Abstract
An outbreak of coronavirus disease 2019 (COVID-19) caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) occurred aboard the Diamond Princess cruise ship between her January 20 departure and late February 2020. Here, we used phylodynamic analyses to investigate the transmission dynamics of SARS-CoV-2 during the outbreak. Using a Bayesian coalescent-based method, the estimated mean nucleotide substitution rate of 240 SARS-CoV-2 whole-genome sequences was approximately 7.13 × 10−4 substitutions per site per year. Population dynamics and the effective reproductive number (Re) of SARS-CoV-2 infections were estimated using a Bayesian framework. The estimated origin of the outbreak was January 21, 2020. The infection spread substantially before quarantine on February 5. The Re peaked at 6.06 on February 4 and gradually declined to 1.51, suggesting that transmission continued slowly even after quarantine. These findings highlight the high transmissibility of SARS-CoV-2 and the need for effective measures to control outbreaks in confined settings.
Abbreviations: COVID-19, Coronavirus disease 2019; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; SARS, severe acute respiratory syndrome; MERS, Middle East respiratory syndrome; RT-PCR, reverse-transcription PCR; R0, basic reproductive number; Re, effective reproductive number; GISAID, Global Initiative on Sharing All Influenza Data; MCMC, Markov chain Monte Carlo; GTR, general time-reversible; ML, maximum likelihood; ESS, effective sample size; HPD, highest posterior density; tMRCA, time to the most recent common ancestor
Keywords: SARS-CoV-2, Nucleotide substitution rate, Outbreak, Population dynamics, Effective reproductive number, Cruise ship, Confined settings
1. Introduction
Coronavirus disease 2019 (COVID-19) is a respiratory infectious disease caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) (Coronaviridae Study Group of the International Committee on Taxonomy of Viruses, 2020, Huang et al., 2020). The initial case was reported in China in December 2019 (Zhou et al., 2020), and the outbreak spread rapidly around the world resulting in an ongoing pandemic (WHO, 2020b). As of September 27, 2020, the total number of confirmed cases had reached 32.7 million, and 991,000 people had died of COVID-19 (WHO, 2020a).
Coronaviruses are a family of enveloped positive-strand RNA viruses that possess a single-stranded, positive-sense RNA genome comprising 26–32 kilobases (kb). Among them, SARS-CoV-2 belongs to the genus Betacoronavirus and has a genome that is approximately 30 kb in length (Lu et al., 2020). Some betacoronaviruses have caused severe disease outbreaks such as severe acute respiratory syndrome (SARS) and the Middle East respiratory syndrome (MERS). The COVID-19 pandemic is the third major outbreak caused by betacoronaviruses in the past two decades. In contrast to SARS and MERS, a substantial proportion of SARS-CoV-2 infections causing COVID-19 have been asymptomatic (He et al., 2020, Mizumoto et al., 2020, Nishiura et al., 2020). Moreover, since asymptomatic and presymptomatic individuals infected with SARS-CoV-2 can transmit the virus (Bai et al., 2020, Ganyani et al., 2020, Rothe et al., 2020), epidemiological data can be unreliable, causing difficulties in understanding transmission dynamics and controlling the spread of infection.
An outbreak of COVID-19 occurred on board the Diamond Princess cruise ship in February 2020. The Diamond Princess sailed from Yokohama on January 20, 2020, arrived at Hong Kong on January 25, and returned to Yokohama on February 3 with 2666 passengers and 1045 crew members. The presumed COVID-19 index case was reportedly a passenger from Hong Kong who boarded the ship in Yokohama on January 20 and developed a mild dry cough on January 23 (Leung et al., 2020). After the passenger returned to Hong Kong and disembarked on January 25, he became feverish on January 29 and was diagnosed with COVID-19 on February 1. After learning that a former passenger had been diagnosed with COVID-19, the Japanese Ministry of Health, Labour, and Welfare quarantined the ship in Yokohama on February 5. Movement of the passengers was restricted, and self-isolation in cabins was implemented. Symptomatic and high-risk individuals were initially tested for SARS-CoV-2 using reverse-transcription polymerase chain reaction (RT-PCR), and then, asymptomatic passengers were tested after February 11 (National institute of infectious diseases, 2020, Nishiura, 2020, Yamahata and Shibata, 2020). Individuals testing positive were transported from the ship to the quarantine location or hospitals (Yamahata and Shibata, 2020). A total of 712 individuals had tested positive among >3600 tests as of August 2020 (Ministry of Health, 2020).
Understanding the patterns of transmission and the dynamics of an infectious agent, among a susceptible population, is essential for establishing effective strategies to control and prevent the spread of infection, particularly when the pathogenic agent is novel (Gardy and Loman, 2018, Ladner et al., 2019). The COVID-19 outbreak on the Diamond Princess cruise ship might provide valuable samples and information about the transmission dynamics of SARS-CoV-2, particularly in a confined setting. In this case, the presumed index case is known, the number of susceptible individuals was constant, timely preventive measures were deployed, and the passengers were extensively tested using RT-PCR (Liu et al., 2020). Although the number of confirmed cases increased soon after returning to port suggesting that the infection had spread among passengers and crew during the voyage, the temporal dynamics of transmission remained elusive. Here, we analyzed the transmission dynamics of SARS-CoV-2 aboard the Diamond Princess using its viral genome sequence in a Bayesian framework. We estimated the effective population size, as well as basic (R0) and effective (Re) reproductive numbers of SARS-CoV-2 during the outbreak and evaluated changes in transmissibility over time.
2. Materials and methods
2.1. SARS-CoV-2 genomic sequence dataset
All publicly available SARS-CoV-2 genome sequences with clinical information as of August 7, 2020 (n = 78,448), were retrieved from the Global Initiative on Sharing All Influenza Data (GISAID) database (Elbe and Buckland-Merrett, 2017). We included sequences with specific collection date known to the day, sampling location, and host information indicating that they had been collected from human hosts. We retained only complete or nearly complete sequences between 29,600 and 31,000 nt that contained <1% ambiguous nucleotides. Sequences were aligned and trimmed according to the reference sequence (NC_045512.2, 29,903 nt) using MAFFT (v.7.453; https://mafft.cbrc.jp/alignment/software/) with the “-addfragments” option. Finally, we excluded sequences with >20 nt gaps, >20 mismatched nucleotides, or concentrated ambiguity calls (>10 in any 50-nt window) as described (Korber et al., 2020) and manually inspected the sequences using AliView (v.1.26; https://ormbunkar.se/aliview/). All retained sequences were randomly resampled up to two per country, year, and month using nextstrain CLI (v.1.16.5) (Hadfield et al., 2018) to maintain the geographic and temporal diversity of the virus. All available isolates from Wuhan sampled in December 2019 were included in the dataset. The first and last 50 nt of the 5′ and 3′ terminals, respectively (1–50 and 29,854–29,903 nt of NC_045512.2), were trimmed as being error-rich. An outlier sequence was removed based on root-to-tip regression analysis using treetime (v.0.7.6; https://github.com/neherlab/treetime), and the resulting global dataset comprised 240 sequences (Supplementary Table 1). Single nucleotide variants were called using snp-sites (v.2.5.1; https://github.com/sanger-pathogens/snp-sites), VEP (v.101.0; https://asia.ensembl.org/info/docs/tools/vep/index.html), and bcftools (v.1.10.2; http://samtools.github.io/bcftools/bcftools.html). Supplementary Fig. 1 shows the characteristics of single nucleotide variants in the reference dataset.
In total, 74 SARS-CoV-2 sequences from infected persons on the Diamond Princess were identified in the GISAID database as of August 7, 2020. Sequences with multiple ambiguous nucleotides (>1%) and long gaps in the coding region were excluded. Finally, 67 sequences were retained and subsequently analyzed (Supplementary Table 2). All sequences from the ship were collected between February 10 and 17, 2020.
2.2. Phylogenetic analyses
The general time-reversible model with invariable sites and γ distribution among site rate variation (GTR+G+I) was selected as the best-fit model for the reference dataset using ModelTest-NG (v.0.1.6; https://github.com/ddarriba/modeltest). Phylogenetic trees were constructed using RAxML (v.8.2.9; https://cme.h-its.org/exelixis/software.html) with 1000 replicates using the maximum likelihood (ML) algorithm. Trees were created using FigTree (v.1.4.4; http://evomics.org/resources/software/molecular-evolution-software/figtree/) and were re-rooted using the earliest sample (IPBCAMS-WH-01, GenBank entry: MT019529.1). Bayesian phylogenetic analysis was conducted using a constant coalescent model and the strict molecular clock model implemented in BEAST (v.1.10.4) (Drummond and Rambaut, 2007).
2.3. Substitution rate estimation
The substitution rate of the SARS-CoV-2 sequences was calculated in a Bayesian framework using the Markov chain Monte Carlo (MCMC) algorithm implemented in BEAST (v.1.10.4). The substitution rate of the global dataset (n = 240) was estimated under constant, exponential, and Bayesian skyline plot coalescent models with a strict molecular clock model and an uncorrelated relaxed molecular clock model with a lognormal distribution. GTR+G+I was used as a nucleotide substitution model. Each MCMC run contained 50 million states with sampling every 10,000 states. The MCMC convergence and effective sample size (ESS) were analyzed using Tracer (v.1.7.1) (Drummond and Rambaut, 2007), and ESS values >200 were accepted after burn-in. The best-fitting model was selected based on marginal likelihood estimation using path sampling and stepping-stone sampling methods (Baele et al., 2012). Root-to-tip regression of the ML tree was analyzed using Tempest (v. 1.5.3). The root was determined according to the coefficient of determination, R2.
2.4. Phylodynamic analyses
The Bayesian skyline plot is a coalescent-based nonparametric method for estimating past population dynamics over time from molecular sequence data (Drummond et al., 2005). The SARS-CoV-2 population dynamics on the Diamond Princess were inferred using a Bayesian skyline plot coalescent model with the strict molecular clock model implemented in BEAST (v.1.10.4). The GTR+G+I substitution model was used, and the prior for the nucleotide substitution rate was set based on the values obtained in the rate estimation. The MCMC run contained 100 million states, with sampling every 10,000 states. We accepted ESS values >200 after a 10% burn-in, and three independent runs were combined using LogCombiner (v. 1.10.4) implemented in the BEAST package. The effective population sizes were plotted using R (v.4.0.2, R Core Team, 2018; https://www.R-project.org/).
2.5. Basic reproductive number estimation
The R0 is the predicted number of secondary cases that one case would generate in a completely susceptible population (Delamater et al., 2019). Generally, an outbreak should continue if R0 has a value >1, but not if R0 is <1. The R0 was estimated using the linear equation, R0 = rD + 1, where r is the exponential growth rate and D is the average duration of infectiousness (Pybus et al., 2001, Wallinga and Lipsitch, 2007). We calculated the exponential growth rate of 67 sequences derived from persons aboard the cruise ship using an exponential coalescent model with the strict molecular clock model implemented in BEAST2 (v.2.6.3). Three independent runs were combined. The average duration of infectiousness was calculated as 9.3 days (95% CI, 7.8–10) (He et al., 2020), and the R0 was calculated using R (v.4.0.2) (Supplementary information). Doubling time was calculated as ln(2)/r. Limitations of this method include biases due to the duration of infectiousness and the estimated substitution rate (Pybus et al., 2001).
2.6. Effective reproductive number estimation
The birth–death skyline model is based on a forward-in-time model of transmission, death/recovery, and sampling (Stadler et al., 2013). Temporal changes in Re during the outbreak were estimated using the birth–death skyline contemporary model implemented in BEAST2 (v.2.6.3). The GTR + G + I substitution and strict molecular clock models were used with nucleotide substitution rate estimates. For the birth–death model analysis, the epidemiological parameters comprising the rate of becoming noninfectious (δ), sampling proportion (ρ), Re, and the origin of the epidemic were established based on previous estimates (Stadler et al., 2013, Byrne et al., 2020, He et al., 2020, Lai et al., 2020a). The rate of becoming noninfectious was calculated as the reciprocal of the average duration of infectiousness (9.3 days, 95%; CI 7.8–10) (He et al., 2020), which corresponds to 39.4/year (95%CI: 36.5–46.8). A gamma prior with parameters α = 1.7 and β = 23 (mean, 39.1 year−1; 95% CI, 3.59–116]) was taken as a less informative prior to allow the broader range of the period. Sampling proportion (ρ) is the proportion of lineages sampled in the analysis. The number of infected individuals up to February 17 was expected to range between 454 and 3711, which corresponds to the cumulative cases up to February 17, and the total number of individuals aboard the ship, respectively. Therefore, we calculated ρ to be between 0.018 and 0.14, and set a beta prior with parameters α = 2 and β = 15 (mean, 0.118; 95% CI, 0.0155–0.302). The Re of SARS-CoV-2 has been reported to be around 2–3 and 11 in the community-based infection and confined settings, respectively. A lognormal prior with a log-mean of 1 and an SD of 1.25 (mean, 5.94; 95% CI, 0.235–31.5) was used in this study. The origin of the epidemic is considered as the time point at which the index case became infected. We used 1/X prior as the origin of the epidemic prior. Three independent runs were combined as described previously herein, and Re was plotted using R (v.4.0.2) with bdskytools (https://github.com/laduplessis/bdskytools).
2.7. Confirmed cases on the Diamond Princess
The numbers of daily confirmed and cumulative cases obtained from the website of the Japanese Ministry of Health, Labour, and Welfare (https://www.mhlw.go.jp/index.html) and previous studies (Mizumoto et al., 2020, Yamahata and Shibata, 2020) were plotted using R (v.4.0.2).
3. Results
3.1. SARS-CoV-2 sequences and phylogenetic analyses
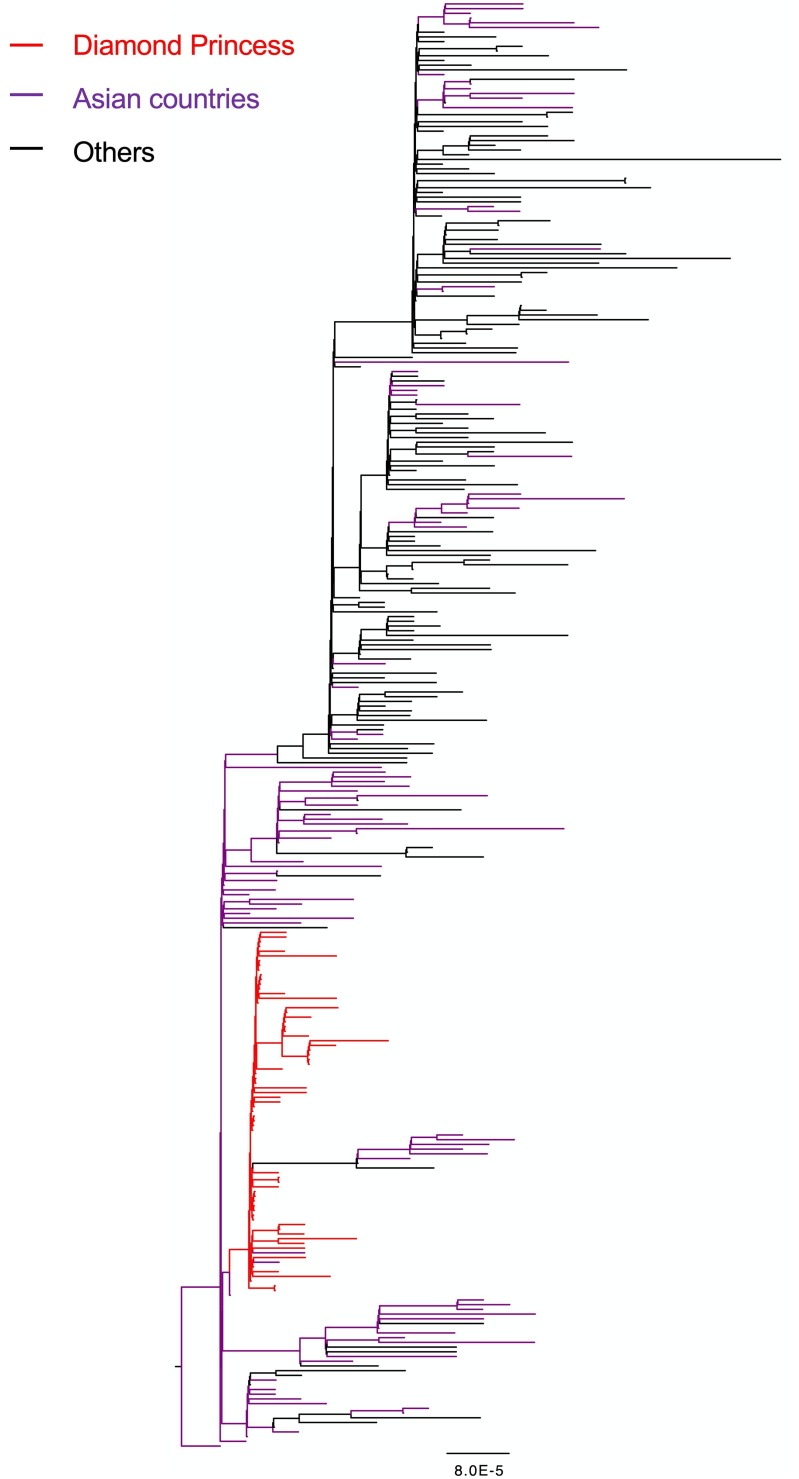
An ML phylogenetic tree was constructed using the SARS-CoV-2 whole-genome sequences from the 67 persons aboard the Diamond Princess and the 240 global reference sequences (Fig. 1 , Supplementary Fig. 2). Although branch bootstrap values were low, all sequences from the cruise ship clustered with some isolates collected in other countries. The Bayesian phylogenetic analysis also showed a similar clustering for samples from the ship (Supplementary Fig. 3), suggesting that the outbreak originated from a single source.
Fig. 1.
Maximum likelihood phylogenetic tree based on SARS-CoV-2 whole-genome sequences of 240 global reference strains and 67 isolates sampled from persons on board the Diamond Princess. Branches of sequences from the Diamond Princess and those from Asian countries are colored red and purple, respectively. The tree was rooted using the earliest sample in the dataset (IPBCAMS-WH-01, GenBank entry: MT019529.1). Branch length reflects units of nucleotide substitutions per site according to the provided scale. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.2. Substitution rate estimation
Using the reference sequence dataset, the substitution rate was estimated under several settings (Supplementary Table 3). However, convergence was not achieved using the Bayesian skyline plot model. We selected strict clock and exponential coalescent models as providing the best fit based on marginal likelihood estimations. The estimated mean substitution rate was 7.13 × 10−4 substitutions per site per year (95% highest posterior density [HPD] intervals, 6.16–8.16 × 10−4). The estimated mean time to the most recent common ancestor (tMRCA) was 2019.80 (95% HPD, 2019.71–2019.88), which corresponded to October 19, 2019 (95% HPD, September 18–November 18, 2019). Root-to-tip regression analysis of the ML phylogenetic trees against sampling time was conducted using Tempest (v. 1.5.3). The estimated mean evolutionary rate was 7.82 × 10−4 substitutions per site per year (Supplementary Fig. 4), and the estimated tMRCA was 2019.83 (November 30, 2019).
3.3. Phylodynamic analyses
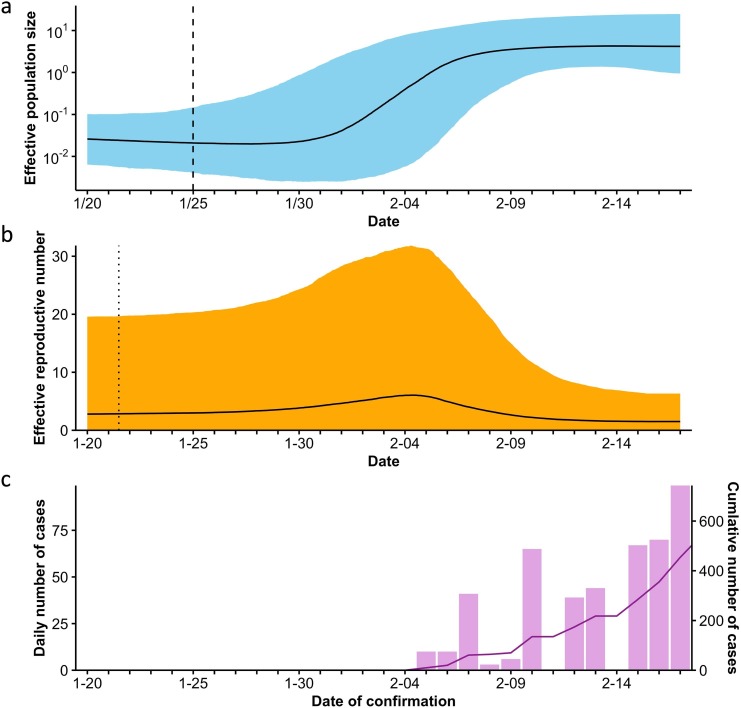
The SARS-CoV-2 population dynamics on the Diamond Princess were inferred using the Bayesian skyline plot coalescent and strict molecular clock models. The Bayesian skyline plot showed that the effective population size began to increase around January 30 and exponentially increased between February 2 and 6, 2020 (Fig. 2 a). Subsequently, the population growth slowed down but continued on a slightly increasing trajectory, followed by a plateau phase from approximately February 10, 2020. The estimated mean tMRCA was 2020.066 (95% HPD, 2020.027–2020.095), corresponding to January 25, 2020 (95% HPD, January 10–February 4).
Fig. 2.
Transmission dynamics of SARS-CoV-2 on board the Diamond Princess. (a) Effective population size of SARS-CoV-2 on board the Diamond Princess over time, (b) effective reproductive number (Re) over time, and (c) daily and cumulative numbers of confirmed cases. Effective population size and Re were estimated from 67 whole-genome sequences sampled from persons on board the Diamond Princess. Thick black line in (a) and (b), median estimate; colored area, 95% highest posterior density (HPD) confidence intervals of the estimate. Dashed (a) and dotted (b) lines, time to the most recent common ancestor and estimated time of outbreak origin, respectively.
3.4. Basic and effective reproductive numbers over time
To estimate R0 and the doubling time of infections, the exponential growth rate for 67 sequences from the cruise ship was calculated under an exponential coalescent model with a strict clock model. The estimated exponential growth rate was 61.5 per year (95% HPD, 1.66–143.5), corresponding to 0.168 per day (95% HPD, 0.00454–0.392). The estimated R0 was 2.56 (95% confidence interval [CI], 1.04–4.65), and the direct estimation of the mean doubling time was 4.11 days (95% CI, 1.76–152.4; Table 1 ).
Table 1.
Estimated epidemiological parameters based on exponential growth rate.
| Mean estimates | 95% CI | |
|---|---|---|
| Basic reproductive number (R0) | 2.56 | 1.04, 4.65 |
| Doubling time (days) | 4.11 | 1.76, 152.4 |
We estimated epidemiological parameters and temporal changes in Re for the outbreak using the birth–death skyline contemporary model (Table 2 , Fig. 2b). The number of mean infected persons was calculated as 682 (95% HPD, 293–17000) based on the estimated sampling proportion (ρ). The Bayesian birth–death skyline plot showed a median Re of 2.84 (95% HPD, 0.0204–19.6) at the origin of the epidemic, which we estimated had been between January 21 and 22, 2020. The Re remained at 2–3 during the cruise, reached a high of 6.06 (95% HPD, 0.0379–31.8) on February 4, and then gradually declined to 1.51 (0.0320–6.33) on February 17, reflecting the effects of placing the ship in quarantine.
Table 2.
Estimated epidemiological parameters using a birth–death model.
| Mean estimates | 95% HPD | |
|---|---|---|
| Origin of the epidemic (y) | 0.0722 (January 21, 2020) | 0.0360, 0.124 (Jan 2–Feb 3, 2020) |
| Rate of becoming noninfectious per year | 30.4 | 4.83, 64.1 |
| Sampling proportion (ρ) | 0.0982 | 3.93 × 10−3, 0.229 |
| Tree root (tMRCA, year) | 0.0573 | 0.0334, 0.0858 |
HPD, highest posterior density; tMRCA, time to the most recent common ancestor.
3.5. Confirmed cases on the Diamond Princess
Fig. 2c shows daily confirmed and cumulative cases over time. Symptomatic and subsequently asymptomatic passengers and crew were tested using RT-PCR when the ship returned to Yokohama on February 3. The first results were obtained on February 5, 2020. The number of confirmed cases increased substantially, and the cumulative number exponentially increased after February 7, which reflected the estimated exponential increase of infections on the cruise ship after February 2.
4. Discussion
The present study investigated transmission dynamics of SARS-CoV-2 on the Diamond Princess cruise ship based on viral genome data using a Bayesian framework. Previous studies have estimated these transmission dynamics on the ship based on mathematical models using the daily number of confirmed cases or the daily number of symptomatic confirmed cases (Mizumoto and Chowell, 2020, Nishiura, 2020, Rocklöv et al., 2020). However, these epidemiological data could have been biased because of the underestimation of infections owing to the limited diagnostic test capacity in the early phase of the outbreak and the limited sensitivity of RT-PCR (Woloshin et al., 2020). In addition, the origin of the outbreak might be difficult to estimate due to the lack of diagnostic tests on the voyage. In contrast, recent advances in genomics and computing have facilitated the extraction of valuable information from viral genome data, and analytic methods have been developed to infer transmission history and past population dynamics of the pathogen with limited epidemiological data (Pybus et al., 2001, Drummond et al., 2005, Stadler et al., 2013). We aimed to elucidate the dynamics of transmission from the perspective of genomic epidemiology, which would contribute to further understanding of the outbreak.
SARS-CoV-2 can spread rapidly via close contact and in confined spaces (Li et al., 2020, Rocklöv and Sjödin, 2020), such as cruise ships (Moriarty et al., 2020, Sando et al., 2020, Sekizuka et al., 2020b) and airplanes (Hoehl et al., 2020, Pavli et al., 2020, Speake et al., 2020). Our results showed the transmission dynamics of the SARS-CoV-2 outbreak in a high-transmission environment. The estimated origin of the epidemic was January 21, which coincided with the date when the presumed index case boarded the ship (Leung et al., 2020). The estimated R0 was 2.56 (95% HPD, 1.04–4.65), which was in the range of the community-level R0 (2.2–2.7) (Li et al., 2020, Wu et al., 2020) and in lower range of previous estimates for the ship (2.28–14.8) (Rocklöv et al., 2020, Zhang et al., 2020b). The effective population size began to increase around January 30 and exponentially surged from February 2, before the start of quarantine on February 5. These findings indicate that most infections had already occurred before quarantine, which is consistent with previous findings (Mizumoto et al., 2020, National institute of infectious diseases, 2020, Nishiura et al., 2020). When we compare our findings with epidemiological data, the exponential increase in confirmed cases after February 7 appears to follow the estimated substantial growth in infections by approximately 5 days. This time interval is in close agreement with the incubation period of the virus, which is on average 5.2 days with a 95th percentile of the distribution at 12.5 days (Li et al., 2020). Although the number of confirmed cases might be affected by the limited capacity of RT-PCR tests or change of test strategies, this paralleled increase confirms that our estimates reasonably reflect the increase in infections. Previous studies also used the number of confirmed febrile cases by date of symptom onset (Mizumoto and Chowell, 2020, National institute of infectious diseases, 2020, Nishiura, 2020, Tsuboi et al., 2020). The number of confirmed febrile cases surged on February 7 and gradually declined afterwards. The increase in febrile cases might reflect the estimated exponential increase in infections in the beginning of February, although the number of confirmed febrile cases might be biased by the distribution of thermometer among all individuals and the establishment of the Fever Call Center on February 7. The interval between the estimated increase in infections and the observed peak of daily febrile cases appears to be relatively close, considering the average incubation period of 5.2 (95% CI: 4.1–7.0) days (Li et al., 2020). However, the incubation period of COVID-19 varies across individuals within a population, and factors such as age and disease severity affect the length of the incubation period (Dai et al., 2020, Kong, 2020, Lai et al., 2020b). Therefore, the interval between our estimates and the number of febrile cases could be explained by the temporal change of infected subpopulations with different backgrounds.
The modern cruise ships carry large numbers of people in limited spaces for a certain period, which facilitate the transmission of infectious diseases and outbreaks (Mitruka and Wheeler, 2008, Kak, 2015). The Diamond Princess had many passengers and crew from various countries, and her cruise lasted more than 2 weeks. The facilities on the ship provided semi-closed spaces for passengers and crew, which contributed to high transmission of SARS-CoV-2. Large cocktail parties during the cruise played an important role in the dissemination of infections among passengers and crew (Yamagishi et al., 2020). From the perspective of virology, SARS-CoV-2 has highly contagious characteristics, including a higher R0, longer incubation period, shorter interval between symptom onset and infectivity, and higher proportions of asymptomatic persons and patients with mild illness, compared to those with influenza, SARS-CoV, and MERS-CoV (Petersen et al., 2020). These environmental and virologic factors might contribute to the outbreak of SARS-CoV-2 onboard the cruise ship.
The Re peaked at 6.06 on February 4 and then decreased to 1.51. The effective population size continued to slightly increase after quarantine and reached a plateau around February 10. Although quarantine greatly reduced the number of infections, our results suggested that the infection spread slowly even after quarantine, which is consistent with previous epidemiological studies using mathematical models (Mizumoto and Chowell, 2020, Nishiura, 2020) and haplotype network analysis using viral genome sequences (Sekizuka et al., 2020a). Strict implementation of quarantine on the cruise ship was encumbered by difficulties in developing effective traffic lines or zones between infectious and noninfectious passengers and in providing daily support and medical procedures for passengers (Yamahata and Shibata, 2020). The former possibly promoted transmission among passengers after quarantine through shared onboard facilities or cabins (Sekizuka et al., 2020a). The latter necessitated activities of the crew, which put members at risk of infection. A previous study estimated that the epidemic peaked among passengers and crew between February 2–4 and February 8–10, respectively (Nishiura, 2020). The epidemic period among the crew coincided with the increase in the effective population size after quarantine, suggesting that the slight increase in infections reflected the spread of infection among crew members. Further investigations are essential to understand the transmission dynamics and mechanism of SARS-CoV-2, including the possibility and effect of airborne transmission (Centers for Disease Control and Prevention, 2020). It is also necessary to develop effective strategies for preventing and controlling outbreaks in confined settings. The development of vaccines and anticoronaviral drugs are urgent for preventing future COVID-19 epidemic (Basu et al., 2020, Shin et al., 2020, Zhang et al., 2020a). Based on lessons learnt from the outbreak aboard the Diamond Princess, appropriate protective measures, and separation of clean and contaminated areas should be implemented comprehensively under the supervision of infectious disease specialists. In addition, activities of staff should be restricted to a minimum under quarantine, and suspected infected staff should be isolated as early as possible (Tokuda et al., 2020). Finally, cooperation of various specialists and an established chain of command are essential for controlling a large-scale outbreak in confined settings (Tokuda et al., 2020). Comprehensive and practical protocols should be designed for future epidemic events.
Our study had certain advantages when compared with previous studies on the transmission dynamics of SARS-CoV-2 aboard the Diamond Princess using mathematical models and the number of laboratory-confirmed febrile cases (Mizumoto and Chowell, 2020, Nishiura, 2020). First, our study takes asymptomatic cases into consideration. Although previous studies focused on symptomatic population, a substantial proportion of SARS-CoV-2 infections is asymptomatic and has a significantly longer duration of viral shedding than that in the symptomatic population (Long et al., 2020, Mizumoto et al., 2020, Yanes-Lane et al., 2020), suggesting a critical role of asymptomatic individuals in the outbreak. In contrast, most sequences in this study were obtained from both symptomatic and asymptomatic patients through extensive tests, enabling our study to reflect the entire dynamics of transmission on the ship. Second, our analysis used objective data such as viral genome sequences and sampling dates. In contrast, the dates of fever onset can be affected by factors such as the background of individuals and testing situation on the ship. For instance, older adults with SARS-CoV-2 infection become more symptomatic and have a longer incubation period (Dai et al., 2020, Yamagishi et al., 2020). In addition, distribution of thermometers among all individuals and the establishment of the Fever Call Center on February 7 could increase the detection of symptoms (Tsuboi et al., 2020). The data used in this study are more robust against these potential biases. Third, our study investigates the origin of the epidemic using viral genomic data. Our results showed that the outbreak had a single origin and that the epidemic started immediately after departure. However, in our study, one of the disadvantages was that the association between transmission dynamics and epidemiological factors could not be analyzed. Factors such as age, sex, location of cabin, and membership might largely affect transmission patterns. It was technically difficult to combine this information with phylogenetic analysis in this study. Therefore, we could not analyze the transmission dynamics in each subpopulation or between subpopulations. Second disadvantage is the potential sampling bias and sequencing errors due to next-generation sequencing techniques. It is difficult to obtain a complete and high-quality viral sequence from a sample with a low viral load. The viral load can be affected by the sampling procedure, immunological characteristics of individuals, and interval between infection and sampling. Therefore, there is a possibility that specific groups were excluded from our study.
This study has several potential limitations. First, we used SARS-CoV-2 sequences sampled over a period of 8 months. The short time range and relatively low diversity of SARS-CoV-2 might have affected the estimation of the substitution rate. To mitigate this challenge, we selected sequences to represent the temporal and geographical diversity of SARS-CoV-2 at the time while minimizing the impact of sampling bias. Epidemiological parameters of SARS-CoV-2 infection are currently limited. Although the prior used herein was set based on available epidemiological data, these settings might need modification based on future data. However, our results were consistent with those of previous epidemiological studies using mathematical models. Regarding the application of the Bayesian Skyline plot for epidemic data, sequences should be sampled sparsely from a large population under the assumption of a coalescent model. Sequences used in this study were less than 10% of the total confirmed cases, which might be sufficiently sparse (Fu, 2006). In addition, the HPD interval under a coalescent model tends to be overconfident when the number of lineages in the tree gets close to population size (Stadler et al., 2013). Therefore, the HPD interval in the early phase of an epidemic should be interpreted with caution. Re was estimated using the birth–death skyline contemporary model in this study. The birth–death model can be biased largely when sampling process is mis-specified in the use of serially-sampled sequences (Volz and Frost, 2014). It was difficult to establish a prior reflecting the complex sampling process on the ship, and convergence was not achieved. Because sequences were obtained almost contemporaneously, we used the birth–death contemporary model with the sampling proportion prior that can be set with reasonable certainty. This model selection could affect the estimated change of Re within approximately 1 day; however, our results would be robust against the irregular sampling process on the ship. Finally, we analyzed sequences as they were sampled from only one group. However, the transmission patterns differed between the passengers and crew, which might have affected our findings. The discrepancies in transmission patterns between passengers and crew can be explained by different living patterns on the cruise ship, locations of cabins in distinct decks, and extent of activity after quarantine (Nishiura, 2020, Tsuboi et al., 2020, Yamagishi et al., 2020). The incorporation of the different transmission patterns into phylogenetic analyses would contribute to more accurate projections of transmission dynamics. However, it is difficult to incorporate phylogenetic analysis with complicated epidemiological models, including age, structure of host contact rates, or multiple host populations with different life history traits (Volz et al., 2013). Alternatively, we could analyze sequences from a specific subpopulation, for instance passengers or crew. We identified four sequences from the crew in our dataset, which were too few to analyze transmission dynamics among the passengers and crew. Future studies involving more genetic and finer epidemiological data could facilitate further understanding of the complexity of SARS-CoV-2 transmission in confined spaces.
In conclusion, we investigated the transmission dynamics associated with the COVID-19 outbreak aboard the Diamond Princess cruise ship. Based on the estimated substitution rate, phylodynamic analysis showed that the outbreak on the cruise ship occurred between late January and early February, before quarantine started on February 5, 2020. Although quarantine was clearly effective, the infection continued to slowly spread, even after the countermeasures were initiated. More effective measures should be considered to control outbreaks, particularly in confined settings.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
CRediT authorship contribution statement
Kunikazu Hoshino: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Project administration, Visualization, Writing - original draft. Tatsuji Maeshiro: Writing - review & editing. Nao Nishida: Writing - review & editing. Masaya Sugiyama: Writing - review & editing. Jiro Fujita: Writing - review & editing. Takashi Gojobori: Conceptualization, Investigation, Supervision, Writing - review & editing. Masashi Mizokami: Conceptualization, Investigation, Supervision, Project administration, Writing - review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We gratefully acknowledge the team at GISAID for creating the SARS-CoV-2 global database and the authors and investigators who submitted the sequences.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.gene.2021.145496.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- Baele G., Lemey P., Bedford T., Rambaut A., Suchard M.A., Alekseyenko A.V. Improving the accuracy of demographic and molecular clock model comparison while accommodating phylogenetic uncertainty. Mol. Biol. Evol. 2012;29:2157–2167. doi: 10.1093/molbev/mss084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai Y., Yao L., Wei T., Tian F., Jin D.Y., Chen L., Wang M. Presumed Asymptomatic Carrier Transmission of COVID-19. JAMA. 2020;323:1406–1407. doi: 10.1001/jama.2020.2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu S., Veeraraghavan B., Ramaiah S., Anbarasu A. Novel cyclohexanone compound as a potential ligand against SARS-CoV-2 main-protease. Microb. Pathog. 2020;149 doi: 10.1016/j.micpath.2020.104546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne A.W., McEvoy D., Collins A.B., Hunt K., Casey M., Barber A., Butler F., Griffin J., Lane E.A., McAloon C., O'Brien K., Wall P., Walsh K.A., More S.J. Inferred duration of infectious period of SARS-CoV-2: rapid scoping review and analysis of available evidence for asymptomatic and symptomatic COVID-19 cases. BMJ Open. 2020;10 doi: 10.1136/bmjopen-2020-039856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention, 2020. Scientific Brief: SARS-CoV-2 and Potential Airborne Transmission. https://www.cdc.gov/coronavirus/2019-ncov/more/scientific-brief-sars-cov-2.html (last accessed: October 10, 2020).
- Coronaviridae Study Group of the International Committee on Taxonomy of Viruses The species Severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020;5:536–544. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai J., Yang L., Zhao J. Probable longer incubation period for elderly COVID-19 cases: analysis of 180 contact tracing data in Hubei Province, China. Risk Manag. Healthc. Policy. 2020;13:1111–1117. doi: 10.2147/RMHP.S257907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delamater P.L., Street E.J., Leslie T.F., Yang Y.T., Jacobsen K.H. Complexity of the Basic Reproduction Number (R0) Emerg. Infect. Dis. 2019;25:1–4. doi: 10.3201/eid2501.171901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond A.J., Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond A.J., Rambaut A., Shapiro B., Pybus O.G. Bayesian coalescent inference of past population dynamics from molecular sequences. Mol. Biol. Evol. 2005;22:1185–1192. doi: 10.1093/molbev/msi103. [DOI] [PubMed] [Google Scholar]
- Elbe S., Buckland-Merrett G. Data, disease and diplomacy: GISAID's innovative contribution to global health. Glob Chall. 2017;1:33–46. doi: 10.1002/gch2.1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y.X. Exact coalescent for the Wright-Fisher model. Theor Popul Biol. 2006;69:385–394. doi: 10.1016/j.tpb.2005.11.005. [DOI] [PubMed] [Google Scholar]
- Ganyani T., Kremer C., Chen D., Torneri A., Faes C., Wallinga J., Hens N. Estimating the generation interval for coronavirus disease (COVID-19) based on symptom onset data, March 2020. Euro Surveill. 2020;25:2000257. doi: 10.2807/1560-7917.ES.2020.25.17.2000257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardy J.L., Loman N.J. Towards a genomics-informed, real-time, global pathogen surveillance system. Nat. Rev. Genet. 2018;19:9–20. doi: 10.1038/nrg.2017.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadfield J., Megill C., Bell S.M., Huddleston J., Potter B., Callender C., Sagulenko P., Bedford T., Neher R.A. Nextstrain: real-time tracking of pathogen evolution. Bioinformatics. 2018;34:4121–4123. doi: 10.1093/bioinformatics/bty407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X., Lau E.H.Y., Wu P., Deng X., Wang J., Hao X., Lau Y.C., Wong J.Y., Guan Y., Tan X., Mo X., Chen Y., Liao B., Chen W., Hu F., Zhang Q., Zhong M., Wu Y., Zhao L., Zhang F., Cowling B.J., Li F., Leung G.M. Temporal dynamics in viral shedding and transmissibility of COVID-19. Nat. Med. 2020;26:672–675. doi: 10.1038/s41591-020-0869-5. [DOI] [PubMed] [Google Scholar]
- Hoehl S., Karaca O., Kohmer N., Westhaus S., Graf J., Goetsch U., Ciesek S. Assessment of SARS-CoV-2 transmission on an international flight and among a tourist group. JAMA Netw. Open. 2020;3 doi: 10.1001/jamanetworkopen.2020.18044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., Cheng Z., Yu T., Xia J., Wei Y., Wu W., Xie X., Yin W., Li H., Liu M., Xiao Y., Gao H., Guo L., Xie J., Wang G., Jiang R., Gao Z., Jin Q., Wang J., Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kak V. Infections on cruise ships. Microbiol. Spectr. 2015;3 doi: 10.1128/microbiolspec.IOL5-0007-2015. [DOI] [PubMed] [Google Scholar]
- Kong T.K. Longer incubation period of coronavirus disease 2019 (COVID-19) in older adults. Aging Med. (Milton) 2020;3:102–109. doi: 10.1002/agm2.12114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korber B., Fischer W.M., Gnanakaran S., Yoon H., Theiler J., Abfalterer W., Hengartner N., Giorgi E.E., Bhattacharya T., Foley B., Hastie K.M., Parker M.D., Partridge D.G., Evans C.M., Freeman T.M., de Silva T.I., Sheffield C.-G.-G., McDanal C., Perez L.G., Tang H., Moon-Walker A., Whelan S.P., LaBranche C.C., Saphire E.O., Montefiori D.C. Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell. 2020;182:812–827. doi: 10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladner J.T., Grubaugh N.D., Pybus O.G., Andersen K.G. Precision epidemiology for infectious disease control. Nat. Med. 2019;25:206–211. doi: 10.1038/s41591-019-0345-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai A., Bergna A., Acciarri C., Galli M., Zehender G. Early phylogenetic estimate of the effective reproduction number of SARS-CoV-2. J. Med. Virol. 2020;92:675–679. doi: 10.1002/jmv.25723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai C., Yu R., Wang M., Xian W., Zhao X., Tang Q., Chen R., Zhou X., Li X., Li Z., Li Z., Deng G., Wang F. Shorter incubation period is associated with severe disease progression in patients with COVID-19. Virulence. 2020;11:1443–1452. doi: 10.1080/21505594.2020.1836894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung W.S., Chan J.M.C., Chik T.S.H., Lau D.P.L., Choi C.Y.C., Lau A.W.T., Tsang O.T.Y. Presumed COVID-19 index case on diamond princess cruise ship and evacuees to Hong Kong. J. Travel Med. 2020;27:taaa073. doi: 10.1093/jtm/taaa073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Guan X., Wu P., Wang X., Zhou L., Tong Y., Ren R., Leung K.S.M., Lau E.H.Y., Wong J.Y., Xing X., Xiang N., Wu Y., Li C., Chen Q., Li D., Liu T., Zhao J., Liu M., Tu W., Chen C., Jin L., Yang R., Wang Q., Zhou S., Wang R., Liu H., Luo Y., Liu Y., Shao G., Li H., Tao Z., Yang Y., Deng Z., Liu B., Ma Z., Zhang Y., Shi G., Lam T.T.Y., Wu J.T., Gao G.F., Cowling B.J., Yang B., Leung G.M., Feng Z. Early Transmission dynamics in Wuhan, China, of novel coronavirus-infected pneumonia. N. Engl. J. Med. 2020;382:1199–1207. doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F., Li X., Zhu G. Using the contact network model and Metropolis-Hastings sampling to reconstruct the COVID-19 spread on the “Diamond Princess”. Sci Bull (Beijing) 2020;65:1297–1305. doi: 10.1016/j.scib.2020.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long Q.X., Tang X.J., Shi Q.L., Li Q., Deng H.J., Yuan J., Hu J.L., Xu W., Zhang Y., Lv F.J., Su K., Zhang F., Gong J., Wu B., Liu X.M., Li J.J., Qiu J.F., Chen J., Huang A.L. Clinical and immunological assessment of asymptomatic SARS-CoV-2 infections. Nat. Med. 2020;26:1200–1204. doi: 10.1038/s41591-020-0965-6. [DOI] [PubMed] [Google Scholar]
- Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., Wang W., Song H., Huang B., Zhu N., Bi Y., Ma X., Zhan F., Wang L., Hu T., Zhou H., Hu Z., Zhou W., Zhao L., Chen J., Meng Y., Wang J., Lin Y., Yuan J., Xie Z., Ma J., Liu W.J., Wang D., Xu W., Holmes E.C., Gao G.F., Wu G., Chen W., Shi W., Tan W. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ministry of Health, Labor and Welfare., The situation report on the novel coronavirus infections, as of 17 August 2020 (in Japanese). https://www.mhlw.go.jp/stf/newpage_13013.html (last accessed: October 10, 2020).
- Mitruka K., Wheeler R.E. Cruise ship travel. Travel Med. 2008:351–360. [Google Scholar]
- Mizumoto K., Chowell G. Transmission potential of the novel coronavirus (COVID-19) onboard the diamond Princess Cruises Ship, 2020. Infect. Dis. Model. 2020;5:264–270. doi: 10.1016/j.idm.2020.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizumoto K., Kagaya K., Zarebski A., Chowell G. Estimating the asymptomatic proportion of coronavirus disease 2019 (COVID-19) cases on board the Diamond Princess cruise ship, Yokohama, Japan, 2020. Euro. Surveill. 2020;25:2000180. doi: 10.2807/1560-7917.ES.2020.25.10.2000180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriarty L.F., Plucinski M.M., Marston B.J., Kurbatova E.V., Knust B., Murray E.L., Pesik N., Rose D., Fitter D., Kobayashi M., Toda M., Cantey P.T., Scheuer T., Halsey E.S., Cohen N.J., Stockman L., Wadford D.A., Medley A.M., Green G., Regan J.J., Tardivel K., White S., Brown C., Morales C., Yen C., Wittry B., Freeland A., Naramore S., Novak R.T., Daigle D., Weinberg M., Acosta A., Herzig C., Kapella B.K., Jacobson K.R., Lamba K., Ishizumi A., Sarisky J., Svendsen E., Blocher T., Wu C., Charles J., Wagner R., Stewart A., Mead P.S., Kurylo E., Campbell S., Murray R., Weidle P., Cetron M., Friedman C.R. Public Health responses to COVID-19 outbreaks on cruise ships - worldwide, February-March 2020. MMWR Morb. Mortal. Wkly Rep. 2020;69:347–352. doi: 10.15585/mmwr.mm6912e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- National institute of infectious diseases, 2020. Field Briefing: Diamond Princess COVID-19 Cases, 20 Feb Update. https://www.niid.go.jp/niid/en/2019-ncov-e/9417-covid-dp-fe-02.html (last accessed: October 10, 2020).
- Nishiura H. Backcalculating the incidence of infection with COVID-19 on the Diamond Princess. J. Clin. Med. 2020;9:657. doi: 10.3390/jcm9030657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiura H., Kobayashi T., Miyama T., Suzuki A., Jung S.M., Hayashi K., Kinoshita R., Yang Y., Yuan B., Akhmetzhanov A.R., Linton N.M. Estimation of the asymptomatic ratio of novel coronavirus infections (COVID-19) Int J Infect Dis. 2020;94:154–155. doi: 10.1016/j.ijid.2020.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavli A., Smeti P., Hadjianastasiou S., Theodoridou K., Spilioti A., Papadima K., Andreopoulou A., Gkolfinopoulou K., Sapounas S., Spanakis N., Tsakris A., Maltezou H.C. In-flight transmission of COVID-19 on flights to Greece: an epidemiological analysis. Travel Med. Infect. Dis. 2020;38 doi: 10.1016/j.tmaid.2020.101882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen E., Koopmans M., Go U., Hamer D.H., Petrosillo N., Castelli F., Storgaard M., Al Khalili S., Simonsen L. Comparing SARS-CoV-2 with SARS-CoV and influenza pandemics. Lancet Infect. Dis. 2020;20:e238–e244. doi: 10.1016/S1473-3099(20)30484-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pybus O.G., Charleston M.A., Gupta S., Rambaut A., Holmes E.C., Harvey P.H. The epidemic behavior of the hepatitis C virus. Science. 2001;292:2323–2325. doi: 10.1126/science.1058321. [DOI] [PubMed] [Google Scholar]
- Rocklöv J., Sjödin H. High population densities catalyse the spread of COVID-19. J. Travel Med. 2020;27:taaa038. doi: 10.1093/jtm/taaa038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocklöv J., Sjödin H., Wilder-Smith A. COVID-19 outbreak on the Diamond Princess cruise ship: estimating the epidemic potential and effectiveness of public health countermeasures. J. Travel Med. 2020;27:taaa030. doi: 10.1093/jtm/taaa030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothe C., Schunk M., Sothmann P., Bretzel G., Froeschl G., Wallrauch C., Zimmer T., Thiel V., Janke C., Guggemos W., Seilmaier M., Drosten C., Vollmar P., Zwirglmaier K., Zange S., Wölfel R., Hoelscher M. Transmission of 2019-nCoV infection from an asymptomatic contact in Germany. N. Engl. J. Med. 2020;382:970–971. doi: 10.1056/NEJMc2001468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sando E., Morimoto K., Narukawa S., Nakata K. COVID-19 outbreak on the Costa Atlantica cruise ship: use of a remote health monitoring system. J. Travel Med. 2020:taaa163. doi: 10.1093/jtm/taaa163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekizuka T., Itokawa K., Kageyama T., Saito S., Takayama I., Asanuma H., Nao N., Tanaka R., Hashino M., Takahashi T., Kamiya H., Yamagishi T., Kakimoto K., Suzuki M., Hasegawa H., Wakita T., Kuroda M. Haplotype networks of SARS-CoV-2 infections in the Diamond Princess cruise ship outbreak. Proc. Natl. Acad. Sci. U S A. 2020;117:20198–20201. doi: 10.1073/pnas.2006824117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekizuka T., Kuramoto S., Nariai E., Taira M., Hachisu Y., Tokaji A., Shinohara M., Kishimoto T., Itokawa K., Kobayashi Y., Kadokura K., Kamiya H., Matsui T., Suzuki M., Kuroda M. SARS-CoV-2 genome analysis of Japanese travelers in Nile River Cruise. Front. Microbiol. 2020;11:1316. doi: 10.3389/fmicb.2020.01316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin M.D., Shukla S., Chung Y.H., Beiss V., Chan S.K., Ortega-Rivera O.A., Wirth D.M., Chen A., Sack M., Pokorski J.K., Steinmetz N.F. COVID-19 vaccine development and a potential nanomaterial path forward. Nat. Nanotechnol. 2020;15:646–655. doi: 10.1038/s41565-020-0737-y. [DOI] [PubMed] [Google Scholar]
- Speake H., Phillips A., Chong T., Sikazwe C., Levy A., Lang J., Scalley B., Speers D.J., Smith D.W., Effler P., McEvoy S.P. Flight-associated transmission of severe acute respiratory syndrome coronavirus 2 corroborated by whole-genome sequencing. Emerg. Infect. Dis. 2020;26:2872–2880. doi: 10.3201/eid2612.203910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadler T., Kuhnert D., Bonhoeffer S., Drummond A.J. Birth-death skyline plot reveals temporal changes of epidemic spread in HIV and hepatitis C virus (HCV) Proc. Natl. Acad. Sci. U S A. 2013;110:228–233. doi: 10.1073/pnas.1207965110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokuda Y., Sakihama T., Aoki M., Taniguchi K., Deshpande G.A., Suzuki S., Uda S., Kurokawa K. COVID-19 outbreak on the Diamond Princess Cruise Ship in February 2020. J. Gen. Fam. Med. 2020;21:95–97. doi: 10.1002/jgf2.326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuboi M., Hachiya M., Noda S., Iso H., Umeda T. Epidemiology and quarantine measures during COVID-19 outbreak on the cruise ship Diamond Princess docked at Yokohama, Japan in 2020: a descriptive analysis. Global Health Med. 2020;2:102–106. doi: 10.35772/ghm.2020.01037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volz E.M., Frost S.D. Sampling through time and phylodynamic inference with coalescent and birth-death models. J. R. Soc. Interface. 2014;11:20140945. doi: 10.1098/rsif.2014.0945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volz E.M., Koelle K., Bedford T. Viral phylodynamics. PLoS Comput. Biol. 2013;9 doi: 10.1371/journal.pcbi.1002947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallinga J., Lipsitch M. How generation intervals shape the relationship between growth rates and reproductive numbers. Proc. Royal Soc. B: Biol. Sci. 2007;274:599–604. doi: 10.1098/rspb.2006.3754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO, 2020a. Coronavirus disease (COVID-19) Weekly Epidemiological Up. Data as received by WHO from national authorities, as of 27 September 2020, 10 am CEST. https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200928-weekly-epi-update.pdf?sfvrsn=9e354665_6 (last accessed: October 10, 2020).
- WHO, 2020b. WHO Director General’s opening remarks at the media briefing on COVID-19-11. March 11, 2020. https://www.who.int/dg/speeches/detail/who-director-general-s-opening-remarks-at-the-media-briefing-on-covid-19---11-march-2020 (last accessed: October 10, 2020).
- Woloshin S., Patel N., Kesselheim A.S. False negative tests for SARS-CoV-2 infection — challenges and implications. N. Engl. J. Med. 2020;383 doi: 10.1056/NEJMp2015897. [DOI] [PubMed] [Google Scholar]
- Wu J.T., Leung K., Leung G.M. Nowcasting and forecasting the potential domestic and international spread of the 2019-nCoV outbreak originating in Wuhan, China: a modelling study. Lancet. 2020;395:689–697. doi: 10.1016/S0140-6736(20)30260-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagishi T., Kamiya H., Kakimoto K., Suzuki M., Wakita T. Descriptive study of COVID-19 outbreak among passengers and crew on Diamond Princess cruise ship, Yokohama Port, Japan, 20 January to 9 February 2020. Euro. Surveill. 2020;25:2000272. doi: 10.2807/1560-7917.ES.2020.25.23.2000272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamahata Y., Shibata A. Preparation for quarantine on the cruise ship Diamond Princess in Japan due to COVID-19. JMIR Public Health Surveill. 2020;6 doi: 10.2196/18821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanes-Lane M., Winters N., Fregonese F., Bastos M., Perlman-Arrow S., Campbell J.R., Menzies D. Proportion of asymptomatic infection among COVID-19 positive persons and their transmission potential: a systematic review and meta-analysis. PLoS ONE. 2020;15 doi: 10.1371/journal.pone.0241536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Lin D., Sun X., Curth U., Drosten C., Sauerhering L., Becker S., Rox K., Hilgenfeld R. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science. 2020;368:409–412. doi: 10.1126/science.abb3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S., Diao M., Yu W., Pei L., Lin Z., Chen D. Estimation of the reproductive number of novel coronavirus (COVID-19) and the probable outbreak size on the Diamond Princess cruise ship: a data-driven analysis. Int. J. Infect. Dis. 2020;93:201–204. doi: 10.1016/j.ijid.2020.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., Chen H.D., Chen J., Luo Y., Guo H., Jiang R.D., Liu M.Q., Chen Y., Shen X.R., Wang X., Zheng X.S., Zhao K., Chen Q.J., Deng F., Liu L.L., Yan B., Zhan F.X., Wang Y.Y., Xiao G.F., Shi Z.L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.