Abstract
Mapping the gene regulatory networks dysregulated in human disease would allow the design of network-correcting therapies that treat the core disease mechanism. However, small molecules are traditionally screened for their effects on one to several outputs at most, biasing discovery and limiting the likelihood of true disease-modifying drug candidates. Here, we developed a machine learning approach to identify small molecules that broadly correct gene networks dysregulated in a human induced pluripotent stem cell (iPSC) disease model of a common form of heart disease involving the aortic valve. Gene network correction by the most efficacious therapeutic candidate, XCT790, generalized to patient-derived primary aortic valve cells and was sufficient to prevent and treat aortic valve disease in vivo in a mouse model. This strategy, made feasible by human iPSC technology, network analysis, and machine learning, may represent an effective path for drug discovery.
One Sentence Summary:
A machine learning approach identifies a therapeutic that corrects dysregulated gene networks and treats cardiac valve disease in vivo.
Main Text:
Determining the gene regulatory networks that drive human disease allows the design of therapies that target the core disease mechanism rather than symptomatic management. However, small molecules utilized as therapeutic agents are traditionally screened for their effects on one to several outputs at most. Their predicted efficacy on the disease is then extrapolated from those narrow data. In principle, mapping the architecture of the dysregulated network to elucidate key nodes could overcome this obstacle by screening for molecules that correct a gene network’s core regulatory elements rather than peripheral downstream effectors that may not be disease-modifying. There has been much interest in cataloging gene network effects of small molecule libraries and integrating effects of a compound with dysregulated pathways in disease for further testing (1, 2). However, those efforts are typically in standard laboratory cell lines that often do not represent the cells affected in the disease of interest. Disease biology is often cell-type specific so approaches harnessing human induced pluripotent stem cell (iPSC) and gene-editing technologies would allow direct screening of libraries for evaluation of gene network correction in human disease-relevant cells. The advent of targeted RNA-seq methods, which focus sequencing on select transcripts of interest (3), could permit cost-effective high-throughput screening for network-correcting molecules by determining their effect on 100–200 signature transcripts within the disease network.
We previously reported that heterozygous loss-of-function NOTCH1 (N1) mutations cause aortic valve (AV) stenosis and calcific AV disease (CAVD) (4), common forms of human heart disease. Genetic disruption of Notch signaling in mouse valve endothelium results in valve thickening and calcification, suggesting endothelial cells (ECs) as the cell type responsible for valve disease caused by insufficient N1 signaling (5). In the disease setting, ECs undergo an abnormal mesenchymal transition into valve interstitial cells, which are susceptible to developing an osteoblast-like state leading to calcification (5). In human iPSC-derived ECs from patients with N1 heterozygosity, similar gene dysregulation was observed with adoption of an osteogenic-like gene program and activation of inflammatory pathways (6). Integrating genome-wide N1 DNA-binding data with epigenetic and transcriptomic changes in N1 heterozygous arterial-like iPSC-derived endothelial cells (ECs) revealed the underlying transcriptional mechanism (6) that results in reprogramming of valve cells into osteoblast-like cells (7).
N1 is a transmembrane receptor that is cleaved in response to fluid shear stress (6), resulting in nuclear translocation of the intracellular domain, which functions as a transcriptional regulator. N1 has been implicated in ~4% of sporadic CAVD cases through discovery of de novo genetic mutations as well as more broadly through transcriptional repression of N1 (8–10). N1 is also associated with a congenital anomaly present in 1–2% of the population where the AV has two, rather than three, leaflets (bicuspid AV (BAV)). Although BAV is typically asymptomatic at birth, most develop AV stenosis with age, usually earlier than those with tricuspid AV (TAV) (11). Despite the discovery of N1 haploinsufficiency as a genetic cause, the only current treatment for AV stenosis and CAVD is surgical valve replacement, necessitating over 100,000 valve replacements annually in the United States alone (4). Given that valve calcification progresses with age, a medical therapy that could arrest or even slow progression would have tremendous impact.
Here, we performed a network-based drug screen to identify small molecules that corrected the gene network dysregulated by N1 haploinsufficiency in isogenic human iPSC-derived ECs from a patient affected by AV stenosis and CAVD. The restorative effect of the top corrector, XCT790, on dysregulated network nodes generalized to a panel of primary AV ECs from patients with sporadic CAVD. Furthermore, XCT790 prevented and treated N1-dependent cardiac valve disease in mice, providing in vivo validation and representing a promising candidate for a medical therapy of cardiac valve disease in humans. Thus, network-based screening can distinguish molecules with broadly restorative effects on gene networks dysregulated in human disease, translating to efficacy in vivo.
Map of gene network dysregulated by N1 haploinsufficiency
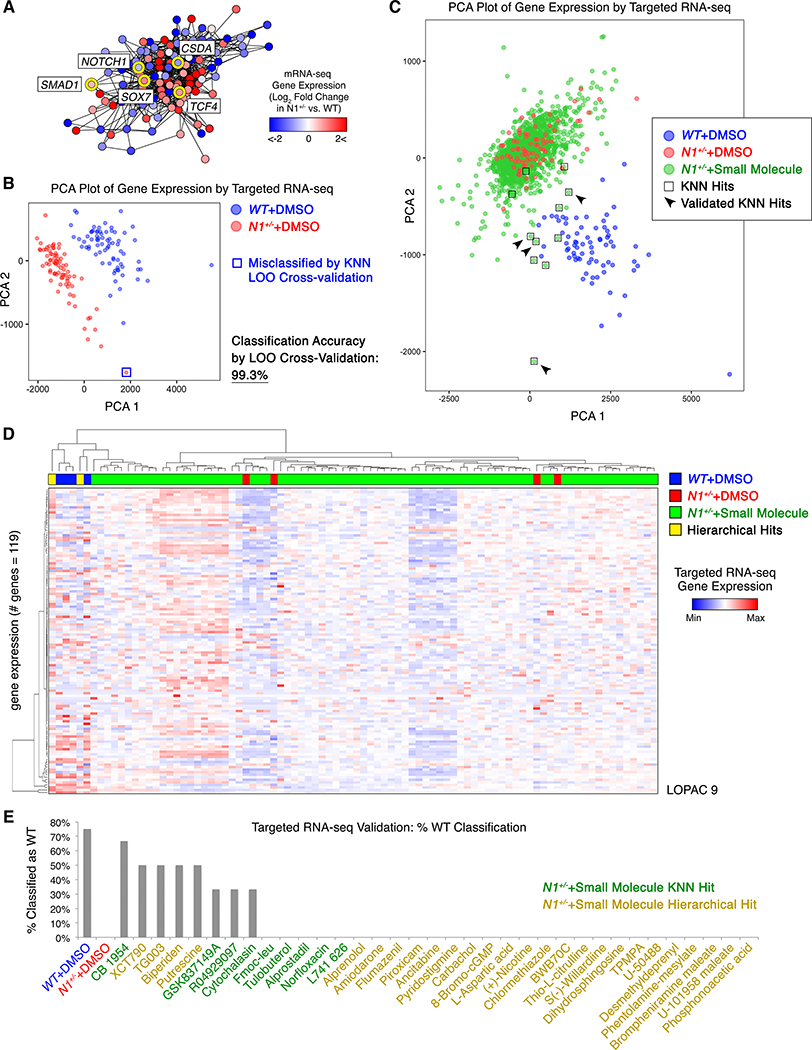
To map the gene network disrupted by N1 haploinsufficiency and identify small molecules that correct the network back towards the normal state, we designed a targeted RNA-seq strategy assaying expression of 119 signature genes in patient-derived N1+/− iPSC-derived ECs or gene-corrected isogenic cells (6, 12), exposed to either DMSO or each of 1595 small molecules (from LOPAC and in-house libraries). The selected 119 target genes were either predicted central regulatory nodes or peripheral genes positioned within varied regions of the N1-dependent network in human iPSC-derived ECs as we previously described by whole transcriptome RNA-seq (6) (Data S1).
The large number of replicates of isogenic WT and N1-haploinsufficient ECs treated with DMSO or small molecules allowed us to map the gene network regulated by N1 to a higher degree of confidence than in previous experiments with fewer observations of the network state. Network inference predicted that SOX7 and the WNT signaling effector TCF4, both of which are upregulated by N1 haploinsufficiency and are potentially pro-osteogenic genes, were highly connected within the dysregulated network, with SOX7 being the third most highly connected gene overall (Fig. 1A, Fig. S1A–B). Concordant with prior perturbation experiments (6), BMP signaling effector SMAD1, which is also upregulated by N1 haploinsufficiency, was predicted to be more peripheral within the network. The most highly interconnected gene was CSDA, which is downregulated by N1 haploinsufficiency and serves as an anti-inflammatory repressor of the GM-CSF promoter (13). The endothelial transcription factors ETS1 and ETS2 were also highly interconnected, as expected given the cell type.
Fig. 1. Targeted RNA-seq and machine learning in human iPSC-derived cells revealed CAVD network-correcting small molecules.
(A) Map of the gene network dysregulated by N1 haploinsufficiency in human iPSC-derived ECs (network inference based on DMSO-treated WT n=72 or N1+/− n=79 and single replicates of N1+/− treated with one of 1595 molecules). (B) Principal component analysis (PCA) plot of gene expression in WT (n=72) or N1+/− (n=79) ECs. LOO=leave-one-out. (C) PCA plot of gene expression in N1+/− ECs treated with one of 1595 molecules (each n=1) compared to DMSO-treated N1+/− (n=79) or WT (n=72) ECs. (D) Example of hierarchical clustering of transcriptional profile of N1+/− ECs treated with each of the small molecules (from LOPAC library plate 9 of 16) compared to DMSO-treated N1+/− or WT ECs (each column n=1). (E) Percent of targeted RNA-seq validation replicates that classified as WT with true identities of DMSO-exposed WT (n=4), DMSO-exposed N1+/− (n=4), or small molecule-treated N1+/− (KNN hits n=3, hierarchical hits n=2). In (A-E): targeted RNA-seq.
Network-based screen identifies network-correcting small molecules
Having mapped the key regulatory nodes within the network dysregulated by N1 haploinsufficiency, we next screened for small molecules that corrected the network back towards the normal state using a machine learning approach. We trained a K-nearest neighbors (KNN) algorithm to classify the network gene expression by targeted RNA-seq as WT or N1+/− based on isogenic ECs of each genotype exposed to DMSO (WT n=72, N1+/− n=79). The KNN algorithm classified ECs as either WT or N1+/− with 99.3% accuracy by leave-one-out cross-validation. The single N1+/− EC replicate that was misclassified as WT appeared near the boundary of the two classes when mapped onto two principal components (Fig. 1B).
When we applied the trained KNN algorithm to N1+/− ECs exposed to each of 1595 small molecules, the vast majority remained classified as N1+/−; however, 11 molecules sufficiently corrected the network gene expression such that the treated N1+/− ECs classified as WT (Fig. 1C, Data S2). To enhance the diversity of algorithms used to select initial candidates, we also applied hierarchical clustering using a complete agglomeration method to identify additional small molecules that shifted gene expression networks such that treated N1-haploinsufficient ECs clustered with WT ECs (Fig. 1D, Fig. S2–5, Data S2). Of the promising candidates identified by the aforementioned methods, eight compounds sufficiently corrected the network gene expression such that one or more replicates of treated N1+/− ECs were classified as WT by the KNN algorithm in validation trials (Fig. 1E). When plotted on two principal components, N1-haploinsufficient ECs treated with the validated molecules localized more closely to WT ECs than those treated with molecules that did not validate (Fig. 1C).
The validated molecules included compounds predicted to inhibit osteogenesis (XCT790) and atherosclerosis (TG003, GSK837149A), both of which may impact CAVD. Statins, which have been shown to be ineffective in preventing CAVD in clinical trials (14), were not identified by either method to be corrective molecules (9 statins tested), serving as validated negative controls. Overall, the network-based screening approach identified molecules that had a broad restorative impact on the gene network dysregulated in N1 haploinsufficient ECs.
Network-correcting molecules, most prominently XCT790, normalize aberrant pathways
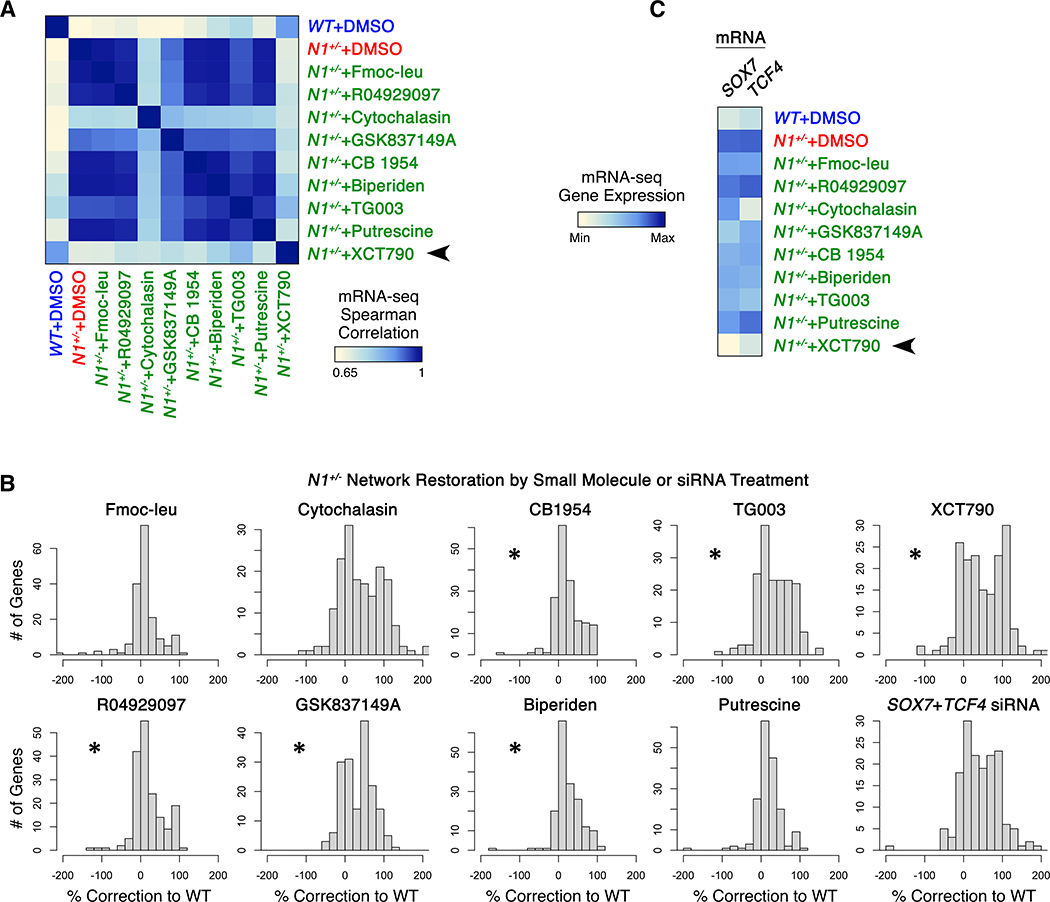
We next performed whole transcriptome RNA-seq in triplicate to determine the effect of each identified network-correcting small molecule on the entire transcriptional landscape (Fig. 2A). We included Fmoc-leu, a molecule that promoted classification of N1+/− ECs as WT in the initial screen but did not validate, as a negative control. XCT790-treated ECs correlated most strongly with the transcriptional profile of WT ECs (Fig. 2A). XCT790 effectively restored the transcription of mesoderm and cell cycle checkpoint genes and downregulated cadherin-11, repression of which prevents CAVD in N1+/− mice exposed to high fat diet (15) (Fig. S6A).
Fig. 2. Network-correcting small molecules broadly restored NOTCH1-dependent transcriptional dysregulation.
(A) Correlation of gene expression in WT ECs with gene expression in N1+/− ECs exposed to indicated molecules. (B) Extent of N1-dependent network restoration in N1+/− ECs treated with indicated molecules or siRNA targeting SOX7 and TCF4 (*p<0.05, one-sided t-test vs. negative control Fmoc-leu, most to least significant: XCT790, TG003, GSK837149A, Biperiden, CB1954, R04929097). Positive values indicate correction towards or past WT expression level. Negative values indicate worsened dysregulation. (C) Gene expression of SOX7 and TCF4 regulatory nodes in response to indicated molecule treatment. In (A-C): whole transcriptome RNA-seq, n=3.
Given SOX7 and TCF4’s predicted role as central regulatory nodes within the network dysregulated by N1 haploinsufficiency, correction of these nodes would be predicted to have a broad restorative effect on the network as a whole. Indeed, siRNA directly targeting these nodes broadly corrected both upregulated and downregulated genes in N1-haploinsufficient ECs back towards their normal state (Fig. 2B, S6B). Concordantly, XCT790, the compound with the strongest restorative effect on these regulatory nodes, drove N1-haploinsufficient ECs to correlate most strongly with WT ECs and corrected the greatest number of genes to the highest degree among the compounds identified in the network-based screen (Fig. 2B–C, S6C). Thus, chemical treatment by XCT790 that corrected the expression of central regulatory nodes SOX7 and TCF4 appeared to be the most effective at restoring the network as a whole.
XCT790 restores pathways dysregulated in primary AV cells from patients with sporadic CAVD
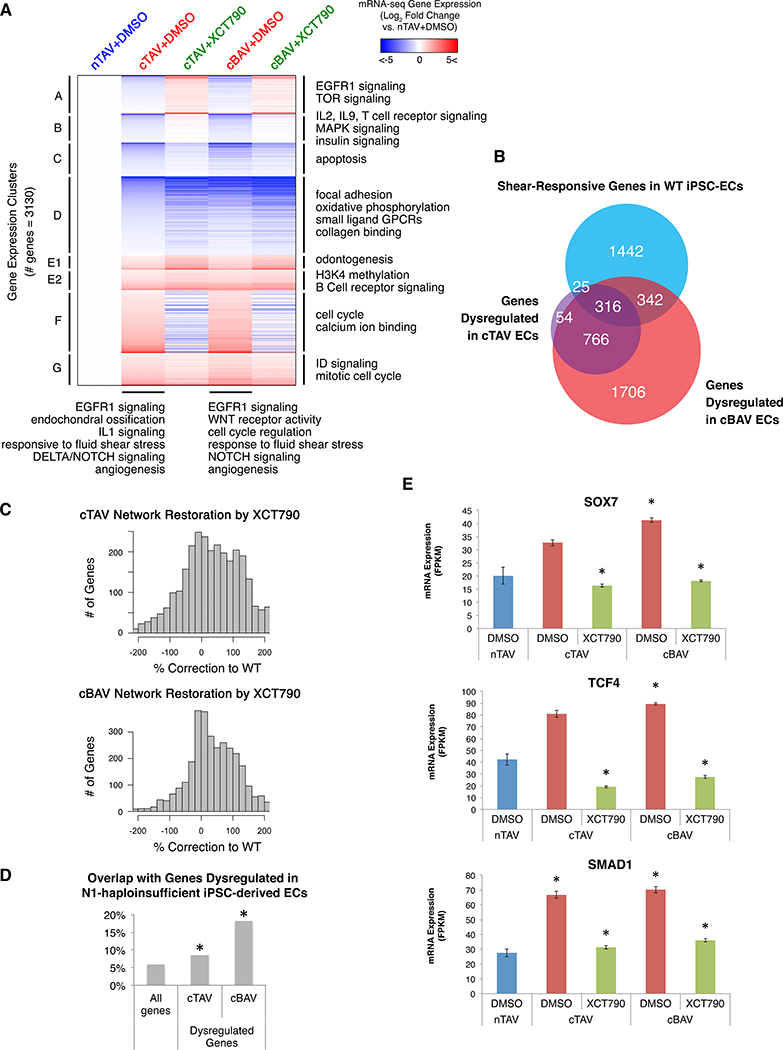
As treatment with XCT790 was sufficient to cluster N1+/− with WT iPSC-derived ECs, we tested whether the effect of XCT790 could generalize to primary AV ECs from multiple patients with sporadic CAVD. We performed RNA-seq in primary human AV ECs cultured from explanted normal tricuspid AVs (nTAVs, n=5), calcified tricuspid AVs (cTAVs, n=9), and calcified bicuspid AVs (cBAVs, n=12) treated with XCT790 or DMSO. While N1 haploinsufficiency in humans is linked to the developmental malformation of BAV, there are individuals with N1 heterozygous mutations that develop AV calcification even with TAV, indicating that the osteogenic fate switch leading to calcification due to N1 haploinsufficiency is not solely linked to the bicuspid malformation (4). 1161 genes were dysregulated in cTAV ECs compared to nTAV ECs, and top dysregulated GO pathways included EGFR1 signaling (previously implicated in both valve development and atherosclerosis (16)), endochondral ossification, IL1 signaling, Delta-Notch signaling, and angiogenesis (Fig. 3A, Data S3–4). Of these genes, 1082 were also dysregulated in cBAV ECs, along with another 2048 genes for a total of 3130 genes dysregulated in cBAV ECs compared to nTAV ECs. Genes dysregulated in cBAV ECs were also enriched for GO pathways including angiogenesis, cell cycle regulation, WNT receptor activity, and EGFR signaling. Genes dysregulated in both cTAV and cBAV ECs were enriched for shear stress-responsive genes, indicating that the response to shear stress may be defective in these valves, similarly to in N1-haploinsufficient ECs (Fig. 3B). There were no genes significantly dysregulated in cBAV ECs compared to cTAV ECs, suggesting an overall common mechanism for calcification in both tricuspid and bicuspid AVs.
Fig. 3. XCT790 alleviates dysregulation in primary ECs from calcified human aortic valves.
(A) mRNA expression by whole transcriptome RNA-seq of genes dysregulated in primary valve ECs from patients with cTAV or cBAV as compared to ECs from patients with nTAV in response to DMSO or XCT790 treatment (p<0.05, negative binomial test, FDR correction). (B) Overlap of genes dysregulated in cTAV or cBAV with previously reported shear-responsive genes in iPSC-derived ECs (6) (p<0.05, X2 test, Bonferroni correction). (C) Extent of dysregulated gene restoration towards nTAV in cTAV or cBAV ECs treated with XCT790. (Positive values indicate correction towards or past nTAV expression level. Negative values indicate worsened dysregulation.) (D) Overlap of genes dysregulated in cTAV or cBAV with genes dysregulated in N1-haploinsufficient iPSC-derived ECs as previously reported in Theodoris et al. 2015 (p<0.05, X2 test, Bonferroni correction). (E) Expression of the key regulatory nodes SOX7, TCF4, and SMAD1 in nTAV, cTAV, or cBAV ECs by RNA-seq (*p<0.05, negative binomial test, FDR correction). In (A-E): nTAV n=5, cTAV n=9, cBAV n=12.
XCT790 was effective in broadly correcting the dysregulated genes back to the normal state in both primary cTAV and cBAV ECs (71% and 69% of genes, respectively), including EGFR signaling, shear-responsive, and cell cycle genes (Fig. 3A, 3C). Overall, there was a significant overlap in genes dysregulated in N1-haploinsufficient ECs with those dysregulated in the same direction in cTAV ECs and cBAV ECs (Fig. 3D). N1 was significantly upregulated in cBAV ECs, potentially reflecting a compensatory response to primary defects in a downstream region of the network, which was significantly relieved by treatment with XCT790 (Fig. S7). NOTCH2, however, was significantly downregulated in both cTAV and cBAV ECs but was also corrected towards normal by XCT790.
SOX7, TCF4, and SMAD1 were key pro-calcific regulatory nodes significantly upregulated in N1-haploinsufficient iPSC-derived ECs. Concordant with prior network inference and perturbation experiments in N1-haploinsufficient iPSC-derived ECs (6), the inferred gene network governing primary AV ECs mapped SOX7 and TCF4 as more central regulatory nodes, with TCF4 being the third most highly connected gene overall, whereas SMAD1 was inferred to be more peripheral within the network (Fig. S8A). Analogously to N1-haploinsufficient iPSC-derived ECs, SOX7 and TCF4 were significantly upregulated in cBAV ECs, showed a trend of upregulation in cTAV ECs, and were corrected towards normal by XCT790 (Fig. 3E). SMAD1 was significantly upregulated in both cTAV and cBAV ECs and was also corrected towards normal by XCT790 (Fig. 3E).
The most highly connected gene within the inferred network in primary AV ECs was the ETS factor ERG, which promotes chondrocyte development as well as endocardial-mesenchymal transformation in cardiac valve morphogenesis (17, 18) (Fig. S8B–C). ERG was significantly upregulated in cTAV and cBAV ECs, and XCT790 significantly normalized ERG expression in both settings (Fig. S7). ELK3, an ETS factor in the TCF subfamily that is also involved in vascular development but later becomes restricted to cartilage, was the fourth most highly connected gene in the inferred network and also was upregulated in cTAV and cBAV ECs but was corrected to normal by XCT790. ERG and ELK3 upregulation potentially indicates a chondrocyte identity switch and promotion of endothelial-mesenchymal transformation, which have both been previously linked to valve calcification and may be alleviated by XCT790 treatment. CEBPB, which was also highly connected within the inferred network, induces cartilage degradation (19) and was significantly downregulated in cBAV ECs. CEBPB expression was upregulated by XCT790 treatment in both cTAV and cBAV ECs, which may protect against valve calcification.
The significant overlap of dysregulated genes including key nodes SOX7, TCF4, and SMAD1 and efficacy of XCT790 in correcting this dysregulation in both settings supports a congruent mechanism of calcification in sporadic cases to that of N1 haploinsufficiency, which may thus represent a model for CAVD where effective compounds generalize to sporadic disease.
Network-correcting molecules reduce AV disease in vivo
Recognizing that mice have much longer telomeres than humans (20), we previously reported that N1+/− mice with telomeres shortened to be more similar to human telomeres develop age-dependent AV thickening, calcification, and stenosis as well as pulmonary valve (PV) stenosis, mimicking the range of human disease caused by N1 haploinsufficiency (7). To determine whether the identified network-correcting molecules were sufficient to prevent cardiac valve disease caused by N1 haploinsufficiency in vivo, we performed a pre-clinical trial in N1+/− mice intercrossed for two generations (G2) with mice lacking telomerase activity due to mutations in the telomerase RNA component (mTR). As the valve phenotype is incompletely penetrant, we tested a total of 6 of the 8 compounds in pilot studies of 40 mice randomized to compound or control solvent, omitting biperiden due to solubility issues and cytochalasin due to known toxicity that would likely be incompatible with chronic use, and adding Fmoc-leu as a negative control. Mice were treated daily for 4 weeks starting at 4 weeks of age with no signs of adverse effects or toxicity. At doses tested representing levels tolerable in mice, compounds Fmoc-leu, putrescine, CB1954, RO4929097, and GSK837149A elicited no improvement in peak velocity of blood flow across the AV or PV by echocardiography, a typical measure of clinical valve stenosis (Fig. S9A–D). TG003 and XCT790 showed promising results in pilot studies of AV peak velocity so were repeated with a second cohort. TG003 trended toward improvement but ultimately did not impact disease progression with statistical significance at the trialed dosages (Fig. S10A–F).
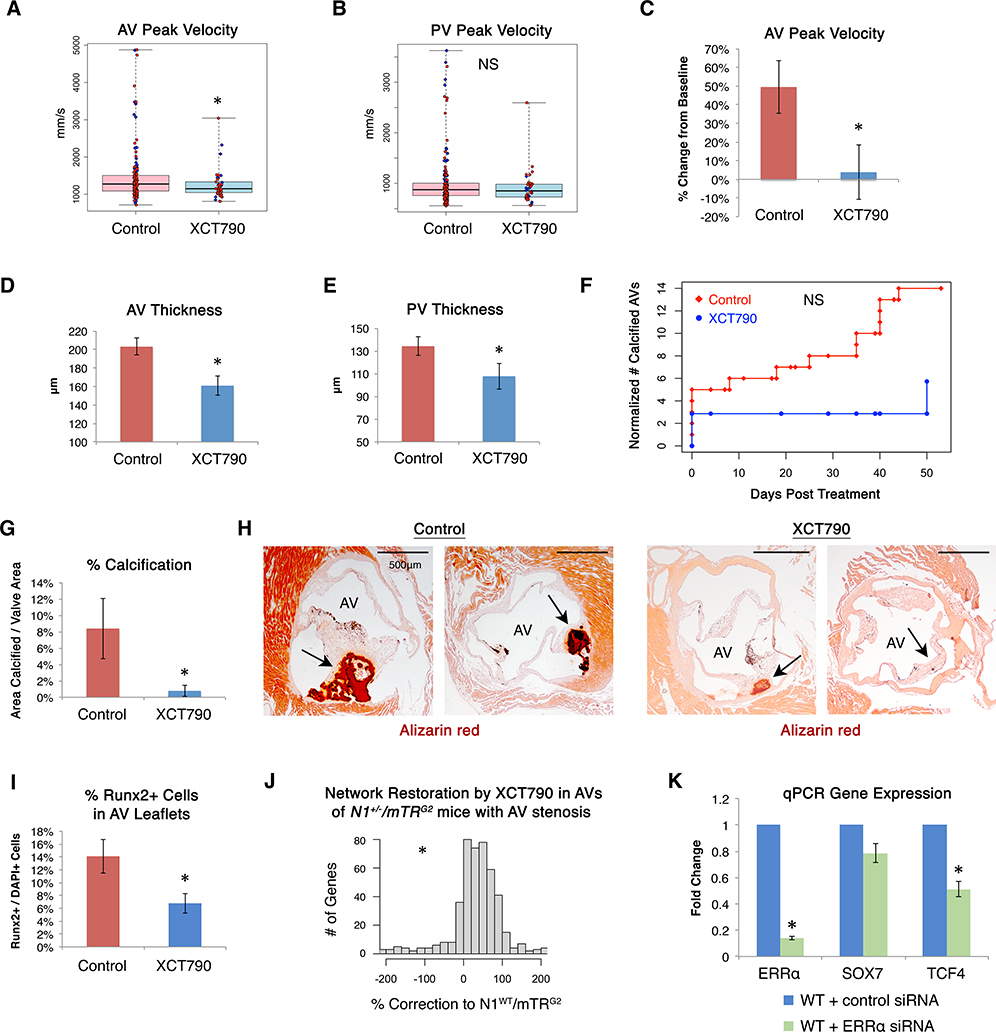
In contrast, XCT790, which most effectively restored the dysregulated gene network in vitro, was sufficient to prevent increased AV peak velocity, a measure of AV stenosis, in vivo by echocardiography (p=0.017) (Fig. 4A). It also showed a trend of reducing PV peak velocity, a measure of PV stenosis (p=0.25) (Fig. 4B). When treating a subset of mice with documented increased AV peak velocity and blood pressure gradient across the valve at 1 month of age with XCT790, we observed arrest of progression and even reversal in some mice within one month of treatment (p=0.033) (Fig. 4C, S10G–I). XCT790 also significantly reduced the thickness of treated AVs (p=0.0018) and PVs (p=0.034) (Fig. 4D–E, Fig. S11A). After the month of treatment, XCT790 showed a trend of reducing the risk of developing any calcification by 59% (Fig. 4F), and of those that did calcify, the extent of calcification was reduced by 91% (p=0.031) (Fig. 4G–H). Furthermore, XCT790 treatment significantly reduced the percentage of cells in AVs expressing the osteoblast transcriptional regulator RUNX2 (21), indicating a reduction in the osteogenic switch underlying CAVD (Fig. 4I, Fig. S11B–C).
Fig. 4. Network-correcting molecule XCT790 prevented and treated aortic valve disease in vivo.
(A) AV or (B) PV peak velocity by echocardiography in N1+/−/mTRG2 mice treated with XCT790 (n=42) or control solvent (n=174) (AV *p=0.017, one-sided Mann-Whitney rank test). (C) AV peak velocity by echocardiography in N1+/−/mTRG2 mice with documented increased AV peak velocity at 4 weeks of age (AV peak velocity higher than any WT/mTRG2 mice) treated with XCT790 or control solvent for 4 weeks (n=4, all male, *p=0.033, one-sided t-test). (D) AV and (E) PV thickness in N1+/−/mTRG2 mice treated with XCT790 (n=20) or control solvent (n=49) (AV *p=0.0018, PV *p=0.034, one-sided t-test). (F) Normalized number of mice with calcified AVs by Alizarin red staining in N1+/−/mTRG2 mice at the indicated number of days after completion of four weeks of treatment with XCT790 (n=37) or control solvent (n=106) to evaluate the sustainability of treatment effects after discontinuation. (G) Of the 106 N1+/−/mTRG2 mice treated with control solvent, 14 mice developed valve calcification (7 females, 7 males). Of the 37 N1+/−/mTRG2 mice treated with XCT790, only 2 developed any valve calcification (1 female, 1 male). Of the N1+/−/mTRG2 mice that developed valve calcification when treated by control solvent (n=14) compared to XCT790 (n=2), those treated with XCT790 had significantly less calcification by Alizarin red staining (*p=0.031, one-sided t-test). (H) Representative AV calcification by Alizarin red staining in mice treated with control solvent or XCT790. “AV”=valve lumen; arrows highlight Alizarin red staining. (I) Percentage of Runx2-positive cells by immunohistochemistry within the AV leaflets of N1+/−/mTRG2 mice treated with XCT790 (n=20) or control solvent (n=19) (*p=0.01, one-sided t-test). (J) Extent of N1-dependent network restoration by whole transcriptome RNA-seq in AVs from N1+/−/mTRG2 mice with increased AV peak velocity at 4 weeks of age treated for 1 month with XCT790 (n=3) compared to control solvent (n=3), where the WT network gene expression is defined by AVs from N1WT/mTRG2 mice with healthy valves at 4 weeks treated with control solvent for 1 month (n=4). All mice were male. Positive values indicate correction towards or past WT expression level. Negative values indicate worsened dysregulation. (*p<0.000001, one-sided Mann-Whitney rank test.) (K) Normalized qPCR gene expression of ERRα, SOX7, or TCF4 in WT ECs exposed to control (n=4) or one of three different ERRα (n=3) siRNAs (*p<0.05 by one-sided t-test). In (A-B): Boxes=interquartile range, whiskers=range, line=median, red circles=female, blue diamonds=male. In (C-E, G, I, K): Errors bars=standard error. NS=non-significant.
Given the incomplete penetrance of AV disease in N1+/−/mTRG2 mice, we compared the gene dysregulation by RNA-seq in N1+/−/mTRG2 AVs, with or without AV disease by echocardiography (increased AV peak velocity and pressure gradient), at one month of age that were then treated for four weeks with control solvent or XCT790 (Fig. 4J, Fig. S11D, Data S5). Compared to N1WT/mTRG2, genes uniquely dysregulated in the AVs of control-treated N1+/−/mTRG2 mice with AV disease (as opposed to N1+/−/mTRG2 mice with healthy AVs) were enriched for GO pathways including steroid hormone receptor activity, bone growth, and WNT signaling. AV transcriptomes from control-treated N1+/−/mTRG2 mice with healthy AVs clustered more closely with N1WT/mTRG2 mice than N1+/−/mTRG2 mice with AV disease. In vivo XCT790 treatment corrected 77% of genes dysregulated in N1+/−/mTRG2 mice with AV disease towards the N1WT/mTRG2 state and promoted clustering with N1WT/mTRG2 and N1+/−/mTRG2 mice with healthy AVs. Although expressed at low baseline levels, among the downregulated genes significantly restored by XCT790 were Sfrp5 and Ace2, both of which have been reported to protect against atherosclerosis, via downregulation of WNT signaling in the case of Sfrp5.
XCT790 is annotated as an inverse agonist of the orphan nuclear receptor ERRα involved in WNT signaling, and indeed XCT790 significantly repressed the canonical ERRα target ACADM (22) in human N1-haploinsufficient iPSC-derived ECs and showed a trend of repressing multiple other downstream targets (Fig. S12A). In N1+/− ECs and primary VECs, ERRα was significantly upregulated at the mRNA level in response to XCT790, likely reflecting a compensatory response to inhibition of ERRα at the protein level (Fig. S7, S12C).
ERRα promotes osteogenesis through WNT signaling specifically in the presence of its coactivator, PPARGC1A (23, 24), which was upregulated in N1-haploinsufficient ECs and reduced by XCT790 (Fig. S12A–B). Concordantly, direct repression of ERRα with three siRNAs in N1+/+ human iPSC-derived ECs also led to a significant reduction in the central regulatory node and WNT signaling effector TCF4, supporting that the mechanism of XCT790 may at least be partially through inhibition of its known target ERRα (Fig. 4K). N1+/− ECs had a greater level of baseline SOX7 and TCF4 upregulation that was not able to be overcome by the ERRα siRNAs tested. However, XCT790 was able to significantly reduce SOX7 and TCF4 even in the N1+/− ECs (Fig. S12C), suggesting that XCT790 may have supplementary effects outside of ERRα inhibition that contribute to its efficacy.
Discussion
This work represents a broad gene network-based drug screen in human disease-relevant iPSC-derived cells that was validated in a range of primary human cells and in a mouse model, identifying potential therapeutic molecules for a target human cardiac disease. Harnessing targeted RNA-seq technology and machine learning enabled the cost-effective interrogation of the effect of nearly 1600 small molecules on a broad panel of signature genes within the disease network as opposed to only a few disease markers. Furthermore, verification that the molecule XCT790 restored key dysregulated nodes in primary ECs cultured from valves of patients with non-familial CAVD validated use of the iPSC-derived EC model and demonstrated extension of findings to sporadic cases. Finally, in vivo testing of molecules discovered using human iPSC-derived ECs revealed that XCT790, the most effective network-correcting molecule in vitro, was sufficient to prevent and treat CAVD in a mouse model of the disease.
XCT790, an inverse agonist of ERRα, is a promising candidate for both N1-dependent familial and sporadic CAVD. Sporadic cases not due to N1 mutations, all of which largely share similar pathology, appear to have comparable gene network dysregulation that ultimately results in calcification, as demonstrated by significant overlap in gene dysregulation including key nodes TCF4, SOX7, and SMAD1 in primary AV ECs. XCT790 corrected gene dysregulation, including in these key nodes, towards normal in primary AV ECs from numerous patients with sporadic CAVD.
Because CAVD is often detected years before intervention and progresses gradually in an age-dependent manner, there is potential for an effective medical therapy to impact the natural history of this disease (11). This is particularly true in the setting of BAV, which is often detected prior to calcification, and is present in 1–2% of the population (11). Simply slowing progression and shifting the age requiring intervention by 5–10 years may avoid tens of thousands of surgical replacements annually. Medical therapy may also impact vascular calcification, which is often associated with CAVD. In addition to reducing calcification, XCT790 also limited thickening of the aortic and pulmonary valves, which are common causes of childhood heart disease requiring intervention (25). The potential of XCT790 to alter the natural progression of childhood, and perhaps even fetal, aortic and pulmonary valve stenosis warrants further study.
In sum, network-based screening, particularly leveraging iPSC and machine learning technologies, is an effective strategy to discover molecules with broadly restorative effects on gene networks dysregulated in human disease that can be validated in vivo. Application of this strategy to other human models of disease may increase the likelihood of identifying disease-modifying candidate therapies that are successful in vivo.
Supplementary Material
Acknowledgements:
We are grateful to Srivastava Lab and Gladstone/UCSF members for scientific discussions. We thank B. Taylor for editorial assistance and manuscript preparation. We thank Gladstone Institutes Histology, Genomics, and Bioinformatics Cores for technical expertise. A Patent Cooperation Treaty patent application was filed in association with this work (PCT/US2019/047295, WO 2020/041333 A1).
Funding:
D.S. was supported by the Winslow Family, L.K. Whittier Foundation, Roddenberry Foundation, Younger Family Fund, California Institute of Regenerative Medicine (DISC2-09098), NHLBI/NIH, and the National Center for Research Resources (U01-HL098179, U01-HL100406, C06-RR018928). S.D. was supported by the Roddenberry Foundation. A.M., V.U., and A.K. were supported by Russian Science Foundation 18-14-00152. C.V.T. was supported by American Heart Association, Roddenberry, Winslow, and UCSF Discovery Fellowships, UCSF DSCB (NIH-T32-HD007470), UCSF Medical Scientist Training Program (NIH-T32-GM007618), Boston Combined Residency Program in Pediatrics, and Harvard Medical School Genetics Residency Program. T.N. was supported by a research fellowship from the Uehara Memorial Foundation. S.S.R. was supported by a training grant from the NHLBI/NIH (T32-HL007544) and a Winslow Fellowship. C.A.G. was supported by a Howard Hughes Medical Institute Fellowship of the Damon Runyon Cancer Research Foundation (DRG-2206-14).
Footnotes
Data and materials availability:
Data is available in the main text, supplementary materials, and Gene Expression Omnibus (GSE125299).
References and Notes:
- 1.Subramanian A et al. , A Next Generation Connectivity Map: L1000 platform and the first 1,000,000 profiles. Cell. 171, 1437 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stathias Vasileios et al. , Drug and disease signature integration identifies synergistic combinations in glioblastoma. Nature Communications. 9 (2018), doi:10.1038/s41467–018-07659-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mercer TR et al. , Targeted RNA sequencing reveals the deep complexity of the human transcriptome. Nat Biotechnol. 30, 99–104 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Garg V et al. , Mutations in NOTCH1 cause aortic valve disease. Nature. 437, 270–274 (2005). [DOI] [PubMed] [Google Scholar]
- 5.Hofmann JJ et al. , Endothelial deletion of murine Jag1 leads to valve calcification and congenital heart defects associated with Alagille syndrome. Development. 139, 4449–4460 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Theodoris CV et al. , Human Disease Modeling Reveals Integrated Transcriptional and Epigenetic Mechanisms of NOTCH1 Haploinsufficiency. Cell. 160, 1072–1086 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Theodoris CV et al. , Long telomeres protect against age-dependent cardiac disease caused by NOTCH1 haploinsufficiency. The Journal of Clinical Investigation. 127, 1683 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mohamed SA et al. , Novel missense mutations (p.T596M and p.P1797H) in NOTCH1 in patients with bicuspid aortic valve. Biochemical and Biophysical Research Communications. 345, 1460–1465 (2006). [DOI] [PubMed] [Google Scholar]
- 9.Foffa I et al. , Sequencing of NOTCH1, GATA5, TGFBR1 and TGFBR2 genes in familial cases of bicuspid aortic valve. BMC Med. Genet. 14, 44 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hadji F et al. , Altered DNA Methylation of Long Noncoding RNA H19 in Calcific Aortic Valve Disease Promotes Mineralization by Silencing NOTCH1. Circulation. 134, 1848–1862 (2016). [DOI] [PubMed] [Google Scholar]
- 11.Otto CM, Calcification of bicuspid aortic valves. Heart. 88, 321–322 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.White MP et al. , Limited Gene Expression Variation in Human Embryonic Stem Cell and Induced Pluripotent Stem Cell Derived Endothelial Cells. Stem Cells. 31, 92–103 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Occhiodoro F, Vadas MA, Shannon MF, Cold shock domain proteins repress transcription from the GM-CSF promoter. Nucleic Acids Research. 24, 2311 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cowell SJ et al. , A Randomized Trial of Intensive Lipid-Lowering Therapy in Calcific Aortic Stenosis. N Engl J Med. 352, 2389–2397 (2005). [DOI] [PubMed] [Google Scholar]
- 15.Clark CR, Bowler MA, Snider JC, Merryman WD, Targeting Cadherin-11 Prevents Notch1-Mediated Calcific Aortic Valve Disease. Circulation. 135, 2448–2450 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Makki N, Thiel K, Miller F Jr., The Epidermal Growth Factor Receptor and Its Ligands in Cardiovascular Disease. IJMS. 14, 20597–20613 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Iwamoto M et al. , Transcription factor ERG and joint and articular cartilage formation during mouse limb and spine skeletogenesis. Dev. Biol. 305, 40–51 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vijayaraj P et al. , Erg is a crucial regulator of endocardial-mesenchymal transformation during cardiac valve morphogenesis. Development. 139, 3973–3985 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hirata M et al. , C/EBPβ and RUNX2 cooperate to degrade cartilage with MMP-13 as the target and HIF-2α as the inducer in chondrocytes. Human Molecular Genetics. 21, 1111–1123 (2011). [DOI] [PubMed] [Google Scholar]
- 20.Kipling D, Cooke HJ, Hypervariable ultra-long telomeres in mice. Nature. 347, 400–402 (1990). [DOI] [PubMed] [Google Scholar]
- 21.Ducy P, Zhang R, Geoffroy V, Ridall AL, Karsenty G, Osf2/Cbfa1: a transcriptional activator of osteoblast differentiation. Cell. 89, 747–754 (1997). [DOI] [PubMed] [Google Scholar]
- 22.Chisamore MJ, Cunningham ME, Flores O, Wilkinson HA, Chen JD, Characterization of a Novel Small Molecule Subtype Specific Estrogen-Related Receptor α Antagonist in MCF-7 Breast Cancer Cells. PLoS ONE. 4, e5624 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Auld KL et al. , Estrogen-related receptor α regulates osteoblast differentiation via Wnt/β-catenin signaling. J. Mol. Endocrinol. 48, 177–191 (2012). [DOI] [PubMed] [Google Scholar]
- 24.Wang H, Wang J, Estrogen-related receptor alpha interacts cooperatively with peroxisome proliferator-activated receptor-gamma coactivator-1alpha to regulate osteocalcin gene expression. Cell Biol. Int. 37, 1259–1265 (2013). [DOI] [PubMed] [Google Scholar]
- 25.Eroğlu AG, Atik SU, Çinar B, Bakar MT, Saltik İL, Echocardiographic Follow-Up of Congenital Aortic Valvular Stenosis II. Pediatr Cardiol. 39, 1547–1553 (2018). [DOI] [PubMed] [Google Scholar]
- 26.Fu Z, Luo B, Li M, Peng B, Wang Z, Effects of Raloxifene on the Proliferation and Apoptosis of Human Aortic Valve Interstitial Cells. Biomed Res Int. 2016, 5473204 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Margolin AA et al. , ARACNE: an algorithm for the reconstruction of gene regulatory networks in a mammalian cellular context. BMC Bioinformatics. 7 Suppl 1, S7 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hagberg AA, Schult DA, Swart PJ, Exploring Network Structure, Dynamics, and Function using NetworkX. SciPy (2008). [Google Scholar]
- 29.Dobin A et al. , STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 29, 15–21 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Trapnell C et al. , Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat Biotechnol. 31, 46–53 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van der Laan MJ, Pollard KS, A new algorithm for hybrid hierarchical clustering with visualization and the bootstrap. stat-www.berkeley.edu, (available at http://stat-www.berkeley.edu/~laan/Research/Research_subpages/Papers/hopach.pdf).
- 32.Chen J, Bardes EE, Aronow BJ, Jegga AG, ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Research. 37, W305–W311 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zambon AC et al. , GO-Elite: a flexible solution for pathway and ontology over-representation. Bioinformatics. 28, 2209–2210 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data is available in the main text, supplementary materials, and Gene Expression Omnibus (GSE125299).