Abstract
The stem cell niche is a specialized environment that dictates the proper functioning of stem cells during normal development and homeostasis. Despite recent progress, the identity of the mammary gland stem cell (MaSC) niche remains largely unknown. Here, we show that Dll1, a Notch pathway ligand, is enriched in MaSCs and mediates critical interactions between these cells and their surrounding niche. Conditional deletion of Dll1 resulted in reduced number of MaSCs and defective ductal morphogenesis in both virgin and pregnant mammary glands. Dll1-mediated Notch signaling maintains the MaSC function via its activation of Notch signaling in stromal macrophages, leading to increased expression of Wnt family ligands such as Wnt3, Wnt10A, and Wnt16, which feed back to Dll1+ mammary stem cells to promote their activity. These findings revealed a functionally important cross talk between MaSCs and their macrophageal niche through Dll1/Notch-mediated signaling.
One sentence summary:
Dll1-mediated crosstalk between mammary stem cells and macrophages is critical for mammary gland development and function
Mammary epithelial cells are composed of two major cell types—the basal cells that rest on the basement membrane, and the luminal cells that face the lumen. These two lineages of mammary epithelial cells are believed to be derived from MaSCs during puberty and each round of pregnancy and lactation cycle (1, 2). The existence of MaSCs that give rise to these different lineages of mammary epithelial cells has been demonstrated by transplantation (3, 4), and more recently, lineage tracing experiments (5, 6). The epithelial cells in the mammary gland is surrounded by various stromal cell types, including adipocytes, fibroblasts, macrophages, endothelial cells and lymphoid cells, that constitute the mammary gland microenvironment (7, 8). Previous studies have indicated the contribution of some of these stromal cells to mammary gland development and homeostasis, including ductal morphogenesis during puberty and pregnancy, and involution after weaning (9–15). However, little is known about the functional involvement and signaling mechanism of the mammary stromal cells in regulating MaSC activity.
The Notch signaling pathway has emerged as a key regulator of several essential developmental processes in the mammary gland, including stem cell maintenance, cell fate decisions, and de-differentiation (16–18). Four Notch receptors (Notch1-Notch4) and five Notch ligands (Delta-like1, Delta-like3, Delta-like4, Jagged1, and Jagged2) have been reported to mediate Notch signaling in mammals. Recently, lineage tracing experiments demonstrated the function of Notch1–3 receptors in luminal progenitor cells during puberty and pregnancy (19, 20), and earlier studies have revealed the importance of Notch1 and Notch4 in basal MaSCs (16, 18). Notably, most of these studies on Notch signaling and mammary gland development have focused on the receptors, while relatively little is known about the role of Notch ligands in regulating MaSC behavior and cell fate. Interestingly, Dll1 has been implicated in intestinal stem cells (21, 22), although its functional importance in MaSC is unknown.
Besides Notch signaling, another crucial regulatory pathway of mammary gland development and tumorigenesis is the Wnt signaling. Wnt signaling is well established for sustaining adult stem cells in many organs (23), including MaSCs (3, 24–27). Binding of the Wnt ligands to receptors leads to nuclear translocation of β-catenin and activation of Tcf/Lef target genes (28). Several studies have showed that Wnt ligands such as Wnt3a and Wnt4 are important for the self-renewal of MaSCs (25, 29); however, these Wnt ligands are not expressed by the basal stem cells, suggesting that an adjacent MaSC niche might be responsible for secretion of the ligands. Indeed, recent studies showed that Wnt4 control MaSC function through luminal-myoepithelial crosstalk (29). It remains unclear whether stromal niche cells can also produce Wnt ligands to regulate MaSC.
In this study, we provide evidence that Dll1 expression is enriched in MaSCs and is essential for their function by engaging Notch signaling in stromal macrophages to induce the expression of Wnt ligands. Our study defines a Dll1-mediated MaSC niche that involves the coupling of Notch and Wnt signaling between MaSCs and macrophages.
Results
Dll1 is required for mammary morphogenesis in virgin and pregnant mammary glands
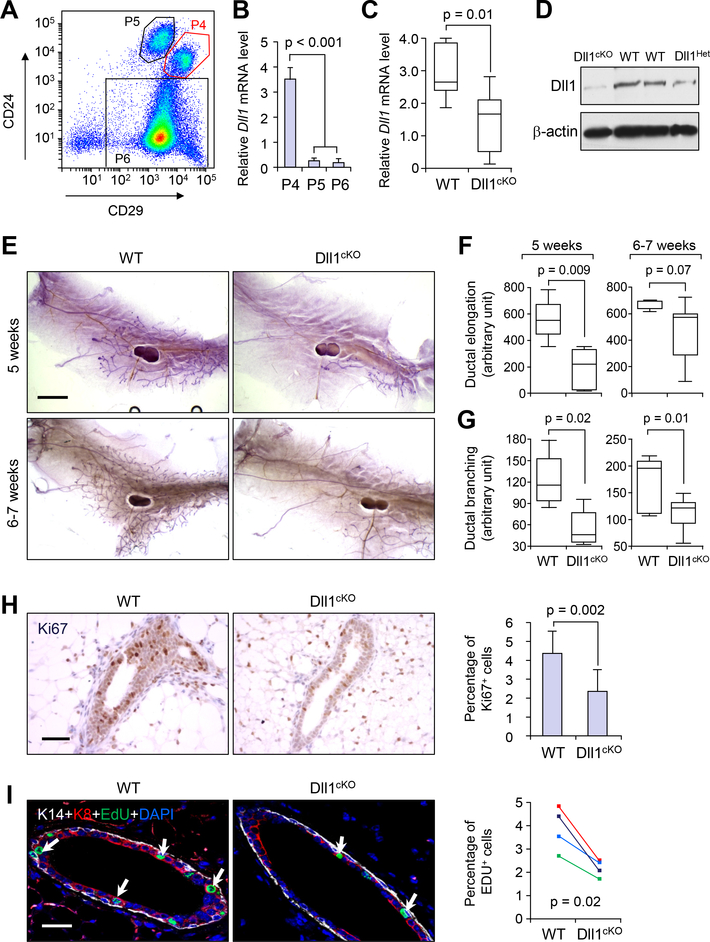
Until recently, most of the Notch signaling studies on normal mammary gland development have focused on the role of the Notch receptors. In our recent gene expression profiling analysis of different populations of cells from the mammary gland (30), Notch ligand Dll1 is found to be predominantly expressed in MaSC-enriched basal cells (P4, Lin−CD24+CD29hi) compared to luminal (P5, Lin−CD24+CD29lo) and stromal-enriched cells (P6, Lin−CD24loCD29lo) (Fig. 1A, B). To investigate the potential role of Dll1 in mammary gland development, we employed the Cre-loxP recombination system to generate K14-Cre mediated Dll1 conditional knockout mice that target Dll1 in both basal (P4) and luminal cells (P5) (31–33). A robust knockout of Dll1 in the mammary gland was confirmed by significant reduction of Dll1 mRNA (Fig. 1C) and Dll1 protein (Fig. 1D) in mammary epithelial cells in MECs of K14-Cre/Dll1f/f (Dll1cKO) mammary glands and by immunofluorescence in P4 cells (fig. S1A). Interestingly, K14-Cre-mediated deletion of Dll1 resulted in no apparent hair formation phenotype (fig. S1B), unlike knockout mice of other Notch ligands such as Jagged1 (34, 35), suggesting that Dll1 is dispensable for hair follicle development. In contrast, whole mount carmine staining of the mammary glands from 5-week and 6–7 week old Dll1cKO mice revealed a significant reduction in mammary ductal elongation and branching compared to the wild-type littermate (Fig. 1E–G).
Fig. 1. Dll1 is required for mammary gland ductal morphogenesis.
(A) FACS profile of different populations of Lin− mammary epithelial cells based on staining of CD24 and CD29. (B) qRT-PCR analysis of Dll1 mRNA expression in different subpopulations of mammary epithelial cells as in (A). qRT-PCR values were normalized to the housekeeping gene Gapdh. n = 5 samples, experiments were performed three times, each with qRT-PCR in technical duplicate, and data are presented as the mean ± SD. * p < 0.001 by Student’s t-test. (C) qRT-PCR analysis of Dll1 mRNA expression in mammary epithelial cells from WT and Dll1 conditional knockout mice (Dll1cKO). The boxes represent the 75th, 50th and 25th percentile of the values. The top and bottom lines represent maximum and minimal data points within the 1.5x IQ (inter quarter) range, respectively. p-value of box plot in c was computed by Mann Whitney U test. n = 7 samples for WT and n = 9 samples for Dll1cKO in (C). (D) Western blot showing Dll1 protein expression in the mammary epithelial cells from WT and Dll1cKO mice. (E) Representative alum carmine stained whole mount mammary outgrowths from WT and Dll1cKO mice at 5 weeks and 6–7 weeks respectively. (F-G) Box plot analyses of ductal elongation and branching in WT and Dll1cKO mice. Quantification of ductal branching (tertiary branch points) was measured in defined area. The boxes represent the 75th, 50th and 25th percentile of the values. The top and bottom lines represent maximum and minimal data points within the 1.5x IQ (inter quarter) range, respectively. p-value computed by Mann-Whitney U test. n = 5 samples per genotype. (H) Ki67 staining in outgrowth sections from WT and Dll1cKO mice mammary gland sections (left panel). Quantification of Ki67+ cell percentage among total epithelial cells in field of view is shown (right panel). p-value was computed by Student’s t-test. n = 7 samples for WT and n = 5 samples for Dll1cKO. (I) Keratin-14 (K14), Keratin-8 (K8) and EdU staining of mammary gland sections of WT and Dll1cKO mice at 5–6 weeks age (left). White arrows indicate EdU+ cells in left panel. Quantification of EdU+ cell percentage among total epithelial cells in field of view is shown in the right panel. p-value computed by paired Student’s t-test as sibling cohorts of mice were used. n = 4 samples for each genotype in the left panel. Size bar, 2 mm in (E), 40 μm (H) and (I) respectively.
Ki67 and EdU staining revealed fewer proliferating cells in mammary epithelial cells from Dll1cKO mammary gland compared to WT littermate control (Fig. 1H, I). To further characterize the cell population in the Dll1cKO mammary epithelium, we used basal (K14) and luminal (K8) cell lineage markers. Examination of the mammary glands with these lineage markers showed no aberrant expression of K14 and K8, suggesting that the cell fate was not significantly altered in the Dll1cKO mammary epithelial cells compared to WT controls (Fig. 1I). Interestingly, the reduced ductal morphogenesis phenotype of Dll1cKO mice persisted into pregnancy, as the alveoli density in Dll1cKO mammary glands was nearly half that of wild-type glands at lactation day 1, leading to reduced survival of pups (fig. S1C, D). 80% of all pups from the Dll1cKO mothers died within two days of birth, regardless of their genotype (fig. S1C). Histological analyses by Hematoxylin and Eosin (H&E) staining indicated that a smaller subset of alveoli from Dll1cKO mammary gland display features of secretory differentiation such as lipid droplets and milk production compared to WT (fig. S1D). Immunohistochemistry with Ki67 antibodies confirmed reduced proliferation in the Dll1cKO mammary glands compared to WT mammary glands during lactaction (fig. S1D). Since the alveoli in the Dll1cKO mammary gland were lacking in lipid droplets and milk secretion, we next tested whether the proliferation defect was associated with secretory differentiation failure by staining for Np2b (Na-Pi cotransporter) protein, whose absence at parturition is indicative of a lack of secretory function (36). Indeed, the apical membranes of secretory alveoli in Dll1cKO mammary gland showed reduced Npt2b staining compared to the wild-type mammary glands that exhibited intense Npt2b staining (fig. S1D). Overall, our data indicates that reduced proliferation and a block in secretory differentiation may both play a critical role in contributing to the defects in lobuloalveolar development of Dll1cKO mice.
Dll1 is critical for maintaining MaSC numbers
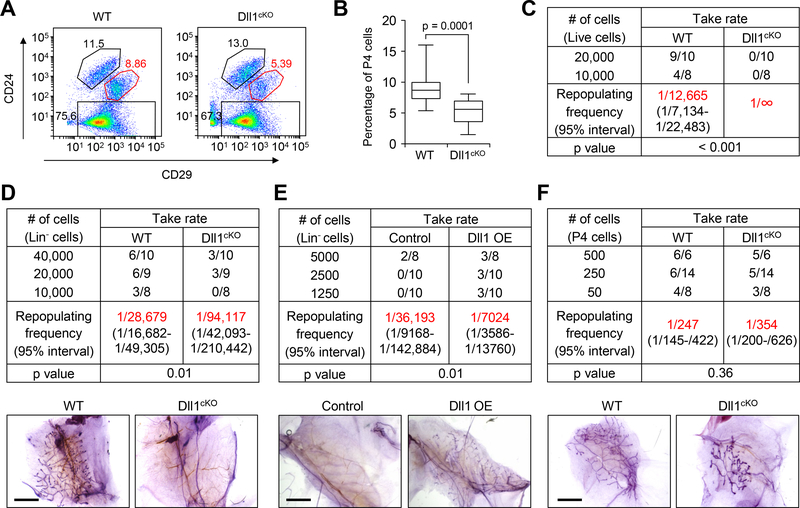
Studies have demonstrated that MaSCs are believed to reside among the basal cells (3). Since Dll1 is predominantly expressed in basal cells, we next probed whether the reduced ductal morphogenesis is associated with altered MaSC number or function. FACS analysis demonstrated a significant decrease of MaSC-enriched Lin−CD24+CD29hi (P4 cells) population in Dll1cKO mice compared to WT mice (Fig. 2A, B and fig. S2A). Limiting dilution mammary cleared fat pad transplantation assay with live cells (propidium iodide-negative) or lineage negative live cells (Lin−, i.e. CD31−, Ter119− and CD45−) revealed a significantly reduced (~3-fold) mammary gland repopulating frequency of mammary epithelial cells (live or Lin−) from Dll1cKO mice compared to WT mice (Fig. 2C, D). Conversely, overexpression of Dll1 in mammary epithelial cells by lentiviral transduction prior to transplantation resulted in an increased MaSC repopulation frequency compared to control cells (Fig. 2E and fig. S2B). Next, we analyzed the repopulating ability of isolated P4 and P5 cells from WT and Dll1cKO mice. As expected, luminal cells (P5) showed no ductal growth from either WT or Dll1cKO mammary glands (fig. S2C, D). Intriguingly, no significant difference between WT and Dll1cKO mice was observed when P4 cells were used for limiting dilution assays (Fig. 2F). Moreover, only a modest difference was observed in the serial transplant take rate of WT and Dll1cKO basal cells (fig. S2E, F). These results indicated that the primary reason for the reduced ductal growth in the Dll1cKO mammary glands is the reduction of MaSC numbers, rather than their cellular properties. This phenotype of reduced basal (P4) population, which harbors MaSCs, was also observed during lactation (fig. S2G, H). Taken together, our studies suggest that Dll1 is a critical regulator of mammary gland development via its role in maintaining MaSC numbers.
Fig. 2. Dll1 is required for maintaining MaSC activity.
(A) Representative FACS profile of Lin− mammary epithelial cells from WT and Dll1cKO mouse at 5–6 weeks of age based on staining with CD24 and CD29. (B) Box plot showing percentage of P4 (basal) cells in WT and Dll1cKO mouse after FACS. n = 18 samples for WT and Dll1cKO. Please see Fig. S2A for individual values of indicated groups. The boxes represent the 75th, 50th and 25th percentile of the values. The top and bottom lines represent maximum and minimal data points within the 1.5x IQ (inter quarter) range, respectively. p value was computed by paired t test. (C) Table showing reconstitution efficiency at limiting dilution of total live cells from WT and Dll1cKO mouse mammary glands injected into cleared mammary fat pads of recipient mice. (D) Table showing reconstitution efficiency at limiting dilution of total lineage negative (Lin−) cells from WT and Dll1cKO mouse mammary glands injected into cleared mammary fat pads of recipient mice. Representative alum carmine stained mammary outgrowths from transplantation with 10,000 Lin− cells are shown in the bottom panel. (E) Table showing reconstitution efficiency at limiting dilution of total lineage negative (Lin−) cells from WT and Dll1-overexpressing (Dll1 OE) mammary gland injected into cleared mammary fat pads of recipient mice. Lin− population was isolated and transduced with vector (control), or Dll1 expressing lentivirus and transplanted into cleared fat pads of recipient mice. Representative alum carmine stained mammary outgrowths from transplantation with 10,000 Lin− cells are shown in the bottom panel. (F) Table showing reconstitution efficiency at limiting dilution of Lin−CD24+CD29hi (P4) cells from WT and Dll1cKO mouse mammary glands injected into cleared mammary fat pads of recipient mice. Representative alum carmine stained mammary outgrowths from transplantation are shown in the bottom panel. n = number of mammary fat pad injections as indicated in the table in (C-F). P value was obtained by Pearson’s Chi-squared test using ELDA software. Size bar, 2 mm in (D), (E) and (F) respectively.
Dll1+ cell population has enriched MaSC activities
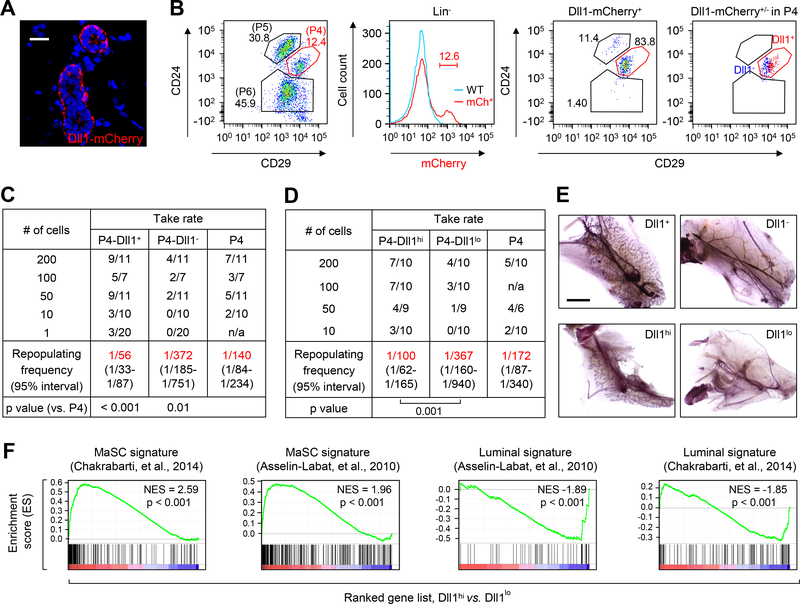
To further characterize the function and expression of Dll1 in mammary gland cells, we used a Dll1-mCherry transgenic mouse model in which the mCherry reporter gene is driven by the Dll1 genomic regulatory sequences. In adult mammary gland of 5–6 week-old mice, Dll1-mCherry reporter is expressed predominantly in basal cells (Fig. 3A), corroborating the Dll1 mRNA data (Fig. 1A, B). Dll1+ cells are a subset of K14+ and ΔNp63+ basal cells, and are negative for K8+ luminal cells (fig. S3A, B). We next tested the expression of Dll1mCherry in different mammary gland developmental stages. Dll1mCherry was expressed in all stages of mammary gland development (virgin and pregnancy) and its expression was predominantly in the basal compartment throughout development (fig. S3C–F). FACS analysis indicated that ~12% of Lin− cells are Dll1mCherry positive in virgin mice (4–6 weeks of age) (Fig. 3B and fig. S3D), and this population increases dramatically during pregnancy and lactation (fig. S3D). Dll1mCherry expression is predominantly enriched in the P4 basal cell population (fig. S3E). The P4-Dll1mCherry+ (Dll1+) and P4-Dll1mCherry− (Dll1−) basal cells were expressed towards the upper right (top) and lower left (bottom) portion of the Lin−CD24+CD29hi (P4) basal population, respectively in all developmental stages (Fig. 3B and fig. S3F).
Fig. 3. Dll1+ population is enriched in cells with MaSC activity.
(A) Immunofluorescence image of Dll1-mCherry reporter mice mammary gland section stained with mCherry antibody. (B) Representative FACS profile of mammary epithelial cells from Dll1-mCherry reporter mice at 6 weeks of age based on staining with CD24 and CD29 (left panel). Middle panel: mCherry+ cells in lineage negative (Lin−) population. Right two panels: distribution of Dll1-mCherry+ cells in different epithelial populations (left), and Dll1-mCherry+ and Dll1-mCherry− cells in the P4 population (right). (C-D) Table showing reconstitution efficiency at limiting dilution of different groups of P4 cells (as indicated in table) from Dll1-mCherry reporter mice mammary glands injected into cleared mammary fat pads of recipient mice. For sorting of P4-Dll1hi and P4-Dll1lo, top and bottom 10–12% of the population were chosen from the P4-Dll1+ cell population respectively. In (C) and (D), n = number of mammary fat pad injections as indicated in the table. p value was obtained by Pearson’s Chi-squared test using ELDA software. (E) Representative alum carmine stained mammary outgrowths from transplantation as shown in (C) and (D). (F) GSEA demonstrating enriched MaSC signatures in P4-Dll1hi population compared to P4-Dll1lo populations. In contrast, luminal progenitor cell signatures are enriched in Dll1lo populations. Size bar, 40 μm in (A) and 2 mm in (F) respectively.
To directly examine the repopulating ability of cell populations with different levels of Dll1, we tested Dll1+ and Dll1− cells from both lineage-negative and basal cells (P4) by transplantation assays to examine their reconstitution potential. We found that Lin−Dll1+ cells generated the mammary outgrowths more efficiently (repopulation frequency 1/164) than the Lin−Dll1− cells (repopulation frequency 1/5435) or the total population of lineage negative cells (repopulation frequency 1/1752) (fig. S4A, B). Similarly, P4-Dll1+ cells had a much higher repopulation frequency (repopulation frequency 1/56) than the P4-Dll1− cells (repopulation frequency 1/372) or the total populations of P4 cells (repopulation frequency 1/140) (Fig. 3C, E), suggesting that the Dll1+ cells contain a MaSC-enriched population. Since a relatively high proportion of basal cells expressed Dll1 (Fig. 3B and fig. S3C), other basal cells besides MaSCs, such as basal progenitors or differentiated basal cells, likely also express certain level of Dll1. We wondered whether the P4-Dll1mCherry-high (Dll1hi) cells with higher level of Dll1 expression are more likely to function as MaSCs compared to the P4-Dll1mCherry-low (Dll1lo) cells. Mammary gland repopulation assay in vivo showed that P4-Dll1hi basal cells indeed have increased reconstitution potential (repopulation frequency 1/100) than the P4-Dll1lo basal cells (repopulation frequency 1/367) (Fig. 3D, E). Corroborating these findings, gene set enrichment analysis (GSEA) confirmed that P4-Dll1hi cells were enriched for MaSC signatures (30, 37), whereas P4-Dll1lo cells were enriched for luminal signatures (Fig. 3F). These data suggests that the Dll1hi cells have enhanced mammary stem cells activity, whereas Dll1lo cells could be the potential basal progenitor cells, which share some gene expression features of luminal cells. Immunostaining with lineage markers K8 and K14 showed no major structural difference between the outgrowths from Dll1+/hi and Dll1−/lo cells (fig. S4C, D). However, outgrowths from Dll1− cells had fewer K14+ basal cells compared to outgrowths from Dll1+ cells and occasionally produced a closed lumen. Finally, in serial transplantation assays, both Dll1+ and Dll1hi cells continued to be more efficient in reconstitution compared to Dll1− and Dll1lo cells, respectively (fig. S4E–G), further supporting the notion that Dll1 is enriched in MaSC population.
Dll1+ enriched MaSCs can give rise to both basal and luminal cells
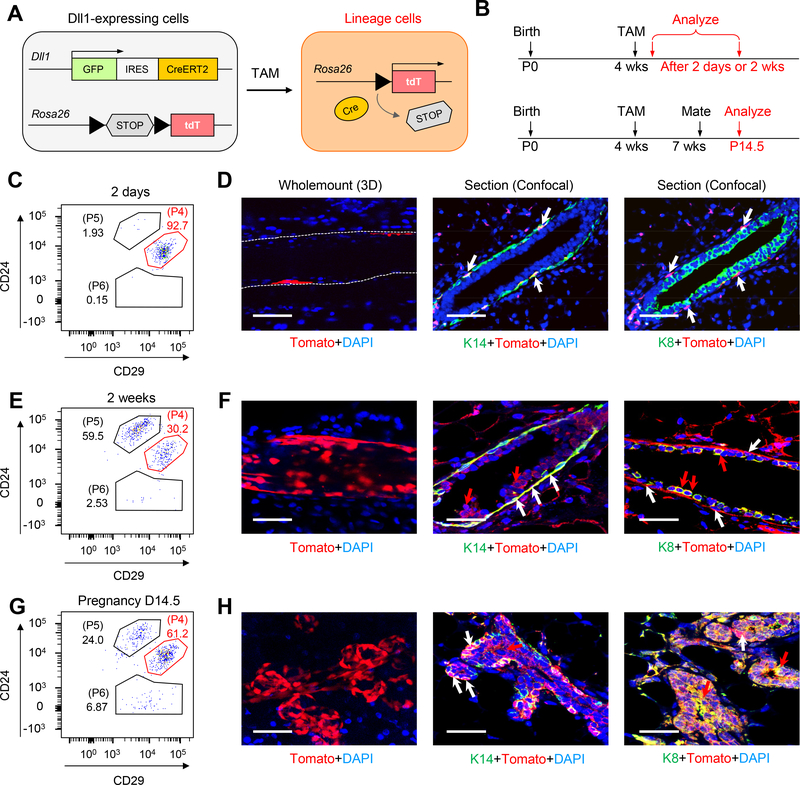
To examine the function of Dll1+ cells during mammary gland development in physiological conditions, we performed a lineage tracing experiment. For this purpose, we used the previously described Dll1-GFP-IRES-Cre-ERT2 knock-in mouse model in which GFP and Cre-ER were bicistronically expressed under the endogenously Dll1 promoter (21). Similar to our observation in the Dll1mCherry mouse model, Dll1GFP was predominantly expressed in basal cells, which are K14+/ΔNp63+ and K8− cells (fig. S5A–D). To trace the fate of Dll1GFP+ cells, Dll1-GFP-IRES-Cre-ERT2 mice were mated with tdTomato reporter mice (Jackson Laboratory) (Fig. 4A, B). The tdTomato reporter expression was traced at different time points after initial induction with tamoxifen at 4 weeks old mice (Fig. 4B). As expected, FACS analysis at early time point (2 day post induction) revealed tdTomato expression to be predominantly in the basal compartment (Fig. 4C). 3D wholemount staining and optical section further confirmed tdTomato expression in basal cells, which are K14+ and K8− (Fig. 4D). At 2-week and 6-week of tracing post induction, tdTomato expression was observed in both basal and luminal cells by FACS analysis, indicating that Dll1+ cells can give rise to both basal and luminal cells (Fig. 4E and fig. S6A). This finding was further confirmed with 3D whole mount mammary gland staining and optical section (Fig. 4F and fig. S6A). Similar to virgin mammary gland, lineage tracing revealed tdTomato+ basal and luminal cells at pregnancy 14 day in both FACS and immunofluorescence staining analyses (Fig. 4G–H). Similar finding was also observed when we performed lineage tracing at the adult stage by inducing the Dll1 mediated tomato expression at 7–8 weeks old mice (fig. S6B–F). FACS analysis and immunofluorescence staining at early time point (2 day post induction) revealed tdTomato expression to be predominantly in the basal compartment and almost none in the luminal compartment (fig. S6C, D). At 4-week of tracing post induction in adult stage, tdTomato expression was observed in both basal and luminal cells by FACS analysis and immunofluorescence staining, indicating that Dll1+ cells can give rise to both basal and luminal cells (fig. S6E, F). Taken together, our data indicates that Dll1+ MaSCs can give rise to both the basal and luminal lineage.
Fig. 4. Lineage tracing shows Dll1+ cells can give rise to both basal and luminal cell populations in mammary gland.
(A-B) Strategy for tamoxifen inducible Cre-mediated cell tracking using Dll1-GFP-IRES-Cre-ERT2; ROSA-tdTomato mice. Red cell indicates Dll1-Cre activated Tomato+ cells, which were used for lineage tracing of Dll1+ cells. (C) FACS plot of mammary epithelial cells from Dll1-GFP-IRES-Cre-ERT2; ROSA-tdTomato mice mammary gland after 2 days of induction with Tamoxifen (TAM), showing the percentage of tdTomato+ cells in various mammary epithelial populations based on staining with CD24 and CD29. (D) Whole mount 3-D staining showing Tomato+ cells in basal cells (left two panels). Staining with K14, K8 and Tomato antibodies on confocal sections on Dll1-GFP-IRES-Cre-ERT2; ROSA-tdTomato mouse mammary glands (right panel) after 2 days of TAM. White arrows indicate Tomato+K14+ basal cells. (E) FACS plot of mammary epithelial cells from Dll1-GFP-IRES-Cre-ERT2; ROSA-tdTomato mouse mammary gland after 2 weeks of induction with Tamoxifen (TAM), showing the percentage of tdTomato+ cells in various mammary epithelial populations based on staining with CD24 and CD29. (F) Whole mount 3-D staining showing Tomato+ cells in both luminal and basal cells suggesting Dll1+ cells can give rise to both basal and luminal cells (left panel). Staining with K14, K8 and Tomato antibodies on confocal sections on Dll1-GFP-IRES-Cre-ERT2; ROSA-tdTomato mouse mammary gland (right two panels) after 2 weeks of TAM. White arrows indicate Tomato+K14+ basal cells and red arrows indicate Tomato+ K8+ luminal cells. (G) FACS plot of mammary epithelial cells from Dll1-GFP-IRES-Cre-ERT2; ROSA-tdTomato mouse mammary gland during pregnancy after induction with Tamoxifen (TAM), showing the percentage of tdTomato+ cells in various mammary epithelial populations based on staining with CD24 and CD29. (H) Whole mount 3-D staining showing Tomato+ cells in both luminal and basal cells suggesting Dll1+ cells can give rise to both basal and luminal cells (left panel). Staining with K14, K8 and Tomato antibodies on confocal sections on Dll1-GFP-IRES-Cre-ERT2; ROSA-tdTomato mouse mammary gland (right two panels) at pregnancy 14.5 days. Size bar: 40μm in (D), (F) and (H) respectively. White arrows indicate Tomato+K14+ basal cells and red arrows indicate Tomato+ K8+ luminal cells. n = 5 samples per developmental stage.
Mammary gland macrophages have unique molecular properties and are regulated by Dll1+ MaSCs
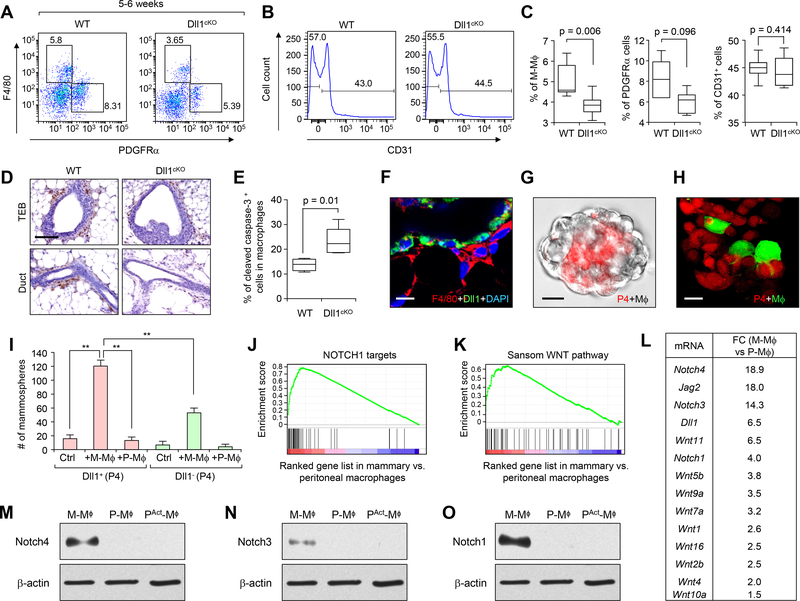
Since Notch signaling requires direct cell-cell contacts, we considered the possibility that Dll1 may regulate MaSC through signaling crosstalk to stromal niche cells. To this end, we first investigated the neighboring stromal populations of Dll1+ basal cells in WT and Dll1cKO mammary glands. FACS analysis with F4/80 (macrophages), CD140a/PDGFRα (fibroblasts) and CD31 (endothelial cells), showed reduced F4/80+ macrophage and moderately reduced PDGFRα+ fibroblasts population in Dll1cKO mammary glands compared to WT mammary glands (Fig. 5A–C). No significant difference was observed for the CD31+ endothelial population between the two groups (Fig. 5B, C). Since macrophages have been shown to be important for mammary gland development (12–15), we next performed immunostaining with F4/80 antibody, which further confirmed reduced F4/80+ macrophages in Dll1cKO TEBs and ducts compared to the WT type mammary glands (Fig. 5D). The notion of crosstalk between basal cells and macrophages were further reinforced by a positive correlation of the number of P4 cells and F4/80+ macrophages in WT and Dll1cKO mammary glands in different developmental stages (fig. S7A–I). Similar to reduction of P4 cells in Dll1cKO mammary glands compared to WT, F4/80+ macrophages also showed a reduction in number in Dll1cKO mammary glands compared to WT (fig. S7A–I). Dll1 mRNA expression shows no significant difference between WT and Dll1cKO macrophages, fibroblasts and endothelial cells (fig. S8A) supporting the activity of K14-Cre specifically in epithelial cells. Furthermore, flow cytometry analysis using a cleaved caspase-3 antibody showed increased apoptotic activity in macrophages from Dll1cKO mammary gland compared to WT (Fig. 5E), suggesting that the reduced number of macrophages in Dll1cKO mammary gland may be caused by decrease survival of macrophages due to reduced Dll1-mediated Notch signaling. Immunofluorescence analysis demonstrated that Dll1+ basal cells were neighboring to the F4/80+ stromal cells in the Dll1mCherry+ mammary gland (Fig. 5F), suggesting the possibility of crosstalk between the two populations via juxtacrine or paracrine signaling. This observation was further confirmed in a novel in vitro mammosphere co-culture assay where MaSC-enriched P4 basal cells (red) were mixed with macrophages (green from GFP mice or no color from WT mice) (Fig. 5G, H). In such 3D co-culture system, the MaSC-enriched cells and macrophages formed a close heterotypic organoid structure where macrophages were juxta-positioned to MaSCs (Fig. 5G, H), as seen in mammary gland architecture in vivo (Fig. 5F). Using this 3-D co-culture assay, we tested the function of macrophages on Dll1+ MaSC activity. Furthermore, we used peritoneal macrophages as a control in this assay to test if macrophages from other tissue besides mammary gland can have same effect on Dll1+ MaSCs. We found that mammary gland macrophages can induce MaSC activity (increased mammospheres) while peritoneal macrophages could not (Fig. 5I). Furthermore, such enhancement of stem cell activity was much more prominent in P4-Dll1+ cells than in P4-Dll1− cells (Fig. 5I), suggesting that MaSCs depends on Dll1 to engage and respond to mammary gland macrophages. The sphere-forming ability of Dll1− cells was modestly increased by co-culture with macrophage derived from WT glands as these macrophages likely maintains some MaSC-supporting properties when they were isolated from WT mammary gland, where they were exposed to in Dll1+ P4 cells.
Fig. 5. Mammary gland macrophages have unique molecular properties and are regulated by Dll1+ MaSCs.
(A) FACS plot of mammary epithelial cells from WT and Dll1cKO mammary gland based on staining with F4/80 (macrophages) and PDGFRα (fibroblasts). (B) Histogram from FACS analyses showing CD31+ endothelial cells in WT and Dll1cKO mammary gland based on staining with CD31 antibody. (C) Box plots showing quantification (percentage) of F4/80+, PDGFRα+ and CD31+ stromal cell populations of WT and Dll1cKO mammary glands based on staining with respective antibodies. The boxes represent the 75th, 50th and 25th percentile of the values. The top and bottom lines represent maximum and minimal data points within the 1.5x IQ (inter quarter) range, respectively. Mann Whitney U test was used for all these analyses. n = 5 samples per genotype. (D) F4/80 antibody staining in WT and Dll1cKO mammary gland sections show reduced F4/80+ cells at TEBs and ducts of Dll1cKO mice compared to WT mammary glands. n = 3 samples per genotype. (E) Box plots showing quantification (percentage) of CD45+ F4/80+ macrophages which are positive for cleaved caspase-3 activity in WT and Dll1cKO mammary glands based on staining with respective antibodies. The boxes represent the 75th, 50th and 25th percentile of the values. The top and bottom lines represent maximum and minimal data points within the 1.5x IQ (inter quarter) range, respectively. Mann Whitney U test was used for all these analyses. n = 5 samples for WT and n = 6 samples for Dll1cKO. (F) Immunofluorescence image of Dll1-mCherry reporter mouse mammary gland section at 6 weeks shows juxtaposition of Dll1-mCherry+ cells (green) with F4/80+ (red) macrophages. Dll1mCherry+ cells were indirectly detected using a secondary antibody for mCherry that was conjugated to Alexa 488 green fluorescent dye. Macrophages were stained with F4/80 antibody, which conjugated to Alexa 568 red fluorescent dye. (G) Mammosphere assay of P4 cells from WT mice (bright field) with macrophages from Actin-dsRED mice (red). (H) Confocal images of mammospheres of P4 cells from Actin-dsRED mice with macrophages from Actin-GFP mice (green) mammary glands showing juxtaposition of basal cells (red) with macrophages (green) in mammospheres. (I) Bar graphs showing number of mammospheres formed by P4-Dll1+ and P4-Dll1− cells with and without macrophages from either mammary gland or peritoneum, respectively. n = 3 samples. Student’s t-test was used for statistical analysis. (J-K), GSEA showing enrichment of Notch and Wnt signaling pathway signature in mammary gland macrophages compared to peritoneal macrophage populations. (L) Fold-change in gene expression of the most differentially expressed Notch and Wnt genes between mammary gland and peritoneal macrophage populations from WT mice. (M-O) Western blot showing Notch4, Notch3 and Notch1 protein expression in the sorted population of mammary resident macrophages (M-Mϕ) (6 weeks old virgin mice were used), resident peritoneal macrophages (P-Mϕ) and activated peritoneal macrophages (PAct-Mϕ) respectively. Size bar: 40 μm in (D) and 20 μm in (F and H) respectively.
Next, we performed gene expression profiling to study the molecular difference between mammary and peritoneal (both resting and activated) macrophages. Unsupervised clustering shows that the overall gene signature of the mammary macrophages is more similar to the resting peritoneal macrophages than activated peritoneal macrophage (fig. S8B). Interestingly, GSEA showed that, compared to peritoneal macrophages, the mammary gland macrophages are enriched for Wnt and Notch related gene signatures (Fig. 5J, K), including several Wnt ligands and Notch receptors (Fig. 5L), which are known for their involvement in stem cell regulation. Elevated expression of Notch 1, 3, and 4 in mammary macrophages compared to peritoneal macrophages was further confirmed by western blot analysis (Fig. 5M–O).
Depletion of macrophages reduces function of Dll1+ MaSCs
We next investigated the Dll1-Notch-dependent function of macrophages as a component of the MaSC niche. First, we used clodronate liposomes (CL) to systemically deplete macrophages in Dll1-mCherry transgenic mice by intra-peritoneal (IP) injection on alternate days for one week starting at 3–4 weeks of age. The clodronate liposomes are non-toxic until ingested by macrophages. Once ingested, they are then broken down by liposomal phospholipases to release the drug that subsequently induces cell death in macrophages by apoptosis (38). Systemic ablation of macrophages decreased Dll1mcherry+ MaSCs (fig. S9A, B), which is consistent with the increased apoptosis of Dll1mcherry+ MaSCs (fig. S9C, D). As a parallel approach, we performed the cleared fat pad transplantation assay with control and CL treated Dll1mcherry+ MaSCs, using a similar method as described in an earlier report (15). We observed a nearly complete inhibition of reconstitution by the CL-treated Dll1mcherry+ MaSCs in contrast to the control cells, indicating the dependence of MaSCs on macrophages (fig. S9E, F).
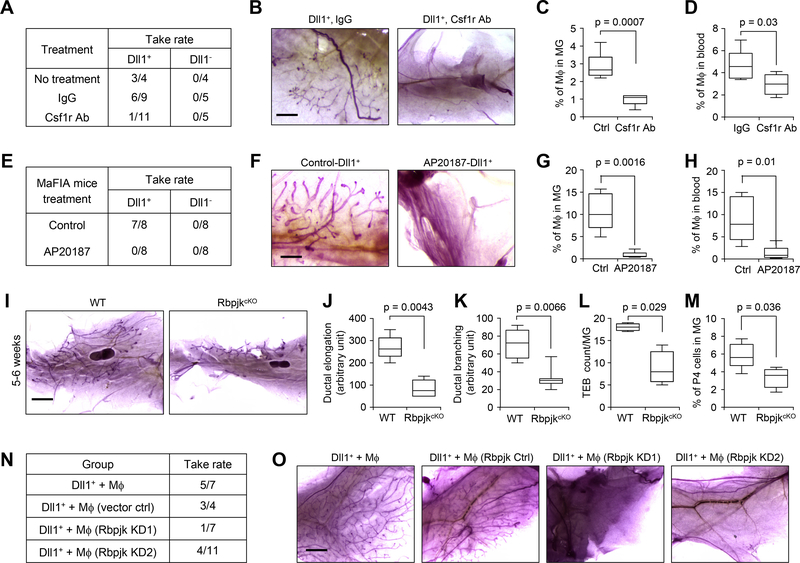
To more specifically test whether macrophages are necessary for Dll1+ MaSC activity, we used two additional approaches to deplete macrophages in vivo: 1) Csf1r blocking antibody treatment (39) (Fig. 6A–D), and 2) Macrophage Fas-Induced Apoptosis (MaFIA) mice (40) (Fig. 6E–H). In MaFIA transgenic mice, the Csf1r promoter was used to drive the expression of a mutant human FK506 binding protein 1A, which preferentially binds the dimerization drug AP20187. Administration of AP20187 causes inducible apoptosis and depletion of macrophages in the MaFIA mice (40). Both Csf1r blocking antibody treatment in WT mice and AP20187 treatment in MaFIA mice significantly reduced the number of macrophages (Fig. 6C, D and Fig. 6G, H) without affecting dendritic cells and neutrophils (fig. S9G), and dramatically blocked the repopulation of the mammary gland by Dll1+ P4 cells (Fig. 6A, B, E, F).
Fig. 6. Depletion of macrophages or genetic knockout of Notch signaling in macrophage reduces stem cell activity of Dll1+ basal cells.
(A) Table shows take rate of transplantation with 200 P4-Dll1+ and P4-Dll1− cells from Dll1-mCherry mouse mammary gland. Recipient mice were treated with IgG (control) and Csf1r antibody (500 μg/mouse) for 4–5 weeks. (B) Representative alum carmine stained whole-mount mammary outgrowths from (A). (C-D) Quantification of macrophages from mammary gland and peripheral blood from control and Csf1r antibody treated mice. FACS was performed using CD45 and F4/80 antibodies to detect macrophages (n= 5 mice per condition). (E) Table shows take rate of transplantation with 500 P4-Dll1+ and P4-Dll1− cells from Dll1-mCherry mice mammary gland into MaFIA recipient mice. Recipient MaFIA mice were treated with either vehicle or AP20187 (5 mg/Kg) for 3 weeks. AP20187 was used to induce depletion of macrophages in MaFIA mice. (F) Representative alum carmine stained whole-mount mammary outgrowths from (E). (G-H) Quantification of macrophages from mammary gland and peripheral blood from control and MaFIA treated mice. FACS was performed using CD45 and F4/80 antibodies to detect macrophages (n= 5 mice per condition). (I) Representative alum carmine stained whole-mount mammary outgrowths from WT and RbpjkcKO (CD11c-Cre; Rbpjkf/f) mice at 5–6 weeks. CD11c-Cre deletes Rbpjk in mammary tissue macrophages (41). (J-L) Box plot analyses of ductal elongation, branching and terminal end buds (TEB) counts in WT and RbpjkcKO mice. Quantification of ductal branching (tertiary branch points) was measured in defined area. (M) Box plot showing percentage of P4 (basal) cells in WT and RpjkcKO mouse after FACS using CD24 and CD29 antibodies in Lin− cells. In (J-M), n = 5 samples for WT and RbpjkcKO. (N) Table shows take rate of transplantation using mixed population of 500 P4-Dll1+ and 2000 mammary macrophages. Mammary macrophages were sorted from Actin-GFP mice using F4/80 and CD140 antibodies and either infected with control lentivirus or Rbpjk shRNAs (KD1 and KD2). P4-Dll1+ cells were obtained from sorting of Dll1-mCherry mouse mammary gland. p=0.0266 by Fisher Exact test when comparing the two control groups vs. the two Rbjp KD groups. Please refer to schematic in fig. S9H for additional details of the experimental design. (O) Representative carmine alum stained images of transplants of different groups from (N). In all box plots, the boxes represent the 75th, 50th and 25th percentile of the values. The top and bottom lines represent maximum and minimal data points within the 1.5x IQ (inter quarter) range, respectively. Mann Whitney U test was used in (C), (D), (G), (H) and (J-M). Size bar, 1 mm in (B), (F) and (O), 2 mm in (I).
We further used two in vivo models to investigate the importance of Notch signaling in macrophage for sustaining MaSC activity. First, we used the previously reported RbpjkcKO (CD11c-Cre; Rbpjk floxed) mouse model (41). In these mice, Rbpjk, a mediator of Notch signaling is conditionally deleted in the macrophage population using CD11c-Cre. Whole-mount carmine staining of the mammary glands from 5–6 week old RbpjkcKO mice revealed a significant reduction in mammary ductal elongation, branching, terminal end buds (TEB) counts compared to the wild-type littermate (Fig. 6I–L). This phenotype of altered branching morphogenesis in RbpjkcKO mice mammary gland is also associated with decreased basal (P4) population (Fig. 6M), phenocopying Dll1cKO mouse model. Next, we used an ex vivo transplant method to test the dependence of MaSC activity of Dll1+ cells on Notch signaling in macrophages (fig. S9H). Macrophages were isolated from mammary gland of Actin-GFP mice (WT), followed by lentiviral knock down of Rbpjk (fig. S9I) using two previously reported shRNAs (32). Rbpjk-KD and control mammary macrophages were then mixed with P4-Dll1+ cells obtained from Dll1-mCherry reporter mouse and transplanted into recipient NSG mice, which have defective macrophages. Take rate of mammary outgrowths showed a significant reduction when Rbpjk KD macrophages were mixed with P4-Dll1mcherry+ cells as compared to control macrophages (Fig. 6N–O and fig. S9H–I). Taken together, these studies demonstrate a functional dependence of Dll1+ MaSCs on mammary macrophages through Notch signaling.
Dll1 regulates Notch signaling in neighboring macrophages
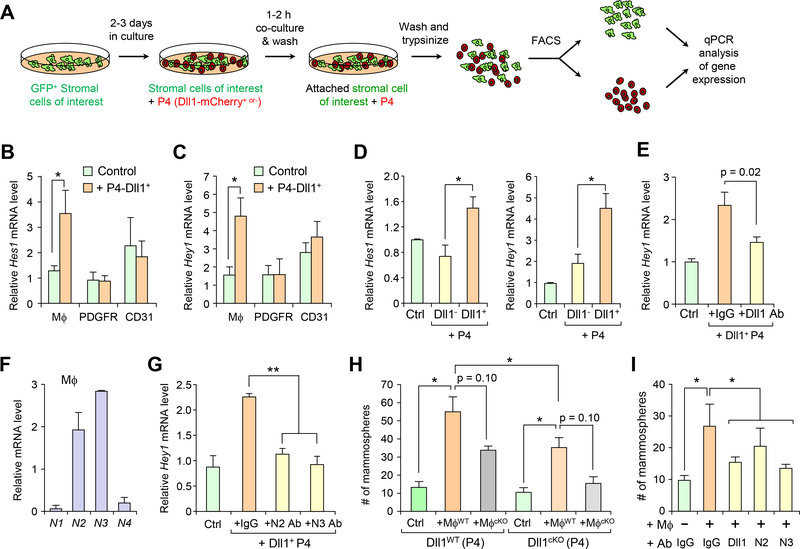
We developed an in vitro co-culture assay to further interrogate the molecular connection between Dll1-mediated Notch signaling between macrophages and Dll1+ MaSCs. Briefly, stromal cells from mammary glands were isolated from GFP transgenic mice by lineage-specific sorting and plated on gelatin-coated plates for 3 days, followed by the addition of P4-Dll1mcherry+ cells for 90 minutes, before sorting the adhered P4 and stromal cells based on their fluorescence marker expression for gene expression analysis by qPCR (Fig. 7A). Notably, this short-term 3–5 day culture of macrophages on gelatin did not significantly alter their characteristics as observed by most cell remains positive for F4/80 expression (fig. S10A). Interestingly, addition of P4-Dll1mcherry+ cells to the culture induced Hes1 and Hey1 expression in only the F4/80+ macrophage population, but not in fibroblasts and endothelial cells (Fig. 7B, C), indicating a functional Notch signaling between Dll1+ MaSCs and their neighboring macrophages. To further confirm the Dll1-mediated specific interaction between MaSCs and macrophages at the MaSC niche, we compared the interaction between P4-Dll1mCherry+ and P4-Dll1mCherry− cells with macrophages using the co-culture system. We observed greater Hes1 and Hey1 expression in Dll1mCherry+ basal cells compared to Dll1mCherry− basal cells when co-cultured with macrophages (Fig. 7D), and such Dll1-dependent gene activation was suppressed with a Dll1-blocking monoclonal antibody (Fig. 7E). These results indicate a Dll1-dependent specific interaction between MaSCs and macrophages, as well as subsequent activation of Notch signaling in the macrophages.
Fig. 7. Dll1 regulates Notch signaling in macrophages as part of the MaSC niche.
(A) Schematic of co-culture assay using Dll1+ and Dll1−basal cells with different mammary gland stromal cells including macrophages. (B-C) qRT-PCR analyses of the expression of Hes1 and Hey1 in respective stromal cells (F4/80+ or PDGFRα+ or CD31+) after co-culture of control (no cell added) or P4-Dll1+ cells for 90 mins. (D) qRT-PCR analyses of the expression of Hes1 and Hey1 in F4/80+ cells after co-culture with P4-Dll1+ and P4-Dll1− cells respectively. (E) qRT-PCR analyses of the expression of Hey1 in F4/80+ cells after co-culture with P4-Dll1+ cells with and without blocking antibody against Dll1. (F) qRT-PCR analyses of the expression of Notch receptors 1–4 (N1, N2, N3, N4) in freshly sorted F4/80+ cells from WT mammary gland. (G) qRT-PCR analyses of the expression of Hey1 in F4/80+ cells after co-culture with P4-Dll1+ cells with or without antibodies against Notch2 and Notch3 receptors. (H) Mammosphere assay with P4 cells from WT and Dll1cKO mammary epithelial cells with and without WT and Dll1cKO macrophages as indicated in the Figure, n = 5 samples. (I) Mammosphere assay with P4-Dll1+ cells with and without macrophages and treatment of antibodies against Dll1, Notch2, and Notch3, as indicated in the Figure, n = 3 samples. Initial sort for stromal cells was performed using an antibody cocktail of F4/80, CD140 and CD31 antibodies. WT mice were used except in (D), where Actin-GFP mice were used to isolate stromal cells (green color). For overlay of P4-Dll1+ cells, Dll1-mCherry reporter mouse mammary gland was used. Combination of lineage, CD24, CD29 and mCherry fluorescence color was used to sort P4-Dll1+ cells or P4-Dll1+ cells. After 90 minutes of co-culture, cells were sorted based on gating on mCherry population as stromal cells were from either WT mice or Actin-GFP mice. All qRT-PCR experiments were performed three times. n = 3 samples, each with qRT-PCR in technical duplicate, and data are presented as the mean ± SD. *p < 0.05, **p < 0.01 by Student’s t-test.
We next evaluated which Notch receptor was responsible for the Dll1-mediated signaling between MaSCs and macrophages. qPCR analysis of F4/80+ macrophages from WT type cells of the mammary gland indicated Notch2 and Notch3 as the most abundantly expressed Notch receptors in macrophages (Fig. 7F). To examine whether Dll1-mediated Notch signaling was through Notch2 or Notch3 receptor, or both, we performed a co-culture experiment between P4-Dll1+ mammary stem cells and macrophages with antibodies blocking either Notch2 or Notch3. qRT-PCR analysis demonstrated reduced Hey1 (Fig. 7G) and Hes1 (data not shown) expression in F4/80+ macrophages co-cultured with Dll1+ MaSCs when the co-culture was treated with the Notch2 or Notch 3 blocking antibody, indicating that Dll1-mediated crosstalk between macrophages and MaSCs is likely mediated by both Notch2 and Notch3 receptors in macrophages.
Next, we examined the functional importance of Dll1-Notch signaling in MaSC-macrophage niche interaction by using mammosphere co-culture assays. No significant difference in mammosphere number was observed between WT and Dll1cKO P4 cells (Fig. 7H), which is consistent with earlier data showing no functional difference of these P4 cells in transplantation assay (Fig. 2F). When WT macrophages were mixed with WT P4 cells, a dramatic increase in mammosphere number was observed compared to the P4 cells alone (Fig. 7H), suggesting a macrophage-mediated effect on MaSCs. This increase in mammosphere number was reduced when Dll1cKO P4 cells were co-cultured with WT macrophages or Dll1cKO macrophages. Interestingly, a trend toward reduced number of mammospheres was also observed when macrophages from Dll1cKO mice (Mϕ cKO) were used in conjunction with WT or Dll1cKO P4 cells, indicating an altered cellular property of the macrophages from Dll1cKO mammary glands. Indeed, Notch1–4 expression was reduced in Dll1cKO macrophages compared to WT macrophages (fig. S10B), in line with similarly lower level of Notch receptor expression in peritoneal macrophages as compared to mammary gland macrophages (Fig. 5L), indicating a Dll1-dependent influence of mammary epithelial cells on the cellular properties of mammary gland macrophages. Consistent with the role of Dll1 and Notch2/3 in mediating the crosstalk between MaSC and macrophage, treatment of the mammosphere co-culture by antibodies against Dll1, Notch 2 or Notch 3 significantly reduced the number of mammospheres (Fig. 7I). Overall, our data indicates a novel Dll1-mediated Notch signaling between MaSCs and macrophages that are crucial for supporting MaSC activity.
Dll1-dependent expression of Wnt ligands in macrophages
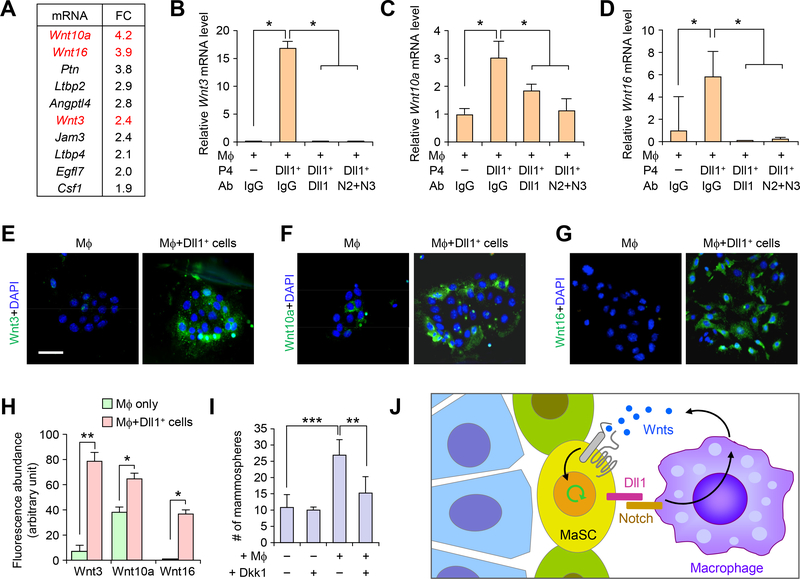
To gain mechanistic insight as to how macrophages dictate the cell fate of MaSCs at the niche, we performed global transcriptomic analysis of F4/80+ macrophages isolated from WT and Dll1cKO mammary glands. Focusing on extracellular secreted factors and cytokines, we found that among the 10 most differentially expressed genes were three Wnt ligands: Wnt10A, Wnt16 and Wnt3 (Fig. 8A), which is consistent with our earlier data (Fig. 5K, L) showing that mammary macrophages are enriched for Wnt signaling genes compared to peritoneal macrophages. To further confirm Dll1-dependent stimulation of Wnt ligand gene expression in macrophages, we performed qRT-PCR analysis of Wnt3, Wnt10a, and Wnt16 in the co-culture system (Fig. 8A). All three Wnt genes were strongly induced in macrophages after co-culture with Dll1+ MaSCs (Fig. 8B–D), which was further confirmed with immunofluorescence analysis of protein expression (Fig. 8E–H). This induction was blocked with antibodies against Dll1 or against both Notch 2 and Notch 3 (Fig. 8B–D). The critical function of Wnt signaling has been well established in MaSCs (25, 26, 28) and increased Wnt signaling has been shown to result in the expansion of the MaSC population (3). Therefore, we next tested the role of Wnt signaling in macrophages-mediated enhancement of MaSC using the mammosphere co-culture assay. Consistent with our earlier data (Fig. 7H–I), MaSC-enriched P4 cells co-cultured with macrophages exhibited a 3-fold increase of mammosphere number (Fig. 8I), an effect that was largely blocked when the co-culture was treated with a Wnt signaling inhibitor Dkk1 (Dickkopf-1) (Fig. 8I). These results indicate that the regulation of the MaSC population by macrophage is likely mediated by increased Wnt ligand production by macrophages in response to Dll1-Notch signaling.
Fig. 8. Dll1 mediated crosstalk between MaSCs and macrophages promote Wnt ligand expression in macrophages to support MaSC activity.
(A) Fold-change in gene expression of the most differentially expressed genes encoding secreted factors or extracellular proteins between WT and Dll1cKO macrophage populations from mammary glands. (B-D) qRT-PCR analyses of expression of Wnt3, Wnt10a and Wnt16 in F4/80+ cells after co-culture with P4-Dll1+ cells with and without blocking antibody against Dll1, Notch2 and Notch3 antibodies. n = 3 samples, each with qRT-PCR in technical duplicate, and data are presented as the mean ± SD. (E-G) Representative IF images of co-culture cells (macrophages cultured for 3 days followed by addition of P4-Dll1+ cells for 5h) stained with Wnt3, Wnt10a and Wnt16 antibodies. Co-culture was washed extensively to remove P4-Dll1+ overlay cells from macrophages in short co-culture system. (H) Quantification of Wnt3, Wnt10a and Wnt16 immunofluorescence intensity in indicated groups from (E-G). Control was macrophage cultured alone without P4-Dll1+ cells. (I) Mammosphere assay of WT P4 cells with and without co-culture with macrophages along with Wnt inhibitor, Dkk1, n = 4 samples. For macrophage isolation, combination of F4/80 and CD140 antibodies were used. ***p < 0.001 and **p < 0.01 by Student’s t-test in (B-D) and (I). Mann Whitney U test was used in (H). Size bar, 10 μm in (E), (F) and (G). (J) Model showing crosstalk of Dll1+ MaSC enriched population with macrophages through Notch and Wnt signaling.
DISCUSSION
The stem cell niche dictates the stem cell number and other key properties such as self-renewal through inter-cellular signaling between stem cells and the niche stromal cells. Unlike other tissues such as intestine, muscle, blood, hair follicle and skin, where the niche has been extensively studied, MaSC niche has not been well characterized. This is mostly due to the lack of insights into the precise localization of MaSCs and their associated stromal niche components in the mammary gland. In our study, we first showed that Dll1 is a marker and crucial regulator of MaSCs. Using transplantation assays, we showed that Dll1+/hi cells are enriched for MaSCs with increased regenerative potential. We further confirmed that Dll1+ basal cells can give rise to both basal and luminal cells by lineage tracing experiment. However, similar to other published models using lineage tracing (27, 42), we cannot completely rule out the possible contribution of a small fraction of Dll1+ luminal cells to the luminal population expansion, although cleared fat pad injection experiments clearly demonstrated very low MaSC activity of the luminal population. Moreover, tomato expression after 2 days of induction shows only a small ~1% tomato+ population in the luminal compartment indicating most of the induced cells are in the basal cell population with increased regenerative potential (fig. S5A and Fig. 4C).
Macrophages have been reported to be components of the spermatogonial and hematopoetic stem cell niches (43, 44). They have also been shown to be important for mammary gland development (15); however, the exact signaling mechanism between macrophages and MaSCs is not known. In our study, we found that the F4/80+ macrophage population was reduced in Dll1cKO mammary gland in different developmental stages, likely due to increased cell death as seen by cleaved caspase-3 activity. It is also possible that lower expression level of Csf1, a critical cytokine for macrophage differentiation, in mammary gland macrophage of Dll1cKO mice as compared to WT mice (Fig. 8A) might also contribute to the lower number of mature macrophage. Interestingly, reduced ductal elongation and branching phenotype of Dll1cKO mammary gland is similar to the mammary gland defects observed in the Csf1 knockout mice with macrophage deficiency (14). Similarly, we also observed that Dll1+ cells could not regenerate mammary gland when macrophages are depleted by clodronate liposomes or Csf1r antibody treatment, or by AP20187 injection in the MaFIA mice. Moreover, using CD11c-Cre;RbpjkcKO mice and lentiviral mediated Rbjpjk knockdown in macrophages, we further showed the dependence of Dll1+ MaSCs on Notch signaling in mammary macrophages. These studies thus delineate macrophages as one of the important components of the mammary stem cell stromal niche. Gene expression analysis identified several Wnt and Notch signaling genes enriched in the mammary macrophage populations compared to peritoneal macrophages, indicating unique nature of mammary gland macrophages to sustain MaSC pool. These results collectively indicate reciprocal interactions between macrophages and MaSCs — Dll1-Notch signaling from MaSCs to macrophages maintains the number and niche-related activity of the macrophages; conversely, the macrophageal niche is crucial for sustaining the MaSC pool.
Consistent with these notions, we established and used a novel 3-D co-culture assay to show that Dll1+ MaSC-enriched basal cells interact with stromal macrophages through Notch2/3 receptors. We would like to point out that this organoid co-culture system aims to mimic the point where there is substantial contact between MaSC and macrophages. Using this system, co-culturing MaSCs with macrophages resulted in significant increase of stem cell activity of Dll1+ cells. Conditional knockout of Dll1 in the mammary epithelial cells not only renders the Dll1cKO basal cells less responsive to macrophage activation, but also reduces the potency (number and fate) of macrophages from Dll1cKO mice in supporting MaSC function. Importantly, macrophages isolated from Dll1cKO mice have reduced expression of several Wnt family ligands, including Wnt3, Wnt10A, Wnt16, suggesting that Dll1-dependent Notch signaling is responsible for promoting the expression of the Wnt ligands in the macrophages that are in close contact with MaSCs.
Although there is strong evidence that Wnt signaling is important for MaSCs, the source of such Wnt ligands is previously unknown. Our study shows that mammary gland macrophages produce Wnt ligands after Notch signaling is activated by Dll1 from MaSCs, which in return induces the MaSC activity. This situation is somewhat reminiscent of the crypt stem cell niche, where Paneth cells produce large amounts of Wnt3 to maintain stem cells, and where stem and Paneth cells communicate through Notch/delta signaling (45, 46). Stroma mediated Wnt/β-catenin signal has also been reported to promote the self-renewal of hematopoietic stem cells (47). Notably, we have previously reported high level of ΔNp63 expression in MaSCs, which transcriptionally activates the expression of Wnt receptor Fzd7 (30). Therefore, ΔNp63 and Dll1 not only are markers of MaSCs, but also functionally support MaSC activity through sustaining a locally enriched Wnt signaling environment.
In conclusion, our study establishes macrophages as important cellular components of the MaSC niche through intercellular coupling of Notch and Wnt signaling (Fig. 8J). It is possible that additional niche stromal cells may also play an important role for MaSC regulation, which requires future exploration. In the context of Dll1 mediated Notch signaling in MaSC-macrophage cross-talk, we found that Dll1 produced from MaSCs activates Notch signaling in macrophages to enhance the expression of Wnt ligands, which in turn supports Wnt signaling in MaSCs to maintain stem cell activity (Fig. 8J). Since Dll1-Notch signaling requires direct cell-cell contact, and Wnt ligands mostly act as short-range intercellular signals, the Dll1-mediated coupling of Notch-Wnt signaling ensures a spatially delimiting mechanism for localized MaSC-macrophageal niche interaction (23). As Notch and Wnt pathways have been reported to be key oncogenic pathways in breast cancer, and macrophages are a major component of the tumor microenvironment, future studies of Notch-Wnt dependent interaction between MaSCs and macrophage may provide novel insights into tumor initiation and progression in breast cancer.
Material and Methods
Animal studies
Animal procedures were conducted in compliance with Institutional Animal Care and Use Committee (IACUC) of Princeton University, University of Pennsylvania and Memorial Sloan Kettering Cancer Center. Dll1 floxed mice (33), Dll1-GFP-IRES-Cre-ERT2 mice (21) and CD11c-Cre; RbpjkcKO mice (41) mice have been described previously. The Dll1-mCherry transgenic mice, which were generated using a genomic BAC clone with mCherry cDNA inserted after the start codon of Dll1, will be described elsewhere (Gao & Aifantis, manuscript in preparation). tdTomato mice (B6;129S6-Gt(ROSA)26Sortm9(CAG-tdTomato)Hze/J), Actin-GFP, Actin-dsRED mice and MaFIA mice were obtained from Jackson Laboratory. For all animal experiments, control littermate animals were utilized. For cleared fat pad injection experiment, C57/B6, athymic nude and NSG mice at 3 weeks old were anaesthetized and a small incision was made to reveal the mammary gland. Mammary epithelial cells (MECs) as specified in each experiment were injected into cleared inguinal (#4) mammary fat pads according to the standard procedures (48).
Limiting dilution assay (LDA)
Single cell suspension of primary MECs from WT and Dll1cKO mammary glands at 5–7 weeks were sorted using the lineage (CD31, Ter119 and CD45), CD24, and CD29 markers to obtain MaSC-enriched P4 population (Lin−CD24+CD29hi), which were then injected into cleared mammary fat pads. The outgrowths were analyzed at 6–8 weeks post-transplantation. Transplantation was performed with indicated number of cells resuspended in 50% Matrigel and 50% PBS. For transplantation assay with clodronate liposomes (CL) treatment, assay was performed following protocol from the previously published work (15). For transplantation assay using Csf1r blocking antibody, mice were pre-treated once with either control IgG or blocking antibody followed by treatment every 3 days with antibodies at a concentration of 500 μg/mice. 200 P4-Dll1+ or P4-Dll1− basal cells were used for transplantation. Treatment continued for 4 weeks and mice were harvested at 6 weeks after injection. For transplantation using MaFIA mice as recipients, 500 P4-Dll1+ or 500 P4-Dll1− basal cells were injected into cleared mammary gland of MaFIA mice and these mice were treated (by IP injection) with AP20187 at 5 mg/Kg (Ariad Pharmaceuticals, Cambridge, MA) every 3 days, which leads to depletion of macrophages. Similar to Csf1r antibody experiment, mice were pre-treated once with the control or drug AP20187 at the concentration of 10 mg/Kg. Treatment continued for 3 weeks and mice were euthanized to examine reconstitution of mammary gland at 5 weeks after injection. For, the ex vivo transplant assay with mixture of mammary macrophages with P4-Dll1mCherry+ cells, please see schematic in Fig. S9H for the detailed process. Frequency of MaSCs in the transplanted cell suspension was calculated using L-calc software (StemCell Technologies) or ELDA (Extreme Limiting Dilution Assay) (49). Single hit model was also tested using ELDA and value of slope was 1. MaSC abundances were assumed to follow a Poisson distribution in LDAs, and generalized linear models utilizing a log-log link function were used to derive repopulation frequency parameters. Self-renewal activity of MaSCs after transplantation was tested by their ability to regenerate functional mammary glands in virgin mouse.
Clondronate liposome (CL) assay
For systemic treatment of Dll1mCherry+ reporter mice, the animals were treated with CL (150–170 μl) at 5 weeks of age (mice body weight ~15–17g) every other day for a week before harvesting the mammary gland. For mammary fat pad clearing assay, P4-Dll1mCherry+ cells were treated with or without CL following the published procedure (15) and then injected into cleared mammary fat pad of C57/B6 mice. Transplants were harvested 6 weeks post injection.
Mammosphere assays
Stem cells from the mammary gland have been successfully maintained and passaged in vitro as spheroids in suspension (50–52). Cells were cultured as previously described (32). For co-culture mammosphere assay with P4 basal cells (WT or Actin-dsRED) and macrophages (WT or Actin-dsRED or Actin-GFP), 5,000 P4 cells were mixed with 20,000 macrophages and grown in low adherent plate in mammosphere media (32). This 1:4 ratio of P4:macrophage co-culture was determined to be the optimal in vitro co-culture condition in which macrophages strongly enhance the mammosphere forming activity of P4 cells (data not shown)
Cloning, viral production and infection
The pLEX plasmid (Open Biosystems) expressing Dll1 cDNAs was generated by routine molecular cloning techniques. All plasmids were packaged into virus using HEK293-T cells as packaging cell lines and helper plasmids VSVG and dR8.9 following standard protocols. Primary cells were spin-infected with virus-containing media supplemented with 2 μg/mL polybrene for 2 hours at 1000 g at 4°C and then transplanted. Rbpjk shRNAs (purchased from Open Biosystems Inc.,Huntsville, AL) were previously validated in our earlier studies (32). Macrophages from Actin-GFP mice were sorted using cocktail of F4/80 and CD140 antibodies and were spin infected similar to mammary epithelial cells using lentivirus.
Co-culture assay
Briefly, various stromal cell populations from WT or Actin-GFP+ mice mammary glands were isolated by sorting and plated on gelatin-coated plates for 3–5 days. Dll1 (0.75 μg/ml) or Notch2 or Notch3 (1.5 μg/ml) blocking antibodies were added alone or in combination followed by the addition of control (no P4 cell), P4- Dll1mCherry+ cells (P4-Dll1+) or P4- Dll1mcherry− cells (P4-Dll1−) for 90 minutes. Cells are then washed, trypsinized and sorted for either mCherry+ and mCherry− population or mCherry+ and GFP+ population followed by RNA isolation for gene expression analysis. For IF with Wnt antibodies, macrophages were co-cultured for 5 hours with P4- Dll1mCherry+ cells. Co-culture was washed extensively to remove P4 cells. Attached macrophages were stained with respective Wnt antibodies.
Protein extraction and western blot analysis
Proteins were extracted from primary epithelial cell cultures and cell lines in RIPA buffer as previously described (31). Western blot analysis was performed using the standard protocol. Antibodies and dilutions used are listed in Supplementary Table S1.
Histological analysis, immunohistochemistry (IHC) and immunofluorescence (IF)
For histological analysis, mammary gland specimens were processed as previously described (32). Antibodies and dilutions used are listed in the Supplementary Table S1. DAPI was used to stain nuclei. Confocal images were taken using a Nikon A1 confocal microscope or Nikon TiE microscope. For immunofluorescence analysis of sorted P4 cells, cells were attached to slides by gentle cytospin followed by immunofluorescence which was performed after fixing and permeabilizing the cells for 20 mins at RT. Dll1 antibody is listed in Supplementary Table S1.
Flow Cytometry/FACS Sorting
Single mammary epithelial cells were obtained from mammary glands following the published protocol (3, 4, 30, 32). Briefly, MECs were stained with a combination of lineage, CD24 and CD29 antibodies (3) for 20–30 minutes on ice following the published protocol. FACS analysis was performed using the LSRII Flow Cytometer (BD Biosciences) and data were analyzed using FlowJo software (TreeStar, Inc). For sorting cells, FACS Vantage or Aria II instruments were used. For cleaved caspase-3 assay, MECs were fixed and then stained with antibodies following manufacturer’s protocol (BD Biosciences). For isolation and or FACS analysis of macrophages from different tissues, either CD140 and F4/80 antibody cocktail or CD45 and F4/80 antibody cocktail was used. For DCs and neutrophils, cocktail of CD45, CD11b, Gr1 and CD11c antibodies were used. Live cells were gated out using either DAPI or PI. Details about all FACS related antibodies are listed in Supplementary Table S2.
EdU assay
Mice were intraperitoneally injected with EdU (0.2 mg per 10 g body weight, Invitrogen) 2h or 12h before mammary gland harvest. EdU was visualized using Click-it Imaging reagents (647 and 488) from Invitrogen following the protocol from manufacturer. For EdU assay in FACS along with other antibodies, samples were first stained with CD24, CD29, Ter119, CD45 and CD31 antibodies, fixed and then stained with EdU following the protocol from manufacturer (Invitrogen). For immunofluorescence, paraffin embedded sections were first rehydrated using standard protocol and then stained with EdU followed by other antibodies following the manufacturer’s instructions.
qRT-PCR analyses
Total RNA was isolated from primary cells using Qiagen RNA extraction kit in accordance with the manufacturer’s instructions. Real-time RT-PCR was performed on an ABI 7900 96 HT series PCR machine (Applied Biosystem) using SYBR Green Supermix (Bio-Rad Laboratories). The gene-specific primer sets were used at a final concentration of 0.2 μM and their sequences are listed in Supplementary Table S3. All qRT-PCR assays were performed in duplicate in at least three independent experiments using three different tissue samples.
Microarray analysis
The P4, P5, P6 subpopulations of mammary epithelial cells (MECs) were isolated from the mammary glands (4 mammary glands from each group) of virgin mice. MECs were isolated using FACS as described in (53). The sorted P4 cells from Dll1-mCherry mice mammary gland or macrophages from WT and Dll1cKO mice (C57/B6 strain) at 5–6 weeks age were prepared as described. For activated peritoneal macrophages, macrophages were activated with Bio-Gel P-100 and obtained from C57/B6 mice. RNA was collected from these samples using the RNAeasy Mini Kit (Qiagen, Valencia VA) according to manufacturer’s instructions. The gene expression profiles of various populations of macrophages from the wild type and Dll1cKO mice or P4 (Dll1hi or Dll1lo) were determined using Agilent mouse GE 8×60k two-color microarrays system (Agilent, G4852A), following the manufacturer’s instructions. Briefly, the RNA samples and universal mouse reference RNA (Agilent 740100) were labeled with CTP-cy5 and CTP-cy3, respectively, using the Agilent Quick Amp Labeling Kit. Labeled testing and reference RNA samples were mixed in equal proportions, and hybridized to the mouse GE 8×60K array. The arrays were scanned with an Agilent G2505C scanner and raw data was extracted using Agilent Feature Extraction software (v11.0). Data was analyzed using the GeneSpring 13 software (Agilent). The expression value of individual probes refers to the Log2(Cy5/Cy3) ratio.
Gene set enrichment analysis (GSEA)
GSEA v2.2.0 was used to perform the GSEA on various functional and/or characteristic gene signatures (54, 55). Normalized microarray expression data were rank-ordered by differential expression between cell populations and/or genetic background as indicated, using the provided ratio of classes (i.e., fold change) metric. Two independent MaSC specific gene signatures were used to characterize MaSC characteristics. Both are defined by significantly upregulated genes (p < 0.05 and FC > 3) in MaSC-enriched subpopulations from MECs of wild type mice. Among which, the “MaSC signature and luminal cell signature from Chakrabarti et al, 2014” is derived from the microarray data collected from our lab as described in previous study (30) (GSE47493). The genes showing >3 folds up-regulation in P4 comparing to both P5 and P6 of wild type mice were included in the MaSC gene set. For luminal signature, the genes showing >3 folds up-regulation in P5 comparing to both P4 and P6 of wild type mice were included in the luminal gene set. The other MaSC and luminal cell signature is derived from published dataset (37). For gene expression in macrophages, the genes showing >3 folds up-regulation in mammary resting macrophages comparing to resting peritoneal macrophage of wild type mice were included.
Lineage tracing
Lineage tracing experiment was performed following protocols previously described (5). In brief, tdTomato reporter expression in Dll1-GFP-IRES-Cre-ERT2/ROSA- tdTomato mice were induced by intraperitoneal injection of 1.5 mg of tamoxifen (75 μl of 20mg/ml) diluted in corn oil (Sigma) at the indicated age during puberty or adulthood and kept for different time points followed by whole mount or FACS analysis.
Statistical Analysis
Results were generally reported as mean ± SD (standard deviation) as indicated in the figure legend. For comparisons of central tendencies, normally distributed datasets were analyzed using unpaired (with the exception of analyses of cellular populations from paired samples) two-sided Student’s t-tests under assumption of equal variance. Non-normally distributed datasets were analyzed using non–parametric Mann-Whitney U tests. To adjust for host effects, paired two-sided Student’s t-tests assuming equal variance were used for experiments in which cellular populations were compared following matched control and experimental cell types (Fig. 1l, 2B, S2F and S9A–C). Statistical analyses specific to Limiting Dilution Assays and Gene Set Enrichment Analysis are described above. All the experiments with representative images (including western blot, FACS plot, histology and immunofluorescence) have been repeated at least thrice and representative images were shown. For animal studies, no statistical test was performed to pre-determine the sample size. Animals were excluded only if they died or had to be sacrificed because of moribund conditions following the IACUC protocol.
Accession numbers for datasets
Microarray data reported herein have been deposited at the NCBI Gene Expression GSE77504.
Supplementary Material
Acknowledgements
This work was supported by a DOD Postdoctoral Fellowship (BC103740) and a NCI-K22 grant to R.C. (K22CA193661), a Susan G. Komen Fellowship to T. C-T (PDF15332075), and grants from the Brewster Foundation, the Breast Cancer Research Foundation, Department of Defense (BC123187), and the National Institutes of Health (R01CA141062) to Y.K and R01 CA198280-01 and P30 CA008748 to M.O.L. This research was also supported by the Genomic Editing and Flow Cytometry Shared Resources of the Cancer Institute of New Jersey (P30CA072720).
Footnotes
Competing interests
The authors declare no potential conflicts of interest.
References
- 1.Visvader JE, Stingl J, Mammary stem cells and the differentiation hierarchy: current status and perspectives. Genes Dev 28, 1143 (June 1, 2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Makarem M et al. , Stem cells and the developing mammary gland. Journal of mammary gland biology and neoplasia 18, 209 (June, 2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shackleton M et al. , Generation of a functional mammary gland from a single stem cell. Nature 439, 84 (January 05, 2006). [DOI] [PubMed] [Google Scholar]
- 4.Stingl J et al. , Purification and unique properties of mammary epithelial stem cells. Nature 439, 993 (February 23, 2006). [DOI] [PubMed] [Google Scholar]
- 5.Rios AC, Fu NY, Lindeman GJ, Visvader JE, In situ identification of bipotent stem cells in the mammary gland. Nature 506, 322 (February 20, 2014). [DOI] [PubMed] [Google Scholar]
- 6.Van Keymeulen A et al. , Distinct stem cells contribute to mammary gland development and maintenance. Nature 479, 189 (November 10, 2011). [DOI] [PubMed] [Google Scholar]
- 7.Bussard KM, Smith GH, The mammary gland microenvironment directs progenitor cell fate in vivo. Int J Cell Biol 2011, 451676 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brisken C, Duss S, Stem cells and the stem cell niche in the breast: an integrated hormonal and developmental perspective. Stem Cell Rev 3, 147 (June, 2007). [DOI] [PubMed] [Google Scholar]
- 9.Hovey RC, Aimo L, Diverse and active roles for adipocytes during mammary gland growth and function. Journal of mammary gland biology and neoplasia 15, 279 (September, 2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gregor MF et al. , The role of adipocyte XBP1 in metabolic regulation during lactation. Cell Rep 3, 1430 (May 30, 2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu X et al. , ROCK inhibitor and feeder cells induce the conditional reprogramming of epithelial cells. Am J Pathol 180, 599 (February, 2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schwertfeger KL, Rosen JM, Cohen DA, Mammary gland macrophages: pleiotropic functions in mammary development. Journal of mammary gland biology and neoplasia 11, 229 (October, 2006). [DOI] [PubMed] [Google Scholar]
- 13.O’Brien J, Martinson H, Durand-Rougely C, Schedin P, Macrophages are crucial for epithelial cell death and adipocyte repopulation during mammary gland involution. Development 139, 269 (January, 2012). [DOI] [PubMed] [Google Scholar]
- 14.Gouon-Evans V, Rothenberg ME, Pollard JW, Postnatal mammary gland development requires macrophages and eosinophils. Development 127, 2269 (June, 2000). [DOI] [PubMed] [Google Scholar]
- 15.Gyorki DE, Asselin-Labat ML, van Rooijen N, Lindeman GJ, Visvader JE, Resident macrophages influence stem cell activity in the mammary gland. Breast Cancer Res 11, R62 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bouras T et al. , Notch signaling regulates mammary stem cell function and luminal cell-fate commitment. Cell Stem Cell 3, 429 (October 9, 2008). [DOI] [PubMed] [Google Scholar]
- 17.Callahan R, Egan SE, Notch signaling in mammary development and oncogenesis. Journal of mammary gland biology and neoplasia 9, 145 (April, 2004). [DOI] [PubMed] [Google Scholar]
- 18.Dontu G et al. , Role of Notch signaling in cell-fate determination of human mammary stem/progenitor cells. Breast Cancer Res 6, R605 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rodilla V et al. , Luminal progenitors restrict their lineage potential during mammary gland development. PLoS Biol 13, e1002069 (February, 2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sale S, Lafkas D, Artavanis-Tsakonas S, Notch2 genetic fate mapping reveals two previously unrecognized mammary epithelial lineages. Nat Cell Biol 15, 451 (May, 2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van Es JH et al. , Dll1+ secretory progenitor cells revert to stem cells upon crypt damage. Nat Cell Biol 14, 1099 (October, 2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pellegrinet L et al. , Dll1- and dll4-mediated notch signaling are required for homeostasis of intestinal stem cells. Gastroenterology 140, 1230 (April, 2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Clevers H, Loh KM, Nusse R, Stem cell signaling. An integral program for tissue renewal and regeneration: Wnt signaling and stem cell control. Science 346, 1248012 (October 3, 2014). [DOI] [PubMed] [Google Scholar]
- 24.Badders NM et al. , The Wnt receptor, Lrp5, is expressed by mouse mammary stem cells and is required to maintain the basal lineage. PLoS One 4, e6594 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zeng YA, Nusse R, Wnt proteins are self-renewal factors for mammary stem cells and promote their long-term expansion in culture. Cell Stem Cell 6, 568 (June 4, 2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van Amerongen R, Bowman AN, Nusse R, Developmental stage and time dictate the fate of Wnt/beta-catenin-responsive stem cells in the mammary gland. Cell Stem Cell 11, 387 (September 7, 2012). [DOI] [PubMed] [Google Scholar]
- 27.Plaks V et al. , Lgr5-expressing cells are sufficient and necessary for postnatal mammary gland organogenesis. Cell Rep 3, 70 (January 31, 2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Clevers H, Nusse R, Wnt/beta-catenin signaling and disease. Cell 149, 1192 (June 8, 2012). [DOI] [PubMed] [Google Scholar]
- 29.Rajaram RD et al. , Progesterone and Wnt4 control mammary stem cells via myoepithelial crosstalk. EMBO J 34, 641 (March 4, 2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chakrabarti R et al. , DeltaNp63 promotes stem cell activity in mammary gland development and basal-like breast cancer by enhancing Fzd7 expression and Wnt signalling. Nat Cell Biol 16, 1004 (October, 2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Choi YS, Chakrabarti R, Escamilla-Hernandez R, Sinha S, Elf5 conditional knockout mice reveal its role as a master regulator in mammary alveolar development: failure of Stat5 activation and functional differentiation in the absence of Elf5. Dev Biol 329, 227 (May 15, 2009). [DOI] [PubMed] [Google Scholar]
- 32.Chakrabarti R et al. , Elf5 regulates mammary gland stem/progenitor cell fate by influencing notch signaling. Stem Cells 30, 1496 (July, 2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hozumi K et al. , Delta-like 1 is necessary for the generation of marginal zone B cells but not T cells in vivo. Nat Immunol 5, 638 (June, 2004). [DOI] [PubMed] [Google Scholar]
- 34.Estrach S, Ambler CA, Lo Celso C, Hozumi K, Watt FM, Jagged 1 is a beta-catenin target gene required for ectopic hair follicle formation in adult epidermis. Development 133, 4427 (November, 2006). [DOI] [PubMed] [Google Scholar]
- 35.Estrach S, Cordes R, Hozumi K, Gossler A, Watt FM, Role of the Notch ligand Delta1 in embryonic and adult mouse epidermis. J Invest Dermatol 128, 825 (April, 2008). [DOI] [PubMed] [Google Scholar]
- 36.Miyoshi K et al. , Signal transducer and activator of transcription (Stat) 5 controls the proliferation and differentiation of mammary alveolar epithelium. J Cell Biol 155, 531 (November 12, 2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Asselin-Labat ML et al. , Control of mammary stem cell function by steroid hormone signalling. Nature 465, 798 (June 10, 2010). [DOI] [PubMed] [Google Scholar]
- 38.van Rooijen N, Hendrikx E, Liposomes for specific depletion of macrophages from organs and tissues. Methods Mol Biol 605, 189 (2010). [DOI] [PubMed] [Google Scholar]
- 39.Ries CH et al. , Targeting tumor-associated macrophages with anti-CSF-1R antibody reveals a strategy for cancer therapy. Cancer Cell 25, 846 (June 16, 2014). [DOI] [PubMed] [Google Scholar]
- 40.Burnett SH et al. , Conditional macrophage ablation in transgenic mice expressing a Fas-based suicide gene. J Leukoc Biol 75, 612 (April, 2004). [DOI] [PubMed] [Google Scholar]
- 41.Franklin RA et al. , The cellular and molecular origin of tumor-associated macrophages. Science 344, 921 (May 23, 2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang D et al. , Identification of multipotent mammary stem cells by protein C receptor expression. Nature 517, 81 (January 1, 2015). [DOI] [PubMed] [Google Scholar]
- 43.Winkler IG et al. , Bone marrow macrophages maintain hematopoietic stem cell (HSC) niches and their depletion mobilizes HSCs. Blood 116, 4815 (December 2, 2010). [DOI] [PubMed] [Google Scholar]
- 44.DeFalco T et al. , Macrophages Contribute to the Spermatogonial Niche in the Adult Testis. Cell Rep 12, 1107 (August 18, 2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Clevers H, The intestinal crypt, a prototype stem cell compartment. Cell 154, 274 (July 18, 2013). [DOI] [PubMed] [Google Scholar]
- 46.Sato T et al. , Paneth cells constitute the niche for Lgr5 stem cells in intestinal crypts. Nature 469, 415 (January 20, 2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim JA et al. , Identification of a stroma-mediated Wnt/beta-catenin signal promoting self-renewal of hematopoietic stem cells in the stem cell niche. Stem Cells 27, 1318 (June, 2009). [DOI] [PubMed] [Google Scholar]
- 48.Deome KB, Faulkin LJ Jr., Bern HA, Blair PB, Development of mammary tumors from hyperplastic alveolar nodules transplanted into gland-free mammary fat pads of female C3H mice. Cancer research 19, 515 (June, 1959). [PubMed] [Google Scholar]
- 49.Lim E et al. , Aberrant luminal progenitors as the candidate target population for basal tumor development in BRCA1 mutation carriers. Nature medicine 15, 907 (August, 2009). [DOI] [PubMed] [Google Scholar]
- 50.Dontu G et al. , In vitro propagation and transcriptional profiling of human mammary stem/progenitor cells. Genes & development 17, 1253 (May 15, 2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Reynolds BA, Weiss S, Clonal and population analyses demonstrate that an EGF-responsive mammalian embryonic CNS precursor is a stem cell. Developmental biology 175, 1 (April 10, 1996). [DOI] [PubMed] [Google Scholar]
- 52.Guo W et al. , Slug and Sox9 cooperatively determine the mammary stem cell state. Cell 148, 1015 (March 2, 2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tiede BJ, Owens LA, Li F, DeCoste C, Kang Y, A novel mouse model for non-invasive single marker tracking of mammary stem cells in vivo reveals stem cell dynamics throughout pregnancy. PloS one 4, e8035 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mootha VK et al. , Integrated analysis of protein composition, tissue diversity, and gene regulation in mouse mitochondria. Cell 115, 629 (November 26, 2003). [DOI] [PubMed] [Google Scholar]
- 55.Subramanian A et al. , Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America 102, 15545 (October 25, 2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.