Abstract
Background
Diabetic osteoporosis (DOP) is a systemic metabolic bone disease caused by diabetes mellitus (DM). Adipose-derived stem cells (ASCs) play an important role in bone regeneration. Our previous study confirmed that ASCs from DOP mice (DOP-ASCs) have a lower osteogenesis potential compared with control ASCs (CON-ASCs). However, the cause of this poor osteogenesis has not been elucidated. Therefore, this study investigated the underlying mechanism of the decline in the osteogenic potential of DOP-ASCs from the perspective of epigenetics and explored methods to enhance their osteogenic capacity.
Methods
The expression level of JNK1-associated membrane protein (JKAMP) and degree of DNA methylation in CON-ASCs and DOP-ASCs were measured by mRNA expression profiling and MeDIP sequencing, respectively. JKAMP small interfering RNA (siRNA) and a Jkamp overexpression plasmid were used to assess the role of JKAMP in osteogenic differentiation of CON-ASCs and DOP-ASCs. Immunofluorescence, qPCR, and western blotting were used to measure changes in expression of Wnt signaling pathway-related genes and osteogenesis-related molecules after osteogenesis induction. Alizarin red and ALP staining was used to confirm the osteogenic potential of stem cells. Bisulfite-specific PCR (BSP) was used to detect JKAMP methylation degree.
Results
Expression of JKAMP and osteogenesis-related molecules (RUNX2 and OPN) in DOP-ASCs was decreased significantly in comparison with CON-ASCs. JKAMP silencing inhibited the Wnt signaling pathway and reduced the osteogenic ability of CON-ASCs. Overexpression of JKAMP in DOP-ASCs rescued the impaired osteogenic capacity caused by DOP. Moreover, JKAMP in DOP-ASCs contained intragenic DNA hypermethylated regions related to the downregulation of JKAMP expression.
Conclusions
Intragenic DNA methylation inhibits the osteogenic ability of DOP-ASCs by suppressing expression of JKAMP and the Wnt signaling pathway. This study shows an epigenetic explanation for the reduced osteogenic ability of DOP-ASCs and provides a potential therapeutic target to prevent and treat osteoporosis.
Keywords: DNA methylation, JKAMP, Wnt signaling pathway, Adipose-derived stem cells, Osteogenic differentiation, Diabetic osteoporosis
Background
Diabetic osteoporosis (DOP) is a serious metabolic complication of diabetes mellitus (DM) in the bone and joint system. Characterized by hyperglycemic microenvironment and systemic impairment of bone microstructure, strength, and mass, patients with DOP are prone to fractures and have difficulty in controlling diabetes, which makes treatment and rehabilitation difficult [1–3]. The incidence of DOP is about 50% among DM patients [4, 5]. DOP patients with fractures or bone defects have reduced bone healing and regeneration, which leads to poor bone healing, nonunion, and bone defects [6, 7]. Studies are investigating targets to prevent or treat DOP. Bone tissue engineering is a promising method to repair bone defects, which consists of seed cells, scaffold materials, and growth factors [8, 9]. Adipose-derived stem cells (ASCs) as seed cells have attracted widespread attention for clinical applications, including reconstruction of bone defects [10]. In our previous study, the osteogenic ability of DOP-ASCs was decreased in comparison to control ASCs (CON-ASCs). However, the regulatory mechanism of osteogenic differentiation of DOP-ASCs has not been elucidated, which has hindered their application to treatment of DOP bone fractures and defects.
DNA methylation is one of the earliest discovered DNA modification pathways. Studies have shown that DNA methylation changes the chromatin structure, DNA conformation, DNA stability, and the manner through which proteins act on DNA [11, 12]. It is generally believed that hypermethylation of gene promoters contributes to gene silencing, and DNA demethylation and hypermethylation have opposite effects [13–15]. DNA methylation is very important for the self-renewal, multi-directional differentiation ability, and aging of embryonic stem cells [16]. The occurrence of osteoporosis, osteoarthritis, and other skeletal diseases has been confirmed to be related to changes in the degree of DNA methylation in stem cells [17, 18]. Thus, investigating DNA methylation of DOP-ASCs may provide a reference for the mechanism of DOP-related bone disease. Our previous study showed that the overall DNA methylation level was significantly increased in DOP-ASCs in comparison with CON-ASCs and the Wnt/β-Catenin signaling pathway related to bone differentiation was inhibited [19]. However, the regulatory mechanism of DNA methylation in differentiation of DOP-ASCs into osteoblasts has not been reported.
JNK1-associated membrane protein (JKAMP) has been shown to be associated with JNK1 through its C-terminal domain, which increases and prolongs JNK1 activity after activation [20]. In particular, c-Jun N-terminal kinases (JNKs)-members of the mitogen-activated protein kinase superfamily play an important role in regulating various cellular processes including differentiation, apoptosis, and proliferation [21]. JNK1 as a critical protein in the Wnt-PCP signaling pathway significantly regulates its downstream signaling molecules and the canonical Wnt signaling pathway [22]. The Wnt signaling pathway is a highly conserved signaling pathway. Abnormal changes in Wnt signaling pathway markers are linked to changes in bone metabolism and the osteogenic ability of mesenchymal stem cells [23–26]. Recent study has shown that JNK1 is essential for osteoblast function in vivo and in vitro [27, 28]. Therefore, JKAMP may be associated with the regulation of the osteogenic ability of mesenchymal stem cells via JNK1 and Wnt signaling pathway. However, the possible role of JKAMP and JNK1 in decline of the osteogenic capacity of DOP-ASCs is unclear.
In this study, we isolated CON-ASCs and DOP-ASCs from C57BL/6 mice and diabetic osteoporosis C57BL/6 mice, respectively. The cells were treated with Si-Jkamp and a plasmid (Jkamp) for silencing and overexpression of JKAMP, respectively. Bisulfite-specific PCR (BSP) was used to detect the methylation degree of JKAMP. Gene function analysis was employed to assess changes in the JKAMP and the related signaling pathways. Thus, we explored the effects of JKAMP on the osteogenic ability of DOP-ASCs through the Wnt signaling pathway and evaluated the role of DNA methylation in this process.
Methods
Establishment of the diabetic osteoporosis animal model
C57BL/6 mice were provided by the Experimental Animal Center of the Department of Basic Medicine, Southwest Medical University. All animal procedures were reviewed and approved by the Ethics Committee of the Southwest Medical University. The animal care and anesthesia were conducted in accordance with the guidelines of the Care and Use of Laboratory Animals (Ministry of Science and Technology of China, 2006). Briefly, 4-week-old male mice were randomly divided into model (DOP) and control (CON) groups by the random number table method. The CON group was fed normal feed and the DOP group was fed high-fat and sugar feed for 4 weeks. We established the DOP animal model by injection of streptozotocin (STZ; Sigma, St Louis, USA). The two groups of mice were fasted for 12 h and then the DOP group received a single intraperitoneal injection of STZ (140 mg/kg). The CON group received a single intraperitoneal injection of citric acid-sodium citrate buffer (140 mL/kg). After the procedure, each mouse was returned to its original cage for normal feeding (room temperature, 20–25 °C; relative humidity, 60%–80%, free access to water and normal feed).
Isolation and culture of CON-ASCs and DOP-ASCs
CON-ASCs and DOP-ASCs were obtained from subcutaneous adipose tissue of the groin in CON and DOP mice. The adipose tissue was cut into pieces and incubated in 0.075% type I collagenase (Sigma-Aldrich, St Louis, USA) for 30 min at 37 °C. The digestion was terminated with 10% α-modified Eagle’s medium (α-MEM, Hyclone, Pittsburgh, USA) containing 10% fetal bovine serum (FBS, Schaumburg, USA). The cells were centrifuged at 200 g for 5 min and the supernatant was discarded. Then, the cells were resuspended in α-MEM containing 10% FBS and 1% penicillin-streptomycin, seeded in 25-cm2 culture flasks, and incubated at 37 °C with 5% CO2. Non-adherent cells were removed by changing the medium every 2 days. At 80% confluence, the cells were passaged. Passage 3 cells were used in experiments.
Cell transfection
JKAMP siRNA was designed and synthesized by GenePharma (Shanghai, China) for gene silencing. Cells were transiently transfected using a riboFECT CP Transfection Kit (RiboBio, Guangzhou, China) in accordance with the manufacturer’s instructions. SiRNA sequences are shown in Table 1.
Table 1.
SiRNA sequences designed for specific gene silencing
| Sequence (5′ → 3′) | ||
|---|---|---|
| SiRNA | Sense | GCACCAUGGCAGCUAUCAUTT |
| Antisense | AUGAUAGCUGCCAUGGUGCTT | |
| SiRNA-NC | Sense | UUCUCCGAACUGGUCACGUTT |
| Antisense | ACGUGACACGUUCGGAGAATT | |
For JKAMP overexpression, JKAMP was amplified and subcloned into the pcDNA3.1 vector. The empty pGFP3.1 vector carrying eGFP was used as a negative control. Plasmids were transfected into cells using an Auto Electroporator (Bimake, TX, USA) in accordance with the manufacturer’s instructions.
Alizarin red and ALP staining
Alizarin red and ALP staining was used to analyze mineralized nodule formation and alkaline phosphatase activity of differentiated CON-ASCs and DOP-ASCs. CON-ASCs and DOP-ASCs were seeded on 12-well plates at 5 × 104 cells per well. The medium was changed to osteogenic induction medium (Cyagen, Guangzhou, China), after CON-ASCs and DOP-ASCs were transfected with Si-Jkamp and plasmid (Jkamp), respectively. The medium was changed every 3 days. After 3 and 5 days of osteoinduction, the osteogenesis induction medium was discarded. Cells were washed twice with PBS and then fixed with 4% neutral buffered formalin for 30 min. ALP activity was detected using an Alkaline Phosphatase Assay Kit (Beyotime, Shanghai, China) in accordance to the manufacturer’s protocol. At 14 days after osteogenesis induction, we performed alizarin red staining (Cyagen) in accordance with the manufacturer’s protocol to assess the formation of calcium nodules. Staining was observed using the DFC 7000T system (Leica, Wetzlar, Germany). Each image was selected to save by a visual camera.
Western blot analysis
Western blotting was used to detect the levels of JKAMP, JNK1, GSK-3β, p-GSK-3β, β-catenin, RUNX2, and OPN. Total protein was isolated from cells using a total protein extraction kit (Keygen Biotech, Nanjing, China). The protein samples were mixed with loading buffer, boiled for 5 min, separated by SDS-PAGE, and transferred to polyvinylidene fluoride membranes [29, 30]. The membranes were blocked with 5% dry skim milk in Tris-buffered saline with 0.05% (v/v) Tween-20 (TBST) for 1 h and then incubated with primary antibodies against JKAMP (NBP2-36446SS) (Novus, Littleton, USA), GAPDH (ab181602), JNK1 (ab110724), β-Catenin (ab32572), RUNX2 (ab92336) and OPN (ab8448) (Abcam, Cambridge, UK), GSK-3β (12456), or p-GSK-3β (5558) (Cell Signaling Technology, Danvers, USA) at 4 °C overnight. The membrane was washed three times with TBST and then incubated with a secondary labeled anti-rabbit or anti-mouse antibody (1:3000) for 1 h. The membrane was then washed three times with TBST and developed using an enhanced chemiluminescence detection system (Bio-Rad, Hercules, USA).
Quantitative polymerase chain reaction
Gene expression levels after 3 and 6 days of osteogenesis induction were measured by qPCR, including JNK1-associated membrane protein (Jkamp), c-Jun-N-terminal-kinase-1 (Jnk1), cadherin-associated protein, delta 1 (β-catenin), runt-related transcription factor 2 (Runx2), and osteopontin (Opn). The primer sequences are shown in Table 2. Briefly, an RNeasy Plus Mini kit (Qiagen, Hilden, Germany) and genomic DNA eliminator was used to isolate and purify total RNA from cells and then cDNA was synthesized using a PrimeScript RT kit with gDNA Eraser (Takara Bio, Tokyo, Japan). qPCR was performed using a PrimeScript RT-PCR Kit (Takara Bio) with the following amplification program: denaturation at 95 °C for 30 s and then 45 cycles of 95 °C for 5 s and 60 °C for 34 s for amplification. Gene expression from was averaged and normalized against Gapdh [31, 32].
Table 2.
Primer sequences for qPCR amplification of specific genes
| Genes | Sequence (5′ → 3′) | |
|---|---|---|
| Gapdh | Forward | GGTGAAGGTCGGTGTGAACG |
| Reverse | CTCGCTCCTGGAAGATGGTG | |
| Jkamp | Forward | CCAATGGCTGTCGATATTCAACC |
| Reverse | CTTGGGCATACCCCACATTCT | |
| Jnk1 | Forward | GTGGAATCAAGCACCTTCACT |
| Reverse | TCCTCGCCAGTCCAAAATCAA | |
| β-Catenin | Forward | GCTGCGTGGACAATGGCTACTC |
| Reverse | AGCGTCAAACTGCGTGGATGG | |
| Runx2 | Forward | GACTGTGGTTACCGTCATGGC |
| Reverse | ACTTGGTTTTTCATAACAGCGGA | |
| Opn | Forward | TCCCTCCCGGTGAAAGTGACTG |
| Reverse | TCCTCGCTCTCTGCATGGTCTC | |
Immunofluorescence staining
ASCs and DOP-ASCs were transfected with Si-Jkamp or a Jkamp plasmid. The cells were then incubated in osteogenic induction medium for 3 days. Subsequently, the cells were fixed with 4% paraformaldehyde at 4 °C for 30 min. After treatment with 0.5% Triton X-100 for 10 min to permeabilize the cell membrane, the cells were incubated with 5% sheep serum for 1 h and then with diluted primary antibodies against RUNX2 or OPN at 4 °C overnight. The samples were rewarmed for 30 min and then incubated with a fluorescent dye-conjugated anti-rabbit secondary antibody (1:500, Invitrogen, CA, USA) for 1 h at 37 °C. The nucleus and cytoskeleton of the cells were stained with DAPI and phalloidin, respectively. Cells were washed with PBS between each step. Finally, images were captured under a confocal laser microscope (Nikon, Tokyo, Japan).
Statistical analysis
SPSS 19.0 software (IBM, NY, USA) was used for statistical analysis. The t test or one-way analysis of variance (ANOVA) was applied to the experimental data to evaluate their reliability. Each experiment was repeated at least three times. The results are expressed as the mean ± standard deviation (SD). Data were considered statistically different at P < 0.05.
Results
JKAMP, the Wnt signaling pathway, and osteogenesis-related molecules are downregulated in DOP-ASCs
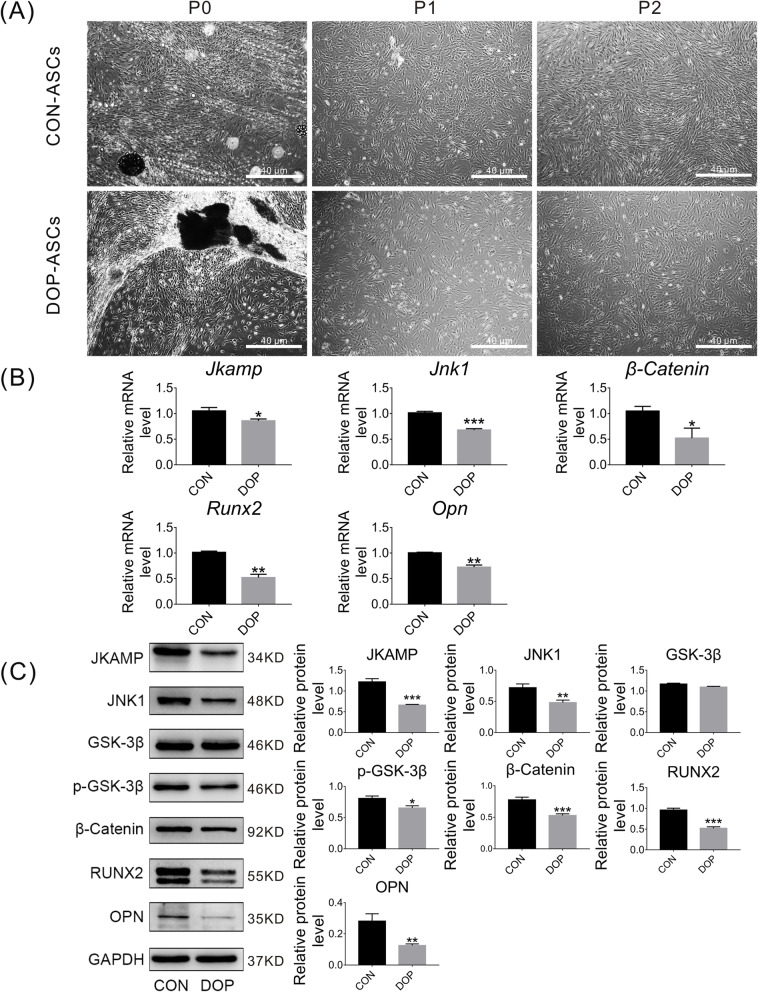
To investigate differences in expression of JKAMP, Wnt signaling pathway markers, and osteogenesis-related molecules in CON-ASCs and DOP-ASCs, we cultured CON-ASCs and DOP-ASCs to passage 2 (Fig. 1a). After 3 days of osteogenesis induction, mRNA levels of Jkamp, Jnk1, β-Catenin, Runx2, and Opn were measured by qPCR. Moreover, western blotting was used to measure the protein levels of JKAMP, JNK1, GSK-3β, p-GSK-3β, β-catenin, RUNX2, and OPN. Compared with CON-ASCs, the gene and protein levels of Wnt signaling pathway markers and downstream osteogenesis-related molecules were reduced significantly in DOP-ASCs (Fig. 1b, c). These results showed that expression of JKAMP, the Wnt signaling pathway, and the osteogenic ability were downregulated in DOP-ASCs.
Fig. 1.
JKAMP, the Wnt signaling pathway, and osteogenesis-related molecules are suppressed in DOP-ASCs. a Normal appearance passage 2 CON-ASCs and DOP-ASCs. b mRNA levels of Jkamp, Jnk1, β-catenin, Runx2, and Opn in CON-ASCs and DOP-ASCs. c Protein levels of JAKMP, Wnt signaling pathway-related molecules, and osteogenesis-related molecules in CON-ASCs and DOP-ASCs. Data represent the mean ± SD of at least three independent experiments, *P < 0.05, **P < 0.01
JKAMP silencing suppresses the Wnt signaling pathway in CON-ASCs
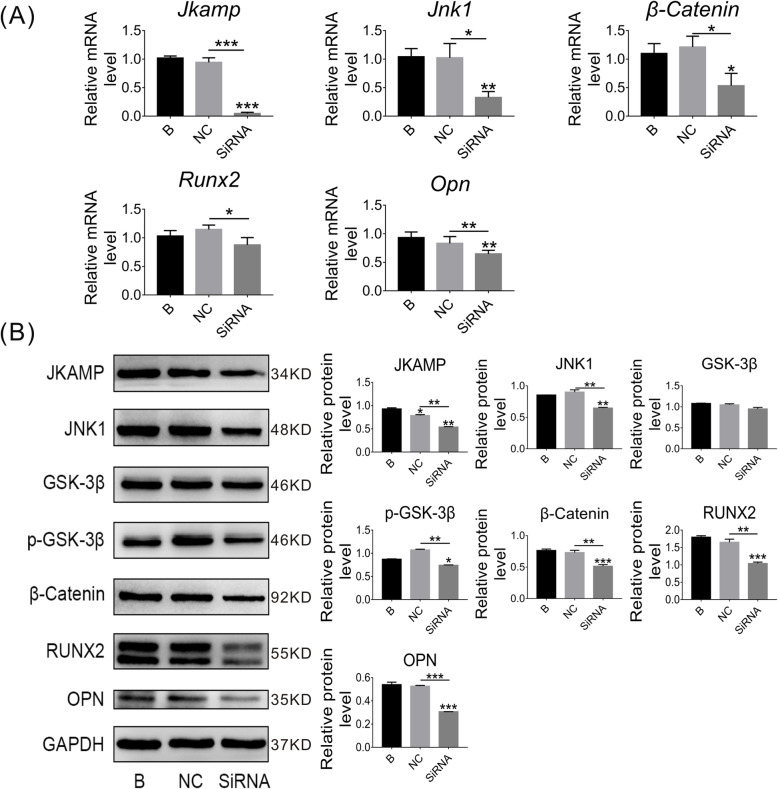
JKAMP was highly expressed in CON-ASCs. However, the relationship between JKAMP and the Wnt signaling pathway in CON-ASCs was unclear. Therefore, we silenced the JKAMP gene in CON-ASCs and then detected expression of related genes and proteins in the Wnt signaling pathway. In the siRNA group, Si-Jkamp was used to silence Jkamp. Negative control (NC) and blank (B) groups were treated with the siRNA negative control and normal medium, respectively. The mRNA and protein levels of Wnt signaling pathway markers and downstream osteogenesis-related molecules in the SiRNA group were reduced significantly compared with B and NC groups after 3 days of osteogenic induction (Fig. 2a, b). On day 6 of osteogenesis induction, we obtained similar results (Fig. 3a, b). These results demonstrated that JKAMP and the Wnt signaling pathway were positively correlated.
Fig. 2.
Si-Jkamp suppresses Wnt signaling and osteogenesis-related molecules in CON-ASCs (osteoinduction for 3 days). a, b Si-Jkamp downregulated the gene and protein levels of Wnt signaling pathway markers and osteogenesis-related molecules in CON-ASCs (osteoinduction for 3 days). Data represent the mean ± SD of at least three independent experiments, *P < 0.05, **P < 0.01
Fig. 3.
Si-Jkamp suppresses Wnt signaling and osteogenesis-related molecules in CON-ASCs (osteoinduction for 6 days). a, b mRNA and protein levels of Wnt signaling pathway markers and osteogenesis-related molecules were downregulated after si-Jkamp transfection into CON-ASCs (osteoinduction for 6 days). Data represent the mean ± SD of three or more independent experiments, *P < 0.05, **P < 0.01
Osteogenesis of CON-ASCs cells decreases after JKAMP silencing
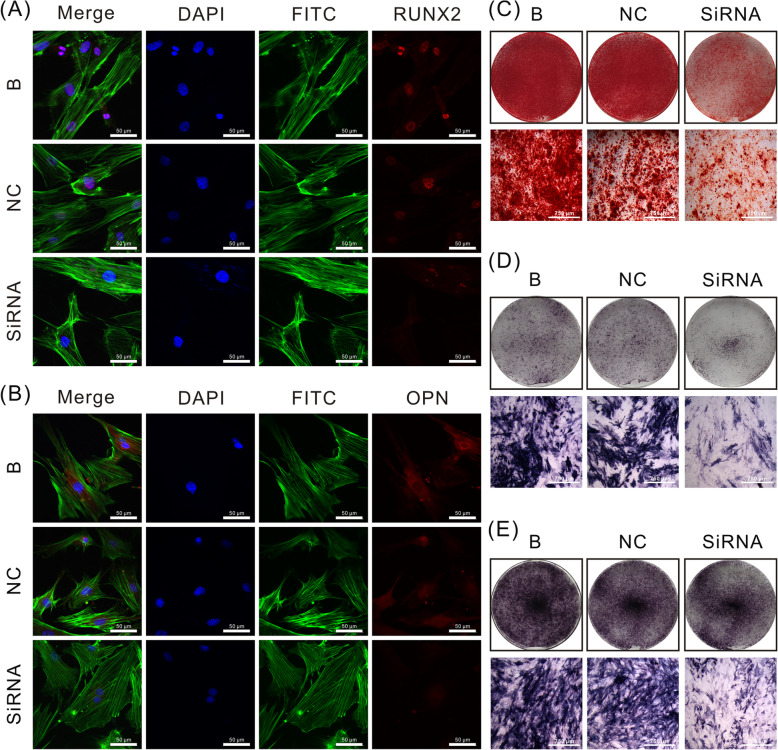
We used immunofluorescence, alizarin red, and ALP staining to determine changes in osteogenesis of CON-ASCs after Jkamp was silenced by Si-Jkamp. Immunofluorescence staining showed that OPN and RUNX2 proteins in the siRNA group were decreased after 3 days of osteogenic induction (Fig. 4a, b). After culturing the cells in osteogenic induction medium for 14 days, alizarin red staining revealed fewer mineralized nodules in the siRNA group compared with B and NC groups (Fig. 4c). ALP staining showed less alkaline phosphatase in the siRNA group compared with the other groups at 3 and 5 days after osteoinduction (Fig. 4d, e). These changes indicated that JKAMP positively regulated the osteogenic ability of CON-ASCs.
Fig. 4.
Si-Jkamp decreases the osteogenic ability of CON-ASCs. a, b Immunofluorescence staining of RUX2 and OPN proteins in CON-ASCs after 3 days after osteoinduction. c Alizarin red staining of CON-ASCs after 14 days of osteoinduction. d, e ALP staining of CON-ASCs after 3 and 5 days of osteoinduction. d Osteoinduction for 3 days. e Osteoinduction for 5 days
JKAMP overexpressing activates the Wnt signaling pathway in DOP-ASCs
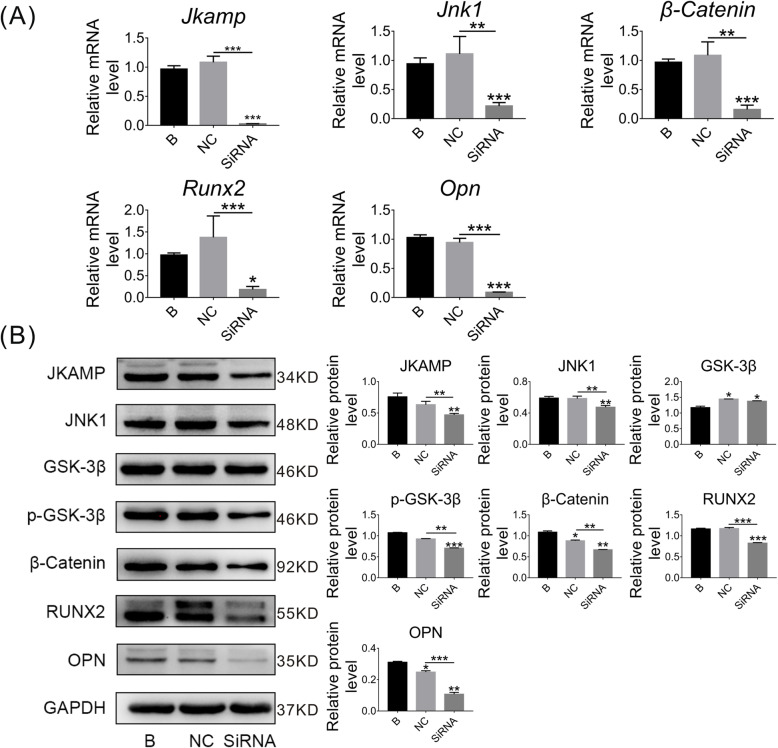
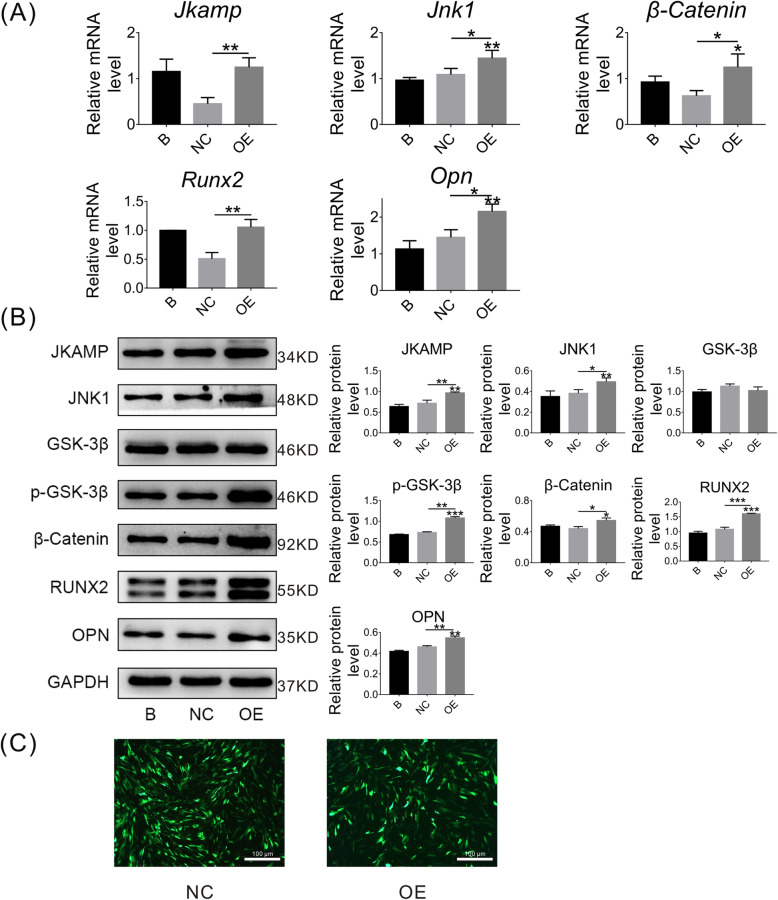
To activate the originally inactive Wnt signaling pathway in DOP-ASCs, we transfected an overexpression plasmid carrying the JKAMP gene into DOP-ASCs (Fig. 5c). Osteogenesis was then induced for 3 days after transfection. Compared with B and NC groups, DOP-ASCs transfected with the Jkamp plasmid (OE group) showed higher mRNA and protein expression of Wnt signaling pathway markers and downstream osteogenesis-related molecules (Fig. 5a, b). After osteogenesis induction for 6 days, similar results were obtained (Fig. 6a, b). These results indicated that overexpression of Jkamp activated the Wnt signaling pathway in DOP-ASCs.
Fig. 5.
Overexpression of Jkamp increases Wnt signaling and osteogenesis-related molecules in DOP-ASCs (osteoinduction for 3 days). a, b The Jkamp plasmid upregulated mRNA and protein levels of Wnt signaling pathway markers and osteogenesis-related molecules in DOP-ASCs (osteoinduction for 3 days). c DOP-ASCs cellular uptake of NC- plasmids and OE- plasmids after treated for 48 h. Data represent the mean ± SD of at least three independent experiments, *P < 0.05, **P < 0.01
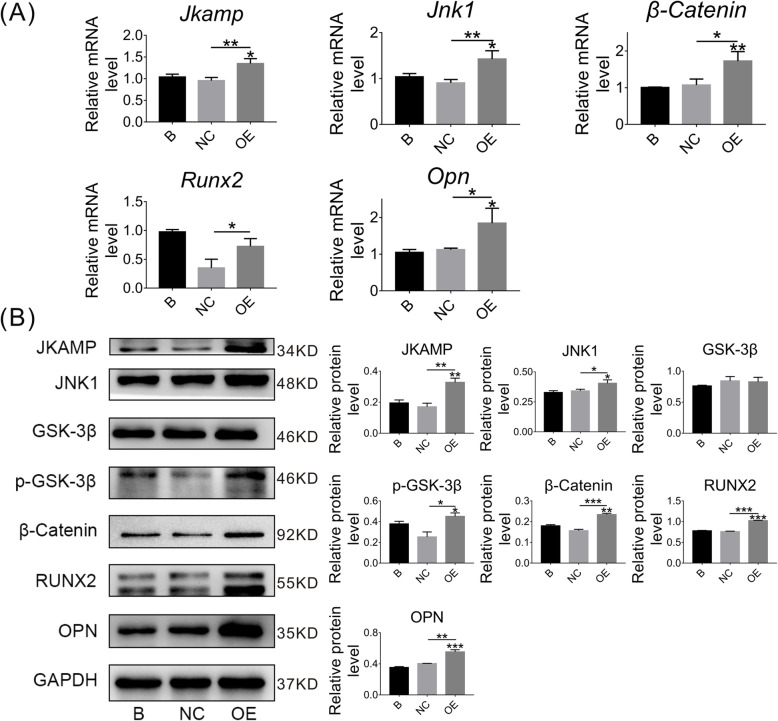
Fig. 6.
Overexpression of Jkamp increases Wnt signaling and osteogenesis-related molecules in DOP-ASCs (osteoinduction for 6 days). a, b mRNA and protein levels of Wnt signaling pathway markers and osteogenesis-related molecules were upregulated after Jkamp plasmid transfection into DOP-ASCs (osteoinduction for 6 days). Data represent the mean ± SD of at least three independent experiments, *P < 0.05, **P < 0.01
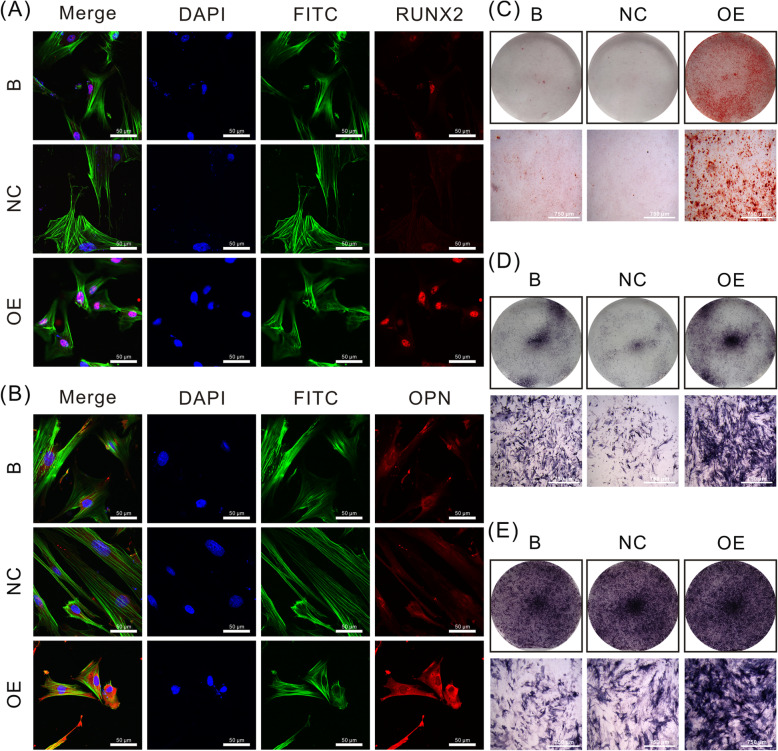
JKAMP overexpression increases osteogenesis of DOP-ASCs
Immunofluorescence, Alizarin red, and ALP staining were used to examine changes in the osteogenic ability of DOP-ASCs after Jkamp overexpression. Immunofluorescence staining showed that OPN and RUNX2 proteins in the OE group had increased compared with those in B and NC groups at 3 days after induction of osteogenesis (Fig. 7a, b). Alizarin red staining at day 14 of osteogenesis induction revealed that the OE group had more mineralized nodules than B and NC groups (Fig. 7c). ALP staining was performed at days 3 and 5 of osteogenesis induction, which showed that the OE group had more alkaline phosphatase activity than B and NC groups (Fig. 7d, e). These results showed that overexpression of Jkamp enhanced the osteogenic ability of DOP-ASCs.
Fig. 7.
Overexpression of Jkamp increases the osteogenic ability of DOP-ASCs. a, b Immunofluorescence staining of RUX2 and OPN proteins in DOP-ASC after 3 days of osteoinduction. c Alizarin red staining of DOP-ASCs after 14 days of osteoinduction. d, e ALP staining of DOP-ASCs after 3 and 5 days of osteoinduction. c Osteoinduction for 3 days. d Osteoinduction for 5 days
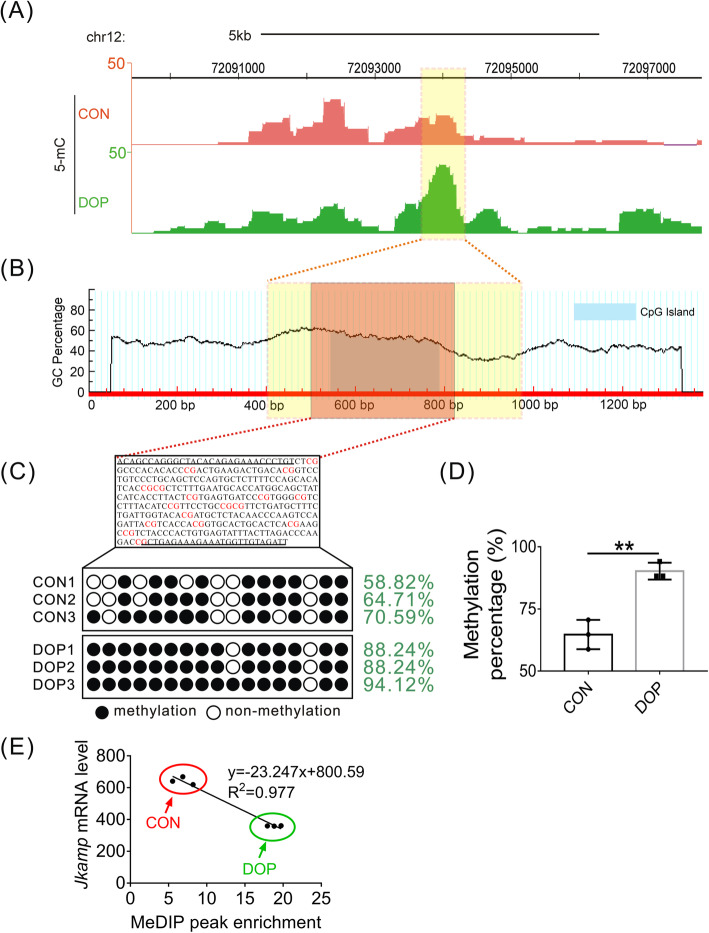
Increased intragenic methylation level of JKAMP in DOP-ASCs
To explore the reasons for the decrease of JKAMP expression in DOP-ASCs, we performed MeDIP sequencing and BSP analyses of JKAMP in CON-ASCs and DOP-ASCs. Interestingly, MeDIP sequencing analysis revealed that the methylation peak of the JKAMP gene in DOP-ASCs was significantly higher than that in CON-ASCs, especially near exon 3/4/5/6/7 regions. However, there was no such difference in the promoter region (Fig. 8a). A recent study confirmed the existence of putative promoters near exons. Similar to promoter DNA methylation, putative promoter DNA methylation also individually regulates the expression of exons [33]. We hypothesized that expression of JKAMP was influenced by the CpG island (CGI) located at the gene body. Therefore, we examined the representative exon 4 region (gene coordinates: chr12, 72093937–72094143). Through calculation of Meth Primer software, we found a large amount of CGI enrichment in the region near exon 4 (genomic coordinate: chr12, 72093357–72094707) (Fig. 8b). BSP results also demonstrated that the degree of methylation of the genomic coordinate chr12 72093857–72094207 in DOP-ASCs was higher than that in CON-ASCs (Fig. 8c, d). Additionally, combined with the results of mRNA expression profiling and MeDIP sequencing, we found a strongly negative relationship between JKAMP expression and the methylation level of the corresponding CGI (Fig. 8e). Taken together, these observations indicated that the decrease in expression of JKAMP in DOP-ASCs was related to increased intragenic methylation.
Fig. 8.
Increased methylation level of JKAMP in DOP-ASCs. a MeDIP sequencing showed that the intragenic methylation peaks of the JKAMP gene in DOP-ASCs were significantly higher than those in CON-ASCs. b Meth Primer software analysis showed that a large amount of CGI was enriched in the region near JKAMP exon 4 (genomic coordinates: chr12, 72093357–72094707). c, d BSP confirmed that the methylation degree of CON-ASCs was lower than that of matched DOP-ASCs in the region near exon 4 of JKAMP (genomic coordinates: chr12, 72093857–72094207). e Correlation between Jkamp mRNA levels and MeDIP peak enrichment near exon 4 of JKAMP (r2 = 0.977). Each data point corresponds to a high-throughput sequencing result, and the data were analyzed for correlation. Data represent the mean ± SD of at least three independent experiments, *P < 0.05, **P < 0.01
Discussion
The hyperglycemic microenvironment caused by diabetes exacerbates damage of osteoblasts. Chronic inflammation in diabetic patients inhibits the activity of stem cells and increases calcium excretion [1, 34, 35]. Additionally, the interaction of hyperglycemia with parathyroid hormone and the vitamin D system weakens bone turnover in diabetic patients and reduces osteocalcin produced by osteoblasts [36, 37]. The diabetic microenvironment makes bone tissue more susceptible to accumulation of microdamage, which leads to increased bone fragility and diabetic osteoporosis [2, 38, 39]. Recent studies have shown that DNA methylation is involved in multiple biological processes of stem cells. Zhang et al. found that transplantation of BMSCs modified by DNA methylation rescues the osteogenic ability in lupus mice [40]. Vera et al. reported that DNA methylation in a hyperglycemic environment changes the expression of CXCR4 receptors and migration of CD34+ stem cells [41]. However, at present, there are no reports of epigenetic changes in stem cells, which cause diabetic bone disease [38]. In this study, we established a mouse model of DOP and revealed the molecular mechanism by which JKAMP affects osteogenesis of DOP-ASCs via intragenic DNA methylation.
JKAMP is a seven-transmembrane protein that is mainly located in the plasma membrane of cells [42]. It provides cells with apoptosis-related signals and affects the activity and duration of JNK1 through competition with mitogen-activated protein kinase phosphatase 5 [20]. Sabapathy et al. reported that the main function of JNK1 is activation of c-Jun after stress, and JNK2 appears to regulate the stability of c-Jun under non-stressed conditions [43]. Boutros et al. reported that JNK1 interacts with Dsh to regulate the Wnt-PCP Pathway [44]. Another study has shown that JNK1 also cooperates with β-catenin to participate in the canonical Wnt pathway [22]. However, there is no study that has directly shown the relationship between JKAMP and the Wnt signaling pathway. In CON-ASCs, we found that silencing Jkamp decreased the expression levels of β-catenin and p-GSK-3β by reducing expression of JNK1. Furthermore, overexpression of Jkamp increased the expression levels of β-catenin and p-GSK-3β in DOP-ASCs by upregulating the expression of JNK1. In general, our results demonstrated that JKAMP positively regulated the Wnt signaling pathway through JNK1.
The Wnt/β-catenin signaling pathway plays an indispensable role in osteogenic differentiation and bone development of stem cells [45, 46]. Ubiquitination and degradation of β-catenin are regulated by the balance of GSK-3β and p-GSK-3β [22]. β-Catenin interacts with TCF/LEF-1 or other transcription coactivators in the nucleus and induces the expression of osteogenesis-related molecules to regulate osteogenesis [24, 25, 47]. Compared with CON-ASCs, DOP-ASCs exhibited relatively downregulated expression of β-catenin, p-GSK-3β, RUNX2, and OPN. These results indicated that the Wnt signaling pathway in DOP-ASCs was inhibited with a subsequent decline in osteogenesis. After silencing Jkamp, expression of Wnt signaling pathway markers in CON-ASCs was decreased, which decreased osteogenesis-related molecules over time. Additionally, after overexpressing Jkamp in DOP-ASCs, the levels of osteogenesis-related molecules were rescued by activating the Wnt signaling pathway. Interestingly, ALP and Alizarin red staining indicated that inhibiting the expression of JKAMP reduced the early and late osteogenic differentiation capacities of CON-ASCs. Conversely, overexpression of JKAMP rescued the early and late osteogenic differentiation capacities of DOP-ASCs. Considering that the changes in the osteogenesis-related molecules via Wnt signaling pathway after silencing or overexpression of Jkamp are consistent with the variations in ALP and Alizarin red staining, we have reason to believe that the osteogenic ability of ASCs was regulated by Jkamp and Wnt signaling pathway. Therefore, JKAMP inhibits the osteogenic capacity of DOP-ASCs by modulating the Wnt signaling pathway.
DNA methylation is covalent addition of a methyl group to the C5 position of a cytosine pyrimidine ring [48]. Generally, in a CpG dinucleotide, hypermethylation of CpG sites in a promoter leads to transcriptional silencing [49]. As an epigenetic marker, the main functions of DNA methylation include gene silencing and maintenance of genomic integrity. Additionally, it plays a vital role in genomic imprinting and suppression of repeated sequences [50–53]. Zhao et al. reported that promoter DNA methylation regulates the osteogenic differentiation ability of BMSCs from osteoporotic mice [54]. Zhang et al. showed that DNA methylation-related enzymes (DNMT1 and DNMT3a) are significantly upregulated in AGE-induced ASCs of diabetic models, which reduces osteogenesis [19]. In recent years, some studies have found that the role of intragenic methylation is seriously underestimated. In fact, intragenic and promoter methylations are both involved in transcriptional regulation of HIV-1 and reduce transcription efficiency of the virus [55]. Ma et al. found that putative promoters may exist in the gene bodies of certain cancer-related genes. DNA methylation of CGI near these putative promoters also silences these genes [32]. Mathios et al. found that intragenic DNA methylation of glioblastoma may also be a regulatory mechanism of ZMIZ1 gene transcription [56]. Interestingly, in our study, we found that the promoter of JKAMP showed no obvious difference in the DNA methylation degree between CON-ASCs and DOP-ASCs. However, the methylation degree of the CON-ASCs group was lower than that of the matched DOP-ASC group in the gene body, especially near exon 3/4/5/6/7 regions. Additionally, correlation analysis determined that the change in the JKAMP mRNA level was related to these intragenic methylations of the corresponding CGIs. These results suggest that the decreased expression of JKAMP in DOP-ASCs was related to intragenic DNA methylations rather than promoter DNA methylation.
Long non-coding RNAs (LncRNAs) are generally recognized as non-coding RNA molecules greater than 200 nucleotides in length [57]. Recent studies have postulated that they have a vital function in a series of important cellular processes [58, 59]. Moreover, growing evidence is supporting the involvement of LncRNAs in influencing gene expression by regulating DNA methylation of specific CGI [60]. Therefore, how LncRNAs interact with intragenic DNA methylations to affect the expression of JKAMP is a potential epigenetic mechanism that needs us to entail. And this research provides an epigenetic explanation for the reduced osteogenic ability of DOP-ASCs and a potential therapeutic target for the prevention and treatment of diabetic osteoporosis.
Conclusions
This study shows that decreased expression of JKAMP reduces the osteogenic potential of DOP-ASCs by inhibiting the Wnt signaling pathway. Overexpression of JKAMP effectively rescues the decline in osteogenesis of DOP-ASCs. Furthermore, intragenic methylation of JKAMP in DOP-ASCs has a strong negative correlation with JKAMP expression, which provides a possible research route for bone tissue regeneration of diabetic osteoporosis.
Acknowledgements
Not applicable.
Abbreviations
- DOP
Diabetic osteoporosis
- DM
Diabetes mellitus
- ASCs
Adipose-derived stem cells
- CON
Control
- BSP
Bisulfite-specific PCR
- PCR
Quantitative real-time polymerase chain reaction
- STZ
Streptozotocin
- ALP
Alkaline phosphatase
- Jnk1
c-Jun-N-terminal-kinase-1
- JKAMP
JNK1\MAPK8-associated membrane protein
- β-Catenin
Cadherin-associated protein
- Runx2
Runt-related transcription factor 2
- Opn
Osteopontin
- GAPDH
Glyceraldehyde phosphate dehydrogenase
- OE
Over-expressed
- B
Blank
- NC
Negative control
- GSK-3β
Glycogen synthase kinase 3 beta
- DNMT1
DNA methyltransferases 1
- DNMT3a
DNA methyltransferases 3a
- CGI
CpG island
- LncRNA
Long non-coding RNA
Authors’ contributions
All authors have made important contributions to this research. Shuanglin Peng conducted in vitro experiments, executed the analysis of the data, and wrote the main manuscript. Sirong Shi conducted in vitro experiments and wrote the main manuscript. Gang Tao reviewed and revised the manuscript. Yanjing Li and Lang Wang collected the data. Dexuan Xiao and Qing He established the animal model of diabetic osteoporosis. Xiaoxiao Cai designed the experimental project, analyzed data, and revised the manuscript. Jingang Xiao conceived and designed the experiment, analyzed data, revised the manuscript, and provided funding. All authors have read and approved the final manuscript.
Funding
This work was funded by the National Natural Science Foundation of China (81870746, 81771125), and the Joint project of Luzhou Municipal People’s Government and Southwest Medical University (2020LZXNYDZ09).
Availability of data and materials
The datasets generated or analyzed during the current study can be obtained from the corresponding author in accordance with reasonable requirements.
Ethics approval and consent to participate
The Ethics Committee of Southwest Medical University reviewed and approved the experimental animal procedures, and we conducted animal care and anesthesia in accordance with the guidelines of the Care and Use of Laboratory Animals (Ministry of Science and Technology of China, 2006).
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Xiaoxiao Cai, Email: dentistcai@hotmail.com.
Jingang Xiao, Email: drxiaojingang@163.com.
References
- 1.Cho J, Scragg R, Petrov MS. Risk of mortality and hospitalization after post-pancreatitis diabetes mellitus vs type 2 diabetes mellitus: a population-based matched cohort study. Am J Gastroenterol. 2019;114(5):804–812. doi: 10.14309/ajg.0000000000000225. [DOI] [PubMed] [Google Scholar]
- 2.Gilbert MP, Pratley RE. The impact of diabetes and diabetes medications on bone health. Endocr Rev. 2015;36(2):194–213. doi: 10.1210/er.2012-1042. [DOI] [PubMed] [Google Scholar]
- 3.Shanbhogue VV, Hansen S, Frost M, Jorgensen NR, Hermann AP, Henriksen JE, Brixen K. Bone geometry, volumetric density, microarchitecture, and estimated bone strength assessed by HR-pQCT in adult patients with type 1 diabetes mellitus. J Bone Miner Res. 2015;30(12):2188–2199. doi: 10.1002/jbmr.2573. [DOI] [PubMed] [Google Scholar]
- 4.Karim L, Bouxsein ML. Effect of type 2 diabetes-related non-enzymatic glycation on bone biomechanical properties. Bone. 2016;82:21–27. doi: 10.1016/j.bone.2015.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Khosla S, Shane E. A crisis in the treatment of osteoporosis. J Bone Miner Res. 2016;31(8):1485–1487. doi: 10.1002/jbmr.2888. [DOI] [PubMed] [Google Scholar]
- 6.Rachner TD, Khosla S, Hofbauer LC. Osteoporosis: now and the future. Lancet. 2011;377(9773):1276–1287. doi: 10.1016/S0140-6736(10)62349-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yamamoto M, Sugimoto T. Advanced glycation end products, diabetes, and bone strength. Curr Osteoporos Rep. 2016;14(6):320–326. doi: 10.1007/s11914-016-0332-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Agarwal R, Garcia AJ. Biomaterial strategies for engineering implants for enhanced osseointegration and bone repair. Adv Drug Deliv Rev. 2015;94:53–62. doi: 10.1016/j.addr.2015.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cox SC, Thornby JA, Gibbons GJ, Williams MA, Mallick KK. 3D printing of porous hydroxyapatite scaffolds intended for use in bone tissue engineering applications. Mater Sci Eng C Mater Biol Appl. 2015;47:237–247. doi: 10.1016/j.msec.2014.11.024. [DOI] [PubMed] [Google Scholar]
- 10.Lin S, Zhang Q, Shao X, Zhang T, Xue C, Shi S, Zhao D, Lin Y. IGF-1 promotes angiogenesis in endothelial cells/adipose-derived stem cells co-culture system with activation of PI3K/Akt signal pathway. Cell Prolif. 2017;50(6):e12390. doi: 10.1111/cpr.12390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hlady RA, Zhao X, Pan X, Yang JD, Ahmed F, Antwi SO, Giama NH, Patel T, Roberts LR, Liu C, Robertson KD. Genome-wide discovery and validation of diagnostic DNA methylation-based biomarkers for hepatocellular cancer detection in circulating cell free DNA. Theranostics. 2019;9(24):7239–7250. doi: 10.7150/thno.35573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hu X, Tang J, Hu X, Bao P, Deng W, Wu J, Liang Y, Chen Z, Gao L, Tang Y. Silencing of long non-coding RNA HOTTIP reduces inflammation in rheumatoid arthritis by demethylation of SFRP1. Mol Ther Nucleic Acids. 2019;19:468–481. doi: 10.1016/j.omtn.2019.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 13.Long J, Chen P, Lin J, Bai Y, Yang X, Bian J, Lin Y, Wang D, Yang X, Zheng Y, Sang X, Zhao H. DNA methylation-driven genes for constructing diagnostic, prognostic, and recurrence models for hepatocellular carcinoma. Theranostics. 2019;9(24):7251–7267. doi: 10.7150/thno.31155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Godler DE, Amor DJ. DNA methylation analysis for screening and diagnostic testing in neurodevelopmental disorders. Essays Biochem. 2019;63(6):785–795. doi: 10.1042/EBC20190056. [DOI] [PubMed] [Google Scholar]
- 15.Chen Q, Yan W, Duan E. Epigenetic inheritance of acquired traits through sperm RNAs and sperm RNA modifications. Nat Rev Genet. 2016;17(12):733–743. doi: 10.1038/nrg.2016.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tang WW, Kobayashi T, Irie N, Dietmann S, Surani MA. Specification and epigenetic programming of the human germ line. Nat Rev Genet. 2016;17(10):585–600. doi: 10.1038/nrg.2016.88. [DOI] [PubMed] [Google Scholar]
- 17.Vrtacnik P, Marc J, Ostanek B. Hypoxia mimetic deferoxamine influences the expression of histone acetylation- and DNA methylation-associated genes in osteoblasts. Connect Tissue Res. 2015;56(3):228–235. doi: 10.3109/03008207.2015.1017573. [DOI] [PubMed] [Google Scholar]
- 18.Ghayor C, Weber FE. Epigenetic regulation of bone remodeling and its impacts in osteoporosis. Int J Mol Sci. 2016;17(9):e1446. doi: 10.3390/ijms17091446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang M, Li Y, Rao P, Huang K, Luo D, Cai X, Xiao J. Blockade of receptors of advanced glycation end products ameliorates diabetic osteogenesis of adipose-derived stem cells through DNA methylation and Wnt signalling pathway. Cell Prolif. 2018;51(5):e12471. doi: 10.1111/cpr.12471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kadoya T, Khurana A, Tcherpakov M, Bromberg KD, Didier C, Broday L, Asahara T, Bhoumik A, Ronai Z. JAMP, a Jun N-terminal kinase 1 (JNK1)-associated membrane protein, regulates duration of JNK activity. Mol Cell Biol. 2005;25(19):8619–8630. doi: 10.1128/MCB.25.19.8619-8630.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Davis R. Signal transduction by the JNK group of MAP kinases. Cell. 2000;103(2):239–252. doi: 10.1016/S0092-8674(00)00116-1. [DOI] [PubMed] [Google Scholar]
- 22.van der Velden JL, Guala AS, Leggett SE, Sluimer J, Badura EC, Janssen-Heininger YM. Induction of a mesenchymal expression program in lung epithelial cells by wingless protein (Wnt)/beta-catenin requires the presence of c-Jun N-terminal kinase-1 (JNK1) Am J Respir Cell Mol Biol. 2012;47(3):306–314. doi: 10.1165/rcmb.2011-0297OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Giuliani C. The flavonoid quercetin induces AP-1 activation in FRTL-5 thyroid cells. Antioxidants (Basel). 2019;8(5):e112. doi: 10.3390/antiox8050112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu AX, Xu ML, Yao P, Kwan KK, Liu YX, Duan R, Dong TT, Ko RK, Tsim KW. Corylin, a flavonoid derived from Psoralea Fructus, induces osteoblastic differentiation via estrogen and Wnt/beta-catenin signaling pathways. FASEB J. 2020. 10.1096/fj.201902319RRR. [DOI] [PubMed]
- 25.Xia K, Cen X, Yu L, Huang X, Sun W, Zhao Z, Liu J. Long noncoding RNA expression profiles during the NEL-like 1 protein–induced osteogenic differentiation. J Cell Physiol. 2020. 10.1002/jcp.29526. [DOI] [PubMed]
- 26.Tao Y, Kang B, Petkovich DA, Bhandari YR, In J, Stein-O'Brien G, Kong X, Xie W, Zachos N, Maegawa S, Vaidya H, Brown S, Chiu Yen RW, Shao X, Thakor J, Lu Z, Cai Y, Zhang Y, Mallona I, Peinado MA, Zahnow CA, Ahuja N, Fertig E, Issa JP, Baylin SB, Easwaran H. Aging-like spontaneous epigenetic silencing facilitates Wnt activation, stemness, and Braf(V600E)-induced tumorigenesis. Cancer Cell. 2019;35(2):315–328. doi: 10.1016/j.ccell.2019.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu R, Zhang C, Shin DY, Kim J-M, Lalani S, Li N, Yang Y-S, Liu Y, Eiseman M, Davis RJ, Shim J-H, Greenblatt MB. C-Jun N-terminal kinases (JNKs) are critical mediators of osteoblast activity In vivo. J Bone Miner Res. 2017;32(9):1811–1815. doi: 10.1002/jbmr.3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ke D, Zhu Y, Zheng W, Fu X, Chen J, Han J. Autophagy mediated by JNK1 resists apoptosis through TRAF3 degradation in osteoclastogenesis. Biochimie. 2019;167:217–227. doi: 10.1016/j.biochi.2019.10.008. [DOI] [PubMed] [Google Scholar]
- 29.Ma W, Shao X, Zhao D, Li Q, Liu M, Zhou T, Xie X, Mao C, Zhang Y, Lin Y. Self-assembled tetrahedral DNA nanostructures promote neural stem cell proliferation and neuronal differentiation. ACS Appl Mater Interfaces. 2018;10(9):7892–7900. doi: 10.1021/acsami.8b00833. [DOI] [PubMed] [Google Scholar]
- 30.Liu N, Zhang X, Li N, Zhou M, Zhang T, Li S, Cai X, Ji P, Lin Y. Tetrahedral framework nucleic acids promote corneal epithelial wound healing in vitro and in vivo. Small. 2019;15(31):e1901907. doi: 10.1002/smll.201901907. [DOI] [PubMed] [Google Scholar]
- 31.Zhan Y, Ma W, Zhang Y, Mao C, Shao X, Xie X, Wang F, Liu X, Li Q, Lin Y. DNA-based nanomedicine with targeting and enhancement of therapeutic efficacy of breast cancer cells. ACS Appl Mater Interfaces. 2019;11(17):15354–15365. doi: 10.1021/acsami.9b03449. [DOI] [PubMed] [Google Scholar]
- 32.Xie X, Shao X, Ma W, Zhao D, Shi S, Li Q, Lin Y. Overcoming drug-resistant lung cancer by paclitaxel loaded tetrahedral DNA nanostructures. Nanoscale. 2018;10(12):5457–5465. doi: 10.1039/C7NR09692E. [DOI] [PubMed] [Google Scholar]
- 33.Ma L, Muhammad T, Wang H, Du G, Sakhawat A, Wei Y, Ali Khan A, Cong X, Huang Y. Putative promoters within gene bodies control exon expression via TET1-mediated H3K36 methylation. J Cell Physiol. 2020;235(10):6711–6724. doi: 10.1002/jcp.29566. [DOI] [PubMed] [Google Scholar]
- 34.Sundbom M, Franzen S, Ottosson J, Svensson AM. Superior socioeconomic status in patients with type 2 diabetes having gastric bypass surgery: a case-control analysis of 10 642 individuals. BMJ Open Diab Res Care. 2020;8(1):e000989. doi: 10.1136/bmjdrc-2019-000989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dong X, Bi L, He S, Meng G, Wei B, Jia S, Liu J. FFAs-ROS-ERK/P38 pathway plays a key role in adipocyte lipotoxicity on osteoblasts in co-culture. Biochimie. 2014;101:123–131. doi: 10.1016/j.biochi.2014.01.002. [DOI] [PubMed] [Google Scholar]
- 36.Hofbauer LC, Brueck CC, Singh SK, Dobnig H. Osteoporosis in patients with diabetes mellitus. J Bone Miner Res. 2007;22(9):1317–1328. doi: 10.1359/jbmr.070510. [DOI] [PubMed] [Google Scholar]
- 37.Madsen JOB, Jorgensen NR, Pociot F, Johannesen J. Bone turnover markers in children and adolescents with type 1 diabetes-a systematic review. Pediatr Diabetes. 2019;20(5):510–522. doi: 10.1111/pedi.12853. [DOI] [PubMed] [Google Scholar]
- 38.Murray CE, Coleman CM. Impact of diabetes mellitus on bone health. Int J Mol Sci. 2019;20(19):e4837. doi: 10.3390/ijms20194873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen S, Jia L, Zhang S, Zheng Y, Zhou Y. DEPTOR regulates osteogenic differentiation via inhibiting MEG3-mediated activation of BMP4 signaling and is involved in osteoporosis. Stem Cell Res Ther. 2018;9(1):185. doi: 10.1186/s13287-018-0935-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu S, Liu D, Chen C, Hamamura K, Moshaverinia A, Yang R, Liu Y, Jin Y, Shi S. MSC transplantation improves osteopenia via epigenetic regulation of notch signaling in lupus. Cell Metab. 2015;22(4):606–618. doi: 10.1016/j.cmet.2015.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vigorelli V, Resta J, Bianchessi V, Lauri A, Bassetti B, Agrifoglio M, Pesce M, Polvani G, Bonalumi G, Cavallotti L, Alamanni F, Genovese S, Pompilio G, Vinci MC, Abnormal DNA. Methylation induced by hyperglycemia reduces CXCR 4 gene expression in CD 34(+) stem cells. J Am Heart Assoc. 2019;8(9):e010012. doi: 10.1161/JAHA.118.010012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roy SJ, Glazkova I, Frechette L, Iorio-Morin C, Binda C, Petrin D, Trieu P, Robitaille M, Angers S, Hebert TE, Parent JL. Novel, gel-free proteomics approach identifies RNF5 and JAMP as modulators of GPCR stability. Mol Endocrinol. 2013;27(8):1245–1266. doi: 10.1210/me.2013-1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sabapathy K, Hochedlinger K, Nam SY, Bauer A, Karin M, Wagner EF. Distinct roles for JNK1 and JNK2 in regulating JNK activity and c-Jun-dependent cell proliferation. Mol Cell. 2004;15(5):713–725. doi: 10.1016/j.molcel.2004.08.028. [DOI] [PubMed] [Google Scholar]
- 44.Boutros M, Paricio N, Strutt D, Mlodzik M. Dishevelled activates JNK and discriminates between JNK pathways in planar polarity and wingless signaling. Cell. 1998;94(1):109–118. doi: 10.1016/S0092-8674(00)81226-X. [DOI] [PubMed] [Google Scholar]
- 45.Baron R, Kneissel M. WNT signaling in bone homeostasis and disease: from human mutations to treatments. Nat Med. 2013;19(2):179–192. doi: 10.1038/nm.3074. [DOI] [PubMed] [Google Scholar]
- 46.Jiang T, Xia C, Chen X, Hu Y, Wang Y, Wu J, Chen S, Gao Y. Melatonin promotes the BMP9-induced osteogenic differentiation of mesenchymal stem cells by activating the AMPK/beta-catenin signalling pathway. Stem Cell Res Ther. 2019;10(1):408. doi: 10.1186/s13287-019-1511-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shen G, Ren H, Shang Q, Zhao W, Zhang Z, Yu X, Tang K, Tang J, Yang Z, Liang JX. Foxf1 knockdown promotes BMSC osteogenesis in part by activating the Wnt/beta-catenin signalling pathway and prevents ovariectomy-induced bone loss. EBioMedicine. 2020;52:102626. doi: 10.1016/j.ebiom.2020.102626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen D, Chao DL, Rocha L, Kolar M, Nguyen Huu VA, Krawczyk M, Dasyani M, Wang T, Jafari M, Jabari M, Ross KD, Saghatelian A, Hamilton BA, Zhang K, Skowronska-Krawczyk D. The lipid elongation enzyme ELOVL2 is a molecular regulator of aging in the retina. Aging Cell. 2020;19(2):e13100. doi: 10.1111/acel.13100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Morgan AE, Davies TJ, Mc Auley MT. The role of DNA methylation in ageing and cancer. Proc Nutr Soc. 2018;77(4):412–422. doi: 10.1017/S0029665118000150. [DOI] [PubMed] [Google Scholar]
- 50.Delaval K, Feil R. Epigenetic regulation of mammalian genomic imprinting. Curr Opin Genet Dev. 2004;14(2):188–195. doi: 10.1016/j.gde.2004.01.005. [DOI] [PubMed] [Google Scholar]
- 51.Goll MG, Bestor TH. Eukaryotic cytosine methyltransferases. Annu Rev Biochem. 2005;74:481–514. doi: 10.1146/annurev.biochem.74.010904.153721. [DOI] [PubMed] [Google Scholar]
- 52.Suzuki MM, Bird A. DNA methylation landscapes: provocative insights from epigenomics. Nat Rev Genet. 2008;9(6):465–476. doi: 10.1038/nrg2341. [DOI] [PubMed] [Google Scholar]
- 53.Wu Y, Chen X, Zhao Y, Wang Y, Li Y, Xiang C. Genome-wide DNA methylation and hydroxymethylation analysis reveal human menstrual blood-derived stem cells inhibit hepatocellular carcinoma growth through oncogenic pathway suppression via regulating 5-hmC in enhancer elements. Stem Cell Res Ther. 2019;10(1):151. doi: 10.1186/s13287-019-1243-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhao QH, Wang SG, Liu SX, Li JP, Zhang YX, Sun ZY, Fan QM, Tian JW. PPARgamma forms a bridge between DNA methylation and histone acetylation at the C/EBPalpha gene promoter to regulate the balance between osteogenesis and adipogenesis of bone marrow stromal cells. FEBS J. 2013;280(22):5801–5814. doi: 10.1111/febs.12500. [DOI] [PubMed] [Google Scholar]
- 55.Kint S, Trypsteen W, De Spiegelaere W, Malatinkova E, Kinloch-de Loes S, De Meyer T, Van Criekinge W, Vandekerckhove L. Underestimated effect of intragenic HIV-1 DNA methylation on viral transcription in infected individuals. Clin Epigenetics. 2020;12(1):36. doi: 10.1186/s13148-020-00829-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mathios D, Hwang T, Xia Y, Phallen J, Rui Y, See AP, Maxwell R, Belcaid Z, Casaos J, Burger PC, McDonald KL, Gallia GL, Cope L, Kai M, Brem H, Pardoll DM, Ha P, Green JJ, Velculescu VE, Bettegowda C, Park CK, Lim M. Genome-wide investigation of intragenic DNA methylation identifies ZMIZ1 gene as a prognostic marker in glioblastoma and multiple cancer types. Int J Cancer. 2019;145(12):3425–3435. doi: 10.1002/ijc.32587. [DOI] [PubMed] [Google Scholar]
- 57.Kamel LM, Atef DM, Mackawy AMH, Shalaby SM, Abdelraheim N. Circulating long non-coding RNA GAS5 and SOX2OT as potential biomarkers for diagnosis and prognosis of non-small cell lung cancer. Biotechnol Appl Biochem. 2019;66(4):634–642. doi: 10.1002/bab.1764. [DOI] [PubMed] [Google Scholar]
- 58.Quinn JJ, Chang HY. Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet. 2016;17(1):47–62. doi: 10.1038/nrg.2015.10. [DOI] [PubMed] [Google Scholar]
- 59.Wang C, Jiang X, Li X, Song S, Meng Q, Wang L, Lu Y, Xin X, Pu H, Gui X, Li T, Lu D. Long noncoding RNA HULC accelerates the growth of human liver cancer stem cells by upregulating CyclinD1 through miR675-PKM2 pathway via autophagy. Stem Cell Res Ther. 2020;11(1):8. doi: 10.1186/s13287-019-1528-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Qi X, Yu XJ, Wang XM, Song TN, Zhang J, Guo XZ, Li GJ, Shao M. Knockdown of KCNQ1OT1 suppresses cell invasion and sensitizes osteosarcoma cells to CDDP by upregulating DNMT1-mediated Kcnq1 expression. Mol Ther Nucleic Acids. 2019;17:804–818. doi: 10.1016/j.omtn.2019.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated or analyzed during the current study can be obtained from the corresponding author in accordance with reasonable requirements.