Abstract
Background
We evaluated the association between the I/D polymorphism in the ACE gene and lung cancer risk by performing a meta-analysis.
Methods
The heterogeneity in the study was tested using the Cochran χ2-based Q statistic test and I2 test, and then the random ratio or fixed effect was utilized to merge the odds ratios (ORs) and 95% confidence intervals (CIs) to estimate the strength of the association between ACE polymorphisms and susceptibility to lung cancer. Sensitivity analysis was also performed. Using funnel plot and Begg’s rank test, we investigated the publication bias. All statistical analyses were performed using Stata 12.0 and RevMan 5.3.
Results
A total of 4307 participants (2181 patients; 2126 controls) were included in the 12 case–control studies. No significant association was found between the ACE I/D polymorphism and lung cancer risk (II vs. ID + DD: OR = 1.22, 95% CI = 0.89–1.68; II + ID vs. DD: OR = 1.21, 95% CI = 0.90–1.63; I vs. D: OR = 1.15, 95% CI = 0.95–1.39). In the subgroup analysis by ethnicity, no significant association between the ACE I/D polymorphism and lung cancer risk was found among Asian and Caucasian populations for the comparisons of II vs. ID + DD, II + ID vs. DD, and I vs. D genetic models.
Conclusion
The ACE I/D polymorphism is not associated with the risk of lung cancer.
Keywords: ACE, Polymorphism, Lung cancer, Meta-analysis
Background
Lung cancer has become the largest malignant tumor in terms of harm to human health and life [1, 2]. The incidence and mortality of lung cancer are also increasing every year, and its proportion in tumor mortality has also expanded [2]. The 5-year survival rate of lung cancer is only approximately 15% [3].
The human angiotensin-converting enzyme (ACE) gene, located on chromosome 17q23, has a length of 21 kb and consists of 26 exons and 25 introns [4]. The biological function of the ACE gene is affected by an insertion/deletion in the 16th intron, producing three genotypes: insertion homozygous (II), deletion homozygous (DD), and insertion/deletion of heterozygous type (ID) [5]. ACE activity in serum is related to the I/D polymorphism of the ACE gene [6]. The II genotype exhibits low activity, the DD gene exhibits high activity, and the heterozygous ID genotype has activity between the two [7].
In recent years, there have been many studies on the role of ACE I/D polymorphism in the risk of lung cancer, but there were some contradictions among the results of these studies. Some studies showed an obvious trend of the ACE ‘II’ genotype with increased risk of lung cancer [8, 9], whereas other studies have shown that the DD genotype of ACE contributes to a higher risk of lung cancer [10–13]. However, another study has shown that the ACE ‘ID’ genotype might increase the risk of lung cancer [14]. In addition, other studies have shown no association between ACE I/D polymorphism and lung cancer [15–19]. To more accurately assess the potential relationship between the ACE I/D polymorphism and the risk of lung cancer, we performed a meta-analysis using all eligible published studies.
Methods
Search strategies
We conducted a comprehensive search of the literature in the Web of Science, PubMed, Cochrane Library, Embase, and China National Knowledge Infrastructure (CNKI) electronic databases, covering relevant studies published as of June 31, 2019. The keywords for the search were as follows: (“angiotensin-converting enzyme” OR “ACE”) AND (“polymorphism” OR “variant” OR “mutation”) AND (“Lung cancer” OR “lung neoplasm”’). The literature on relevant data was searched in English and Chinese. In addition, retrieved articles and references were manually searched. Referring to the Preferred Reporting Project (PRISMA) Guide for Systematic Evaluation and Meta-Analysis [20], an information flow diagram related to the final eligibility data was constructed by screening all retrieved studies.
Inclusion and exclusion criteria
Screening for the studies of the relationship between ACE I/D polymorphism and the risk of lung cancer was performed according to the following inclusion criteria: (1) the design of the study was case–control; (2) the full text can be found; (3) the genotype information of the ACE I/D polymorphism was available; (4) the relationship of the ACE I/D polymorphism and the risk of lung cancer was evaluated. The major exclusion criteria were: (1) not a case–control study; (2) repeating early publications (studies used in different publications for the same sample data, including only the most complete samples after careful review); (3) unpublished articles, conference papers, meta-analysis, and systematic reviews; (4) family-based pedigree research. This meta-analysis strictly followed the requirements of PRISMA [20].
Data extraction
The data of the selected studies were independently extracted by two researchers using standard data collection forms. The information extracted from the literature was as follows: first author, year of publication, country of origin, mean age and gender in cases and controls, number of cases and controls, Hardy-Weinberg equilibrium, genotyping method, source of controls, and available genotype frequency information for ACE I/D. If the same sample data appeared in multiple publications, only publications with the largest sample size were included in the study. The differences between the two investigators were resolved through discussion. If the discussion could not resolve the objection between the two researchers, the objection was judged by a third investigator. All data were obtained from the full text of the published research, and the authors were not contacted for further information. All information on the participants in the selected studies is presented in Table 1.
Table 1.
Characteristic of studies included in the meta-analysis
| Author | year | country | Ethnicity | Age group | Genotype Methods |
Source of control | NOS score | HWE |
|---|---|---|---|---|---|---|---|---|
| Peddireddy et al [14] | 2018 | South Indian | Asia | Adult | PCR | PB | 8 | 0.726 |
| Phukan et al [9] | 2014 | Northeast India | Asia | Adult | PCR | PB | 8 | 0.227 |
| Ozen et al [16] | 2013 | Turkey | Caucasians | Adult | PCR | PB | 7 | 0.920 |
| Shi et al [12] | 2014 | China | Asia | Adult | PCR-SSP | PB | 6 | 0.308 |
| Cheon et al [15] | 2000 | Korea | Asia | Adult | PCR | – | 6 | 0.133 |
| Yaren et al [17] | 2008 | Turkey | Caucasians | Adult | PCR | – | 7 | 0.470 |
| Nacak et al [8] | 2010 | Turkey | Caucasians | Adult | PCR | PB | 8 | 0.268 |
| Wang et al [13, 21] | 2000 | China | Asia | Adult | PCR | – | 6 | 0.861 |
| Zhang et al [18] | 2005 | China | Asia | Adult | PCR | HB | 7 | 0.109 |
| Gao et al [11] | 2012 | China | Asia | Adult | PCR | HB | 6 | 0.018 |
| Devic Pavlic et al [10] | 2012 | Croatia | Caucasians | Adult | PCR | HB | 7 | 0.909 |
| Ding et al [19] | 2008 | China | Asia | Adult | PCR | HB | 7 | 0.175 |
Study quality assessment
Two evaluators evaluated the quality of the included studies according to the Newcastle-Ottawa Scale (NOS) [22], which is applicable to the quality assessment of observational studies. The difference between the two evaluators was reported and resolved by a third evaluator. The scores of research quality included mainly the following three aspects: (1) selection of the case groups and control groups (4 stars); (2) quality of confounding factor correction in the case and control populations (2 stars); and (3) determination of the exposure of interest in the studies (3 stars). For each item numbered in the selection and exposure categories, one study can be rated as up to one star, and comparability can be assigned up to two stars. Higher scores indicated an increase in the quality of the study. Studies with scores equal to or higher than six were considered high-quality studies.
Data analysis
The heterogeneity in the study was tested using the Cochran χ2-based Q statistic test and I2 test, and then the random ratio or fixed effect was utilized to merge the odds ratios (ORs) and 95% confidence intervals (CIs). The significance of the pooled OR was analyzed by the Z-test (P < 0.05, judged statistically significant). To estimate the strength of the association between ACE polymorphisms and susceptibility to lung cancer, we performed a sensitivity analysis. Using funnel plot and Begg’s rank test, publication bias was investigated. All statistical analyses were performed using Stata 12.0 (Stata Corp, College Station, TX, USA) and RevMan 5.3.
Results
Literature search and study characteristics
A flow chart of the literature search is shown in Fig. 1. A total of 279 potentially relevant articles were selected for the preliminary online search. After verifying and deleting 104 duplicate articles, 175 articles were included for further consideration. Through a review of the titles and abstracts, 15 articles were included for full-text review. Finally, 12 articles were included in the final analysis. These studies were published between 2005 and 2018, and included 2181 patients with lung cancer and 2126 controls. Except for one study, the distribution of genotypes in the controls followed the Hardy–Weinberg equilibrium (HWE). In addition, the NOS scores for all the studies ranged from 6 to 8 points; thus, the selected articles were considered to have high methodological quality. The relevant information of the included articles is shown in Tables 1 and 2.
Fig. 1.
The flow sheet of identification of eligible studies
Table 2.
The genotype distribution of ACE I/ D
| Author | Sample size (case/control) |
Female (%) (case/control) |
case | control | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| I/I | I/D | D/D | I | D | I/I | I/D | D/D | I | D | |||
| Peddireddy et al [14] | 246/250 | 28.0/28.0 | 48 | 161 | 37 | 257 | 235 | 111 | 113 | 26 | 335 | 165 |
| Phukan et al [9] | 151/151 | 45.7 /45.7 | 61 | 62 | 28 | 184 | 118 | 44 | 68 | 39 | 156 | 146 |
| Ozen et al [16] | 52/212 | 11.5/− | 10 | 30 | 12 | 50 | 54 | 67 | 105 | 40 | 239 | 185 |
| Shi et al [12] | 120/62 | 39.2/− | 47 | 49 | 24 | 143 | 97 | 26 | 31 | 5 | 83 | 41 |
| Cheon et al [15] | 218/121 | 26.6/42.1 | 72 | 116 | 31 | 260 | 178 | 48 | 50 | 23 | 146 | 96 |
| Yaren et al [17] | 75/85 | 8.0/9.4 | 4 | 39 | 32 | 47 | 103 | 14 | 37 | 34 | 65 | 105 |
| Nacak et al [8] | 125/165 | 12.0/48.5 | 37 | 50 | 38 | 124 | 126 | 29 | 72 | 64 | 130 | 200 |
| Wang et al [13, 21] | 34/38 | 23.5/44.7 | 10 | 6 | 18 | 26 | 42 | 13 | 18 | 7 | 44 | 32 |
| Zhang et al [18] | 47/54 | 14.9 /29.6 | 21 | 21 | 5 | 63 | 31 | 20 | 30 | 4 | 70 | 38 |
| Gao et al [11] | 684/602 | 27.2/33.6 | 351 | 271 | 62 | 973 | 395 | 320 | 253 | 29 | 893 | 311 |
| Devic Pavlic et al [10] | 308/353 | 29.5/38.5 | 64 | 148 | 96 | 276 | 340 | 78 | 177 | 98 | 333 | 373 |
| Ding et al [19] | 121/33 | 31.4/30.3 | 55 | 56 | 10 | 166 | 76 | 19 | 10 | 4 | 48 | 18 |
Meta-analysis results
The heterogeneity of the three genetic models was determined using the Q test and I squared statistics. As shown in Fig. 2, there was significant heterogeneity in the three models (II vs. ID + DD: P < 0.001, I2 = 77.9%; II + ID vs. DD: P = 0.002, I2 = 61.7%; I vs. D: P < 0.001, I2 = 73.0%); thus, the random-effect model was employed in the analysis of the three models. Our results revealed that there were no significant associations between the ACE I/D polymorphism and lung cancer in the II vs. ID + DD (OR = 1.22, 95% CI = 0.89–1.68, P = 0.22), II + ID vs. DD (OR = 1.21, 95% CI = 0.90–1.63, P = 0.21), and I vs. D (OR = 1.15, 95% CI = 0.95–1.39, P = 0.15). In the subgroup analysis by ethnicity, no significant association was found among the three models in both Caucasian and Asian populations (Table 3). Sensitivity analysis was used to assess the impact of each individual study on the pooled OR by sequentially removing each eligible study. Our results suggest that none of the studies affected the overall outcome of the pooled OR (Fig. 3). Begg’s funnel plot was used to assess publication bias, and the results showed that publication bias was not reflected in the three genetic models. (II vs. ID + DD: P = 0.41; II + ID vs. DD: P = 0.34; I vs. D: P = 0.89) (Fig. 4).
Fig. 2.
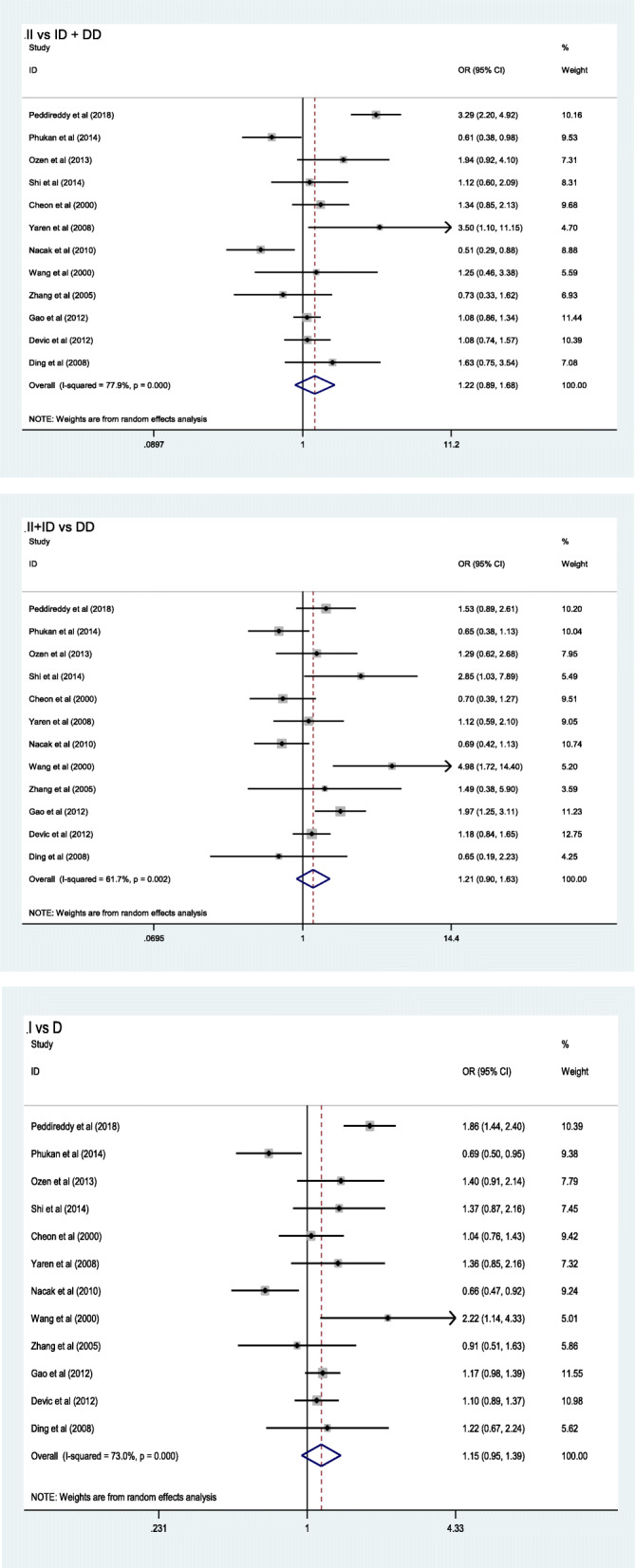
Forest plot results of meta- analysis for association between the ACE I/D polymorphism and lung cancer under different genetic models (II vs ID+DD; II + ID vs DD; I vs D)
Table 3.
Summary of pooled OR in different ethnicities
| Genetic model | group | Pooled OR (95% CI) | Heterogeneity | Test for overall effect | ||
|---|---|---|---|---|---|---|
| P | I2 | Z | P | |||
| II vs ID+DD | Caucasians | 1.22 (0.75–2.00) | 0.007 | 71.9% | 0.80 | 0.423 |
| Asia | 1.21 (0.76–1.94) | < 0.01 | 82.9% | 0.81 | 0.419 | |
| II + ID vs DD | Caucasians | 0.98 (0.79–1.23) | 0.284 | 20.5% | 0.15 | 0.881 |
| Asia | 1.56 (0.94–2.59) | 0.005 | 67.3% | 1.71 | 0.088 | |
| I vs D | Caucasians | 1.05 (0.82–1.34) | < 0.01 | 73.0% | 0.39 | 0.694 |
| Asia | 1.24 (0.92–1.66) | < 0.01 | 77.9% | 1.41 | 0.159 | |
Fig. 3.
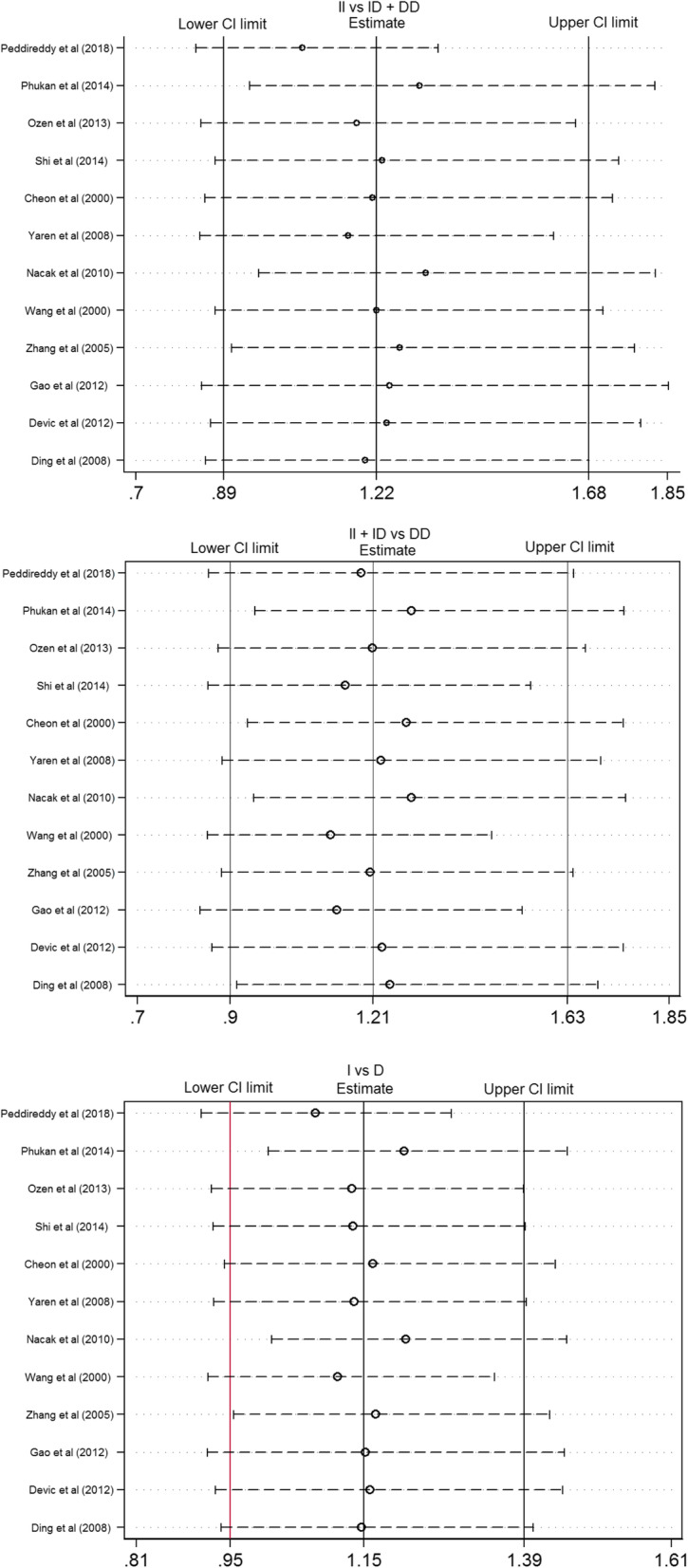
Sensitivity analysis examining the association between the ACE I/D polymorphism and risk of lung cancer under three model (II vs ID+DD, II + ID vs DD, I vs D)
Fig. 4.
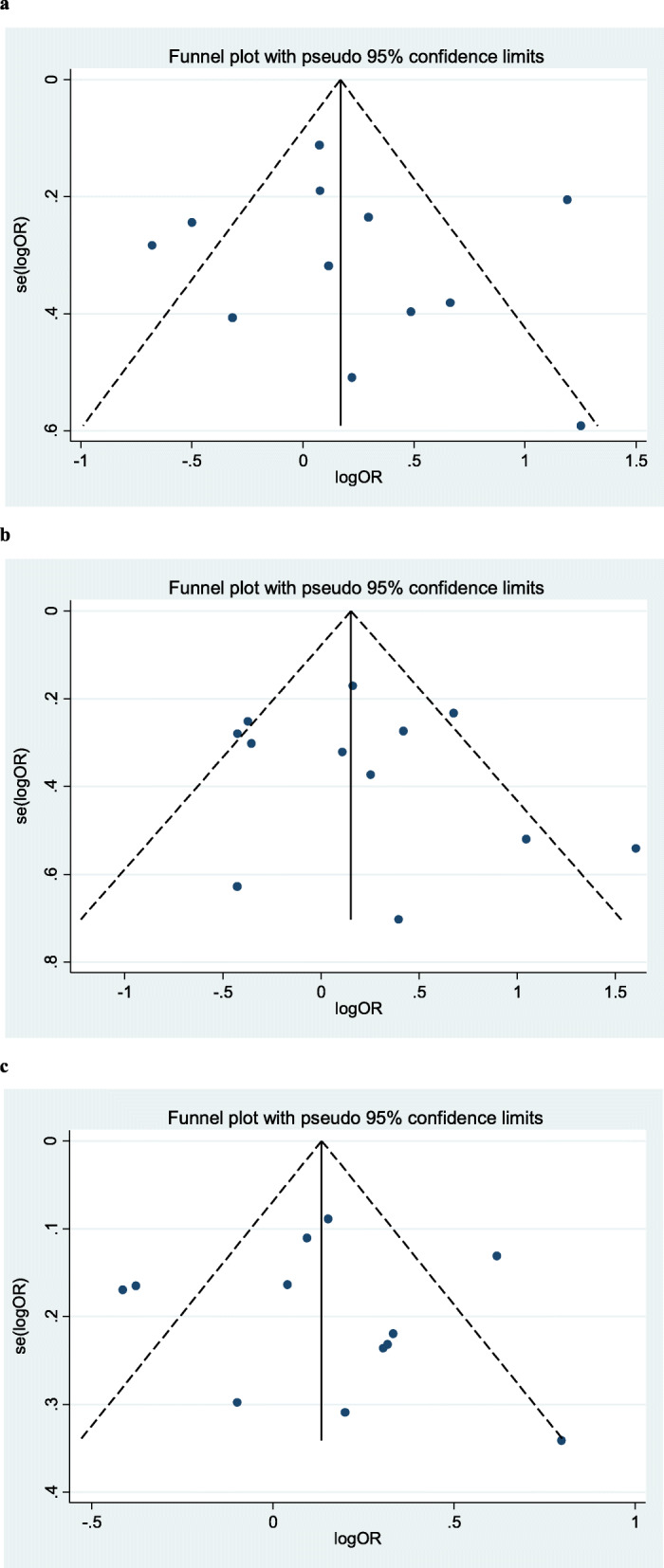
Begg’s funnel plot for publication bias analysis. a is the model of II vs ID+DD; b is the model of II + ID vs DD; c is the model of I vs D
Discussion
Recent studies have reported that ACE may be involved in the development of tumors [23–25]. ACE is a key enzyme in the renin-angiotensin system, which converts angiotensin I to angiotensin II and inactivates bradykinin. The mechanism of action may be due to the fact that angiotensin II can stimulate the synthesis of DNA and protein in vascular smooth muscle cells and promote the synthesis and secretion of vascular endothelial growth factor [26]. In addition, it may be involved in tumor development through bradykinin that can increase the permeability of the cell membrane to electrolytes and peptides. ACE can inactivate bradykinin [27]; however, low levels of ACE in tumor tissues can promote the invasive growth of malignant tumors. An increasing number of studies have investigated the association between ACE I/D polymorphism and lung cancer risk; however, there are inconsistencies and conflicting results. To further assess the association between ACE I/D polymorphism and lung cancer risk, we performed a meta-analysis of 12 case–control studies, including 2181 cases and 2126 controls.
The results of this meta-analysis showed no significant association between the three genetic models and the development of lung cancer. Although previous studies have revealed that ACE may have a certain effect on the etiology of lung cancer, our results suggest that these effects may not be caused by ACE gene mutations. The exact pathogenetic role of ACE in the etiology of lung cancer remains unclear. Our results suggest that the ACE I/D polymorphism does not affect cancer risk. Moreover, considering that this polymorphism may affect serum ACE levels and ACE levels may affect the risk of lung cancer, the risk for lung cancer is not directly caused by ACE gene mutations. Therefore, future research is necessary to determine the association between ACE polymorphism, ACE levels, and cancer risk.
Previously, a meta-analysis of eight published studies [8, 10, 11, 13, 15, 17–19] performed by Cheng et al. [28], including 1612 cases and 1442 controls, showed that the ACE I/D polymorphism is not associated with lung cancer. Wang et al. [21] have also conducted a meta-analysis of six published studies with 807 cases and 816 controls [8, 10, 13, 15, 18], and the results also showed that the ACE I/D polymorphism may not be associated with lung cancer risk. In our meta-analysis, 12 studies were included with 2181 cases and 2126 controls. Therefore, the statistical power of the current analysis is better than those of the two previous meta-analyses. Compared with the other studies, this study is more comprehensive regarding the relationship between ACE I/D polymorphism and lung cancer risk. Despite the differences between the studies included in the analysis, the results of our study suggest that the ACE I/D polymorphism may not lead to cancer risk, which is consistent with the findings of Cheng and Wang.
However, there are certain limitations to our study. First, databases that include studies published only in Chinese and English language were selected for analysis, and studies in other languages or unpublished potential research are missing. Second, due to the lack of raw data, we were unable to assess the potential gene–gene and gene–environment interactions. Third, the meta-analysis included data from Europeans and Asians, so the results apply only to these two ethnic groups. Fourth, among the three models, heterogeneity may greatly influence the conclusions of the meta-analysis. Lastly, the power of our statistical analysis with the current sample size was not high enough, so the results obtained from this study should be verified in the future.
Conclusion
In summary, our study showed that ACE I/D polymorphism did not increase or decrease the risk to lung cancer. Further well-designed studies should be conducted to confirm our findings in different populations and age groups, such as different races in Asia or Europe or other populations or children and adults. Future research will also need to explore the possible role of ACE I/D gene–gene and gene–environment interactions in the susceptibility to lung cancer.
Acknowledgements
Not applicable.
Abbreviations
- OR
Odds ratios
- CI
Confidence interval
- ACE
Angiotensin-converting enzyme
- CNKI
China Nationa Knowledge Infrastructure
- NOS
Newcastle-Ottawa Scale
- HWE
Hardy Weinberg equilibrium
Authors’ contributions
JC and RL: Conceptualization, data curation, formal analysis, methodology, writing and review and editing. MS and MZ: Software, supervision, validation, visualization. All authors have read and approved the manuscript.
Funding
Not applicable.
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Yokota J, Shiraishi K, Kohno T. Genetic basis for susceptibility to lung cancer: recent progress and future directions. Adv Cancer Res. 2010;109:51–72. doi: 10.1016/B978-0-12-380890-5.00002-8. [DOI] [PubMed] [Google Scholar]
- 2.Schwartz AG, Cote ML. Epidemiology of lung Cancer. Adv Exp Med Biol. 2016;893:21–41. doi: 10.1007/978-3-319-24223-1_2. [DOI] [PubMed] [Google Scholar]
- 3.Sculier JP, Berghmans T, Meert AP. Advances in target therapy in lung cancer. Eur Respir Rev. 2015;24(135):23–29. doi: 10.1183/09059180.00011014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Castellon R, Hamdi HK. Demystifying the ACE polymorphism: from genetics to biology. Curr Pharm Des. 2007;13(12):1191–1198. doi: 10.2174/138161207780618902. [DOI] [PubMed] [Google Scholar]
- 5.Harrap SB, Davidson HR, Connor JM, Soubrier F, Corvol P, Fraser R, Foy CJ, Watt GC. The angiotensin I converting enzyme gene and predisposition to high blood pressure. Hypertension. 1993;21(4):455–460. doi: 10.1161/01.HYP.21.4.455. [DOI] [PubMed] [Google Scholar]
- 6.Rocken C, Lendeckel U, Dierkes J, et al. The number of lymph node metastases in gastric cancer correlates with the angiotensin I-converting enzyme gene insertion/deletion polymorphism. Clin Cancer Res. 2005;11(7):2526–2530. doi: 10.1158/1078-0432.CCR-04-1922. [DOI] [PubMed] [Google Scholar]
- 7.Rigat B, Hubert C, Alhenc-Gelas F, Cambien F, Corvol P, Soubrier F. An insertion/deletion polymorphism in the angiotensin I-converting enzyme gene accounting for half the variance of serum enzyme levels. J Clin Investig. 1990;86(4):1343–1346. doi: 10.1172/JCI114844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nacak M, Nacak I, Sanli M, Ozkur M, Pektas M, Aynacioglu AS. Association of angiotensin converting enzyme gene insertion/deletion polymorphism with lung cancer in Turkey. Cancer Genet Cytogenet. 2010;198(1):22–26. doi: 10.1016/j.cancergencyto.2009.11.019. [DOI] [PubMed] [Google Scholar]
- 9.Phukan RK, Borah PK, Saikia BJ, Das M, Sekhon GS, Mahanta J. Interaction of tobacco smoking and chewing with angiotensin converting enzyme (insertion/deletion) gene polymorphisms and risk of lung cancer in a high risk area from Northeast India. Asian Pac J Cancer Prev. 2014;15(24):10691–10695. doi: 10.7314/APJCP.2014.15.24.10691. [DOI] [PubMed] [Google Scholar]
- 10.Devic Pavlic S, Ristic S, Flego V, Kapovic M, Radojcic BA. Angiotensin-converting enzyme insertion/deletion gene polymorphism in lung cancer patients. Genet Test Mol Biomarkers. 2012;16(7):722–725. doi: 10.1089/gtmb.2011.0306. [DOI] [PubMed] [Google Scholar]
- 11.Gao M, Wang Y, Shi Y, et al. The relationship between three well-characterized polymorphisms of the angiotensin converting enzyme gene and lung cancer risk: a case-control study and a meta-analysis. J Renin-Angiotensin-Aldosterone Syst. 2012;13(4):455–460. doi: 10.1177/1470320312443912. [DOI] [PubMed] [Google Scholar]
- 12.Shi G, Sun Y, Wu ED, Liang Z, Zhang B. Association of genetic polymorphism of angiotensin converting enzyme gene with lung cancer. China Medical Herald. 2014;11(15):43–46. [Google Scholar]
- 13.Wang H, Nie ZH, Duan Y, et al. Angiotensin -converting enzyme gene polymorphism in patients with lung carcinoma. J Navy Med. 2000;04:319–322. [Google Scholar]
- 14.Peddireddy V, Badabagni SP, Gundimeda SD, Mundluru HP. Association of eNOS and ACE gene polymorphisms and plasma nitric oxide with risk of non-small cell lung cancer in South India. Clin Respir J. 2018;12(1):207–217. doi: 10.1111/crj.12517. [DOI] [PubMed] [Google Scholar]
- 15.Cheon KT, Choi KH, Lee HB, Park SK, Rhee YK, Lee YC. Gene polymorphisms of endothelial nitric oxide synthase and angiotensin-converting enzyme in patients with lung cancer. Lung. 2000;178(6):351–360. doi: 10.1007/s004080000039. [DOI] [PubMed] [Google Scholar]
- 16.Ozen F, Polat F, Arslan S, Ozdemir O. Combined germline variations of thrombophilic genes promote genesis of lung cancer. Asian Pac J Cancer Prev. 2013;14(9):5449–5454. doi: 10.7314/APJCP.2013.14.9.5449. [DOI] [PubMed] [Google Scholar]
- 17.Yaren A, Oztop I, Turgut S, Turgut G, Degirmencioglu S, Demirpence M. Angiotensin-converting enzyme gene polymorphism is associated with anemia in non small-cell lung cancer. Exp Biol Med (Maywood, NJ) 2008;233(1):32–37. doi: 10.3181/0705-RM-141. [DOI] [PubMed] [Google Scholar]
- 18.Zhang Q, Liu X, Zhang Z, Wang L. Analysis of the relationship between polymorphism of angiotensin-converting enzyme gene and lung cancer. Chin J Lung Cancer. 2005;8(3):211–214. doi: 10.3779/j.issn.1009-3419.2005.03.11. [DOI] [PubMed] [Google Scholar]
- 19.Ding X. Analysis of the relationship between the polymorphism of angiotensin- converting enzyme gene and Lung Cancer. Suzhou: Soochow University; 2008.
- 20.Moher D, Liberati A, Tetzlaff J, Altman DG. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. Ann Intern Med. 2009;151(4):264–269. doi: 10.7326/0003-4819-151-4-200908180-00135. [DOI] [PubMed] [Google Scholar]
- 21.Wang N, Yang D, Ji B, Li J. Angiotensin-converting enzyme insertion/deletion gene polymorphism and lung cancer risk: a meta-analysis. J Renin Angiotensin Aldosterone Syst. 2015;16(1):189–194. doi: 10.1177/1470320314552310. [DOI] [PubMed] [Google Scholar]
- 22.Stang A. Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur J Epidemiol. 2010;25(9):603–605. doi: 10.1007/s10654-010-9491-z. [DOI] [PubMed] [Google Scholar]
- 23.Coulson R, Liew SH, Connelly AA, et al. The angiotensin receptor blocker, losartan, inhibits mammary tumor development and progression to invasive carcinoma. Oncotarget. 2017;8(12):18640–18656. doi: 10.18632/oncotarget.15553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pandith AA, Qasim I, Zahoor W, Shah P, Bhat AR. ACE I/D sequence variants but not MTHFR C677T, is strongly linked to malignant glioma risk and its variant DD genotype may act as a promising predictive biomarker for overall survival of glioma patients. Gene. 2018;639:62–68. doi: 10.1016/j.gene.2017.10.013. [DOI] [PubMed] [Google Scholar]
- 25.Sugimoto M, Furuta T, Shirai N, Ikuma M, Sugimura H, Hishida A. Influences of chymase and angiotensin I-converting enzyme gene polymorphisms on gastric cancer risks in Japan. Cancer Epidemiol Biomarkers Prev. 2006;15(10):1929–1934. doi: 10.1158/1055-9965.EPI-06-0339. [DOI] [PubMed] [Google Scholar]
- 26.Walther T, Menrad A, Orzechowski HD, Siemeister G, Paul M, Schirner M. Differential regulation of in vivo angiogenesis by angiotensin II receptors. FASEB J. 2003;17(14):2061–2067. doi: 10.1096/fj.03-0129com. [DOI] [PubMed] [Google Scholar]
- 27.Yang HY, Erdos EG, Levin Y. A dipeptidyl carboxypeptidase that converts angiotensin I and inactivates bradykinin. Biochim Biophys Acta. 1970;214(2):374–376. doi: 10.1016/0005-2795(70)90017-6. [DOI] [PubMed] [Google Scholar]
- 28.Cheng Z, Ma R, Tan W, Zhang L, Tan Q. Lack of association between ACE insertion/deletion polymorphism and lung cancer: a meta-analysis. J Renin Angiotensin Aldosterone Syst. 2015;16(2):453–458. doi: 10.1177/1470320314544163. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.