Abstract
Colorectal cancer (CRC) is a heterogeneous disease with various genetic and epigenetic factors leading to difficulties in response to both the therapy and drug resistance. Moreover, even in tumors with similar histopathological characteristics, different responses and molecular features could be observed because of the genetic basis and its interactions with the living environment. Through personalized medicine, we can classify patients into separate groups according to their genetic and epigenetic features and their susceptibility for a specific disease which could help with choosing the best therapeutic approach. In this review, genetic and epigenetic factors that cause heterogeneity in colorectal cancer are evaluated and proper drug administration in both chemotherapy and target therapy are suggested.
Key Words: Colorectal cancer, Personalized medicine, Biomarker, Targeted therapy, Chemotherapy
Introduction
Medical and biological sciences have shown that despite similar phenotype and pathology of a certain disease, etiology is different in patients and patients might respond differently to a single treatment. Regarding this, the variable molecular basis that contributes to different outcomes was detected. Notably, only 25% of cancer patients have responded to selective treatments. Genetic and epigenetic alterations are the main cause of hereditary and sporadic form of CRCs(1). Factors involved in personalized medicine of colorectal cancer include patient’s characteristics and unique properties of tumors that provide information about the prognosis. Source of high heterogeneity in CRC patients is genetic and epigenetic alterations such as CIN, MSI and CIMP. These changes could occur individually or concurrently , which brings about inconsistency in tumors. Another source of heterogeneity in CRC is the signaling pathways. Change in Wnt, RAS-MAPK, OI3K, TGF-β, and P53 in the marker of mutations in critical genes in CRC. Host-tumor interactions are additional source of heterogeneity in individuals affected by this disease. It is believed that these interactions are highly dependent on the genetic composition of normal cells. Therefore, clinical manifestations will occur as the result of the genetic background and genomic changes. In addition to all these factors, tumor microenvironment, background diseases, hormonal alterations, stress, nutritional diet, and lifestyle impact disease heterogeneity.
CRC Heterogeneity
Two types of heterogeneity including inter-tumor (between the subtypes of CRC) and intra-tumor (between the tumors of one person with cellular heterogeneity) lead into different prognosis, drug resistance, and challenges in selecting the optimum treatment (2-7). Heterogeneity in CRC has been discussed in terms of local and systemic variables. Host-tumor interactions play a role as a key variable not only in the tumor but also in the whole organism. These interactions are strongly related to the genetic composition of a normal cell, hence different clinical manifestations are seen in the affected person, which is hidden in their genetic background(8). Tumor’s micro environment with variable cells are major variables between the patients(9). Immunological heterogeneity in CRC is an important issue that more than one decay is in challenge and different criteria until now was approved i.e., MHC antigens alteration enable colorectal cancerous cells to be hidden from immune cells(10) or another achievement was the effectives of anti PDL1 drug? on the specific type of patients, MSI subtype(11). Metastatic patients also show all these heterogeneities(12). Cell-to-cell heterogeneity in one tumor probably has a developmental-like mechanism or occurs in response to an exogenous factor like a drug. Researches have shown that this type of heterogeneity plays a vital role in drug resistance and disease occurrence, and relapse as a consequence(13). Association between heterogeneity and genetic variation changed the normal biological process and signaling pathways in cancerous cells. The main signaling pathway in normal colorectal tissue is Wnt associate with promoting cell proliferation and mutation in downstream molecules. It may cause the inactivation of APC and activation of β-Catenin and enter the cell to tumorigenesis process(14). Wnt pathway with about 93% change undergoes most changes in this disease and tumors have shown disorders in this pathway, regardless of their mutation rate (low or high)(9). Carcinogenesis process of colorectal tissue promote by different bypass pathways that cell growth, proliferation and differentiation. They are receptor tyrosine kinase (RTK) signaling, TGFβ and TNF-α signaling. Receptor or downstream molecules in these pathways is targeted for therapies(15). Besides, tumors with high mutation rate are rich in genetic alterations in TGF-β pathway, while tumors with low mutation level are primarily affected in the P53 pathway (9). Based on transcription profile, tumors are divided into three groups of MSI/CIMP, CIN, and aggressive mesenchymal phenotype. Importantly, primary pathways of MSI and CIMP are dependent on epigenetic changes. Thus, epigenetics is one of the causes of heterogeneity in CRC(6). MicroRNA molecules are the other regulatory biomarkers reported in angiogenesis, disease relapse, and mortality rate. They could work as a reliable tool for diagnosis and therapy (16, 17), i.e., association between miR-19a expression level and poor prognosis or significant correlation of cluster miR-17-92a expression level with relapses in CRC patients (18).
Prognostic and predictive biomarker in CRC
Prognostic and predictive biomarkers are defined as patient selection factor for treatment type and evaluation of the treatment on patients, respectively (19). The most important causes of heterogeneity in CRC used as molecular markers to optimized and tailored treatment regimens and predict the prognosis i.e. MSI and KRAS hopefully become biological prognostic and/or predictive markers. Most diagnosis criteria do not have adequate sensitivity and accuracy regarding multiple studies on the role of KRAS demonstrated that the prognostic indicator of specific mutation type of KRAS or detected only in some stages or recurrence with other abnormalities like p53 mutation (20-22). With conflicting results of conventional biomarkers in CRC, new studies are needed to select the best therapy or predict prognosis . In addition to many detected biomarkers detected, some of which applied in clinic, there is a large number of biomarkers to be approved in CRC. Recently, great efforts have been made to identify novel biomarkers for more effective treatment of the colorectal cancer patients. Three of these markers are telomerase length, telomerase activity and micronuclei frequency(23) which have been proposed as new potential biomarkers to ascertain the prognosis of the disease and predictive respond to treatments. The critical domain in personalized medicine is disease relapse prediction. Several studies have been conducted to find biomarkers for CRC relapse prediction. The expression level of a gene such as MACC1 or micro-RNAs like miR-19a might probably be helpful for this prediction (18), which require further studies. Table 1 demonstrates prognostic and predictive biomarkers in CRC.
Table 1.
Prognostic and predictive biomarkers in CRC
| Tissue derived | Blood derived | |
|---|---|---|
| Prognostic biomarkers | MUC2, SATB2, CK20/CDX2 | preoperative CEA |
| VEGF, Imp3, TNIK, KRAS, NRAS | postoperative CEA | |
| P53, BRAF, miRNAs, MSI | CA19-9, CTC | |
| Predictive biomarkers | KRAS/NRAS, BRAF, PI3K | MSI, CD133 |
Drugs
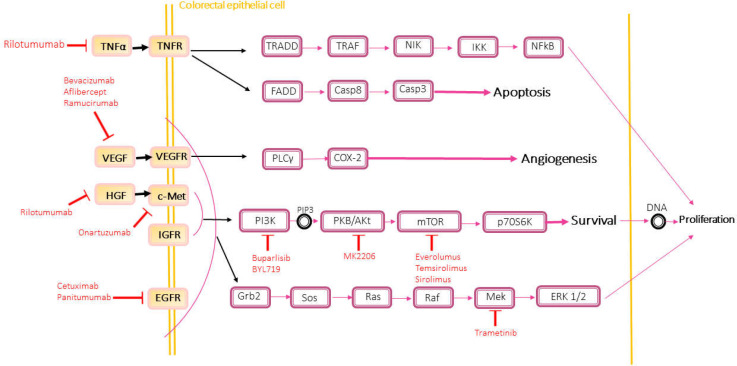
Personalized medicine offers the most efficient therapeutic process for the patients. All the targeted therapy programs presented for CRC refer to the association of critical genes to the central mechanisms of disease progression, as represented in Figure1, all pathways that could be repressed by the targeted drugs to inhibit the proliferation and invasion. However, more research is needed to investigate drug resistance mechanisms in distinct groups of CRC to understand probable prognosis in different categories of the patients. Chemotherapy compositions have been regarded to have had more challenges in recent years. Because of the judgment of CRC patients’ prognosis nowadays is restricted to the limited information based on the high or low frequency of MSI, patient’s MSS status, mutations in BRAF or PIK3CA genes, all of which influence in the identification of the potential tumors’ invasion rate and differentiation, good or poor prognosis, and the degree of progression, and sometimes the tumor’s location. Here two categories of treatment in CRC will explain in details the pharmacogenetics used in clinical trials or in researches.
Figure 1.
All cell signaling pathways blocked by targeted drugs such as Cetuximab which is a monoclonal antibody for blocking epidermal growth factor receptor. Thus, survival, proliferation, migration, invasion, and angiogenesis of cancer cells can be inhibited. Resistance to these drugs is as a result of a mutation in one of the involved genes in these cell signaling
Drugs Related to Targeted Therapies
1.Bevacizumab: this drug is a monoclonal, recombinant human IgG1-antibody that acts against the VEGF-A ligand and is related to VEGFR1 and 2 receptors. In 2006, FDA approved Bevacizumab combinatorial drug based on 5-Fluorouracil as the second-line treatment for mCRC. This drug is prescribed because of the significant results of OS measure improvement in patients receiving Bevacizumab in addition to FOLFOX4. Furthermore, US food and drug administration approved Bevacizumab in combination with Fluoropyrimidine-Irinotecan or Fluoropyrimidine-Oxaliplatin for mCRC patients who have received Bevacizumab in the first-line of treatment, and had progressive disease. Thus, patients were given a new regimen depending on their previous treatment regimen. A significant improvement was seen in OS and PFS indicators of patients who had received Bevacizumab along with the described chemotherapies (24). In addition to metastatic colorectal cancer, this drug is used to treat lung cancers, glioblastoma, and renal cells carcinoma(25).
Variations in VEGFA gene are shown to alter sensitivity to bevacizumab. rs2305948 SNP in VEGFR2 has been associated with diminished bevacizumab treatment outcome. In a study on cancer mCRC patients who were treated with bevacizumab, researchers observed increased PFS/OS associated with rs699947, rs833061, rs2010963, rs3025039 SNPs in VEGFA(26). A large cohort study on mCRC patients revealed an increased risk toxicities associated with a treatment regimen based on first-line 5-flourouracil and irinotecan plus bevacizumab(27). Recently, it has been determined that bevacizumab and alternative splicing of VEGFA could be predictive and prognostic biomarkers respectively, in patients treated with irinotecan-based chemotherapy and bevacizumab respectively (28).
2. Ramucirumab: This drug is also a monoclonal human IgG1 antibody similar to Bevacizumab which is classified in anti-VEGFR drugs group, and inhibits VGFR2 thereby preventing the progress of signaling pathways for angiogenesis, division, and survival(29). In 2015, FDA approved this drug in the second-line treatment in combination with FOLFIRI for treating mCRC patients. The mentioned patients have received Bevacizumab with Oxaliplatin and Fluoropyrimidine in the first-line treatment but were still experiencing disease progression. The treatment regimen of Ramucirumab accompanied by FOLFIRI leads to the improved OS and PFS rates (30). Unfortunately, no biomarker has yet been identified for choosing the patients to respond to this drug. This drug is also used for gastric and lung cancers, and since CRC treatment with this drug does not last long, no unique mechanism is found for the resistance to this drug(31).
3. Aflibercept: This drug which is also known as Zaltrap is an inhibitor of the receptor protein bound to angiogenesis ligands including VEGF-A, VEGF-B, and PLGF to prevent the progression of angiogenesis signaling pathways. In 2012, Zaltrap regimen along with FOLFIRI was approved by the US for patients who were resistant to regimens containing Oxaliplatin, because the research emphasized improved OS in patients accepting this strategy(32). Mechanism of resistance to this drug and other anti-VEGFR drugs are alike and still unknown. As shown in the researches, resistance to VEGF-Trap treatment leads to an increased expression of VEGF-C which bestows the improvement of alternate angiogenesis pathways for adaptation against the drugs(33).
4. Regorafenib: This drug acts by targeting several protein kinases in angiogenesis pathway associated with VEGFR1, VEGFR3, VEGFR2, and TIE2. Its oncogenic targets include KIT, RET, BRAF, and BRAF-V600E. PDGFRα and β are tumor microenvironment targets andp38 MAP kinase, FGFR1 and 2 are other tyrosine kinases that are targets for this drug(34). This drug was approved in 2012 for mCRC patients who had previously used regimens based on Oxaliplatin, Irinotecan, or Fluoropyrimidine or had received anti-VEGFR and anti-EGFR regimens(35). Since the extent of inhibition is immense, the resistance mechanism is too complicated and still unexplained. Some researchers state that overcoming the resistance requires extensive changes. Hence, the Notch signaling pathway has attracted them. This pathway regulates different cellular processes such as proliferation, differentiation, and apoptosis so that it could be a way for resistance to various targeted treatments. In research on colorectal cancer cells resistant to Regorafenib, a high level of Notch-1 expression and transcription factors like HEY1 and HES1 has been observed. Invasiveness of those colorectal cancer cells resistant to Regorafenib reduced the following Notch-1 knockdown (36).
5. Cetuximab: This drug belongs to the anti-EGFR treatments group and is a human recombinant monoclonal IgG1 antibody that acts by blocking the epidermal growth factor receptor. Blockage of this receptor prevents progression of signaling pathways involved in cellular survival, angiogenesis, and invasion. Several studies issued in US food and drug administration approved this drug. It is utilized either in a combinatorial form or as an individual drug(37).
Somatic mutation in EGFR is one of the resistance mechanisms, in which the replacement of serine by arginine in codon 492 (S492R) affects the external domain of this receptor, and causes resistance to Cetuximab drug(38). Researches have shown that this somatic mutation is acquired after receiving anti-EGFR drugs(39). According to molecular analyses, the mutation in BRAF and KRAS genes leads to resistance to anti-EGF agents. In addition to these genes, the mutation in NRAS (a membrane GTPase enzyme which is mutant in 2-5% of the mCRC patients) is effective in causing resistance to Cetuximab and its peer, Panitumumab(22). However, some patients are still resistant to anti-EGFR drugs without mutation in KRAS and BRAF genes. Several experiments have been performed to clarify this issue. For example, it was observed that those individuals who have a mutation in exon 20 of their PIK3CA gene imply resistant to Cetuximab(40). On the other hand, some researches mention mutation, lack of expression or hyper methylation of the PTEN gene linked to unresponsiveness to Cetuximab(41). It is also stated that patients without a mutation in KRAS and BRAF genes encounter enhanced HER2 gene in their samples. Researchers believe that anti-HER2 method could be beneficial for this subtype which is also known as HER2-therapy(42). Another extensive research on over 3000 mCRC patients has also confirmed that more investigations are required to prove the correlation between HER2 overexpression and resistance to anti-EGFR therapeutic strategy or disease relapse(43). MET or hepatocyte growth factor receptor (a tyrosine kinase involved in cell division and apoptosis) could be mentioned among resistance factors to anti-EGFR strategy. It is suggested that MET has the potential to activate PIK3/PKB pathway independent from RAS and could interfere with anti-EGFR strategy(44). High IGF1R expression has also been recognized in 50 to 90% of CRC patients and plays a vital role in oncogenic changes and cancerous cells growth and survival, and is indicative of poor prognosis and resistance to anti-EGFR(45, 46).
6. Panitumumab: This drug is also among epidermal growth factor receptor inhibitors like Cetuximab. It belongs to the human monoclonal IgG2 antibodies. The effect of this drug on OS and PFS rates is similar to Cetuximab; furthermore, the failing of its effects when patients encounter with a mutation in the KRAS gene have been ascertained(47). Mechanism of resistance to Panitumumab is comparable to resistance mechanisms for Cetuximab; the only difference is the observation of acquired mutations in EGFR after treatment with Cetuximab but not for Panitumumab(39).
Targeted drugs related to epithelial cell signaling pathway from CRC mentioned above are shown in Figure 1.
B) Chemotherapies
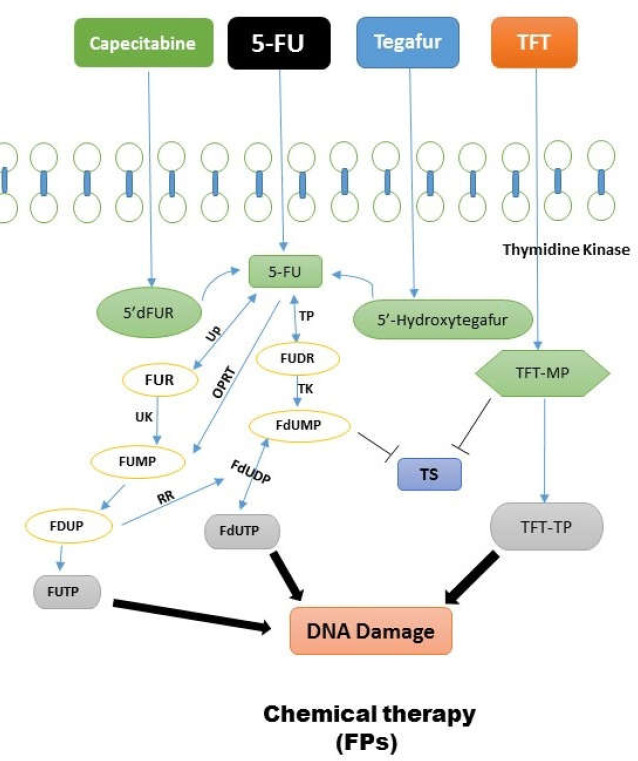
1. Fluoropyrimidines (FPs) family: 5-FU and Capecitabine are part of this group. These drugs provoke cellular death through inducing disturbance in DNA duplication and transcription. Mutation in MMRs and hyper methylation of promoters of mentioned genes could make resistance to this group of drugs(48). 5-FU is a drug from FPs class that has extensive application in colon cancer treatment. This compound as a synthetic fluorinated pyrimidine analog is administered through venous injection. It has several metabolites including FdUMP, 5-FdUTP, and 5-FUTP. This drug tends to produce fluorinated nucleotides by different mechanisms and inserts them in DNA structure instead of thymidine, thereby disrupting duplication and ultimately causing cellular death (see Figure 2). The primary active metabolite of this drug, FdUMP, inhibits TS enzyme, hence preventing dUMP conversion into dTMP and interfering with cell cycle in the cancerous cell(49). TS enzyme is coded by TYMS gene, and the studies designated that an increase in the expression of this gene reduces the effects of this drug or drives resistance to 5-FU(50). In some other studies, it has been mentioned that diminished expression of MTHFR enzyme which is the regulator of cell folate results in elevated inhibition of TS enzyme, enhancing 5-FU drug efficacy(51). Therefore, the SNPs of this enzyme that reduce its activity and causes drug sensitivity were evaluated. In fact, this issue is still under study and has not been approved entirely. Converting 5-FU to the active metabolite needs 5-FdUMP, TP enzymes which are encoded by the TYMP gene. Some investigations showed that high TP levels lead to sensitivity to this drug. Besides, it is declared that this enzyme is involved in metastasis and angiogenesis enhancement(52). OPRT is also another enzyme that plays an essential role in converting 5-FU into the active metabolite, and it is encoded by the UPMS gene. High expression of this gene and the subsequent elevation in OPRT enzyme level is accompanied by sensitivity to 5-FU. However, more studies are required to prove these findings(53).
Figure 2.
Fluoropyrimidines family cause cellular death through inducing disturbance in DNA duplication and transcription
Some in vitro studies on malignant cells detected high expression of an enzyme encoded by the DPYD gene which is responsible for 5-FU catabolism. Indeed, this mechanism leads to the elimination of drug efficacy. Choosing this enzyme as a marker for resistance to 5-FU requires more clinical evidence. Generally, it can be stated that alterations in DPYD, MTHFR, TYMS, and UPMS genes could be potential targets for combatting resistance to 5-FU(52-54). Capecitabine belongs to FPs family and is administered orally. This drug finally converts into 5-FU in tumor cell through specific steps. Thus, it seems that the resistance mechanism for this drug is like 5-FU; high DPD expression is related to resistance to this drug and raised TP expression causes more sensitivity to it(48). Tegafur is also an oral FP like Capecitabine and finally changes into Fluorouracil. Cytochrome P-450 is required to metabolize this drug and is coded by CYP2A6. Researches show that patients whose CYP2A6 gene is wild-type were more sensitive to the drug. Polymorphisms probably involved in the reduced metabolization of the drug are also under examination(55).
Trifluridine/Tipiracil or TAS-102: In 2015, this compound drug earned approval for metastatic colorectal cancer patients. These patients had already used anti-EGFR or anti-VEGFR drugs or chemotherapy regimens containing Irinotecan, Oxaliplatin or Fluoropyrimidine. Both OS and PFS values improved after taking this drug. It is administered orally and contains a Fluoropyrimidine part. Phosphorylation of this part (TFT) by thymidine kinase inhibits TS enzyme similar to other FPs.
Moreover, TFT with three phosphates (TFT-TP) is inserted into DNA structure and disturbs duplication. It has probably a resistance mechanism similar to FPs. Resistance mechanism for this drug has not currently been defined(48).
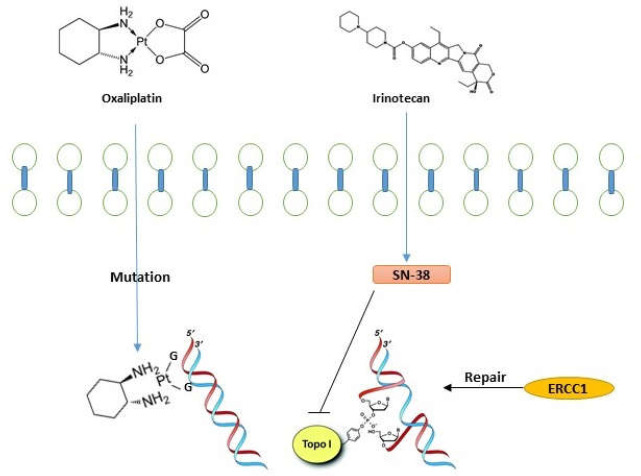
2. Oxaliplatin: This drug is a platinum compound that produces cross-linkages in DNA by DNA-Pt adduct (see Figure. 3).
Figure 3.
Oxaliplatin is a compound that produces cross-linkages in DNA. This defect can be repaired by MMR system and NER mechanism. Irinotecan effect on duplication and transcription by inhibiting topoisomerase I enzyme
Afterwards, the MMR system identifies these defects and induces apoptosis in cells. Repair of DNA damages through the NER mechanism could explain chemotherapy failure. High expression of ERCC1 and ERCC2 proteins which are involved in this mechanism is seen followed by a weak response to Oxaliplatin-based regimen. Furthermore, numerous polymorphisms in ERCC1 gene could be a suitable prediction candidate for response to treatment(52, 56). On the other hand, BRCA1 protein has an essential role in repairing the DNA cross-links, and any kind of change in this gene or its epigenetic knockdown which rarely occurs in CRC is indicative of sensitivity to Oxaliplatin. The interaction between BRCA1 gene products and SRBC gene determines the role of this factor in responding to this drug, because it has been perceived that elevated SRBC expression accompanies with sensitivity to the drug. Besides, it seems that SRBC is involved in trafficking of intracellular vesicles (57, 58). Efflux of drug from the cell is another theory for explaining drug resistance. Combination of Platinum with glutathione (GSH), an antioxidant molecule which prevents nucleic acid oxidative damages, causes drug efflux from the cell by ABC transporter proteins. Investigations showed that high GSH expression levels in tumors induce resistance to Platinum(59).
3. Irinotecan: This drug interferes with DNA duplication by inhibiting topoisomerase I enzyme, thereby causing cell death (see Figure 3). This drug acts better when combined with FPs and produces a remarkable improvement in OS and PFS values. The active metabolite of this drug (SN-38) reversibly forms a complex with TOPO I and DNA. Any alteration in the activity of this complex decreased TOPO I expression, or low SN-38 level could make resistance to Irinotecan. Cytochrome P-450 enzymes convert Irinotecan into an inactive metabolite, and then carboxylesterase 1 and 2 produce SN-38 with hydrolyzing. Some studies revealed that carboxylesterase activity is correlated with drug sensitivity(60). Besides, drug efflux by ABC proteins creates resistance to this drug. Polymorphisms in transporter proteins explain a variety of toxicity in patients(61, 62). Researches have shown that TOPO 1 expression rate or copy numbers could be useful in drug resistance, meaning that response rate to the mentioned drug is directly correlated with TOPO 1 gene copy numbers. Point mutations in TOPO1 gene which damage its binding sites with SN-38 also cause drug resistance(63, 64). The major enzyme responsible for glucuronidation of SN-38 is UGT1A1 which metabolizes and detoxifies irinotecan. FDA of the US has approved this as a method for the prediction of irinotecan-related acute diarrhea and neutropenia.
To increase the efficacy and patient’s safety, UGT1A1 genotyping was initiated for dose-escalated irinotecan in mCRC. Patients with homozygous UGT1A1*28/*28 are more vulnerable to irinotecan, while patients with heterozygous UGT1A1*1/*28) encounter are exposed to an increased risk of irinotecan toxicity and patients with the homozygous UGT1A1*1/*1 genotype are more resistant to irinotecan(65).
Future perspective
Since personalized medicine works on three subjects of determining disease indices in people, choosing the best therapeutic method and predicting disease relapse, it seems that regarding colorectal cancer, more researches are required in order to achieve favorable results. For determining the indices of familial colorectal cancer, familial background can be traced. Considering the most common inherited colon cancer which is known as Lynch syndrome, the presence of people affected by this disease in the family and follow-up of PMS2, MSH2, MSH6, and MLH1 genes might be helpful. The index for familial adenomatous polyposis (FAP) is also a mutation in the APC gene. Reports of the possible role of other genes such as KIF23, CENPE, MUTYH, POLE, and POLD1 in genetic predisposition for CRC and presence of a mutation in specific genes in populations affected by CRC are all suggestive of unidentified features of personalized medicine for colorectal cancer. Much needs to be clarified regarding the indices of sporadic colorectal cancer that may firstly result in early diagnosis of the disease and then the patients’ targeted therapy. Although many mutations and signaling pathways involved in the disease have been identified, an efficient index for predicting the sporadic type of this disease has not yet been introduced. The reason for this is the variety in genetic and molecular details of the mentioned disease. The role of studies based on personalized medicine for this disease is irrefutable. The overall results of these researches could help efficient classification of CRC patients because the existing classifications have several deficits.
Regarding the variety in response to drugs for colorectal cancer, the need for more researches about patient’s classification is being felt. Various drug resistances in these patients demonstrate that molecular information available about this disease is insufficient for understanding its complexities. Consequently, additional studies about all the individual mentioned subjects in the drug resistance factors could be carried out. Extensive studies on personalized medicine in breast cancer treatment have led into specific classification for this disease and treatments are more targeted and efficient than before. Thus, mortality rate and drug resistance during treatment have reached the minimum levels. Systemic and comprehensive genomics, proteomics, and metabolomics studies along with bioinformatics evaluations are of great benefit.
Patient derived xenografts (PDX) are models of cancer applied clinically and do not determine all limitations, but the number of them is the insufficient amount of fresh tissue in some case or the model not established for all patients. Data analysis from samples used for drug screening by PDX and tumor organoid represents a similar result. Genomic analysis between native tumor tissue and both models show nearly complete concordance.
This study sheds light on the integrated data obtained from methods, including genetic screening, bioinformatics algorithms and PDX, and organoid models might be helpful in diagnosis and proper drug administration in both chemotherapy and target therapy to predict the prognosis.
Conclusion
With all chemotherapies or targeted therapies in colorectal cancer, the most critical factor in treating patients is to focus on early diagnosis and development of studies that work on early diagnosis biomarkers. Pharmacogenomic and pharmacogenetics discoveries have recently begun necessitating the inclusion of various populations. All of which requires the development of molecular methods, availability, and low cost. On the other hand, regardless of sophisticated molecular methods, it is possible to find the effective drug and the appropriate dosage for the patient's tumor spheroids, solely by cellular methods and focusing on heterozygous tumor cells in the presence of various therapeutic regimens in the laboratory.
Acknowledgment
This study was part of a Ph.D. thesis of student Padina Vaseghi Maghvan, which is financially supported by Proteomics research Centre of Shahid Beheshti University of Medical Sciences (grant NO. 5283).
Conflict of interests
The authors declare that they have no conflict of interest.
References
- 1.Safaei A, Sobhi S, Rezaei-Tavirani M, Zali MR. Genomic and epigenetic instability in colorectal cancer. Iran J Cancer Prev. 2013;6:54–63. [PMC free article] [PubMed] [Google Scholar]
- 2.Blanco-Calvo M, Concha Á, Figueroa A, Garrido F, Valladares-Ayerbes M. Colorectal cancer classification and cell heterogeneity: a systems oncology approach. Int J Mol Sci. 2015;16:13610–32. doi: 10.3390/ijms160613610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rezaie-Tavirani M, Fayazfar S, Heydari-Keshel S, Rezaee MB, Zamanian-Azodi M, Rezaei-Tavirani M, et al. Effect of essential oil of Rosa Damascena on human colon cancer cell line SW742. Gastroenterol Hepatol Bed Bench. 2013;6:25. [PMC free article] [PubMed] [Google Scholar]
- 4.Markowitz SD, Bertagnolli MM. Molecular basis of colorectal cancer. N Engl J Med. 2009;361:2449–60. doi: 10.1056/NEJMra0804588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lao VV, Grady WM. Epigenetics and colorectal cancer. Nature reviews Gastroenterol Hepatol. 2011;8:686. doi: 10.1038/nrgastro.2011.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Herrera M, Islam AB, Herrera A, Martín P, García V, Silva J, et al. Functional heterogeneity of cancer-associated fibroblasts from human colon tumors shows specific prognostic gene expression signature. Clin Cancer Res. 2013;19:5914–26. doi: 10.1158/1078-0432.CCR-13-0694. [DOI] [PubMed] [Google Scholar]
- 7.Siena S, Sartore-Bianchi A, Di Nicolantonio F, Balfour J, Bardelli A. Biomarkers predicting clinical outcome of epidermal growth factor receptor–targeted therapy in metastatic colorectal cancer. J Natl Cancer Inst. 2009;101:1308–24. doi: 10.1093/jnci/djp280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berdiel-Acer M, Sanz-Pamplona R, Calon A, Cuadras D, Berenguer A, Sanjuan X, et al. Differences between CAFs and their paired NCF from adjacent colonic mucosa reveal functional heterogeneity of CAFs, providing prognostic information. Mol Oncol. 2014;8:1290–305. doi: 10.1016/j.molonc.2014.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vermeulen L, Felipe De Sousa EM, Van Der Heijden M, Cameron K, De Jong JH, Borovski T, et al. Wnt activity defines colon cancer stem cells and is regulated by the microenvironment. Nat Cell Biol. 2010;12:468. doi: 10.1038/ncb2048. [DOI] [PubMed] [Google Scholar]
- 10.Masoodi M, Zali MR, Ehsani-Ardakani MJ, Mohammad-Alizadeh AH, Aiassofi K, Aghazadeh R, et al. Abdominal pain due to lead-contaminated opium: a new source of inorganic lead poisoning in Iran. Arch Iran Med. 2006;9:72–5. [PubMed] [Google Scholar]
- 11.Llosa NJ, Cruise M, Tam A, Wicks EC, Hechenbleikner EM, Taube JM, et al. The vigorous immune microenvironment of microsatellite instable colon cancer is balanced by multiple counter-inhibitory checkpoints. Cancer Discov. 2015;5:43–51. doi: 10.1158/2159-8290.CD-14-0863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van Zijl F, Krupitza G, Mikulits W. Initial steps of metastasis: cell invasion and endothelial transmigration. Mutati Res. 2011;728:23–34. doi: 10.1016/j.mrrev.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gillies RJ, Verduzco D, Gatenby RA. Evolutionary dynamics unifies carcinogenesis and cancer therapy. Nat Rev Cancer. 2012;12:487. doi: 10.1038/nrc3298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Segditsas S, Tomlinson I. Colorectal cancer and genetic alterations in the Wnt pathway. Oncogene. 2006;25:7531. doi: 10.1038/sj.onc.1210059. [DOI] [PubMed] [Google Scholar]
- 15.Barzi A, Lenz AM, Labonte MJ, Lenz H-J. Molecular pathways: estrogen pathway in colorectal cancer. Clin Cancer Res. 2013;19:5842–8. doi: 10.1158/1078-0432.CCR-13-0325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Popat S, Hubner R, Houlston R. Systematic review of microsatellite instability and colorectal cancer prognosis. J Clin Oncol. 2005;23:609–18. doi: 10.1200/JCO.2005.01.086. [DOI] [PubMed] [Google Scholar]
- 17.Deyati A, Bagewadi S, Senger P, Hofmann-Apitius M, Novac N. Systems approach for the selection of micro-RNAs as therapeutic biomarkers of anti-EGFR monoclonal antibody treatment in colorectal cancer. Sci Rep. 2015;5:8013. doi: 10.1038/srep08013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matsumura T, Sugimachi K, Iinuma H, Takahashi Y, Kurashige J, Sawada G, et al. Exosomal microRNA in serum is a novel biomarker of recurrence in human colorectal cancer. Br J Cancer. 2015;113 doi: 10.1038/bjc.2015.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oldenhuis C, Oosting S, Gietema J, De Vries E. Prognostic versus predictive value of biomarkers in oncology. Eur J Cancer. 2008;44:946–53. doi: 10.1016/j.ejca.2008.03.006. [DOI] [PubMed] [Google Scholar]
- 20.Itzkowitz SH, Yio X. Inflammation and cancer IV Colorectal cancer in inflammatory bowel disease: the role of inflammation. Am J Physiol Gastrointest Liver Physiol. 2004;287:G7–17. doi: 10.1152/ajpgi.00079.2004. [DOI] [PubMed] [Google Scholar]
- 21.Roth AD, Tejpar S, Delorenzi M, Yan P, Fiocca R, Klingbiel D, et al. Prognostic role of KRAS and BRAF in stage II and III resected colon cancer: results of the translational study on the PETACC-3, EORTC 40993, SAKK 60-00 trial. J Clin Oncol. 2009;28:466–74. doi: 10.1200/JCO.2009.23.3452. [DOI] [PubMed] [Google Scholar]
- 22.Therkildsen C, Bergmann TK, Henrichsen-Schnack T, Ladelund S, Nilbert M. The predictive value of KRAS, NRAS, BRAF, PIK3CA and PTEN for anti-EGFR treatment in metastatic colorectal cancer: a systematic review and meta-analysis. Acta Oncol. 2014;53:852–64. doi: 10.3109/0284186X.2014.895036. [DOI] [PubMed] [Google Scholar]
- 23.Nikolouzakis TK, Vassilopoulou L, Fragkiadaki P, Mariolis Sapsakos T, Papadakis GZ, Spandidos DA, et al. Improving diagnosis, prognosis and prediction by using biomarkers in CRC patients. Oncol Rep. 2018;39:2455–72. doi: 10.3892/or.2018.6330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bennouna J, Sastre J, Arnold D, Österlund P, Greil R, Van Cutsem E, et al. Continuation of bevacizumab after first progression in metastatic colorectal cancer (ML18147): a randomised phase 3 trial. Lancet Oncol. 2013;14:29–37. doi: 10.1016/S1470-2045(12)70477-1. [DOI] [PubMed] [Google Scholar]
- 25.Wahid M, Mandal RK, Dar SA, Jawed A, Lohani M, Areeshi MY, et al. Therapeutic potential and critical analysis of trastuzumab and bevacizumab in combination with different chemotherapeutic agents against metastatic breast/colorectal cancer affecting various endpoints. Crit Rev Oncol Hematol. 2016;104:124–30. doi: 10.1016/j.critrevonc.2016.06.009. [DOI] [PubMed] [Google Scholar]
- 26.Lambrechts D, Lenz H-J, de Haas S, Carmeliet P, Scherer SJ. Markers of response for the antiangiogenic agent bevacizumab. J Clin Oncol. 2013;31:1219–30. doi: 10.1200/JCO.2012.46.2762. [DOI] [PubMed] [Google Scholar]
- 27.Cremolini C, Del Re M, Antoniotti C, Lonardi S, Bergamo F, Loupakis F, et al. DPYD and UGT1A1 genotyping to predict adverse events during first-line FOLFIRI or FOLFOXIRI plus bevacizumab in metastatic colorectal cancer. Oncotarget. 2018;9:7859. doi: 10.18632/oncotarget.23559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pentheroudakis G, Mavroeidis L, Papadopoulou K, Koliou G-A, Bamia C, Chatzopoulos K, et al. Angiogenic and Antiangiogenic VEGFA Splice Variants in Colorectal Cancer: Prospective Retrospective Cohort Study in Patients Treated With Irinotecan-Based Chemotherapy and Bevacizumab. Clin Colorectal Cancer. 2019;18:e370–84. doi: 10.1016/j.clcc.2019.07.007. [DOI] [PubMed] [Google Scholar]
- 29.Aprile G, Bonotto M, Ongaro E, Pozzo C, Giuliani F. Critical appraisal of ramucirumab (IMC-1121B) for cancer treatment: from benchside to clinical use. Drugs. 2013;73:2003–15. doi: 10.1007/s40265-013-0154-8. [DOI] [PubMed] [Google Scholar]
- 30.Tabernero J, Yoshino T, Cohn AL, Obermannova R, Bodoky G, Garcia-Carbonero R, et al. Ramucirumab versus placebo in combination with second-line FOLFIRI in patients with metastatic colorectal carcinoma that progressed during or after first-line therapy with bevacizumab, oxaliplatin, and a fluoropyrimidine (RAISE): a randomised, double-blind, multicentre, phase 3 study. Lancet Oncol. 2015;16:499–508. doi: 10.1016/S1470-2045(15)70127-0. [DOI] [PubMed] [Google Scholar]
- 31.Obermannová R, Lordick F. Insights into next developments in advanced gastric cancer. Curr Opin Oncol. 2016;28:367–75. doi: 10.1097/CCO.0000000000000289. [DOI] [PubMed] [Google Scholar]
- 32.Van Cutsem E, Tabernero J, Lakomy R, Prenen H, Prausová J, Macarulla T, et al. Addition of aflibercept to fluorouracil, leucovorin, and irinotecan improves survival in a phase III randomized trial in patients with metastatic colorectal cancer previously treated with an oxaliplatin-based regimen. J Clin Oncol. 2012;30:3499–506. doi: 10.1200/JCO.2012.42.8201. [DOI] [PubMed] [Google Scholar]
- 33.Li D, Xie K, Ding G, Li J, Chen K, Li H, et al. Tumor resistance to anti-VEGF therapy through up-regulation of VEGF-C expression. Cancer Lett. 2014;346:45–52. doi: 10.1016/j.canlet.2013.12.004. [DOI] [PubMed] [Google Scholar]
- 34.Majithia N, Grothey A. Regorafenib in the treatment of colorectal cancer. Expert Opin Pharmacother. 2016;17:137–45. doi: 10.1517/14656566.2016.1118054. [DOI] [PubMed] [Google Scholar]
- 35.Grothey A, Van Cutsem E, Sobrero A, Siena S, Falcone A, Ychou M, et al. Regorafenib monotherapy for previously treated metastatic colorectal cancer (CORRECT): an international, multicentre, randomised, placebo-controlled, phase 3 trial. Lancet. 2013;381:303–12. doi: 10.1016/S0140-6736(12)61900-X. [DOI] [PubMed] [Google Scholar]
- 36.Mirone G, Perna S, Shukla A, Marfe G. Involvement of Notch‐1 in Resistance to Regorafenib in Colon Cancer Cells. J Cell Physiol. 2016;231:1097–105. doi: 10.1002/jcp.25206. [DOI] [PubMed] [Google Scholar]
- 37.Van Cutsem E, Köhne C-H, Hitre E, Zaluski J, Chang Chien CR, Makhson A, et al. Cetuximab and chemotherapy as initial treatment for metastatic colorectal cancer. N Engl J Med. 2009;360:1408–17. doi: 10.1056/NEJMoa0805019. [DOI] [PubMed] [Google Scholar]
- 38.Van Emburgh BO, Sartore-Bianchi A, Di Nicolantonio F, Siena S, Bardelli A. Acquired resistance to EGFR-targeted therapies in colorectal cancer. Mol Oncol. 2014;8:1084–94. doi: 10.1016/j.molonc.2014.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Price TJ, Peeters M, Kim TW, Li J, Cascinu S, Ruff P, et al. Panitumumab versus cetuximab in patients with chemotherapy-refractory wild-type KRAS exon 2 metastatic colorectal cancer (ASPECCT): a randomised, multicentre, open-label, non-inferiority phase 3 study. Lancet Oncol. 2014;15:569–79. doi: 10.1016/S1470-2045(14)70118-4. [DOI] [PubMed] [Google Scholar]
- 40.De Roock W, Claes B, Bernasconi D, De Schutter J, Biesmans B, Fountzilas G, et al. Effects of KRAS, BRAF, NRAS, and PIK3CA mutations on the efficacy of cetuximab plus chemotherapy in chemotherapy-refractory metastatic colorectal cancer: a retrospective consortium analysis. Lancet Oncol. 2010;11:753–62. doi: 10.1016/S1470-2045(10)70130-3. [DOI] [PubMed] [Google Scholar]
- 41.Sood A, McClain D, Maitra R, Basu-Mallick A, Seetharam R, Kaubisch A, et al. PTEN gene expression and mutations in the PIK3CA gene as predictors of clinical benefit to anti-epidermal growth factor receptor antibody therapy in patients with KRAS wild-type metastatic colorectal cancer. Clin Colorectal Cancer. 2012;11:143–50. doi: 10.1016/j.clcc.2011.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pohl M, Schmiegel W. Therapeutic strategies in diseases of the digestive tract-2015 and beyond targeted therapies in colon cancer today and tomorrow. Dig Dis. 2016;34:574–9. doi: 10.1159/000445267. [DOI] [PubMed] [Google Scholar]
- 43.Richman SD, Southward K, Chambers P, Cross D, Barrett J, Hemmings G, et al. HER2 overexpression and amplification as a potential therapeutic target in colorectal cancer: analysis of 3256 patients enrolled in the QUASAR, FOCUS and PICCOLO colorectal cancer trials. J Pathol. 2016;238:562–70. doi: 10.1002/path.4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Inno A, Di Salvatore M, Cenci T, Martini M, Orlandi A, Strippoli A, et al. Is there a role for IGF1R and c-MET pathways in resistance to cetuximab in metastatic colorectal cancer? Clin Colorectal Cancer. 2011;10:325–32. doi: 10.1016/j.clcc.2011.03.028. [DOI] [PubMed] [Google Scholar]
- 45.Abdollahpour-Alitappeh M, Lotfinia M, Gharibi T, Mardaneh J, Farhadihosseinabadi B, Larki P, et al. Antibody-drug conjugates (ADCs) for cancer therapy: Strategies, challenges, and successes. J Cell Physiol. 2019;234:5628–5642. doi: 10.1002/jcp.27419. [DOI] [PubMed] [Google Scholar]
- 46.Rezaei-Tavirani M, Safaei A, Zali MR. The association between polymorphismsin insulin and obesity related genesand risk of colorectal cancer. Iran J Cancer Prev. 2013;6:179. [PMC free article] [PubMed] [Google Scholar]
- 47.Douillard J-Y, Siena S, Cassidy J, Tabernero J, Burkes R, Barugel M, et al. Randomized, phase III trial of panitumumab with infusional fluorouracil, leucovorin, and oxaliplatin (FOLFOX4) versus FOLFOX4 alone as first-line treatment in patients with previously untreated metastatic colorectal cancer: the PRIME study. J Clini Oncol. 2010;28:4697–705. doi: 10.1200/JCO.2009.27.4860. [DOI] [PubMed] [Google Scholar]
- 48.Wilson PM, Danenberg PV, Johnston PG, Lenz H-J, Ladner RD. Standing the test of time: targeting thymidylate biosynthesis in cancer therapy. Nat Rev Clin Oncol. 2014;11:282. doi: 10.1038/nrclinonc.2014.51. [DOI] [PubMed] [Google Scholar]
- 49.Longley DB, Harkin DP, Johnston PG. 5-fluorouracil: mechanisms of action and clinical strategies. Nat Rev Cancer. 2003;3:330. doi: 10.1038/nrc1074. [DOI] [PubMed] [Google Scholar]
- 50.Qiu LX, Tang QY, Bai JL, Qian XP, Li RT, Liu BR, et al. Predictive value of thymidylate synthase expression in advanced colorectal cancer patients receiving fluoropyrimidine‐based chemotherapy: evidence from 24 studies. Int J Cancer. 2008;123:2384–9. doi: 10.1002/ijc.23822. [DOI] [PubMed] [Google Scholar]
- 51.Etienne‐Grimaldi MC, Milano G, Maindrault‐Gœbel F, Chibaudel B, Formento JL, Francoual M, et al. Methylenetetrahydrofolate reductase (MTHFR) gene polymorphisms and FOLFOX response in colorectal cancer patients. Br J Clin Pharmacol. 2010;69:58–66. doi: 10.1111/j.1365-2125.2009.03556.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Panczyk M. Pharmacogenetics research on chemotherapy resistance in colorectal cancer over the last 20 years. World J Gastroenterol. 2014;20:9775. doi: 10.3748/wjg.v20.i29.9775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Koopman M, Venderbosch S, van Tinteren H, Ligtenberg MJ, Nagtegaal I, Van Krieken JH, et al. Predictive and prognostic markers for the outcome of chemotherapy in advanced colorectal cancer, a retrospective analysis of the phase III randomised CAIRO study. Eur J Cancer. 2009;45:1999–2006. doi: 10.1016/j.ejca.2009.04.017. [DOI] [PubMed] [Google Scholar]
- 54.Vallböhmer D, Kuramochi H, Shimizu D, Danenberg KD, Lindebjerg J, Nielsen JN, et al. Molecular factors of 5-fluorouracil metabolism in colorectal cancer: analysis of primary tumor and lymph node metastasis. Int J Oncol. 2006;28:527–33. doi: 10.3892/ijo.28.2.527. [DOI] [PubMed] [Google Scholar]
- 55.Kim S, Hong Y, Shim E, Kong S, Shin A, Baek J, et al. S-1 plus irinotecan and oxaliplatin for the first-line treatment of patients with metastatic colorectal cancer: a prospective phase II study and pharmacogenetic analysis. Br J Cancer. 2013;109:1420. doi: 10.1038/bjc.2013.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Baba H, Watanabe M, Okabe H, Miyamoto Y, Sakamoto Y, Baba Y, et al. Upregulation of ERCC1 and DPD expressions after oxaliplatin-based first-line chemotherapy for metastatic colorectal cancer. Br J Cancer. 2012;107:1950. doi: 10.1038/bjc.2012.502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Moutinho C, Martinez-Cardús A, Santos C, Navarro-Pérez V, Martínez-Balibrea E, Musulen E, et al. Epigenetic inactivation of the BRCA1 interactor SRBC and resistance to oxaliplatin in colorectal cancer. J Natl Cancer Inst. 2014:106. doi: 10.1093/jnci/djt322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zamanian-Azodi M, Rezaei-Tavirani M, Rahmati-Rad S, Hasanzadeh H, Tavirani MR, Seyyedi SS. Protein-Protein Interaction Network could reveal the relationship between the breast and colon cancer. Gastroenterol Hepatol Bed Bench. 2015;8:215. [PMC free article] [PubMed] [Google Scholar]
- 59.Goudarzi M, Goudarzi H, Alebouyeh M, Azimi Rad M, Shayegan Mehr FS, Zali MR, et al. Antimicrobial susceptibility of clostridium difficile clinical isolates in iran. Iran Red Crescent Med J. 2013;15:704–11. doi: 10.5812/ircmj.5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Boyer J, McLean EG, Aroori S, Wilson P, McCulla A, Carey PD, et al. Characterization of p53 wild-type and null isogenic colorectal cancer cell lines resistant to 5-fluorouracil, oxaliplatin, and irinotecan. Clin Cancer Res. 2004;10:2158–67. doi: 10.1158/1078-0432.ccr-03-0362. [DOI] [PubMed] [Google Scholar]
- 61.Longley D, Johnston P. Molecular mechanisms of drug resistance. J Pathol. 2005;205:275–92. doi: 10.1002/path.1706. [DOI] [PubMed] [Google Scholar]
- 62.Zhao J, Li W, Zhu D, Yu Q, Zhang Z, Sun M, et al. Association of single nucleotide polymorphisms in MTHFR and ABCG2 with the different efficacy of first-line chemotherapy in metastatic colorectal cancer. Med Oncol. 2014;31:802. doi: 10.1007/s12032-013-0802-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nygård SB, Christensen IJ, Nielsen SL, Nielsen HJ, Brünner N, Spindler K-LG. Assessment of the topoisomerase I gene copy number as a predictive biomarker of objective response to irinotecan in metastatic colorectal cancer. Scand J Gastroenterol. 2013;49:84–91. doi: 10.3109/00365521.2013.856464. [DOI] [PubMed] [Google Scholar]
- 64.Gongora C, Vezzio-Vie N, Tuduri S, Denis V, Causse A, Auzanneau C, et al. New Topoisomerase I mutations are associated with resistance to camptothecin. Mol Cancer. 2011;10:64. doi: 10.1186/1476-4598-10-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ma CJ, Huang CW, Chang TK, Tsai HL, Su WC, Yeh YS, et al. Oncologic Outcomes in Metastatic Colorectal Cancer with Regorafenib with FOLFIRI as a Third-or Fourth-Line Setting. Transl Oncol. 2019;12:502–12. doi: 10.1016/j.tranon.2018.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]