Summary
The host response to SARS-CoV-2, the etiologic agent of the COVID-19 pandemic, demonstrates significant interindividual variability. In addition to showing more disease in males, the elderly, and individuals with underlying comorbidities, SARS-CoV-2 can seemingly afflict healthy individuals with profound clinical complications. We hypothesize that, in addition to viral load and host antibody repertoire, host genetic variants influence vulnerability to infection. Here we apply human induced pluripotent stem cell (hiPSC)-based models and CRISPR engineering to explore the host genetics of SARS-CoV-2. We demonstrate that a single-nucleotide polymorphism (rs4702), common in the population and located in the 3′ UTR of the protease FURIN, influences alveolar and neuron infection by SARS-CoV-2 in vitro. Thus, we provide a proof-of-principle finding that common genetic variation can have an impact on viral infection and thus contribute to clinical heterogeneity in COVID-19. Ongoing genetic studies will help to identify high-risk individuals, predict clinical complications, and facilitate the discovery of drugs.
Keywords: SARS-CoV-2, human induced pluripotent stem cell, host genetics, brain, neurons, FURIN rs4702
Graphical abstract

Highlights
-
•
hiPSC-derived model of SARS-CoV-2 infection in human neurons, lung, and intestinal cells
-
•
Infection of post-mitotic human neurons by SARS-CoV-2
-
•
Effect of host gene expression (ACE2, FURIN, BSG, and TMPRSS2) on SARS-CoV-2 infection
-
•
Impact of host genotype with common SNP rs4702 on SARS-CoV-2 infection
Brennand, Akbarian, and colleagues combine human induced pluripotent stem cell-based models and CRISPR engineering to explore the host genetics of SARS-CoV-2 in alveolar cells, intestinal cells, and neurons. A SNP(rs4702) located in the 3′ UTR of the protease FURIN influences alveolar and neuron infection by SARS-CoV-2 in vitro.
Introduction
A growing number of human genetic variants have been identified that contribute to enhanced susceptibility or resistance to viral diseases (Kenney et al., 2017). Genetic discoveries related to virus-host interactions implicate genes encoding virus receptors, receptor-modifying enzymes, and a wide variety of innate and adaptive immunity-related proteins in disease outcome. Such insights have been made across a range of pathogenic viruses, including influenza A virus (Ciancanelli et al., 2015), norovirus (Lindesmith et al., 2003), and human immunodeficiency virus (Samson et al., 1996).
There is marked variability between individuals in response to SARS-related coronavirus 2 (SARS-CoV-2) infection, with outcomes ranging from asymptomatic to critical (6.1%) (Guan et al., 2020). There is an urgent need to explore the molecular mechanisms underlying this unexpected clinical heterogeneity. A range of factors in addition to viral load can have an impact on outcomes, from genetic predisposition (e.g., blood type, Ellinghaus et al., 2020; mutations in immune signaling receptors, van der Made et al., 2020; or inborn errors of type I interferon immunity, Zhang et al., 2020) to immune repertoire (e.g., cross-reacting antibodies from other coronaviruses, Yuan et al., 2020, or autoantibodies to interferon, Bastard et al., 2020). Although the host response to SARS-CoV-2 has been defined in lung cultures and tissue (Karczewski et al., 2020), it remains unclear the full extent to which this varies across cell types and donors. Given the complicated host immune response to SARS-CoV-2 (Blanco-Melo et al., 2020), the expression of a number of human genes likely modulates infection.
In an effort to determine whether host variants that may influence viral entry might contribute to the heterogeneity of COVID-19 symptoms, we assessed human variants of FURIN. The spike glycoprotein that resides on the surface of the SARS-CoV-2 virion facilitates viral entry into target cells by engaging host angiotensin-converting enzyme 2 (ACE2) as the entry receptor and host cellular serine protease TMPRSS2 for spike protein priming (Hoffmann et al., 2020; Yan et al., 2020). More controversial is the extent to which host BSG, a transmembrane glycoprotein, serves as a secondary entry receptor (Shilts et al., 2021; Wang et al., 2020). The SARS-CoV-2 spike protein incorporates a four amino acid insertion that introduces a proposed cleavage site for FURIN, a host membrane-bound pro-protease convertase, potentially resulting in priming of spike protein before viral exit from the cell (Coutard et al., 2020; Wrapp et al., 2020). In contrast to SARS-CoV-2, the SARS-CoV spike protein lacks this FURIN cleavage site, thus requiring cleavage to facilitate subsequent cell entry (Walls et al., 2020). This theoretical hijacking of host FURIN activity is one possible explanation for the increased infectivity of SARS-CoV-2. Not only do these host genes show tissue-specific and cell-type-specific expression patterns, but each is associated with non-coding common genetic variants thought to regulate the expression of each gene (GTEx Consortium et al., 2017). Here we test the hypothesis that variability in the expression of host genes between cell types and individuals predicts susceptibility to infection. Given that CRISPR (clusters of regularly interspaced short palindromic repeats)-based allelic conversion of rs4702 in human induced pluripotent stem cells (hiPSCs) from AA to GG decreased FURIN mRNA levels, reduced neurite outgrowth, and altered neuronal activity in induced glutamatergic neurons (Schrode et al., 2019), here we consider the impact of FURIN expression and genotype on SARS-CoV-2 infection across hiPSC-derived lung, intestinal, and brain models. Our findings suggest that uncovering the genetic underpinnings of SARS-CoV-2 outcomes may help predict susceptibility to COVID-19, as well as facilitating precision treatment and prevention approaches.
Results
Infection of neurons by SARS-CoV-2 in vivo and in vitro
A subset of COVID-19 patients present with neurological symptoms (Mao et al., 2020). COVID-19 has been associated with acute disseminated encephalomyelitis (Pilotto et al., 2020); mid- and long-term sequelae, including classical Guillain-Barré syndrome (Toscano et al., 2020); and COVID-19-associated delirium (McLoughlin et al., 2020). After hospital discharge, an alarmingly high fraction of patients, as high as 33%, suffer from a dysexecutive syndrome consisting of inattention, disorientation, or poorly organized movements in response to command (Helms et al., 2020). It is critical to resolve whether potential SARS-CoV-2 pathological mechanisms include direct infection of brain cells, consistent with reports of neurotropism and transsynaptic spread by other coronaviruses (Li et al., 2020), or simply reflect endothelial injury, vascular coagulopathy, and/or diffuse neuroinflammatory processes.
A variety of studies reveal that human brain tissue and cultured neurons express critical host genes required for viral entry. RNA transcripts and protein for ACE2, FURIN, and BSG were detected in post-mortem adult human brain (prefrontal cortex and midbrain substantia nigra) (Figures 1A and 1B; Figure S1). Likewise, RNA-sequencing data from hiPSC-derived neural progenitor cells (NPCs) (Hoffman et al., 2017), NGN2-induced glutamatergic neurons (Zhang et al., 2013), ASCL1/DLX2-induced GABAergic neurons (Yang et al., 2017), and ASCL1/NURR1/LMX1A-induced dopaminergic neurons (Dobrindt et al., 2020), as well as differentiated forebrain neurons (comprising a mixture of neurons and astrocytes), confirmed expression of many candidate host genes (Figure 1A). Although induced GABAergic and dopaminergic neurons expressed higher levels of some host genes, NGN2-induced glutamatergic neurons were used for all neuronal infections because the expression quantitative trait loci (eQTL) effect associated with the common variant rs4702 is best characterized in NGN2-induced glutamatergic neurons (Schrode et al., 2019). Expression of FURIN, ACE2, BSG, and TMPRSS2 in NGN2-induced glutamatergic neurons, in mock- (uninfected) and SARS-CoV-2-infected populations, was confirmed by qPCR, normalized to lung alveolosphere samples to facilitate inter-cell-type comparisons of expression, with FURIN and BSG being most prominent and ACE2 being very low and almost undetectable (Figure 1C).
Figure 1.
Infection of neurons by SARS-CoV-2 in vitro
(A) Expression of host genes associated with SARS-CoV-2 infection (Gordon et al., 2020; Ou et al., 2020; Wruck and Adjaye, 2020) examined by RNA sequencing (log2 RNA-seq, CPM medians) of post-mortem human prefrontal cortex (PFC), human induced pluripotent stem cells (IPSC), neural progenitor cells (NPC), and four hiPSC-derived neuronal cell types: induced glutamatergic (GLUT), differentiated forebrain (FB) populations (comprising glutamatergic neurons, GABAergic neurons, and astrocytes), induced GABAergic (GABA), and induced dopaminergic (DOPA).
(B) Immunoblot showing ACE2, FURIN, and BSG protein in post-mortem human adult substantia nigra (SN) and PFC.
(C) Baseline host gene expression in day 21 NGN2-induced glutamatergic neurons, uninfected or infected with SARS-CoV-2 (MOI 0.5), normalized relative to alveolospheres, to ensure comparability across cell types. Data are presented as mean ± SEM from two independent donors from two independent experiments, one or two replicates each.
(D) qPCR analysis of relative change in subgenomic vRNA nucleocapsid (N) transcript and interferon β (IFNB1) in mock- and SARS-CoV-2-infected day 21 NGN2-induced glutamatergic neurons (MOI of 0.05, 24 h). Data are presented as mean ± SEM from two donors from two independent experiments, one or two replicates each. ∗∗∗∗p < 0.0001.
(E) Representative immunostaining for SARS-CoV-2 N protein (magenta), neuronal marker β-III-tubulin (green), and DAPI (blue) in day 7 NGN2-induced glutamatergic neurons infected with SARS-CoV-2 (MOI of 0.05, 48 h). Scale bar: 50 μm.
(F) Immunoblot against SARS-CoV-2 N protein shows replication in NGN2-induced glutamatergic neurons at an MOI of 1 after 24 h.
To determine neuronal susceptibility to SARS-CoV-2 virus in vitro, human NGN2-induced glutamatergic neurons derived from two independent donors were infected with SARS-CoV-2 virus at low multiplicity of infection (MOI 0.05). Glutamatergic neurons were susceptible to SARS-CoV-2 virus infection, rapidly increasing expression of the subgenomic viral RNA (vRNA) nucleocapsid (N) transcript by qPCR (148,124-fold, p < 0.0001) within 24 h, as well as increasing interferon β (IFNB1), a type I interferon canonical of the innate immune response to viral infection (Figure 1D). In addition, protein immunohistochemistry for SARS-CoV-2 N was detected within 48 h of infection (MOI of 0.05) (Figure 1E). It is important to note that the qPCR primers used assay for subgenomic transcript sequences only and do not detect the initial inoculum (see Experimental procedures for explanation). At higher titers (MOI 1, 24 h), an immunoblot against N protein was consistent with SARS-CoV-2 replication in neurons (Figure 1F).
To further explore the susceptibility of the in vivo brain to SARS-CoV-2, we analyzed RNA-sequencing datasets of brains of 3–5 week old male Golden Syrian hamsters infected intranasally with SARS-CoV-2 or a phosphate-buffered saline control (GSE161200) (Hoagland et al., 2021). By RNA sequencing, we identified high levels of SARS-CoV-2 host gene expression in hamster brain tissue, with notably high levels of FURIN and BSG/CD147 (Figure 2A). SARS-CoV-2 infection resulted in widespread up- and downregulation of transcripts (Figure 2B), notably upregulation of interferon-associated genes within 24 h, an effect that subsided somewhat by day 8 (Figure 2C). Gene set enrichment analysis of differentially expressed genes was performed across a collection of 698 neural-themed gene sets subdivided into eight categories (Schrode et al., 2019). Notably, significantly upregulated gene sets (false discovery rate < 5%) included those related to abnormal neuronal morphology and development, membrane trafficking, and abnormal synaptic transmission (clustered hierarchically by significance in Figure 2D), while downregulated gene sets included ion channel and neurotransmitter signaling pathways (Figure 2E). Nonetheless, alignment of the transcriptomic data to the SARS-CoV-2 genome failed to detect reads for virus beyond background, suggesting that inflammatory changes in gene expression persist with SARS-CoV-2 exposure despite a lack of robust viral replication in vivo.
Figure 2.
RNA-sequencing analysis of expression changes in hamster brain after SARS-CoV-2 infection in vivo
(A) Expression of relevant host genes associated with SARS-CoV-2 infection in hamster brain after mock infection, 1 or 8 days following SARS-CoV-2 infection. Data are derived from at least two animals each.
(B) Volcano plot highlighting the top 50 significantly differentially expressed genes after a high plaque-forming unit challenge with SARS-CoV-2.
(C) Interferon response over the course of infection, expression shown in log2 CPM, gene set from Blanco-Melo et al. (2020).
(D) Gene set enrichment for upregulated pathways using the MAGMA gene set.
(E) Gene set enrichment for downregulated pathways using curated gene sets (Schrode et al., 2019).
Altogether, these analyses suggest that although neuronal infection by SARS-CoV-2 can occur in response to high MOI in vitro, it may occur less frequently under clinically relevant conditions in vivo, but might still lead to upregulation of inflammatory pathways.
Host-gene-dependent infection of brain, lung, and intestinal cells by SARS-CoV-2
SARS-CoV-2 infects hiPSC-derived lung alveolospheres (Abo et al., 2020; Han et al., 2021; Huang et al., 2020) and intestinal organoids (Lamers et al., 2020; Yang et al., 2020; Zhou et al., 2020), particularly after dissociation; here we similarly apply published stepwise differentiation protocols for distal lung alveolar type 2-like epithelium (hereafter “alveolar cells”) (Jacob et al., 2019) and intestinal organoids (Koike et al., 2019; Zhang et al., 2018).
Alveolospheres formed after 30 days (Figure 3A) robustly express the endothelial marker EPCAM as well as prosurfactant protein C (SFTPC) and NKX2-1 (Figures 3B and 3D). Alveolar cells were also shown to express host genes associated with SARS-CoV-2 infection (Figure 3C), including ACE2 (Figure 3B). Consistent with other studies, we observed infection by SARS-CoV-2 of EPCAM- and NKX2-1-positive lung alveolospheres derived from two donors across three infections on the protein level (MOI 0.5, 24 h) (Figure 3D) and on the RNA level (MOI 0.1 and 0.5, 24 h) (Figure 3E). Dissociated alveolar cells showed detectable SARS-CoV-2 N protein (MOI of 0.1, 24 h) (Figure 3F). A plaque assay confirms effective viral production from the alveolospheres 24 h post-infection at an MOI of 0.1 (Figure S2).
Figure 3.
Infection of lung alveolospheres by SARS-CoV-2 in vitro
(A) Representative bright-field image of alveolosphere cultures on day 30. Scale bar: 100 μm.
(B) Representative immunofluorescence staining against surfactant protein C (SFTPC) and ACE2. Scale bars: 50 μm.
(C) Baseline SARS-CoV-2 host gene expression in alveolospheres. Data are presented as mean ± SEM from two donors, from three independent experiments, with approximately 15 organoids per sample.
(D) Representative immunofluorescence staining against SARS-CoV-2 nucleocapsid (N) (magenta), epithelial marker EPCAM (green), NKX2-1 (blue, left), and DAPI (blue, right) in alveolospheres with mock or SARS-CoV-2 infection (MOI 0.5, 24 h). Scale bar: 100 μm.
(E) qPCR analysis of relative change in subgenomic vRNA N transcript in mock- and SARS-CoV-2-infected day 34 alveolospheres from two donors (MOI of 0.1 or 0.5, 24 h). Data are presented as mean ± SEM from two donors, from three independent experiments, with approximately 15 organoids per sample. t test, relative to respective mock, ∗p < 0.05, ∗∗p < 0.01.
(F) Dissociated alveolar cells after SARS-CoV-2 infection on day 34 are positive for SARS-CoV-2 N protein (magenta). Scale bar: 100 μm.
Intestinal organoids form within 24 days and express CDX2 and KRT20 protein, as well as ACE2 (Figure 4A). Similar to alveolospheres, EPCAM-positive intestinal organoids (one donor) also showed SARS-CoV-2 infection on the protein level (MOI 0.5 and 1, 48 h) (Figure 4B) and on the RNA level (MOI 0.05 and 0.5, 24 h) (Figure 4D). Intestinal organoids express high levels of FURIN, ACE2, and BSG, but lower levels of TMPRSS2, relative to alveolospheres (Figure 4C). Dissociated intestinal cells showed detectable SARS-CoV-2 N protein (MOI of 0.05, 48 h) (Figure 4E).
Figure 4.
Infection of intestinal organoids by SARS-CoV-2 in vitro
(A) Representative immunofluorescence staining of intestinal organoids for the intestinal markers CDX2 (red) and KRT20 (green) and the SARS-CoV-2 host factor ACE2. Scale bar: 100 μm.
(B) Immunofluorescence staining of intestinal organoids against SARS-CoV-2 nucleocapsid (N) (red), the epithelial marker EPCAM (green), and DAPI (blue), following mock or SARS-CoV-2 infection (MOI of 0.5 or 1, 48 h). Scale bar: 100 μm.
(C) Baseline SARS-CoV-2 host gene expression in mock-treated intestinal organoids days 24 and 50. Data are presented as mean ± SEM from one donor, from two independent experiments, with approximately 15 organoids per sample.
(D) qPCR analysis of relative change in subgenomic vRNA N transcript in mock- and SARS-CoV-2-infected intestinal organoids (MOI of 0.05 or 0.5, 24 h). Data are presented as mean ± SEM from one donor, from two independent experiments, with approximately 15 organoids per sample. t test, relative to mock, ∗p < 0.05, ∗∗p < 0.01.
(E) Representative immunofluorescence staining of dissociated intestinal cells after SARS-CoV-2 infection on day 25 (MOI of 0.05, 48 h), positive for SARS-CoV-2 N protein (red), EPCAM (green), and DAPI (blue). Scale bar: 50 μm.
We optimized MOIs (neuron, 0.05; alveolosphere, 0.1; intestine, 0.05) and exposure times (neuron, 24 h for RNA, 48 h for protein; alveolosphere, 24 h; intestine, 24 h for RNA, 48 h for protein) for subsequent studies of host gene and variant effects, toward maximizing cell viability and detection of SARS-CoV-2 N protein.
To test the functional impact of decreasing expression of critical host genes, we applied a short hairpin RNA (shRNA) strategy to knock down expression of ACE2, FURIN, BSG, and TMPRSS2 in neurons (Figures 5A and 5B: FURIN expression reduced to 56.9%, p < 0.00045; ACE2 expression reduced to 20%, p = 0.0001; BSG expression reduced to 37.8%, p < 0.0001; and TMPRSS2 expression reduced to 32.9%, p = 0.002374), lung alveolar cells (Figure 5D: FURIN expression reduced to 14.5%, p = 0.0208; ACE2 expression reduced to 21.1%, p = 0.002; BSG expression reduced to 16.5%, p < 0.0001; and TMPRSS2 expression reduced to 13.8%, p < 0.0001), and intestinal cells (Figure 5G: FURIN expression reduced to 27.6%, p = 0.0063; ACE2 expression reduced to 28.4%, p = 0.0323; and BSG expression reduced to 13.4%, p = 0.0434). TMPRSS2 knockdown in intestinal cells was inconsistent due to extremely low host transcript expression and could not be included. Otherwise, observed knockdown did not significantly differ across cell types or donors. Moreover, susceptibility to infection was quantified by qPCR for subgenomic vRNA N transcript and immunocytochemistry with high-content imaging for N protein. By comparing vRNA N transcript expression in mock- and SARS-CoV-2-infected neurons (Figure 5C), dissociated alveolosphere cells (Figure 5E), and dissociated intestinal organoid cells (Figure 5H) expressing shRNAs against ACE2, TMPRSS2, BSG, and FURIN, as well as a scrambled control, we demonstrate non-cell-type-specific requirements for ACE2, BSG, and FURIN in neurons (Figure 5C: FURIN, vRNA expression reduced to 37.7%, p = 0.0005; ACE2, vRNA expression reduced to 26.6%, p = 0.0011; BSG, vRNA expression reduced to 48.4%, p = 0.0177; and TMPRSS2, vRNA expression reduced to 38.7%, p = 0.0053), lung alveolar cells (Figure 5E: FURIN, vRNA expression reduced to 29.3%, p = 0.0055; ACE2, vRNA expression reduced to 12.8%, p = 0.0008; BSG, vRNA expression reduced to 17.2%, p = 0.0009; and TMPRSS2, vRNA expression reduced to 26.8%, p = 0.0067), and intestinal cells (Figure 5H: FURIN, vRNA expression reduced to 49.7%, p < 0.0001; ACE2, expression reduced to 33.3%, p < 0.0001; and BSG, expression reduced to 11.1%, p < 0.0001). These findings were confirmed through quantification of fluorescence via high-content imaging in dissociated alveolar cells for viral N protein, which revealed decreased total infected cells compared with scrambled control (MOI of 0.1, 24 h): shFURIN, N protein reduced to 50.7%, p < 0.0001; shACE2, N protein reduced to 10.7%, p < 0.0001; shBSG, N protein reduced to 20.1%, p < 0.0001; and TMPRSS2, N protein reduced to 63.2%, p < 0.0001 (Figure 5F). Consistent with this, knockdown of FURIN, ACE2, and BSG in dissociated organoid intestinal cells led to a decreased number of N protein-positive SARS-CoV-2-infected cells through quantification of fluorescence via high-content imaging (MOI 0.5 after 48 h): shFURIN, N protein reduced to 0.42%, p < 0.0001; shACE2, N protein reduced to 10.05%, p < 0.0001; and shBSG, N protein reduced to 12.87%, p < 0.0001 (Figure 5I). It is possible that the reduced percentage of N-positive cells in the scrambled controls with higher SARS-CoV-2 infection (MOI 0.5) resulted from increased cell death in alveolar cells, perhaps due to increased cellular stress from the combination of SARS-CoV-2 infection, shRNA transduction, and antibiotic selection. The SARS-CoV-2 alone control (not shown) did indeed exhibit a higher infection rate at MOI 0.5. Since the infection rate of neurons by immunocytochemistry was extremely low, high-content imaging analysis of neurons could not be included.
Figure 5.
Downregulation of SARS-CoV-2 host genes and the impact on infection
(A) Schematic of the experimental setup (created with BioRender.com).
(B) shRNA knockdown of FURIN, ACE2, BSG, and TMPRSS2 host genes, in day 7 NGN2-induced glutamatergic neurons, normalized to 18S levels and scrambled.
(C) Subgenomic vRNA nucleocapsid (N) transcript in infected NGN2-induced glutamatergic neurons (MOI of 0.05 for 24 h) treated with shRNAs against SARS-CoV-2 receptors. t test, relative to scrambled shRNA, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001. Data are presented as mean ± SEM from two donors, from three independent experiments, one or two replicates each.
(D) shRNA knockdown of FURIN, ACE2, BSG, and TMPRSS2 host genes in dissociated alveolar cells, normalized to 18S levels and scrambled.
(E) qPCR analysis of relative change in subgenomic vRNA N transcript in mock- and SARS-CoV-2-infected dissociated alveolar cells (MOI 0.1, 24 h) treated with shRNAs against SARS-CoV-2 receptorstt. t test relative to scrambled shRNA, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001. Data are presented as mean ± SEM from two donors, from two independent experiments, with one or two replicates each.
(F) Quantification of high-content imaging for SARS-CoV-2 N protein in mock- and SARS-CoV-2-infected dissociated alveolar cells infected at MOI of 0.1 and 0.5 for 24 h. Each data point represents the mean from at least 100 cells from an individual well, from two independent experiments. Two-way ANOVA, relative to scrambled shRNA; ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
(G) shRNA knockdown of FURIN, ACE2, and BSG host genes in dissociated intestinal cells, normalized to 18S levels and infected scrambled control.
(H) qPCR analysis of relative change in subgenomic vRNA N transcript in mock- and SARS-CoV-2-infected dissociated intestinal cells (MOI 0.05, 24 h) treated with shRNAs against SARS-CoV-2 receptors. t test, relative to scrambled shRNA, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗∗p < 0.0001. Data are presented as mean ± SEM from one donor, from three independent experiments, one or two replicates each.
(I) Quantification of high-content imaging for SARS-CoV-2 N protein in mock- and SARS-CoV-2-infected dissociated intestinal cells infected at MOI of 0.05 and 0.5 for 48 h. Each data point represents an independent well presented as mean, from two independent experiments. Two-way ANOVA, relative to respective scrambled shRNA control; ∗∗∗∗p < 0.0001.
Critically, shRNA-mediated knockdown of host genes resulted in large changes in their endogenous expression, which may not well model subtle changes in gene expression resulting from common variation between individual people but can serve here as positive controls to negatively affect SARS-CoV-2 entry and replication.
Host FURIN rs4702 genotype infection by SARS-CoV-2
To achieve more physiologically relevant changes in gene expression, CRISPR-based allelic conversion was used to generate isogenic hiPSCs that differed at a single non-coding regulatory single-nucleotide polymorphism (SNP) at the FURIN locus (rs4702, NC_000015.10:g.90883330G>A). This SNP was predicted to be an eQTL in the human brain (Fromer et al., 2016) and in NGN2-induced glutamatergic neurons (Forrest et al., 2017), and fine-mapping analysis identified a single putative causal cis-eQTL (p = 0.94) (Schrode et al., 2019). rs4702 was empirically validated to regulate FURIN expression, neuronal outgrowth, and neuronal activity following allelic conversion from AA to GG in NGN2-induced glutamatergic neurons (Schrode et al., 2019). Here we evaluate the regulatory effect of this common SNP, determining the extent to which rs4702 regulates FURIN expression and SARS-CoV-2 infection in lung cells and neurons (Figure 6).
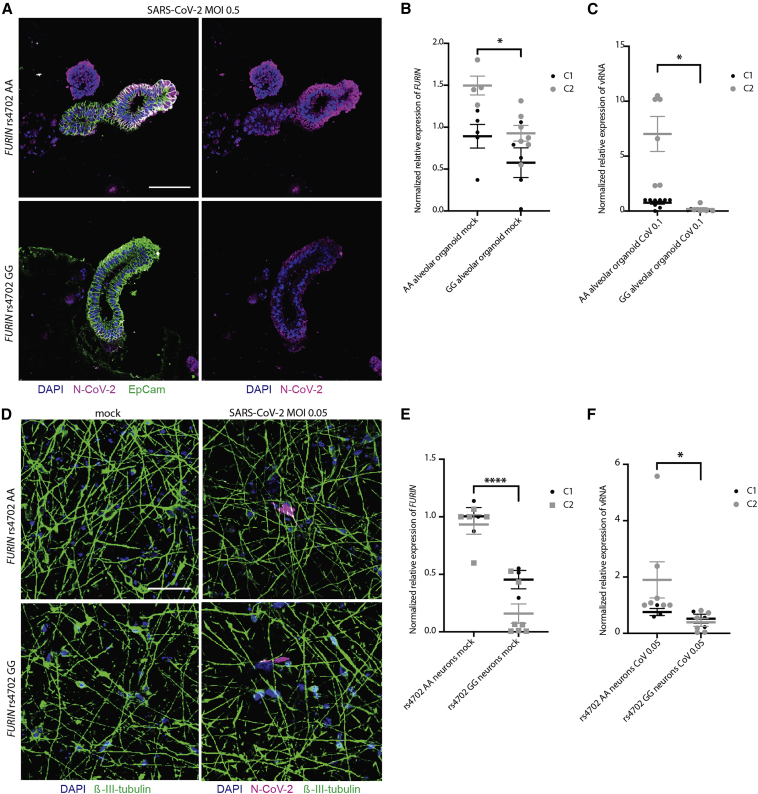
Figure 6.
Allelic conversion at FURIN rs4702 in alveolospheres and neurons influences SARS-CoV-2 infection
(A) Representative immunofluorescence staining against SARS-CoV-2 nucleocapsid (N) protein (magenta), epithelial marker EPCAM (green), and DAPI (blue). Alveolospheres were generated from C2 FURIN rs4702 AA and GG lines and infected with mock or an MOI of 0.5 SARS-CoV-2 for 24 h. Scale bar: 100 μm.
(B) FURIN RNA expression in rs4702 alveolospheres, relative to isogenic AA alveolospheres, in two control donors. Unpaired t test (−0.327-fold, p = 0.0347), ∗p < 0.05. Data are presented as mean ± SEM from three independent experiments, with approximately 15 organoids per sample.
(C) qPCR analysis of relative change in subgenomic vRNA N transcript in mock- and SARS-CoV-2-infected GG and AA alveolospheres from two control donors. Unpaired t test (−0.95-fold, p = 0.0215), ∗p < 0.05. Data are presented as mean ± SEM from three independent experiments, approximately 15 organoids per sample.
(D) Representative immunofluorescence staining of NGN2-induced glutamatergic FURIN rs4702 AA and GG neurons against N protein (magenta), β-III-tubulin (green), and DAPI (blue), after infection with SARS-CoV-2 (MOI of 0.05, 48 h). Scale bar: 50 μm.
(E) FURIN RNA expression in day 7 rs4702 GG NGN2-induced neurons, relative to isogenic AA neurons. Unpaired t test (−0.74-fold, p < 0.0001), ∗∗∗∗p < 0.0001. Data are presented as mean ± SEM from three independent experiments, with two or three replicates each.
(F) qPCR analysis of relative change in subgenomic vRNA N transcript in mock- and SARS-CoV-2-infected day 7 NGN2-induced neurons (MOI of 0.05, 24 h). Unpaired t test (−0.72-fold, p = 0.0416), ∗p < 0.05. Data are presented as mean ± SEM from three independent experiments, with two or three replicates each.
rs4702 GG alveolospheres exhibited a reduced FURIN expression (−0.327-fold, p = 0.0347) (Figure 6B) and demonstrated reduced genotype-dependent SARS-CoV-2 infection across both donors (−0.95-fold, p = 0.0215) (Figures 6A and 6C). In 7 day old CRISPR-edited rs4702 GG NGN2-induced glutamatergic neurons, we observed decreased FURIN expression relative to AA neurons, from two independent donors (−0.74-fold, p < 0.0001) (Figure 6E), consistent with Schrode et al. (2019). Both rs4702 AA and rs4702 GG neurons were infected by SARS-CoV-2 (MOI 0.05) at 48 h (Figure 6D), trending toward decreased SARS-CoV-2 infection in GG neurons by qPCR for subgenomic vRNA N transcript after 24 h (−0.72-fold, p = 0.0416) (Figure 6F). Altogether, these results indicate that reduced FURIN levels mediated by rs4702 result in decreased SARS-CoV-2 infection.
Overall, these findings suggest not only that gross manipulations of host genes can have an impact on SARS-CoV-2 infection in lung and brain cells, but also that the more subtle gene expression changes associated with genetic common variants are sufficient to influence viral infection levels.
Discussion
We applied an hiPSC-based model to explore the genetics of cell-type-specific host response to SARS-CoV-2. First, we reported infection of post-mitotic human neurons by SARS-CoV-2. Second, we further demonstrated functional validation of the impact of host genes (ACE2, FURIN, BSG, and TMPRSS2) and genetic variation (rs4702) on SARS-CoV-2 infection in human neurons, lung, and intestinal cells. Our results highlight the importance of FURIN as a mediator for SARS-CoV-2 infection and further demonstrate that a common variant, rs4702, located in the 3′ UTR of the FURIN gene and with allele frequency estimates from G = 0.34 to G = 0.45 (Tryka et al., 2014), is capable of influencing SARS-CoV-2 infection in vitro. More specifically, CRISPR-Cas9-mediated allelic conversion (from AA to GG) at the common variant rs4702 resulted in decreased neuronal and alveolar expression of the cis-gene target FURIN, and reduced SARS-CoV-2 infection. Our isogenic hiPSC-based strategy provides a proof-of-principle demonstration that common human genetic variation may directly affect SARS-CoV-2 infection.
The ability of SARS-CoV-2 to infect neurons observed here is consistent with other studies, which include reports of infection of human neurons and astrocytes using monolayer and organoid approaches (Jacob et al., 2020; Ramani et al., 2020; Song et al., 2021), highlighting greater and productive infection in choroid plexus epithelial cells (Jacob et al., 2020) and less infection in NPCs (Ramani et al., 2020; Song et al., 2021). One study comparing different hiPSC-derived cell types was able to detect infection in dopaminergic, but not cortical, neurons, using an MOI of 0.01 on the protein level (Yang et al., 2020), thus a lower titer and a less sensitive detection method than used in our study, which are potentially less clinically relevant. Although it remains possible that neurons and/or glia in the brain do become infected in vivo, we see no evidence that infected brain cells consistently produce mature virions in our hamster model. The inflammatory signature we observe might be triggered by viral products and debris that make their way into the circulatory system and/or systemic cytokines. The consequences of neuronal infection include transcriptional dysregulation indicative of an inflammatory response (Jacob et al., 2020), missorted and phosphorylated Tau (Ramani et al., 2020), and increased cell death (Jacob et al., 2020; Ramani et al., 2020; Song et al., 2021).
Large-scale international consortia have assembled to identify host-genetic associations with COVID-19 clinical outcomes (Initiative, 2020; Shelton et al., 2020; Tanigawa and Rivas, 2020), although cohorts remain underpowered by genome-wide association study (GWAS) standards. To date, such studies have found only a handful of loci to be genome-wide significant (Initiative, 2020; Kachuri et al., 2020; Ramlall et al., 2020). The largest data freeze to date (https://www.covid19hg.org) does not include rs4702 within its summary statistics; however, we note an excess of nominally significant genetic variants within the FURIN cis-region (Table S1; binomial test p = 3.57 × 10−31) comparing individuals hospitalized from COVID-19 with the general population (Initiative, 2020). This indicates the necessity of expanding GWAS to more comprehensively explore the polygenic architecture for COVID-19 clinical outcome. In addition to FURIN and components of the immune system, variants in ACE2, TMPRSS2, and BSG harbor genetic variants that are significantly enriched in COVID-19 patients and are predicted to influence susceptibility to SARS-CoV-2 infection (Benetti et al., 2020; Pairo-Castineira et al., 2020; Russo et al., 2020). Such discoveries could help to generate hypotheses for drug repurposing, identify individuals at unusually high or low risk, and contribute to the global knowledge of the biology of SARS-CoV-2 infection and disease. Discovery-based approaches to broadly identify all genes required for host infection and survival across tissues and cell types are also necessary to inform in silico drug-repurposing screens (Han et al., 2021). Integration of ongoing clinical GWASs with in vitro CRISPR-based forward genetic screening (e.g., Perturb-seq, Dixit et al., 2016) and pooled eQTL screens (e.g., crisprQTL mapping, Gasperini et al., 2019, and Census-seq, Mitchell et al., 2020) approaches will more rapidly identify host variants that have an impact on the entry, replication, and egress of SARS-CoV-2, cellular survival, and host immune response.
This study focused only on host genes known to mediate SARS-CoV-2 infection. The broad requirement of ACE2 and TMPRSS2 expression for SARS-CoV-2 infection across cell types was consistent with established literature (Hoffmann et al., 2020), whereas the observed impact of BSG expression was more surprising, given recent evidence suggesting that BSG is incapable of binding SARS-CoV-2 (Shilts et al., 2021). Together with the data shown here, this suggests that BSG protein may enhance viral entry by means other than directly binding to or modifying SARS-CoV-2, that it may require a secondary cofactor, and/or that BSG has cell-type-specific functions in vivo. The impact of FURIN expression was not unexpected (Coutard et al., 2020; Wrapp et al., 2020); however, because rs4702 lies in the binding site for microRNA-338 (Hou et al., 2018), this is a context-dependent eQTL and so dependent upon cell-type-specific and donor-dependent expression of miR-338 (Schrode et al., 2019). Overall, it remains possible that our in vitro findings will not predict clinical response because of the influence of other genetic and/or environmental factors. For example, SARS-CoV-2 antagonism of the immune response has been substantially defined (Konno et al., 2020; Lei et al., 2020; Thoms et al., 2020; Xia et al., 2020), and so genetic variants that result in decreased replication efficiency of SARS-CoV-2 in vitro may ultimately prove advantageous for the virus in vivo, permitting low levels of virus to spread successfully while remaining undetected by the immune system.
The expected clinical response to SARS-CoV-2 infection in the donors from whom we derived hiPSCs is not known; therefore, we cannot explore the extent to which in vitro findings recapitulate clinical outcomes. Moreover, hiPSC-derived cells of most (if not all) lineages are immature relative to those in adult humans (Hoffman et al., 2017; Jacob et al., 2019; Pavlovic et al., 2018), which may affect SARS-CoV-2 infection/response, particularly since clinical outcomes in children are generally mild (Ludvigsson, 2020). Given that clinical evidence suggests that SARS-CoV-2 infection can lead to heterogeneous acute and chronic outcomes across a range of tissues, future work should explore the wider range of cell types, both the cell autonomous response to infection and the non-cell-autonomous effects resulting from subsequent cytokine storms that occur as part of the response to infection. Moving forward, we should strive toward an expanded and unbiased isogenic strategy that will prove more broadly useful in exploring the impact of host genome on SARS-CoV-2 infection across diverse cell types and tissues.
In summary, we demonstrate that a single non-coding SNP is sufficient to influence SARS-CoV-2 infection in human neurons and alveolar cells. This work supports ongoing efforts to discover host genes associated with SARS-CoV-2 infection, both in vitro and in the clinic. Our hope is that such efforts might better predict clinical outcomes before the onset of symptoms and facilitate the discovery of drugs that might prevent or treat COVID-19 disease.
Experimental procedures
See also the supplemental information.
Donors
hiPSC lines and culture
hiPSCs were cultured in StemFlex medium (Gibco, A3349401) and passaged with EDTA (Life Technologies, 15575-020). All wild-type and shRNA experiments in lung alveolosphere cells, as well as all CRISPR-engineered AA and GG experiments, were conducted in NSB3113, here C1 (female, 18 years of age, European descent), and NSB3188, here C2 (female, 14 years of age, European descent) (Schrode et al., 2019). shRNA experiments in neurons were conducted in control NSB553 (male, 31 years of age, European descent) and NSB2607 (male, 15 years of age, European descent) hiPSC-derived NPCs (Hoffman et al., 2017); wild-type and shRNA intestinal cell experiments were conducted in 1383D6 (male, 36 years of age, Asian descent) (Takayama et al., 2017).
Cell culture and differentiations
See the supplemental information.
SARS-CoV-2 virus propagation and infections
SARS-CoV-2, isolate USA-WA1/2020 (NR-52281), was deposited by the Centers for Disease Control and Prevention and obtained through BEI Resources, NIAID, NIH. SARS-CoV-2 was propagated in Vero E6 cells in DMEM supplemented with 2% FBS, 4.5 g/L D-glucose, 4 mM L-glutamine, 10 mM non-essential amino acids, 1 mM sodium pyruvate, and 10 mM HEPES. Virus stock was filtered by centrifugation using an Amicon Ultra-15 centrifugal filter unit (Sigma, UFC910096) and resuspended in viral propagation medium. All infections were performed with passage 3 or 4 SARS-CoV-2. Infectious titers of SARS-CoV-2 were determined by plaque assay in Vero E6 cells in Minimum Essential Medium supplemented with 4 mM L-glutamine, 0.2% BSA, 10 mM HEPES, and 0.12% NaHCO3 and 0.7% Oxoid agar (OXLP0028B). All SARS-CoV-2 infections were performed in the CDC/USDA-approved BSL-3 facility of the Global Health and Emerging Pathogens Institute at the Icahn School of Medicine at Mount Sinai in accordance with institutional biosafety requirements.
Molecular and biochemical analysis
Additional details in supplemental information.
SARS-CoV-2 qPCR
Primers were used as in the supplemental experimental procedures. The SARS-CoV-2 genome comprises two long open reading frames that encode non-structural polyproteins, followed by a series of largely structural genes. For these latter genes to be produced in the quantities necessary to manufacture mature virions, the replicase/transcriptase complex that is formed by the protein products of the long polyproteins recombines the "leader sequence," which initiates transcription with different lengths of the 3′ end of the genome, generating "subgenomic" vRNA. The N primer sequences were designed so that the forward primer is specific to the leader sequence and the reverse primer is specific to N. In this way, the primers should amplify only subgenomic vRNA, which should be detected only when the viral replication cycle has been initiated, as it is not present in packaged virions.
Data analysis
Data from all the phenotypic assays above were first organized in a Microsoft Excel spreadsheet and analyzed using GraphPad PRISM 8 software or R. For qPCR data analysis, values are expressed as the mean ± SEM. Statistical significance was tested using either one-sided Student’s t test or two-way ANOVA with Tukey's post hoc test for comparison of all sample means.
Data and code availability
All donor-derived and CRISPR-edited hiPSCs are available at the NIMH repository at RUCDR. Hamster brain SARS-CoV-2 RNA-sequencing datasets are available at the Gene Expression Omnibus: GSE161200.
Author contributions
K.J.B., S.A., and B.T. conceptualized this collaborative approach. K.J.B., S.A., B.T., K.D., and D.A.H. contributed to the experimental design and wrote the manuscript. K.D. coordinated all hiPSC-based studies and led the neuron differentiation, B.K. carried out lung differentiation, and C.S. carried out intestinal differentiations. D.K. provided critical expertise in lung organoid differentiation and K.I. and T.T. provided expertise not just in intestinal organoids, but also in frozen differentiated intestinal organoids. D.A.H. coordinated all SARS-CoV-2 infections, assisted by R.M. and K.D. qPCR experiments were supported by C.O., M.I., B.K., and M.F.G. Post-mortem brain studies were done by B.J., C.P., J.F.C., and S.A. High-content imaging was carried out by K.D. shRNA virus production was carried out by P.J.M.D. and K.D. Additional neuronal inductions, viral production, and organoid sectioning and staining were supported by A.M. and M.B.F. Bioinformatics of hiPSC neurons was done by S.P. and that of hamster brains by C.S.
Declaration of interests
The authors declare no competing interests.
Acknowledgments
This work was supported by R56 MH101454 (K.J.B.), R01MH106056 (K.J.B., S.A.), 1U01DA048279-01 (S.A.), the NIH Director's New Innovator Award (DP2 DK128799-01) (T.T.), and the New York Stem Cell Foundation (K.J.B and T.T.), as well as COVID-19 seed fund 0285VV12 from the Icahn School of Medicine at Mount Sinai. All hiPSC research was conducted under the oversight of the Institutional Review Board (IRB) and Embryonic Stem Cell Research Overview (ESCRO) committees at the Icahn School of Medicine at Mount Sinai (ISSMS). Informed consent was obtained from all skin cell donors as part of a study directed by Judith Rapoport MD at the National Institute of Mental Health (NIMH).
Published: March 9, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.stemcr.2021.02.010.
Contributor Information
Schahram Akbarian, Email: kristen.brennand@mssm.edu.
Kristen J. Brennand, Email: schahram.akbarian@mssm.edu.
Supplemental information
References
- Abo K.M., Ma L., Matte T., Huang J., Alysandratos K.D., Werder R.B., Mithal A., Beermann M.L., Lindstrom-Vautrin J., Mostoslavsky G. Human iPSC-derived alveolar and airway epithelial cells can be cultured at air-liquid interface and express SARS-CoV-2 host factors. bioRxiv. 2020 doi: 10.1101/2020.06.03.132639. [DOI] [Google Scholar]
- Bastard P., Rosen L.B., Zhang Q., Michailidis E., Hoffmann H.H., Zhang Y., Dorgham K., Philippot Q., Rosain J., Beziat V. Autoantibodies against type I IFNs in patients with life-threatening COVID-19. Science. 2020;370:eabd4585. doi: 10.1126/science.abd4585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benetti E., Tita R., Spiga O., Ciolfi A., Birolo G., Bruselles A., Doddato G., Giliberti A., Marconi C., Musacchia F. ACE2 gene variants may underlie interindividual variability and susceptibility to COVID-19 in the Italian population. Eur. J. Hum. Genet. 2020;28:1602–1614. doi: 10.1038/s41431-020-0691-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco-Melo D., Nilsson-Payant B.E., Liu W.C., Uhl S., Hoagland D., Moller R., Jordan T.X., Oishi K., Panis M., Sachs D. Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell. 2020;181:1036–1045.e9. doi: 10.1016/j.cell.2020.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciancanelli M.J., Huang S.X., Luthra P., Garner H., Itan Y., Volpi S., Lafaille F.G., Trouillet C., Schmolke M., Albrecht R.A. Infectious disease. Life-threatening influenza and impaired interferon amplification in human IRF7 deficiency. Science. 2015;348:448–453. doi: 10.1126/science.aaa1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coutard B., Valle C., de Lamballerie X., Canard B., Seidah N.G., Decroly E. The spike glycoprotein of the new coronavirus 2019-nCoV contains a furin-like cleavage site absent in CoV of the same clade. Antivir. Res. 2020;176:104742. doi: 10.1016/j.antiviral.2020.104742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixit A., Parnas O., Li B., Chen J., Fulco C.P., Jerby-Arnon L., Marjanovic N.D., Dionne D., Burks T., Raychowdhury R. Perturb-seq: dissecting molecular circuits with scalable single-cell RNA profiling of pooled genetic screens. Cell. 2016;167:1853–1866.e17. doi: 10.1016/j.cell.2016.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobrindt K., Zhang H., Das D., Abdollahi S., Prorok T., Ghosh S., Weintraub S., Genovese G., Powell S.K., Lund A. Publicly available hiPSC lines with extreme polygenic risk scores for modeling schizophrenia. Complex Psychiatr. 2020;6:68–82. doi: 10.1159/000512716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellinghaus D., Degenhardt F., Bujanda L., Buti M., Albillos A., Invernizzi P., Fernandez J., Prati D., Baselli G., Asselta R. Genomewide association study of severe covid-19 with respiratory failure. N. Engl. J. Med. 2020;383:1522–1534. doi: 10.1056/NEJMoa2020283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forrest M.P., Zhang H., Moy W., McGowan H., Leites C., Dionisio L.E., Xu Z., Shi J., Sanders A.R., Greenleaf W.J. Open chromatin profiling in hiPSC-derived neurons prioritizes functional noncoding psychiatric risk variants and highlights neurodevelopmental loci. Cell Stem Cell. 2017 doi: 10.1016/j.stem.2017.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromer M., Roussos P., Sieberts S.K., Johnson J.S., Kavanagh D.H., Perumal T.M., Ruderfer D.M., Oh E.C., Topol A., Shah H.R. Gene expression elucidates functional impact of polygenic risk for schizophrenia. Nat. Neurosci. 2016;19:1442–1453. doi: 10.1038/nn.4399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasperini M., Hill A.J., McFaline-Figueroa J.L., Martin B., Kim S., Zhang M.D., Jackson D., Leith A., Schreiber J., Noble W.S. A genome-wide framework for mapping gene regulation via cellular genetic screens. Cell. 2019;176:377–390 e319. doi: 10.1016/j.cell.2018.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon D.E., Jang G.M., Bouhaddou M., Xu J., Obernier K., White K.M., O'Meara M.J., Rezelj V.V., Guo J.Z., Swaney D.L. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020 doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan W.J., Ni Z.Y., Hu Y., Liang W.H., Ou C.Q., He J.X., Liu L., Shan H., Lei C.L., Hui D.S.C. Clinical characteristics of coronavirus disease 2019 in China. N. Engl. J. Med. 2020;40:275–280. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GTEx Consortium, Battle A., Brown C.D., Engelhardt B.E., Montgomery S.B. Genetic effects on gene expression across human tissues. Nature. 2017;550:204–213. doi: 10.1038/nature24277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y., Duan X., Yang L., Nilsson-Payant B.E., Wang P., Duan F., Tang X., Yaron T.M., Zhang T., Uhl S. Identification of SARS-CoV-2 inhibitors using lung and colonic organoids. Nature. 2021;589:270–275. doi: 10.1038/s41586-020-2901-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helms J., Kremer S., Merdji H., Clere-Jehl R., Schenck M., Kummerlen C., Collange O., Boulay C., Fafi-Kremer S., Ohana M. Neurologic features in severe SARS-CoV-2 infection. N. Engl. J. Med. 2020;382:2268–2270. doi: 10.1056/NEJMc2008597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoagland D.A., Møller R., Uhl S.A., Oishi K., Frere J., Golynker I., Horiuchi S., Panis M., Blanco-Melo D., Sachs D. Leveraging the antiviral type-I interferon system as a first line defense against SARS-CoV-2 pathogenicity. Immunity. 2021 doi: 10.1016/j.immuni.2021.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman G.E., Hartley B.J., Flaherty E., Ladran I., Gochman P., Ruderfer D.M., Stahl E.A., Rapoport J., Sklar P., Brennand K.J. Transcriptional signatures of schizophrenia in hiPSC-derived NPCs and neurons are concordant with post-mortem adult brains. Nat. Commun. 2017;8:2225. doi: 10.1038/s41467-017-02330-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Kruger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Y., Liang W., Zhang J., Li Q., Ou H., Wang Z., Li S., Huang X., Zhao C. Schizophrenia-associated rs4702 G allele-specific downregulation of FURIN expression by miR-338-3p reduces BDNF production. Schizophr Res. 2018;199:176–180. doi: 10.1016/j.schres.2018.02.040. [DOI] [PubMed] [Google Scholar]
- Huang J., Hume A.J., Abo K.M., Werder R.B., Villacorta-Martin C., Alysandratos K.D., Beermann M.L., Simone-Roach C., Lindstrom-Vautrin J., Olejnik J. SARS-CoV-2 infection of pluripotent stem cell-derived human lung alveolar type 2 cells elicits a rapid epithelial-intrinsic inflammatory response. Cell Stem Cell. 2020;27:962–973.e7. doi: 10.1016/j.stem.2020.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Initiative C.-H.G. The COVID-19 Host Genetics Initiative, a global initiative to elucidate the role of host genetic factors in susceptibility and severity of the SARS-CoV-2 virus pandemic. Eur. J. Hum. Genet. 2020;28:715–718. doi: 10.1038/s41431-020-0636-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob A., Vedaie M., Roberts D.A., Thomas D.C., Villacorta-Martin C., Alysandratos K.D., Hawkins F., Kotton D.N. Derivation of self-renewing lung alveolar epithelial type II cells from human pluripotent stem cells. Nat. Protoc. 2019;14:3303–3332. doi: 10.1038/s41596-019-0220-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob F., Pather S.R., Huang W.K., Zhang F., Wong S.Z.H., Zhou H., Cubitt B., Fan W., Chen C.Z., Xu M. Human pluripotent stem cell-derived neural cells and brain organoids reveal SARS-CoV-2 neurotropism predominates in choroid plexus epithelium. Cell stem cell. 2020;27:937–950.e9. doi: 10.1016/j.stem.2020.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kachuri L., Francis S.S., Morrison M.L., Wendt G.A., Bosse Y., Cavazos T.B., Rashkin S.R., Ziv E., Witte J.S. The landscape of host genetic factors involved in immune response to common viral infections. Genome Med. 2020;12:93. doi: 10.1186/s13073-020-00790-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karczewski K.J., Francioli L.C., Tiao G., Cummings B.B., Alfoldi J., Wang Q., Collins R.L., Laricchia K.M., Ganna A., Birnbaum D.P. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581:434–443. doi: 10.1038/s41586-020-2308-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenney A.D., Dowdle J.A., Bozzacco L., McMichael T.M., St Gelais C., Panfil A.R., Sun Y., Schlesinger L.S., Anderson M.Z., Green P.L. Human genetic determinants of viral diseases. Annu. Rev. Genet. 2017;51:241–263. doi: 10.1146/annurev-genet-120116-023425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koike H., Iwasawa K., Ouchi R., Maezawa M., Giesbrecht K., Saiki N., Ferguson A., Kimura M., Thompson W.L., Wells J.M. Modelling human hepato-biliary-pancreatic organogenesis from the foregut-midgut boundary. Nature. 2019;574:112–116. doi: 10.1038/s41586-019-1598-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konno Y., Kimura I., Uriu K., Fukushi M., Irie T., Koyanagi Y., Sauter D., Gifford R.J., Consortium U.-C., Nakagawa S. SARS-CoV-2 ORF3b is a potent interferon antagonist whose activity is increased by a naturally occurring elongation variant. Cell Rep. 2020:108185. doi: 10.1016/j.celrep.2020.108185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamers M.M., Beumer J., van der Vaart J., Knoops K., Puschhof J., Breugem T.I., Ravelli R.B.G., Paul van Schayck J., Mykytyn A.Z., Duimel H.Q. SARS-CoV-2 productively infects human gut enterocytes. Science. 2020;369:50–54. doi: 10.1126/science.abc1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei X., Dong X., Ma R., Wang W., Xiao X., Tian Z., Wang C., Wang Y., Li L., Ren L. Activation and evasion of type I interferon responses by SARS-CoV-2. Nat. Commun. 2020;11:3810. doi: 10.1038/s41467-020-17665-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y.C., Bai W.Z., Hashikawa T. The neuroinvasive potential of SARS-CoV2 may play a role in the respiratory failure of COVID-19 patients. J. Med. Virol. 2020;92:552–555. doi: 10.1002/jmv.25728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindesmith L., Moe C., Marionneau S., Ruvoen N., Jiang X., Lindblad L., Stewart P., LePendu J., Baric R. Human susceptibility and resistance to Norwalk virus infection. Nat. Med. 2003;9:548–553. doi: 10.1038/nm860. [DOI] [PubMed] [Google Scholar]
- Ludvigsson J.F. Systematic review of COVID-19 in children shows milder cases and a better prognosis than adults. Acta Paediatr. 2020;109:1088–1095. doi: 10.1111/apa.15270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Made C.I., Simons A., Schuurs-Hoeijmakers J., van den Heuvel G., Mantere T., Kersten S., van Deuren R.C., Steehouwer M., van Reijmersdal S.V., Jaeger M. Presence of genetic variants among young men with severe COVID-19. JAMA. 2020;324:1–11. doi: 10.1001/jama.2020.13719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao L., Jin H., Wang M., Hu Y., Chen S., He Q., Chang J., Hong C., Zhou Y., Wang D. Neurologic manifestations of hospitalized patients with coronavirus disease 2019 in Wuhan, China. JAMA Neurol. 2020;77:683–690. doi: 10.1001/jamaneurol.2020.1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLoughlin B.C., Miles A., Webb T.E., Knopp P., Eyres C., Fabbri A., Humphries F., Davis D. Functional and cognitive outcomes after COVID-19 delirium. Eur. Geriatr. Med. 2020;11:857–862. doi: 10.1007/s41999-020-00353-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell J.M., Nemesh J., Ghosh S., Handsaker R.E., Mello C., Meyer D., Raghunathan K., de Rivera H., Tegtmeyer M., Hawes D. Mapping genetic effects on cellular phenotypes with "cell villages. bioRxiv. 2020 doi: 10.1101/2020.06.29.174383. [DOI] [Google Scholar]
- Ou X., Liu Y., Lei X., Li P., Mi D., Ren L., Guo L., Guo R., Chen T., Hu J. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat. Commun. 2020;11:1620. doi: 10.1038/s41467-020-15562-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pairo-Castineira E., Clohisey S., Klaric L., Bretherick A.D., Rawlik K., Pasko D., Walker S., Parkinson N., Fourman M.H., Russell C.D. Genetic mechanisms of critical illness in Covid-19. Nature. 2020 doi: 10.1038/s41586-020-03065-y. [DOI] [PubMed] [Google Scholar]
- Pavlovic B.J., Blake L.E., Roux J., Chavarria C., Gilad Y. A comparative assessment of human and chimpanzee iPSC-derived cardiomyocytes with primary heart tissues. Scientific Rep. 2018;8:15312. doi: 10.1038/s41598-018-33478-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilotto A., Odolini S., Masciocchi S., Comelli A., Volonghi I., Gazzina S., Nocivelli S., Pezzini A., Foca E., Caruso A. Steroid-responsive encephalitis in coronavirus disease 2019. Ann. Neurol. 2020;88:423–427. doi: 10.1002/ana.25783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramani A., Muller L., Ostermann P.N., Gabriel E., Abida-Islam P., Muller-Schiffmann A., Mariappan A., Goureau O., Gruell H., Walker A. SARS-CoV-2 targets neurons of 3D human brain organoids. EMBO J. 2020;39:e106230. doi: 10.15252/embj.2020106230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramlall V., Thangaraj P., Meydan C., Foox J., Butler D., May B., de Freitas J., Glicksberg B.S., Mason C., Tatonetti N.P. Identification of immune complement function as a determinant of adverse SARS-CoV-2 infection outcome. medRxiv. 2020 doi: 10.1101/2020.05.05.20092452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo R., Andolfo I., Lasorsa V.A., Iolascon A., Capasso M. Genetic analysis of the coronavirus SARS-CoV-2 host protease TMPRSS2 in different populations. Front. Genet. 2020;11:872. doi: 10.3389/fgene.2020.00872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samson M., Libert F., Doranz B.J., Rucker J., Liesnard C., Farber C.M., Saragosti S., Lapoumeroulie C., Cognaux J., Forceille C. Resistance to HIV-1 infection in caucasian individuals bearing mutant alleles of the CCR-5 chemokine receptor gene. Nature. 1996;382:722–725. doi: 10.1038/382722a0. [DOI] [PubMed] [Google Scholar]
- Schrode N., Ho S.M., Yamamuro K., Dobbyn A., Huckins L., Matos M.R., Cheng E., Deans P.J.M., Flaherty E., Barretto N. Synergistic effects of common schizophrenia risk variants. Nat. Genet. 2019;51:1475–1485. doi: 10.1038/s41588-019-0497-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelton J.F., Shastri A.J., Ye C., Weldon C.H., Filshtein-Somnez T., Coker D., Symons A., Esparza-Gordillo J., Aslibekyan S., Auton A. Trans-ethnic analysis reveals genetic and non-genetic associations with COVID-19 susceptibility and severity. medRxiv. 2020 doi: 10.1101/2020.09.04.20188318. [DOI] [PubMed] [Google Scholar]
- Shilts J., Crozier T.W.M., Greenwood E.J.D., Lehner P.J., Wright G.J. No evidence for basigin/CD147 as a direct SARS-CoV-2 spike binding receptor. Sci. Rep. 2021;11:413. doi: 10.1038/s41598-020-80464-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song E., Zhang C., Israelow B., Lu-Culligan A., Prado A.V., Skriabine S., Lu P., Weizman O.E., Liu F., Dai Y. Neuroinvasion of SARS-CoV-2 in human and mouse brain. J. Exp. Med. 2021;218:e20202135. doi: 10.1084/jem.20202135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takayama K., Akita N., Mimura N., Akahira R., Taniguchi Y., Ikeda M., Sakurai F., Ohara O., Morio T., Sekiguchi K. Generation of safe and therapeutically effective human induced pluripotent stem cell-derived hepatocyte-like cells for regenerative medicine. Hepatol. Commun. 2017;1:1058–1069. doi: 10.1002/hep4.1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanigawa Y., Rivas M. Initial review and analysis of COVID-19 host genetics and associated phenotypes. Preprints. 2020 doi: 10.20944/preprints202003.0356.v1. [DOI] [Google Scholar]
- Thoms M., Buschauer R., Ameismeier M., Koepke L., Denk T., Hirschenberger M., Kratzat H., Hayn M., Mackens-Kiani T., Cheng J. Structural basis for translational shutdown and immune evasion by the Nsp1 protein of SARS-CoV-2. Science. 2020;369:1249–1255. doi: 10.1126/science.abc8665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toscano G., Palmerini F., Ravaglia S., Ruiz L., Invernizzi P., Cuzzoni M.G., Franciotta D., Baldanti F., Daturi R., Postorino P. Guillain-barre syndrome associated with SARS-CoV-2. N. Engl. J. Med. 2020;382:2574–2576. doi: 10.1056/NEJMc2009191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tryka K.A., Hao L., Sturcke A., Jin Y., Wang Z.Y., Ziyabari L., Lee M., Popova N., Sharopova N., Kimura M. NCBI's database of genotypes and phenotypes: dbGaP. Nucleic Acids Res. 2014;42:D975–D979. doi: 10.1093/nar/gkt1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181:281–292.e6. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K., Chen W., Zhang Z., Deng Y., Lian J.Q., Du P., Wei D., Zhang Y., Sun X.X., Gong L. CD147-spike protein is a novel route for SARS-CoV-2 infection to host cells. Signal. Transduct. Target Ther. 2020;5:283. doi: 10.1038/s41392-020-00426-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wruck W., Adjaye J. SARS-CoV-2 receptor ACE2 is co-expressed with genes related to transmembrane serine proteases, viral entry, immunity and cellular stress. Sci. Rep. 2020;10:21415. doi: 10.1038/s41598-020-78402-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia H., Cao Z., Xie X., Zhang X., Chen J.Y., Wang H., Menachery V.D., Rajsbaum R., Shi P.Y. Evasion of type I interferon by SARS-CoV-2. Cell Rep. 2020;33:108234. doi: 10.1016/j.celrep.2020.108234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan R., Zhang Y., Li Y., Xia L., Guo Y., Zhou Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;367:1444–1448. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang N., Chanda S., Marro S., Ng Y.H., Janas J.A., Haag D., Ang C.E., Tang Y., Flores Q., Mall M. Generation of pure GABAergic neurons by transcription factor programming. Nat. Methods. 2017;14:621–628. doi: 10.1038/nmeth.4291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L., Han Y., Nilsson-Payant B.E., Gupta V., Wang P., Duan X., Tang X., Zhu J., Zhao Z., Jaffre F. A human pluripotent stem cell-based platform to study SARS-CoV-2 tropism and model virus infection in human cells and organoids. Cell Stem Cell. 2020;27:125–136.e7. doi: 10.1016/j.stem.2020.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan M., Wu N.C., Zhu X., Lee C.-C.D., So R.T.Y., Lv H., Mok C.K.P., Wilson I.A. A highly conserved cryptic epitope in the receptor-binding domains of SARS-CoV-2 and SARS-CoV. Science. 2020:eabb7269. doi: 10.1126/science.abb7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Pak C., Han Y., Ahlenius H., Zhang Z., Chanda S., Marro S., Patzke C., Acuna C., Covy J. Rapid single-step induction of functional neurons from human pluripotent stem cells. Neuron. 2013;78:785–798. doi: 10.1016/j.neuron.2013.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R.R., Koido M., Tadokoro T., Ouchi R., Matsuno T., Ueno Y., Sekine K., Takebe T., Taniguchi H. Human iPSC-derived posterior gut progenitors are expandable and capable of forming gut and liver organoids. Stem Cell Reports. 2018;10:780–793. doi: 10.1016/j.stemcr.2018.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Bastard P., Liu Z., Le Pen J., Moncada-Velez M., Chen J., Ogishi M., Sabli I.K.D., Hodeib S., Korol C. Inborn errors of type I IFN immunity in patients with life-threatening COVID-19. Science. 2020;370:eabd4570. doi: 10.1126/science.abd4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Li C., Liu X., Chiu M.C., Zhao X., Wang D., Wei Y., Lee A., Zhang A.J., Chu H. Infection of bat and human intestinal organoids by SARS-CoV-2. Nat. Med. 2020;26:1077–1083. doi: 10.1038/s41591-020-0912-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All donor-derived and CRISPR-edited hiPSCs are available at the NIMH repository at RUCDR. Hamster brain SARS-CoV-2 RNA-sequencing datasets are available at the Gene Expression Omnibus: GSE161200.