deficiency virus (HIV) and hepatitis C virus (HCV) infections, targeting the viral proteases were able to provide several anti-viral agents which were effective against those infections [99,100]. Moreover, the HIV protease inhibitors lopinavir and ritonavir were effective against SARS-CoV and MERS-CoV and were also tested clinically against COVID-19 [4,6,101]. Hence, the proteases of SARS-CoV-2 are druggable targets to develop anti-viral agents and have higher odds to achieve the desirable anti-viral agent for COVID-19 treatment.
Meanwhile, desirable pharmacokinetic characteristics are very important for drug discovery and development. Reports already supported that macromolecular and/or peptidomimetic compounds are advantageous over the low molecular weight compounds in terms of Mpro and PLpro inhibitory potency as well as selectivity [104]. However, these types of compound may exhibit ADME issues. An analysis of the reported SARS-CoV Mpro and PLpro inhibitors for their drug likeliness properties has revealed that compounds A002-A003, A005-A006, A008, A019, A022-A023, A024, A026, A027, A032-A033, A037-A038, A042-A043 failed to pass Lipinski rule of five principle [105] due to combinations of different factors, e.g. AlogP: 5, molecular weight: 500, number of hydrogen bond acceptors: 10, number of hydrogen bond donor: 5 [106]. In this scenario, small molecule based lead optimization strategies may be beneficial. In our previous study [107], we have shown that baicalein (MW 270.24 Da) based lead optimization by taking into account different good fragments can generate potential inhibitors and may effectively block the SARS-CoV-2 Mpro target. The design of protease inhibitors should also take into account the PAINS (Pan-assay interference compounds) alert as these compounds can bind nonspecifically with different target. An analysis of the SARS-CoV Mpro and PLpro inhibitors by SwissADME [108] has revealed that compounds A006, B015–B021 have possible PAINS alert to be avoided in the lead optimization of protease inhibitors.
In addition to this, there is a high possibility of nonsense mutation to be introduced in the viral genome of SARS-CoV and SARS-CoV-2 due to the higher replication rate as well as mutation frequency of the virus. In this case, rational drug design of protease inhibitors targeting the conserved catalytic residues in the active site will be a preferred strategy [109]. Among the two proteases, drug discovery against PLpro target is challenging due to interference of host-cell deubiquitinases [110]. However, Mpro can be an ideal target of reduced off-target effects [111,112] as no human host-cell proteases are known with similar substrate specificity.
Declaration of competing interest
The authors have no conflict of interests.
Acknowledgment
Sk. Abdul Amin sincerely acknowledges Council of Scientific & Industrial Research (CSIR), New Delhi, India for awarding the Senior Research Fellowship [FILE NO.: 09/096(0967)/2019-EMR-I, Dated: 01-04-2019]. Suvankar Banerjee and Tarun Jha are thankful for the financial support from RUSA 2.0 of UGC, New Delhi, India to Jadavpur University, Kolkata, India. We are very much thankful to the Department of Pharmaceutical Technology, Jadavpur University, Kolkata, India and Department of Pharmaceutical Sciences, Dr. Harisingh Gour University, India for providing the research facilities.
Biographies
Sk. Abdul Amin is a Council of Scientific & Industrial Research (CSIR)-Senior Research Fellow at Department of Pharmaceutical Technology, Jadavpur University, Kolkata, India. His research area includes design and synthesis of small molecules with anti-cancer and anti-viral properties, computational chemistry, and large-scale structure-activity relationship analysis. He has published seventy four research/review articles in different reputed peer-reviewed journals and four book chapters. His SCOPUS h-index is 17 (till 15th Feb 2020). Apart from that he is a heritage enthusiast and travel writer. His interests are history through the lens of Art, culture, and religion. He enjoys a good conversation on science, regional history, contemporary art and books.
Suvankar Banerjee is a Research Scholar at Department of Pharmaceutical Technology, Jadavpur University, Kolkata, India. He is working under the guidance of Tarun Jha. His research area includes design of small molecules with anti-cancer and anti-viral properties. He has published twelve articles in different reputed peer-reviewed journals.
Shovanlal Gayen, is an Assistant Professor at Department of Pharmaceutical Sciences, Dr. Harisingh Gour University, Sagar, India. He is actively involved in different drug design and discovery projects. He has published more than ninety research articles in different reputed peer-reviewed journals and has filed two Indian patents.
Tarun Jha, a Professor at Department of Pharmaceutical Technology, Jadavpur University, Kolkata, India, has supervised 16 Ph.D. students and guided nine research projects funded by different organizations. His research area includes design and synthesis of anti-cancer small molecules. He has published more than 175 research articles in different reputed peer-reviewed journals. Prof. Jha is a member of the Academic Advisory Committee of National Board of Accreditation (NBA), New Delhi, India.
Abbreviations used
- 3CLpro
3C-like protease or main protease
- CoV
coronavirus
- COVID-19
coronavirus disease 2019
- E protein
envelope protein
- EBOV
Ebola virus
- Mpro
main protease
- M protein
membrane protein
- MERS-CoV
Middle East respiratory syndrome coronavirus
- N protein
nucleocapsid protein
- Nsp
non-structural proteins
- NTD
N-terminal domain
- ORF
open reading frame
- PLpro
papain-like protease
- QSAR
Quantitative structure-activity relationship
- RdRp
RNA-dependent RNA polymerase
- S protein
spike protein
- SAR
Structure-activity relationship
- SARS-CoV
severe acute respiratory syndrome coronavirus
- SARS-CoV-2
severe acute respiratory syndrome coronavirus 2
- SPCI
Structural and physico-chemical interpretation
- WHO
World Health Organization
References
- 1.Tay M.Z., Poh C.M., Rénia L., A MacAry P., P Ng L.F. The trinity of COVID-19: immunity, inflammation and intervention, Nat. Rev. Immunol. 2020;20:363–374. doi: 10.1038/s41577-020-0311-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Estrada E. Topological analysis of SARS CoV-2 main protease. Chaos. 2020:30. doi: 10.1063/5.0013029. 061102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang L., Lin D., Sun X., Curth U., Drosten C., Sauerhering L., Becker S., Rox K., Hilgenfeld R. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved alpha-ketoamide inhibitors. Science. 2020;368:409–412. doi: 10.1126/science.abb3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dai W., Zhang B., Jiang X.-M., Su H., Li J., Zhao Y., Xie X., Jin Z., Peng J., Liu F., Li C., Li Y., Bai F., Wang H., Cheng X., Cen X., Hu S., Yang X., Wang J., Liu X., Xiao G., Jiang H., Rao Z., Zhang L.-K., Xu Y., Yang H., Liu H. Structure-based design of antiviral drug candidates targeting the SARS-CoV-2 main protease. Science. 2020;368:1331–1335. doi: 10.1126/science.abb4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.WHO Coronavirus Disease (Covid-19) Dashboard https://covid19.who.int/ [accessed on 2nd Feb., 2021]
- 6.Florindo H.F., Kleiner R., Vaskovich-Koubi D., Acúrcio R.C., Carreira B., Yeini E., Tiram G., Liubomirski Y., Satchi-Fainaro R. Immune-mediated approaches against COVID-19. Nat. Nanotechnol. 2020;15:630–645. doi: 10.1038/s41565-020-0732-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Amin S.A., Jha T. Fight against novel coronavirus: a perspective of medicinal chemists, Eur. J. Med. Chem. 2020;201:112559. doi: 10.1016/j.ejmech.2020.112559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choudhury C. Fragment tailoring strategy to design novel chemical entities as potential binders of novel corona virus main protease. J. Biomol. Struct. Dyn. 2020 doi: 10.1080/07391102.2020.1771424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Douangamath A., Fearon D., Gehrtz P., Krojer T., Lukacik P., Owen C.D., Resnick E., Strain-Damerell C., Aimon A., Ábrányi-Balogh P., Brandão-Neto J., Carbery A., Davison G., Dias A., Downes T.D., Dunnett L., Fairhead M., Firth J.D., Jones S.P., Keeley A., Keserü G.M., Klein H.F., Martin M.P., Noble M.E.M., O’Brien P., Powell A., Reddi R.N., Skyner R., Snee M., Waring M.J., Wild C., London N., von Delft F., Walsh M.A. Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease. Nat. Commun. 2020;11:5047. doi: 10.1038/s41467-020-18709-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rut W., Lv Z., Zmudzinski M., Patchett S., Nayak D., Snipas S.J., El Oualid F., Huang T.T., Bekes M., Drag M., Olsen S.K. Activity profiling and crystal structures of inhibitor-bound SARS-CoV-2 papain-like protease: a framework for anti-COVID-19 drug design. Sci. Adv. 2020:6. doi: 10.1126/sciadv.abd4596. eabd4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McClain C.B., Vabret N. SARS-CoV-2: the many pros of targeting PLpro, Signal Transduct. Target Ther. 2020;5:223. doi: 10.1038/s41392-020-00335-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Derosa G., Maffioli P., D’Angelo A., Di Pierro F. A role for quercetin in coronavirus disease 2019 (COVID-19) Phytother Res. 2020 doi: 10.1002/ptr.6887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gao X., Qin B., Chen P., Zhu K., Hou P., Wojdyla J.A., Wang M., Cui S. Crystal structure of SARS-CoV-2 papain-like protease. Acta Pharm. Sin. B. 2020 doi: 10.1016/j.apsb.2020.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vuong W., Khan M.B., Fischer C., Arutyunova E., Lamer T., Shields J., Saffran H.A., McKay R.T., van Belkum M.J., Joyce M.A., Young H.S., Tyrrell D.L., Vederas J.C., Lemieux M.J. Feline coronavirus drug inhibits the main protease of SARS-CoV-2 and blocks virus replication. Nat. Commun. 2020;11:4282. doi: 10.1038/s41467-020-18096-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhu W., Xu M., Chen C.Z., Guo H., Shen M., Hu X., Shinn P., Klumpp-Thomas C., Michael S.G., Zheng W. Identification of SARS-CoV-2 3CL protease inhibitors by a quantitative high-throughput screening, ACS Pharmacol. Transl. Sci. 2020;3:1008–1016. doi: 10.1021/acsptsci.0c00108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Loffredo M., Lucero H., Chen D.-Y., O’Connell A., Bergqvist S., Munawar A., Bandara A., De Graef S., Weeks S.D., Douam F., Saeed M., Munawar A.H. The effect of famotidine on SARS-CoV-2 proteases and virus replication. bioRxiv. 2020 doi: 10.1101/2020.07.15.203059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li Q., Kang C. Progress in developing inhibitors of SARS-CoV-2 3C-like protease. Microorganisms. 2020;8:1250. doi: 10.3390/microorganisms8081250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ullrich S., Nitsche C. The SARS-CoV-2 main protease as drug target, Bioorg. Med. Chem. Lett. 2020;30:127377. doi: 10.1016/j.bmcl.2020.127377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ghosh A.K., Brindisi M., Shahabi D., Chapman M.E., Mesecar A.D. Drug development and medicinal chemistry efforts toward SARS-coronavirus and Covid-19 therapeutics. ChemMedChem. 2020;15:907–932. doi: 10.1002/cmdc.202000223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Petushkova A.I., Zamyatnin A.A., Jr. Papain-like proteases as coronaviral drug targets: current inhibitors, opportunities, and limitations. Pharmaceuticals. 2020:13. doi: 10.3390/ph13100277. Basel. E277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lu I.L., Mahindroo N., Liang P.H., Peng Y.H., Kuo C.J., Tsai K.C., Hsieh H.P., Chao Y.S., Wu S.Y. Structure-based drug design and structural biology study of novel nonpeptide inhibitors of severe acute respiratory syndrome coronavirus main protease. J. Med. Chem. 2006;49:5154–5161. doi: 10.1021/jm060207o. [DOI] [PubMed] [Google Scholar]
- 22.Shie J.J., Fang J.M., Kuo T.H., Kuo C.J., Liang P.H., Huang H.J., Wu Y.T., Jan J.T., Cheng Y.S., Wong C.H. Inhibition of the severe acute respiratory syndrome 3CL protease by peptidomimetic alpha,beta-unsaturated esters. Bioorg. Med. Chem. 2005;13:5240–5252. doi: 10.1016/j.bmc.2005.05.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shie J.J., Fang J.M., Kuo C.J., Kuo T.H., Liang P.H., Huang H.J., Yang W.B., Lin C.H., Chen J.L., Wu Y.T., Wong C.H. Discovery of potent anilide inhibitors against the severe acute respiratory syndrome 3CL protease. J. Med. Chem. 2005;48:4469–4473. doi: 10.1021/jm050184y. [DOI] [PubMed] [Google Scholar]
- 24.Wu C.Y., King K.Y., Kuo C.J., Fang J.M., Wu Y.T., Ho M.Y., Liao C.L., Shie J.J., Liang P.H., Wong C.H. Stable benzotriazole esters as mechanism-based inactivators of the severe acute respiratory syndrome 3CL protease. Chem. Biol. 2006;13:261–268. doi: 10.1016/j.chembiol.2005.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tsai K.C., Chen S.Y., Liang P.H., Lu I.L., Mahindroo N., Hsieh H.P., Chao Y.S., Liu L., Liu D., Lien W., Lin T.H., Wu S.Y. Discovery of a novel family of SARS-CoV protease inhibitors by virtual screening and 3D-QSAR studies. J. Med. Chem. 2006;49:3485–3495. doi: 10.1021/jm050852f. [DOI] [PubMed] [Google Scholar]
- 26.Mukherjee P., Desai P., Ross L., White E.L., Avery M.A. Structure-based virtual screening against SARS-3CL(pro) to identify novel non-peptidic hits. Bioorg. Med. Chem. 2008;16:4138–4149. doi: 10.1016/j.bmc.2008.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Blanchard J.E., Elowe N.H., Huitema C., Fortin P.D., Cechetto J.D., Eltis L.D., Brown E.D. High-throughput screening identifies inhibitors of the SARS coronavirus main proteinase. Chem. Biol. 2004;11:1445–1453. doi: 10.1016/j.chembiol.2004.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jacobs J., Grum-Tokars V., Zhou Y., Turlington M., Saldanha S.A., Chase P., Eggler A., Dawson E.S., Baez-Santos Y.M., Tomar S., Mielech A.M., Baker S.C., Lindsley C.W., Hodder P., Mesecar A., Stauffer S.R. Discovery, synthesis, and structure-based optimization of a series of N-(tert-butyl)-2-(N-arylamido)-2-(pyridin-3-yl) acetamides (ML188) as potent noncovalent small molecule inhibitors of the severe acute respiratory syndrome coronavirus (SARS-CoV) 3CL protease. J. Med. Chem. 2013;56:534–546. doi: 10.1021/jm301580n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nguyen T.T., Ryu H.J., Lee S.H., Hwang S., Breton V., Rhee J.H., Kim D. Virtual screening identification of novel severe acute respiratory syndrome 3C-like protease inhibitors and in vitro confirmation. Bioorg. Med. Chem. Lett. 2011;21:3088–3091. doi: 10.1016/j.bmcl.2011.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kumar V., Tan K.P., Wang Y.M., Lin S.W., Liang P.H. Identification, synthesis and evaluation of SARS-CoV and MERS-CoV 3C-like protease inhibitors. Bioorg. Med. Chem. 2016;24:3035–3042. doi: 10.1016/j.bmc.2016.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kumar V., Shin J.S., Shie J.J., Ku K.B., Kim C., Go Y.Y., Huang K.F., Kim M., Liang P.H. Identification and evaluation of potent Middle East respiratory syndrome coronavirus (MERS-CoV) 3CLPro inhibitors. Antivir. Res. 2017;141:101–106. doi: 10.1016/j.antiviral.2017.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lin M.H., Moses D.C., Hsieh C.H., Cheng S.C., Chen Y.H., Sun C.Y., Chou C.Y. Disulfiram can inhibit MERS and SARS coronavirus papain-like proteases via different modes. Antivir. Res. 2018;150:155–163. doi: 10.1016/j.antiviral.2017.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ratia K., Pegan S., Takayama J., Sleeman K., Coughlin M., Baliji S., Chaudhuri R., Fu W., Prabhakar B.S., Johnson M.E., Baker S.C., Ghosh A.K., Mesecar A.D. A noncovalent class of papain-like protease/deubiquitinase inhibitors blocks SARS virus replication, Proc. Natl. Acad. Sci. U. S. A. 2008;105:16119–16124. doi: 10.1073/pnas.0805240105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Báez-Santos Y.M., Barraza S.J., Wilson M.W., Agius M.P., Mielech A.M., Davis N.M., Baker S.C., Larsen S.D., Mesecar A.D. X-ray structural and biological evaluation of a series of potent and highly selective inhibitors of human coronavirus papain-like proteases. J. Med. Chem. 2014;57:2393–2412. doi: 10.1021/jm401712t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ghosh A.K., Takayama J., Rao K.V., Ratia K., Chaudhuri R., Mulhearn D.C., Lee H., Nichols D.B., Baliji S., Baker S.C., Johnson M.E., Mesecar A.D. Severe acute respiratory syndrome coronavirus papain-like novel protease inhibitors: design, synthesis, protein-ligand X-ray structure and biological evaluation. J. Med. Chem. 2010;53:4968–4979. doi: 10.1021/jm1004489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ghosh A.K., Takayama J., Aubin Y., Ratia K., Chaudhuri R., Baez Y., Sleeman K., Coughlin M., Nichols D.B., Mulhearn D.C., Prabhakar B.S., Baker S.C., Johnson M.E., Mesecar A.D. Structure-based design, synthesis, and biological evaluation of a series of novel and reversible inhibitors for the severe acute respiratory syndrome-coronavirus papain-like protease. J. Med. Chem. 2009;52:5228–5240. doi: 10.1021/jm900611t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Park J.Y., Kim J.H., Kim Y.M., Jeong H.J., Kim D.W., Park K.H., Kwon H.J., Park S.J., Lee W.S., Ryu Y.B. Tanshinones as selective and slow-binding inhibitors for SARS-CoV cysteine proteases. Bioorg. Med. Chem. 2012;20:5928–5935. doi: 10.1016/j.bmc.2012.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chou C.Y., Chien C.H., Han Y.S., Prebanda M.T., Hsieh H.P., Turk B., Chang G.G., Chen X. Thiopurine analogues inhibit papain-like protease of severe acute respiratory syndrome coronavirus. Biochem. Pharmacol. 2008;75:1601–1609. doi: 10.1016/j.bcp.2008.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cheng K.W., Cheng S.C., Chen W.Y., Lin M.H., Chuang S.J., Cheng I.H., Sun C.Y., Chou C.Y. Thiopurineanalogs and mycophenolic acid synergistically inhibit the papain-like protease of Middle East respiratory syndrome coronavirus. Antivir. Res. 2015;115:9–16. doi: 10.1016/j.antiviral.2014.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Park J.Y., Jeong H.J., Kim J.H., Kim Y.M., Park S.J., Kim D., Park K.H., Lee W.S., Ryu Y.B. Diarylheptanoids from Alnus japonica inhibit papain-like protease of severe acute respiratory syndrome coronavirus. Biol. Pharm. Bull. 2012;35:2036–2042. doi: 10.1248/bpb.b12-00623. [DOI] [PubMed] [Google Scholar]
- 41.Cho J.K., Curtis-Long M.J., Lee K.H., Kim D.W., Ryu H.W., Yuk H.J., Park K.H. Geranylated flavonoids displaying SARS-CoV papain-like protease inhibition from the fruits of Paulownia tomentosa. Bioorg. Med. Chem. 2013;21:3051–3057. doi: 10.1016/j.bmc.2013.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee H., Lei H., Santarsiero B.D., Gatuz J.L., Cao S., Rice A.J., Patel K., Szypulinski M.Z., Ojeda I., Ghosh A.K., Johnson M.E. Inhibitor recognition specificity of MERS-CoV papain-like protease may differ from that of SARS-CoV. ACS Chem. Biol. 2015;10:1456–1465. doi: 10.1021/cb500917m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ghosh A.K., Xi K., Ratia K., Santarsiero B.D., Fu W., Harcourt B.H., Rota P.A., Baker S.C., Johnson M.E., Mesecar A.D. Design and synthesis of peptidomimetic severe acute respiratory syndrome chymotrypsin-like protease inhibitors. J. Med. Chem. 2005;48:6767–6771. doi: 10.1021/jm050548m. [DOI] [PubMed] [Google Scholar]
- 44.Lee T.W., Cherney M.M., Huitema C., Liu J., James K.E., Powers J.C., Eltis L.D., James M.N. Crystal structures of the main peptidase from the SARS coronavirus inhibited by a substrate-like aza-peptide epoxide. J. Mol. Biol. 2005;353:1137–1151. doi: 10.1016/j.jmb.2005.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang S., Chen S.J., Hsu M.F., Wu J.D., Tseng C.T., Liu Y.F., Chen H.C., Kuo C.W., Wu C.S., Chang L.W., Chen W.C., Liao S.Y., Chang T.Y., Hung H.H., Shr H.L., Liu C.Y., Huang Y.A., Chang L.Y., Hsu J.C., Peters C.J., Hsu M.C. Synthesis, crystal structure, structure-activity relationships, and antiviral activity of a potent SARS coronavirus 3CL protease inhibitor. J. Med. Chem. 2006;49:4971–4980. doi: 10.1021/jm0603926. [DOI] [PubMed] [Google Scholar]
- 46.Ghosh A.K., Xi K., Grum-Tokars V., Xu X., Ratia K., Fu W., Houser K.V., Baker S.C., Johnson M.E., Mesecar A.D. Structure-based design, synthesis, and biological evaluation of peptidomimetic SARS-CoV 3CLpro inhibitors. Bioorg. Med. Chem. Lett. 2007;17:5876–5880. doi: 10.1016/j.bmcl.2007.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ghosh A.K., Gong G., Grum-Tokars V., Mulhearn D.C., Baker S.C., Coughlin M., Prabhakar B.S., Sleeman K., Johnson M.E., Mesecar A.D. Design, synthesis and antiviral efficacy of a series of potent chloropyridyl ester-derived SARS-CoV 3CLpro inhibitors. Bioorg. Med. Chem. Lett. 2008;18:5684–5688. doi: 10.1016/j.bmcl.2008.08.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Regnier T., Sarma D., Hidaka K., Bacha U., Freire E., Hayashi Y., Kiso Y. New developments for the design, synthesis and biological evaluation of potent SARS-CoV3CL(pro) inhibitors. Bioorg. Med. Chem. Lett. 2009;19:2722–2727. doi: 10.1016/j.bmcl.2009.03.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Konno S., Thanigaimalai P., Yamamoto T., Nakada K., Kakiuchi R., Takayama K., Yamazaki Y., Yakushiji F., Akaji K., Kiso Y., Kawasaki Y., Chen S.E., Freire E., Hayashi Y. Design and synthesis of new tripeptide-type SARS-CoV 3CL protease inhibitors containing an electrophilic arylketone moiety. Bioorg. Med. Chem. Lett. 2013;21:412–424. doi: 10.1016/j.bmc.2012.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Thanigaimalai P., Konno S., Yamamoto T., Koiwai Y., Taguchi A., Takayama K., Yakushiji F., Akaji K., Kiso Y., Kawasaki Y., Chen S.E., Naser-Tavakolian A., Schön A., Freire E., Hayashi Y. Design, synthesis, and biological evaluation of novel dipeptide-type SARS-CoV 3CL protease inhibitors: structure-activity relationship study. Eur. J. Med. Chem. 2013;65:436–447. doi: 10.1016/j.ejmech.2013.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Thanigaimalai P., Konno S., Yamamoto T., Koiwai Y., Taguchi A., Takayama K., Yakushiji F., Akaji K., Chen S.E., Naser-Tavakolian A., Schön A., Freire E., Hayashi Y. Development of potent dipeptide-type SARS-CoV 3CL protease inhibitors with novel P3 scaffolds: design, synthesis, biological evaluation, and docking studies. Eur. J. Med. Chem. 2013;68:372–384. doi: 10.1016/j.ejmech.2013.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jain R.P., Pettersson H.I., Zhang J., Aull K.D., Fortin P.D., Huitema C., Eltis L.D., Parrish J.C., James M.N., Wishart D.S., Vederas J.C. Synthesis and evaluation of keto-glutamine analogues as potent inhibitors of severe acute respiratory syndrome 3CLpro. J. Med. Chem. 2004;47:6113–6116. doi: 10.1021/jm0494873. [DOI] [PubMed] [Google Scholar]
- 53.Wu C.Y., Jan J.T., Ma S.H., Kuo C.J., Juan H.F., Cheng Y.S., Hsu H.H., Huang H.C., Wu D., Brik A., Liang F.S., Liu R.S., Fang J.M., Chen S.T., Liang P.H., Wong C.H. Small molecules targeting severe acute respiratory syndrome human coronavirus. Proc. Natl. Acad. Sci. U. S. A. 2004;101:10012–10017. doi: 10.1073/pnas.0403596101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kao R.Y., Tsui W.H., Lee T.S., Tanner J.A., Watt R.M., Huang J.D., Hu L., Chen G., Chen Z., Zhang L., He T., Chan K.H., Tse H., To A.P., Ng L.W., Wong B.C., Tsoi H.W., Yang D., Ho D.D., Yuen K.Y. Identification of novel small-molecule inhibitors of severe acute respiratory syndrome-associated coronavirus by chemical genetics. Chem. Biol. 2004;11:1293–1299. doi: 10.1016/j.chembiol.2004.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhou L., Liu Y., Zhang W., Wei P., Huang C., Pei J., Yuan Y., Lai L. Isatin compounds as noncovalent SARS coronavirus 3C-like protease inhibitors. J. Med. Chem. 2006;49:3440–3443. doi: 10.1021/jm0602357. [DOI] [PubMed] [Google Scholar]
- 56.Zhang H.Z., Zhang H., Kemnitzer W., Tseng B., Cinatl J., Jr., Michaelis M., Doerr H.W., Cai S.X. Design and synthesis of dipeptidylglutaminylfluoromethyl ketones as potent severe acute respiratory syndrome coronovirus (SARS-CoV) inhibitors. J. Med. Chem. 2006;49:1198–1201. doi: 10.1021/jm0507678. [DOI] [PubMed] [Google Scholar]
- 57.Shao Y.M., Yang W.B., Peng H.P., Hsu M.F., Tsai K.C., Kuo T.H., Wang A.H., Liang P.H., Lin C.H., Yang A.S., Wong C.H. Structure-based design and synthesis of highly potent SARS-CoV 3CL protease inhibitors. Chembiochem. 2007;8:1654–1657. doi: 10.1002/cbic.200700254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wen C.C., Kuo Y.H., Jan J.T., Liang P.H., Wang S.Y., Liu H.G., Lee C.K., Chang S.T., Kuo C.J., Lee S.S., Hou C.C., Hsiao P.W., Chien S.C., Shyur L.F., Yang N.S. Specific plant terpenoids and lignoids possess potent antiviral activities against severe acute respiratory syndrome coronavirus. J. Med. Chem. 2007;50:4087–4095. doi: 10.1021/jm070295s. [DOI] [PubMed] [Google Scholar]
- 59.Ramajayam R., Tan K.P., Liu H.G., Liang P.H. Synthesis and evaluation of pyrazolone compounds as SARS-coronavirus 3C-like protease inhibitors. Bioorg. Med. Chem. 2010;18:7849–7854. doi: 10.1016/j.bmc.2010.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ramajayam R., Tan K.P., Liu H.G., Liang P.H. Synthesis, docking studies, and evaluation of pyrimidines as inhibitors of SARS-CoV 3CL protease. Bioorg. Med. Chem. Lett. 2010;20:3569–3572. doi: 10.1016/j.bmcl.2010.04.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ryu Y.B., Jeong H.J., Kim J.H., Kim Y.M., Park J.Y., Kim D., Nguyen T.T., Park S.J., Chang J.S., Park K.H., Rho M.C., Lee W.S. Biflavonoids from Torreyanucifera displaying SARS-CoV 3CL(pro) inhibition. Bioorg. Med. Chem. 2010;18:7940–7947. doi: 10.1016/j.bmc.2010.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ryu Y.B., Park S.J., Kim Y.M., Lee J.Y., Seo W.D., Chang J.S., Park K.H., Rho M.C., Lee W.S. SARS-CoV 3CLpro inhibitory effects of quinone-methidetriterpenes from Tripterygiumregelii. Bioorg. Med. Chem. Lett. 2010;20:1873–1876. doi: 10.1016/j.bmcl.2010.01.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Akaji K., Konno H., Mitsui H., Teruya K., Shimamoto Y., Hattori Y., Ozaki T., Kusunoki M., Sanjoh A. Structure-based design, synthesis, and evaluation of peptide-mimetic SARS 3CL protease inhibitors. J. Med. Chem. 2011;54:7962–7973. doi: 10.1021/jm200870n. [DOI] [PubMed] [Google Scholar]
- 64.Turlington M., Chun A., Tomar S., Eggler A., Grum-Tokars V., Jacobs J., Daniels J.S., Dawson E., Saldanha A., Chase P., Baez-Santos Y.M., Lindsley C.W., Hodder P., Mesecar A.D., Stauffer S.R. Discovery of N-(benzo[1,2,3]triazol-1-yl)-N-(benzyl)acetamido)phenyl) carboxamides as severe acute respiratory syndrome coronavirus (SARS-CoV) 3CLpro inhibitors: identification of ML300 and noncovalentnanomolar inhibitors with an induced-fit binding. Bioorg. Med. Chem. Lett. 2013;23:6172–6177. doi: 10.1016/j.bmcl.2013.08.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liu W., Zhu H.M., Niu G.J., Shi E.Z., Chen J., Sun B., Chen W.Q., Zhou H.G., Yang C. Synthesis, modification and docking studies of 5-sulfonyl isatin derivatives as SARS-CoV 3C-like protease inhibitors. Bioorg. Med. Chem. 2014;22:292–302. doi: 10.1016/j.bmc.2013.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lee H., Mittal A., Patel K., Gatuz J.L., Truong L., Torres J., Mulhearn D.C., Johnson M.E. Identification of novel drug scaffolds for inhibition of SARS-CoV 3-Chymotrypsin-like protease using virtual and high-throughput screenings. Bioorg. Med. Chem. 2014;22:167–177. doi: 10.1016/j.bmc.2013.11.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shimamoto Y., Hattori Y., Kobayashi K., Teruya K., Sanjoh A., Nakagawa A., Yamashita E., Akaji K. Fused-ring structure of decahydroisoquinolin as a novel scaffold for SARS 3CL protease inhibitors. Bioorg. Med. Chem. 2015;23:876–890. doi: 10.1016/j.bmc.2014.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yoshizawa S.I., Hattori Y., Kobayashi K., Akaji K. Evaluation of an octahydroisochromene scaffold used as a novel SARS 3CL protease inhibitor. Bioorg. Med. Chem. 2020;28:115273. doi: 10.1016/j.bmc.2019.115273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Amin S.A., Ghosh K., Gayen S., Jha T. Chemical-informatics approach to COVID-19 drug discovery: Monte Carlo based QSAR, virtual screening and molecular docking study of some in-house molecules as papain-like protease (PLpro) inhibitors. J. Biomol. Struct. Dyn. 2020 doi: 10.1080/07391102.2020.1780946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pillaiyar T., Meenakshisundaram S., Manickam M. Recent discovery and development of inhibitors targeting coronaviruses, Drug Discov. Today Off. 2020;25:668–688. doi: 10.1016/j.drudis.2020.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.2021. http://www.who.int/csr/sars/archive/2003_05_07a/en as accessed on 4th Feb.
- 72.2021. http://www.who.int/csr/sars/country/en/country2003_08_15.pdf as accessed on 4th Feb.
- 73.Li F. Structure, function, and evolution of coronavirus spike proteins. Annu. Rev. Virol. 2020;3:237–261. doi: 10.1146/annurev-virology-110615-042301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hamre D., Procknow J.J. A new virus isolated from the human respiratory tract, Proc. Soc. Exp. Biol. Med. 1966;121:190–193. doi: 10.3181/00379727-121-30734. [DOI] [PubMed] [Google Scholar]
- 75.McIntosh K., Dees J.H., Becker W.B., Kapikian A.Z., Chanock R.M. Recovery in tracheal organ cultures of novel viruses from patients with respiratory disease. Proc. Natl. Acad. Sci. U. S. A. 1967;57:933. doi: 10.1073/pnas.57.4.933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ghosh K., Amin S.A., Gayen S., Jha T. Chemical-informatics approach to COVID-19 drug discovery: exploration of important fragments and data mining based prediction of some hits from natural origins as main protease (Mpro) inhibitors. J. Mol. Struct. 2020;1224:129026. doi: 10.1016/j.molstruc.2020.129026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Khailany R.A., Safdar M., Ozaslan M. Genomic characterization of a novel SARS-CoV-2. Gene Rep. 2020;16:100682. doi: 10.1016/j.genrep.2020.100682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yu R., Chen L., Lan R. R.Shen, P. Li, Computational screening of antagonist against the SARS-CoV-2 (COVID-19) coronavirus by molecular docking. Int. J. Antimicrob. Agents. 2020;56:106012. doi: 10.1016/j.ijantimicag.2020.106012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhou Y., Hou Y., Shen J., Huang Y., Martin W., Cheng F. Network-based drug repurposing for novel coronavirus 2019-nCoV/SARS-CoV-2. Cell Discov. 2020;6:1–8. doi: 10.1038/s41421-020-0153-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zumla A., Chan J.F., Azhar E.I., Hui D.S., Yuen K.Y. Coronaviruses - drug discovery and therapeutic options. Nat. Rev. Drug Discov. 2016;15:327–347. doi: 10.1038/nrd.2015.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Perlman S., Netland J. Coronaviruses post-SARS: update on replication and pathogenesis. Nat. Rev. Microbiol. 2009;7:439–450. doi: 10.1038/nrmicro2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Báez-Santos Y.M., John S.E., Mesecar A.D. The SARS-coronavirus papain-like protease: structure, function and inhibition by designed antiviral compounds. Antivir. Res. 2015;115:21–38. doi: 10.1016/j.antiviral.2014.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Jin Z., Du X., Xu Y., Deng Y., Liu M., Zhao Y., Zhang B.L., Zhang X.L., Peng C., Duan Y., Yu J., Wang L., Yang K., Liu F., Jiang R., Yang X., You T., Liu X., Yang X., Bai F., Liu H., Liu X., Guddat L.W., Xu W., Xiao G., Qin C., Shi Z., Jiang H., Rao Z., Yang H. Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors. Nature. 2020;582:289–293. doi: 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- 84.Su H., Yao S., Zhao W., Li M., Liu J., Shang W., Xie H., Ke C., Gao M., Yu K., Liu H., Shen J., Tang W., Zhang L., Zuo J., Jiang H., Bai F., Wu Y., Ye Y., Xu Y. Discovery of baicalin and baicalein as novel, natural product inhibitors of SARS-CoV-2 3CL protease in vitro. BioRxiv. 2020 doi: 10.1101/2020.04.13.038687. [DOI] [Google Scholar]
- 85.Ma C., Sacco M.D., Hurst B., Townsend J.A., Hu Y., Szeto T., Zhang X., Tarbet B., Marty M.T., Chen Y., Wang J. Boceprevir, GC-376, and calpain inhibitors II, XII inhibit SARS-CoV-2 viral replication by targeting the viral main protease. BioRxiv. 2020 doi: 10.1101/2020.04.20.051581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Goyal B., Goyal D. Targeting the dimerization of the main protease of coronaviruses: a potential broad-spectrum therapeutic strategy, ACS Comb. Sci. 2020;22:297–305. doi: 10.1021/acscombsci.0c00058. [DOI] [PubMed] [Google Scholar]
- 87.Gimeno A., Mestres-Truyol J., Ojeda-Montes M.J., Macip G., Saldivar-Espinoza B., Cereto-Massagué A., Pujadas G., Garcia-Vallvé S. Prediction of novel inhibitors of the main protease (M-pro) of SARS-CoV-2 through consensus docking and drug reposition. Int. J. Mol. Sci. 2020;21:3793. doi: 10.3390/ijms21113793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tang B., He F., Liu D., Fang M., Wu Z., Xu D. AI-aided design of novel targeted covalent inhibitors against SARS-CoV-2. BioRxiv. 2020 doi: 10.1101/2020.03.03.972133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Adhikari N., Baidya S.K., Saha A., Jha T. In: Viral Proteases and Their Inhibitors. Gupta S.P., editor. Academic Press; USA: 2017. Structural insight into the viral 3C-like protease inhibitors: comparative SAR/QSAR approaches. [Google Scholar]
- 90.Harcourt B.H., Jukneliene D., Kanjanahaluethai A., Bechill J., Severson K.M., Smith C.M., Rota P.A., Baker S.C. Identification of severe acute respiratory syndrome coronavirus replicase products and characterization of papain-like protease activity. J. Virol. 2004;78:13600–13612. doi: 10.1128/JVI.78.24.13600-13612.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Chen X., Chou C.Y., Chang G.G. Thiopurine analogue inhibitors of severe acute respiratory syndrome-coronavirus papain-like protease, a deubiquitinating and deISGylating enzyme. Antivir. Chem. Chemother. 2009;19:151–156. doi: 10.1177/095632020901900402. [DOI] [PubMed] [Google Scholar]
- 92.Clementz M.A., Chen Z., Banach B.S., Wang Y., Sun L., Ratia K., Baez-Santos Y.M., Wang J., Takayama J., Ghosh A.K., Li K., Mesecar A.D., Baker S.C. Deubiquitinating and interferon antagonism activities of coronavirus papain-like proteases. J. Virol. 2010;84:4619–4629. doi: 10.1128/JVI.02406-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Katsila T., Spyroulias G.A., Patrinos G.P., Matsoukas M.-T. Computational approaches in target identification and drug discovery, Comput. Struct. Biochem. J. 2016;14:177–184. doi: 10.1016/j.csbj.2016.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bandyopadhyay D., Kreatsoulas C., Brady P.G., Boyer J., He Z., Scavello G., Jr., Peryea T., Jadhav A., Nguyen D.-T., Guha R. Scaffold-based analytics: enabling hit-to-lead decisions by visualizing chemical series linked across large datasets. J. Chem. Inf. Model. 2019;59:4880–4892. doi: 10.1021/acs.jcim.9b00243. [DOI] [PubMed] [Google Scholar]
- 95.Murray C.W., Verdonk M.L., Rees D.C. Experiences in fragment-based drug discovery. Trends Pharmacol. Sci. 2012;33:224–232. doi: 10.1016/j.tips.2012.02.006. [DOI] [PubMed] [Google Scholar]
- 96.Kodadek T. The rise, fall and reinvention of combinatorial chemistry. Chem. Commun. 2011;47:9757–9763. doi: 10.1039/c1cc12102b. [DOI] [PubMed] [Google Scholar]
- 97.van Hilten N., Chevillard F., Kolb P. Virtual compound libraries in computer-assisted drug discovery. J. Chem. Inf. Model. 2019;59:644–651. doi: 10.1021/acs.jcim.8b00737. [DOI] [PubMed] [Google Scholar]
- 98.Romasanta A.K.S., van der Sijde P., Hellsten I., Hubbard R.E., Keseru G.M., van Muijlwijk-Koezen J., de Esch I.J.P. When fragments link: a bibliometric perspective on the development of fragment-based drug discovery, Drug Discov. Today Off. 2018;23:1596–1609. doi: 10.1016/j.drudis.2018.05.004. [DOI] [PubMed] [Google Scholar]
- 99.Turk B. Targeting proteases: successes, failures and future prospects. Nat. Rev. Drug Discov. 2006;5:785–799. doi: 10.1038/nrd2092. [DOI] [PubMed] [Google Scholar]
- 100.Drag M., Salvesen G.S. Emerging principles in protease-based drug discovery. Nat. Rev. Drug Discov. 2010;9:690–701. doi: 10.1038/nrd3053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Amanat F., Krammer F. SARS-CoV-2 vaccines: status report. Immunity. 2020;52:583–589. doi: 10.1016/j.immuni.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Unpublished data.
- 103.Freitas B.T., Durie I.A., Murray J., Longo J.E., Miller H.C., Crich D., Hogan R.J., Tripp R.A., Pegan S.D. Characterization and noncovalent inhibition of the deubiquitinase and deISGylase activity of SARS-CoV-2 papain-like protease, ACS Infect. Dis. 2020;6:2099–2109. doi: 10.1021/acsinfecdis.0c00168. [DOI] [PubMed] [Google Scholar]
- 104.Amin S.A., Banerjee S., Ghosh K., Gayen S., Jha T. Protease targeted COVID-19 drug discovery and its challenges: insight into viral main protease (Mpro) and papain-like protease (PLpro) inhibitors. Bioorg. Med. Chem. 2021;29:115860. doi: 10.1016/j.bmc.2020.115860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Lipinski C.A., Lombardo F., Dominy B.W., Feeney P.J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 2001;46:3–26. doi: 10.1016/s0169-409x(00)00129-0. [DOI] [PubMed] [Google Scholar]
- 106.Biovia D.S. Biovia; San Diego: 2016. Discovery Studio. [Google Scholar]
- 107.Amin S.A., Banerjee S., Singh S., Qureshi I.A., Gayen S., Jha T. First structure-activity relationship analysis of SARS-CoV-2 virus main protease (Mpro) inhibitors: an endeavor on COVID-19 drug discovery. Mol. Divers. 2021 doi: 10.1007/s11030-020-10166-3. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Daina A., Michielin O., Zoete V. SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017;7:42717. doi: 10.1038/srep42717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Zhou J., Fang L., Yang Z., Xu S., Lv M., Sun Z., Chen J., Wang D., Gao J., Xiao S. Identification of novel proteolytically inactive mutations in coronavirus 3C-like protease using a combined approach. Faseb. J. 2019;33:14575–14587. doi: 10.1096/fj.201901624RR. [DOI] [PubMed] [Google Scholar]
- 110.Ullrich S., Nitsche C. The SARS-CoV-2 main protease as drug target, Bioorg. Med. Chem. Lett. 2020;30:127377. doi: 10.1016/j.bmcl.2020.127377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Hilgenfeld R. From SARS to MERS: crystallographic studies on coronaviral proteases enable antiviral drug design. FEBS J. 2014;281:4085–4096. doi: 10.1111/febs.12936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Zhang L., Lin D., Kusov Y. α-Ketoamides as broad-spectrum inhibitors of coronavirus and enterovirus replication: structure-based design, synthesis, and activity assessment. J. Med. Chem. 2020;63:4562–4578. doi: 10.1021/acs.jmedchem.9b01828. [DOI] [PubMed] [Google Scholar]