Abstract
Expression of the andrgogen receptor splice variant 7 (AR-V7) is frequently detected in castrate resistant prostate cancer and associated with resistance to AR-targeted therapies. While we have previously noted that homodimerization is required for the transcriptional activity of AR-V7 and that AR-V7 can also form heterodimers with the full-length AR (AR-FL), there are still many gaps of knowledge in AR-V7 stepwise activation. In the present study, we show that neither AR-V7 homodimerization nor AR-V7/AR-FL heterodimerization requires cofactors or DNA binding. AR-V7 can enter the nucleus as a monomer and drive a transcriptional program and DNA-damage repair as a homodimer. While forming a heterodimer with AR-FL to induce nuclear localization of unliganded AR-FL, AR-V7 does not need to interact with AR-FL to drive gene transcription or DNA-damage repair in prostate cancer cells that co-express AR-V7 and AR-FL. These data indicate that AR-V7 can function independently of its interaction with AR-FL in the true castrate state or “absence of ligand”, providing support for the utility of targeting AR-V7 in improving outcomes of patients with castrate resistant prostate cancer.
Keywords: Androgen receptor, splice variant, dimerization, nuclear localization, DNA damage response, transactivation, prostate cancer
Introduction
The androgen receptor (AR) is a potent ligand-regulated transcription factor that regulates a comprehensive transcriptional program in prostate cancer cells. AR-targeted therapies either block the production of the ligand [1] or compete with the ligand for binding to AR [2-4]. Despite the advent of second-generation AR-targeted therapies, such as enzalutamide, apalutamide, and abiraterone, disease progression is common with maintained AR signaling [5].
Multiple mechanisms that contribute to continued AR activity in castrate resistant prostate cancer (CRPC) have been uncovered, including intratumoral androgen biosynthesis, AR gene and enhancer amplification, gain-of-function AR mutations, aberrant activation of AR with alternate drivers, and the emergence of constitutively active truncated AR splice variants (AR-Vs) that lack a functional ligand-binding domain (LBD) [5, 6]. Over 20 AR-Vs have been detected in clinical specimens [7, 8]. To date, AR splice variant-7 (AR-V7) has been studied in greatest detail owing to its relative abundance, frequency of detection [8, 9], and potential clinical utility as a marker for treatment selection in men with metastatic CRPC [10-15].
The AR-V7 protein is rarely expressed in primary prostate cancer but is frequently detected following androgen deprivation therapy, with further increase in expression following abiraterone or enzalutamide therapy [9, 15-19]. In CRPC, AR-V7 expression is predominantly nuclear, and its expression is correlated with androgen-independent cell proliferation and disease progression after abiraterone or enzalutamide therapy [10, 20-23]. AR-V7 expression in circulating tumor cells has been shown to be a negative predictive biomarker of response to AR-targeted therapies [10, 11, 14]. These studies establish that AR-V7 expression is an adaptive response to AR-targeted therapies and associated with endocrine resistance.
Structurally, AR-V7 contains the AR N-terminal transactivation domain, the DNA-binding domain, and a unique C-terminal 16-amino-acid sequence, resulting from the inclusion of a cryptic exon [7]. Since AR-Vs have the same DNA-binding domain and N-terminal transactivation domain as the full-length AR (AR-FL), they are able to bind to DNA and drive a transcriptional program, albeit in a ligand-independent manner [24-29]. We and others have shown that, while driving a similar transcriptional program and mediating DNA-damage repair following radiation as AR-FL, AR-Vs also regulate a distinct set of genes/pathways [18, 21, 24, 26-35]. In addition, we have noted that AR-V homodimerization is required for the transcriptional activity of AR-Vs [26, 36, 37]. AR-Vs can also form heterodimers with AR-FL, and nuclear-localized AR-Vs, such as AR-V7 and ARv567es, can induce nuclear localization of unliganded AR-FL [20, 36]. However, there are still many gaps of knowledge in AR-V stepwise activation, i.e., whether AR-V/AR-V homodimerization and/or AR-V/AR-FL heterodimerization requires other factors or DNA binding, whether AR-V/AR-V homodimerization is required for AR-V to translocate to the nucleus or to mediate DNA-damage repair following radiation, whether AR-V/AR-FL heterodimerization impedes AR-FL ligand binding, and whether heterodimerization with AR-FL is required for AR-Vs to induce nuclear localization of unliganded AR-FL. Filling in these gaps of knowledge will allow a better understanding of the mechanism of action of AR-Vs in therapy-resistant prostate cancer.
In addition to a detailed characterization of AR-V stepwise activation, the present study tackled another unresolved critical issue in prostate cancer research - whether AR-Vs can function independently of AR-FL. In AR-null cell models and/or models with depleted AR-FL, many AR-Vs have been shown to have the ability to regulate gene transcription in the absence of AR-FL [17-19, 25-27, 30, 34, 38-42]. In contrast, in models that express AR-FL, AR-V7 activity has been indicated to be largely dependent on AR-FL [29, 43]. For example, the expression of AR-V7 in AR-FL-overexpressing LNCaP xenograft tumors did not affect the growth-inhibitory efficacy of the AR-FL antagonist enzalutamide [43]. Moreover, in LNCaP95 cells that express both AR-FL and AR-V7, AR-V7 was shown to interact with AR-FL on the DNA to repress the transcription of a set of growth-suppressive genes [29]. Since AR-Vs are almost always co-expressed with AR-FL in the clinical setting, dissecting AR-FL-dependent and/or –independent functions of AR-Vs has significant clinical implications. We tackled this issue here using a mutant AR-V7 that is unable to dimerize with AR-FL. We show that AR-V7 can regulate transcriptional programs and mediate DNA damage repair after irradiation independently of its interaction with AR-FL.
Results
AR-V7/AR-FL and AR-V7/AR-V7 dimerization do not require other co-factors or DNA binding.
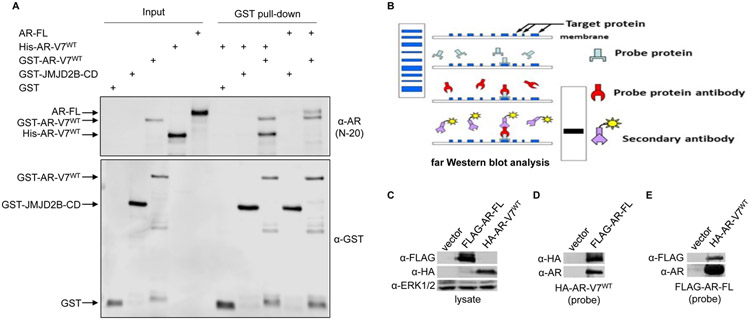
We first used purified recombinant proteins in an in vitro pull-down assay to assess whether AR-V7/AR-FL and/or AR-V7/AR-V7 dimerization requires other factors. We observed that purified AR-FL and His-tagged AR-V7 proteins were readily pulled down by GST-fused AR-V7, but not GST alone or GST-fused catalytic domain of JMJD2B (Fig. 1A). Consistent with the pull-down data, FLAG-tagged AR-FL and HA-tagged AR-V7 immuno-affinity purified from 293T cells hybridized with each other on far Western blots (Fig. 1B-E). Of note, the affinity-purified proteins were extensively washed to remove most of the proteins that were in complex. Moreover, the target protein was run on a denaturing gel, which would disrupt the interactions with any residual proteins. The target protein was renatured after being transferred to a PVDF membrane, allowing it to interact with the probe protein. With reciprocal binding of AR-FL and AR-V7 as either the target or the probe protein, the far Western results strongly support their direct interactions. Together, the data from these two assays indicate for the first time that AR-V7/AR-FL heterodimerization and AR-V7 homodimerization can occur independently of cofactors and DNA binding.
Figure 1. AR-V7/-FL or AR-V7/-V7 dimerization does not require other factors or DNA-binding.
A. In vitro pull-down assay with purified proteins demonstrating direct interactions of GST-fused AR-V7 with His-tagged AR-V7 and with AR-FL. His-tagged AR-V7 and AR-FL recombinant proteins were pulled down using glutathione-bead-immobilized GST alone or GST-JMJD2B-catalytic-domain (GST-JMJD2B-CD) or GST-AR-V7 fusion protein and subjected to Western blotting with an anti-AR or anti-GST antibody. B. Schematic of far Western blot analysis. C-E. Reciprocal far Western blot analysis with purified proteins supporting direct interaction between HA-tagged AR-V7 and FLAG-tagged AR-FL. C: Input showing the expression of FLAG-tagged AR-FL and HA-tagged AR-V7. D&E: Extensively washed FLAG-tagged AR-FL (D) or HA-tagged AR-V7 (E) was run on a denaturing gel, transferred to a PVDF membrane, renatured, and hybridized to extensively-washed HA-tagged AR-V7 (D) or FLAG-tagged AR-FL (E) probe protein. The interactions were detected using anti-HA and anti-AR antibodies (D) or anti-FLAG and anti-AR antibodies (E).
AR-V7 does not affect androgen binding to AR-FL.
To evaluate whether the interaction with AR-V7 impacts the ability of AR-FL to bind to androgens, we performed a ligand-binding assay in cumate-inducible AR-V7-expressing LNCaP cells. Scatchard analysis showed that induction of AR-V7 expression did not alter the ligand-binding affinity of AR-FL (Fig. 2, KD). The ligand-binding capacity of AR-FL, however, displayed a trend of reduction after AR-V7 expression, although the reduction was not statistically significant (Fig. 2, Bmax). These data indicate that AR-V7 expression has minimal impact on androgen binding to AR-FL.
Figure 2. AR-V7 does not significantly affect AR-FL ligand binding:
A. Scatchard analysis of binding of [3H]R1881 with AR-FL in LNCaP extracts with or without cumate-induced AR-V7 expression in three independent experiments. B. KD and Bmax values calculated from the Scatchard analysis showing that AR-V7 expression does not have a significant impact on AR-FL ligand-binding affinity or capacity, although the capacity shows a trend of decrease. C. Western blotting confirming AR-V7 induction by cumate in these replicates.
FXXLF-motif is critical for AR-V7/-FL heterodimerization.
We have previously used the bimolecular fluorescence complementation (BiFC) and bioluminescence resonance energy transfer (BRET) assays to characterize AR-V7 and AR-FL dimerization [36]. Both assays entail the expression of AR-V7 and AR-FL proteins fused with a bulky fluorescent and/or bioluminescent protein in AR-null cell models. To further delineate AR-V7 and AR-FL interactions, we performed proximity ligation assays (PLA) with endogenous AR-FL and inducibly expressed AR-V7 fused with a small FLAG tag in LNCaP cells. Consistent with our previous findings [36], robust androgen-independent interactions between AR-V7 and AR-FL were evident from the PLA results (Fig. 3). The interactions were detected in both the nucleus and the cytoplasm, although the majority of the signal was observed in the nucleus, possibly due to nuclear translocation of AR-V7 (Supplementary Fig. S1). Interestingly, addition of 10 nM dihydrotestosterone (DHT) only modestly enhanced their interactions (Supplementary Fig. S2A).
Figure 3. FXXLF motif is critical for AR-V7/-FL heterodimerization.
LNCaP cells stably expressing cumate-inducible FLAG-tagged AR-V7WT or AR-V7MT were evaluated for interaction between AR-FL and AR-V7 using proximity ligation assays (PLA) under conditions of vehicle control or cumate stimulation in androgen-deprived condition. An AR C-terminal antibody and a FLAG-targeted antibody were used to track AR-FL and AR-V7, respectively. DAPI was used to stain the nucleus. These data indicate that AR-V7WT, AR-V7D, and AR-V7P, but not AR-V7F or AR-V7FD, can interact with AR-FL. The enlarged images in the inset show the interactions in a representative single cell. *, P < 0.001 from control.
Using the BiFC assay, we previously showed that the AR-V7 FXXLF motif and dimerization box (D-box) together mediate AR-V7/AR-FL dimerization [36]. To validate these dimerization interfaces, we generated LNCaP cells with cumate inducible expression of a mutant AR-V7 (AR-V7MT) with point mutations in either the FXXLF motif (F23A/F27A/L26A AR-V7F), the D-box (A596T/S597T, AR-V7D), both the FXXLF motif and D-box (F23A/F27A/L26A/A596T/S597T, AR-V7FD) [36, 44, 45], or the P-box (R585K, AR-V7P), which mediates AR interaction with DNA [46]. We then evaluated AR-FL interactions with these AR-V mutants by PLA studies. While AR-V7D and AR-V7P retained the ability to interact with AR-FL, AR-V7F and AR-V7FD were unable to interact with AR-FL (Fig. 3). The expression of AR-V7MT was confirmed by Western blotting (Supplementary Fig. S2B). While these PLA studies indicate that mutating the FXXLF motif alone is sufficient to abolish AR-V7/AR-FL heterodimerization, the finding differs from our previous observations in the BiFC assay, where mutating both the FXXLF motif and the D-box was needed for disrupting AR-V7/AR-FL interactions [36]. The influence of the bulky fluorescent protein fused to AR-V7 and AR-FL in the BiFC assay may account for the discrepancy, and the PLA studies are more likely to be representative of AR-V7 and AR-FL interactions within a prostate cancer cell. This was supported by the inability of AR-V7F to form a complex with AR-FL in co-immunoprecipitation assay (Supplementary Fig. S3).
While AR-V7 can enter the nucleus as a monomer, heterodimerization with AR-FL is required for AR-V7 to mediate nuclear localization of unliganded AR-FL.
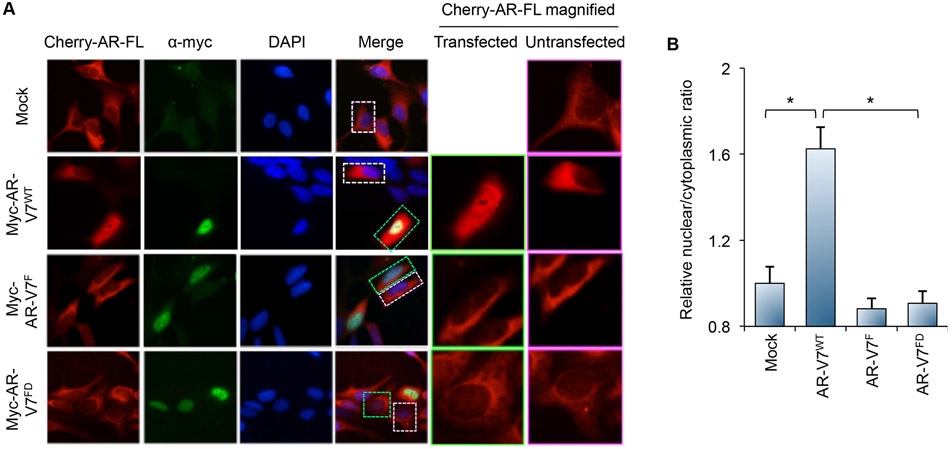
We have previously shown that AR-V7 induces AR-FL nuclear localization in the absence of androgen [20]. To determine whether the induced AR-FL nuclear localization is mediated by heterodimerization with AR-V7, we created LNCaP cells stably expressing cherry-tagged AR-FL and transiently transfected with either the mock vector or myc-tagged AR-V7WT or AR-V7MT (Western blots in Supplementary Fig. S4). Thus, while all cells express AR-FL, only a subset expresses AR-V7 (due to transfection efficiency), enabling direct comparisons between cells expressing AR-FL alone and cells expressing both AR-FL and AR-V7. The cells were cultured under androgen-deprived condition. Cherry-tagged AR-FL was largely cytoplasmic in the mock-transfected cells (Fig. 4). Transfection of AR-V7WT significantly induced nuclear localization of unliganded cherry-tagged AR-FL (Fig. 4) in the transfected cells that express AR-V7WT (magnified image in Column 5). In adjacent untransfected cells that do not express AR-V7WT, cherry-tagged AR-FL is largely cytoplasmic (magnified image in Column 6). In contrast, transfection of AR-V7F or AR-V7FD did not impact the subcellular localization of cherry-tagged AR-FL (Fig. 4). These data were validated in 293T cells stably transfected with a GFP-tagged AR-FL and transiently transfected with these myc-tagged AR-V7 expression plasmids (Supplementary Fig. S5). It is of note that the differences observed were not due to altered subcellular localization of AR-V7MT, as AR-V7F and AR-V7FD remained predominantly nuclear, no matter whether they are tagged (Fig. 4A and Supplementary Figs. S1 and S5) or untagged (Fig. 5). Together, these data indicate that heterodimerization with AR-V7 mediates nuclear localization of unliganded AR-FL.
Figure 4. Dimerization with AR-FL is required for AR-V7 to induce nuclear localization of unliganded AR-FL.
A. LNCaP cells stably expressing cherry-AR-FL were evaluated using confocal microscopy for AR-FL distribution following transfection with the indicated myc-AR-V7 plasmid under androgen-deprived condition (first column). The second column shows the distribution of the transfected myc-AR-V7 protein detected by immunofluorescence staining using an anti-myc antibody. The third column shows DAPI-stained nuclei of the cells. The fourth column is merged images of the first 3 columns. The fifth column is an enlarged view of a single transfected cell from the image in the first column, corresponding to the cell in the green box in the merge column. The sixth column is an enlarged view of a single untransfected cell from the image in the first column, corresponding to the cell in the white box in the merge column. B. Quantification of AR-FL nuclear and cytoplasmic signals in confocal images showing that only AR-V7WT is able to induce nuclear localization of unliganded AR-FL. *, P < 0.001.
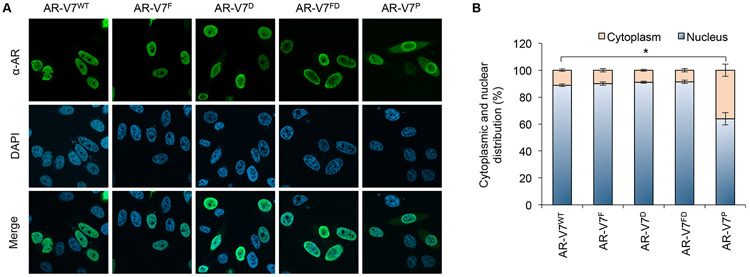
Figure 5. Subcellular localization of AR-V7 mutants.
A. Confocal microscopy images of PC-3 cells transfected with the indicated AR-V7WT or AR-V7MT plasmid under androgen-deprived condition. The cells were immunofluorescently stained with a pan-AR antibody or stained with DAPI for visualization of nucleus. B. Quantification of AR-V7 cytoplasmic and nuclear distribution showing that AR-V7WT, AR-V7F. AR-V7D, and AR-V7FD are largely nuclear, whereas AR-V7P exhibits some cytoplasmic distribution. *, P < 0.001.
We also assessed the subcellular distribution of untagged AR-V7D and AR-V7P. While AR-V7D was primarily nuclear, AR-V7P was more evenly distributed between the nucleus and the cytoplasm (Fig. 5). Since AR-V7D does not homodimerize [36] and the P-box mutation abolishes AR DNA binding [46], these data indicate that AR-V7 can enter the nucleus as a monomer and that interacting with DNA helps AR-V7 stay in the nucleus. Interestingly, co-expression of AR-V7P with AR-FL or other AR-Vs, AR-V4 and AR-V6, which we previously showed to heterodimerize with AR-V7 [37], did not alter the mixed nuclear and cytoplasmic distribution of AR-V7P (Supplementary Figs. S1 and S6), indicating that DNA binding by its heterodimerization partner may not have a significant impact on AR-V7 nuclear localization.
AR-V7 can drive gene transcription independently of interaction with AR-FL.
To assess whether heterodimerization with AR-FL is required for the trans-activating activity of AR-V7 in prostate cancer cells, we co-transfected PC-3 and LNCaP cells with the AR-V7WT or AR-V7MT plasmid with a luciferase reporter construct driven by three copies of androgen response element (ARE-luc). Transfection of AR-V7WT or AR-V7F induced robust luciferase expression in both cell lines under androgen-deprived condition (Fig. 6A-B). In contrast, AR-V7D, AR-V7P, and AR-V7FD were not able to induce luciferase expression. Since mutating the D-box and the P-box, respectively, abolishes AR-V7 homodimerization [36] and DNA binding (Supplementary Fig. S7), these data support a critical role of homodimerization [36] and/or DNA binding in AR-V7 transactivation (Fig. 6A-B). Notably, in addition to losing the ability to homodimerize, AR-V7D was unable to bind to DNA (Supplementary Fig. S7). Since AR-V7 homodimerization does not require DNA binding (Fig. 1), it is likely that homodimerization is a prerequisite for AR-V7 DNA binding and thereby transactivation.
Figure 6. AR-V7 retains the ability to drive transcription independently of heterodimerization with AR-FL.
A & B. Luciferase assays in PC-3 (A) or LNCaP cells (B) co-transfected with an ARE-driven luciferase reporter construct and mock control or indicated AR-V7 expression plasmid showing that, in contrast to AR-V7D, AR-V7FD, and AR-V7P, AR-V7F retains the ability to transactivate. *, P < 0.001 from mock. C & D. RNA-seq analyses of LNCaP cells stably expressing cumate-inducible AR-V7WT or AR-V7MT showing that the transcriptional program induced by AR-V7WT is in general maintained in AR-V7F-expressing cells, but not in AR-V7FD-expressing cells. C: Clustering analysis was performed using genes that were differentially expressed between AR-V7WT-expressing cells and the mock control cells. D: Relative expression levels from the RNA-seq data showing that AR-V7F retains the ability to induce the expression of KLK3, FKBP5, and TMPRSS2 but loses the ability to induce GNMT and FASN expression. *, FDR < 0.01 from mock. E. ChIP assay showing that the similarity and/or difference between the wildtype and mutant AR-V7 in regulating target genes can be largely explained by their ability/inability to bind to the 5’-regulatory region of these genes. The results were from 3 independent experiments. *, P < 0.01 from mock.
In order to further elucidate the role of heterodimerization with AR-FL in AR-V7 transactivation, we performed RNA-sequencing (RNA-seq) analyses of cumate-treated mock and cumate-inducible AR-V7WT-, AR-V7F-, or AR-V7FD-expressing LNCaP cells under androgen-deprived condition from replicate experiments. The analyses identified 140 genes that were upregulated and 31 genes that were downregulated upon AR-V7WT expression (Fig. 6C and Supplementary Fig. S8, FDR < 0.05, listed in Supplementary Table S1). Comparison of this list of genes with published AR-V7-associated targets in LNCaP95 cells [29] and gene expression changes caused by knocking down all AR-Vs in AR-FL-deleted CWR22Rv1-AR-EK cells [47] demonstrated 45% and 39% overlaps respectively, suggesting significant conservation of AR-V function in different cell models. Gene Set Enrichment Analysis (GSEA) demonstrated that androgen response pathway was the most enriched pathway in AR-V7WT-expressing cells, followed by Myc, E2F, and G2M checkpoint pathways (Supplementary Fig. S9). In line with the reporter gene studies, the vast majority of these AR-V7WT-regulated genes were similarly modulated by AR-V7F, but not AR-V7FD (Fig. 6C). AR-V7WT and AR-V7F-induced expression clustered together in hierarchical clustering and principal component analyses (Fig. 6C and Supplementary Fig. S10A). For example, canonical AR-regulated genes such as KLK3, FKBP5, and TMPRSS2 were similarly upregulated in AR-V7WT- and AR-V7F-expressing cells but not in AR-V7FD-expressing cells (Fig. 6D). These observations were validated by RT-qPCR (Supplementary Fig. S10B) and by AR-V7 chromatin immunoprecipitation (ChIP) analyses (Fig. 6E), indicating that transcription and AR promoter occupancy of these genes were similarly regulated by AR-V7WT and AR-V7F but not modulated by AR-V7FD. A small subset (10) of the 140 upregulated genes such as GNMT and FASN were differentially regulated by AR-V7WT and AR-V7F, at both the DNA binding and expression level (Fig. 6D & E). Taken together, these data provide support that heterodimerization with AR-FL is not required for a majority of AR-V7 cistromic and transcriptomic activity in AR-FL-expressing prostate cancer cells.
Homodimerization and DNA binding, but not heterodimerization with AR-FL, is required for AR-V7 to mediate DNA-damage repair.
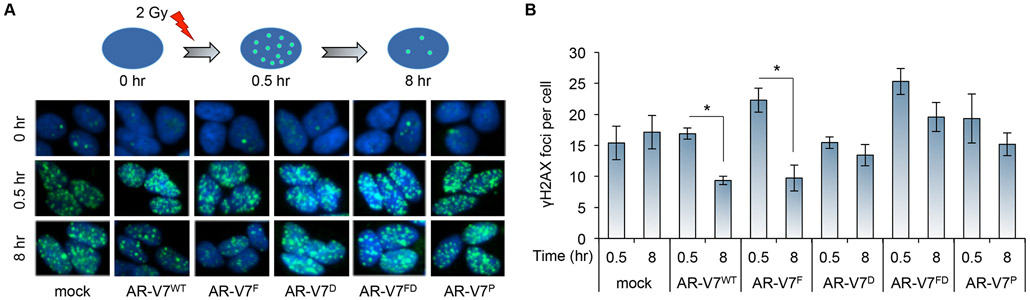
We and others recently demonstrated that AR-Vs could orchestrate DNA-damage repair after irradiation [35, 47]. In support of this notion, functional annotation of the 140 genes that were upregulated upon AR-V7WT expression in LNCaP cells showed that 6 of these genes regulate DNA damage response (CUL4B, NBN, NKX3-1, PIK3R1, PLK1, and USP43). To evaluate the importance of homodimerization, DNA binding, and/or heterodimerization with AR-FL to the DNA-damage repair function of AR-V7, we performed DNA-damage resolution studies in cumate-inducible AR-V7WT- or AR-V7MT-expressing LNCaP cells (Fig. 7A). We noted that γ-H2Ax foci (within 0.5 hour) were induced by 2Gy radiation treatment in androgen-deprived mock control cells and had minimal resolution by 8 hours (Fig. 7A & 7B), suggesting that unliganded AR-FL alone in these cells is unable to mediate DNA-damage repair. Under these conditions, expression of AR-V7WT or AR-V7F enabled DNA-damage repair, as shown by resolution of γ-H2Ax foci within 8 hours (Fig. 7). However, the AR-V7D, AR-V7FD, and AR-V7P mutants are unable to mediate DNA-damage repair (Fig. 7). These data indicate that homodimerization and DNA binding, but not interaction with AR-FL, is critical for AR-V7 to mediate DNA-damage repair. Supplementary Fig. S11 provides a summary of the impact of different mutations on various functions of AR-V7. Through inducible expression of these mutants in AR-FL-expressing cells, we show that, in prostate cancer cells that co-express AR-V7 and AR-FL, AR-V7 can largely function independently of the interaction with AR-FL in terms of regulating gene expression and inducing DNA-damage repair.
Figure 7. Homodimerization and DNA binding, but not dimerization with AR-FL, is required for AR-V7 to mediate DNA-damage repair.
Evaluation for double-stranded DNA breaks using γ-H2AX foci in LNCaP cells stably expressing cumate-inducible FLAG-tagged AR-V7WT or AR-V7MT construct at 0, 0.5, or 8 hr after 2 Gy ionizing irradiation under androgen-deprived condition showing that AR-V7F, but not AR-V7D, AR-V7FD, or AR-V7P, retains the ability to mediate DNA-damage repair. A. Schematic of the experiment and images of γ-H2AX foci within the cells. B. Quantification of more than 50 nuclei per condition for the average number of γ-H2AX foci per cell. *, P < 0.01.
Discussion
Our data, for the first time, show that neither AR-V7 homodimerization nor AR-V7/AR-FL heterodimerization requires cofactors or DNA binding. Together with our previous studies [26, 36], we have employed 5 different approaches, in vitro pull-down, far Western, BiFC, BRET, and PLA, to test AR-V homo- and/or hetero-dimerization. These approaches, through the use of recombinant proteins, transfected fusion proteins, to almost equivalent to endogenous proteins, have unequivocally established the direct interactions of AR-V7 with itself and with AR-FL. Further, using luciferase assay, RT-qPCR, DNA pull-down, electrophoretic mobility shift assay, ChIP, and DNA damage resolution assay in this and previous studies [26, 35, 36], we demonstrate that, although interacting with AR-FL is not required for AR-V7 to transactivate or to mediate DNA-damage repair, homodimerization and DNA binding are imperative for these functions of AR-V7.
Additionally, we note that mutating the dimerization interface of AR-V7 does not impact its nuclear localization, indicating that AR-V7 can enter the nucleus as a monomer, similar to AR-FL nuclear translocation upon androgen binding [45]. We also show that AR-V7-induced AR-FL nuclear localization depends on their heterodimerization. This may be caused by either AR-V7 dimerizing with AR-FL in the cytoplasm and bringing AR-FL to the nucleus or their dimerization in the nucleus preventing cytoplasmic export of AR-FL. Based on the finding that both AR-V7 and androgen-bound AR-FL enter the nucleus as a monomer, it is tempting to speculate that the latter scenario might be more likely. In fact, our data on the lack of significant impact of AR-V7 on AR-FL ligand binding can be easily explained by unliganded AR-FL remaining as a monomer in the cytoplasm even in the presence of AR-V7, although other mechanisms are also plausible.
Cistromic studies have indicated that AR-V7 and AR-FL can bind to the same sites on the chromatin [18, 21, 24, 26-34]. Since AR-Vs are almost always co-expressed with AR-FL in the clinical setting, one of the key unanswered questions in the field was whether AR-Vs need the interaction with AR-FL to function. Using AR-V7 point mutations that disrupt its interaction with AR-FL, we show that, when AR-V7 and AR-FL are co-expressed in LNCaP cells under androgen-deprived condition, their heterodimerization results in altered AR-V7 promoter occupancy and transcriptional regulation of only a small number of genes of unclear biological significance. AR-V7 regulation of the vast majority of the genes does not require interaction with AR-FL. This is also the case for AR-V7 to mediate DNA-damage repair following radiation under androgen-deprived condition. While requiring AR-V7 homodimerization and DNA binding, the DNA-damage repair function of AR-V7 does not rely on interaction with AR-FL in cells co-expressing AR-V7 and AR-FL. Together, our data indicate that the transcriptional and DNA-damage repair functions of AR-V7 are largely independent of its interaction with AR-FL in the true castrate state or “absence of ligand”.
Intratumoral androgen biogenesis has been shown to be one mechanism of CRPC progression after AR-targeted therapy, leading to sufficient intraprostatic androgen levels to activate AR-FL [48]. Our finding that AR-V7 expression does not significantly affect AR-FL ligand binding indicates that, in these CRPCs, AR-FL maintains the ability to bind to androgens despite the expression of AR-V7. In the PLA studies, we note that AR-V7 heterodimerizes with AR-FL regardless of the presence or absence of androgen. These data suggest that AR-V7 is likely to interact with both unliganded and liganded AR-FL. Interestingly, the transcriptional activity of liganded AR-FL has been shown to rely on its interaction with AR-V7 in LNCaP95 cells, the growth of which is known to be suppressed by androgens [29]. Future studies in CRPC cells that are growth-sustained by androgens are necessary to further delineate the role of AR-V7 in ligand-induced transcription driven by the AR-FL, since the progression of a significant proportion of CRPCs is driven by intratumoral androgens.
Overall, the present study helps generate a blueprint of AR-V7 stepwise activation. AR-V7 can enter the nucleus as a monomer and drive a transcriptional program and DNA-damage repair as a dimer. AR-V7 dimerization does not require other factors or DNA binding. While forming a heterodimer with AR-FL to induce nuclear localization of unliganded AR-FL, AR-V7 does not need to interact with AR-FL to drive gene transcription or DNA-damage repair. CRPC evolves a mechanism to make sure that the AR-driven transcriptional program and DNA-damage repair are maintained. As an adaptive response to AR-targeted therapies, the expression of AR-V7 appears to be sufficient to drive AR function within CRPC cells. Thus, therapeutic strategies to target AR-V7 could be of utility in combatting CRPC.
Materials and Methods
Cell culture.
LNCaP, PC-3, and 293T cells were obtained from ATCC. LNCaP and PC-3 cells were grown in RPMI-1640 medium (Hyclone) with 10% fetal bovine serum (FBS, Gibco). 293T cells were cultured in DMEM medium (Corning) with 10% FBS. Cells used in all experiments were within 3 months of resuscitation of frozen cell stocks established within 3 passages after receipt of the cells. Cell authentication was performed at the Genetica DNA Laboratories, and cells were evaluated monthly for mycoplasma contamination.
In vitro pull-down assay using recombinant proteins.
To generate the GST or His fusion construct of AR-V7, we cloned PCR-amplified coding region of AR-V7 into the pGEX-6p-1 or pET-28a vector (Millipore Sigma). These constructs were transformed individually into E. coli strain BL-21, and protein expression was induced using 0.4 mM IPTG. Fusion proteins were purified using glutathione-agarose beads (GE Life Sciences) or Ni-NTA resin (Thermo Fisher). For pull-down assays, 2 μg of His-AR-V7 or AR-FL recombinant protein (Creative Biomart) was incubated with 2 μg of GST-AR-V7 protein, GST alone, or GST-fused catalytic domain of JMJD2B that was immobilized on glutathione beads in 50 μl of binding buffer (25mM Tris, 0.1% TX-100, 0.5 M NaCl in water, and proteinase inhibitor cocktail) for 2 hr at 4°C on a rotating platform. After centrifugation at 5000xg for 15 sec, the agarose beads were washed 4 times with the binding buffer. The proteins were then subjected to SDS-PAGE and Western blotting.
Far Western Blot Analysis.
The analysis was performed as detailed in [49]. In brief, 293T cells were transfected with FLAG-AR-FL or HA-AR-V7 using Fugene 6. At 48 hr following transfection, AR was immuno-affinity purified with an anti-FLAG or anti-HA antibody and extensively washed. Proteins were resolved by SDS-PAGE, transferred to PVDF membrane, and denatured and renatured. After blocking, membranes were incubated with probes, washed, fixed, and quenched. Membranes were then blotted for an HA, FLAG, AR, or ERK1/2 antibody.
Plasmids.
The cumate-inducible 3xFLAG-tagged AR-V7 expression construct, in the pCDH-Cuo-MCS-EF1α-CymR-T2A-Puro backbone (System Biosciences), was kindly provided by Dr. Stephen Plymate (University of Washington, Seattle, WA). The constitutive AR-V7 expression lentiviral construct was generated by cloning PCR-amplified coding region of AR-V7 into the pGEM-T Easy vector (Promega), followed by subcloning into the pLVX-puro lentiviral vector (Clontech). The AR-V7MT constructs were generated by site-directed mutagenesis. AR-V7 and mutants were subcloned into the pcDNA3.1-Myc plasmid to generate the Myc-tagged AR-V7 versions.
Cumate-inducible AR-V7-expressing LNCaP cells.
The cumate-inducible wildtype or mutant AR-V7 expression constructs were individually packaged into lentiviral particles in 293T cells, and the lentivirus particles were used to transduce LNCaP cells as described previously [50]. At 48 hr after transduction, 1 μg/ml puromycin was added to the medium to start selection, and the selection continued for two weeks to obtain polyclonal colonies of resistant cells. AR-V7 was expressed by exposing the cells to 1x cumate (System Biosciences).
Ligand-binding assay.
3H-Methyltrienolone R1881 (PerkinElmer) was added to cells grown on a 6-well plate in phenol red-free medium with 10% CSS to final concentrations of 0.25, 0.5, 1, 2.5, 5, 7.5 nM in the presence or absence of 200-fold excess unlabeled R1881 for 1 hr at 37°C in a CO2 incubator. Cells were washed three times with ice-cold PBS, and the rinse solution was completely removed after each wash. 700 μl absolute ethanol was added to each well, and the plates were rocked for 30 min at room temperature. The cell lysates were collected and transferred to scintillation vials with 4 ml scintillation fluid for radioactivity counting.
PLA.
Cells grown on coverslips were fixed with 10% formalin for 20 min and permeabilized with 0.5% Triton X-100 for 15 min at room temperature. AR-V7 and AR-FL were detected using an anti-FLAG (Sigma-Aldrich, F3165) and anti-AR-FL antibody (C19, Santa Cruz; against AR C-terminus), respectively. In situ proximity ligation was performed using a Duolink Detection Kit (Sigma-Aldrich). Nuclear foci were imaged using a Lionheart FX microscope (×40 magnification). The number of nuclear foci/cell was quantified using ImageJ with >50 cells analyzed per experiment per condition.
AR-FL localization assessment.
LNCaP cells stably expressing Cherry-AR-FL or 293T cells stably expressing GFP-AR-FL grown on coverslips were transiently transfected with pcDNA-Myc AR-V7 wildtype or mutants and continued to grow for 48 hr in phenol-red free RPMI-1640 with 3-5% CSS. Coverslips were fixed with methanol for 10 min at −20°C and permeabilized with 0.2% TX-100 for 30 min at room temperature. Cells were stained with an anti-Myc tag antibody (Cell Signaling, 9B11) and counterstained with DAPI. Cells were imaged using a widefield Delta Vision Fluorescence microscope, and nuclear/cytoplasmic localization of cherry-AR-FL or GFP-AR-FL was quantified using ImageJ with >50 cells analyzed per experiment per condition.
Immunofluorescence staining and confocal fluorescence microscopy of AR-V7 localization.
After fixation with 4% paraformaldehyde, cells were incubated with an anti-FLAG (for cumate-inducible AR-V7-expressing LNCaP cells; Sigma-Aldrich, F3165) or AR-441 pan-AR antibody (for PC-3 cells transfected with AR-V7; Santa Cruz Biotechnology, sc-7305) overnight at 4°C and then washed and incubated with Alexa Fluor 488-conjugated secondary antibody (Thermo Fisher, A-11001) for 1 hr at room temperature in the dark and counterstained with DAPI. An average of 10 fields were captured for each group using a Nikon A1r confocal fluorescence microscope. The intensity ratios of nuclear and cytoplasmic fluorescence signals were quantitated using ImageJ.
Plasmid transfection and reporter-gene assay.
LNCaP, PC-3, and 293T cells were transfected by using the Lipofectamine 3000 (Thermo Fisher), TransIT-2020 (Mirus Bio LLC), and TurboFect (Thermo Fisher) reagent, respectively. Reporter gene assay was performed as previously described [51] with ARE-luc plasmid. To ensure even transfection efficiency, we conducted the transfection in bulk and then split the transfected cells for luciferase assay.
RNA-seq analysis.
Total RNA isolated using the TRIzol reagent (Thermo Fisher) was sent to Beijing Genomics Institute (BGI) for polyA-selected, 100-base paired-end RNA sequencing using an Illumina HiSeq 4000 system. Data analysis was performed using RSEM [52]. The transcripts per million (TPM) values from the RSEM output were used for transcript abundance measurement. Differential expression was assessed using the DESeq package [53] with R 3.6.0. Genes were defined as differentially expressed between groups if having FDR < 0.05. The list of genes that were differentially expressed between the AR-V7-expressing group and the mock control group were used in hierarchical clustering analysis and principal component analysis to elucidate similarity among wildtype AR-V7 and AR-V7 mutants in ability to regulate target genes. GSEA was performed on compiled normalized count matrix with 1000 permutations, the gene-set permutation type, and weighted enrichment statistics. The genes were ranked using the Signal2Noise matrix, and the data were run against the hallmark gene sets in the Molecular Signatures Database.
ChIP-qPCR.
ChIP analysis was performed as previously described [35] using an anti-FLAG (Sigma-Aldrich, F3165) or anti-AR-FL antibody (C19, Santa Cruz) or normal rabbit IgG (Abcam). Purified ChIP and input DNA were analyzed by qPCR.
RT-qPCR and Western blotting analyses.
cDNA synthesis and qPCR analysis were performed as described [54, 55], and the primer-probe sets were from IDT. The Western blotting procedure was described previously [55].
Double-strand break repair assay.
At each time point after 2 Gy radiation, cells were subjected to γH2AX immunofluorescence as previously described [35], and the number of γ-H2AX foci per cell was quantified using ImageJ. More than 200 γ-H2AX–positive cells were analyzed per experiment per condition.
Statistical analysis.
Sample size was selected based on the ability to achieve an overall significance level of P = 0.05 and 80% power. The Student two-tailed t test was used to determine the mean differences between two groups. P < 0.05 is considered significant. Data are presented as mean ± SEM.
Supplementary Material
Acknowledgments
We are grateful to Dr. Stephen Plymate at the University of Washington for providing cumate-inducible AR-V7 expression construct. We appreciate the support from the Tulane Cancer Next Generation Sequence Analysis core for utilization of resources and expertise for this work.
Competing interests
This work was funded by grants from the Department of Defense (W81XWH-19-1-0363, W81XWH-17-1-0674), Prostate Cancer Foundation Challenge Award, Mimi and John Cole Foundation and Charles Y. Pak grant to GVR, from the National Institutes of Health (R01CA188609) and the Department of Defense (W81XWH-15-1-0439 and W81XWH-16-1-0317) to YD, and from the National Institutes of Health (R01 CA178338) to DG. The content is solely the responsibility of the authors and does not necessarily represent the official views of the funding agencies.
Footnotes
Dedication
This paper is dedicated to the memory of Amy Rui Li, lab manager of the Raj laboratory, who lost her battle with cancer.
References
- 1.de Bono J, Eisenberger M, Sartor O. Abiraterone in Metastatic Prostate Cancer. N Engl J Med 2017; 377: 1694–1695. [DOI] [PubMed] [Google Scholar]
- 2.Davis ID, Martin AJ, Stockler MR, Begbie S, Chi KN, Chowdhury S et al. Enzalutamide with Standard First-Line Therapy in Metastatic Prostate Cancer. N Engl J Med 2019; 381: 121–131. [DOI] [PubMed] [Google Scholar]
- 3.Hussain M, Fizazi K, Saad F, Rathenborg P, Shore N, Ferreira U et al. Enzalutamide in Men with Nonmetastatic, Castration-Resistant Prostate Cancer. N Engl J Med 2018; 378: 2465–2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Loriot Y, Fizazi K, de Bono JS, Forer D, Hirmand M, Scher HI. Enzalutamide in castration-resistant prostate cancer patients with visceral disease in the liver and/or lung: Outcomes from the randomized controlled phase 3 AFFIRM trial. Cancer 2017; 123: 253–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sartor O, de Bono JS. Metastatic Prostate Cancer. N Engl J Med 2018; 378: 1653–1654. [DOI] [PubMed] [Google Scholar]
- 6.Yuan X, Cai C, Chen S, Chen S, Yu Z, Balk SP. Androgen receptor functions in castration-resistant prostate cancer and mechanisms of resistance to new agents targeting the androgen axis. Oncogene 2014; 33: 2815–2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cao S, Zhan Y, Dong Y. Emerging data on androgen receptor splice variants in prostate cancer. Endocr Relat Cancer 2016; 23: T199–T210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Robinson D, Van Allen EM, Wu YM, Schultz N, Lonigro RJ, Mosquera JM et al. Integrative clinical genomics of advanced prostate cancer. Cell 2015; 161: 1215–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sharp A, Coleman I, Yuan W, Sprenger C, Dolling D, Rodrigues DN et al. Androgen receptor splice variant-7 expression emerges with castration resistance in prostate cancer. J Clin Invest 2019; 129: 192–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Antonarakis ES, Lu C, Wang H, Luber B, Nakazawa M, Roeser JC et al. AR-V7 and resistance to enzalutamide and abiraterone in prostate cancer. N Engl J Med 2014; 371: 1028–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Scher HI, Lu D, Schreiber NA, Louw J, Graf RP, Vargas HA et al. Association of AR-V7 on Circulating Tumor Cells as a Treatment-Specific Biomarker With Outcomes and Survival in Castration-Resistant Prostate Cancer. JAMA Oncol 2016; 2: 1441–1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Todenhofer T, Azad A, Stewart C, Gao J, Eigl BJ, Gleave ME et al. AR-V7 Transcripts in Whole Blood RNA of Patients with Metastatic Castration Resistant Prostate Cancer Correlate with Response to Abiraterone Acetate. J Urol 2017; 197: 135–142. [DOI] [PubMed] [Google Scholar]
- 13.Welti J, Rodrigues DN, Sharp A, Sun S, Lorente D, Riisnaes R et al. Analytical Validation and Clinical Qualification of a New Immunohistochemical Assay for Androgen Receptor Splice Variant-7 Protein Expression in Metastatic Castration-resistant Prostate Cancer. Eur Urol 2016; 70: 599–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sharp A, Welti JC, Lambros MBK, Dolling D, Rodrigues DN, Pope L et al. Clinical Utility of Circulating Tumour Cell Androgen Receptor Splice Variant-7 Status in Metastatic Castration-resistant Prostate Cancer. Eur Urol 2019; 76: 676–685. [DOI] [PubMed] [Google Scholar]
- 15.Hornberg E, Ylitalo EB, Crnalic S, Antti H, Stattin P, Widmark A et al. Expression of androgen receptor splice variants in prostate cancer bone metastases is associated with castration-resistance and short survival. PLoS One 2011; 6: e19059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dehm SM, Schmidt LJ, Heemers HV, Vessella RL, Tindall DJ. Splicing of a novel androgen receptor exon generates a constitutively active androgen receptor that mediates prostate cancer therapy resistance. Cancer Res 2008; 68: 5469–5477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sun S, Sprenger CC, Vessella RL, Haugk K, Soriano K, Mostaghel EA et al. Castration resistance in human prostate cancer is conferred by a frequently occurring androgen receptor splice variant. J Clin Invest 2010; 120: 2715–2730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guo Z, Yang X, Sun F, Jiang R, Linn DE, Chen H et al. A novel androgen receptor splice variant is up-regulated during prostate cancer progression and promotes androgen depletion-resistant growth. Cancer Res 2009; 69: 2305–2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hu R, Dunn TA, Wei S, Isharwal S, Veltri RW, Humphreys E et al. Ligand-independent androgen receptor variants derived from splicing of cryptic exons signify hormone-refractory prostate cancer. Cancer Res 2009; 69: 16–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cao B, Qi Y, Zhang G, Xu D, Zhan Y, Alvarez X et al. Androgen receptor splice variants activating the full-length receptor in mediating resistance to androgen-directed therapy. Oncotarget (1802 pii) 2014; 5: 1646–1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li Y, Chan SC, Brand LJ, Hwang TH, Silverstein KA, Dehm SM. Androgen receptor splice variants mediate enzalutamide resistance in castration-resistant prostate cancer cell lines. Cancer Res 2013; 73: 483–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mostaghel EA, Marck BT, Plymate SR, Vessella RL, Balk S, Matsumoto AM et al. Resistance to CYP17A1 Inhibition with Abiraterone in Castration-Resistant Prostate Cancer: Induction of Steroidogenesis and Androgen Receptor Splice Variants. Clinical Cancer Research 2011; 17: 5913–5925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nadiminty N, Tummala R, Liu C, Yang J, Lou W, Evans CP et al. NF-kappaB2/p52 induces resistance to enzalutamide in prostate cancer: role of androgen receptor and its variants. Mol Cancer Ther 2013; 12: 1629–1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krause WC, Shafi AA, Nakka M, Weigel NL. Androgen receptor and its splice variant, AR-V7, differentially regulate FOXA1 sensitive genes in LNCaP prostate cancer cells. Int J Biochem Cell Biol 2014; 54: 49–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lu J, Lonergan PE, Nacusi LP, Wang L, Schmidt LJ, Sun Z et al. The cistrome and gene signature of androgen receptor splice variants in castration resistant prostate cancer cells. J Urol 2015; 193: 690–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chan SC, Selth LA, Li Y, Nyquist MD, Miao L, Bradner JE et al. Targeting chromatin binding regulation of constitutively active AR variants to overcome prostate cancer resistance to endocrine-based therapies. Nucleic Acids Res 2015; 43: 5880–5897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.He Y, Lu J, Ye Z, Hao S, Wang L, Kohli M et al. Androgen receptor splice variants bind to constitutively open chromatin and promote abiraterone-resistant growth of prostate cancer. Nucleic Acids Res 2018; 46: 1895–1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen Z, Wu D, Thomas-Ahner JM, Lu C, Zhao P, Zhang Q et al. Diverse AR-V7 cistromes in castration-resistant prostate cancer are governed by HoxB13. Proc Natl Acad Sci U S A 2018; 115: 6810–6815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cato L, de Tribolet-Hardy J, Lee I, Rottenberg JT, Coleman I, Melchers D et al. ARv7 Represses Tumor-Suppressor Genes in Castration-Resistant Prostate Cancer. Cancer Cell 2019; 35: 401–413 e406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hu R, Lu C, Mostaghel EA, Yegnasubramanian S, Gurel M, Tannahill C et al. Distinct transcriptional programs mediated by the ligand-dependent full-length androgen receptor and its splice variants in castration-resistant prostate cancer. Cancer Res 2012; 72: 3457–3462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lu J, Lonergan PE, Nacusi LP, Wang L, Schmidt LJ, Sun Z et al. The cistrome and gene signature of androgen receptor splice variants in castration-resistant prostate cancer cells. J Urol 2014; 193:690–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shafi AA, Putluri V, Arnold JM, Tsouko E, Maity S, Roberts JM et al. Differential regulation of metabolic pathways by androgen receptor (AR) and its constitutively active splice variant, AR-V7, in prostate cancer cells. Oncotarget 2015; 6: 31997–32012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Magani F, Bray ER, Martinez MJ, Zhao N, Copello VA, Heidman L et al. Identification of an oncogenic network with prognostic and therapeutic value in prostate cancer. Mol Syst Biol 2018; 14: e8202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cai L, Tsai YH, Wang P, Wang J, Li D, Fan H et al. ZFX Mediates Non-canonical Oncogenic Functions of the Androgen Receptor Splice Variant 7 in Castrate-Resistant Prostate Cancer. Mol Cell 2018; 72: 341–354 e346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yin Y, Li R, Xu K, Ding S, Li J, Baek G et al. Androgen Receptor Variants Mediate DNA Repair after Prostate Cancer Irradiation. Cancer Res 2017; 77: 4745–4754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu D, Zhan Y, Qi Y, Cao B, Bai S, Xu W et al. Androgen Receptor Splice Variants Dimerize to Transactivate Target Genes. Cancer Res 2015; 75: 3663–3671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhan Y, Zhang G, Wang X, Qi Y, Bai S, Li D et al. Interplay between Cytoplasmic and Nuclear Androgen Receptor Splice Variants Mediates Castration Resistance. Mol Cancer Res 2017; 15: 59–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Uo T, Dvinge H, Sprenger CC, Bradley RK, Nelson PS, Plymate SR. Systematic and functional characterization of novel androgen receptor variants arising from alternative splicing in the ligand-binding domain. Oncogene 2017; 36: 1440–1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hu R, Isaacs WB, Luo J. A snapshot of the expression signature of androgen receptor splicing variants and their distinctive transcriptional activities. The Prostate 2011; 71: 1656–1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chan SC, Li Y, Dehm SM. Androgen receptor splice variants activate androgen receptor target genes and support aberrant prostate cancer cell growth independent of canonical androgen receptor nuclear localization signal. J Biol Chem 2012; 287: 19736–19749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kohli M, Ho Y, Hillman DW, Van Etten JL, Henzler C, Yang R et al. Androgen Receptor Variant AR-V9 Is Coexpressed with AR-V7 in Prostate Cancer Metastases and Predicts Abiraterone Resistance. Clin Cancer Res 2017; 23: 4704–4715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nyquist MD, Li Y, Hwang TH, Manlove LS, Vessella RL, Silverstein KA et al. TALEN-engineered AR gene rearrangements reveal endocrine uncoupling of androgen receptor in prostate cancer. Proc Natl Acad Sci USA 2013; 110: 17492–17497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Watson PA, Chen YF, Balbas MD, Wongvipat J, Socci ND, Viale A et al. Constitutively active androgen receptor splice variants expressed in castration-resistant prostate cancer require full-length androgen receptor. Proc Natl Acad Sci U S A 2010; 107: 16759–16765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dubbink HJ, Hersmus R, Verma CS, van der Korput HA, Berrevoets CA, van Tol J et al. Distinct recognition modes of FXXLF and LXXLL motifs by the androgen receptor. Mol Endocrinol 2004; 18: 2132–2150. [DOI] [PubMed] [Google Scholar]
- 45.van Royen ME, van Cappellen WA, de VC, Houtsmuller AB, Trapman J. Stepwise androgen receptor dimerization. JCell Sci 2012; 125: 1970–1979. [DOI] [PubMed] [Google Scholar]
- 46.Shaffer PL, Jivan A, Dollins DE, Claessens F, Gewirth DT. Structural basis of androgen receptor binding to selective androgen response elements. Proc Natl Acad Sci U S A 2004; 101: 4758–4763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kounatidou E, Nakjang S, McCracken SRC, Dehm SM, Robson CN, Jones D et al. A novel CRISPR-engineered prostate cancer cell line defines the AR-V transcriptome and identifies PARP inhibitor sensitivities. Nucleic Acids Research 2019; 47: 5634–5647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cai C, Balk SP. Intratumoral androgen biosynthesis in prostate cancer pathogenesis and response to therapy. EndocrRelat Cancer 2011; 18: R175–R182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ta HQ, Dworak N, Ivey ML, Roller DG, Gioeli D. AR phosphorylation and CHK2 kinase activity regulates IR-stabilized AR–CHK2 interaction and prostate cancer survival. eLife 2020; 9: e51378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bai S, Cao S, Jin L, Kobelski M, Schouest B, Wang X et al. A positive role of c-Myc in regulating androgen receptor and its splice variants in prostate cancer. Oncogene 2019; 38: 4977–4989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dong Y, Zhang H, Gao AC, Marshall JR, Ip C. Androgen receptor signaling intensity is a key factor in determining the sensitivity of prostate cancer cells to selenium inhibition of growth and cancer-specific biomarkers. Mol Cancer Ther 2005; 4: 1047–1055. [DOI] [PubMed] [Google Scholar]
- 52.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 2011; 12: 323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol 2010; 11: R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cao S, Ma T, Ungerleider N, Roberts C, Kobelski M, Jin L et al. Circular RNAs add diversity to androgen receptor isoform repertoire in castration-resistant prostate cancer. Oncogene 2019; 38: 7060–7072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dong Y, Lee SO, Zhang HT, Marshall J, Gao AC, Ip C. Prostate specific antigen expression is down-regulated by selenium through disruption of androgen receptor signaling. Cancer Research 2004; 64: 19–22. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.