Abstract
Knowledge of viral load is essential to formulate strategies for antiviral treatment, vaccination, and epidemiological control of COVID-19. Moreover, identification of patients with high viral loads can also be useful to understand risk factors such as age, comorbidities, severity of symptoms and hypoxia, to decide on the need for hospitalization. Several ongoing studies are analyzing viral load in different types of samples and evaluating its relationship with clinical outcomes and viral transmission pathways. However, in a great number of emerging studies, cycle threshold (Ct) values alone are often used as viral load indicators, which may be a mistake. In this study, we compared tracheal aspirate with nasopharyngeal swab samples obtained from critically ill COVID-19 patients and here we report how the raw Ct can lead to misinterpretation of results. Furthermore, based on analysis of nasopharyngeal swab samples we propose a method to reduce evaluation errors that could occur from using raw Ct data. Based on these findings, we show the impact that normalization of Ct values has on interpretation of SARS-CoV-2 viral load from different biological samples.
Keywords: SARS-CoV-2, COVID-19, Viral load, Clinical outcomes, Ct values
1. Introduction
Besides investigating risk factors for mortality in hospitalized patients with coronavirus disease 2019 (COVID-19), such as older age, obesity, comorbidities, C-reactive protein (CRP) and inflammatory cytokines, the impact of SARS-CoV-2 viral load in clinical outcomes also is extremely important (Dietz and Santos-Burgoa, 2020; Huang et al., 2020; Wang et al., 2020b). Moreover, reliable data on viral load are needed to guide antiviral treatment, infection control and epidemiological metrics. Several types of biological samples have been analyzed for the presence of SARS-CoV-2 viral RNA, such as nasal swabs, throat swabs, sputum, rectal swabs, vaginal swabs, blood, placenta, human breast milk and urine, among others (Vivanti et al., 2020; Wang et al., 2020c). Although in most of these samples, the SARS-CoV-2 RNA was detectable, the pattern of viral load in these samples is not yet clear.
The differential expression of SARS-CoV-2 viral RNA among patient groups is a current topic of interest, and viral load has been associated with a diversity of outcomes (Heald-Sargent et al., 2020; Liu et al., 2020; Magleby et al., 2020; Vivanti et al., 2020). The gold standard method to detect SARS-CoV-2 infection is reverse-transcription quantitative PCR (RT-qPCR), which is based on the amplification of regions of viral RNA that have been reverse transcribed in each cycle of the reaction (WHO, 2020). The earlier the cycle when the fluorescent signal is detectable above the threshold, known as cycle threshold (Ct), indicates a higher concentration of the target gene in the sample. In a great number of studies of the new coronavirus, the Ct value by itself is often used as a viral load indicator. For example, raw Ct values have been used to correlate viral load with a higher risk of intubation (Magleby et al., 2020), to compare viral load between nasopharyngeal swab (NPS) and oropharyngeal swab (OPS) samples (Wang et al., 2020a), and to investigate the relationship between viral load and age range (Heald-Sargent et al., 2020). These types of applications to evaluate viral load of different types of viruses are commonly used, but high variance has been reported, often due to different devices, PCR reagents and standards used (Hayden et al., 2012).
Moreover, the type of biological material has a strong impact when evaluating viral load. For instance, if serum is used, viral load can be expressed per mL for each sample. However, SARS-CoV-2 viral load is often evaluated in NPS samples, where the amount of biological material retrieved by the swab can vary depending on the quality of the collection. This makes normalization by using a reference gene important when interpreting the results of RT-qPCR (Guest et al., 2020). In the present work, we compared tracheal aspirate (TA) with NPS samples obtained from critically ill COVID-19 patients. We compared the raw Ct values and ΔCt and found that the ΔCt value provides better accuracy. Furthermore, we analyzed SARS-CoV-2 positive NPS samples and we proposed a method to reduce the error that can occur from using raw Ct values. Application of a normalization method for the Ct values of RT-qPCR can improve the interpretation of studies that use viral load of SARS-CoV-2.
2. Materials and methods
2.1. Samples
In this study, RT-qPCR data were obtained from 138 patients that tested positive for SARS-CoV-2. In total, there were 138 NPS samples, one from each patient, and 21 TA samples from intubated patients who were admitted to the intensive care unit, at Instituto Estadual do Cérebro Paulo Niemeyer, Rio de Janeiro, Brazil. TA samples were collected on the same day as NPS samples from each patient. The study involving human participants was reviewed and approved by the institute’s ethics committee (protocol number 3.997.619).
2.2. RT-qPCR
The TaqMan™ RT-qPCR assays were performed in a QuantStudio 7™ Flex Real-Time PCR System (Applied Biosystems, Foster City, CA, USA), directed to the nucleocapsid N gene regions (N1 and N2) of SARS-CoV-2 viral RNA (CDC assays for SARS-CoV-2 detection, manufactured by Integrated DNA Technologies, Iowa, USA). We used a final volume of 20 μL for RT-qPCR reactions: 5 μL of RNA, 0.4 μL of GoScript™ RT Mix for 1-Step RT-qPCR 50x (Promega, Madison, WI, USA), 9.98 μL of GoTaq® qPCR Master Mix 2x (with CXR reference dye), 0.02 μL of CXR reference dye, 3.1 μL nuclease-free water, and 1.5 μL of TaqMan™ RT-qPCR assay. Thermal cycling was performed at 45 °C for 15 min for reverse transcription, followed by 95 °C for 2 min and then 45 cycles at 95 °C for 3 s and 55 °C for 30 s. A cycle threshold value less than 40 was interpreted as positive for SARS-CoV-2 RNA. In this assay, a RNase P gene region is used as an endogenous internal control for the analysis of biological samples. It is normally used to ensure the quality of the test, by excluding the possibility of false negatives due to the presence of inhibitors or the low quality and integrity of RNA samples (Guest et al., 2020). However, all human cells have a single copy of the RNase P gene, which encodes the mRNA moiety for the RNAse P enzyme. Therefore, their Ct values are associated with a range of input cell numbers in the RNA extraction (Fernandes-Monteiro et al., 2015). In order to evaluate possible variability in the amount of material retrieved from NPS and other specimen types, we utilized RNase P as reference gene to normalize the input data.
Standard curves were plotted by using serial 10-fold dilutions of standard synthetic RNA transcripts of the SARS-CoV-2 N1 and N2 genes, ranging from 1 × 106 to 1 copies/ reaction (IDT, USA). We evaluated the amplification efficiency of both assays using standard curve analysis, since even though the reported efficiency is close to 100 %, it is of utmost importance to validate this with the particular laboratory setup (Vogels et al., 2020). The assay with best efficiency was used for further analysis and the experimental limit of detection (LoD) was determined.
2.3. RT-qPCR normalization
When performing relative gene expression analysis of qPCR data, first we calculate the Delta Ct (ΔCt). This is obtained by subtracting the reference gene Ct from the target gene Ct, to account for the input fluctuation that can occur (Livak and Schmittgen, 2001). For this measurement to be accurate, one needs to assume that amplification efficiency is 100 %. We obtained ΔCt from our samples using RNaseP as a reference gene (ΔCt = CtN1-CtRNaseP). When comparing different sample types (TA and NPS), we used Ct and ΔCt from paired samples to check whether there was a difference in viral RNA load or in the amount of biological material. When evaluating RT-qPCR data of swabs, we compared Ct and ΔCt and proposed a method to reduce the error that can occur from using raw Ct values. We applied a formula that corrects the Ct values to achieve the closest relation to ΔCt values. This is a simple correction based on the formula proposed by Duchamp and collaborators (Duchamp et al., 2010). They used this formula to correct influenza A viral load per sample, calculating a Ct value modified according to the ratio of sample RNase P and mean RNase P Ct values ([sample influenza A Ct value x sample RNaseP Ct value/mean RNaseP Ct value]).
2.4. Statistical analysis
All data analysis was performed with GraphPad Prism 6 (GraphPad Software Inc., USA). Data were expressed as mean ± standard deviation. The Student t-test was used for comparison between two groups. Spearman correlation was used to compare the relationship between N1 Ct and ΔCt. Differences were considered to be significant at a level of P < 0.05.
3. Results
3.1. Uncorrected Ct values and misinterpretation of viral load
Before analyzing the results, we evaluated the efficiency levels of the TaqMan™ assay from the CDC kit: efficiency of 100.177 % for the N1 assay (R2 = 0.999, slope = -3318, error = 0.03); 98.322 % for the N2 assay (R2 = 0.997, slope = -3363, error = 0.045); and 107.274 % (R2 = 0.997, slope = -3159, error = 0.045) for the RNase P assay. Then we performed the following tests using only N1 as a viral target since it had the best efficiency. Considering our experimental setup, the LoD was 10 copies per reaction (10 out of 10 positives), whereas at the final concentration tested (1 copy per reaction), we observed only 2 out of 10 positives. These results are in accordance with data previously reported by Lu et al. from CDC, where they concluded that the LoD of the N1 assay was 5 copies/reaction (Lu et al., 2020).
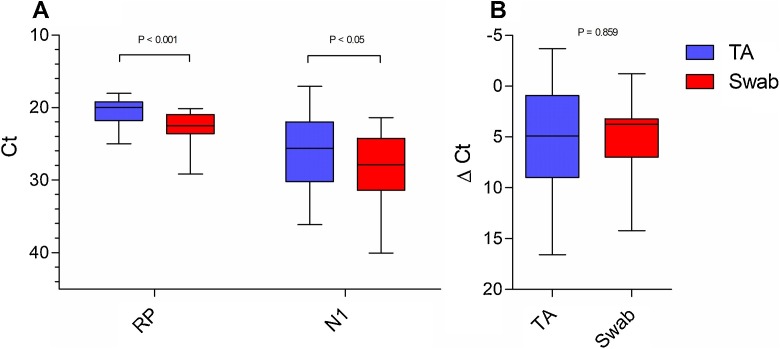
When comparing 21 paired samples of TA and NPS, the TA samples had a lower N1 Ct value than the NPS samples (P < 0.05), and also had lower RNase P Ct values (P < 0.001) (Fig. 1 A). However, when the ΔCt values were compared, there was no difference between TA and NPS samples (P = 0.859) (Fig. 1B). It is important to note that for one patient, the NPS sample was negative for SARS-CoV-2 and the TA sample was positive (N1 Ct = 34). The difference in Ct values having similar ΔCt values indicated that the higher concentration of viral RNA in TA samples was possibly a consequence of a higher concentration of total RNA. When RNA concentrations of both types of samples were quantified using a NanoDrop Lite® spectrophotometer (Thermo Fischer, Wilmington, DE, USA), we observed that the median of TA samples’ RNA concentration was 373.0 ng/μL (IQR = 136.7–862.0), as opposed to 121.3 ng/μL (IQR = 56.9–196.4) for NPS samples.
Fig. 1.
Comparison between nasopharyngeal swab and tracheal aspirate samples for SARS-CoV-2 detection. (A) N1 and RP Ct values of NPS X TA samples (N1: P < 0.05, TA = 25.6 (17.04 – 36.11) / 26.13 ± 4.99 and NPS = 27.87 (21.37 – 31.36) / 28.22 ± 4.54; RP: P*< 0.001, TA = 19.94 (18.02 – 24.98) / 20.49 ± 1.76 and NPS = 22.47 (20.11-29.16) / 22.61 ± 2.09). (B) ΔCt (N1 – RP) of NPS X TA samples (P = 0.859, TA = 4.90 (-3.71 – 16.59) / 5.64 ± 5.65 and NPS = 3.74 (-1.21 – 14.21) / 5.06 ± 3.91). Data are expressed as median (min – max)/ mean ± standard deviation. Statistical difference was evaluated by the paired T-test. (RP = RNAse P, NPS = nasopharyngeal swab, TA = tracheal aspirate).
3.2. Discrepancy between uncorrected Ct values and ΔCt values
In order to demonstrate the discrepancy that can arise when comparing results of N1 Ct and ΔCt, we plotted those values obtained from the 138 NPS positive samples. The summary of statistics is as follows: N1, mean = 25.31/ StdD = 5.47; RP, mean = 24.99/ StdD = 2.10; and ΔCt, mean = 0.32/ StdD = 5.31. Even though we found a correlation between those values (R = 0.94), this could provide a misleading result. On the X-axis, a variation of 1 ΔCt from -3 to -2 included 11 samples that had N1 Ct values ranging from 18.61 to 25.5. Interestingly, when analyzing the corresponding ΔCt values of these min and max N1 Ct values (18.61 and 25.5), we obtained -2.10 and -2.21, respectively (Fig. 2 A). When using uncorrected Ct values as a measure of viral load difference between those samples, we obtained a difference of 6.89 cycles, which corresponded roughly to a difference of 118 times more viral RNA present in the sample with lower Ct. Meanwhile, when we applied the fold change formula (2−ΔΔCt) to compare the same samples, we obtained a fold change of 1.08.
Fig. 2.
N1 Ct X ΔCt using different corrections. (A) No correction. (B)Correction proposed by Duchamp et al. (2010): Ct = CtN1* Sample CtRNaseP/mean CtRNaseP.(C)Modification of the method proposed in B: Ct = CtN1* mean CtRNaseP/Sample CtRNaseP.(D)Method with direct relation to ΔCt variation: Ct = CtN1– (Sample CtRNaseP– mean CtRNaseP).
3.3. Reducing the discrepancy between Ct and ΔCt Ct
We then applied a formula to correct Ct values based on the RNase P mean Ct (CtN1* sample CtRNaseP /mean CtRNaseP), as proposed by Duchamp et al. (2010). However, as can be observed in Fig. 2B, this method further increased the distance in Ct values of samples that had similar ΔCt values (a difference of cutoff cycle threshold values of 12.70), which is undesirable. We also observed a decrease in the correlation between those values (R = 0.76). So we modified this formula, trying to decrease the discrepancy of original Ct values of samples with similar ΔCt values, since this is a result of the differences in the amount of biological material used as input. A modification of the formula was used (CtN1* mean CtRNaseP/sample CtRNaseP). After this adjustment, the minimum Ct value increased from 18.61 to 22.9 and the maximum declined from 25.5 to 23.7, now with a difference of 0.8 cycle, indicating around 1.74 times more viral RNA. This adjusted Ct value had an even stronger correlation with the original ΔCt value, achieving a Spearman rank correlation of 0.99 (Fig. 2C).
Even after reducing the discrepancy and increasing the correlation, we observed that Ct values <20 and >30 were not adjusted in a similar way to intermediate values. We then applied a third formula, where we obtained the difference between sample RNase P Ct and mean RNase P Ct, and then subtracted it from sample N1 Ct (CtN1 – (Sample CtRNaseP -mean CtRNaseP)). With this, all Ct values became directly related with ΔCt values, yielding a correlation value of R = 1 (Fig. 2D).
4. Discussion and conclusion
The impact of the pandemic on society has increased the demand for quick responses and solutions, promoting the adaptation of sample collection sources due to shortage of materials, like the use of nasopharyngeal swabs or oropharyngeal swabs (LeBlanc et al., 2020). Unfortunately, the use of raw Ct data, inappropriate references for normalization, or even non-standardization are being widely considered. Consequently, the molecular data being used to infer SARS-CoV-2 viral load and correlate it with different outcomes can lead to inaccurate conclusions. Due to the diversity of sample types, the quantities of starting material for RT-qPCR vary. Moreover, there are differences among commercial detection kits, experimental conditions, and real-time equipment for COVID-19 diagnosis. Therefore, when data from real-time RT-qPCR are used for quantitative purposes, it is helpful to have a reference gene for normalization, to know assay efficiency and validate assay sensitivity.
In our study, we demonstrated that when Ct values were used without any correction, the test results indicated that TA samples had significantly more SARS-CoV-2 viral RNA than NPS samples (P < 0.05). However, we clearly noted that RNAse P Ct values were significantly different (P < 0.001), indicating that TA have higher amounts of biological material than NPS. This was confirmed when RNA concentration was evaluated in the samples. In short, when we performed the extraction of total RNA from, for example, 200 μL of tracheal aspirate, the result did not correspond to that of 200 μL of swab material. Even though this method did not provide actual viral RNA quantification, it was sufficient to show how the use of raw Ct values can be misleading, and it was easy to apply even in a diagnostic setup. The study of Liu et al. (2020) is one of the few that have used ΔCt values. They observed that the ΔCt values of severe cases were significantly lower than those of mild cases at the time of admission. They found that mean viral load of severe cases was around 60 times higher than that of mild cases, suggesting that higher viral loads might be associated with severe clinical outcomes. However, one of the most cited studies on viral load (>1500 citations) used only raw Ct data. The authors observed that the viral load in asymptomatic patients was similar to that in symptomatic patients, and that viral load was higher in the nose than in the throat (Zou et al., 2020).
Pujadas et al. (2020) reported an independent relation between high viral load and mortality. They argued that transforming qualitative testing into a quantitative measurement of viral load would assist clinicians in risk-stratifying patients and choosing among available therapies and trials. On the other hand, Wang et al. (2020a, 2020b, 2020c) evaluated nasopharyngeal (NPS) and oropharyngeal swab (OPS) samples collected from 120 patients with confirmed COVID-19. They found mean Ct value (uncorrected) for NPS of 37.8, significantly lower than that of OPS (39.4), indicating that the SARS-CoV-2 load was significantly higher in NPS than OPS samples. If sample concentration were taken into account, a different conclusion could have been drawn from such comparison. Thus, it is extremely important to have an internal control for a human reference gene when comparing samples.
Heald-Sargent et al. (2020) described that levels of SARS-CoV-2 viral nucleic acid in NPS are significantly greater in young children (younger than 5 years) than in older children (aged 5–17 years). They reported that young children and older children had median Ct values of 6.5 and 11, respectively. However, we found that within a range as far as 7 cycles in Ct for a viral target, samples could actually have a difference of only 0.1 cycle when ΔCt was taken into consideration. A multicentric study demonstrated that viral load estimations for several viruses can vary considerably between different laboratories, since there are no standardized required resources (Hayden et al., 2012). Fernandes-Monteiro and collaborators reported that serum samples tested for yellow fever had small variation in RNase P, even though there was significant difference in viral load between samples (Fernandes-Monteiro et al., 2015). For other sample types, like NPS, RNase P Ct varied depending on the quality of sample and efficiency of acquisition (Guest et al., 2020).
Wang et al. (2020a, 2020b, 2020c) investigated the biodistribution of viral RNA among different types of biological samples, including bronchoalveolar lavage fluid, fibrobronchoscope brush biopsy, sputum, feces, blood and urine, among others. They evaluated 1070 specimens collected from 205 patients with COVID-19 and observed that Ct values (uncorrected) of all specimen types were higher than 30, except for nasal swabs, with a mean Ct value of 24.3 (range of 16.9–38.4). However, without a correction in the Ct values, it is not possible to confirm these differences. Vivanti et al. (2020) demonstrated the transplacental transmission of SARS-CoV-2 in a neonate born to a mother infected in the last trimester, causing neurological compromise. The authors detected SARS-CoV-2 RNA in amniotic fluid, vaginal and rectal swab material, blood and NPS and called attention to the very high viral load in the placenta. However, an important point is that different types of biological samples have different concentrations in number of cells and particles. Moreover, the complex composition of some sample types includes proteins, fats, humic acid, phytic acid, immunoglobulin G, bile, calcium chloride, EDTA, heparin and ferric chloride, many of which have been recognized as PCR inhibitors (Nolan et al., 2006).
Real-time RT-PCR has become a common technique, and indeed in many cases is the main method to measure the presence of viral RNA, due to its sensitivity and high potential for accurate quantification. Despite RT-qPCR’s inability to differentiate between infective and non infective (antibody-neutralized or dead) viruses, using an estimative of viral RNA load remains plausible for clinical hypothesis formulation. The evaluation of infectiousness of a sample is not a simple procedure, since SARS-CoV-2 virus isolation in cell cultures need to be conducted in Biosafety Level 3 (BSL-3) laboratories (WHO, 2020). To achieve this, however, appropriate normalization strategies are required to control for variations that can arise from processing RNA from biological samples. We agree that the ideal approach is to use the Standard Curve Method with an endogenous control. In this method, standard curves are constructed for both the target and the endogenous reference. For each experimental sample, the amounts of target and endogenous reference are determined from the appropriate standard curve. However, this method has high cost, since standard curves need to be plotted in all experiments.
Here we propose a formula that can achieve perfect correlation between the corrected Ct values and ΔCt values, allowing researchers to use these corrected Ct values to calculate the number of viral copies. Even though experiments were conducted using only N1 as a target, Vogels et al. performed comparisons of RT-qPCR analytical efficiency and sensitivity of assays used to detect SARS-CoV-2 viral RNA. They included US CDC assay targets (N1, N2 and N3) and concluded that all assays, except RdRp-SARSr (Charité), had similar amplification efficiency and limit of detection (Vogels et al., 2020).
In conclusion, we found that in general, TA samples had more total RNA than NPS samples, even though there was no difference in viral load. Thus, if a reference gene is taken into consideration when analyzing NPS, samples initially considered to have different viral loads by raw Ct comparison actually had the same viral load. Thus, when comparing samples, the use of a reference gene is extremely important before drawing conclusions related to COVID-19 viral load.
Author contributions
RLM, AG and CHAL worked directly with samples, performed the literature search, prepared the figures, interpreted the data and wrote the manuscript draft; MG and PNF participated in the experimental design, discussion of results and manuscript preparation. All authors critically reviewed the manuscript for intellectual content and approved it in its final version.
Declaration of Competing Interest
The authors report no declarations of interest.
Acknowledgement
This work was supported by grants from the Rio de Janeiro State Research Foundation (FAPERJ).
References
- Dietz W., Santos-Burgoa C. Obesity and its implications for COVID-19 mortality. Obesity (Silver Spring) 2020 doi: 10.1002/oby.22818. [DOI] [PubMed] [Google Scholar]
- Duchamp M.B., Casalegno J.S., Gillet Y., Frobert E., Bernard E., Escuret V., Billaud G., Valette M., Javouhey E., Lina B., Floret D., Morfin F. Pandemic A(H1N1)2009 influenza virus detection by real time RT-PCR : is viral quantification useful? Clin. Microbiol. Infect. 2010;16:317–321. doi: 10.1111/j.1469-0691.2010.03169.x. [DOI] [PubMed] [Google Scholar]
- Fernandes-Monteiro A.G., Trindade G.F., Yamamura A.M.Y., Moreira O.C., de Paula V.S., Duarte A.C.M., Britto C., Lima S.M.B. New approaches for the standardization and validation of a real-time qPCR assay using TaqMan probes for quantification of yellow fever virus on clinical samples with high quality parameters. Hum. Vaccines Immunother. 2015;11:1865–1871. doi: 10.4161/21645515.2014.990854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guest J.L., Sullivan P.S., Valentine-Graves M., Valencia R., Adam E., Luisi N., Nakano M., Guarner J., del Rio C., Sailey C., Goedecke Z., Siegler A.J., Sanchez T.H. Suitability and sufficiency of telehealth clinician-observed, participant-collected samples for SARS-CoV-2 testing: the iCollect cohort pilot study. JMIR Public Heal. Surveill. 2020;6 doi: 10.2196/19731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayden R.T., Yan X., Wick M.T., Rodriguez A.B., Xiong X., Ginocchio C.C., Mitchell M.J., Caliendo A.M. Factors contributing to variability of quantitative viral PCR results in proficiency testing samples: a multivariate analysis. J. Clin. Microbiol. 2012;50:337–345. doi: 10.1128/JCM.01287-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heald-Sargent T., Muller W.J., Zheng X., Rippe J., Patel A.B., Kociolek L.K. Age-related differences in nasopharyngeal severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) levels in patients with mild to moderate coronavirus disease 2019 (COVID-19) JAMA Pediatr. 2020 doi: 10.1001/jamapediatrics.2020.3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., Cheng Z., Yu T., Xia J., Wei Y., Wu W., Xie X., Yin W., Li H., Liu M., Xiao Y., Gao H., Guo L., Xie J., Wang G., Jiang R., Gao Z., Jin Q., Wang J., Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020 doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeBlanc J.J., Heinstein C., MacDonald J., Pettipas J., Hatchette T.F., Patriquin G. A combined oropharyngeal/nares swab is a suitable alternative to nasopharyngeal swabs for the detection of SARS-CoV-2. J. Clin. Virol. 2020;128 doi: 10.1016/j.jcv.2020.104442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Yan L.M., Wan L., Xiang T.X., Le A., Liu J.M., Peiris M., Poon L.L.M., Zhang W. Viral dynamics in mild and severe cases of COVID-19. Lancet Infect. Dis. 2020;20:656–657. doi: 10.1016/S1473-3099(20)30232-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lu X., Wang L., Sakthivel S.K., Whitaker B., Murray J., Kamili S., Lynch B., Malapati L., Burke S.A., Harcourt J., Tamin A., Thornburg N.J., Villanueva J.M., Lindstrom S. US CDC real-time reverse transcription PCR panel for detection of severe acute respiratory syndrome coronavirus 2. Emerg. Infect. Dis. 2020;26:1654–1665. doi: 10.3201/eid2608.201246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magleby R., Westblade L.F., Trzebucki A., Simon M.S., Rajan M., Park J., Goyal P., Safford M.M., Satlin M.J. Impact of SARS-CoV-2 viral load on risk of intubation and mortality among hospitalized patients with coronavirus disease 2019. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolan T., Hands R.E., Bustin S.A. Quantification of mRNA using real-time RT-PCR. Nat. Protoc. 2006;1:1559–1582. doi: 10.1038/nprot.2006.236. [DOI] [PubMed] [Google Scholar]
- Pujadas E., Chaudhry F., McBride R., Richter F., Zhao S., Wajnberg A., Nadkarni G., Glicksberg B.S., Houldsworth J., Cordon-Cardo C. SARS-CoV-2 viral load predicts COVID-19 mortality. Lancet Respir. Med. 2020 doi: 10.1016/S2213-2600(20)30354-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vivanti A.J., Vauloup-Fellous C., Prevot S., Zupan V., Suffee C., Do Cao J., Benachi A., De Luca D. Transplacental transmission of SARS-CoV-2 infection. Nat. Commun. 2020;11:3572. doi: 10.1038/s41467-020-17436-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogels C.B.F., Brito A.F., Wyllie A.L., Fauver J.R., Ott I.M., Kalinich C.C., Petrone M.E., Casanovas-Massana A., Catherine Muenker M., Moore A.J., Klein J., Lu P., Lu-Culligan A., Jiang X., Kim D.J., Kudo E., Mao T., Moriyama M., Oh J.E., Park A., Silva J., Song E., Takahashi T., Taura M., Tokuyama M., Venkataraman A., Weizman O.-E., Wong P., Yang Y., Cheemarla N.R., White E.B., Lapidus S., Earnest R., Geng B., Vijayakumar P., Odio C., Fournier J., Bermejo S., Farhadian S., Dela Cruz C.S., Iwasaki A., Ko A.I., Landry M.L., Foxman E.F., Grubaugh N.D. Analytical sensitivity and efficiency comparisons of SARS-CoV-2 RT–qPCR primer–probe sets. Nat. Microbiol. 2020 doi: 10.1038/s41564-020-0761-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Liu Q., Hu J., Zhou M., Yu M.Q., Li K.Y., Xu D., Xiao Y., Yang J.Y., Lu Y.J., Wang F., Yin P., Xu S.Y. Nasopharyngeal swabs are more sensitive than oropharyngeal swabs for COVID-19 diagnosis and monitoring the SARS-CoV-2 load. Front. Med. 2020;7:1–8. doi: 10.3389/fmed.2020.00334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T., Du Z., Zhu F., Cao Z., An Y., Gao Y., Jiang B. Comorbidities and multi-organ injuries in the treatment of COVID-19. Lancet. 2020 doi: 10.1016/S0140-6736(20)30558-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Xu Y., Gao R., Lu R., Han K., Wu G., Tan W. Detection of SARS-CoV-2 in different types of clinical specimens. JAMA. 2020 doi: 10.1001/jama.2020.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO . 2020. Laboratory Testing for 2019 Novel Coronavirus (2019-nCoV) in Suspected Human Cases. [WWW Document]https://www.who.int/publications-detail/laboratory-testing-for-2019-novel-coronavirus-in-suspected-human-cases-20200117 URL. [Google Scholar]
- Zou L., Ruan F., Huang M., Liang L., Huang H., Hong Z., Yu J., Kang M., Song Y., Xia J., Guo Q., Song T., He J., Yen H.-L., Peiris M., Wu J. SARS-CoV-2 viral load in upper respiratory specimens of infected patients. N. Engl. J. Med. 2020;382:1177–1179. doi: 10.1056/NEJMc2001737. [DOI] [PMC free article] [PubMed] [Google Scholar]