Abstract
Despite major advances in our understanding of players and mechanisms involved in peroxisome biogenesis and peroxisome degradation, very few studies have focused on unraveling the multi-layered connections between, and the coordination of, these two opposing processes that regulate peroxisome homeostasis. The intersection between these processes also provides exciting avenues for future research. This review highlights the links between peroxisome biogenesis and degradation, incorporating an integrative approach that is critical not only for a mechanistic understanding, but also for manipulating the balance between these processes in relevant disease models.
Overview of the Players and Mechanisms Involved in Peroxisome Biogenesis
Peroxisomes are ubiquitous, single membrane organelles, that play an essential role in lipid metabolism and redox homeostasis [1]. They are essential in humans and deficiencies in their biogenesis manifest as peroxisome biogenesis disorders (PBDs), that can be debilitating and fatal [1]. Their biogenesis has been studied in many model organisms ranging from single-celled yeasts to multi-cellular mammals. However, because these biogenesis pathways have been reviewed in depth elsewhere [2–4], an overview is only provided here, as the context for subsequent discussion of peroxisome homeostasis.
Matrix and membrane proteins destined for peroxisomes rely on peroxisome targeting signals (PTSs), called PTS1 (C-terminal SKL tripeptide or its conserved variants), or PTS2 (internal sequence with the consensus (R/K)(L/V/I/Q)XX(L/V/I/H/Q)(L/S/G/A/K)X(H/Q)(L/A/F)) for peroxisomal matrix proteins, and mPTSs (membrane PTSs lacking a consensus sequence) for peroxisomal membrane proteins (PMPs) [3] (Figure 1). Following synthesis of peroxisomal matrix proteins in the cytosol, they are recognized by specific PTS receptors, such as Pex5 (nomenclature for yeast and mammalian components is Pex5 and PEX5, respectively), and Pex7, which interact with PTS1 and PTS2 cargoes, respectively. Yeasts have a PTS2 coreceptor, called Pex18, Pex20, or Pex21, while mammalian PEX5 has two isoforms that differ by alternative splicing and the longer one, PEX5L, also binds PEX7.
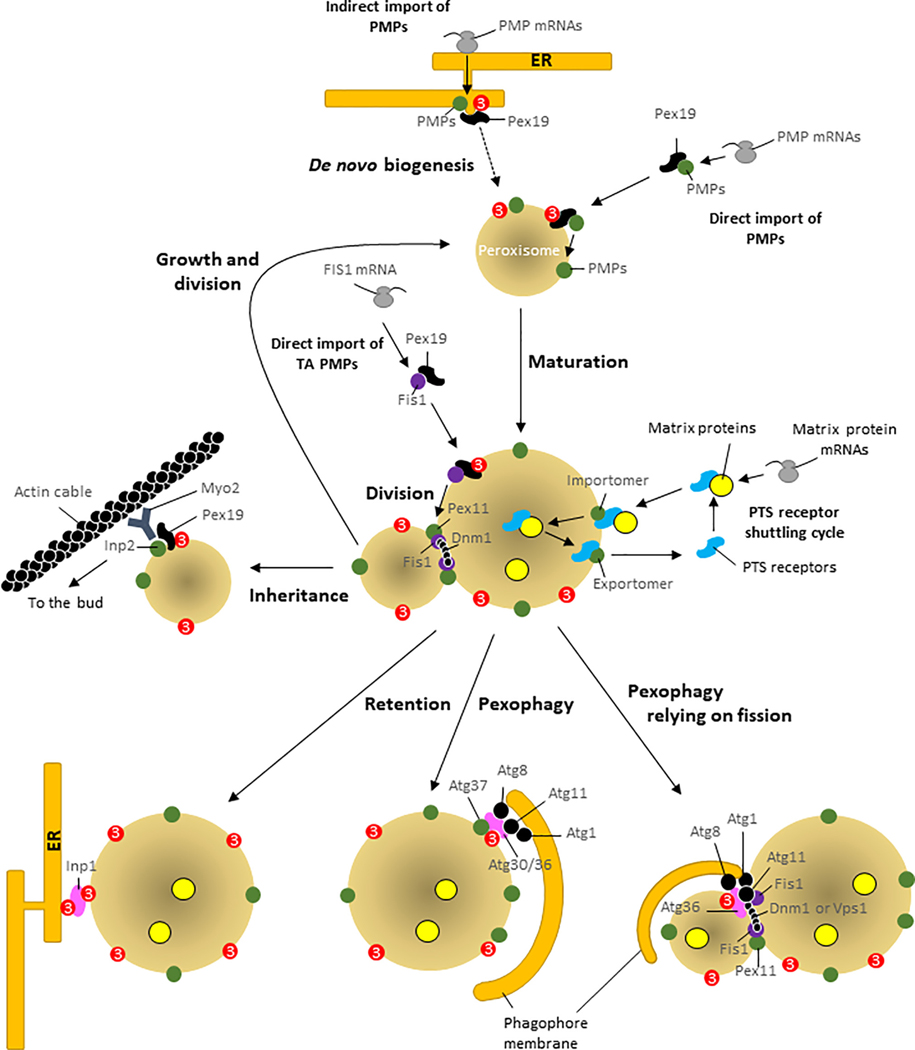
Figure 1. Peroxisome Homeostasis and Key Roles of Pex3 in the Fate of Peroxisomes.
Direct and indirect import of PMPs: direct PMP import to the membrane requires Pex19, which docks with Pex3 and inserts PMPs into the membrane (growth and division model). Alternatively, PMPs may be imported indirectly via the ER, wherein PMPs bud from the ER using Pex3 and Pex19, and the resulting vesicles fuse with pre-existing peroxisomes. In cells lacking peroxisomes, the vesicles containing PMPs fuse and grow to form new peroxisomes (de novo biogenesis model). Direct import of TA proteins: these (e.g., Fis1) are imported post-translationally to the peroxisome membrane using Pex19 and Pex3. Maturation and PTS receptor shuttling cycle: proteins destined for the peroxisomal matrix are translocated by PTS receptors across the importomer. Following cargo release in the matrix, PTS receptors recycle back to the cytosol, via the exportomer. Division: another common mode of peroxisome proliferation is by growth and division of pre-existing peroxisomes, during which daughter peroxisomes bud from a mother peroxisome, using the division machinery comprised of Pex11, Fis1, and Dnm1 in yeast. Inheritance and Retention: during cytokinesis in yeast, daughter peroxisomes are inherited using the Myo2 motor interacting with peroxisomal Inp2 and actin filaments, whereas the peroxisome in the mother cell is retained by the interaction of Inp1 with Pex3 at the cortical ER. Pexophagy: requires SARs (Atg30, Atg36) and a SAR regulator (Atg37 in P. pastoris), which associate with, or are inserted into, the peroxisome membrane by Pex3. SARs activated by environmental cues engage the autophagy machinery (Atg8, Atg11, and Atg1 kinase) allowing phagophore membranes to engulf peroxisomes. Pexophagy relying on fission: in S. cerevisiae, pexophagy requires peroxisome division. Abbreviations: ER, endoplasmic reticulum; PMPs, peroxisomal membrane proteins; PTS, peroxisomal targeting signal; TA, tail-anchored; SARs, selective autophagy receptors.
The formation of a cytosolic PTS receptor/cargo complex is followed by its interaction with a peroxisome membrane docking complex, comprised of Pex13, Pex14, and Pex17 in yeast, and PEX13 and PEX14 in mammals (Figures 1 and 2). Pex14 and the PTS receptors form the minimal translocon (part of the importomer complex) for transport of matrix proteins across the peroxisomal membrane [5,6]. Following receptor/cargo translocation across the peroxisome membrane, cargo is released in the peroxisome lumen. Next, the receptors (such as PEX5) or co-receptors (Pex20) are monoubiquitylated on Cys residues near the N-termini of these proteins, rather than on Lys residues, and recycled to the cytosol by an exportomer complex comprised of PEX1 and PEX6, and tethered to peroxisomes via PEX26 (in humans) [7,8]. Once in the cytosol, deubiquitylating (DUB) enzymes (Ubp15 in yeast and USP9X in mammals), remove the ubiquitin from Pex5 [9,10]. This deubiquitylation step is necessary for peroxisomal matrix protein import in Saccharomyces cerevisiae [11].
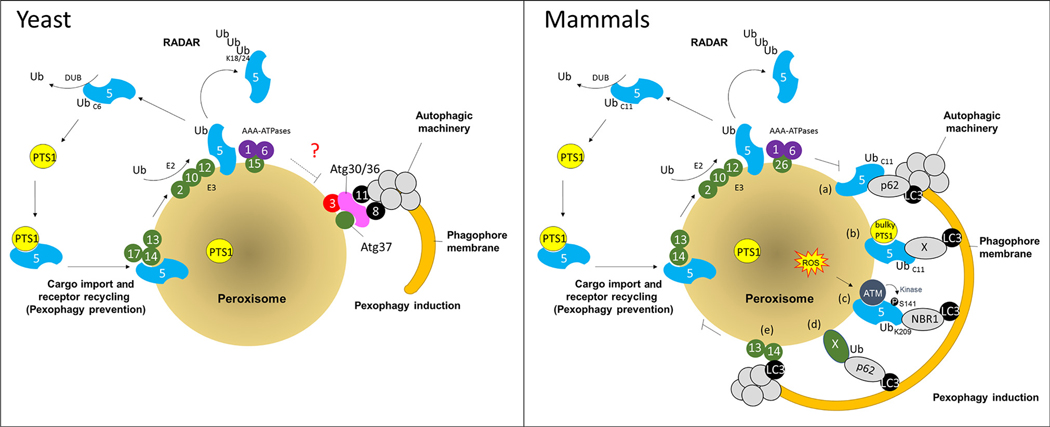
Figure 2. Role of Pex5 in Biogenesis and Pexophagy in Yeast and Mammalian Cells.
During peroxisome biogenesis in both yeast (left panel) and mammals (right panel) Pex5 recognizes the PTS1-containing cargoes, and transports them into the peroxisome matrix after interacting with the importomer proteins (conserved components are Pex13 and Pex14). After cargo release, Pex5 recycles back to the cytoplasm using exportomer components, Pex1 and Pex6, anchored at the peroxisome membrane through Pex15 in yeast and PEX26 in mammals. Receptor recycling requires Pex5 mono-ubiquitylation (at residue C6 in yeast and C11 in mammals), via an E2 enzyme (Pex4 in yeast and UbcH5 in mammals) and RING E3 ligases (Pex2, Pex10, and Pex12). If Pex5 recycling is blocked, then it can be polyubiquitylated and targeted for UPS-mediated turnover (RADAR pathway). Alternatively, when peroxisomes are damaged or redundant, pexophagy is activated. In yeast, a block in receptor recycling triggers pexophagy. It is still unclear (marked as ‘?’) how the yeast the AAA-ATPases regulate pexophagy. In mammals, either (a) the accumulation of mono-ubiquitylated PEX5 at the peroxisome membrane, or (b) the presence of a bulky PTS1 that cannot be released form PEX5, can activate pexophagy [24]. Alternatively (c), as a part of quality control, under conditions of oxidative stress, the ATM kinase phosphorylates PEX5 at S141, leading to its subsequent ubiquitylation by the RING E3 complex at K209, and activation of pexophagy [25]. Pexophagy can also be triggered in mammals in a PEX5-independent manner, either (d) by ubiquitination of PMPs (marked by ‘X’), such as PEX3 and PMP70, which are then recognized by autophagy adaptor proteins NBR1, and/or p62 [27], or (e) by direct binding of PEX14 to LC3 [26]. Abbreviations: ATM, ataxia-telangiectasia mutated; DUB, deubiquitylating enzyme; PTS1, peroxisomal targeting signal 1; RADAR, receptor accumulation and degradation in the absence of recycling; Ub, ubiquitin; UPS, ubiquitin-proteasome system; ROS, reactive oxygen species.
When PTS receptor recycling from the peroxisome membrane is either delayed or blocked, the PTS receptor is polyubiquitylated on Lys residues and extracted from the peroxisome membrane by unknown proteins distinct from two AAA ATPases (ATPases Associated with diverse cellular Activities), Pex1, and Pex6 involved in PTS receptor recycling. This is followed by degradation of the PTS receptor via the ubiquitin-proteasome system (UPS) and a process called RADAR (receptor accumulation and degradation in the absence of recycling) in yeast, but not so well-characterized in mammals (Figure 2) [12–14]. This constitutes a quality control system, to prevent potential traffic jams caused by blocked receptor recycling, from the peroxisome to the cytosol.
PMPs with their mPTSs can be targeted to the peroxisome membrane either directly from the cytosol, or indirectly via the endoplasmic reticulum (ER), using the proteins PEX3, PEX16, and PEX19 (or their yeast homologs) (Figure 1) [3].
Peroxisome inheritance, studied mostly in yeast, depends on the proteins, Inp1 and Inp2 [15,16] (Figure 1). Inp1 maintains peroxisomes in mother cells (retention) by interactions with the PMP, Pex3, at the cortical ER, whereas the cell cycle regulated protein, Inp2, is responsible for the movement of daughter peroxisomes (inheritance) from the mother to the newly-budded cells [15].
Peroxisome division and number are regulated by many proteins in yeast. However, the key conserved proteins involved in peroxisome division are yeast Pex11 or mammalian PEX11 (and its isoforms), and a conserved division machinery comprised of FIS1, MFF1, either dynamin 1 or a dynamin-like protein (DLP1) in yeast, or a dynamin-related protein (DRP1) in mammals (Figure 1), as well as glutathione-S-transferase, GDAP1 (Ganglioside induced differentiation associated protein 1), and a nucleoside diphosphate kinase, called DYNAMO1 (dynamin-based ring motive-force organizer 1). All these, except PEX11, are used both for peroxisomal and mitochondrial division [17,18].
Mitochondrial Rho GTPases, Miro1 and Miro2, are important for the microtubule-dependent motility of mitochondria, while also being responsible for the mitochondrial shape transition, thereby regulating mitophagy. However, recent reports show that these proteins are also associated with peroxisomes [19]. While their roles in peroxisome motility is unclear, they inhibit peroxisome fission by negatively regulating the recruitment of DRP1 to both peroxisomes and mitochondria, thereby inhibiting the common DRP1-dependent fission of these organelles, and promoting elongation of peroxisomes [20].
Peroxisome division can occur both during the growth and division of pre-existing peroxisomes, as well as during de novo biogenesis involving the budding of pre-peroxisomal vesicles from the ER (Figure 1) [2].
Brief Summary of Pexophagy
Three mechanisms exist for mammalian peroxisome degradation, including selective autophagy (pexophagy), proteolysis by peroxisomal Lon protease 2, and 15-lipoxygenase-1-mediated autolysis [21]. The role of pexophagy is in focus here [22], because 70–80% of peroxisomes are degraded by this mechanism [21].
Both general and selective autophagy have been reviewed elsewhere [23], therefore herein summarized, are only the key features of pexophagy, as a conserved form of selective autophagy that is involved in peroxisome homeostasis.
The core autophagy machinery, generally activated by starvation, is involved in the ‘self-digestion’ and recycling of unnecessary, damaged, or redundant cellular proteins and organelles in the lysosomes (or vacuoles in yeast) [23]. Cargoes destined for autophagy are captured, sequestered into double-membrane vesicular autophagosomes, and delivered to lysosomes for degradation and recycling. The process relies on autophagy-related (ATG) genes. Superimposed on the core autophagy genes that are required, common for most general and selective autophagy pathways, are selective autophagy genes. The latter encode selective autophagy receptors (SARs), regulators, scaffold proteins, and signaling components, that engage selective cargo on the one hand, with the core autophagic machinery on the other [22].
General autophagy involves five sequential steps [23]: (i) initiation; (ii) elongation; (iii) maturation; (iv) fusion; and (v) degradation. In steps (i)–(iii), cargoes are sequestered by a growing double membrane, called the phagophore membrane (Figures 1 and 2), that expands and fuses to form an autophagosome. In step (iv), the autophagosome fuses with lysosomes to form autolysosomes, followed by step (v), where the cargoes are degraded by lysosomal hydrolases and their constituent building blocks (amino acids, sugars, etc.) are recycled back to cytoplasm by lysosomal transporters.
Pexophagy requires SARs, such as Atg30 in Pichia pastoris, Atg36 in S. cerevisiae, as well as the autophagy adaptors, NBR1 and p62, in mammals (Figures 1 and 2). In mammalian cells, both PEX5-dependent [24,25] and PEX5-independent [26,27] pexophagy have been described, where the latter involves other proteins, such as PEX3, PMP70, and PEX14 (Figure 2). Additional requirements may include a SAR regulator, such as Atg37 in P. pastoris (whose mammalian counterpart is ACBD5, but its role in pexophagy is controversial [28,29]); scaffold proteins, such as Atg11 (FIP200 in mammals), and Atg17. Also required for pexophagy are signaling proteins, such as Hrr25 and other kinases in yeast, that phosphorylate and activate the SAR to engage the peroxisome cargo with the core autophagy machinery, such as Atg8 (mammalian counterparts are LC3 and GABARAP proteins), required for phagophore elongation, and Atg1 (mammalian ULK1) kinase, necessary for phagophore expansion [22,30–32].
A Balancing Act Between the Opposing Processes of Peroxisome Biogenesis and Pexophagy Maintains Homeostasis
Described next, is the extensive interdependency between the many components of the peroxisome biogenesis machinery, peroxisomal metabolites, and the proteins involved in pexophagy (Table 1). Knowledge of the intricate connections between these two opposing processes is critical for a comprehensive understanding of peroxisome homeostasis.
Table 1.
Cellular Components Influencing Both Peroxisome Biogenesis and Pexophagy.
| Proteins/Signaling moleculesa | Role in peroxisome biogenesisa | Role in peroxisome degradationa | Refs |
|---|---|---|---|
| I. Scaffolding and Signaling platforms | |||
| Yeast Pex3 | PMP import into peroxisome membranes in yeast and mammals, and intra-ER trafficking of Pex2 and Pex11C in P. pastoris | Required for the formation of the pexophagic RPC in P. pastoris and for phosphorylation of Atg30 that enables Atg11 and Atg8 recruitment | [28,32,33–35,37] |
| Mammalian PEX3 | Overexpression of PEX3 triggers pexophagy | [27,34,35] | |
| II. Translocon components | |||
| Mammalian PEX5 | PTS receptor | Target of ubiquitylation during pexophagy | [3,25] |
| P. pastoris Pex14 | Importomer component that recruits cargo-bound PTS receptors to the peroxisomal membrane and facilitates matrix protein import | Interacts with Atg30 and required for its peroxisomal localization | [3,37] |
| H. polymorpha Pex14 | Required for pexophagy | [3,39] | |
| Mammalian PEX14 | Binds directly to LC3-II and is implicated in pexophagy | [3,26] | |
| III. AAA ATPases | |||
| Yeast Pex1 and Pex6 | Import of peroxisome matrix proteins and mediates the recycling of Pex5 from the peroxisome membrane to the cytosol during matrix protein import in yeast and mammals. Also implicated in fusion of pre-peroxisomal vesicles that bud from yeast ER | Pex1 and Pex6 repress pexophagy in yeast and mammalian cells | [7,34,41–43,92] |
| Mammalian PEX1 and PEX6 | |||
| IV. Ubiquitylation/Deubiquitylation | |||
| Yeast Pex2 | Part of E3 ligase complex (with Pex10 and Pex12) required for PTS receptor ubiquitylation during matrix protein import in yeast and mammals | In H. polymorpha, it ubiquitylates Pex3 after its dissociation from peroxisomes for subsequent degradation by UPS, which is required for pexophagy | [12,92,93] |
| Mammalian PEX2 | PEX2 is the E3 ligase that ubiquitylates PEX5 and PMP70 to target peroxisomes for starvation-induced pexophagy | [12,92,45] | |
| Mammalian UPS30, USP9x | USP9x deubiquitinase removes Ub from recycled mono-ubiquitylated PEX5 to allow PEX5 participation in another round of import | USP30 localizes to peroxisomes and counteracts the effects of PEX2 (above) by deubiquitylating PEX5 to allow its recycling from the peroxisome membrane, thus inhibiting pexophagy | [46–48] |
| V. Cytoskeletal proteins | |||
| S. cerevisiae Myo2 and actin filaments | Peroxisome transport from mother to bud on actin cables using the Myo2 motor, via interaction with Inp2, Pex19, and Pex3 during cell division | Autophagosome mobilization towards vacuoles and the PAS require actin and myosin. | [15,17,62,63] |
| Mammalian Kinesin, Dynein and microtubules | Peroxisome transport towards cell periphery via Kinesin-1, KifC3, and towards the cell interior via dynein-dynactin complex | Autophagosome transport to the cell periphery and interior (lysosome) requires Kinesin and Dynein/dynactin motors, respectively; membrane vesicle delivery to omegasome requires Myosin II and actin. | [59,64–67,94] |
| VI. Peroxisomal metabolites | |||
| ROS/RNS | Presence of ROS reduces the ubiquitylation of Cys11 on PEX5 thus hindering PTS1-specific import | ROS accumulation leads to phosphorylation of PEX5, leading to its ubiquitylation, eventually causing activation of pexophagy | [50,53–55] |
Abbreviations: ER, endoplasmic reticulum; PAS, pre-autophagosomal structure; PMPs, peroxisomal membrane proteins; PTS, peroxisomal targeting signal; RNS, reactive nitrogen species; RPC, receptor protein complex; ROS, reactive oxygen species; Ub, ubiquitin.
Common Platforms for Peroxisome Homeostasis
Two peroxins, Pex3 and Pex14, central for the import of peroxisomal matrix proteins and PMPs, also play critical roles in pexophagy.
Pex3 is a common scaffold for peroxisome biogenesis, inheritance, and pexophagy, and is likely to play a role in coordinating all these processes directly or indirectly (Figure 1) [33]. Pex3, along with Pex19 (and PEX16 in mammals), is necessary for the post-translational import of PMPs into the peroxisome membrane [34], and along with Pex19 and Pex36 (a P. pastoris homolog of mammalian PEX16 and S. cerevisiae Pex34) is also required for the intra-ER trafficking of some P. pastoris RING-domain proteins (Pex2 and Pex11C) [34,35].
During cytokinesis in yeast, Inp1 binds Pex3 and bridges pools of Pex3 located at the peroxisomes and the cortical ER [36], thereby facilitating the retention of peroxisomes in the mother cell (Figure 1).
Pex3 is necessary for pexophagy, by being indispensable for the import and targeting of certain pexophagy components [28,33]. In P. pastoris, Atg30 and Atg37 are the pexophagy receptor and regulator, respectively, which together assemble the selective autophagy receptor protein complex (RPC) to initiate pexophagy [28]. Because Atg37 is a PMP, its targeting to the peroxisome membrane depends on Pex3 and Pex19. In contrast, Atg30 is only peroxisome associated, facilitated by its interactions with Pex3 and Pex14 [33,37].
Pex3 is also critical for the phosphorylation of Atg30, a step necessary for its activation during pexophagy [32,33]. Hypophosphorylated Atg30 cannot associate with its normal partners, Atg11 and Atg37 [32,33]. The proper localization of Pex3 on peroxisomes (but not the targeting of Pex3 itself to peroxisomes) depends partially on Atg37 and Atg30 [32]. However, neither Atg30 nor Atg37 is required for the targeting and import of PMPs and peroxisomal matrix proteins, or for peroxisome assembly [28,37].
PEX3 may also regulate mammalian pexophagy. Its overexpression induces pexophagy [27]. However, the expression of a PEX3 mutant with substitution of all lysine and cysteine residues by arginine and alanine, respectively, also induces peroxisome ubiquitylation and degradation, suggesting that another endogenous, unidentified peroxisomal protein, is ubiquitylated to activate pexophagy [27]. In contrast, studies in Hansenula polymorpha suggest that pexophagy requires the removal and proteasomal degradation of peroxisomal Pex3, before the completion of pexophagy [38].
In H. polymorpha, Pex14 is required critically, for both peroxisome biogenesis and pexophagy [39]. Mammalian PEX14 is also implicated in pexophagy and in CHO-KI cells it interacts directly with the microtubule-associated protein I light chain 3 (LC3), a homolog of yeast Atg8 [26] (Figure 2). Since both PEX5 and LC3 compete for PEX14, their competitive binding might modulate the balance between biogenesis and pexophagy in response to nutritional conditions, but this remains to be tested. Although, we still do not know if Pex14 has a similar ability to bind Atg8 in yeast, its requirement for Atg30 recruitment to peroxisomes in P. pastoris implicates Pex14 in pexophagy as well.
Regulation of Pexophagy by Other Peroxisome Biogenesis Proteins
Other peroxins regulating pexophagy include Pex1 and Pex6, which alternate to form a hetero-hexameric double ring, and play a role in the recycling of Pex5 from peroxisomes back to the cytosol, after each round of matrix protein import (Figure 2) [40]. In support of this role, both proteins in the mammalian PEX1/PEX6 complex bind mono-ubiquitylated PEX5 in the peroxisome membrane, and the hydrolysis of ATP then drives the unfolding and extraction of mono-ubiquitylated PEX5 from the peroxisome membrane during PEX5 recycling to the cytosol [7]. This recognition of ubiquitylated Pex5 by the AAA ATPases also extends to yeast [41]. In addition to this function, Pex1, Pex6, and Pex15 in yeast, repress pexophagy [42], as do their mammalian counterparts PEX1, PEX6, and PEX26 (a homolog of yeast Pex15) [43].
Both Peroxisome Biogenesis and Pexophagy are Regulated by Ubiquitylation/Deubiquitylation
In PEX5-dependent mammalian pexophagy, peroxisomes are targeted by ubiquitylation of PEX5 either at K209 or at C11, depending on the cell type or signals (Figure 2) [44]. The E3 ligase that ubiquitylates PEX5 (K209) is PEX2 [45]. Consequently, overexpression of PEX2 induces pexophagy in an NBR1-dependent manner. PEX2, and its interacting RING-domain proteins, PEX10 and PEX12, are also E3 ligases for peroxisome biogenesis [12,13].
The mammalian DUB USP30, reverses PEX5 ubiquitylation and inhibits pexophagy [46,47], whereas deubiquitylation (by USP9X in mammalian cells) is necessary for PEX5 recycling during peroxisome biogenesis (Figure 2) [48].
Peroxisomal Metabolites Regulate Pexophagy
Peroxisomal metabolism requires proper biogenesis of PMPs, and peroxisomal matrix proteins, that perform metabolic functions. Peroxisomal enzymes catalyze oxidative reactions that use oxygen (O2) to produce hydrogen peroxide (H2O2), which is then degraded by peroxisomal catalase. Additionally, peroxisomal metabolic pathways also produce reactive oxygen species (ROS)/reactive nitrogen species (RNS), and peroxisomes also have antioxidant proteins, such as catalase, peroxiredoxins, superoxide dismutase, glutathione-S-transferases, and epoxide hydrolases [49]. Peroxisomes in rat liver may be responsible for as much as 20% of the O2 consumption, and 35% of the H2O2 production [49].
In mammalian cells, ROS triggers pexophagy [50], as does hypoxia, which activates pexophagy by inducing the hypoxia-inducible factor, HIF-2α [51,52]. Peroxisomally-generated ROS was shown to activate ataxia-telangiectasia mutated (ATM) kinase, which translocates to peroxisome membranes in a PEX5-dependent manner, and phosphorylates peroxisome membrane-associated PEX5 at Ser141 (Figure 2). This phosphorylation is required for a subsequent monoubiquitylation at K209, which is then recognized by the pexophagy machinery to cause peroxisome turnover [25]. In view of this result, it is not surprising that the inhibition of catalase, which degrades peroxisomal H2O2, causes ROS accumulation and induces pexophagy [53].
Common Intracellular Signals That Impact Peroxisome Biogenesis and Pexophagy
Interestingly, the ROS generated by peroxisomes not only induces pexophagy, but also impairs peroxisome biogenesis. This is exemplified by the redox regulation of the activity of Pex5, which, as explained earlier, has a Cys near the N-terminus that is necessary for receptor recycling to the cytosol [48], but this residue is also sensitive to redox regulation, so that in the presence of ROS, PEX5 function is dampened and peroxisome biogenesis is inhibited [54,55]. Since ROS activates pexophagy in mammalian cells, it would make physiological sense to inhibit peroxisome biogenesis under such conditions.
Pexophagy Depends on the Peroxisome Division Machinery Necessary for Biogenesis
In S. cerevisiae, peroxisome division is necessary for pexophagy (Figure 1) [56]. During peroxisome proliferation in yeast, peroxisomes grow fairly large, which influences the specific proteins required for pexophagy [57]. Thus, in P. pastoris, where the average peroxisome size is even larger than that in S. cerevisiae, the requirement for pexophagy proteins, such as Atg26 (and Atg11), is higher. These two proteins are required for the formation of a membranous structure called the micropexophagic membrane apparatus (MIPA), which is necessary for the engulfment of large peroxisomes [58]. In S. cerevisiae, although the ATG26 gene is conserved, it is not required for pexophagy, possibly because the specific role of Atg26 in engulfing large peroxisomes is not conserved. Instead, in S. cerevisiae, peroxisomes must divide to become smaller before they can be captured into pexophagosomes [56]. However, the generality of this mechanism is unclear because in P. pastoris, there is no impairment in pexophagy in peroxisome division mutants, such as pex11 cells.
Some insight into how pexophagy components and peroxisome division influence each other, comes from the finding that in S. cerevisiae, both the pexophagy receptor Atg36, and Atg11 the scaffold protein for selective autophagy, interact with two dynamin-like proteins, Dnm1 and Vps1, which play roles in peroxisome division (Figure 1) [17].
Common Intracellular Movement Highways Used by Peroxisomes and Autophagosomes
In yeast, peroxisome movement is dependent on the actin-myosin system [17], whereas in mammalian cells, it is kinesin/dynein and microtubule dependent (Figure 3) [59].
Figure 3. Peroxisome and Pexophagosome Movement Along the Cytoskeleton in Yeast and Mammals.
In yeast (left panel), newly-divided peroxisomes containing Inp2, interacting with Pex19 (which is bound to Pex3), engage the Myo2 motor on actin filaments to move peroxisomes to the bud. During pexophagy, the phagophore membrane expands by the delivery of vesicles to the PAS (site of initiation of pexophagy) using the actin cytoskeleton. The resulting pexophagosome moves to the vacuole along the actin cytoskeleton using unknown receptors on the pexophagosome. In mammals (right panel), peroxisomes move using kinesin or dynein motors, in the anterograde (cell periphery) and retrograde (cell interior) directions, respectively. These motors engage with microtubules. Phagophore formation relies on the delivery of membrane vesicles from different sources to the omegasome (site of initiation of pexophagy), using the Myosin II motor moving along the actin cytoskeleton network. Pexophagy uses PEX5-dependent or -independent SARs on peroxisomes to engage selective autophagy adaptors (NBR1 or p62). The pexophagosome resulting from peroxisome engulfment by the phagophore membrane also uses kinesin and dynein motors moving along microtubules. Abbreviations: PAS, pre-autophagosomal structure; PMPs, peroxisomal membrane proteins; SARs, selective autophagy receptors.
In yeast, the myosin, Myo2, interacts with Inp2, Pex19, and Pex3, and drives peroxisome movement on the actin cytoskeleton [17], during peroxisome inheritance from mother to daughter cell following cytokinesis (Figure 3) [15].
Drosophila [60] and mammalian [59] peroxisomes are evenly distributed in the cytoplasm of most cells, however in certain cell types, such as neurons and kidney proximal tubule cells, peroxisomes are unevenly distributed. Peroxisome distribution is achieved by the association of peroxisomes with microtubules, and their bidirectional transport by microtubule motor proteins, kinesin-1 (Kif5 and KifC3), and cytoplasmic dynein/dynactin (Figure 3). KifC3 co-immunoprecipitates with PEX1, and interacts with it in a yeast two-hybrid screen. RNAi knockdown of KifC3, results in an increase of cells with perinuclear-clustered peroxisomes, indicating enhanced minus-end directed motility of peroxisomes. KifC3 may play a regulatory role by competing with, and thereby reducing, the motor function of dynein, which controls minus-end directed peroxisomal transport [59]. In human cells, PEX14 is also involved in microtubule binding and microtubule-based peroxisome motility (Figure 3) [61].
Pexophagy also depends on similar motors and cytoskeletal elements. For example, in yeast, autophagosome formation around peroxisomes and the movement of the autophagosomes to the vacuole depend on actin [62], and only by inference, myosin [63]. In mammalian cells, the retrograde movement of autophagosomes is dependent on microtubules and dynein/dynactin for retrograde motility [64–66], whereas the anterograde movement is dependent on kinesin motors [67].
Therefore, the same cytoskeletal and motor proteins are involved in both peroxisome biogenesis (movement and inheritance), as well as autophagosome and membrane vesicle movement during pexophagy.
Tilting the Homeostatic Balance of Peroxisomes in Disease States
Peroxisomes as Signaling Centers
Peroxisomes, in addition to mitochondria, serve as intracellular signaling centers during viral infections and contribute to innate immunity [68]. MAVS, a protein involved in mitochondrial anti-viral signaling (MAVS) is located both on mitochondria and peroxisomes. Upon viral infection, certain cytosolic proteins detect the presence of the virus and bind to, and activate, MAVS. This causes the virally-infected cell to secrete cytokines, which activate a cellular immune response that, in addition to other effects, kills virus-infected cells, resulting in viral clearance. Not surprisingly, as part of the evolutionary arms race between host immunity and viruses, the latter have found evasion mechanisms that compromise peroxisome homeostasis.
MAVS is a tail-anchored protein (Figure 1) that targets to both peroxisomes and mitochondria in human and mouse cells, like other such proteins (e.g., FIS1 and MFF1, which are involved in the division of both organelles). A major site of MAVS signaling is the mitochondria-associated membrane (MAM) of the ER. However, MAVS-dependent signaling emanates from both peroxisomes and mitochondria, with the initial interferon (IFN)-independent response being coordinated from the peroxisomal MAVS, followed later and synergistically, by an IFN-dependent signaling from mitochondrial MAVS [68,69]. However, another study revealed differences in the peroxisomal and mitochondrial MAVS signaling routes [70], suggesting that these details need to be studied further. Retinoic acid-inducible gene-I (RIG-I)-like receptors (RLRs), involved in innate immunity, respond to RNAs from many viruses such as human immunodeficiency virus (HIV), hepatitis C, dengue (DENV), and Zika [70–72], as well as DNA from human cytomegalovirus (HCMV) and herpes simplex virus 1 (HSV 1) [73,74]. Upon reovirus infection, activation of peroxisomal MAVS causes peroxisomes to tubulate and aggregate [68]. During RNA virus infection, RIG-I is recruited to the MAM to bind MAVS protein. Dynamic MAM tethering to mitochondria and peroxisomes then coordinates MAVS localization to form a signaling synapse between membranes [75].
Viral Mechanisms to Subvert MAVS Signaling
The NS3–4A protease of hepatitis C virus cleaves MAVS protein localized on peroxisomes and mitochondria [70], whereas the nsp1 protein of porcine diarrhea virus reduces type III IFN induction, in part by reducing peroxisome pools via an unknown mechanism [76].
HCMV hijacks PEX19 to transport its own protein, vMIA (viral mitochondria-localized inhibitor of apoptosis), to the peroxisome membrane in HepG2 and human foreskin fibroblast cells, where it inhibits peroxisomal MAVS [73].
Viral Strategies to Influence Peroxisome Homeostasis
Viruses use two independent strategies to influence this homeostasis, culminating either in the reduction or increase in the peroxisome number. For example, the Vpu protein of HIV-1 downregulates peroxisome biogenesis to evade signaling from the peroxisomal MAVS [77]. It achieves this by inducing the expression of four microRNAs that target mRNAs encoding peroxins (PEX2, PEX7, PEX11, and PEX13). Several other viruses target the peroxisome biogenesis factor, PEX19, and these are seen during infection of cells with West Nile virus (WNV) and DENV, wherein PEX19 is selectively degraded [78]. The WNV and DENV capsid proteins target PEX19 and cause reduced levels of peroxisomes, and a suppressed type III IFN response. Zika virus infection also causes PEX19 loss, but this can be countered by overexpression of PEX11Β, which causes peroxisome proliferation [72].
Plant tombusviruses, such as cucumber necrosis virus, replicate on peroxisomes, so they induce peroxisome formation in multivesicular bodies [79]. Infection of cells with HCMV and HSV1, whose replication requires peroxisomes [80], also induces the peroxisomal proteome and peroxisome biogenesis by growth and division.
In addition to directly interfering with the RIG-I signaling pathway, the reduction of peroxisomal MAVS signaling could be achieved either by impairing biogenesis, or by inducing pexophagy.
While no reports exist currently regarding this second strategy, there is precedent that a viral protein, BHRF1, an Epstein-Barr virus (EBV) homolog of cellular BCL2, activates mitophagy by stimulating the mitochondrial division machinery that is also used for peroxisome division [81]. However, because general autophagy, whose machinery is shared by selective autophagy pathways, acts as part of the innate immunity pathway against viruses, and is also involved in the presentation of viral antigens for the cellular adaptive immunity responses, there could be an evolutionary selection against viral mechanisms that activate pexophagy. Only further research will reveal whether this is true or not.
Relevance of Peroxisome Homeostasis and Its Manipulation to Human Diseases
Mutations in a dozen PEX genes are responsible for the PBDs. While homozygous loss of function phenotypes caused by mutations in these genes can be severe, including premature death of the patient, there are many cases with milder phenotypes that have been attributed to missense, temperature-sensitive mutations, in several PEX genes, including PEX1, PEX2, PEX6, PEX13, and PEX26 [82,83]. In fact, all Infantile Refsum disease cell lines, and some neonatal adrenoleukodystrophy lines belonging to several different complementation groups, are temperature sensitive for peroxisome biogenesis [82]. The reason for the milder phenotypes is that these proteins have some residual function, such that their required levels of activity could, in principle, be restored sufficiently either by boosting peroxisome biogenesis or by reducing pexophagy, using either small molecules that help stabilize misfolded mutant proteins [84], or modulate these processes [85].
Mutations in many core autophagy genes, as well in the gene encoding p62, are associated with a direct or indirect impairment of selective autophagy [86]. In this context, small molecules, such as 1,10-phenanthroline and dimethyloxalylglycine, do induce pexophagy in cell lines [52,87], but have not been tested for their therapeutic potential in disease states.
Conversely, excessive pexophagy could also be associated with Parkinson’s disease [88] and certain cancers [51], but a causative link remains to be established. The study of over 200 clear cell renal cell carcinoma (ccRCC) tissues revealed that peroxisome abundance is reduced in VHL-deficient ccRCC characterized by high HIF-2α levels, suggesting that HIF-2α-mediated pexophagy is relevant to human disease. Small molecule antagonists have been described for HIF-2α [89], but these have not been tested for their ability to inhibit baseline pexophagy.
Recently, microRNAs have also been found to regulate both peroxisome biogenesis [77] and pexophagy [90], and could offer new tools for shifting the balance between the biogenesis and degradative arms for peroxisomes.
Concluding Remarks
The use of common molecular platforms, components, metabolites, division proteins, post-translational modifications, and cytoskeletal machinery for peroxisome biogenesis and pexophagy, highlights the intricate connections that must exist to coordinate peroxisome homeostasis. Research on these connections is necessary, for deeper knowledge and mechanistic insights into the regulatory mechanisms involved, and related areas that are ripe for the next generation of scientists to delve into (see Outstanding Questions).
Outstanding Questions.
Recognizing that peroxisome biogenesis and pexophagy have to be maintained in the right balance in cells and tissues, what are the molecular mechanisms that alter this balance?
What are the signaling events that determine whether common molecular platforms used by peroxisome biogenesis and pexophagy, are used for one or the other process?
If peroxisome division is necessary for pexophagy generally, then how do the pexophagy signals activate peroxisome division?
How do the AAA ATPases, Pex1 and Pex6, which are required for peroxisome matrix protein import, also repress pexophagy?
How is the activity of the peroxisomal RING E3 ligase complex regulated to facilitate peroxisome biogenesis or turnover?
Which metabolites generated by peroxisomes, regulate peroxisome biogenesis and pexophagy, and what are the mechanisms?
Given that peroxisomes and pexophagosomes use the same cytoskeletal elements for their intracellular transport, what factors control these movements without creating a traffic jam?
How do viral proteins downregulate peroxisome biogenesis and innate immunity, and are these used physiologically in uninfected cells?
What small molecules modulate peroxisome homeostasis, and are these therapeutically useful for human PBDs?
What diseases are associated with defective or excessive pexophagy?
From a disease standpoint, mutations in over a dozen PEX genes causing PBDs and cellular microRNAs, have been shown to affect peroxisome homeostasis. Additionally, because peroxisomes are intracellular platforms for anti-viral signaling during innate immunity, and several enveloped viruses require peroxisomal lipids [91] or membranes [80], many insights into the regulation of peroxisome homeostasis are emerging from virology. These early studies are likely to shed more light, not only into mechanisms by which viruses perturb peroxisome homeostasis, but also into cellular mechanisms that regulate them. Ultimately, this knowledge will become useful for therapeutic interventions that could be used to modulate peroxisome homeostasis in disease states.
Highlights.
Emerging studies on the integration and coordination of the opposing processes of peroxisome biogenesis and pexophagy present new vistas for research.
Physiologically, the processes of organelle biogenesis and turnover must be interconnected and regulated. For example, some peroxisomal metabolites could act as crucial signaling cues to inhibit biogenesis and trigger pexophagy to achieve a coordinated response to a changing cellular state. Moreover, signals that induce peroxisome biogenesis must keep pexophagy in check and vice versa.
Multiple common molecular platforms, representing nodes for regulation, are used to balance peroxisome biogenesis and pexophagy.
Because peroxisomes are essential in humans and there are disease states associated with peroxisomes and their functions, an understanding of the two arms of homeostasis may help in therapeutic interventions.
Studies of peroxisome homeostasis will also shed light on principles involved in the homeostasis of other organelles.
Acknowledgements
This work was supported by an NIH grant (DK41737) to S.S., who holds a Tata Chancellor’s Endowed Professorship in Molecular Biology.
References
- 1.Argyriou C. et al. (2016) Peroxisome biogenesis disorders. Transl. Sci. Rare Dis. 1, 111–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Agrawal G and Subramani S. (2016) De novo peroxisome biogenesis: evolving concepts and conundrums. Biochim. Biophys. Acta 1863, 892–901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Farre JC et al. (2019) Peroxisome biogenesis, membrane contact sites, and quality control. EMBO Rep. 20, e46864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hettema EH et al. (2014) Evolving models for peroxisome biogenesis. Curr. Opin. Cell Biol. 29, 25–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Meinecke M et al. (2010) The peroxisomal importomer constitutes a large and highly dynamic pore. Nat. Cell Biol. 12, 273–277 [DOI] [PubMed] [Google Scholar]
- 6.Ma C et al. (2009) The peroxisomal matrix import of Pex8p requires only PTS receptors and Pex14p. Mol. Biol. Cell 20, 3680–3689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pedrosa AG et al. (2018) Peroxisomal monoubiquitinated PEX5 interacts with the AAA ATPases PEX1 and PEX6 and is unfolded during its dislocation into the cytosol. J. Biol. Chem. 293, 11553–11563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grimm I et al. (2016) Role of AAA(+)-proteins in peroxisome biogenesis and function. Biochim. Biophys. Acta 1863, 828–837 [DOI] [PubMed] [Google Scholar]
- 9.Debelyy MO et al. (2011) Ubp15p, a ubiquitin hydrolase associated with the peroxisomal export machinery. J. Biol. Chem. 286, 28223–28234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grou CP et al. (2012) Identification of ubiquitin-specific protease 9X (USP9X) as a deubiquitinase acting on ubiquitin-peroxin 5 (PEX5) thioester conjugate. J. Biol. Chem. 287, 12815–12827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.El Magraoui F et al. (2019) The deubiquitination of the PTS1-import receptor Pex5p is required for peroxisomal matrix protein import. Biochim. Biophys. Acta, Mol. Cell Res. 1866, 199–213 [DOI] [PubMed] [Google Scholar]
- 12.Liu X and Subramani S. (2013) Unique requirements for monoand polyubiquitination of the peroxisomal targeting signal coreceptor, Pex20. J. Biol. Chem. 288, 7230–7240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Platta HW et al. (2007) Ubiquitination of the peroxisomal import receptor Pex5p is required for its recycling. J. Cell Biol. 177, 197–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leon S et al. (2006) Dynamics of the peroxisomal import cycle of PpPex20p: ubiquitin-dependent localization and regulation. J. Cell Biol. 172, 67–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Knoblach B and Rachubinski RA (2016) How peroxisomes partition between cells. A story of yeast, mammals and filamentous fungi. Curr. Opin. Cell Biol. 41, 73–80 [DOI] [PubMed] [Google Scholar]
- 16.Purnell BA (2017) Peroxisome inheritance and differentiation. Science 355, 490–492 [DOI] [PubMed] [Google Scholar]
- 17.Kuravi K et al. (2006) Dynamin-related proteins Vps1p and Dnm1p control peroxisome abundance in Saccharomyces cerevisiae. J. Cell Sci. 119, 3994–4001 [DOI] [PubMed] [Google Scholar]
- 18.Honsho M et al. (2016) Peroxisome homeostasis: mechanisms of division and selective degradation of peroxisomes in mammals. Biochim. Biophys. Acta 1863, 984–991 [DOI] [PubMed] [Google Scholar]
- 19.Castro IG et al. (2018) A role for mitochondrial Rho GTPase 1 (MIRO1) in motility and membrane dynamics of peroxisomes. Traffic 19, 229–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Covill-Cooke C et al. (2020) Peroxisomal fission is modulated by the mitochondrial Rho-GTPases, Miro1, and Miro2. EMBO Rep. 21, e49865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Till A et al. (2012) Pexophagy: the selective degradation of peroxisomes. Int. J. Cell Biol. 2012, 512721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Germain K and Kim PK (2020) Pexophagy: a model for selective autophagy. Int. J. Mol. Sci. 21, 578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yin Z et al. (2016) Autophagy: machinery and regulation. Microb. Cell 3, 588–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nordgren M et al. (2015) Export-deficient monoubiquitinated PEX5 triggers peroxisome removal in SV40 large T antigen-transformed mouse embryonic fibroblasts. Autophagy 11, 1326–1340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang J et al. (2015) ATM functions at the peroxisome to induce pexophagy in response to ROS. Nat. Cell Biol. 17, 1259–1269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jiang L et al. (2015) Peroxin Pex14p is the key component for coordinated autophagic degradation of mammalian peroxisomes by direct binding to LC3-II. Genes Cells 20, 36–49 [DOI] [PubMed] [Google Scholar]
- 27.Yamashita S et al. (2014) The membrane peroxin PEX3 induces peroxisome-ubiquitination-linked pexophagy. Autophagy 10, 1549–1564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nazarko TY et al. (2014) Peroxisomal Atg37 binds Atg30 or palmitoyl-CoA to regulate phagophore formation during pexophagy. J. Cell Biol. 204, 541–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ferdinandusse S et al. (2017) ACBD5 deficiency causes a defect in peroxisomal very long-chain fatty acid metabolism. J. Med. Genet. 54, 330–337 [DOI] [PubMed] [Google Scholar]
- 30.Farre JC et al. (2013) Phosphorylation of mitophagy and pexophagy receptors coordinates their interaction with Atg8 and Atg11. EMBO Rep. 14, 441–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nakatogawa H. (2015) Hrr25: an emerging major player in selective autophagy regulation in Saccharomyces cerevisiae. Autophagy 11, 432–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zientara-Rytter K et al. (2018) Pex3 and Atg37 compete to regulate the interaction between the pexophagy receptor, Atg30, and the Hrr25 kinase. Autophagy 14, 368–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Burnett SF et al. (2015) Peroxisomal Pex3 activates selective autophagy of peroxisomes via interaction with the pexophagy receptor Atg30. J. Biol. Chem. 290, 8623–8631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Agrawal G et al. (2016) Distinct requirements for intra-ER sorting and budding of peroxisomal membrane proteins from the ER. J. Cell Biol. 212, 335–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Farre JC et al. (2017) A new yeast peroxin, Pex36, a functional homolog of mammalian PEX16, functions in the ER-to-peroxisome traffic of peroxisomal membrane proteins. J. Mol. Biol. 429, 3743–3762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Knoblach B et al. (2013) An ER-peroxisome tether exerts peroxisome population control in yeast. EMBO J. 32, 2439–2453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Farre JC et al. (2008) PpAtg30 tags peroxisomes for turnover by selective autophagy. Dev. Cell 14, 365–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bellu AR et al. (2002) Removal of Pex3p is an important initial stage in selective peroxisome degradation in Hansenula polymorpha. J. Biol. Chem. 277, 42875–42880 [DOI] [PubMed] [Google Scholar]
- 39.van Zutphen T et al. (2008) Pex14 is the sole component of the peroxisomal translocon that is required for pexophagy. Autophagy 4, 63–66 [DOI] [PubMed] [Google Scholar]
- 40.Blok NB et al. (2015) Unique double-ring structure of the peroxisomal Pex1/Pex6 ATPase complex revealed by cryo-electron microscopy. Proc. Natl. Acad. Sci. U. S. A. 112, E4017–E4025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schwerter D et al. (2018) Receptor recognition by the peroxisomal AAA complex depends on the presence of the ubiquitin moiety and is mediated by Pex1p. J. Biol. Chem. 293, 15458–15470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nuttall JM et al. (2014) Deficiency of the exportomer components Pex1, Pex6, and Pex15 causes enhanced pexophagy in Saccharomyces cerevisiae. Autophagy 10, 835–845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Law KB et al. (2017) The peroxisomal AAA ATPase complex prevents pexophagy and development of peroxisome biogenesis disorders. Autophagy 13, 868–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Subramani S. (2015) A mammalian pexophagy target. Nat. Cell Biol. 17, 1371–1373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sargent G et al. (2016) PEX2 is the E3 ubiquitin ligase required for pexophagy during starvation. J. Cell Biol. 214, 677–690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Riccio V et al. (2019) Deubiquitinating enzyme USP30 maintains basal peroxisome abundance by regulating pexophagy. J. Cell Biol. 218, 798–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Marcassa E et al. (2018) Dual role of USP30 in controlling basal pexophagy and mitophagy. EMBO Rep. 19, e45595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang W and Subramani S. (2017) Role of PEX5 ubiquitination in maintaining peroxisome dynamics and homeostasis. Cell Cycle 16, 2037–2045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fransen M et al. (2012) Role of peroxisomes in ROS/RNS-metabolism: implications for human disease. Biochim. Biophys. Acta 1822, 1363–1373 [DOI] [PubMed] [Google Scholar]
- 50.Tripathi DN and Walker CL (2016) The peroxisome as a cell signaling organelle. Curr. Opin. Cell Biol. 39, 109–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Walter KM et al. (2014) Hif-2α promotes degradation of mammalian peroxisomes by selective autophagy. Cell Metab. 20, 882–897 [DOI] [PubMed] [Google Scholar]
- 52.Mu Y et al. (2020) Dimethyloxaloylglycine induces pexophagy in a HIF-2α dependent manner involving autophagy receptor p62. Biochem. Biophys. Res. Commun. Published online February 16, 2020 10.1016/j.bbrc.2020.02.051 [DOI] [PubMed] [Google Scholar]
- 53.Lee JN et al. (2018) Catalase inhibition induces pexophagy through ROS accumulation. Biochem. Biophys. Res. Commun. 501, 696–702 [DOI] [PubMed] [Google Scholar]
- 54.Ma C et al. (2013) Redox-regulated cargo binding and release by the peroxisomal targeting signal receptor, Pex5. J. Biol. Chem. 288, 27220–27231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Apanasets O et al. (2014) PEX5, the shuttling import receptor for peroxisomal matrix proteins, is a redox-sensitive protein. Traffic 15, 94–103 [DOI] [PubMed] [Google Scholar]
- 56.Mao K et al. (2014) The progression of peroxisomal degradation through autophagy requires peroxisomal division. Autophagy 10, 652–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nazarko TY et al. (2009) Peroxisome size provides insights into the function of autophagy-related proteins. Mol. Biol. Cell 20, 3828–3839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yamashita S et al. (2006) PI4P-signaling pathway for the synthesis of a nascent membrane structure in selective autophagy. J. Cell Biol. 173, 709–717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dietrich D et al. (2013) Identification of the kinesin KifC3 as a new player for positioning of peroxisomes and other organelles in mammalian cells. Biochim. Biophys. Acta 1833, 3013–3024 [DOI] [PubMed] [Google Scholar]
- 60.Kural C et al. (2005) Kinesin and dynein move a peroxisome in vivo: a tug-of-war or coordinated movement? Science 308, 1469–1472 [DOI] [PubMed] [Google Scholar]
- 61.Bharti P et al. (2011) PEX14 is required for microtubule-based peroxisome motility in human cells. J. Cell Sci. 124, 1759–1768 [DOI] [PubMed] [Google Scholar]
- 62.Reggiori F et al. (2005) The actin cytoskeleton is required for selective types of autophagy, but not nonspecific autophagy, in the yeast Saccharomyces cerevisiae. Mol. Biol. Cell 16, 5843–5856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Monastyrska I et al. (2006) Atg11 directs autophagosome cargoes to the PAS along actin cables. Autophagy 2, 119–121 [DOI] [PubMed] [Google Scholar]
- 64.Yang Y et al. (2011) Microtubule and kinesin/dynein-dependent, bi-directional transport of autolysosomes in neurites of PC12 cells. Int. J. Biochem. Cell Biol. 43, 1147–1156 [DOI] [PubMed] [Google Scholar]
- 65.Katsumata K et al. (2010) Dynein- and activity-dependent retrograde transport of autophagosomes in neuronal axons. Autophagy 6, 378–385 [DOI] [PubMed] [Google Scholar]
- 66.Kimura S et al. (2008) Dynein-dependent movement of autophagosomes mediates efficient encounters with lysosomes. Cell Struct. Funct. 33, 109–122 [DOI] [PubMed] [Google Scholar]
- 67.Maday S et al. (2012) Autophagosomes initiate distally and mature during transport toward the cell soma in primary neurons. J. Cell Biol. 196, 407–417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dixit E et al. (2010) Peroxisomes are signaling platforms for antiviral innate immunity. Cell 141, 668–681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Odendall C et al. (2014) Diverse intracellular pathogens activate type III interferon expression from peroxisomes. Nat. Immunol. 15, 717–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bender S et al. (2015) Activation of type I and III interferon response by mitochondrial and peroxisomal MAVS and inhibition by Hepatitis C virus. PLoS Pathog. 11, e1005264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Berg RK et al. (2012) Genomic HIV RNA induces innate immune responses through RIG-I-dependent sensing of secondary-structured RNA. PLoS One 7, e29291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wong CP et al. (2019) Interplay between Zika virus and peroxisomes during infection. Cells 8, 725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Magalhaes AC et al. (2016) Peroxisomes are platforms for cytomegalovirus’ evasion from the cellular immune response. Sci. Rep. 6, 26028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zheng C and Su C. (2017) Herpes simplex virus 1 infection dampens the immediate early antiviral innate immunity signaling from peroxisomes by tegument protein VP16. Virol. J. 14, 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Horner SM et al. (2011) Mitochondrial-associated endoplasmic reticulum membranes (MAM) form innate immune synapses and are targeted by hepatitis C virus. Proc. Natl. Acad. Sci. U. S. A. 108, 14590–14595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhang Q et al. (2018) Type III interferon restriction by porcine epidemic diarrhea virus and the role of viral protein nsp1 in IRF1 signaling. J. Virol. 92, e01677–e01717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Xu Z et al. (2020) The HIV-1 accessory protein Vpu downregulates peroxisome biogenesis. mBio 11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.You J et al. (2015) Flavivirus infection impairs peroxisome biogenesis and early antiviral signaling. J. Virol. 89, 12349–12361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Rochon D et al. (2014) The p33 auxiliary replicase protein of Cucumber necrosis virus targets peroxisomes and infection induces de novo peroxisome formation from the endoplasmic reticulum. Virology 452–453, 133–142 [DOI] [PubMed] [Google Scholar]
- 80.Jean Beltran PM et al. (2018) Infection-induced peroxisome biogenesis is a metabolic strategy for herpesvirus replication. Cell Host Microbe 24, 526–541.e7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Vilmen G et al. (2020) BHRF1, a BCL2 viral homolog, disturbs mitochondrial dynamics and stimulates mitophagy to dampen type I IFN induction. Autophagy Published online May 13, 2020. 10.1080/15548627.2020.1758416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hashimoto K et al. (2005) Molecular mechanism of a temperature-sensitive phenotype in peroxisomal biogenesis disorder. Pediatr. Res. 58, 263–269 [DOI] [PubMed] [Google Scholar]
- 83.Tanaka AJ et al. (2019) A newly identified mutation in the PEX26 gene is associated with a milder form of Zellweger spectrum disorder. Cold Spring Harb. Mol. Case Stud. 5, a003483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhang R et al. (2010) Recovery of PEX1-Gly843Asp peroxisome dysfunction by small-molecule compounds. Proc. Natl. Acad. Sci. U. S. A. 107, 5569–5574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Weng H et al. (2015) Induction of peroxisomes by butyrateproducing probiotics. PLoS One 10, e0117851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Stamatakou E et al. (2020) Mendelian neurodegenerative disease genes involved in autophagy. Cell Discov. 6, 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Jo DS et al. (2015) Pexophagy is induced by increasing peroxisomal reactive oxygen species in 1’10-phenanthroline-treated cells. Biochem. Biophys. Res. Commun. 467, 354–360 [DOI] [PubMed] [Google Scholar]
- 88.Jo DS et al. (2020) Loss of HSPA9 induces peroxisomal degradation by increasing pexophagy. Autophagy Published online January 22, 2020. 10.1080/15548627.2020.1712812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wallace EM et al. (2016) A small-molecule antagonist of HIF2α is efficacious in preclinical models of renal cell carcinoma. Cancer Res. 76, 5491–5500 [DOI] [PubMed] [Google Scholar]
- 90.Houri K et al. (2020) miR-142 induces accumulation of reactive oxygen species (ROS) by inhibiting pexophagy in aged bone marrow mesenchymal stem cells. Sci. Rep. 10, 3735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Tanner LB et al. (2014) Lipidomics identifies a requirement for peroxisomal function during influenza virus replication. J. Lipid Res. 55, 1357–1365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Platta HW et al. (2016) Regulation of peroxisomal matrix protein import by ubiquitination. Biochim. Biophys. Acta 1863, 838–849 [DOI] [PubMed] [Google Scholar]
- 93.Williams C and van der Klei IJ (2013) Pexophagy-linked degradation of the peroxisomal membrane protein Pex3p involves the ubiquitin-proteasome system. Biochem. Biophys. Res. Commun. 438, 395–401 [DOI] [PubMed] [Google Scholar]
- 94.Tang HW et al. (2011) Atg1-mediated myosin II activation regulates autophagosome formation during starvation-induced autophagy. EMBO J. 30, 636–651 [DOI] [PMC free article] [PubMed] [Google Scholar]