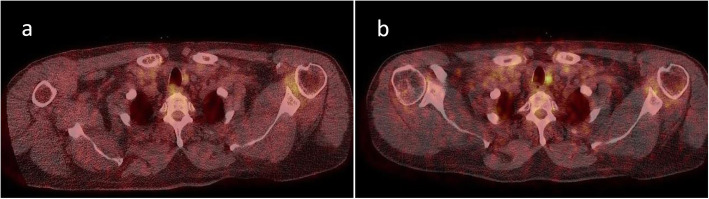Abstract
Background
Expression of CXCR4, a chemokine (C-X-C motif) receptor that plays a central role in tumor growth and metastasis of circulating tumor cells, has been described in a variety of solid tumors. A high expression of CXCR4 has a prognostic significance with regard to overall and progression-free survival and offers a starting point for targeted therapies. In this context, [68]Ga-Pentixafor-Positron Emission Tomography/Computer Tomography (PET/CT) offers promising possibility of imaging the CXCR4 expression profile. We set out to compare a [18F] fluorodeoxyglucose (FDG)-PET/CT and a [68Ga]Pentixafor-PET/CT in (re-)staging and radiation planning of patients with localized esophageal cancer.
Materials and methods
In this retrospective analysis, ten patients, with adeno- or squamous cell carcinoma of the esophagus (n = 3 and n = 7, respectively), which were scheduled for radio (chemo) therapy, were imaged using both Pentixafor and FDG PET/CT examinations. All lesions were visually rated as Pentixafor and FDG positive or negative. For both tracers, SUVmax was measured all lesions and compared to background. Additionally, immunohistochemistry of CXCR4 was obtained in patients undergoing surgery.
Results
FDG-positive tumor-suspicious lesions were detected in all patients and a total of 26 lesions were counted. The lesion-based analysis brought equal status in 14 lesions which were positive for both tracers while five lesions were FDG positive and Pentixafor negative and seven lesions were FDG negative, but Pentixafor positive. Histopathologic correlation was available in seven patients. The CXCR4 expression of four non-pretreated tumour lesion samples was confirmed immunohistochemically.
Conclusion
Our data shows that additional PET/CT imaging with Pentixafor for imaging the CXCR4 chemokine receptor is feasible but heterogeneous in both newly diagnosed and pretreated recurrent esophageal cancer. In addition, the Pentixafor PET/CT may serve as complementary tool for radiation field expansion in radiooncology.
Supplementary Information
The online version contains supplementary material available at 10.1186/s40644-021-00391-w.
Keywords: CXCR4, Pentixafor, Esophageal cancer, Molecular imaging, PET/CT, Radiotherapy
Introduction
In 2018, there were 572,034 new diagnoses of esophageal cancer and 508,585 deaths caused by the disease worldwide [1]. Malignancies of the esophagus cause about 3.5% of all cancer deaths in men and 1.2% in women in Germany [2]. Histopathologically, esophageal carcinoma usually manifests as adenocarcinoma or squamous cell carcinoma. Curative treatment with operation alone in T1–2a tumors and tri-modal treatment with radio-chemotherapy and surgery in locally advanced stages is restricted to patients with no distant metastasis. Since most patients present with advanced disease given late onset of symptoms, they will be directed to non-surgical treatment such as radio- and/or chemotherapy, leading to an expected 5-year survival of less than 15% [3]. As a result, new treatment options including personalized medicine targeting specific molecular markers, are urgently needed.
Positron emission tomography–computed tomography (PET/CT) using fluorodeoxyglucose (FDG) has become an established tool for staging patients with esophageal cancer, since it has the highest sensitivity for the evaluation of distant metastasis [4]. Furthermore FDG PET/CT is used in esophageal cancer to predict response to chemotherapy at an early stage of treatment. Here, FDG PET/CT has shown its predictive value concerning treatment outcome [5].
Chemokine (C-X-C motif) receptor 4 (CXCR4) and its ligand, the alpha-chemokine CXCL12, has been described to play a central role in tumor growth and progression, tumor invasiveness and metastasis [6]. Overexpression of this receptor has been reported in more than 30 different types of cancer [7–10]. Importantly, the CXCL12/CXCR4 axis has been identified as a target for drugs in human tumors due to its critical role in promoting and maintaining cancer stem cells [11]. The CXCL12/CXCR4 axis mobilizes heterogeneous signaling pathways that foster adhesion, chemotaxis, migration, cell proliferation and survival [12].
In primary esophageal cancer, overexpression of CXCR4 was found to be associated with clinicopathological features e.g. gender, histological differentiation, tumor depth, and status of lymph node metastasis, and poor prognosis [13]. Probably CXCR4 expression can be expected in more than 50% of esophageal cancer patients [14]. Wang et al. could show that the ability of esophageal cancer stem cells to spread and metastasize through ERK1/2 signaling pathway could be inhibited by blockage of CXCR4 with inhibitors or shRNA approaches both in vivo and in vitro studies [15]. Zhang et al. suggest that miR-302b, a small non-coding RNA, may be a novel cancer-related inflammation (CRI) regulating miRNA [16]. It inhibits CRI critical pathway and downstream cytokines expression through targeting CXCR4 amongst others, resulting in decrease of tumor growth.
In 2011, a radiolabelled CXCR4-ligand ([68Ga] Pentixafor) for PET imaging has been developed promising diagnostic improvement and targeted treatment [17–19]. Herrmann et al. showed first results of CXCR4-targeted radiotherapy with Lu-marked CXCR4 specific agent pentixather [20].
We hypothesize that additional PET/CT imaging with Pentixafor to visualize the chemokine receptor CXCR4 is feasible in both newly diagnosed and pretreated recurrent esophageal cancer and gives complementary information to FDG PET/CT. Radiolabeled chemokine ligands could contribute as additive imaging for in vivo identification and non-invasive tumor characterization. In this context, they could extend the staging information and possibly enable better patient stratification.
The aim of this analysis is to report on a direct comparison Pentixafor PET/CT and FDG PET/CT in patients with esophageal cancer in (primary) staging as well as part of planning PET CT prior to radio-chemotherapy as a feasible option, as in order to evaluate further treatment options for selected patients.
Materials and methods
Patients
Both FDG and Pentixafor PET/CT were performed for clinical use. In this retrospective analysis we included ten adult esophageal cancer patients suffering of advanced or relapsed disease, and in whom radio (chemo) therapy was planned. We restricted our analyses to PET/CT scans which were performed between November 2014 and March 2015. All consecutive patients underwent an additional Pentixafor PET/CT within a mean period of 8 days (range 2–35) in order to further characterize their CXCR4 expression and to evaluate potential CXCR4-related treatment options.
PET/CT imaging protocol
All PET/CT examinations were performed on a Biograph mCT Flow – Edge 128 PET/CT system (Siemens Medical Solutions) with a 128-slice spiral CT component from the base of the skull to the mid-thigh after patients had fasted for 6 hours. The CT scan for the attenuation correction was performed as a native non-diagnostic scan with a tube current of 30 mAs and a maximum voltage power of 120 kVp. The CT scan was followed by a PET emission scan. FDG was synthesized in house as previously described [21]. Injection activities for FDG were the following: mean 299.2 MBq (range 242–394 MBq). Uptake time between injection and scan was mean 01:11 min (range 00:57–01:37 min). To meet the criteria for European Association of Nuclear Medicine and its Research Ltd. (EARL) certification, reconstruction was performed via ordered subset expectation maximization (OSEM) algorithm (four iterations and twelve subsets), followed by an intrinsic 5 mm Gaussian filter in all directions. Pentixafor was prepared in a fully automated procedure using a module equipped with a single-use cassette kit (Scintomics GmbH, Germany; ABX, Germany) [12]. For radiolabelling, 25 μg of the precursor CPCR4.2 (ABX, Germany) were used. Per patient, approximately 8.3 μg of the [68]Ga-Pentixafor was then applied. Injection activities for Pentixafor were the following: mean 174.5 MBq (range 129–226 MBq). Uptake time between injection and scan was mean 01:04 min (range 00:59–01:11 min).
All patients underwent the PET/CT examinations as part of the clinical workup in order to potentially optimize their individual treatment and with diagnostic intent. All patients signed informed consent in regard of the scientific evaluation of their data. The retrospective evaluation of the data was approved by our ethics committee and conformed to the provisions of the Helsinki Declaration.
Image analysis
PET images were independently analyzed with reference to the contrast CT images by two experienced nuclear medicine specialists. All differences of opinion in interpretation were resolved by consensus. For all image analyses OSEM reconstruction was used. For visual analysis, uptake of all lesions (reference regions) was assessed for both FDG and Pentixafor as positive or negative taking the mediastinal blood pool (MBP) as a reference region. A lesion was considered Pentixafor- or FDG-positive when measured ≥ MBP. In addition, we also considered the liver as reference [22]. Tumor to background ratios were calculated as SUVmax of tumor divided by SUVmean of background (Table 2). For quantitative evaluation the maximal standardized uptake value (SUVmax) was measured in all the lesions, too. The mean standardized uptake value (SUVmean) in the reference regions were determined by placing a sphere with a diameter of 2 cm in the upper right part of the liver, in the mediastinum (refers to ascending aorta), in the spleen, in the bone marrow and in the brain.
Table 2.
Intraindividual comparison of lesion uptake FDG vs. Pentixafor
| Patient no. | Localization of the lesion | Quantitative analyses | Visual analyses | Adeno/SCC | CXCR4 expressionb | ||
|---|---|---|---|---|---|---|---|
| FDG SUVmax | Pentixafor SUVmax | FDGa | Pentixafora | ||||
| #1 | paratracheal left | 2.2 | 5.2 | 0 | 2 | SCC | 1 |
| #2 | mediastinal | 10.1 | 3.2 | 2 | 0 | SCC | 4 |
| infraclavicular | 3.4 | 4.5 | 2 | 1 | |||
| #3 | hip | 7.1 | 2.7 | 2 | 2 | Adeno | 1 |
| thigh | 10.5 | 3.9 | 2 | 2 | |||
| adrenal gland left | 3.3 | 2.9 | 2 | 2 | |||
| left paraaortal lymph node | 3.3 | 1.3 | 1 | 0 | |||
| #4 | esophagus | 7.3 | 5.9 | 2 | 2 | SCC | 3 |
| coeliacal | 9,0 | 5,2 | 2 | 2 | |||
| axillary right | 0,85 | 1,73 | 0 | 2 | |||
| hilary right | 2,0 | 3,5 | 0 | 2 | |||
| gastric curvature | 5,6 | 4,1 | 2 | 2 | |||
| #5 | esophagus | 6.6 | 6.5 | 2 | 2 | Adeno | 0 |
| #6 | mediastinal | 14.4 | 4.2 | 2 | 0 | Adeno | 2 |
| #7 | mediastinal | 15.8 | 4.8 | 2 | 2 | SCC | 3 |
| #8 | adrenal gland left | 2.5 | 5.0 | 2 | 1 | SCC | 4 |
| esophagus | 3.3 | 3.9 | 1 | 0 | |||
| lung upper lobe | 4.9 | 1.5 | 2 | 0 | |||
| #9 | adrenal gland left | 2.7 | 12.0 | 0 | 2 | SCC | 2 |
| #10 | infracarinal | 2.7 | 9.1 | 0 | 2 | SCC | 4 |
| mediastinal | 4.1 | 9.8 | 1 | 2 | |||
| supra right | 2.2 | 5.4 | 0 | 2 | |||
| supra left | 2.8 | 6.6 | 0 | 2 | |||
| esophagus | 14.4 | 3.7 | 2 | 1 | |||
| rib left | 9.2 | 3.8 | 2 | 1 | |||
| cervical left | 13.7 | 4.1 | 2 | 1 | |||
| SUM | 26 | ||||||
Adeno Adenocarcinoma, MPB Mediastinal blood pool, SCC Squamous cell cancer
a 0= < MBP, 1 ≥ MBP, 2 ≤ Liver
b 0 = no CXCR4; 1 = low CXCR4; 2 = intermediate; 3 = strong; 4 = not available
Immunohistochemistry
The reference pathology was performed by one experienced pathologist; again, uncertainties in interpretation were resolved by consensus in cooperation with another pathological specialist with extensive expertise. To exclude a possible influence of concomitant therapy on receptor surface expression from the beginning, biopsies (oesophagogastroduodenoscopy (OGD), n = 4) were performed shortly after PET imaging and before starting treatment. Immunohistochemistry (IHC) was performed on formalin-fixed and paraffin embedded material using the rabbit monoclonal CXCR4 antibody from Cell Signaling Technology (clone D4Z7W; order code 97680; EDTA-buffer, dilution 1:800) on the automated Leica Bond stainer. The CXCR4 expression, which means CXCR4-positive inflammatory cells in the tissue, will be given in a scale range from 0 to 4 (0 = no CXCR4 (0–3 CXCR4 positive cells/high power field (HPF); 1 = low CXCR4 (4–10 CXCR4 positive cells/HPF); 2 = intermediate (11–25 CXCR4 positive cells/HPF); 3 = strong (> 26 CXCR4 positive cells/ HPF); 4 = not available).
Statistics
Quantitative measurements are presented using descriptive statistics. SUVs in the lesions, background regions and their ratios were compared using Wilcoxon matched-pair signed-rank (2 samples) test. Statistical analyses were performed using IBM SPSS statistics (version 22, IBM SPSS Statistics, Armonk, NY, USA) and R Core team (2017, version 4.0.3, Vienna, Austria).
Results
Clinical findings
Eight patients presented with advanced stage disease (infiltration of the tunica adventitia or neighboring structures (T3–4 stage)), two subjects with local disease (infiltration into the muscularis propria layer (T2 stage). Two patients underwent Pentixafor PET/CT as part of restaging after radiotherapy (#8) or radiochemotherapy (#9); the therapies were completed 11 and 9 months ago. Patient #4 was treated with platinum-containing postoperative radiochemotherapy for first-line laryngeal cancer 24 months prior to diagnosis of esophagus cancer. In the majority of cases there was no intervention (e.g. chemotherapy, surgery) done between the scans; however, patient #6 received three fractions, patient #7 one fraction of a radiotherapy in 1.8 Gray single dose before Pentixafor PET/CT (fractions of radiotherapy is equivalent to days before PET/CT). In patients #1,4,6 and 7, CXCR4 expression was determined in cell sample obtained by OGD. In the remaining patients either no cell material was present (n = 3) or the expression was determined after previous radiotherapy ± chemotherapy (n = 4).
Detailed patient characteristics (eight males, mean age 67, standard deviation 8, range 53–76 years) are given in Table 1. In order to optimise the radio-oncological treatment of all patients, the suspicious lesions resulting from the additional information obtained with the Pentixafor PET/CT (compare Figs. 1 and 2) were included in clinical staging. An oncological upstaging was defined for patient #10 in consensus. In four patients (#1, #4, #9, #10) we found additional lesions in Pentixafor PET/CT as compared to FDG PET/CT. We included lesions in the irradiation volume if we found them Pentixafor PET-positive. By definition, most lesions were already located in the area to be irradiated (e.g. lymph drainage area). The additional lesion in the left rib at #10 did not lead to an expansion of the irradiation field. A follow-up was intended here. Consequently, the irradiation field was adjusted based on the gain in information from the Pentixafor PET/CT in one patient (#4, lesion near the right hilus; FDG negative, Pentixafor positive; Fig. 3).
Table 1.
Demographic and clinical characteristics of participants
| Patient no. | Age | Gender | Grading | UICC TNM | Sample origin | Histology |
|---|---|---|---|---|---|---|
| #1 | 61 | m | GX | cT2 cN1 cM0 | I | SCC |
| #2 | 72 | m | G2 | cT4 cN2 cM0 | III | SCC |
| #3 | 76 | m | G3 | cT2 cN1 cM0 | II | Adeno |
| #4 | 58 | m | G3 | uT3 cN2 cM0 | I | SCC |
| #5 | 70 | m | G2 | uT3 cN0 cM0 | II | Adeno |
| #6 | 75 | m | G2 | uT3 cN+ cM0 | I | Adeno |
| #7 | 53 | m | G2 | cT3–4 cN2 cM0 | I | SCC |
| #8 | 74 | w | GX | uT3 cN+ cM0 | III | SCC |
| #9 | 70 | w | GX | uT3 cN1 cM0 | II | SCC |
| #10 | 61 | m | G2 | cT3–4 cN2 cM1 | III | SCC |
Adeno Adenocarcinoma, SCC Squamous cell cancer, UICC TNM The Union for International Cancer Control Tumor Node Metastasis Classification of Malignant Tumours, 7th Edition, I collection by oesophago-gastroduodenoscopy, II collection by surgery, III no sample available for CXCR4 expression determination
Fig. 1.
Esophageal carcinoma patient (#1) with (a) positive FDG-uptake and a suspicious lesion enhanced by Pentixafor PET/CT (b). Arrow indicates the lesion
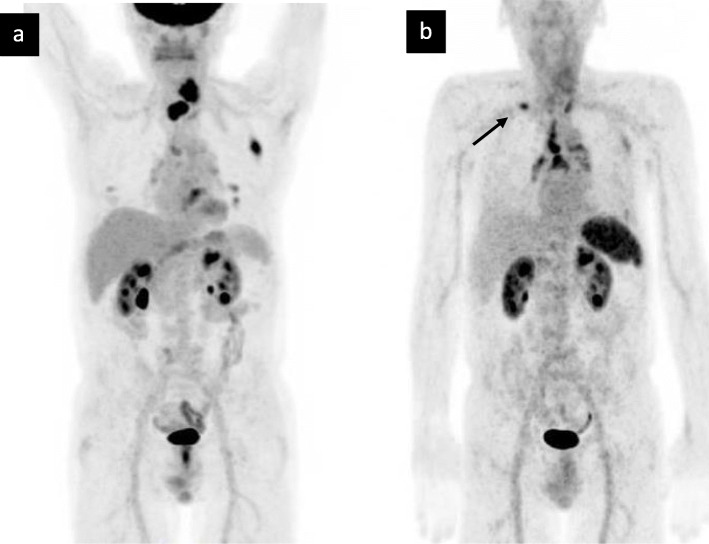
Fig. 2.
Key lesion rather peripher located to the primary tumor. Disconcordance of (a) FDG PET/CT and (b) non-invasive Pentixafor PET/CT imaging in one patient suffering from esophageal cancer. The lesion demonstrates high CXCR4 expression; local response was able to be shown in re-staging PET/CT after radiochemotherapy. Arrow indicates the lesion
Fig. 3.
Key lesion with close relation to the primary tumor: Esophageal carcinoma patient (#4) with (a) negative FDG-uptake and an additional suspicious lesion hilary right enhanced by Pentixafor PET/CT (b). The abdominal uptake is caused by an inserted gastric feeding tube. Arrow indicates the hilary lesion
SUVs in background for FDG and Pentixafor
Mean SUVmax in the tumor lesions were 6.9 ± 4.6 for FDG and 4.7 ± 2.5 for Pentixafor respectively. The mean SUVmean in the reference regions for FDG and Pentixafor were 2.5 ± 0.4/1.4 ± 0.3 in the liver, 1.7 ± 0.5/1.7 ± 0.4 in the mediastinum, 1.8 ± 0.3/5.6 ± 1.0 in the spleen, 1.3 ± 0.5/1.6 ± 0.8 in the bone marrow and 7.7 ± 2.0/0.2 ± 0.1 in the brain (supplementary Table 1). As we suspected, the results from supplementary Table 1 show that FDG and CXCR4 have different biodistributions. We have recognized the SUV on Pentixafor as feasible; however, the SUV must be treated with caution as it is not validated or standardized yet.
Visual analysis
A total of 26 lesions were counted. Patient-based analysis FDG and Pentixafor revealed comparable results in 4/10 patients. Lesion-based analysis showed equal results in 14 lesions which were positive for both tracers while 5 lesions were FDG positive and Pentixafor negative and 7 lesions were FDG negative but Pentixafor positive (Table 2).
Quantitative analysis
Mean intensity of uptake in the lesion was higher, but not statistically significant, for FDG, which is reflected by the mean SUVmax in the lesions 6.9 for FDG and 4.2 for Pentixafor (p = 0.075). For mean SUVmean in the reference regions for FDG and Pentixafor were 2.5 and 1.4 in the liver (p < 0.001), 1.7 and 1.7 in the mediastinum (p = 0.635), 1.8 and 5.6 in the spleen (p < 0.001), 1.3 and 1.6 in the bone marrow (p = 0.4316), and 6.1 and 0.2 in the brain (p = 0.0039), reflecting the higher physiological uptake of FDG in the brain and the liver, and the higher affinity of Pentixafor to the spleen. When choosing the liver as reference region the mean ratio of the SUVmax in the lesion to the SUVmean within the liver was 3.0 for FDG and 3.8 for Pentixafor (p = 0.25) (Table 3).
Table 3.
Tumor to background ratio (TBR) liver/mediastinum for FDG and Pentixafor
| Patient no. | Localization of the lesion | FDG | Pentixafor | ||
|---|---|---|---|---|---|
| TBR liver | TBR MBP | TBR liver | TBR MBP | ||
| #1 | paratracheal left | 1,0 | 1,3 | 2,6 | 3,5 |
| #2 | mediastinal | 5,1 | 6,7 | 2,9 | 2,1 |
| infraclavicular | 1,7 | 2,3 | 4,1 | 3,0 | |
| #3 | hip | 2,2 | 3,4 | 2,5 | 1,1 |
| thigh | 3,2 | 5,0 | 3,5 | 1,6 | |
| adrenal gland left | 1,4 | 2,1 | 2,6 | 1,2 | |
| left paraaortal lymph node | 1,0 | 1,6 | 1,2 | 0,5 | |
| #4 | esophagus | 2,5 | 3,7 | 4,9 | 3,7 |
| coeliacal | 3,1 | 4,5 | 4,3 | 3,25 | |
| axillary right | 0,3 | 0,4 | 1,4 | 1,1 | |
| hilary right | 0,7 | 1 | 2,9 | 2,2 | |
| gastric curvature | 1,9 | 2,8 | 3,4 | 2,6 | |
| #5 | esophagus | 2,8 | 4,1 | 4,1 | 3,4 |
| #6 | mediastinal | 5,1 | 6,3 | 3,0 | 2,5 |
| #7 | mediastinal | 6,6 | 22,6 | 4,0 | 5,3 |
| #8 | adrenal gland left | 0,9 | 1,4 | 4,2 | 2,9 |
| esophagus | 1,2 | 1,8 | 4,2 | 2,9 | |
| lung upper lobe | 1,8 | 2,7 | 1,3 | 0,9 | |
| #9 | adrenal gland left | 1,2 | 1,5 | 12,0 | 6,0 |
| #10 | infracarinal | 1,2 | 1,9 | 5,4 | 4,8 |
| mediastinal | 1,8 | 2,9 | 5,8 | 5,2 | |
| supra right | 1,0 | 1,6 | 3,2 | 2,8 | |
| supra left | 1,2 | 2,0 | 3,9 | 3,5 | |
| esophagus | 6,3 | 10,3 | 2,2 | 1,9 | |
| rib left | 4,0 | 6,6 | 2,2 | 2,0 | |
| cervical left | 6,0 | 9,8 | 2,4 | 2,2 | |
MPB Mediastinal blood pool
Immunohistochemistry
We were able to make semi-quantitative statements about the quantity and number of CXCR4-positive inflammatory cells in the tissue. In 7/10 patients imaging results could be compared to immunohistological staining for CXCR4 derived from surgical specimens (n = 3) as well as biopsies from the primary (n = 4) (Table 1). Regarding the histological evaluation of CXCR4 expression, 2/10 samples (#1, #3) were rated “low” (4–10 CXCR4 positive cells/HPF), 2/10 “intermediate” (11–25 CXCR4 positive cells/HPF) (#6, #9) and 2/10 “strong” (> 26 CXCR4 positive cells/HPF) (#4, #7) positive. Patient #5 was scored negative (0–3 CXCR4 positive cells/HPF) and 3/10 samples (patients #2, #8 and #10) were not scored at all (compare Fig. 4).
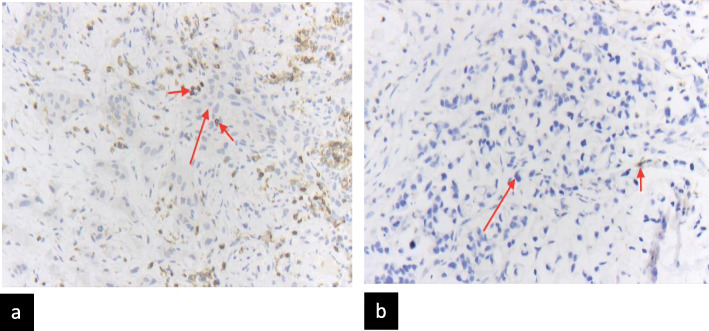
Fig. 4.
Immunohistochemistry: CXCR4 (both photos in same magnification: 200x) coloring in (a) squamous cell carcinoma: (long arrow) is strong positive for CXCR4 (CXCR4 positive inflammatory cells marked with short arrow); and (b) adenocarcinoma (with signet ring cell features; long arrow) scored as negative (less than three CXCR4 positive cells in high power field/HPF; one CXCR4 positive cell marked with short arrow)
Discussion
Our observation of in vivo imaging of CXCR4 expression in humans with both newly diagnosed as well as pre-treated, recurrent esophageal cancer suggest that chemokine receptor 4 (CXCR4) expression in esophageal cancer is not an unusual condition and can be assessed non-invasively by PET/CT and the CXCR4-directed radiopharmaceutical [68Ga]Pentixafor. However, it should be highlighted that CXCR4 expression is not specific for esophageal cancer.
Nevertheless, for almost all tumor lesions we could not find any significant exceptions for high tumor background ratio compared to FDG PET/CT as a reference. This underlines that CXCR4 may prove to be a promising target for endoradiotherapy in selected cases, as recently proposed e.g. by Lapa et al. [23–25]. Recent reports evaluating biopsy samples of esophageal cancer tumors demonstrated a high intensity of CXCR4 receptor expression [26, 27]. Additionally, chemokine receptor expression was a predictor of poor recurrence-free and overall survival [28–31].
The preliminary results of this retrospective analysis indicate a heterogeneity of CXCR4 expression and imply that the information may be considered complementary to Computerized Tomography (CT), endoscopic ultrasound (EUS) and FDG PET/CT within three-modality staging [32–34]. In case of overexpression of CXCR4 in esophageal tumours or their metastases, an additional Pentixafor PET/CT could detect further localisations not visible in FDG PET/CT. As an example, Philipp-Abbrederis et al. demonstrated the benefits of Pentixafor PET imaging in a subset of multiple myeloma patients where specificity and contrast were superior to [18F]FDG [35]. Particularly in view of the low to moderate sensitivity of FDG-PET/CT for lymphonodal staging and the distinction between active tumor tissue and local esophagitis [36].
As Werner et al. noted, future attempts for possible applications of CXCR4-directed imaging could focus on the characterization of lesional heterogeneity by conducting dual radiotracer studies (in coexistence with [18F]FDG) to visualize different levels of tumor differentiation and predict metastases with prognostic relevance. For example, a CXCR4-directed PET/CT could help to visualize receptor-positive cancer stem cells that are considered to be in particular resistant to radiation or chemotherapy [37].
Detailed information of the exact tumor stage is essential for decision making in oncological treatment strategies. Since the treatment of esophageal cancer is a stadium-adapted therapy, accurate and correct staging is of particular importance. In the case of neoadjuvant radiotherapy, the size of the RT field varies depending on the lymph node involvement. Pentixafor PET/CT could have therapeutic impact, e.g. on the size of the field to be irradiated in patients with esophageal cancer. In addition, and of course, the detection of distant metastases would lead to a change in the therapeutic goal towards palliative treatment. Considering the small number of our cases, it appears that a high CXCR4 expression, e.g. samples #4 and #7, is accompanied by a higher T-stage of disease. However, it must be taken into account - which has already been critically demonstrated by Lapa et al. - that receptor presentation on the tumor cell surface appears to be highly dynamic and is influenced by a variety of factors, including previous therapy [38].
In the majority of studies, overexpression of CXCR4 had been investigated in esophageal tumors or its metastases. Łukaszewicz-Zając et al. examined the serum concentrations (SC) of chemokine CXCL12 in patients with esophageal cancer compared to a healthy control group. CXCL12 SC were significantly higher, as those of its receptor CXCR4. This suggests the possibility that CXCR4 could become a prognostic factor in a combined analysis with classical tumor markers such as carcinoembryonic antigen and C-reactive protein levels [39].
In addition, Koishi et al. were able to show that persistence of positive CXCR4 expression is implicated in tumor aggressiveness and poor prognosis in esophageal (squamous cell) cancer (ESCC) after neoadjuvant chemoradiotherapy (nCRT) [40]. Furthermore they showed that nCRT may improve the prognosis of ESCC via CXCL12-CXCR4 signaling pathway [41]. Sasaki et al. demonstrated that positive CXCL12 expression was closely related to tumor development [42]. We can therefore expect that patients, including those with advanced diseases, can be monitored by CXCL12 and thus the effectiveness of multimodal therapies can be observed. At this time, it remains unclear whether chemoradiotherapy could affect CXCL12-CXCR4 signaling or not, since the status of CXCR4 expression after chemoradiotherapy was not available in our cohort.
Nevertheless, our data suggests that a noticeable CXCR4-positive immune or epithelial cell population might accumulate in tumors and we have been able to demonstrate a major fraction of tumor cells to be CXCR4-positive. Our samples #3, #5 and #6 – all examined as adenocarcinoma, all got treated with radiochemotherapy protocol according to CROSS trial, CXCR4 expression was determined in the surgical specimen – ranged from none to intermediate expression, as well as the squamous cell cancer (SCC), which ranged up to strong expression (#4, #7) fixed in OGD sampling [40]. SCC sample #9 received CROSS protocol before determination in surgical specimen, too and was rated intermediate. Of course, it must be critically considered that CXCR4 expression can be up- or underregulated by chemo- and radiotherapy and should therefore be considered with caution [43–45].
Still, there are differences in regard of in vitro and in vivo distribution, when it comes to CXCR4 overexpression. Vag et al. observed that the reported in vitro evidence of CXCR4 overexpression in malignancies such as pancreatic cancer, non-small cell lung cancer, prostate cancer, melanoma, breast cancer, hepatocellular carcinoma, glioblastoma, and sarcoma does not depict the in vivo distribution revealed by Pentixafor PET/CT [46]. These results could potentially differ because 68Ga-Pentixafor PET binds to membrane-associated chemokine receptors. CXCR4 expression levels determined by either transcript or whole-cell protein level analysis is not necessarily representative of the CXCR4 expression level on the cell surface [47, 48]. Therefore, there could be a significant discrepancy between CXCR4 expression profiles determined by analysis of transcript or whole-cell protein protein level analysis and by in vivo quantification of CXCR4 using PET probes. Shim et al. demonstrated that CXCR4 expression in lymph node metastases in breast cancer originates mainly in the cytoplasm. Other relevant factors in this context could be the overexpression of CXCR4 on cancer stem cells, which are believed to represent a drug-resistant cell population.
Even after definitive treatment, esophageal cancer features a high risk of early recurrence after definitive therapy [ 49, 50]. Several series have documented that most recurrences occur in the first 2 years after completion of treatment [51, 52]. Tabouret et al. demonstrated a switch in patients with Glioblastoma multiforme from VEGF pathway to CXCL12 /CXCR4 pathway [53–55]. Potentially, this mechanism could apply for esophageal cancer with early recurrence [56].
Taking again into account the small number of cases in this retrospective analysis, histopathological status seems to be no predictor for CXCR4 expression, in line with results of Kaifi et al. and Gockel et al. [14, 57] It is difficult to state beyond doubt that Pentixafor PET/CT results correlate with immunohistochemistry at n = 10. However, in patients #4 and #9, the IHC measured CXCR4 expression (strong respectively intermediate) was consistent with a coelic lymph node conglomeration (= metastasis) either an adrenal metastasis on the left side. Patient #4 during initial staging; patient #9 for re-staging after radiochemotherapy, which is why the results should at least be considered an exciting trend. For comparative assessment of FDG and Pentixafor uptake, the mediastinum may represent a suitable reference region. High tumor-uptake and low CXCR4 expression in non-tumor regions indicate promising preconditions for a CXCR4 specific radionuclide therapy.
Despite promising results, this pilot research work comes with limitations. First, only a limited number of patients could be included in the investigation. Secondly, both immunohistochemistry was not available in all cases and histological sampling was performed in three patients after nCRT. Third, it is a retrospective analysis, with inherent bias, although our institutional database is managed prospectively with strict tracking of all patients. As a consequence, a controlled study is now needed to shed more light on the potential diagnostic and therapeutic benefits of Pentixafor PET/CT in esophageal cancer.
Conclusion
Our data shows that additional PET/CT imaging with Pentixafor for imaging the CXCR4 chemokine receptor is feasible but heterogeneous in both newly diagnosed and pretreated recurrent esophageal cancer. Therefore, preliminary results of this retrospective analysis imply that information should be considered complementary to CT, EUS and FDG-PET within three-modality (re-)staging. Of note, CXCR4 could provide an additional marker for metastatic tendency in adenocarcinoma or squamous cell carcinoma. In addition, the Pentixafor PET/CT may offer a diagnostic tool for radiation field expansion in radiooncology and thus possibly additional clinical benefit with regard to the oncological outcome.
Supplementary Information
Additional file 1: Supplementary Table 1. SUVmean in background for FDG and Pentixafor.
Acknowledgements
Not applicable.
Abbreviations
- Adeno
Adenocarcinoma
- CXCR4
Chemokine (C-X-C motif) Receptor 4
- CT
Computerized Tomography
- EARL
European Association of Nuclear Medicine and its Research Ltd.
- ESCC
Esophageal (Squamous Cell) Cancer
- EUS
Endoscopic Ultrasound
- FDG
Fluorodeoxyglucose
- IHC
Immunohistochemistry
- MBP
Mediastinal Blood Pool
- nCRT
Neoadjuvant Chemoradiotherapy
- OGD
Oesophagogastroduodenoscopy
- OSEM
Ordered Subset Expectation Maximization
- PET/CT
Positron Emission Tomography/Computer Tomography
- SC
Serum Concentrations
- SCC
Squamous Cell Cancer
- SUV
Standardized Uptake Value
- UICC TNM
The Union for International Cancer Control Tumor Node Metastasis Classification of Malignant Tumours
Authors’ contributions
All authors have read and approved the final manuscript. CK, CB and PL designed the present analysis. CK and PL conducted the statistical analyses of the data. AQ performed the histopathological examinations. All authors contributed to the management, analysis and interpretation of the data.
Funding
There was no funding obtained for this retrospective analysis.
Availability of data and materials
The datasets supporting the conclusions of this article are included within the article (and its additional files).
Ethics approval and consent to participate
A corresponding waiver declaration from the responsible ethics committee is available. All clinical investigations were conducted in accordance with the principles of the Declaration of Helsinki. All procedures were administered in accordance with the regulations of the competent local authorities (District Government of Cologne, Germany) and in accordance with the requirements of the local Institutional Review Board. All patients gave written informed consent to PET imaging with the sole purpose of diagnosis and inclusion of their data in retrospective scientific analysis.
Consent for publication
All patients signed informed consent in regard of the scientific evaluation of their data.
Competing interests
HJW is founder and shareholder of Scintomics. The authors declare that they do not have any competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Various: Krebs in Deutschland für 2015/2016. Gemeinsame Publikation des Zentrums für Krebsregisterdaten und der Gesellschaft der epidemiologischen Krebsregister in Deutschland e.V, vol. 12. Berlin: Robert Koch-Institut Berlin; 2019.
- 3.Zhang Y. Epidemiology of esophageal cancer. World J Gastroenterol. 2013;19(34):5598–5606. doi: 10.3748/wjg.v19.i34.5598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van Vliet EP, Heijenbrok-Kal MH, Hunink MG, et al. Staging investigations for oesophageal cancer: a meta-analysis. Br J Cancer. 2008;98(3):547–557. doi: 10.1038/sj.bjc.6604200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weber WA. Use of PET for monitoring cancer therapy and for predicting outcome. J Nucl Med. 2005;46(6):983–995. [PubMed] [Google Scholar]
- 6.Domanska UM, Kruizinga RC, Nagengast WB, et al. A review on CXCR4/CXCL12 axis in oncology: no place to hide. Eur J Cancer. 2013;49:219–230. doi: 10.1016/j.ejca.2012.05.005. [DOI] [PubMed] [Google Scholar]
- 7.Burger JA, Kipps TJ. CXCR4: a key receptor in the crosstalk between tumor cells and their microenvironment. Blood. 2006;107:1761–1767. doi: 10.1182/blood-2005-08-3182. [DOI] [PubMed] [Google Scholar]
- 8.Burger JA, Peled A. CXCR4 antagonists: targeting the microenvironment in leukemia and other cancers. Leukemia. 2009;23:43–52. doi: 10.1038/leu.2008.299. [DOI] [PubMed] [Google Scholar]
- 9.Muller A, Homey B, Soto H, et al. Involvement of chemokine receptors in breast cancer metastasis. Nature. 2001;410:50–56. doi: 10.1038/35065016. [DOI] [PubMed] [Google Scholar]
- 10.Zhang B, Wu T, Wang Z, et al. p38MAPK activation mediates tumor necrosis factor-alpha-induced apoptosis in glioma cells. Mol Med Rep. 2015;11:3101–3107. doi: 10.3892/mmr.2014.3002. [DOI] [PubMed] [Google Scholar]
- 11.Busillo JM, Benovic JL. Regulation of CXCR4 signaling. Biochim Biophys Acta. 2007;1768(4):952–963. doi: 10.1016/j.bbamem.2006.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dewan MZ, Ahmed S, Iwasaki Y, et al. Stromal cell-derived factor-1 and CXCR4 receptor interaction in tumor growth and metastasis of breast cancer. Biomed Pharmacother. 2006;60(6):273–276. doi: 10.1016/j.biopha.2006.06.004. [DOI] [PubMed] [Google Scholar]
- 13.Wu J, Wu X, Liang W, et al. Clinicopathological and prognostic significance of chemokine receptor CXCR4 overexpression in patients with esophageal cancer: a meta-analysis. Tumor Biol. 2014;353:3709–3715. doi: 10.1007/s13277-013-1490-8. [DOI] [PubMed] [Google Scholar]
- 14.Kaifi JT, Yekebas EF, Schurr P, et al. Tumor-cell homing to lymph nodes and bone marrow and CXCR4 expression in esophageal cancer. J Natl Cancer Inst. 2005;97(24):1840–1847. doi: 10.1093/jnci/dji431. [DOI] [PubMed] [Google Scholar]
- 15.Wang X, Cao Y, Zhang S, et al. Stem cell autocrine CXCL12/CXCR4 stimulates invasion and metastasis of esophageal cancer. Oncotarget. 2017;8(22):36149–36160. doi: 10.18632/oncotarget.15254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang M, Zhang L, Cui M, et al. miR-302b inhibits cancer-related inflammation by targeting ERBB4, IRF2 and CXCR4 in esophageal cancer. Oncotarget. 2017;8(30):49053–49063. doi: 10.18632/oncotarget.17041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Demmer O, Gourni E, Schumacher U, et al. PET imaging of CXCR4 receptors in cancer by a new optimized ligand. ChemMedChem. 2011;6:1789–1791. doi: 10.1002/cmdc.201100320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wester HJ, Keller U, Schottelius M, et al. Disclosing the CXCR4 expression in lymphoproliferative diseases by targeted molecular imaging. Theranostics. 2015;5:618–630. doi: 10.7150/thno.11251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gourni E, Demmer O, Schottelius M, et al. PET of CXCR4 expression by a (68)Ga-labeled highly specific targeted contrast agent. J Nucl Med. 2011;52(11):1803–1810. doi: 10.2967/jnumed.111.098798. [DOI] [PubMed] [Google Scholar]
- 20.Herrmann K, Schottelius M, Lapa C, et al. First-in-human experience of CXCR4-directed Endoradiotherapy with 177Lu- and 90Y-labeled Pentixather in advanced-stage multiple myeloma with extensive intra- and Extramedullary disease. J Nucl Med. 2016;57(2):248–251. doi: 10.2967/jnumed.115.167361. [DOI] [PubMed] [Google Scholar]
- 21.Boellaard R, Delgado-Bolton R, Oyen WJ, et al. FDG PET/CT: EANM procedure guidelines for tumour imaging: version 2.0. Eur J Nucl Med Mol Imaging. 2015;42(2):328–354. doi: 10.1007/s00259-014-2961-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barrington SF, Mikhaeel NG, Kostakoglu L, et al. Role of imaging in the staging and response assessment of lymphoma: consensus of the international conference on malignant lymphomas imaging working group. J Clin Oncol. 2014;32:3048–3058. doi: 10.1200/JCO.2013.53.5229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lapa C, Lückerath K, Rudelius M, et al. [68Ga]Pentixafor-PET/CT for imaging of chemokine receptor 4 expression in small cell lung cancer—initial experience. Oncotarget. 2016;7(8):9288–9295. doi: 10.18632/oncotarget.7063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lapa C, Herrmann K, Schirbel A, et al. CXCR4-directed endoradiotherapy induces high response rates in extramedullary relapsed multiple myeloma. Theranostics. 2017;7:1589–1597. doi: 10.7150/thno.19050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lapa C, Hanscheid H, Kircher M, et al. Feasibility of CXCR4-directed radioligand therapy in advanced diffuse large B cell lymphoma. J Nucl Med. 2018;60:60–64. doi: 10.2967/jnumed.118.210997. [DOI] [PubMed] [Google Scholar]
- 26.Fang HY, Münch NS, Quante M, et al. CXCR4 is a potential target for diagnostic PET/CT imaging in Barrett's dysplasia and esophageal adenocarcinoma. Clin Cancer Res. 2018;24(5):1048–1061. doi: 10.1158/1078-0432.CCR-17-1756. [DOI] [PubMed] [Google Scholar]
- 27.Goto M, Liu M. Chemokines and their receptors as biomarkers in esophageal cancer. Esophagus. 2019. 10.1007/s10388-019-00706-8 [Epub ahead of print]. [DOI] [PubMed]
- 28.Jiang Q, Sun Y, Liu X. CXCR4 as a prognostic biomarker in gastrointestinal cancer: a meta-analysis. Biomarkers. 2019;24(6):510–516. doi: 10.1080/1354750X.2019.1637941. [DOI] [PubMed] [Google Scholar]
- 29.Uchi Y, Takeuchi H, Matsuda S, et al. CXCL12 expression promotes esophageal squamous cell carcinoma proliferation and worsens the prognosis. BMC Cancer. 2016;16:514. doi: 10.1186/s12885-016-2555-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goto M, Yoshida T, Yamamoto Y, et al. CXCR4 expression is associated with poor prognosis in patients with esophageal squamous cell carcinoma. Ann Surg Oncol. 2017;24(3):832–840. doi: 10.1245/s10434-015-4974-5. [DOI] [PubMed] [Google Scholar]
- 31.Schimanski CC, Galle PR, Moehler M. Chemokine receptor CXCR4-prognostic factor for gastrointestinal tumors. World J Gastroenterol. 2008;14(30):4721–4724. doi: 10.3748/wjg.14.4721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schreurs LMA, Janssens ACJW, Groen H, et al. Value of EUS in determining curative resectability in reference to CT and FDG-PET: the optimal sequence in preoperative staging of esophageal cancer? Ann Surg Oncol. 2016;23:1021–1028. doi: 10.1245/s10434-011-1738-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lordick F, Mariette C, Haustermans K, et al. Oesophageal cancer: ESMO clinical practice guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2016;27(suppl 5):v50–v57. doi: 10.1093/annonc/mdw329. [DOI] [PubMed] [Google Scholar]
- 34.Mantziari S, Pomoni A, Prior JO, et al. 18F- FDG PET/CT-derived parameters predict clinical stage and prognosis of esophageal cancer. BMC Med Imaging. 2020;20:7. doi: 10.1186/s12880-019-0401-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Philipp-Abbrederis K, Herrmann K, Knop S, et al. In vivo molecular imaging of chemokine receptor CXCR4 expression in patients with advanced multiple myeloma. EMBO Mol Med. 2015;7(4):477–487. doi: 10.15252/emmm.201404698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kratochwil C, Flechsig P, Giesel FL, et al. 68Ga-FAPI PET/CT: tracer uptake in 28 different kinds of Cancer. J Nucl Med. 2019;60(6):801–805. doi: 10.2967/jnumed.119.227967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Werner RA, Kircher S, Higuchi T, et al. CXCR4-directed imaging in solid tumors. Front Oncol. 2019;9:770. doi: 10.3389/fonc.2019.00770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lapa C, Luckerath K, Kircher S, et al. Potential influence of concomitant chemotherapy on CXCR4 expression in receptor directed endoradiotherapy. Br J Haematol. 2019;184:440–443. doi: 10.1111/bjh.15096. [DOI] [PubMed] [Google Scholar]
- 39.Łukaszewicz-Zając M, Mroczko B, Kozłowski M, et al. The serum concentrations of chemokine CXCL12 and its specific receptor CXCR4 in patients with esophageal Cancer. Dis Markers. 2016;2016:7963895. doi: 10.1155/2016/7963895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van Heijl M, van Lanschot JJ, Koppert LB, et al. Neoadjuvant chemoradiation followed by surgery versus surgery alone for patients with adenocarcinoma or squamous cell carcinoma of the esophagus (CROSS) BMC Surg. 2008;8:21. doi: 10.1186/1471-2482-8-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Koishi K, Yoshikawa R, Tsujimura T, et al. Persistent CXCR4 expression after preoperative chemoradiotherapy predicts early recurrence and poor prognosis in esophageal cancer. World J Gastroenterol. 2006;12(47):7585–7590. doi: 10.3748/wjg.v12.i47.7585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sasaki K, Natsugoe S, Ishigami S, et al. Expression of CXCL12 and its receptor CXCR4 in esophageal squamous cell carcinoma. Oncol Rep. 2009;21(1):65–71. [PubMed] [Google Scholar]
- 43.Li J, Jiang K, Qiu X, et al. Overexpression of CXCR4 is significantly associated with cisplatin-based chemotherapy resistance and can be a prognostic factor in epithelial ovarian cancer. BMB Rep. 2014;47(1):33–38. doi: 10.5483/BMBRep.2014.47.1.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Willers H, Azzoli CG, Santivasi WL, et al. Basic mechanisms of therapeutic resistance to radiation and chemotherapy in lung cancer. Cancer J. 2013;19:200–207. doi: 10.1097/PPO.0b013e318292e4e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sison EA, McIntyre E, Magoon D, et al. Dynamic chemotherapy-induced upregulation of CXCR4 expression: a mechanism of therapeutic resistance in pediatric AML. Mol Cancer Res. 2013;11(9):1004–1016. doi: 10.1158/1541-7786.MCR-13-0114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vag T, Gerngross C, Herhaus P, et al. First experience with chemokine receptor CXCR4-targeted PET imaging of patients with solid cancers. J Nucl Med. 2016;57(5):741–746. doi: 10.2967/jnumed.115.161034. [DOI] [PubMed] [Google Scholar]
- 47.Teicher BA, Fricker SP. CXCL12 (SDF-1)/CXCR4 pathway in cancer. Clin Cancer Res. 2010;16(11):2927–2931. doi: 10.1158/1078-0432.CCR-09-2329. [DOI] [PubMed] [Google Scholar]
- 48.Shim H, Lau SK, Devi S, et al. Lower expression of CXCR4 in lymph node metastases than in primary breast cancers: potential regulation by ligand-dependent degradation and HIF-1alpha. Biochem Biophys Res Commun. 2006;346(1):252–258. doi: 10.1016/j.bbrc.2006.05.110. [DOI] [PubMed] [Google Scholar]
- 49.Lou F, Sima CS, Adusumilli PS, et al. Esophageal cancer recurrence patterns and implications for surveillance. J Thorac Oncol. 2013;8(12):1558–1562. doi: 10.1097/01.JTO.0000437420.38972.fb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhou ZG, Zhen CJ, Bai WW, et al. Salvage radiotherapy in patients with local recurrent esophageal cancer after radical radiochemotherapy. Radiat Oncol. 2015;10:54. doi: 10.1186/s13014-015-0358-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mariette C, Balon JM, Piessen G, et al. Pattern of recurrence following complete resection of esophageal carcinoma and factors predictive of recurrence disease. Cancer. 2003;97:1616–1623. doi: 10.1002/cncr.11228. [DOI] [PubMed] [Google Scholar]
- 52.Abate E, DeMeester ST, Zehetner J, et al. Recurrence after esophagectomy for adenocarcinoma: defining optimal follow-up intervals and testing. J Am Coll Surg. 2010;210:428–435. doi: 10.1016/j.jamcollsurg.2010.01.006. [DOI] [PubMed] [Google Scholar]
- 53.Sneed PK, Gutin PH, Larson DA, et al. Patterns of recurrence of glioblastoma multiforme after external irradiation followed by implant boost. Int J Radiat Oncol Biol Phys. 1994;29:719–727. doi: 10.1016/0360-3016(94)90559-2. [DOI] [PubMed] [Google Scholar]
- 54.Tabouret E, Tchoghandjian A, Denicolai E, et al. Recurrence of glioblastoma after radio-chemotherapy is associated with an angiogenic switch to the CXCL12-CXCR4 pathway. Oncotarget. 2015;6(13):11664–11675. doi: 10.18632/oncotarget.3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Plate KH, Breier G, Weich HA, et al. Vascular endothelial growth factor is a potential tumour angiogenesis factor in human gliomas in vivo. Nature. 1992;359(6398):845–848. doi: 10.1038/359845a0. [DOI] [PubMed] [Google Scholar]
- 56.Zhou W, Guo S, Liu M, et al. Targeting CXCL12/CXCR4 Axis in tumor immunotherapy. Curr Med Chem. 2019;26(17):3026–3041. doi: 10.2174/0929867324666170830111531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gockel I, Schimanski CC, Heinrich C, et al. Expression of chemokine receptor CXCR4 in esophageal squamous cell and adenocarcinoma. BMC Cancer. 2006;6:290. doi: 10.1186/1471-2407-6-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Supplementary Table 1. SUVmean in background for FDG and Pentixafor.
Data Availability Statement
The datasets supporting the conclusions of this article are included within the article (and its additional files).