Abstract
Background
Genetic association of uncoupling proteins (UCPs) variants with the susceptibility of diabetic retinopathy (DR) in diabetes mellitus (DM) patients has been reported but with controversy. Here we aimed to conduct a meta-analysis to confirm the association of different UCPs variants with DR.
Methods
Three databases (Medline Ovid, Embase Ovid and CENTRAL) were applied in the literature search. Five genetic models, including allelic, homozygous, heterozygous, dominant and recessive models, were evaluated. Odds ratios (OR) were estimated under the random or fixed-effects models. Subgroup analyses, publication bias and sensitivity analyses were also conducted.
Results
Eleven studies on 2 UCPs variants (UCP1 rs1800592 and UCP2 rs659366) were included. Our meta-analysis showed that UCP1 rs1800592 was not associated with DR in type-2 DM patients, and UCP2 rs659366 also showed no association with DR. In the subgroup analyses on the stage of DR, allele G of UCP1 rs1800592 significantly increased the susceptibility of proliferative diabetic retinopathy (PDR) in type-2 DM patients in the allelic (OR = 1.26, P = 0.03) and homozygous models (OR = 1.60, P = 0.04). Subgroup analysis on ethnicity did not found any significant association of rs1800592 and rs659366 with DR.
Conclusion
Our meta-analysis confirmed the association of UCP1 rs1800592 variant with PDR in patients with type-2 DM, suggesting its potential as a genetic marker for PDR prediction in population screening.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12886-021-01838-1.
Keywords: Diabetic retinopathy, Uncoupling proteins, Variants, Meta-analysis
Background
Diabetic retinopathy (DR), a common sight-threatening microvascular complication among patients with diabetes mellitus (DM), is the major cause of irreversible blindness and visual impairment in working-age adults [1]. Though the pathophysiological mechanisms of DR remain elusive, increasing evidence suggests that long duration of DM, poor control of blood glucose and high blood pressure mainly contribute to the pathogenesis and development of DR [2]. However, DR could also occur in patients with short duration of DM, well control of blood glucose and normal blood pressure. Besides, epidemiological studies revealed familial inheritance and ethnic variations in DR [3], indicating that genetic factors could play a role in the pathogenesis and development of DR [4].
The elevation of oxidative stress has been suggested contributing to the development of DM complications [5], which is caused by reactive oxygen species (ROS) overproduction, mainly the mitochondrial ROS [5–7]. Excessive ROS resulted from hyperglycemia causes retinal mitochondrial dysfunction with serious damage to the oxidative phosphorylation complexes and abolished adenosine triphosphate biosynthesis [8], and induces capillary endothelial cell apoptosis, which subsequently leads to the dysregulation of the angiogenesis-related genes [9] and diabetic microvascular complications, including DR [10, 11]. Ion channels physiologically play a role in signal transmission and visual processing, but also link to induced oxidative stress and significantly contribute to a wide spectrum of ocular diseases [12]. Uncoupling proteins (UCPs) belong to a group of proton carrier transporters (H+) in the inner membrane of mitochondria [13]. UCPs are able to uncouple the oxidized substrates and dissipate the potential energy on the inner membrane as heat to reduce ROS overproduction from mitochondria [14–17]. The overproduced ROS could cause increases proton conductance by UCP1–3, leading to decease in superoxide radicals through the mitochondria respiratory chain reaction [18]. In human genome, there were five different UCPs, named UCP1 to 5, with various tissue distributions and functions [19]. Uncoupling protein 1 (UCP1) gene is located on chromosome 4q31.1 and found to be expressed in brown adipose tissue, endothelial cells and pericytes of retina [20]. UCP1 mainly plays a role in the maintenance of body temperature in a cold environment through non-shivering thermogenesis [17]. It has been shown that elevated of glucose levels upregulates UCP1 expression, protecting cells from glucose-induced ROS damage [21]. Uncoupling protein 2 (UCP2) and 3 (UCP3) genes are both located in the same cluster on chromosome 11q13.4. UCP2 is ubiquitously expressed across different tissues in the body, whereas UCP3 is mainly expressed in the skeletal muscle tissue [22]. In UCP2 knockout mice, ROS production increases in macrophages and pancreatic islets [23, 24], whereas overexpression of UCP2 inhibits mitochondrial death pathway in cardiomyocytes [25], indicating that UCP2 could be involved in cell protection from ROS damage. UCP2 and UCP3, together with SLC25A27 (UCP4) and BMCP1 (UCP5), exert cytoprotective effects by reducing oxidative stress under certain conditions [22].
Since UCPs are involved in the pathophysiology of glucose-related ROS cell damage, it is reasonable to hypothesize that the UCPs variants could be related to the susceptibility of DR. Yet, inconsistent results have been reported on the association analysis of UCPs variants with the risk of DR [26–28]. Herein, we aimed to conduct a meta-analysis to clarify the association of different UCPs variants with the susceptibility of DR.
Methods
Study design
The protocol of this meta-analysis has been registered in the international prospective register of systematic reviews (PROSPERO protocol CRD42020173510; available at https://www.crd.york.ac.uk/prospero/).
Searching strategies and selection criteria
Three databases, including Medline Ovid, Embase Ovid and CENTRAL, were applied in the literature search for the potential studies. The eligible studies related to the susceptibility of DR and UCPs variants would be included in this meta-analysis. The following terms were used in this search: “diabetic retinopathy”, “uncoupling protein”, and “polymorphisms OR variants”. The detail search strategies and results were shown in Supplementary document.
Literature language was not limited to English. For languages other than Chinese and English, Google Translate (http://translate.google.com/) was used to translate the full text. The bibliographies of the screened articles have been carefully browsed to identify the omitted relevant studies.
The inclusion criteria included: 1) studies on the analysis of the association of UCPs variants with DR; 2) the recruited participants were independent and unrelated to each other; 3) sufficient genotype data for the calculation of odd ratio (OR) with 95% confidence interval (C.I.); 4) participants diagnosed with diabetes without retinopathy (DWR) would be served as the control subjects for the Hardy-Weinberg equilibrium (HWE) analysis or the data provided should be able to calculate HWE of the control group; and 5) the type of diabetes in the participants was clearly provided, including type-1 and type-2 DM. The exclusion criteria included: 1) the genotype distributions of the control subjects did not follow HWE (PHWE < 0.05); and 2) the variants reported only by one study would not be included in this analysis.
Data extraction
Two researchers (X.L. and Z.J.) independently extracted and assessed the full-text reports for all potentially eligible studies. The included studies were evaluated by the Newcastle-Ottawa quality assessment scale (NOS). The extracted items include: first author, year of publication, region of study, ethnicity, number of cases and controls, diagnostic criteria, allele or genotype frequency, Hardy-Weinberg equilibrium (HWE) status, and genotyping method. If there was any disagreement regarding to the eligibility, scores of NOS and extracted items, the judicator (Z.W.) would make the final decision. If any full-text reports have been rejected, the reasons for the rejection would be given. The data extraction form included the following data: 1) the first author and the year of publication; 2) the country and the ethnicity of the studied subjects; 3) the methodology of genotyping; 4) the methodology of DR diagnosis; 5) P-value of HWE in the control group; 6) The genotypic count of each variant in the patient and control groups.
Non-proliferative diabetic retinopathy (NPDR) and proliferative diabetic retinopathy (PDR) are regarded as different stages of DR, which shows different pathology and pathophysiology. This meta-analysis evaluated three types of case groups: (1) DR, (2) only PDR, and (3) combined NPDR, PDR and DR.
Statistical analysis
A publicly available program (https://ihg.gsf.de/cgi-bin/hw/hwa1.pl) was used to estimate the HWE of the included studies. HWE in the control subjects was evaluated by χ2 test, and P < 0.05 was considered as deviation from HWE.
The association of UCPs variants with DR was evaluated by five genetic models, including the allelic (reference allele versus variant allele), homozygous (homozygous reference genotype versus homozygous variant genotype), heterozygous (homozygous reference genotype versus heterozygous genotype), dominant (homozygous reference genotype versus homozygous variant and heterozygous genotypes), and recessive models (homozygous reference and heterozygous genotypes versus homozygous variant genotype). Subgroup analyses were also conducted based on the stage of DR and the ethnicity.
Heterogeneity was examined by the Q statistic (significance defined as P < 0.1) and the I2 statistic (significant inconsistency defined as I2 > 50%) [29]. If heterogeneity test showed significance (P < 0.1 or I2 > 50%), the random-effect model was selected to measure the pooled effect value (DerSimonian and Laird method) [30]; otherwise, the fixed-effect model was applied (Mantel-Haenszel method) [31]. The pooled odds ratio (OR) with 95% confidence intervals (C.I.) was calculated to measure the strength of association between the UCPs variants and DR, which was assessed by the Z test (significance defined as P < 0.05). Sensitivity analysis was used to measure the stability of the results by excluding one study at a time when there were more than two studies. Egger’s test was used to quantitatively evaluate the potential publication bias. All statistical analysis was calculated by the STATA software (version 14.0; STATA Corporation, College Station, TX).
Results
Studies characteristics
Forty studies were resulted and retrieved from the literature search in the 3 databases. After screening on the abstracts and full-text reports, 11 studies met with the inclusion criteria (Brondani et al. 2012; Jin et al. 2017; Jin et al. 2020; Montesanto et al. 2018; Rudofsky et al. 2007; Shen et al. 2014; Zhang et al. 2014; Zhou et al. 2018; Zietz et al. 2006) [20, 26, 27, 32–37]. Jin et al. 2020 is comprised of 2 datasets, of which dataset 1 has been reported in Jin et al. 2017 [34, 35]. For Jin et al. 2020 and Montesanto et al. 2018, only the allelic data was able to be extracted [35, 36]. Therefore, dataset 2 of Jin et al. 2020 as the data for Jin 2020 and Montesanto et al. 2018 were used to calculate the pooled effect in the allelic model. The quality of the included studies was evaluated by NOS, ranging from 5 to 8, and the overall quality was moderate. The flow chart of the study selection was presented in Fig. 1.
Fig. 1.
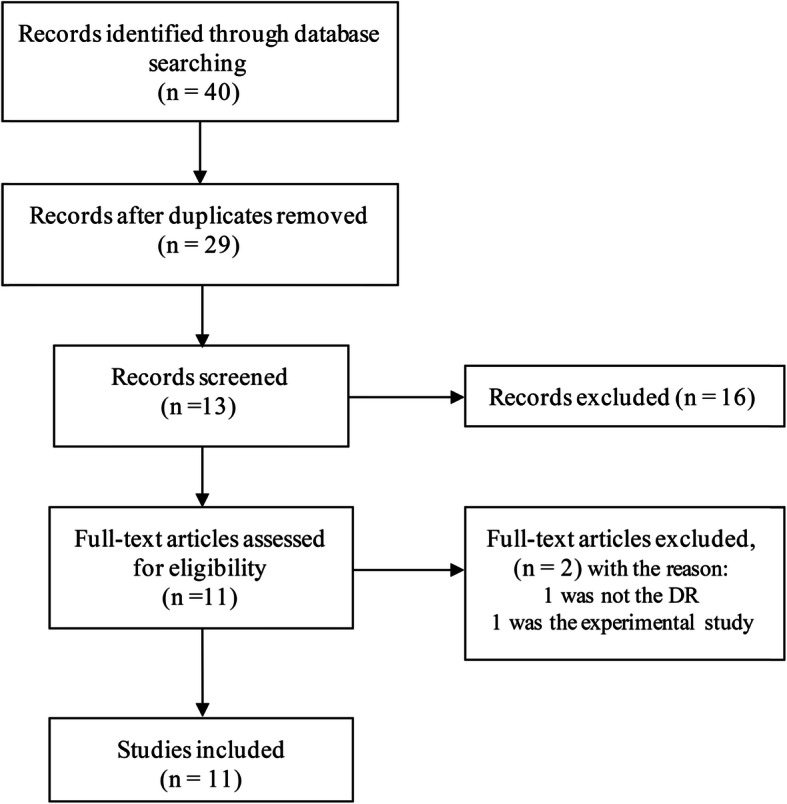
Flow chart for the inclusion and exclusion of the studies in this meta-analysis. n = number
A total of 5 UCPs variants, UCP1 rs1800592, UCP2 rs659366, UCP2 rs660339 (p.A55V), UCP2 45-bp Ins/Del and UCP2 rs1800849, were identified from the literature search (Table 1 and Supplementary Table 1). Yet, only rs1800592 and rs659366 were subjected to further analysis as they were reported in more than 3 studies. Eleven studies for rs1800592 and rs659366 from 4 countries, including China (n = 5), Brazil (n = 2), Germany (n = 3), Brazil (n = 2) and Italy (n = 1), were included. The extracted data (except for Jin et al. 2020 and Montesanto et al. 2018), HWE and minor allele frequency (MAF) were listed in Table 2. For Montesanto et al. 2018, the actual PHWE value was not presented but mentioned all variants with PHWE > 0.05. In addition, Jin et al. 2020 only presented HWE and MAF of the two datasets (rs1800592: MAF = 0.499, PHWE > 0.999 and rs659366: MAF = 0.437, PHWE = 0.640).
Table 1.
Characteristics of the included studies
| Variant | Study | Country | Ethnicity | n (cases/control) | Source of controls | Type of diabetes | Genotyping methods | DMC | Method of DR acertainment | NOS |
|---|---|---|---|---|---|---|---|---|---|---|
| UCP1 rs1800592 | Jin 2017 | China | Han Chinese | 1875 (530/1345) | Population-based study | T2DM | MassARRAY | DR | Direct ophthalmoscopy, fundus fluorescein angiography and OCT | 7 |
| Jin 2020 | China | Han Chinese | 1235 (134/1101) | Hospital-based case-control | T2DM |
Affymetrix Genome-Wide Human SNP Array 6.0 |
DR | Direct ophthalmoscopy, fundus fluorescein angiography and OCT | 7 | |
| Montesanto 2018 | Italy | European | 940 (435/505) | Population-based study | T2DM | MassARRAY | DR | Fundoscopy through dilated pupils and/or fluorescein angiography | 6 | |
| Rudofsky 2007 | Germany | Caucasian | 517 (128/389) | Hospital-based case-control | T2DM | PCR-RFLPs | DR | Fundoscopy, indirect ophthalmoscopy and fundus fluorescein angiography | 8 | |
| Zhang 2014 | China | Han Chinese | 792 (488/344) | Hospital-based case-control | T2DM | PCR-LDR | NPDR, PDR | Fundoscopy and fundus fluorescein angiography | 8 | |
| Zietz 2006 | Germany | Caucasian | 509 (194/315) | Population-based study | T2DM | PCR-RFLPs | NPDR, PDR | Fundus photograpy | 5 | |
| Brondani 2012 | Brazil | European | 257 (154/103) | Hospital-based case-control | T1DM | PCR-RFLPs | DR | Fundoscopy through dilated pupils | 6 | |
| Rudofsky 2006 | Germany | Caucasian | 227 (64/163) | Hospital-based case-control | T1DM | PCR-RFLPs | DR | Ophthalmoscopic examination | 7 | |
| UCP2 rs659366 | Crispim 2010 | Brazil | European |
501 (242/259) 196 (85/111) |
Hospital-based case-control | T1DM, T2DM | PCR-RFLPs | PDR | Fundoscopy through dilated pupils | 7 |
| Jin 2017 | China | Han Chinese | 1875 (530/1344) | Population-based study | T2DM | MassARRAY | DR | Direct ophthalmoscopy, fundus fluorescein angiography and OCT | 7 | |
| Jin 2020 | China | Han Chinese | 1235 (134/1101) | Hospital-based case-control | T2DM |
Affymetrix Genome-Wide Human SNP Array 6.0 |
DR | Direct ophthalmoscopy, fundus fluorescein angiography and OCT | 7 | |
| Rudofsky 2007 | Germany | Caucasian | 645 (128/517) | Hospital-based case-control | T2DM | PCR-RFLPs | DR | Fundoscopy, indirect ophthalmoscopy and fundus fluorescein angiography | 8 | |
| Shen 2014 | China | Han Chinese | 472 (317/155) | Hospital-based case-control | T2DM | ABI 3100 genetic analyzer | NPDR, PDR | Fundoscopy through dilated pupils | 5 | |
| Zhou 2018 | China | Han Chinese | 408 (209/199) | Hospital-based case-control | T2DM | ABI 3730 genetic analyzer | NPDR, PDR | Fundoscopy and fundus photograpy | 6 | |
| Rudofsky 2006 | Germany | Caucasian | 227 (64/163) | Hospital-based case-control | T1DM | PCR-RFLPs | DR | Ophthalmoscopic examination | 7 |
UCP uncoupling protein, T1DM type 1 diabetes mellitus, T2DM type 2 diabetes mellitus, DMC diabetes mellitus complications, DR diabetic retinopathy, PDR proliferative diabetic retinopathy, NPDR non-proliferative diabetic retinopathy, NOS Newcastle-Ottawa quality assessment scale, n: number
Table 2.
Extracted genotype data from the included studies
| Variant | Controls (n) | Cases (n) | Study | Cases | Controls | PHWE | MAF | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| UCP1 rs1800592 | GG | GA | AA | G | A | GG | GA | AA | G | A | |||||
| DWR (2260) | DR (1386) | Jin 2017 | 140 | 251 | 139 | 531 | 529 | 338 | 669 | 338 | 1345 | 1345 | 0.85 | 50% | |
| Rudofsky 2007 | 5 | 63 | 60 | 73 | 183 | 18 | 173 | 198 | 209 | 569 | 0.01 | 27% | |||
| Zhang 2014 | 122 | 217 | 105 | 461 | 427 | 79 | 161 | 94 | 319 | 349 | 0.53 | 50% | |||
| Zietz 2006 | 8 | 72 | 114 | 88 | 300 | 15 | 117 | 183 | 147 | 483 | 0.50 | 23% | |||
| DWR (649) | NPDR (366) | Zhang 2014 | 57 | 110 | 60 | 224 | 230 | 79 | 161 | 94 | 319 | 349 | 0.53 | 48% | |
| Zietz 2006 | 6 | 49 | 84 | 61 | 217 | 15 | 117 | 183 | 147 | 483 | 0.50 | 23% | |||
| DWR (649) | PDR (272) | Zhang 2014 | 65 | 107 | 45 | 237 | 197 | 79 | 161 | 94 | 319 | 349 | 0.53 | 50% | |
| Zietz 2006 | 2 | 23 | 30 | 27 | 83 | 15 | 117 | 183 | 147 | 483 | 0.50 | 24% | |||
| UCP2 rs659366 | AA | AG | GG | A | G | AA | AG | GG | A | G | |||||
| DWR (2244) | DR (1236) | Jin 2017 | 68 | 261 | 201 | 397 | 663 | 177 | 654 | 513 | 1008 | 1680 | 0.16 | 37% | |
| Rudofsky 2007 | 17 | 62 | 49 | 96 | 160 | 49 | 186 | 154 | 284 | 494 | 0.54 | 37% | |||
| Shen 2014 | 59 | 144 | 102 | 262 | 348 | 38 | 73 | 38 | 149 | 149 | 0.81 | 45% | |||
| Zhou 2018 | 57 | 122 | 30 | 236 | 182 | 33 | 110 | 56 | 176 | 222 | 0.09 | 50% | |||
| DWR (348) | NPDR (194) | Shen 2014 | 23 | 54 | 25 | 100 | 104 | 38 | 73 | 38 | 149 | 149 | 0.81 | 50% | |
| Zhou 2018 | 25 | 61 | 6 | 111 | 73 | 33 | 110 | 56 | 176 | 222 | 0.09 | 49% | |||
| DWR (606) | PDR (560) | Crispim 2010 | 45 | 131 | 64 | 221 | 259 | 30 | 116 | 112 | 176 | 340 | 1.00 | 40% | |
| Shen 2014 | 36 | 90 | 77 | 162 | 244 | 38 | 73 | 38 | 149 | 149 | 0.81 | 44% | |||
| Zhou 2018 | 32 | 61 | 24 | 125 | 109 | 33 | 110 | 56 | 176 | 222 | 0.09 | 48% | |||
DWR diabetes without retinopathy, DR diabetic retinopathy, PDR proliferative diabetic retinopathy, NPDR non-proliferative diabetic retinopathy, HWE Hardy-Weinberg equilibrium, MAF minor allele frequency, n: number
Data analysis
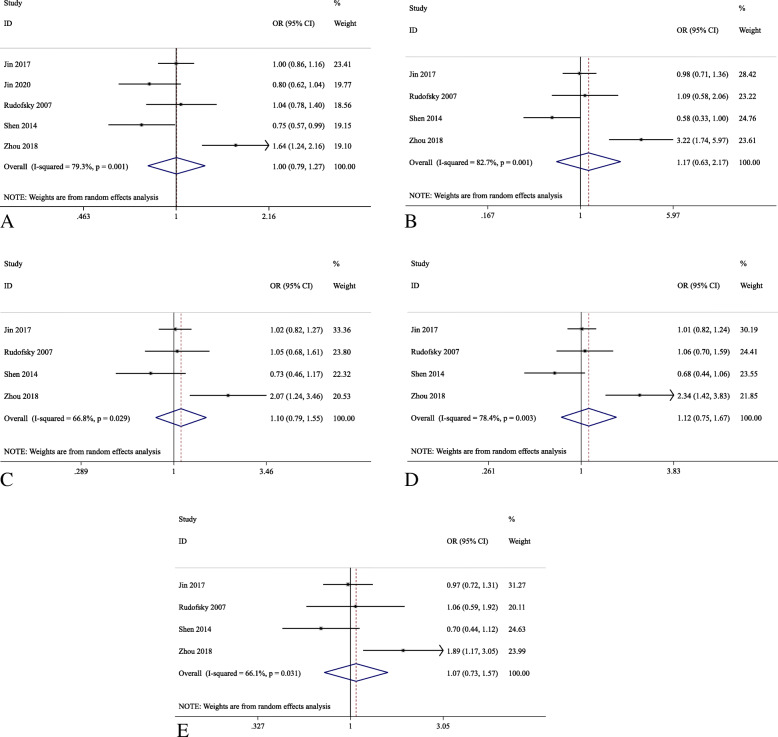
Five studies were identified for the investigation of UCP1 rs1800592, among which Rudofsky et al. 2007 was not included in the pooled effects analysis as its PHWE in the control subjects was less than 0.05 [26]. Only 2 studies, Brondani et al. 2012 and Crispim et al. 2010, included the patients with type-1 DM [20, 28]. Hence, rs1800592 and rs659366 were further analyzed only with patients in type-2 DM. For UCP1 rs1800592, a total of 1781 patients as cases (DR) and 3610 patients as control (DWR) was used for the meta-analysis in the allelic model, and the number of cases and controls in other models were 1212 and 2004, respectively. Similarly, for UCP2 rs659366, the number of cases and controls in the allelic model were 1318 and 3316, respectively, and 1318 patients as case and 3316 patients as control in other models. The pooled effect analysis of UCP1 rs1800592 showed no significant association in type-2 DM patients for all five genetic models (Table 3 and Fig. 2). Similarly, for UCP2 rs659366, the pooled effect analysis showed no statistically significant association with DR in type-2 DM patients for all five genetic models (Table 3 and Fig. 3). In the subgroup analyses, UCP1 rs1800592 showed statistically significant association with PDR in type-2 DM patients for the allelic (G allele versus A allele: OR = 1.26, 95% C.I.: 1.02–1.56, P = 0.035), homozygous model (GG versus AA: OR = 1.60, 95% C.I.: 1.01–2.52, P = 0.044), but not for the heterozygous, dominant and recessive models. (P > 0.05; Table 4 and Fig. 4). However, for other subgroup analyses, no statistically significant association was found in the stage of DR and ethnicity (Supplementary figure 1, 2, 3).
Table 3.
Pooled effect on the association of UCPs variants with DR in type-2 DM patients
| Comparison | Variant | Model | Effect | Pooled OR (95% C.I.) | Z | Pz | I2 | P for heterogeneity |
|---|---|---|---|---|---|---|---|---|
| DR vs DWR | UCP1 rs1800592 | Allelic (G vs A) | Fixed | 1.03 (0.93, 1.13) | 0.51 | 0.609 | 0.0% | 0.533 |
| Homozygous (GG vs AA) | Fixed | 1.10 (0.88, 1.37) | 0.85 | 0.398 | 0.0% | 0.375 | ||
| Heterozygous (AG vs AA) | Fixed | 1.00 (0.84, 1.19) | 0.01 | 0.992 | 0.0% | 0.431 | ||
| Dominant (GG + AG vs AA) | Fixed | 1.03 (0.87, 1.21) | 0.31 | 0.760 | 8.6% | 0.335 | ||
| Recessive (GG vs AG + AA) | Fixed | 1.11 (0.92, 1.33) | 1.07 | 0.284 | 0.0% | 0.684 | ||
| DR vs DWR | UCP2 rs659366 | Allelic (A vs G) | Random | 1.00 (0.79, 1.27) | 0.02 | 0.983 | 79.3% | 0.001 |
| Homozygous (AA vs GG) | Random | 1.17 (0.63, 2.17) | 0.49 | 0.623 | 82.7% | 0.001 | ||
| Heterozygous (GA vs GG) | Random | 1.10 (0.79, 1.55) | 0.56 | 0.572 | 66.8% | 0.029 | ||
| Dominant (AA+GA vs GG) | Random | 1.12 (0.75, 1.67) | 0.55 | 0.584 | 78.4% | 0.003 | ||
| Recessive (AA vs GA + GG) | Random | 1.07 (0.73, 1.57) | 0.35 | 0.73 | 66.1% | 0.031 |
DM diabetes mellitus, DWR diabetes without retinopathy, DR diabetic retinopathy, PDR proliferative diabetic retinopathy, NPDR non-proliferative diabetic retinopathy, C.I. confident interval
Fig. 2.
Forest plots for the association analysis between the UCP1 rs1800592 variant and diabetic retinopathy in type-2 diabetes mellitus patients in five genetic models. Reference allele: A; variant allele: G. a Allelic (G vs A); b Homozygous (GG vs AA); c Heterozygous (AG V. AA); d Dominant (GG + AG vs AA); e Recessive (GG vs AG + AA) models
Fig. 3.
Forest plots for the association analysis of UCP2 rs659366 variant with diabetic retinopathy in type-2 diabetes mellitus patients in five genetic models
Reference allele: G; variant allele: A. a Allelic (A vs G); b Homozygous (AA vs GG); c Heterozygous (GA vs GG); d Dominant (AA+GA vs GG); e Recessive (AA vs GA + GG) models.
Table 4.
Subgroup analysis on subtype of DR and ethnicity
| Variant | Cases vs Controls | Ethnicity | Allelic | Homozygous | Heterozygous | Dominant | Recessive |
|---|---|---|---|---|---|---|---|
| OR (95% C.I.) | OR (95% C.I.) | OR (95% C.I.) | OR (95% C.I.) | OR (95% C.I.) | |||
| UCP1 rs1800592 | G vs A | GG vs AA | AG vs AA | GG + AG vs AA | GG vs AG + AA | ||
| NPDR vs DWR | 1.02 (0.84,1.23) | 1.08 (0.71,1.64) | 0.99 (0.74,1.33) | 1.00 (0.76,1.32) | 1.05 (0.73,1.52) | ||
| PDR vs DWR | 1.26 (1.02,1.56)* | 1.60 (1.01,2.52)* | 1.32 (0.93, 1.87) | 1.38 (0.99,1.92) | 1.32 (0.91,1.92) | ||
| UCP2 rs659366 | A vs G | AA vs GG | GA vs GG | AA+GA vs GG | AA vs GA + GG | ||
| NPDR vs DWR | 1.36 (0.69,2.67) | 2.48 (0.33, 18.37) | 2.33 (0.51,10.57) | 2.35 (0.45, 12.43) | 1.26 (0.58,2.75) | ||
| PDR vs DWR | 1.17 (0.67,2.05) | 1.40 (0.46,4.24) | 1.17 (0.57,2.40) | 1.22 (0.54,2.77) | 1.28 (0.63,2.56) | ||
| UCP1 rs1800592 | G vs A | GG vs AA | AG vs AA | GG + AG vs AA | GG vs AG + AA | ||
| DR vs DWR | Chinese | 1.05 (0.94, 1.16) | 1.12 (0.89,1.40) | 1.00 (0.82,1.22) | 1.04 (0.86,1.26) | 1.12 (0.93,1.35) | |
| Total | 1.03 (0.93, 1.13) | 1.10 (0.88,1.37) | 1.00 (0.84,1.19) | 1.03 (0.87,1.21) | 1.11 (0.92, 1.33) | ||
| UCP2 rs659366 | A vs G | AA vs GG | GA vs GG | AA+GA vs GG | AA vs GA + GG | ||
| DR vs DWR | Chinese | 0.99 (0.74, 1.34) | 1.20 (0.52,2.76) | 1.13 (0.70,1.83) | 1.15 (0.65,2.03) | 1.08 (0.65,1.77) | |
| Total | 1.00 (0.79,1.27) | 1.17 (0.63,2.17) | 1.10 (0.79,1.55) | 1.12 (0.75,1.67) | 1.07 (0.73,1.57) |
*P < 0.05, DWR diabetes without retinopathy, DR diabetic retinopathy, PDR proliferative diabetic retinopathy, NPDR non-proliferative diabetic retinopathy, C.I. confident interval
Fig. 4.
Forest plots for the subgroup analysis of UCP1 rs1800592 variant with the stage of diabetic retinopathy in type-2 diabetes mellitus patients in five genetic models. Reference allele: A; variant allele: G. a Allelic (G vs A); b Homozygous (GG vs AA); c Heterozygous (AG V. AA); d Dominant (GG + AG vs AA); e Recessive (GG vs AG + AA) models
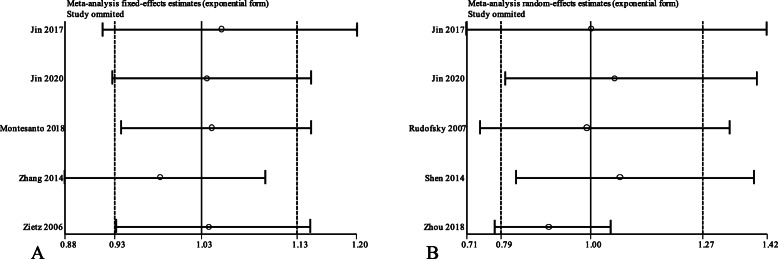
Evaluation of publication bias and sensitivity analyses
The Egger’s test, which was used to quantitatively measure the publication bias, showed no statistically significant publication bias (P > 0.05; Table 5). The results of the sensitivity analysis showed that the pooled OR lied within the 95% C.I. of the total pooled OR (Fig. 5).
Table 5.
Evaluation of publication bias by Egger’s test
| Variant | Cases vs Controls | Allelic | Homozygous | Heterozygous | Dominant | Recessive |
|---|---|---|---|---|---|---|
| UCP1 rs1800592 | DR vs DWR | 0.482 | 0.970 | 0.520 | 0.657 | 0.763 |
| UCP2 rs659366 | DR vs DWR | 0.944 | 0.687 | 0.710 | 0.702 | 0.770 |
α = 0.1
Fig. 5.
Sensitivity analysis of UCP1 rs1800592 and UCP2 rs659366 variants in allelic model. a rs1800592 (G vs A); b rs659366 (A vs G)
Discussion
DR is one of the most common microvascular complications in DM patients. The hyperglycemia-induced ROS is considered as one of the initial and major pathways causing the damage to the endothelial cells. The UCPs are anion carrier proteins in mitochondrial inner membrane. UCPs function to reduce mitochondrial ROS, especially hyperglycemia-induced oxidative stress, and protect endothelial cells from oxidative stress by balancing the proton motive force across the mitochondrial inner membrane [22]. Therefore, UCPs could possibly participate in the development and pathogenesis of DR.
This meta-analysis verified the association of the reported UCPs variants with the susceptibility of DR. Our results showed that UCP1 rs1800592 variant was not significantly associated with DR in type-2 DM patients in the pooled effects analysis (Table 3 and Fig. 2); yet, in the subgroup analysis, UCP1 rs1800592 was significantly associated with PDR in type-2 DM patients in the allelic and homozygous models (Table 4 and Fig. 4). The patients carrying allele G of UCP1 rs1800592 variant have 26% higher risk developing PDR than those carrying allele A. A previous study demonstrated that the carriers of rs1800592 GG genotype exhibited higher UCP1 gene expression than those with AA genotype in the retina samples [20]. This could suggest that higher UCP1 expression in retina by allele G of rs1800592 variant could be associated with higher susceptibility of PDR. Conversely, UCP1 expression was lower in carriers of GG genotype than those with AA genotype in intraperitoneal adipose cells, indicated the tissue-specific effect of rs1800592 on UCP1 expression activity [38]. Moreover, allele G of UCP1 rs1800592 also showed elevated expression of MnSOD2 gene, which is another major scavenger for mitochondrial ROS [20, 39]. Our discovery was resulted from 2 reported studies, and our approach is similar to that from our previous studies [40–42]. Nevertheless, further studies with larger cohorts in different populations are needed to verify its association with PDR.
UCP2 is the most widely distributed uncoupling protein and most frequently studied in DM and DR, and itis associated with the increased oxidative stress and negatively regulates the insulin secretion [43, 44]. Total 4 UCP2 variants, UCP2 rs659366, UCP2 rs660339 (p.A55V), UCP2 45-bp Ins/Del and UCP2 rs1800849, were reported in the association analysis with DR; however, only UCP2 rs659366 variant comprised enough studies for the meta-analysis, and other UCP2 variants have not been further analyzed in this study. UCP2 rs659366 has been reported to be associated with type-2 DM [45]; however, in this meta-analysis, we demonstrated that UCP2 rs659366 variant showed no pooled association with DR in the type-2 DM patients (Table 3 and Fig. 3). The elevation of UCP2 expression could be induced by high glucose treatment in epithelial cell of human vein, and the A allele of UCP2 rs659366 increases promoter activity as compared to the G allele, which can be exacerbated under hyperglycemic condition to exert a protective effect [46]. The negative association of UCP2 rs659366 variant with DR in this meta-analysis might indicated that UCP2 gene variation may not be contributed to the development of DR. Nevertheless, it is of worth to note that, in the F-SNP database analyses, UCP2 rs660339 is strongly linked with UCP2 rs659366, and partially linked with UCP2 45-bp Ins/Del variant [28]. One report showed that the haplotype of 3 differentUCP2 variants [Ins (45-bp Ins/Del), A (rs659366) and Ala (rs660339)] is associated with the decreased UCP2 gene expression in human retina [47]. This could be an independent risk factor for PDR in both type-1 and 2 DM patients [28]. Additional association studies are necessary in order to confirm the association of all 4 UCP2 variants with DR in different ethnic groups.
We conducted the subgroup analyses on ethnicity in this meta-analysis. There was no significant association in different ethnic group, which could be due to the limited and sample sizes after stratification. Thus, the ethnicity-specific effects of these variants need to be determined with larger sample sizes in additional cohort studies.
There are several limitations in this meta-analysis. First, the number of reported studies for each UCPs variant was still limited. Second, the lack of original clinical information would be difficult to adjust the relevant variables, such as duration of diabetes, medications and other chronic diseases.
Conclusions
In summary, our meta-analysis revealed no significant pooled association of UCP1 rs1800592 and UCP2 rs659366 with DR in DM patients; yet allele G of UCP1 rs1800592 variant could be associated with the increased risk of PDR in type-2 DM patients. Our results suggest that UCP1 rs1800592 variant could be clinically applied as a genetic marker for PDR prediction and risk analysis in DM clinics.
Supplementary Information
Additional file 4: Table S1. Characteristics of other UCPs variants. Table S2. Analysis of the included studies by Newcastle-Ottawa quality assessment scale. Figure S1. Subgroup analysis of UCP1 rs1800592 by ethnicity. Figure S2. Subgroup analysis of UCP2 rs659366 by ethnicity. Figure S3. Subgroup analysis of UCP2 rs659366 by stage of DR. Figure S4. Sensitivity analyses of UCP1 rs1800592 in all genetic models. Figure S5. Sensitivity analyses of UCP2 rs659366 in all genetic models
Acknowledgments
Not applicable.
Abbreviations
- UCPs
Uncoupling proteins
- DR
Diabetic retinopathy
- DM
Diabetes mellites
- OR
Odds ratios
- PDR
Proliferative diabetic retinopathy
- C.I.
Confidence interval
- DWR
Diabetes without retinopathy
- HWE
Hardy-Weinberg equilibrium
- NOS
Newcastle-Ottawa quality assessment scale
- MAF
Minor allele frequency
Authors’ contributions
Conceived and designed the study: X.L., T.K.N and Z.W.; Data acquisition: X.L. and Z.J.; Data analysis and Interpretation: X.L.; Manuscript writing: X.L.; Critical revision of the manuscript: T.K.N, G.Z. and Z.W. All authors made substantial contribution to this manuscript meeting authorship criteria, agreed to be accountable for all aspects of the work and have read and approved the final version.
Funding
This work was supported by the Shantou Medical Health, Science and Technology Project Fund (project code:190716155262406 to G.Z.) and the Special Fund for Science and Technology of Guangdong Province (project code: 2019ST017 to Z.W.) and the Grant for Key Disciplinary Project of Clinical Medicine under the Guangdong High-level University Development Program, China. Funding body did not involve in the study and collection, analysis, and interpretation of data and in writing the manuscript.
Availability of data and materials
All the data supporting our findings is contained within the manuscript.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Fong DS, Aiello LP, Ferris FL, 3rd, Klein R. Diabetic retinopathy. Diabetes Care. 2004;27(10):2540–2553. doi: 10.2337/diacare.27.10.2540. [DOI] [PubMed] [Google Scholar]
- 2.Wong TY, Cheung CM, Larsen M, Sharma S, Simo R. Diabetic retinopathy. Nat Rev Dis Primers. 2016;2:16012. doi: 10.1038/nrdp.2016.12. [DOI] [PubMed] [Google Scholar]
- 3.Liew G, Klein R, Wong TY. The role of genetics in susceptibility to diabetic retinopathy. Int Ophthalmol Clin. 2009;49(2):35–52. doi: 10.1097/IIO.0b013e31819fd5d7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Warpeha KM, Chakravarthy U. Molecular genetics of microvascular disease in diabetic retinopathy. Eye (Lond) 2003;17(3):305–311. doi: 10.1038/sj.eye.6700348. [DOI] [PubMed] [Google Scholar]
- 5.Kowluru RA, Chan PS. Oxidative stress and diabetic retinopathy. Exp Diabetes Res. 2007;2007:43603. doi: 10.1155/2007/43603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Golbidi S, Badran M, Laher I. Antioxidant and anti-inflammatory effects of exercise in diabetic patients. Exp Diabetes Res. 2012;2012:941868. doi: 10.1155/2012/941868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ejaz S, Chekarova I, Ejaz A, Sohail A, Lim CW. Importance of pericytes and mechanisms of pericyte loss during diabetes retinopathy. Diabetes Obes Metab. 2008;10(1):53–63. doi: 10.1111/j.1463-1326.2007.00795.x. [DOI] [PubMed] [Google Scholar]
- 8.Donato L, Scimone C, Alibrandi S, Pitruzzella A, Scalia F, D'Angelo R, Sidoti A. Possible A2E Mutagenic Effects on RPE Mitochondrial DNA from Innovative RNA-Seq Bioinformatics Pipeline. Antioxidants (Basel) 2020;9:11. doi: 10.3390/antiox9111158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Scimone C, Alibrandi S, Scalinci SZ, Trovato Battagliola E, D'Angelo R, Sidoti A, Donato L. Expression of Pro-Angiogenic Markers Is Enhanced by Blue Light in Human RPE Cells. Antioxidants (Basel) 2020;9:11. doi: 10.3390/antiox9111154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Santos JM, Tewari S, Kowluru RA. A compensatory mechanism protects retinal mitochondria from initial insult in diabetic retinopathy. Free Radic Biol Med. 2012;53(9):1729–1737. doi: 10.1016/j.freeradbiomed.2012.08.588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brownlee M. Biochemistry and molecular cell biology of diabetic complications. Nature. 2001;414(6865):813–820. doi: 10.1038/414813a. [DOI] [PubMed] [Google Scholar]
- 12.Donato L, Scimone C, Alibrandi S, Abdalla EM, Nabil KM, D'Angelo R, Sidoti A. New Omics-Derived Perspectives on Retinal Dystrophies: Could Ion Channels-Encoding or Related Genes Act as Modifier of Pathological Phenotype? Int J Mol Sci. 2020;22:1. doi: 10.3390/ijms22010070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dalgaard LT, Pedersen O. Uncoupling proteins: functional characteristics and role in the pathogenesis of obesity and type II diabetes. Diabetologia. 2001;44(8):946–965. doi: 10.1007/s001250100596. [DOI] [PubMed] [Google Scholar]
- 14.Jezek P. Possible physiological roles of mitochondrial uncoupling proteins--UCPn. Int J Biochem Cell Biol. 2002;34(10):1190–1206. doi: 10.1016/S1357-2725(02)00061-4. [DOI] [PubMed] [Google Scholar]
- 15.Azzu V, Brand MD. The on-off switches of the mitochondrial uncoupling proteins. Trends Biochem Sci. 2010;35(5):298–307. doi: 10.1016/j.tibs.2009.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cannon B, Shabalina IG, Kramarova TV, Petrovic N, Nedergaard J. Uncoupling proteins: a role in protection against reactive oxygen species--or not? Biochim Biophys Acta. 2006;1757(5–6):449–458. doi: 10.1016/j.bbabio.2006.05.016. [DOI] [PubMed] [Google Scholar]
- 17.Rousset S, Alves-Guerra MC, Mozo J, Miroux B, Cassard-Doulcier AM, Bouillaud F, Ricquier D. The biology of mitochondrial uncoupling proteins. Diabetes. 2004;53(Suppl 1):S130–S135. doi: 10.2337/diabetes.53.2007.S130. [DOI] [PubMed] [Google Scholar]
- 18.Echtay KS, Roussel D, St-Pierre J, Jekabsons MB, Cadenas S, Stuart JA, Harper JA, Roebuck SJ, Morrison A, Pickering S, et al. Superoxide activates mitochondrial uncoupling proteins. Nature. 2002;415(6867):96–99. doi: 10.1038/415096a. [DOI] [PubMed] [Google Scholar]
- 19.Krauss S, Zhang CY, Lowell BB. The mitochondrial uncoupling-protein homologues. Nat Rev Mol Cell Biol. 2005;6(3):248–261. doi: 10.1038/nrm1592. [DOI] [PubMed] [Google Scholar]
- 20.Brondani LA, de Souza BM, Duarte GCK, Kliemann LM, Esteves JF, Marcon AS, Gross JL, Canani LH, Crispim D. The UCP1 -3826A/G polymorphism is associated with diabetic retinopathy and increased UCP1 and MnSOD2 gene expression in human retina. Invest Ophthalmol Vis Sci. 2012;53(12):7449–7457. doi: 10.1167/iovs.12-10660. [DOI] [PubMed] [Google Scholar]
- 21.Cui Y, Xu X, Bi H, Zhu Q, Wu J, Xia X, Qiushi R, Ho PC. Expression modification of uncoupling proteins and MnSOD in retinal endothelial cells and pericytes induced by high glucose: the role of reactive oxygen species in diabetic retinopathy. Exp Eye Res. 2006;83(4):807–816. doi: 10.1016/j.exer.2006.03.024. [DOI] [PubMed] [Google Scholar]
- 22.Liu J, Li J, Li WJ, Wang CM. The role of uncoupling proteins in diabetes mellitus. Journal of Diabetes Research. 2013;2013:585897. doi: 10.1155/2013/585897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krauss S, Zhang CY, Scorrano L, Dalgaard LT, St-Pierre J, Grey ST, Lowell BB. Superoxide-mediated activation of uncoupling protein 2 causes pancreatic beta cell dysfunction. J Clin Invest. 2003;112(12):1831–1842. doi: 10.1172/JCI200319774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Arsenijevic D, Onuma H, Pecqueur C, Raimbault S, Manning BS, Miroux B, Couplan E, Alves-Guerra MC, Goubern M, Surwit R, et al. Disruption of the uncoupling protein-2 gene in mice reveals a role in immunity and reactive oxygen species production. Nat Genet. 2000;26(4):435–439. doi: 10.1038/82565. [DOI] [PubMed] [Google Scholar]
- 25.Teshima Y, Akao M, Jones SP, Marban E. Uncoupling protein-2 overexpression inhibits mitochondrial death pathway in cardiomyocytes. Circ Res. 2003;93(3):192–200. doi: 10.1161/01.RES.0000085581.60197.4D. [DOI] [PubMed] [Google Scholar]
- 26.Rudofsky G, Jr, Schrodter A, Voron'ko OE, Schlotterer A, Humpert PM, Tafel J, Nawroth PP, Bierhaus A, Hamann A. Promoter polymorphisms of UCP1, UCP2, and UCP3 are not associated with diabetic microvascular complications in type 2 diabetes. Horm Metab Res. 2007;39(4):306–309. doi: 10.1055/s-2007-973816. [DOI] [PubMed] [Google Scholar]
- 27.Zhang Y, Meng N, Lv Z, Li H, Qu Y. The gene polymorphisms of UCP1 but not PPAR gamma and TCF7L2 are associated with diabetic retinopathy in Chinese type 2 diabetes mellitus cases. Acta Ophthalmol. 2015;93(3):e223–e229. doi: 10.1111/aos.12542. [DOI] [PubMed] [Google Scholar]
- 28.Crispim D, Fagundes NJ, dos Santos KG, Rheinheimer J, Boucas AP, de Souza BM, Macedo GS, Leiria LB, Gross JL, Canani LH. Polymorphisms of the UCP2 gene are associated with proliferative diabetic retinopathy in patients with diabetes mellitus. Clin Endocrinol. 2010;72(5):612–619. doi: 10.1111/j.1365-2265.2009.03684.x. [DOI] [PubMed] [Google Scholar]
- 29.Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med. 2002;21(11):1539–1558. doi: 10.1002/sim.1186. [DOI] [PubMed] [Google Scholar]
- 30.DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7(3):177–188. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 31.Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst. 1959;22(4):719–748. [PubMed] [Google Scholar]
- 32.Zhou T-C, Yang L, Liu Y-Y, Qin Y, Li Y-P, Zhang L, Yang K, Yang Y. Polymorphisms in the uncoupling protein 2 gene are associated with diabetic retinopathy in Han Chinese patients with type 2 diabetes. Genetic testing and molecular biomarkers. 2018;22(11):637–643. doi: 10.1089/gtmb.2018.0115. [DOI] [PubMed] [Google Scholar]
- 33.Shen Y, Wen Z, Wang N, Zheng Z, Liu K, Xia X, Gu Q, Shi Y, Xu X. Investigation of variants in UCP2 in Chinese type 2 diabetes and diabetic retinopathy. PLoS One. 2014;9(11):e112670. doi: 10.1371/journal.pone.0112670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jin P, Li Z, He J, Chen J, Xu X, Du X, Bai X, Zhang B, He X, Lu L, et al. Associations of single nucleotide polymorphisms of UCPgenes with diabetic retinopathy in Chinese Han population. Zhonghua Shiyan Yanke Zazhi/Chinese Journal of Experimental Ophthalmology. 2017;35(8):737–742. [Google Scholar]
- 35.Jin P, Li Z, He J, Chen J, Xu X, Du X, Bai X, Zhang B, He X, Lu L, et al. Analysis of association between common variants of uncoupling proteins genes and diabetic retinopathy in a Chinese population. BMC Medical Genetics. 2020;21(1):25. doi: 10.1186/s12881-020-0956-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Montesanto A, Bonfigli AR, Crocco P, Garagnani P, De Luca M, Boemi M, Marasco E, Pirazzini C, Giuliani C, Franceschi C, et al. Genes associated with type 2 diabetes and vascular complications. Aging. 2018;10(2):178–196. doi: 10.18632/aging.101375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zietz B, Leonhardt K. Schaffler a: [candidate genes and polymorphism analysis in type 2 diabetes mellitus] Med Klin (Munich) 2006;101(8):605–616. doi: 10.1007/s00063-006-1090-0. [DOI] [PubMed] [Google Scholar]
- 38.Rose G, Crocco P, D'Aquila P, Montesanto A, Bellizzi D, Passarino G. Two variants located in the upstream enhancer region of human UCP1 gene affect gene expression and are correlated with human longevity. Exp Gerontol. 2011;46(11):897–904. doi: 10.1016/j.exger.2011.07.011. [DOI] [PubMed] [Google Scholar]
- 39.Kanwar M, Chan PS, Kern TS, Kowluru RA. Oxidative damage in the retinal mitochondria of diabetic mice: possible protection by superoxide dismutase. Invest Ophthalmol Vis Sci. 2007;48(8):3805–3811. doi: 10.1167/iovs.06-1280. [DOI] [PubMed] [Google Scholar]
- 40.Lu SY, Rong SS, Wu Z, Huang C, Matsushita K, Ng TK, Leung CKS, Kawashima R, Usui S, Tam POS, et al. Association of the CAV1-CAV2 locus with normal-tension glaucoma in Chinese and Japanese. Clin Exp Ophthalmol. 2020;48(5):658–665. doi: 10.1111/ceo.13744. [DOI] [PubMed] [Google Scholar]
- 41.Ng TK, Liang XY, Lai TY, Ma L, Tam PO, Wang JX, Chen LJ, Chen H, Pang CP. HTRA1 promoter variant differentiates polypoidal choroidal vasculopathy from exudative age-related macular degeneration. Sci Rep. 2016;6:28639. doi: 10.1038/srep28639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wong KH, Rong SS, Chong KK, Young AL, Pang CP, Chen LJ. Genetic associations of interleukin-related genes with Graves' Ophthalmopathy: a systematic review and meta-analysis. Sci Rep. 2015;5:16672. doi: 10.1038/srep16672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pi J, Bai Y, Daniel KW, Liu D, Lyght O, Edelstein D, Brownlee M, Corkey BE, Collins S. Persistent oxidative stress due to absence of uncoupling protein 2 associated with impaired pancreatic beta-cell function. Endocrinology. 2009;150(7):3040–3048. doi: 10.1210/en.2008-1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang CY, Baffy G, Perret P, Krauss S, Peroni O, Grujic D, Hagen T, Vidal-Puig AJ, Boss O, Kim YB, et al. Uncoupling protein-2 negatively regulates insulin secretion and is a major link between obesity, beta cell dysfunction, and type 2 diabetes. Cell. 2001;105(6):745–755. doi: 10.1016/S0092-8674(01)00378-6. [DOI] [PubMed] [Google Scholar]
- 45.Bulotta A, Ludovico O, Coco A, Di Paola R, Quattrone A, Carella M, Pellegrini F, Prudente S, Trischitta V. The common -866G/a polymorphism in the promoter region of the UCP-2 gene is associated with reduced risk of type 2 diabetes in Caucasians from Italy. J Clin Endocrinol Metab. 2005;90(2):1176–1180. doi: 10.1210/jc.2004-1072. [DOI] [PubMed] [Google Scholar]
- 46.Crispim D, Rodrigues M, da Silva LPA, Boucas AP, Canani LH, Carlessi R, de Souza BM. The A allele of the UCP2 -866G/A polymorphism changes UCP2 promoter activity in HUVECs treated with high glucose. Mol Biol Rep. 2019;46(5):4735–4741. doi: 10.1007/s11033-019-04918-0. [DOI] [PubMed] [Google Scholar]
- 47.de Souza BM, Assmann TS, Kliemann LM, Marcon AS, Gross JL, Canani LH, Crispim D. The presence of the -866A/55Val/ins haplotype in the uncoupling protein 2 (UCP2) gene is associated with decreased UCP2 gene expression in human retina. Exp Eye Res. 2012;94(1):49–55. doi: 10.1016/j.exer.2011.11.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 4: Table S1. Characteristics of other UCPs variants. Table S2. Analysis of the included studies by Newcastle-Ottawa quality assessment scale. Figure S1. Subgroup analysis of UCP1 rs1800592 by ethnicity. Figure S2. Subgroup analysis of UCP2 rs659366 by ethnicity. Figure S3. Subgroup analysis of UCP2 rs659366 by stage of DR. Figure S4. Sensitivity analyses of UCP1 rs1800592 in all genetic models. Figure S5. Sensitivity analyses of UCP2 rs659366 in all genetic models
Data Availability Statement
All the data supporting our findings is contained within the manuscript.