Abstract
The inflammasome NLRP6 plays a crucial role in regulating inflammation and host defense against microorganisms in the intestine. However, the molecular mechanisms by which NLRP6 function is inhibited to prevent excessive inflammation remain unclear. Here, we demonstrate that the deubiquitinase Cyld prevents excessive interleukin 18 (IL-18) production in the colonic mucosa by deubiquitinating NLRP6. We show that deubiquitination inhibited the NLRP6–ASC inflammasome complex and regulated the maturation of IL-18. Cyld deficiency in mice resulted in elevated levels of active IL-18 and severe colonic inflammation following Citrobacter rodentium infection. Further, in patients with ulcerative colitis, the concentration of active IL-18 was inversely correlated with CYLD expression. Thus, we have identified a novel regulatory mechanism that inhibits the NLRP6-IL-18 pathway in intestinal inflammation.
Inflammatory bowel diseases (IBDs), including ulcerative colitis (UC) and Crohn’s disease, affect about 1.6 million people in the United States of America. Most patients do not respond to current therapies and experience a loss of efficacy over time, or experience adverse events and require a colectomy1. Therefore, there is an unmet and urgent need to understand the mechanism of chronic gastrointestinal inflammation so that more specific targeted therapies can be developed.
Inflammasomes are cytoplasmic multiprotein complexes that are composed of nucleotide-binding domain and leucine-rich repeat receptor (NLR) proteins, such as NLRP1, NLRP3, NLRC4 and NLRP6, which function as innate sensors of endogenous or exogenous stress or damage-associated molecular patterns2–4. NLRP6 is highly expressed in the intestine and plays a critical role in maintaining gut homeostasis5,6. NLRP6 forms a complex with ASC and caspase-1 and facilitates the cleavage of pro-IL-18 into biologically active mature IL-18 (refs. 7,8). However, the molecular mechanism by which NLRP6 function is regulated to prevent excesive inflammation remains unclear.
Post-translational modification of proteins by covalent conjugation of ubiquitin (Ub) plays a crucial role in regulating inflammation9. Ub contains seven lysine residues, but its linkage to adjacent Ub molecules in a Ub chain generally occurs through either lysine 48 (K48) or lysine 63 (K63). K48-linked polyubiquitination predominantly targets proteins for proteasomal degradation, whereas K63-linked polyubiquitination results in non-proteasomal modifications, such as changes in subcellular localization or protein—protein interactions9. Ubiquitination is reversible and removal of Ub molecules is mediated by deubiquitinating enzymes such as Cyld (ref. 10).
In this study, we have discovered that the activity of the NLRP6—ASC inflammasome complex is regulated by Cyld-mediated deubiquitination preventing excessive IL-18 in the intestinal mucosa, which could be exploited therapeutically.
Results
Citrobacter rodentium induces severe inflammation in Cyld-/- mice.
Genome-wide association studies have shown a link between single nucleotide polymorphism and dysregulated expression of the CYLD gene in patients with IBD11,12. In line with these reports, CYLD expression was found to be reduced in the mucosal samples of patients with UC compared to that in control samples (Extended Data Fig. 1). To investigate the contribution of the defect in Cyld expression to intestinal inflammation, we inoculated Cyld-/- mice and littermate wild-type control mice with Citrobacter rodentium. Cyld-/- mice exhibited a significant decrease in body weight and severe diarrhea compared to wild-type mice (Fig. 1a,b). The lengths of the colons were significantly shortened in the C. rodentium– infected Cyld-/- mice compared to wild-type mice (Fig. 1c and Extended Data Fig. 2). Cyld-/- mice exhibited greater submucosal edema, more extensive damage to the surface mucosa and ulceration, suggesting severe colonic inflammation in Cyld-/- mice (Fig. 1d,e). Similar bacterial numbers in the feces of wild-type and Cyld-/- mice were observed following infection, suggesting that Cyld deficiency did not affect initial bacterial colonization. However, from day 8, bacterial numbers increased in the feces of Cyld-/- mice, suggesting that Cyld deficiency resulted in failure to control the C. rodentium infection (Fig. 1f). Cyld-/- mice also displayed enhanced bacterial dissemination to peripheral organs, including the spleen and liver, suggesting disruption of the epithelial barrier (Fig. 1g). To gain further evidence for the disruption of the epithelial barrier, we gavaged mice with fluorescein isothiocyanate (FITC)–dextran and monitored its translocation to the serum13. A markedly elevated level of FITC–dextran was observed in the serum of Cyld-/- mice compared to that in wild-type mice (Fig. 1h). These data collectively suggested that Cyld deficiency leads to severe colonic inflammation and increased disruption of the epithelial barrier.
Fig. 1 |. Cyld−/− mice show severe colitis induced by C. rodentium.
a–c, Body weight change (NSP=0.0965, ***P<0.0001; a), diarrheas core (***P<0.0001; b) and colon length (NSP=0.6847, ***P=0.0004; c) of wild-type (WT) versus Cyld−/− mice (n=10 mice per group) infected (inf) with C. rodentium. d,e, H&E-stained colon tissue sections from C. rodentium–infected wild-type and Cyld−/− mice (scale bars, 100 μm; d) and histology scores (NSP=0.3434, ***P<0.0001; e) of the sections (n=10 mice per group). f, Colony-forming units (CFUs) from the stool (NSP=0.7766, NSP=0.7577, NSP=0.06, **P=0.0037, ***P<0.0001, left to right). g, Quantification of CFUs from the organ culture (n=10 mice per group) (**P =0.0053, ***P=0.0003). h, The mean fluorescence intensity (MFI) showing the translocation of FITC–dextran assayed using serum 4h after administration (n=10 mice per group, NSP=0.2432, ***P<0.0001). The statistics are shown as mean ± s.d., with P values determined by Student’s t-test (two tail). The data are from one experiment representative of three independent experiments with similar results.
Cyld binds to NLRP6.
Next, we sought to identify the dysregulated pathways that lead to severe intestinal inflammation in Cyld−/− mice by a proteomics approach. We performed pulldown assays of His—Cyld protein added to cell lysates of inflamed colonic mucosa of Cyld−/− mice. The precipitated proteins were identified by mass spectrometry (MS) analysis. By this approach, we identified NLRP6 as a Cyld-binding protein in precipitates of His—Cyld. Six specific peptides (Table 1) corresponding to mouse NLRP6 were detected, as illustrated by a representative MS/MS spectrum corresponding to 17ELLMATLEELSQEQLKR33 (Fig. 2a).
Table 1 |.
Representative peptide sequences corresponding to mouse NLRP6, detected in MS/MS analysis
| No. | Sequence of peptide | Start position | End position |
|---|---|---|---|
| 1 | ELLMATLEELSQEQLKR | 17 | 33 |
| 2 | LIEFYEPVPAVEMTR | 65 | 79 |
| 3 | LGPTSVLLSVSAFK | 102 | 115 |
| 4 | LLIAPGTGAVEDELLGPLGEPEPER | 146 | 170 |
| 5 | TPGTPAGGVQK | 516 | 526 |
| 6 | FLFGLLNTEGLR | 544 | 555 |
Fig. 2 |. NLRP6 directly interacts with Cyld.
a, The proteins from the inflamed colonic mucosa of Cyld−/− mice were precipitated using His-tagged Cyld protein and subjected to MS analysis. The MS/MS spectrum corresponding to 17ELLMATLEELSQEQLKR33 is shown. Observed b- and y-ion series are indicated. b, 293T cells were cotransfected with Flag-NLRP6 and Myc-Cyld expression vectors. The cell lysates were immunoprecipitated using anti-Flag and anti-Myc and immunoblotted with anti-Myc and anti-Flag. c, His-tagged NLRP6 protein was expressed in BL21(DE3) Escherichia coli and pulled down using GST-tagged Cyld protein. The precipitated proteins were separated on SDS–polyacrylamide gel electrophoresis (PAGE) and immunoblotted with anti-His and anti-GST. d, Colonic lamina propria lysates from uninfected and infected C57BL6 mice were subjected to immunoprecipitation with IgG, anti-NLRP6 or anti-Cyld and immunoblotted with anti-NLRP6 or anti-Cyld antibody. The data are representative of three or more independent experiments. Uncropped blots (b–d) are shown in the source data.
Next, we immunoprecipitated NLRP6 using anti-NLRP6 and the lysate of inflamed colonic mucosa from wild-type mice, which was followed by MS analysis. In line with the His—Cyld pulldown results, ten specific peptides (Table 2) corresponding to Cyld were found in the immunoprecipitates of NLRP6. A representative MS/MS spectrum corresponding to 157TVSGIFFGVELLEEGR172 is shown in Supplementary Fig. 1a.
Table 2 |.
Representative peptide sequences corresponding to mouse Cyld, detected in MS/MS analysis
| No. | Sequence of peptide | Start position | End position |
|---|---|---|---|
| 1 | VTSPYWEERIFYLLLQECSVTDK | 11 | 33 |
| 2 | FTELLLAITNCEER | 96 | 109 |
| 3 | GLQVDVGSPVK | 122 | 132 |
| 4 | TVSGIFFGVELLEEGR | 157 | 172 |
| 5 | GQGFTDGVYQGK | 173 | 184 |
| 6 | WIGQPPGLSDVLAGLELEDECAGCTDGTFR | 489 | 518 |
| 7 | EKNDIEYYSETQELLR | 625 | 640 |
| 8 | KIFPSLELNITDLLEDTPR | 767 | 785 |
| 9 | DLPDWDWR | 842 | 849 |
| 10 | MSLEDLHSLDSR | 917 | 928 |
To confirm our MS findings, we transfected 293T cells with Myc—Cyld and Flag—NLRP6 expression constructs. Cell lysates were immunoprecipitated using either anti-Flag or anti-Myc. Anti-Myc immunoprecipitated Flag—NLRP6 and anti-Flag immunoprecipitated Myc—Cyld, suggesting that Cyld and NLRP6 physically interact (Fig. 2b). To test whether the Cyld—NLRP6 interaction was direct, we performed pulldown assays using GST—Cyld and His—NLRP6. GST—Cyld but not GST alone precipitated His—NLRP6, confirming a direct Cyld—NLRP6 interaction (Fig. 2c). To determine whether this interaction occurs endogenously, we performed co-immunoprecipitation experiments with anti-Cyld and anti-NLRP6 using the colonic mucosal lysates of C. rodentium—infected mice, which showed an interaction between Cyld and NLRP6 (Fig. 2d). Cyld interaction with NLRP6 was specific, as Cyld did not associate with other members of the NLR family such as NLRP3 and NLRP12 (Supplementary Fig. 1b,c), which are also expressed in the colon. These results suggested that Cyld associates with NLRP6 during an inflammatory response.
Cyld cleaves K63-linked ubiquitination of NLRP6.
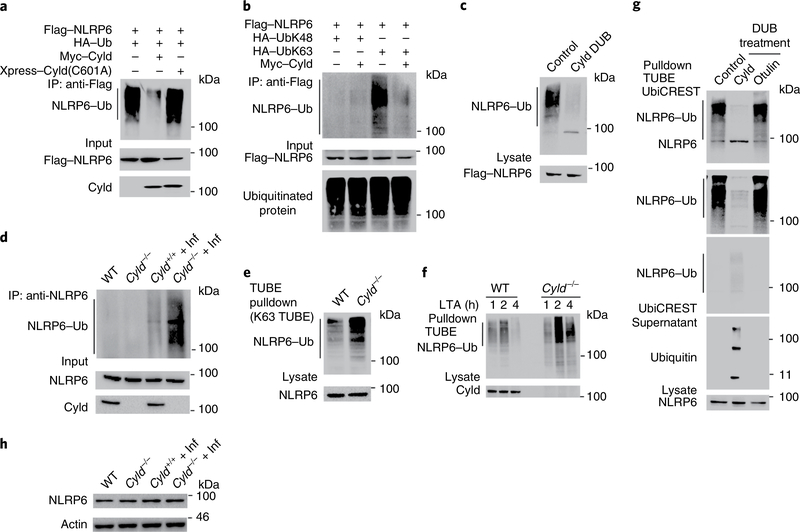
To investigate whether Cyld regulates NLRP6 via deubiquitination, we transfected 293T cells with Flag—NLRP6, Myc—Cyld and hemagglutinin (HA)—Ub. We precipitated NLRP6 using anti-Flag and immunoblotted the membranes with anti-HA. We observed slow-migrating, high-molecular-weight, polyubiquitinated forms of NLRP6 (Fig. 3a, lane 1) only when NLRP6 was coexpressed with Ub. NLRP6 ubiquitination was diminished in the presence of wild-type Cyld but not with the Cyld(C601A) deubiquitinase-defective mutant, suggesting that Cyld deubiquitinates NLRP6 (Fig. 3a).
Fig. 3 |. Cyld controls the deubiquitination of the NLRP6 protein.
a, Immunoassay of 293T cells transfected with expression vectors for various combinations (upper lanes) of Flag-NLRP6, Myc–Cyld, Xpress-Cyld(C601A) and HA-Ub(wild type), followed by immunoprecipitation of lysates with anti-Flag and immunoblot analysis with anti-HA, anti-Flag and anti-Myc. b, 293T cells were transfected with Flag-tagged NLRP6 and the combination of HA-UbK48, HA-UbK63 and Myc-Cyld followed by immunoprecipitation of lysates with anti-Flag and immunoblot analysis with anti-HA and anti-Flag. c, 293T cells were transfected with Flag-tagged NLRP6 and HA-UbK63 followed by immunoprecipitation of lysates with anti-Flag. Immunoprecipitates purified by re-immunoprecipitation and incubated with purified His-Cyld and incubated immunoprecipitates were examined by immunoblotting with anti-HA and anti-Flag. d, Colonic mucosal lysates from uninfected and inflamed wild-type and Cyld−/− mice were immunoprecipitated with anti-NLRP6 and immunoblotted against anti-UbK63. e, Colonic epithelial cells from wild-type and Cyld−/− mice were stimulated with LTA by transfection. The cells were lysed and proteins were pulled down using Flag-tagged K63 TUBEs. Pulled-down proteins were immunoblotted with anti-NLRP6. f, Wild-type and Cyld−/− colonic epithelial cells were stimulated with LTA. The cells were lysed and proteins were pulled down using Flag-tagged K63 TUBEs. Pulled-down proteins were immunoblotted with anti-NLRP6. g, UbiCREST analysis of ubiquitinated NLRP6 isolated with Pan TUBE from LTA-transfected Cyld−/− colonic epithelial cells. h, The lysates of colonic mucosa from uninfected and infected wild-type and Cyld−/− mice were immunoblotted with anti-NLRP6 and anti-actin. The data are representative of three or more independent experiments. Uncropped blots (a–h) are shown in the source data.
To test whether Cyld cleaves K48- or K63-linked ubiquitination, we coexpressed Flag—NLRP6, Myc—Cyld and HA—UbK48 (in which all of the lysine residues within Ub except K48 were mutated to arginine) or HA—UbK63 (in which all of the lysine residues within Ub except K63 were mutated to arginine). Coexpression of Cyld with NLRP6 and UbK63 markedly inhibited ubiquitination of NLRP6 (Fig. 3b, lane 4), suggesting that Cyld cleaves K63-linked Ub chains. In addition to the conventional K48 and K63 polyubiquitination, other forms of polyubiquitination are known to occur9,14. Therefore we tested the possibility of NLRP6 ubiquitination by other linkages (UbK6, UbK11, UbK27, UbK29 and UbK33). However, our results show that ubiquitination of NLRP6 was predominantly K63-linked (Supplementary Fig. 2a).
To confirm that Cyld directly deubiquitinates NLRP6, we coexpressed Flag—NLRP6 with Ub. Ubiquitinated NLRP6 was precipitated using Flag beads followed using by elution with glycine buffer. The eluate was sequentially immunoprecipitated using Flag beads, followed by incubation with Cyld protein. Cyld deubiquitinated NLRP6, suggesting that Cyld directly deubiquitinates NLRP6 (Fig. 3c).
To test whether deubiquitination of NLRP6 was defective in Cyld−/− mice, we immunoprecipitated NLRP6 from wild-type and Cyld−/− mice that were infected with C. rodentium. The immunoprecipitates were then immunoblotted with anti-UbK63. As expected, we found increased NLRP6 ubiquitination in Cyld−/− mice compared to wild-type mice (Fig. 3d). Further, in a similar experiment using tandem ubiquitin-binding entities (TUBEs), we confirmed increased K63-linked ubiquitinated NLRP6 in Cyld−/− mice (Fig. 3e).
Recently, it has been demonstrated that lipoteichoic acid (LTA), a microbial product, activates the NLRP6 inflammasome8. To investigate whether LTA induced K63-linked ubiquitination of NLRP6, we stimulated wild-type and Cyld−/− colonic epithelial cells with LTA. LTA stimulation induced K63-linked ubiquitination, and as expected, ubiquitination of NLRP6 was sustained in Cyld−/− cells compared to wild-type cells (Fig. 3f). Since Cyld can also cleave methionine 1 (M1)-linked Ub chains15,16, we next tested whether M1 ubiquitination occurs on NLRP6. However, M1 TUBEs failed to enrich ubiquitinated NLRP6, suggesting that NLRP6 was not M1-ubiquitinated (Supplementary Fig. 2b). We further confirmed that Cyld cleaves K63- but not M1-linked Ub chains from NLRP6 by UbiCREST analysis using Cyld and otulin (an M1-specific deubiquitinase). Cyld cleaved Ub chains from NLRP6 (Fig. 3g, top blot, lane 2) and released Ub (Fig. 3g, bottom blot, lane 2), whereas otulin failed to deubiquitinate NLRP6 (Fig. 3g, top blot, lane 3). Taken together, these data suggest that Cyld cleaves K63-linked Ub chains on NLRP6.
To investigate whether Cyld-mediated deubiquitination regulates NLRP6 protein stability, we lysed the colon tissues of wild-type and Cyld−/− mice and immunoblotted the lysates using anti-NLRP6. The level of NLRP6 was comparable in wild-type and Cyld−/− tissues (Fig. 3h and Supplementary Fig. 3a), which excluded the possibility of regulation of NLRP6 protein turnover via proteasome or autophagy-mediated proteolysis. We also tested whether the abundance of NLRP6 was altered in colon tissues of patients with UC. In line with our data from mouse models, the abundance of NLRP6 was comparable in samples from unaffected individuals and patients with UC (Supplementary Fig. 3b). These data suggested that Cyld-mediated regulation of NLRP6 is independent of proteolysis.
K63-linked ubiquitination of NLRP6 enhances its interaction With ASC.
NLRP6 has been shown to form an ASC-dependent inflammasome complex via its PYRIN domain to generate active IL-18 (refs. 7,17). To test Cyld-mediated regulation of NLRP6 via non-proteolytic mechanisms, the association of NLRP6 and ASC in the colonic mucosal tissue samples of wild-type and Cyld−/− mice was analyzed. In line with previous reports7, NLRP6 and ASC were found to interact in the colon tissues of wild-type mice. Interestingly, the association of NLRP6 with ASC was pronounced in Cyld−/− tissues (Fig. 4a). To investigate whether deubiquitination of NLRP6 negatively regulates the association of NLRP6 with ASC, we transiently transfected 293T cells with NLRP6, ASC, UbK63 and Cyld. The association of NLRP6 with ASC was markedly enhanced by the overexpression of UbK63 (Fig. 4b). We also found that Cyld-mediated deubiquitination attenuated this association. To further investigate K63-linked ubiquitination on NLRP6 inflammasome activation, the ASC oligomerization was analyzed (Fig. 4c). Our results show that ASC oligomerization was pronounced in K63-linked ubiquitination, and Cyld substantially diminished this oligomerization. These data suggest that K63-mediated ubiquitination promotes an NLRP6—ASC interaction and that Cyld-mediated deubiquitination negatively regulates this association.
Fig. 4 |. NLRP6 association with ASC is pronounced in the absence of Cyld.
a, Colonic mucosal lysates from infected wild-type and Cyld−/− mice were immunoprecipitated with anti-NLRP6 and immunoblotted against anti-ASC and anti-NLRP6. b, Immunoassay of 293T cells transfected with expression vectors for various combinations of Flag-NLRP6, Xpress–ASC, HA–UbK63 and Myc–Cyld, followed by immunoprecipitation of lysates with anti-Flag and immunoblot analysis with anti-Xpress and anti-HA. c, 293T cells were transfected with expression vectors for various combinations of Flag-NLRP6 and Xpress-ASC in the presence and absence of HA-UbK63 and Myc-Cyld. The cells were lysed and the pellet was treated with disuccinimydyl suberate followed by immunoblotting with anti-Xpress, anti-Flag and anti-Myc. d, Colonic mucosal lysates from uninfected and inflamed wild-type and Cyld−/− mice were subjected to SDS-PAGE. The amounts of pro-caspase-1 and cleaved caspase-1 were assessed by immunoblotting with anti-caspase-1. e, Caspase-1 activity assay performed on uninfected and infected wild-type and Cyld−/− mice colon lysates (n=5 mice per group, NSP=0.0856, ***P=0.0006). The statistics are shown as mean ± s.d., with P values determined by Student’s t-test (two tail). The data are from one experiment representative of three independent experiments with similar results. Uncropped blots (a–d) are shown in the source data.
Oligomerization of ASC recruits pro-caspase-1 to the complex, where it is converted into active caspase-1 (ref. 2). To investigate whether the increased NLRP6—ASC association resulted in increased activation of caspase-1, we used the lysate of colonic mucosa from wild-type and Cyld−/− mice that had been infected with C. rodentium. We immunoblotted with anti-caspase-1 and found increased amounts of the cleaved form of caspase-1 in Cyld−/− mice compared with wild-type mice (Fig. 4d). The enzymatic activity of caspase-1 in colonic lysates was determined by the cleavage of the fluorescent caspase-1 substrate, YVAD-AFC (ref. 18). As for the cleaved form of caspase-1, the lysates from Cyld−/− mice had significantly higher caspase-1 enzymatic activity compared to lysates from wild-type mice (Fig. 4e). Recently, NLRP6 was also shown to associate with caspase-11 (ref. 8). Therefore, we tested and show that caspase-11 activation was affected in Cyld−/− mice, similar to caspase-1 (Supplementary Fig. 3c). These data collectively suggest that a defect in deubiquitination of NLRP6 results in increased activation of caspase-1 and caspase-11 in the colonic mucosa of Cyld−/− mice.
Active IL-18 is elevated in Cyld-/- mice.
Next, we analyzed the concentrations of IL-18 in the serum of Cyld−/− and littermate wild-type control mice infected with C. rodentium, which showed a substantially higher amount of IL-18 in Cyld−/− mice compared to wild-type mice (Fig. 5a). Similarly, the concentration of IL-18 was significantly higher in culture supernatants of Cyld−/− colon explants compared to wild-type explants (Fig. 5b). To test the possibility of this elevated IL-18 production being due to an increase in Il18 messenger RNA expression, we performed real-time PCR experiments using gene-specific primers on RNA isolated from the colon tissues. However, the abundance of Il18 mRNA was comparable between littermate wild-type and Cyld−/− tissues (Fig. 5c), which suggested post-translational regulation of IL-18. To further confirm this finding, we performed immunoblotting analysis of colonic mucosal tissue to detect cleaved and uncleaved forms of IL-18. The level of cleaved IL-18 was substantially higher in Cyld−/− tissues (Fig. 5d and Supplementary Fig. 3d). We also measured the expression of Il6, Il22 and Il17a in Cyld−/− mice and wild-type littermate controls infected with C. rodentium. We found a marginal increase in the abundance of Il6 mRNA, whereas the abundance of Il22 and Il17a was comparable between wild-type and Cyld−/− mice (Supplementary Fig. 4a–c). We also analyzed the expression of type I interferons (IFNs) in the C. rodentium—infected wild-type and Cyld−/− mice. Only a marginal increase in Ifn-α was observed and there was no significant difference in Ifn-β expression (Supplementary Fig. 4d,e). The aforementioned results suggest that these cytokines play a minor role in severe colonic inflammation associated with Cyld deficiency.
Fig. 5 |. Increased IL-18 production in Cyld−/− mice following C. rodentium infection.
a, Serum was collected from age-matched uninfected and infected wild-type and Cyld−/− mice (n=8 mice per group). The concentration of IL-18 was measured by ELISA (NSP=0.2103, **P=0.0016). b, Colon tissues from uninfected and infected wild-type and Cyld−/− mice (n=8 mice per group) were cultured for 24h, supernatants were collected, and the IL-18 concentration was measured by ELISA and normalized to colon weight (NSP=0.3657, ***P<0.0001). c, The expression of Il18 was quantified (NSP=0.9792) by real-time PCR on mRNA from colons of age- and sex-matched infected wild-type and Cyld−/− mice (n=8 mice per group). d, Colonic mucosal lysates from uninfected and infected wild-type and Cyld−/− mice were subjected to SDS-PAGE. The amounts of pro-IL-18 and mature IL-18 were assessed by immunoblotting with anti-IL-18. e–g, Body weight change (NSP=0.0520, ***P=0.0001; e), diarrhea score (NSP=0.8683, ***P<0.0001; f) and colon length (***P<0.0001; g) of Cyldfl/fl versus epithelial-specific Cyld−/− (IEC- Cyld(γ9)) mice (n=10 mice per group) infected (inf) with C. rodentium, treated with anti-IL-18 or control IgG. h, The mean fluorescence intensity showing the translocation of FITC-dextran assayed using serum 4h after administration (n=10 mice per group, ***P<0.0001). i,j, H&E-stained colonic sections from infected Cyldfl/fl and IEC-Cyld(Δ9) mice treated with control IgG or anti-IL-18 (scale bars, 50 μm; i) and the histology scores of the sections (n=10 mice per group, ***P<0.0001, **P=0.0011; j). The statistics are shown as mean ± s.d., with P values determined by Student’s t-test (two tail). The data are from one experiment representative of three independent experiments with similar results. Uncropped blots (d) are shown in the source data.
We also induced colitis using 2,4,6-trinitrobenzene sulfonic acid (TNBS)19, in which IL-18 plays a predominant role20,21. We found increased body weight loss, diarrhea and colon shortening, suggesting severe TNBS-induced colitis in Cyld−/− mice (Extended Data Fig. 3a–e). Histological analysis of hematoxylin and eosin (H&E)-stained sections of colon tissue showed massive infiltration and crypt erosion in Cyld−/− mice, suggesting severe inflammation. The concentration of IL-18 was significantly higher in the supernatant of explant cultures (Extended Data Fig. 3f). However, there was no significant difference in the abundance of Il18 mRNA (Extended Data Fig. 3g). Further, we found increased amounts of cleaved biologically active IL-18 in Cyld−/− mice (Extended Data Fig. 3h,i). These data collectively suggest that Cyld plays a critical role in the regulation of colonic inflammation by modulating the maturation of IL-18.
NLRP6 is predominantly expressed in intestinal epithelial cells (IECs)22, whereas Cyld is ubiquitously expressed10. To investigate whether Cyld and NLRP6 actin the same cell type, we crossed Cyldfl/fl mice23 with villin-Cre mice and generated intestinal epithelial-specific conditional Cyld−/− mice (IEC-Cyld(Δ9)). Colitis was induced using C. rodentium. IEC-Cyld(Δ9) mice developed severe colitis compared to control Cyldfl/fl mice (Extended Data Fig. 4). The level of biologically active IL-18 was substantially higher in IEC-Cyld(Δ9) mice compared to Cyldfl/fl mice (Extended Data Fig. 4h). These results collectively suggest that Cyld regulates the conversion of pro-IL-18 to active IL-18 in the gut epithelial cells by deubiquitination of NLRP6.
Neutralization of IL-18 rescues Cyld-/- mice from severe colitis.
Next, we sought to investigate whether neutralizing IL-18 would rescue IEC-Cyld(Δ9) mice from developing severe colitis. Anti-IL-18 treatment markedly improved the loss of body weight and diarrhea score compared to control IgG-treated mice (Fig. 5e,f). We observed less colonic thickening in mice treated with anti-IL-18 than in those treated with control antibody. Anti-IL-18 treatment significantly improved colon thickening and barrier integrity (Fig. 5g–i). Similarly, an improved histological score was observed in mice treated with anti-IL-18 compared to control mice (Fig. 5j). These data indicate that the severe colitis in Cyld−/− mice depends primarily on elevated production of IL-18.
Cyld is essential for regulation of IL-18 abundance.
To investigate whether genetic deletion of Il18 would rescue Cyld−/− mice from severe colitis, we crossed Cyld−/− mice with Il18−/− mice and generated Cyld−/−Il18−/− mice. The Cyld−/−Il18−/− mice did not show any marked change in the primary or secondary lymphoid organs (Supplementary Fig. 5a,b) and colonic architecture (Supplementary Fig. 5c). We induced colitis in Cyld−/−Il18−/− mice using C. rodentium. Real-time PCR and ELISA results showed no Il18 expression in the colonic mucosa of Cyld−/−Il18−/− mice (Fig. 6a,b). However, in line with previously published reports suggesting that a basal amount of IL-18 is essential for preventing colonic inflammation24,25, Il18−/− mice were found to exhibit increased body weight loss and diarrhea compared to littermate wild-type control mice (Extended Data Fig. 5a,b). Our results also showed that colitis severity in Cyld−/−Il18−/− mice was comparable to that in Il18−/− mice (Extended Data Fig. 5a,b).
Fig. 6 |. Deletion of IL-18 and NLRP6 rescues elevated IL-18 levels in colon tissues of Cyld−/− mice ex vivo.
a, mRNA from colonic mucosa of age- and sex-matched C. rodentium-infected wild-type, Cyld−/−, Il18−/− and Cyld−/−Il18−/− mice (n=8 mice per group) was isolated, and the expression of Il18 was quantified by real-time PCR (NSP=0.3390). b, Colon tissues from C. rodentium–infected wild-type, Cyld−/−, Il18−/− and Cyld−/−Il18−/− mice were cultured for 24h (n=8 mice per group), and the IL-18 concentration in the supernatant was measured by ELISA and normalized to colon weight (***P<0.0001). c, mRNA from colonic mucosa of age- and sex-matched C. rodentium-infected wild-type, Cyld−/−, Nlrp6−/− and Cyld−/−Nlrp6−/− mice was isolated (n=8 mice per group), and the level of Il18 was quantified by real-time PCR (NSP=0.9748, NSP=0.2241, NSP=0.5841, left to right). d, Colon tissues from C. rodentium-infected wild-type, Cyld−/−, Nlrp6−/− and Cyld−/−Nlrp6−/− mice (n=8 mice per group) were cultured for 24h, and the IL-18 concentration in the supernatant was measured by ELISA and normalized to colon weight (***P=0.0001; NS, not significant). The statistics are given as mean ± s.d., with P values determined by Student’s t-test (two tail). The data are from one experiment representative of three independent experiments with similar results.
Since a basal level of IL-18 is essential to maintain the epithelial barrier24,25, we investigated whether injection of recombinant IL-18 could rescue the Cyld−/−Il18−/− mice from severe colitis. Injection of IL-18 rescued Il18−/− and Cyld−/−Il18−/− mice but not Cyld−/− mice from body weight loss and diarrhea (Extended Data Fig. 5c,d). Examination of colon length and histological analysis further suggested that injection of recombinant IL-18 rescues Il18−/− and Cyld−/−Il18−/− mice but not Cyld−/− mice (Extended Data Fig. 5e–g). This result suggests that Cyld plays a critical role in regulating the optimum level of IL-18 to prevent colitis.
Further, to investigate the role of NLRP6 and Cyld interaction in regulating IL-18, we induced colitis in Nlrp6−/−, Cyld−/− and Cyld−/−Nlrp6−/− mice using C. rodentium. The abundance of Il18 mRNA in the colonic mucosa was comparable; however, the concentration of IL-18 in colon explant cultures from Cyld−/−Nlrp6−/− mice was substantially reduced (Fig. 6c,d). Furthermore, in line with previous reports, Nlrp6−/− mice exhibited increased body weight loss compared to littermate wild-type controls due to lack of IL-18 (ref. 25). The body weight loss and diarrhea score among Cyld−/−Nlrp6−/− mice were comparable to those of Nlrp6−/− mice (Supplementary Fig. 6a,b). Moreover, injection of recombinant IL-18 rescued Nlrp6−/− and Cyld−/−Nlrp6−/− mice (Supplementary Fig. 6c–g) but not Cyld−/− mice from body weight loss and diarrhea (Extended Data Fig. 5c,d). These data clearly suggest that Cyld plays an essential role in maintaining the optimum level of IL-18 via regulation of NLRP6 to prevent colitis.
The complete deficiency of IL-18 in mice has been shown to result in worse outcomes following C. rodentium infection26. On the contrary, increased secretion of mature IL-18 in the colonic mucosa also promotes severe colitis in mice and humans25,27,28. Our results and published data25 highlight this conundrum of IL-18 being important for the integrity of the colonic epithelium on the one hand as well as being responsible for Cyld-dependent inflammation on the other. To further investigate this hypothesis, we tested whether deletion of one allele of either Il18 or Nlrp6 could rescue Cyld−/− mice from colitis severity. We induced colitis in Cyld−/−Il18+/− and Cyld−/−Nlrp6+/− mice and compared disease severity to that of littermate wild-type and Cyld−/− controls. Interestingly, body weight loss and diarrhea among Cyld−/−Il18+/− and Cyld−/−Nlrp6+/− mice were markedly reduced compared to those of Cyld−/− mice (Fig. 7). Histological examination of the H&E-stained sections of colon tissue further suggested that colitis severity in Cyld−/−Nlrp6+/− and Cyld−/−Il18+/− mice was markedly reduced (Fig. 7).
Fig. 7 |. Heterozygous deletion of Il18 or Nlrp6 rescues Cyld−/− mice.
a–c, Body weight change (**P=0.006, NSP=0.2268; a), diarrhea score (***P<0.0001, NSP=0.2208; b) and colon length (***P<0.0001; c) of wild-type, Cyld−/−, Il18−/−, Il18+/− and Cyld−/−Il18+/− mice (n=10 mice per group) infected (inf) with C. rodentium. d,e, H&E-stained colonic sections from C. rodentium-infected wild-type, Cyld−/− and Cyld−/−Il18+/− mice (scale bars, 50 μm; d) and the histology scores of the colon sections (n=10 mice per group, ***P <0.0001; e). f–h, Body weight change (**P=0.0019, NSP=0.2322; f), diarrhea score (***P<0.0001, NSP=0.1211; g) and colon length (***P<0.0001; h) of wild-type, Cyld−/−, Nlrp6−/−, Nlrp6+/− and Cyld−/−Nlrp6+/− mice (n=8 mice per group) infected (inf) with C. rodentium. i,j, H&E-stained colonic sections from C. rodentium-infected wild-type, Cyld−/− and Cyld−/−Nlrp6+/− mice (scale bars, 50 μm; i ) and the histology scores of the colon sections (**P=0.0011, ***P=0.0006; j). The statistics are given as mean ± s.d., with P values determined by Student’s t-test (two tail). The data are representative of three or more independent experiments.
Next, we reconstituted Cyld−/− colonic epithelial cells with either wild-type Cyld or Cyld(C601A) mutant29 that lacks the deubiquitination activity. The cells were stimulated with LTA and maturation of IL-18 was analyzed by immunoblotting the cell lysate with anti-IL-18 and by ELISA using culture supernatants. In line with a previously published report8, stimulation of epithelial cells with LTA transfection resulted in cleavage of pro-IL-18 into mature IL-18 (Supplementary Fig. 3d), and reconstitution of Cyld−/− epithelial cells with wild-type Cyld but not the Cyld(C601A) mutant normalized the processing of IL-18 (Fig. 8a,b). These data confirm that Cyld-mediated deubiquitination is essential to prevent excessive IL-18 production.
Fig. 8 |. Cyld regulates maturation of IL-18 in colonic epithelial cells and the level of active IL-18 in the colonic mucosa of patients with UC.
Colonic epithelial cells from either wild-type or Cyld−/− mice were transduced with a lentivirus encoding either V5-tagged wild-type Cyld or Cyld(C601A). The cells were then stimulated overnight by transfection with LTA. a, Cells were lysed and immunoblotted with anti-V5, anti-Cyld and anti-IL-18. b, The IL-18 concentration of the supernatant was measured by ELISA (n=8 mice per group, NSP=0.0946, ***P<0.0001, ***P<0.0001, NSP=0.0578, left to right). c, CYLD mRNA expression from the colonic mucosa of patients with UC (n=23 samples from human patients with UC) was measured by real-time PCR and normalized to that of control samples from healthy individuals. The IL-18 concentration was measured from colon mucosal tissue extracts by ELISA. A regression curve was generated by plotting relative CYLD mRNA expression against IL-18 concentration from the corresponding patient’s samples. d, The Il18 mRNA levels were measured using real-time PCR of the RNA from the colonic mucosa of patients. A regression curve was generated by plotting relative Il18 mRNA against relative CYLD mRNA of patients. The statistics (b) are given as mean ± s.d., with P values determined by Student’s t-test (two tail). The data are representative of three or more independent experiments. The statistics (c,d) were measured by the Pearson correlation coefficient. The data are representative of three or more independent experiments.
IL-18 correlates with CYLD expression in patients with UC.
Previous studies have demonstrated elevated amounts of IL-18 in the mucosa of patients with IBD30–33, and this increase was shown to contribute to mucosal inflammation34. Since our results showed that NLRP6-mediated IL-18 activity is inhibited via Cyld-mediated deubiquitination, we investigated whether CYLD expression was correlated with active IL-18 in patients with UC. We observed that CYLD mRNA expression was low in a substantial number of patients. However, in some patients, the level of CYLD mRNA expression was high and was comparable to that of healthy controls (Supplementary Fig. 1a). Next, we tested whether the abundance of IL-18 protein correlated with CYLD mRNA expression in these patients. There was a strong inverse correlation (Pearson coefficient, P=−0.85) between CYLD mRNA expression and active IL-18 production, suggesting that IL-18 production correlates with CYLD gene expression (Fig. 8c). However, there was no significant correlation (Pearson coefficient, P=−0.087) between CYLD and IL18 mRNA abundance (Fig. 8d), suggesting that CYLD gene expression does not correlate with IL18 mRNA expression. Similarly, no significant correlation was observed between CYLD expression and expression of TNF-α and IL6 (Supplementary Fig. 7). These data strongly suggest the clinical relevance of Cyld-mediated regulation of the level of active IL-18 in patients with UC.
Discussion
In the gastrointestinal mucosa, the host is separated from an immense microbial ecosystem by a single layer of epithelial cells. The innate receptors such as NLRs that are expressed by the hematopoietic and epithelial cells recognize the microbe-associated molecular patterns and initiate an inflammatory response2,4. On sensing the relevant signal, NLRs form a complex with ASC. These complexes govern caspase-1 activation and subsequent cleavage of effector cytokines such as IL-1β and IL-18 (ref. 2). To avoid persistent inflammatory reactions, inflammasome-mediated proinflammatory cytokine production must be tightly regulated. A breakdown in these mechanisms will lead to chronic pathologies of the gastrointestinal tract, including IBDs35. In this report, we demonstrate a novel mechanism by which excessive IL-18 response is prevented by inhibition of the NLRP6—ASC complex via its deubiquitination.
Previous studies have shown that inflammasome activity is regulated by autophagy-mediated degradation36. The membrane-associated ring finger 7 (MARCH7) was shown to ubiquitinate NLRP3 and target it for degradation by autophagy37. Another report showed that NLRP3 was constitutively ubiquitinated by the Skp, Cullin, F-box-containing complex subunit F-box L2 (SCFFBXL2), targeting it for degradation by the proteasome38. Deubiquitination of NLRP3 by the BRCA1/ BRCA2-containing complex subunit 3 (BRCC3) promoted NLRP3 inflammasome activity by preventing its degradation39. In contrast to the regulation of NLRP3, ubiquitination of NLRP6 was found to enhance its interaction with ASC and caspase-1 activation. This difference could be due to K63-linked ubiquitination of NLRP6 compared to the K48/K63-linked mixed ubiquitination observed in NLRP3.
The composition and the mechanism of activation of the NLRP6 inflammasome remain poorly understood. The bacterial component LTA and microbial metabolites promote the formation of an NLRP6—ASC—caspase complex leading to maturation of IL-18 (refs. 7,8). Cryogenic electron microscopy and the crystal structures of the NLRP6 pyrin domain (PYD) suggested that NLRP6 assembles into filamentous structures accompanied by large conformational changes and recruits the ASC adaptor using PYD—PYD interactions25. The NLRP6 full-length protein was less efficient compared to NLRP6-PYD and NLRP6-PYD+NBD (nucleotide binding domain), which suggests that monomeric NLRP6 remains in an auto-inhibited form via the C-terminal LRR domain25,40. It is possible that ligands binding to NLRP6 (ref. 8) promote K63-linked ubiquitination, resulting in a conformational change that overcomes the auto-inhibition, leading to recruitment of ASC and inflammasome activation.
Despite extensive studies on IL-18, understanding its contribution to gut homeostasis and colonic inflammation and its relevance to IBD remains controversial. Complete loss of IL-18, the IL-18 receptor or components of the inflammasome predisposes mice to increased epithelial damage41,42. However, conditional deletion of Il18 in IECs or myeloid cells results in increased severity in intestinal inflammation25. This dual role of IL-18 correlates with the clinical observation that increased expression and bioactivity of IL-18 correlate with disease severity in patients with IBD32,33. Our studies identify Cyld-mediated deubiquitination of NLRP6 as a means of regulating this delicate balance of IL-18 in the colonic mucosa to prevent pathogenic inflammation. Further, the abundance of IL-18 in the colonic mucosa negatively correlates with CYLD expression in patients with IBD, suggesting that reduced CYLD expression results in elevated IL-18 and contributes to the pathogenesis of IBDs.
In summary, our studies have uncovered a novel mechanism by which NLRP6-mediated IL-18 maturation is modulated to prevent colonic inflammation. These new findings could pave the way to identify new therapeutic approaches for the prevention and treatment of IBDs.
Online content
Any methods, additional references, Nature Research reporting summaries, source data, extended data, supplementary information, acknowledgements, peer review information; details of author contributions and competing interests; and statements of data and code availability are available at https://doi.org/10.1038/s41590-020-0681-x.
Methods
Mice.
Wild-type, Cyld−/−, Nlrp6−/− and Il18−/− mice were bred in-house at the Baylor Research Institute and the University of Texas Southwestern Medical Center animal facility. Cyld−/− mice were generated by crossing Cyld+/− male mice with Cyld+/− female mice and the wild-type mice generated were used as control mice. Similarly, Il18−/− and Nlrp6−/− mice6 were generated from Il18+/− and Nlrp6+/− mice, respectively. Mice with loxP sites flanking exon 9 of the Cyld locus (Cyldflx9 mice)23 were crossed with villin-Cre mice and generated IEC-specific conditional Cyld−/− mice (IEC-Cyld(Δ9)). Cyld−/−Il18+/−, Cyld−/−Nlrp6+/−, Cyld−/−Il18−/− (double knockout) and Cyld−/−Nlrp6−/− (double knockout) mice were generated by crossing Cyld−/− with Il18−/− and Nlrp6−/− mice, respectively. All experiments were performed using cohoused littermate control mice. All mice were housed in micro-isolator cages in the barrier facility of Baylor Institute for Immunology Research and University of Texas Southwestern Medical Center. All experiments were performed in accordance with the guidelines of the Institutional Animal Care and Use Committee of Baylor Research Institute and University of Texas Southwestern Medical Center.
Antibodies and reagents.
The following antibodies were used in this study: anti-c-Myc (Santa Cruz), anti-Flag (M2, Sigma), anti-Xpress (Invitrogen), anti-V5 (Invitrogen), anti-caspase-1 (Santa Cruz), anti-Cyld (Cell Signaling), anti-β-actin (Sigma), anti-NLRP6 (Sigma), anti-HA (Santa Cruz), anti-His (GE Healthcare), anti-GST (Santa Cruz), anti-IL-18 (Santa Cruz), anti-ASC (Santa Cruz), human anti-NLRP6 (NSJ Bioreagents), InVivoMAb anti-mouse IL-18 (BioXcell), normal rabbit IgG (Santa Cruz) and normal mouse IgG (Santa Cruz). The following reagents were used: mouse rIL-18 (MBL International), picrylsulfonic acid solution (TNBS; Sigma), disuccinimidyl suberate (Thermo Scientific), N-methylmaleimide (Sigma), Pan TUBEs (Life Sensors), K63 TUBEs (Life Sensors), M1 TUBEs (Life Sensors), UbiCREST enzyme kit (Boston Biochem), Flag-tagged agarose beads (Sigma), glutathione beads (GE Healthcare Life Sciences).
C. rodentium-induced colitis model.
The C. rodentium used in this study was purchased from ATCC (51459). Mice (wild-type, Cyld-/-, Cyld-/-Il18+/-, Cyld-/-Nlrp6+/-, Cyld-/-Il18-/-, Cyld-/- Nlrp6-/-, IEC-Cyld(δ9)) were inoculated by gavage with C. rodentium. For inoculations, bacteria were grown overnight in Luria broth. Bacteria were diluted with PBS (pH 7.2) to about 2 × 109 colony-forming units per milliliter and delivered to mice in 100 μl. Mice were killed at 10 d post challenge, and tissues were snap frozen in liquid nitrogen and stored at -80 °C for further analysis.
In vivo permeability assay.
Barrier function was assessed using the permeability tracer FITC-labeled dextran. Wild-type and Cyld−/− mice were deprived of food and water for 4 h and then gavaged with 4 kDa FITC-labeled dextran (Sigma-Aldrich) at 0.6 mg per kg body weight. Blood was retro-orbitally collected after 4 h, and the fluorescence intensity in the serum was measured (excitation, 492 nm; emission, 525 nm). The FITC—dextran concentrations were determined from a standard curve generated by serial dilutions of FITC—dextran.
Protein identification by liquid chromatography-tandem MS.
Inflamed colonic mucosa was isolated from Cyld−/− mice and lysed. A pulldown assay was performed using His-tagged Cyld protein using Ni-NTA beads. The beads were resuspended in 50 μl of 0.1 M ammonium bicarbonate buffer (pH ≈ 8). Cysteines were reduced by adding 50 μl of 10 mM dithiothreitol and incubating at 45 °C for 30 min. Samples were cooled to 30 °C and alkylation of cysteines was achieved by incubating with 65 mM 2-chloroacetamide, under darkness, for 30 min at 30 °C. An overnight digestion with 1 μg of sequencing-grade, modified trypsin was carried out at 37 °C with constant shaking in a Thermomixer. Digestion was stopped by acidification, and peptides were desalted using SepPak C18 cartridges using the manufacturer’s protocol (Waters). Samples were completely dried using a vacufuge. Resulting peptides were dissolved in 8 μl of 0.1% formic acid/2% acetonitrile solution and 2 μl of the peptide solution was resolved on a nano-capillary reverse-phase column (Acclaim PepMap C18, 2 μm, 50 cm, Thermo Scientific) using a 0.1% formic acid/2% acetonitrile (buffer A) and 0.1% formic acid/95% acetonitrile (buffer B) gradient at 300 nl min−1 over a period of 180 min (2–25% buffer B in 110 min, 25–40% in 20 min, 40–90% in 5 min followed by holding at 90% buffer B for 10 min and requilibration with buffer A for 30 min). Eluent was directly introduced into a Q exactive HF mass spectrometer (Thermo Scientific) using an EasySpray source. MS1 scans were acquired at 60-K resolution (AGC target = 3 × 106; max IT = 50 ms). Data-dependent collision-induced dissociation MS/MS spectra were acquired using the top-speed method (3 s) following each MS1 scan (NCE ≈ 28%; 15-K resolution; AGC target = 1 × 105; max IT = 45 ms).
Proteins were identified by searching the MS/MS data against Mus musculus (UniProt, v2016–4–13; 24,861 entries) using Proteome Discoverer (v2.1, Thermo Scientific). Search parameters included an MS1 mass tolerance of 10 ppm and a fragment tolerance of 0.2 Da; two missed cleavages were allowed; carbamidimethylation of cysteine was considered a fixed modification, and oxidation of methionine and deamidation of asparagine and glutamine were considered as potential modifications. The false discovery rate was determined using Percolator and proteins/peptides with a false discovery rate of ≤1% were retained for further analysis.
Plasmid construction and cell transfection.
Myc—Cyld, HA—Ub(wild type), Xpress–Cyld(C601A), HA—UbK63, HA—UbK48, pLenti6.2-Cyld(wild type) and pLenti6.2-Cyld(C601A) were described previously29. Xpress—Pycard (ASC) was amplified from mouse complementary DNA and cloned between the BamHI and NotI sites in the pEF4/HisC plasmid. Nlrp6 (NM_133946.2) was amplified from mouse colonic cDNA and cloned at the EcoRI and XhoI sites of the pCMV-Tag2B vector to generate the Flag-tagged NLRP6. To generate His—NLRP6, the PCR-amplified fragment of mouse Nlrp6 was cloned at the BamHI and NotI sites of the pET-30a(+) vector. GST—Cyld was cloned between the BamHI and NotI sites of the pGEX-4T2 vector. Transient transfection of 293T cells was performed using Lipofectamine 2000 (Invitrogen), according to the manufacturer’s instructions. Epithelial cells isolated from wild-type and Cyld−/− mice were transfected with LTA (Invivogen) using Lipofectamine 2000 as described previously8,43. Lentiviruses were produced in 293FT cells according to the manufacturer’s instructions (ViraPower Lentiviral Expression Systems, ThermoFisher Scientific).
Immunoprecipitation and immunoblot analysis.
Cells were isolated from colonic mucosa of wild-type and Cyld−/− mice after homogenizing in cell lysis buffer. Cell lysis was performed using NP-40 lysis buffer containing 50 mM Tris pH 8.0, 150 mM NaCl, 1% NP-40, EDTA-free protease inhibitor cocktail (Roche), 2 mM sodium fluoride (Boston Bioproducts), 10 mM glycerophosphate (Boston Bioproducts) and 1 mM sodium orthovanadate (Sigma). Protein estimations were performed using the Pierce BCA protein assay kit according to the manufacturer’s protocol. Whole-cell lysates were precleared with 20 μl of Protein A/G plus agarose beads (Santa Cruz) for 1 h at 4 °C. Lysates were then incubated with 1 μg of the desired antibody overnight at 4 °C, followed by a further 1 h incubation at 4 °C with 25 μl of Protein A/G beads. The immunocomplexes were washed five times with NP-40 lysis buffer, denatured using 6× Laemmli buffer, separated by SDS—PAGE, transferred to polyvinylidene difluoride membranes, immunoblotted with the indicated antibodies and visualized with an Amersham ECL prime immunoblot analysis detection system (GE) on a Bio-Rad ChemiDoc. For reprobing, membranes were stripped by incubation in a stripping buffer (62.5 mM Tris-HCl, pH 6.7; 100 mM 2-mercaptoethanol and 2% SDS) at 70 °C for 45 min and washed thoroughly before reprobing.
GST pulldown assay.
GST—Cyld and GST protein were both individually expressed in BL21(DE3) E. coli. Cleared lysates were prepared and the soluble fusion proteins were purified on glutathione Sepharose 4B GST-tagged protein purification resin (GE Healthcare Life Sciences,) and tested by immunoblot using monoclonal anti-GST (Cell Signaling Technology). His-tagged NLRP6 expressed by the pET-30a(+) vector was transformed into BL21(DE3) E. coli and soluble fusion proteins were purified on Ni-NTA His-binding resin (Thermo Fisher Scientific). Purified GST—Cyld and GST were individually incubated with glutathione beads for approximately 4 h. Then the beads were collected by centrifugation, and washed five times with GST pulldown lysis buffer and TBS (1:1). Then, purified His—NLRP6 protein was added to glutathione beads and incubated at 4 °C overnight with gentle rocking. The beads were washed five times and resuspended in SDS—SDS—PAGE sample buffer. After boiling, the bound proteins were analyzed on 12% SDS—PAGE gel followed by immunoblotting using anti-His. His—NLRP6 protein was used as a control.
Ubiquitination assay.
HEK293T cells were transfected with vectors encoding NLRP6 and various constructs as indicated. Cells were washed three times with PBS and lysed in NP-40 lysis buffer containing N-ethylmaleimide (NEM). Immunoprecipitation was performed using anti-Flag. NLRP6-associated HA-tagged Ub was analyzed by immunoblot using antibody to HA, as we described previously13.
TUBE pulldown assay.
The pulldown of Pan-, M1- and K63-linked ubiquitinated proteins was performed with GST and FLAG-tagged TUBE, following the manufacturer’s instructions (Life Sensors). Briefly, for Pan–Ub, cells were collected, washed twice with PBS and lysed with TUBE buffer supplemented with protease inhibitors (Pierce) and 5 mM NEM. Clarified lysates were incubated for 2 h on equilibrated glutathione Sepharose-GST—TUBE under rotation. The resin was washed 3 times with TBS-T (200 mM Tris-HCl pH 8.0, 0.15 M NaCl, 0.1% Tween-20) and resolved by SDS—PAGE. For M1 and K63 TUBE, cells were collected, washed twice in PBS and lysed with TUBE lysis buffer supplemented with protease inhibitors (Pierce), 5 mM NEM and 250 nM Flag—TUBE K63 or 250 nM Flag—TUBE M1. Clarified lysates were diluted with reaction buffer, incubated on ice for 1 h and added to equilibrated FLAG M2 Affinity Resin (Sigma) for 2 h under rotation. The resin was washed three times with wash buffer and resolved by SDS—PAGE.
DUB assay.
HEK293T cells were transfected with a plasmid encoding for NLRP6—Flag and UbK63—HA for 36 h, lysed and pulled down using anti-Flag. The precipitate was subjected to glycine buffer (0.2 M) separation and re-immunoprecipitation was performed using Flag beads to obtain the purified immunoprecipitated product. Ub conjugates from LTA-treated cells were isolated by Flag—TUBE K63, Flag—TUBE M1 or GST—Pan TUBE as described for the TUBE pulldown assay. In vitro DUB assay was performed as described previously44. Beads were resuspended in DUB buffer (50 mM HEPES (pH 7.5), 100 mM NaCl, 2 mM dithiothreitol, 1 mM MnCl2 and 0.01% Brij-35). Ub conjugates were incubated with or without the DUBs Cyld or otulin. Buffers and enzyme were supplied by Boston Biochem (catalog no. K-400) and the Cyld enzyme was purified by us. The enzyme reaction was performed according to the manufacturer’s protocol. Ub conjugates were incubated with DUB enzymes in a 37 °C water bath for 30 min, and the supernatants were collected after brief low-speed centrifugation, boiled with loading buffer and analyzed by immunoblot.
Human samples.
A total of 27 samples from patients with UC and 5 samples from non-inflamed controls were included in the study. Surgically resected samples were collected from patients with UC who were admitted to Baylor University Medical Center at Dallas for therapeutic bowel resection. All human samples were collected and processed in accordance with guidelines and approved protocols of the Baylor Research Institute (IRB 014–117). Informed consent was obtained from the patients.
Real-time PCR analysis.
Total RNA was prepared using an RNeasy Mini Kit (QIAGEN), followed by cDNA synthesis using a Verso cDNA synthesis Kit (Thermo Scientific). Quantitative real-time PCR was performed on a Mastercycler Realplex (Eppendorf). LightCycler 480 SYBR Green I Master reaction mix (Roche) was used in a 20-μl reaction volume. The expression of individual genes was normalized to the expression of β-actin. The cycling conditions were 95 °C for 2 min, followed by 50 cycles of 95 °C for 15 s, 55 °C for 15 s and 72 °C for 20 s (except for human IL-18). For human IL-18, the conitions were Mgcl2 4 mmoll−1 (denaturing 95 °C for 15 s, annealing 55 °C for 10 s, extension 72 °C for 14 s) × 45 cycles. The primer sequences for the genes were as follows: mouse Nlrp6 forward primer, 5'-GCAGACGAGCTGCCTACTTT-3'; mouse Nlrp6 reverse primer, 5'-GCTCCTGGTAACAGCTCCTG-3'; mouse Il18 forward primer, 5'-GCCATGT CAGAAGACTCTTGCGTC-3'; mouse Il18 reverse primer, 5'-GTACAGTGAAG TCGGCCAAAGTTGTC-3'; mouse Il6 forward primer, 5'-TAGTCCTTCCTACC CCAATTTCC-3'; mouse Il6 reverse primer, 5'-TTGGTCCTTAGCCACTCCT TC-3'; mouse Il22 forward primer, 5'-CTGCCTGCTTCTCATTGCCCT-3'; mouse Il22 reverse primer, 5'-CAAGTCTACCTCTGGTCTCAT-3'; mouse Il17A forward primer, 5'-CTGGAGGATAACACTGTGAGAGT-3'; mouse Il17A reverse primer, 5'-TGCTGAATGGCGACGGAGTTC-3'; mouse Ifn-α forward ACAGCCTTGCATT-3’; mouse Ifn-β forward primer, 5’-CCCTATGGAGATGA CGGAGA-3’; mouse Ifn-β reverse primer, 5’-ACCCAGTGCTGGAGAAATTG-3’; mouse Actb forward primer, 5'-GAAATCGTGCGTGACATCAAAG-3'; mouse Actb reverse primer, 5'-TGTAGTTTCATGGATGCCACAG-3'; human CYLD forward primer, 5'TGCCTTCCAACTCTCGTCTT-3'; human CYLD reverse primer, 5'AATCCGCTCTTCCCAGTAG-3'; human IL18 forward primer, 5'-GCTTGAATCTAAATTATCAGTC-3'; human IL18 reverse primer, 5'-GAAGA TTCAAATTGCATCTTAT-3'; human IL6 forward primer, 5'-GGTACATCCTCG ACGGCATCT-3'; human IL6 reverse primer, 5'-GTGCCTCTTTGCTGCTTT CAC-3'; human TNF-α forward primer, 5'-CCCTCACACTCAGATCATCTT CT-3'; human TNF-α reverse primer, 5'-GCTACGACGTGGGCTACAG-3'; human ACTB forward primer, 5'-ACCAACTGGACATGGAGAAA-3'; human ACTB reverse primer, 5'-TAGCACAGCCTGGATAGCAACGTA-3'.
IL-18 cytokine concentrations by ELISA
IL-18 concentrations from serum and colon tissue, colonic epithelial cell culture supernatants of mice and colon tissue lysates of humans were measured using commercially available kits mouse IL-18 platinum ELISA kit (eBioscience) and human IL-18 PicoKine ELISA kit (Bosterbio) according to the manufacturers’ protocols.
Colon tissue culture supernatant was prepared as described previously13. Briefly, 1-cm sections of the proximal colon were cut, washed extensively to remove feces, and then cut in half longitudinally. The colon sections were placed into culture in RPMI-1640 medium with 1% penicillin and streptomycin and cultured at 37 °C with 5% CO2. Supernatants were collected after 24 h, and ELISA was used to determine the cytokine concentrations.
Colonic epithelial cells were prepared and transduced with lentiviral particles encoding either wild-type Cyld or the C601A mutant of Cyld as described previously45. In brief, 5 × 105 cells in each well were cultured with a multiplicity of infection of 10 of the corresponding viral particles for 6 h and the medium was then replaced with fresh medium and incubated for 36 h at 37 °C. LTA treatment was performed for a further 12 h, after which supernatants were collected by a brief centrifugation and ELISA was used to determine the cytokine concentrations.
IL-18 from human tissue was measured according to a previously described protocol46. Briefly, colon samples collected from patients with UC were kept at −80 °C before preparation of crude aqueous extracts. The tissues were weighed, suspended in cell lysis buffer and homogenized. The homogenates were left on ice for 15 min and centrifuged at 14,000 r.p.m. for 15 min at 4 °C. The supernatants were stored at −20 °C until use. A 100-μg quantity of total protein was analyzed for the presence of human IL-18 using two commercial ELISA kits.
Caspase-1 activity assay.
The activity of caspase-1 was determined in colon homogenates by a commercial kit (Abcam, ab39412) according to the manufacturer’s instructions. Briefly, fresh frozen sections of colons were homogenized with lysis buffer (provided in the kit) using a homogenizer, and then the supernatant was separated by centrifugation at 10,000g for 10 min.
Histology scoring.
Colons were isolated from mice, cleaned of feces, fixed with formalin and paraffin-embedded. For H&E staining, 5-μm sections were used. Histologic assessment was performed as described previously. In brief, to denote the severity of inflammation, a three- to four-point scale was used: inflammation (0, none; 1, mild; 2, moderate; and 3, severe), the level of involvement (0, none; 1, mucosa; 2, mucosa and submucosa; and 3, transmural) and the extent of epithelial/ crypt damage (0, none; 1, basal 1/3; 2, basal 2/3; 3, crypt loss; and 4, crypt and surface epithelial destruction).
Densitometry analysis.
The intensities of pro-caspase-1, cleaved caspase-1, cleaved IL-18 and human NLRP6, and the intensities of the pro-caspase-1, cleaved caspase-1, cleaved IL-18, human NLRP6 and actin bands were quantified by densitometry using ImageJ version 1.37a software (National Institutes of Health, Bethesda, MD, USA). The pro-caspase-1 and cleaved-caspase-1 band intensities were normalized to that of the actin band.
Statistical analysis.
The data were analyzed with GraphPad Prism 8.3.1 software to determine the statistical significance using a paired Student’s t-test. The data are expressed as mean ± s.d. Probability (P) values of <0.05 were considered significant: *P<0.05, **P<0.01 and ***P<0.001; NS, not significant.
Reporting Summary.
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Extended Data
Extended Data Fig. 1 |. CYLD mRNA expression in human colonic mucosa.
CYLD mRNA expression from the colonic mucosa of UC patients (n=10 human UC patient samples) and control healthy samples (n=5 healthy control) was measured by real-time PCR and compared (**P=0.0012). Statistics are shown as mean ± SD, with P values determined by Student’s t test (two tail). Data are from one experiment representative of three Experiment with the same samples.
Extended Data Fig. 2 |. Increased colon length shortening in Cyld-/- mice after C. rodentium infection.
Colons of wild-type vs. Cyld-/- mice infected (inf) with C. rodentium for 10 days. Data are from one experiment representative of three independent experiments with similar results.
Extended Data Fig. 3 |. Cyld–/– mice exhibit severe TNBS-induced colitis.
(a–c) (a) Body weight change (nsP=0.9666, ***P<0.0001), (b) diarrhea score (***P<0.0001), and (c) colon length (nsP=0.7766, ***P<0.0001) of wild-type vs. Cyld-/- mice (n = 10 mice per group) given intrarectal administration of TNBS. (d) H&E-stained colon sections from TNBS treated wild-type and Cyld-/- mice (scale bars, 50 μm) and (e) Histology scores of the H&E stained sections (n=10 mice per group, nsP=0.7895, ***P<0.0001). (f) Colon tissues from untreated and TNBS treated wild-type and Cyld–/– mice (n=10 mice per group) were cultured and supernatant IL-18 concentrations measured by ELISA and normalized by colon weight (nsP=0.0573, ***P<0.0001). (g) mRNA from colonic mucosa of age- and sex-matched TNBS treated wild-type and Cyld–/– mice (n=10 mice per group) were isolated, and the expression of Il18 was quantified by real-time PCR (nsP=0.7252). (h) Colonic mucosal extracts from TNBS treated wild-type and Cyld–/– mice was subjected to SDS-PAGE. The amounts of pro-IL-18 and mature IL-18 were assessed by immunoblotting with anti-IL-18 antibody. (i) Densitometric analysis of mature IL-18 expression from wild-type and Cyld–/– mice (n=5 mice per group) colon tissue after immunoblotting (***P=0.0009). a,b,c,e,f,g,i Statistics are mean ± SD and P values were determined by Student’s t test (two tail). Data are from one experiment representative of three independent experiments with similar results. Uncropped blots (h) are shown in the Source Data.
Extended Data Fig. 4 |. Epithelial specific Cyld–/– mice exhibit severe C. rodentium induced colitis.
(a–c) (a) Body weight change (nsP=0.6251, **P=0.0029), (b) diarrhea score (***P=0.0005), and (c) colon length (nsP=0.6433, **P=0.0046) of Cyldfl/fl vs. IEC-Cyld (δ9) (Epithelial specific cyld knockout mice) mice (n = 5 mice per group) infected (inf) with C. rodentium. (d, e) H&E-stained colonic sections from C. rodentium–infected Cyldfl/fl and IEC-Cyld (δ9) mice (scale bars, 50 μm) and histology scores (n = 5 mice per group, nsP=0.99, **P=0.0028). (f) CFU from the organ culture (n = 5 mice per group, *P=0.0241, **P=0.0029, **P=0.0056). (g) Colon tissues from uninfected and infected Cyldfl/fl and IEC-Cyld (δ9) mice were cultured, IL-18 was measured from the supernatant by ELISA and normalized by colon weight (n = 5 mice per group, nsP=0.2755, **P=0.0013). (h) Colonic mucosal lysates from infected Cyldfl/fl and IEC-Cyld (δ9) was subjected to SDS-PAGE. The amounts of pro-IL-18 and mature IL-18 were assessed by immunoblotting with anti–IL-18 antibody. a,b,c,e,f,g Statistics are mean ± SD and P values were determined by Student’s t test (Two tail). Data are from one experiment representative of three independent experiments with similar results. Uncropped blots (h) are shown in the Source Data.
Extended Data Fig. 5 |. Recombinant IL-18 treatment rescues Cyld-/-Il18-/- mice but not Cyld-/- mice from severe colitis.
(a) Body weight change (***P<0.0001) and (b) diarrhea score (***P<0.0001) of wild-type, Cyld-/-, Il18-/-, and Cyld-/-Il18-/- mice (n=8 mice per group) infected (inf) with C. rodentium. (c), Body weight change (***P<0.0001), (d) diarrhea score (***P<0.0001), and (e) colon length (***P<0.0001, **P=0.0039, ***P<0.0001, ***P<0.0001) of wild-type, Cyld-/-, Il18-/- and Cyld-/-Il18-/- mice (n=8 mice per group) infected (inf) with C. rodentium and along with rIL-18. (f) H&E-stained colonic sections from C. rodentium–infected and rIL-18 -treated wild-type, Cyld-/-, Il18-/- and Cyld-/-Il18-/- mice (scale bars, 50 μm) and g, histology scores of the sections (n=8 mice per group, ***P=0.0002, ***P=0.0001). Statistics are mean ± SD, with P values determined by Student’s t test (two tail). Data are from one experiment representative of three independent experiments with similar results.
Supplementary Material
Acknowledgements
We thank E. Burstein for helpful discussions and W. Lancaster for critical reading of the manuscript. This work was supported by the National Institutes of Health (grant R01-DK115668-01), the Cancer Prevention Research Institute of Texas (grants RP160577 and RP190527) and the Baylor Charles A. Sammons Cancer Center and BSWRI-TGEN collaborative grants to K.V.
Footnotes
Competing interests
The authors declare no competing interests.
Additional information
Extended data is available for this paper at https://doi.org/10.1038/s41590-020-0681-x.
Supplementary information is available for this paper at https://doi.org/10.1038/s41590-020-0681-x.
Peer review information L. A. Dempsey was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Data availability
The data that support the findings of this study are available from the corresponding author upon request. Source data for Figs. 1–8 and Extended Data Figs. 1 and 3–5 are provided with the paper.
References
- 1.Neurath MF Cytokines in inflammatory bowel disease. Nat. Rev. Immunol. 14, 329–342 (2014). [DOI] [PubMed] [Google Scholar]
- 2.Broz P & Dixit VM Inflammasomes: mechanism of assembly, regulation and signalling. Nat. Rev. Immunol. 16, 407–420 (2016). [DOI] [PubMed] [Google Scholar]
- 3.Davis BK, Wen H & Ting JP The inflammasome NLRs in immunity, inflammation, and associated diseases. Annu. Rev. Immunol. 29, 707–735 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kanneganti TD, Lamkanfi M & Nunez G Intracellular NOD-like receptors in host defense and disease. Immunity 27, 549–559 (2007). [DOI] [PubMed] [Google Scholar]
- 5.Chen GY, Liu M, Wang F, Bertin J & Nunez G A functional role for Nlrp6 in intestinal inflammation and tumorigenesis. J. Immunol. 186, 7187–7194 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Elinav E et al. NLRP6 inflammasome regulates colonic microbial ecology and risk for colitis. Cell 145, 745–757 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Levy M et al. Microbiota-modulated metabolites shape the intestinal microenvironment by regulating NLRP6 inflammasome signaling. Cell 163, 1428–1443 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hara H et al. The NLRP6 inflammasome recognizes lipoteichoic acid and regulates Gram-positive pathogen infection. Cell 175, 1651–1664.e14 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Venuprasad K, Zeng M, Baughan SL & Massoumi R Multifaceted role of the ubiquitin ligase Itch in immune regulation. Immunol. Cell Biol. 93, 452–460 (2015). [DOI] [PubMed] [Google Scholar]
- 10.Massoumi R Ubiquitin chain cleavage: CYLD at work. Trends Biochem. Sci. 35, 392–399 (2010). [DOI] [PubMed] [Google Scholar]
- 11.Costello CM et al. Dissection of the inflammatory bowel disease transcriptome using genome-wide cDNA microarrays. PLoS Med. 2, e199 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cleynen I et al. Genetic and microbial factors modulating the ubiquitin proteasome system in inflammatory bowel disease.Gut 63, 1265–1274 (2014). [DOI] [PubMed] [Google Scholar]
- 13.Kathania M et al. Itch inhibits IL-17-mediated colon inflammation and tumorigenesis by ROR-γt ubiquitination. Nat. Immunol. 17, 997–1004 (2016). [DOI] [PubMed] [Google Scholar]
- 14.Peng DJ et al. Noncanonical K27-linked polyubiquitination of TIEG1 regulates Foxp3 expression and tumor growth. J. Immunol. 186, 5638–5647 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mevissen TE et al. OTU deubiquitinases reveal mechanisms of linkage specificity and enable ubiquitin chain restriction analysis. Cell 154, 169–184 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hrdinka M et al. CYLD limits Lys63- and Met1-linked ubiquitin at receptor complexes to regulate innate immune signaling. Cell Rep. 14, 2846–2858 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shen C et al. Molecular mechanism for NLRP6 inflammasome assembly and activation. Proc. Natl Acad. Sci. USA 116, 2052–2057 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dixon LJ, Berk M, Thapaliya S, Papouchado BG & Feldstein AE Caspase-1-mediated regulation of fibrogenesis in diet-induced steatohepatitis. Lab. Investig. 92, 713–723 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Singh AK et al. SUMOylation of ROR-γ inhibits IL-17 expression and inflammation via HDAC2. Nat. Commun. 9, 4515 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ten Hove T et al. Blockade of endogenous IL-18 ameliorates TNBS-induced colitis by decreasing local TNF-α production in mice. Gastroenterology 121, 1372–1379 (2001). [DOI] [PubMed] [Google Scholar]
- 21.Kanai T et al. Macrophage-derived IL-18-mediated intestinal inflammation in the murine model of Crohn’s disease. Gastroenterology 121, 875–888 (2001). [DOI] [PubMed] [Google Scholar]
- 22.Wlodarska M et al. NLRP6 inflammasome orchestrates the colonic host-microbial interface by regulating goblet cell mucus secretion. Cell 156, 1045–1059 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Karatzas DN et al. Inactivation of CYLD in intestinal epithelial cells exacerbates colitis-associated colorectal carcinogenesis - a short report. Cell Oncol. 39, 287–293 (2016). [DOI] [PubMed] [Google Scholar]
- 24.Siegmund B Interleukin-18 in intestinal inflammation: friend and foe? Immunity 32, 300–302 (2010). [DOI] [PubMed] [Google Scholar]
- 25.Nowarski R et al. Epithelial IL-18 equilibrium controls barrier function in colitis. Cell 163, 1444–1456 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu Z et al. Role of inflammasomes in host defense against Citrobacter rodentium infection. J. Biol. Chem. 287, 16955–16964 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lochner M & Forster I Anti-interleukin-18 therapy in murine models of inflammatory bowel disease. Pathobiology 70, 164–169 (2002). [DOI] [PubMed] [Google Scholar]
- 28.Siegmund B et al. Neutralization of interleukin-18 reduces severity in murine colitis and intestinal IFN-γ and TNF-α production. Am. J. Physiol. Regul. Integr. Comp. Physiol. 281, R1264–R1273 (2001). [DOI] [PubMed] [Google Scholar]
- 29.Ahmed N et al. The E3 ligase Itch and deubiquitinase Cyld act together to regulate Tak1 and inflammation. Nat. Immunol. 12, 1176–1183 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leach ST et al. Local and systemic interleukin-18 and interleukin-18-binding protein in children with inflammatory bowel disease. Inflamm. Bowel Dis. 14, 68–74 (2008). [DOI] [PubMed] [Google Scholar]
- 31.Leon AJ et al. High levels of proinflammatory cytokines, but not markers of tissue injury, in unaffected intestinal areas from patients with IBD. Mediators Inflamm. 2009, 580450 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Monteleone G et al. Bioactive IL-18 expression is up-regulated in Crohn’s disease. J. Immunol. 163, 143–147 (1999). [PubMed] [Google Scholar]
- 33.Pizarro TT et al. IL-18, a novel immunoregulatory cytokine, is up-regulated in Crohn’s disease: expression and localization in intestinal mucosal cells. J. Immunol. 162, 6829–6835 (1999). [PubMed] [Google Scholar]
- 34.Reuter BK & Pizarro TT Commentary: the role of the IL-18 system and other members of the IL-1R/TLR superfamily in innate mucosal immunity and the pathogenesis of inflammatory bowel disease: friend or foe? Eur. J. Immunol. 34, 2347–2355 (2004). [DOI] [PubMed] [Google Scholar]
- 35.Man SM Inflammasomes in the gastrointestinal tract: infection, cancer and gut microbiota homeostasis. Nat. Rev. Gastroenterol. Hepatol. 15, 721–737 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Deretic V, Saitoh T & Akira S Autophagy in infection, inflammation and immunity. Nat. Rev. Immunol. 13, 722–737 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yan Y et al. Dopamine controls systemic inflammation through inhibition of NLRP3 inflammasome. Cell 160, 62–73 (2015). [DOI] [PubMed] [Google Scholar]
- 38.Han S et al. Lipopolysaccharide primes the NALP3 inflammasome by inhibiting its ubiquitination and degradation mediated by the SCFFBXL2 E3 ligase. J. Biol. Chem. 290, 18124–18133 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Py BF, Kim MS, Vakifahmetoglu-Norberg H & Yuan J Deubiquitination of NLRP3 by BRCC3 critically regulates inflammasome activity. Mol. Cell 49, 331–338 (2013). [DOI] [PubMed] [Google Scholar]
- 40.Hu Z et al. Crystal structure of NLRC4 reveals its autoinhibition mechanism. Science 341, 172–175 (2013). [DOI] [PubMed] [Google Scholar]
- 41.Salcedo R et al. MyD88-mediated signaling prevents development of adenocarcinomas of the colon: role of interleukin 18. J. Exp. Med. 207, 1625–1636 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Takagi H et al. Contrasting action of IL-12 and IL-18 in the development of dextran sodium sulphate colitis in mice. Scand. J. Gastroenterol. 38, 837–844 (2003). [DOI] [PubMed] [Google Scholar]
- 43.Tucker TA et al. Transient transfection of polarized epithelial monolayers with CFTR and reporter genes using efficacious lipids. Am. J. Physiol. Cell Physiol. 284, C791–C804 (2003). [DOI] [PubMed] [Google Scholar]
- 44.Komander D et al. The structure of the CYLD USP domain explains its specificity for Lys63-linked polyubiquitin and reveals a B box module. Mol. Cell 29, 451–464 (2008). [DOI] [PubMed] [Google Scholar]
- 45.Iggo R & Richard E Lentiviral transduction of mammary epithelial cells. Methods Mol. Biol. 1293, 137–160 (2015). [DOI] [PubMed] [Google Scholar]
- 46.Ohta Y, Hamada Y & Katsuoka K Expression of IL-18 in psoriasis. Arch. Dermatol. Res. 293, 334–342 (2001). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon request. Source data for Figs. 1–8 and Extended Data Figs. 1 and 3–5 are provided with the paper.