Abstract
Purpose
To analyze the characteristics and trends of drug resistance for Klebsiella pneumoniae (K. pneumoniae), isolated from urinary tract infections (UTIs), to common antibiotics used in clinics.
Methods
This retrospective study was conducted in a teaching hospital in Chongqing from 2011 to 2019. Laboratory data of isolated bacteria were collected and analyzed.
Results
Among the 17,966 non-repetitive strains isolated from the urine sample, a total of 1543 K. pneumoniae isolates were identified, with an isolation frequency secondary only to Escherichia coli (E. coli) and there was a peak in the K. pneumoniae isolates in the year 2013. During the period, the rate of extended-spectrum β-lactamase (ESBL)-producing K. pneumoniae fell from 48.4% in 2011 to 32.9% in 2019, and a marked jump of resistance was seen in carbapenems from 2.2% to 18.0%. The peak of carbapenem resistance rate (22.6%) to K. pneumoniae was observed in 2017 along with a low ESBL-producing rate (30.9%). Piperacillin/tazobactam and cefepime resistance levels went up from 4.4% to 25.7% and from 18.2% to 30.5%, respectively. Moreover, the K. pneumoniae isolates resistance rate to carbapenems and amikacin gradually grew up, showing their peaks in 2017, and then dropped year by year. However, ceftazidime and aztreonam resistance levels were relatively stable, fluctuating between 21.8% and 35.6%, 32.2% and 39.4%, respectively.
Conclusion
There is a significant upward tendency in carbapenem resistance rate and a downward tendency in ESBL-production rate in K. pneumoniae isolates from UTIs, and continuous surveillance is necessary in the future.
Keywords: urinary tract infections, Klebsiella pneumoniae, carbapenem resistance, extended-spectrum β-lactamase
Introduction
Urinary tract infections (UTIs) are the most frequent bacterial infection in primary care, affecting 150 million people per year worldwide.1 UTI is an inflammatory reaction caused by the invasion of pathogenic microorganisms in the urinary tract, which is a common infection among the inpatients in recent years, and the inappropriate use and abuse of broad-spectrum antibiotics for decolonization not only results in a decrease in the effectiveness of standard treatments, but also leads to the emergence of multidrug resistance.2,3 A modelling study showed that about 3 million severe infections were caused by carbapenem-resistant Escherichia coli (E. coli) and Klebsiella pneumoniae (K. pneumoniae) worldwide.4
Accelerated emergence and effective propagation of carbapenem-resistant K. pneumoniae (CR-KPN) across the world have become a prominent public health challenge due to high mortality rate in healthcare-associated nosocomial infections.5,6 Approximately 60%-80% of UTIs are caused by E. coli and 3%-10% by K. pneumoniae.7–9 This last is an important pathogen that causes a wide range of infections such as pneumonia and bloodstream infections, commonly in neonates and patients admitted in intensive care units. Carbapenems is the last defense against extended-spectrum β-lactamase (ESBL)-producing K. pneumoniae and their frequent use has resulted in the generation and spread of CR-KPN, a clinically significant carbapenem-resistant Enterobacteriales (CRE).10 Since outbreak of CR-KPN in New York hospitals in the early 2000s, this pathogen has spread throughout the United States and worldwide.11,12 In China, CR-KPN infections prevailed from 2.9% in 2005 to 40% in 2017 and the distribution differed greatly by region, with the lowest in northeast and the highest in eastern region of China.13–15 Here, we investigated the distribution and drug resistance trends of K. pneumoniae causing UTIs in a tertiary teaching hospital from 2011 to 2019 to provide reference for rational use of antibiotics.
Materials and Methods
Data Collection
This retrospective study data were collected in the microbiology laboratory of the First Affiliated Hospital of Chongqing Medical University using WHONET software. Strains were isolated from various patients’ urine culture routinely according to the hospital laboratory procedure, and only the first isolate was included in our study. Duplicate isolates, defined as the same bacterial species from the same inpatient during the same inpatient stay, were excluded from analysis.
Antimicrobial Susceptibility Testing
A total of 1543 K. pneumoniae isolates recovered from urine were correctly identified by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) (bioMérieux, MO, France), assisted with Vitek-2 compact system (bioMérieux, MO, France) according to the manufacturer’s instructions. All the isolates were sub-cultured on blood agar and MacConkey agar plates for purity check to confirm species. The susceptibility testing was performed according to the Clinical and Laboratory Standards Institute (CLSI) criteria using the automated Vitek-2 compact system, and further on Mueller Hinton’s agar by the Kirby-Bauer Disk Diffusion method for the following antimicrobials: aztreonam (30μg), meropenem (10μg), ciprofloxacin (5μg) and gentamicin (30μg). The ESBL test was identified by clavulanic acid synergy test, which is included in the AST-GN334 card (bioMérieux, Marcy l’Etoile, France).16 Isolates were defined as CR-KPN if they were resistant to any one of carbapenems tested. MIC results were interpreted according to the CLSI guidelines.17 Quality control was performed with each run using the strains Enterobacter hormaechei ATCC 700323 and Escherichia coli ATCC 25922, to ensure reproducibility of the antibiotic susceptibility testing procedure.
Results
Distribution of Strains
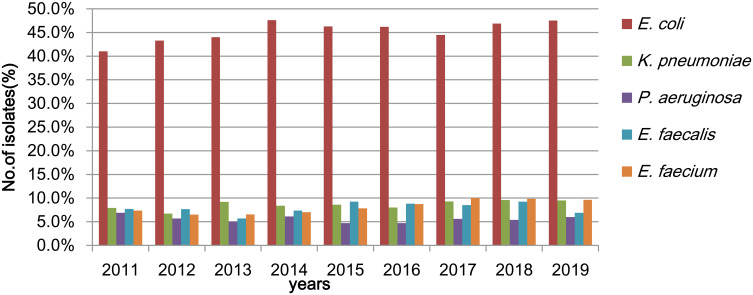
A total of 17,966 strains were isolated from urine culture during the 9 years of study period in our hospital in Chongqing, Southwest China. The top five species among the total identified isolates were E. coli, K. pneumoniae, Enterococcus faecium, Enterococcus faecalis and Pseudomonas aeruginosa. The annual isolation number of K. pneumoniae ranged from 91 to 205 during the 9 years, with 91 (7.9%) strains in the year 2011, 137 (6.7%) in 2012, 205 (9.2%) in 2013, 189 (8.4%) in 2014, 203 (8.6%) in 2015, 177 (8.0%) in 2016, 191 (9.3%) in 2017, 182 (9.6%) in 2018, and 167 (9.5%) in 2019. There was a peak in the K. pneumoniae isolates in the year 2018, accounting for up to 9.6% of the total isolates (Figure 1). Of note, the vast majority of the strains were isolated from the hospitalized patients (data not shown).
Figure 1.
Distribution of strains isolated from urine for the 9-year study.
Abbreviations:E. coli, Escherichia coli; K. pneumoniae, Klebsiella pneumoniae; P. aeruginosa, Pseudomonas aeruginosa; E. faecalis, Enterococcus faecalis; E. faecium, Enterococcus faecium.
Characteristics of K. pneumoniae
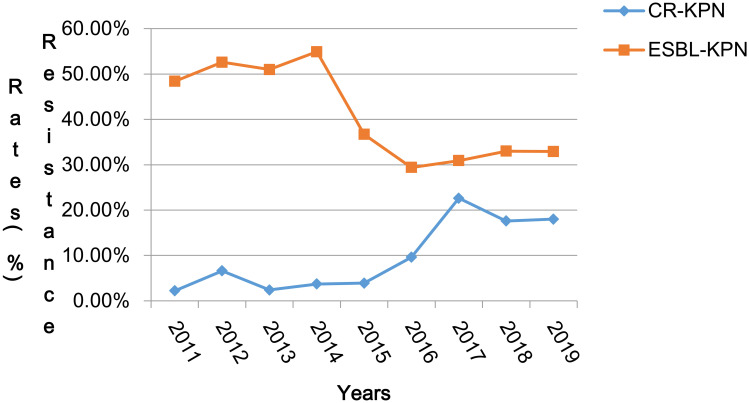
Temporal trends and changes in both the rates of ESBL-production and carbapenem-resistance, and antimicrobial resistance levels in urinary K. pneumoniae isolates over the study period are presented in Figure 2 and Table 1. During this period, the rate of ESBL-producing K. pneumoniae reduced from 48.4% in 2011 to 32.9% in 2019, but a marked increase of resistance against carbapenems was seen from 2.2 to 18.0%. The peak of carbapenem resistance (22.6%) was observed in 2017 along with the low ESBL-production rate (30.9%) in K. Pneumoniae. (Figure 2)
Figure 2.
Rates of ESBL-producing and Carbapenem resistant-K. pneumoniae from 2011 to 2019.
Abbreviations: CR-KPN, carbapenem resistant-K. pneumoniae; ESBL-KPN, ESBL-producing K. pneumoniae.
Table 1.
Resistance Rates (%) of Urine K. pneumoniae Isolates to Antimicrobial Agents
| Antibiotics | 2012 | 2013 | 2014 | 2015 | 2016 | 2017 | 2018 | 2019 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| n = 137 | n = 205 | n = 189 | n = 203 | n = 177 | n = 191 | n = 182 | n = 167 | |||||||||
| n | R% | n | R% | n | R% | n | R% | n | R% | n | R% | n | R% | n | R% | |
| SAM | 137 | 62.0 | 205 | 56.1 | 183 | 59.0 | 188 | 49.5 | 172 | 55.8 | 188 | 60.6 | 182 | 50.5 | / | / |
| TZP | 137 | 4.4 | 205 | 1.5 | 183 | 2.2 | 188 | 5.9 | 172 | 11.6 | 188 | 23.4 | 182 | 13.7 | 167 | 25.7 |
| CAZ | 137 | 31.4 | 205 | 28.8 | 183 | 26.8 | 188 | 21.8 | 172 | 27.3 | 188 | 35.6 | 182 | 28.0 | 167 | 29.3 |
| CRO | 137 | 56.9 | 205 | 56.6 | 183 | 53.0 | 188 | 43.1 | 172 | 43.6 | 190 | 53.7 | 182 | 45.6 | 167 | 46.7 |
| FEP | 137 | 18.2 | 205 | 17.6 | 183 | 15.3 | 188 | 11.2 | 172 | 19.2 | 188 | 30.9 | 182 | 22.0 | 167 | 30.5 |
| CTT | 137 | 2.9 | 205 | 0.5 | 183 | 2.7 | 188 | 4.3 | 172 | 11.0 | 188 | 19.1 | 182 | 15.4 | / | / |
| FOX | / | / | / | / | / | / | 155 | 13.5 | 158 | 24.7 | 180 | 28.9 | 181 | 28.7 | 136 | 28.7 |
| ATM | 137 | 35.8 | 205 | 36.6 | 183 | 36.6 | 188 | 32.4 | 172 | 32.6 | 188 | 39.4 | 180 | 32.2 | 145 | 34.5* |
| ETP | 133 | 6.0 | 200 | 2.0 | 182 | 3.3 | 182 | 4.4 | 171 | 9.9 | 190 | 22.6 | 182 | 17.0 | 167 | 16.8 |
| IPM | 137 | 1.5 | 205 | 0 | 183 | 0.5 | 188 | 4.3 | 172 | 8.7 | 190 | 20.0 | 182 | 13.7 | 167 | 11.4 |
| MEM | / | / | / | / | / | / | 163 | 4.3* | 159 | 8.2* | 184 | 18.5* | 181 | 13.8* | 153 | 11.1* |
| AMK | 137 | 7.3 | 205 | 6.8 | 175 | 6.3 | 175 | 6.9 | 164 | 10.4 | 181 | 21.5 | 182 | 15.4 | 167 | 10.8 |
| GEN | 137 | 42.3 | 205 | 51.7 | 183 | 41.5 | 188 | 33.5 | 175 | 38.3 | 190 | 45.8 | 182 | 34.1 | 51 | 23.5 |
| TOB | 137 | 14.6 | 204 | 21.1 | 183 | 22.4 | 188 | 14.9 | 172 | 16.9 | 188 | 33.0 | 182 | 19.8 | / | / |
| CIP | 137 | 55.5 | 205 | 51.2 | 183 | 59.0 | 188 | 48.9 | 172 | 54.1 | 188 | 58.5 | 182 | 52.2 | / | / |
| LVX | 137 | 38.0 | 205 | 35.6 | 183 | 41.0 | 188 | 37.8 | 172 | 39.0 | 188 | 38.3 | 182 | 40.1 | 167 | 36.5 |
| SXT | 50 | 54.0 | 205 | 53.2 | 183 | 60.1 | 187 | 46.0 | 172 | 54.7 | 188 | 50.5 | 181 | 41.4 | 167 | 43.1 |
Notes: *The data are results using Kirby-Bauer method; /, the data are not detected.
Abbreviations: SAM, ampicillin/sulbactam; TZP, piperacillin/tazobactam; CAZ, ceftazidime; CRO, ceftriaxone; FEP, cefepime; CTT, cefotetan; FOX, cefoxitin; ATM, aztreonam; ETP, ertapenem; IPM, imipenem; MEM, meropenem; AMK, amikacin; GEN, gentamicin; TOB, tobramycin; CIP, ciprofloxacin; LVX, levofloxacin; SXT, sulfamethoxazole/trimethoprim.
Antibioresistence Profiles of K. pneumoniae
Antimicrobial agents checked included quinolones (ciprofloxacin and levofloxacin), sulfonamides (cotrimoxazole), third-generation cephalosporins (ceftazidime and ceftriaxone), fourth-generation cephalosporins (cefepime), cephamycins (cefoxitin and cefotetan), other penicillins (piperacillin/tazobactam), carbapenems (ertapenem, imipenem and meropenem), aminoglycosides (gentamicin, amikacin and tobramycin) and monocyclic β-lactam antibiotics (aztreonam). With regard to drug susceptibility of K. pneumoniae, ESBL producers accounted from 48.4% in 2011 to 32.9% in 2019 (Figure 2). ESBL-producing K. pneumoniae showed a high resistance rate to the antibiotics except cefoxitin, cefotetan, piperacillin/tazobactam, amikacin and carbapenems, while CRKP was highly resistant to all antibiotics (Table 2).
Table 2.
Resistance Rates of Urine K. pneumoniae Isolates with Diverse Characteristics to Antimicrobial Agents
| Antibiotics | ESBL(+)-KPN | ESBL(-)-KPN | CR-KPN | CSE-KPN | ||||
|---|---|---|---|---|---|---|---|---|
| R (%) | S (%) | R (%) | S (%) | R (%) | S (%) | R (%) | S (%) | |
| SAM | 85.3 (480/563) | 2.8 (16/563) | 34.5 (268/777) | 60.6 (471/777) | 100.0 (122/122) | 0 (0/122) | 50.8 (599/1180) | 40.6 (480/1180) |
| TZP | 6.6 (41/617) | 83.6 (516/617) | 13.6 (120/884) | 82.4 (728/884) | 79.2 (118/149) | 9.4 (14/149) | 3.2 (42/1314) | 91.2 (1198/1314) |
| CAZ | 43.6 (269/617) | 50.2 (310/617) | 18.3 (162/885) | 80.5 (712/885) | 96.6 (144/149) | 3.4 (5/149) | 20.5 (269/1314) | 75.9 (997/1314) |
| CRO | 95.8 (591/617) | 3.4 (21/617) | 17.8 (158/886) | 81.6 (723/886) | 98.7 (147/149) | 1.3 (2/149) | 44.3 (583/1316) | 55.1 (725/1316) |
| FEP | 25.6 (158/617) | 70.5 (435/617) | 12.2 (108/885) | 86.9 (769/885) | 85.9 (128/149) | 8.7 (13/149) | 10.3 (135/1314) | 88.1 (1158/1314) |
| CTT | 1.8 (10/563) | 96.4 (543/563) | 11.7 (91/777) | 86.5 (672/777) | 76.2 (93/122) | 14.8 (18/122) | 0.5 (6/1180) | 98.6 (1163/1180) |
| FOX | 22.9 (64/279) | 71.3 (199/279) | 25.8 (138/534) | 71.2 (380/534) | 99.1 (111/112) | 0.9 (1/112) | 12.8 (85/664) | 82.8 (550/664) |
| ATM | 62.6 (382/610) | 35.7 (218/610) | 15.0 (131/872) | 84.5 (737/872) | 92.3 (132/143) | 7.0 (10/143) | 28.0 (364/1300) | 71.0 (923/1300) |
| ETP | 4.2 (26/615) | 94.6 (582/615) | 13.5 (119/880) | 85.8 (755/880) | 100.0 (149/149) | 0 (0/149) | 0 (0/1316) | 100.0 (1316/1316) |
| IPM | 0.6 (4/617) | 98.9 (610/617) | 11.5 (102/886) | 88.0 (780/886) | 71.8 (107/149) | 22.8 (34/149) | 0 (0/1316) | 100.0 (1316/1316) |
| MEM | 1.4 (4/287) | 97.2 (279/287) | 16.2 (90/557) | 83.5 (465/557) | 80.0 (92/115) | 15.7 (18/115) | 0 (0/686) | 100 (686/686) |
| AMK | 8.2 (49/597) | 91.6 (547/597) | 12.2 (106/868) | 87.8 (762/868) | 66.9 (97/145) | 33.1 (48/145) | 4.3 (55/1284) | 95.6 (1228/1284) |
| GEN | 64.8 (378/583) | 34.1 (199/583) | 23.0 (190/826) | 76.3 (630/826) | 76.3 (100/131) | 22.9 (30/131) | 36.5 (451/1235) | 62.9 (777/1235) |
| TOB | 26.3 (148/563) | 29.3 (165/563) | 16.1 (125/777) | 71.9 (559/777) | 76.2 (93/122) | 10.7 (13/122) | 14.6 (172/1180) | 58.8 (694/1180) |
| CIP | 51.1 (313/613) | 40.6 (249/613) | 27.5 (241/876) | 70.2 (615/876) | 91.7 (133/145) | 6.9 (10/145) | 30.7 (400/1305) | 64.2 (838/1305) |
| LVX | 38.9 (240/617) | 54.0 (333/617) | 24.1 (213/884) | 73.0 (645/884) | 87.2 (130/149) | 11.4 (17/149) | 23.2 (305/1314) | 71.6 (941/1314) |
| SXT | 80.4 (456/567) | 19.6 (111/567) | 32.8 (277/846) | 67.2 (569/846) | 60.6 (87/143) | 39.4 (56/143) | 51.0 (630/1236) | 49.0 (606/1236) |
Abbreviations: ESBL(+)-KPN, ESBL-producing K. pneumoniae; ESBL(-)-KPN, non ESBL-producing K. pneumoniae; CR-KPN, carbapenem-resistant K. pneumoniae; CS-KPN, carbapenem-sensitive K. pneumoniae; SAM, ampicillin/sulbactam; TZP, piperacillin/tazobactam; CAZ, ceftazidime; CRO, ceftriaxone; FEP, cefepime; CTT, cefotetan; FOX, cefoxitin; ATM, aztreonam; ETP, ertapenem; IPM, imipenem; MEM, meropenem; AMK, amikacin; GEN, gentamicin; TOB, tobramycin; CIP, ciprofloxacin; LVX, levofloxacin; SXT, sulfamethoxazole/trimethoprim.
Moreover, piperacillin/tazobactam and cefepime resistance levels increased from 4.4% to 25.7% and from 18.2% to 30.5%, respectively (Table 1). The resistance rate of carbapenems and amikacin gradually rose from 2011 to 2017, reaching its peaking in 2017, and then went down year by year. However, ceftazidime and aztreonam resistance levels were stable, fluctuating between 21.8% and 35.6%, 32.2% and 39.4%, respectively (Table 1).
Discussion
Our study highlights the decreasing detection rate of ESBL-producing coupled with rising resistance rates of K. pneumoniae to carbapenems. Isolates of K. pneumoniae are becoming increasingly resistant to antibiotics and subsequently may become even more difficult to be treated.18 Overall, we found a significant reduction of ESBL detection rate for the K. pneumoniae in UTIs from 2011 to 2019. Fortunately, from 2011 onwards, there is a requirement for all hospitals in China to form an antibiotic administrative group with the aim to enforce formulary restrictions and total inpatient consumption of antibiotics (>40 defined daily doses/100 inpatient days).19 The outbreaks of CR-KPN have become increasingly common in China,20,21 and we found a growing prevalence of CR-KPN in the UTIs through retrospective analysis, consistent with findings from previous large surveillance studies.13
We found that the detection of CR-KPN isolates from UTI patients reached its peak in 2017, accompanied by the high resistance rates to carbapenems and amikacin. While piperacillin/tazobactam and cefepime resistance levels increased year after year, ceftazidime and aztreonam resistance levels were relatively stable. Of note, CR-KPN was almost resistant to all the antibiotics tested. High rates of bacterial resistance are often correlated with high rates of antibiotic use and intra- and inter-hospital spread of antibiotic-resistant bacteria.22 In addition, increasing consumption of β-lactam/β-lactamase inhibitors may also increase the frequency of carbapenem-resistant Klebsiella spp.23–25
K. pneumoniae can cause serious, life-threatening infections in humans in endemic and epidemic settings.26 Resistance to carbapenems in Enterobacteriales is primarily linked to different mechanisms, in particular the production of carbapenemases.10 Carbapenems are the main defense against ESBL-producing pathogens, the overuse of which has resulted in the generation and spread of CR-KPN, which is a clinically significant CRE. A series of molecular epidemiology studies on CRE in China have shown that the outbreaks caused by KPC-2-producing K. pneumoniae in these regions were due to the spread of the predominant clone ST11, which is a variant of international clone ST258.26–30 ST258 is strongly associated with KPC production and multidrug resistance, but it may have inherent traits which are responsible for its high rate of transmissibility that results from two distinct genetic clades with a hot spot for DNA recombination.31
There are some limitations of this work. This study is retrospective in nature, so some potential confounders, such as the length of stay, patients from different wards, and time of collection of urine samples, could not be ascertained while observing the trends. At the same time, ESBLs production was not detected by molecular biological methods, so there may be lower detection rates due to the influence of carbapenem production in the CR-KPN isolates on ESBLs detection.
Conclusion
Taken together, our data demonstrated that there was a significant rise in the prevalence of carbapenem resistance and a decline in ESBL production in K. pneumoniae isolates from the patients with UTIs in our hospital. Further work and continuous surveillance are needed to advance the rational and judicious use of antimicrobial agents.
Acknowledgments
The authors would like to thank Xiaofei Xi (an English teacher in international Curriculum Centre of High School Affiliated to South West University, Chongqing, China) for the effort to make the language more accurate and more appropriate.
Funding Statement
This study was supported by the National Natural Science Foundation of China (Grant No. 82072349), and the Medical Research Program of Chongqing Health and Family Planning Commission (No. 2018MSXM009).
Data Sharing Statement
The data sets used and analyzed during the current study are available from the corresponding author on reasonable request.
Ethical Approval
The samples were part of the routine hospital laboratory procedure, not collected for this study, so an institutional review board will not be required.
Author Contributions
All authors read and approved the final manuscript. All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Flores-Mireles AL, Walker JN, Caparon M, Hultgren SJ. Urinary tract infections: epidemiology, mechanisms of infection and treatment options. Nat Rev Microbiol. 2015;13(5):269–284. doi: 10.1038/nrmicro3432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ikram R, Psutka R, Carter A, Priest P. An outbreak of multi-drug resistant Escherichia coli urinary tract infection in an elderly population: a case-control study of risk factors. BMC Infect Dis. 2015;15:224–230. doi: 10.1186/s12879-015-0974-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sun JD, Huang SF, Yang SS, Pu SL, Zhang CM, Zhang LP. Impact of carbapenem heteroresistance among clinical isolates of invasive Escherichia coli in Chongqing, southwestern China. Clin Microbiol Infect. 2015;21(5):469.e1–e10. doi: 10.1016/j.cmi.2014.12.013 [DOI] [PubMed] [Google Scholar]
- 4.Temkin E, Fallach N, Almagor J, et al. Estimating the number of infections caused by antibiotic-resistant Escherichia coli and Klebsiella pneumoniae in 2014: a modelling study. Lancet Glob Health. 2018;6(9):e969–e979. doi: 10.1016/S2214-109X(18)30278-X [DOI] [PubMed] [Google Scholar]
- 5.Pitout JDD, Nordmann P, Poirel L. Carbapenemase-producing Klebsiella pneumoniae, a key pathogen set for global nosocomial dominance. Antimicrob Agents Chemother. 2015;59(10):5873–5884. doi: 10.1128/AAC.01019-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davarci I, Senbayrak S, Aksaray S, Kocoglu ME, Kuskucu MA, Samasti M. Molecular epidemiology of carbapenem-resistant Klebsiella pneumoniae isolates. Anadolu Klinigi. 2019;24(1):1–7. doi: 10.21673/anadoluklin.423081 [DOI] [Google Scholar]
- 7.Koeijers JJ, Verbon A, Kessels AGH, et al. Urinary tract infection in male general practice patients: uropathogens and antibiotic susceptibility. Urology. 2010;76(2):336–340. doi: 10.1016/j.urology.2010.02.052 [DOI] [PubMed] [Google Scholar]
- 8.Sokhn ES, Salami A, Roz AEI, Salloum L, Bahmad HF, Ghssein G. Antimicrobial susceptibilities and laboratory profiles of Escherichia coli, Klebsiella pneumoniae, and Proteus mirabilis isolates as agents of urinary tract infection in Lebanon: paving the way for better diagnostics. Med Sci (Basel). 2020;8(3):32–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Van Driel AA, Notermans DW, Meima A, et al. Antibiotic resistance of Escherichia coli isolated from uncomplicated UTI in general practice patients over a 10-year period. Eur J Clin Microbiol Infect Dis. 2019;38(11):2151–2158. doi: 10.1007/s10096-019-03655-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ravi Shankar P, Balasubramanium R. World Health Organization antimicrobial resistance: global report on surveillance 2014 (Book Review). Australas Med J. 2014;7(5):238–239. [Google Scholar]
- 11.Arnold RS, Thom KA, Sharma S, Phillips M, Kristie Johnson J, Morgan DJ. Emergence of Klebsiella pneumoniae carbapenemase-producing bacteria. South Med J. 2011;104(1):40–45. doi: 10.1097/SMJ.0b013e3181fd7d5a [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guh AY, Limbago BM, Kallen AJ. Epidemiology and prevention of carbapenem-resistant Enterobacteriaceae in the United States. Expert Rev Anti Infect Ther. 2014;12(5):565–580. doi: 10.1586/14787210.2014.902306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hu F-P, Guo Y, Zhu D-M, et al. Resistance trends among clinical isolates in China reported from CHINET surveillance of bacterial resistance, 2005–2014. Clin Microbiol Infect. 2016;22(Suppl 1):S9–S14. doi: 10.1016/j.cmi.2016.01.001 [DOI] [PubMed] [Google Scholar]
- 14.Cusack TP, Ashley EA, Ling CL, et al. Time to switch from CLSI to EUCAST? A Southeast Asian perspective. Clin Microbiol Infect. 2019;25(7):782–785. doi: 10.1016/j.cmi.2019.03.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu A, Zheng B, Xu Y-C, Huang Z-G, Zhong N-S, Zhuo C. National epidemiology of carbapenem-resistant and extensively drug-resistant Gram-negative bacteria isolated from blood samples in China in 2013. Clin Microbiol Infect. 2016;22(Suppl 1):S1–S8. doi: 10.1016/j.cmi.2015.09.015 [DOI] [PubMed] [Google Scholar]
- 16.VITEK®2 (AST-N334) [package insert]. Marcy I’Etoile, France: bioMérieux SA; 2018. [Google Scholar]
- 17.Clinical and Laboratory Standards Institute (CLSI). Performance Standards for Antimicrobial Susceptibility Testing. 29th ed. CLSI supplement M100 Wayne, PA: CLSI; 2019 [Google Scholar]
- 18.Khan F, Siddiqui N, Sultan A, Rizvi M, Shukla I, Khan HM. Seasonal variation in Klebsiella pneumoniae blood stream infection: a five year study. Clin Microbiol. 2016;5(2):247. doi: 10.4172/2327-5073.1000247 [DOI] [Google Scholar]
- 19.Xiao Y, Zhang J, Zheng B, Zhao L, Li S, Li. L. Changes in Chinese policies to promote the rational use of antibiotics. PLoS Med. 2013;10(11):e1001556. doi: 10.1371/journal.pmed.1001556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang X, Xu X, Li Z, et al. An outbreak of a nosocomial NDM-1-producing Klebsiella pneumoniae ST147 at a teaching hospital in Mainland China. Microb Drug Resist. 2014;20(2):144–149. doi: 10.1089/mdr.2013.0100 [DOI] [PubMed] [Google Scholar]
- 21.Zhou J, Li G, Ma X, Yang Q, Yi J. Outbreak of colonization by carbapenemase-producing Klebsiella pneumoniae in a neonatal intensive care unit: investigation, control measures and assessment. Am J Infect Control. 2015;43(10):1122–1124. doi: 10.1016/j.ajic.2015.05.038 [DOI] [PubMed] [Google Scholar]
- 22.Agodi A, Barchitta M, Quattrocchi A, et al. Antibiotic trends of Klebsiella pneumoniae and Acinetobacter baumannii resistance indicators in an intensive care unit of Southern Italy, 2008–2013. Antimicrob Resist Infect Control. 2015;4:43–49. doi: 10.1186/s13756-015-0087-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Federico MP, Furtado GH. Immediate and later impacts of antimicrobial consumption on carbapenem-resistant Acinetobacter spp., Pseudomonas aeruginosa, and Klebsiella spp. in a teaching hospital in Brazil: a 10-year trend study. Eur J Clin Microbiol Infect Dis. 2018;37(11):2153–2158. doi: 10.1007/s10096-018-3352-1 [DOI] [PubMed] [Google Scholar]
- 24.Candevir Ulu A, Kurtaran B, Inal AS, et al. Risk factors of carbapenem-resistant Klebsiella pneumoniae infection: a serious threat in ICUs. Med Sci Monit. 2015;21:219–224. doi: 10.12659/MSM.892516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qu X, Wang H, Chen C, et al. Surveillance of carbapenem-resistant Klebsiella pneumoniae in Chinese hospitals - a five-year retrospective study. J Infect Dev Ctries. 2019;13(12):1101–1107. doi: 10.3855/jidc.11798 [DOI] [PubMed] [Google Scholar]
- 26.Jiang Y, Wei Z, Wang Y, Hua X, Feng Y, Yu Y. Tracking a hospital outbreak of KPC-producing ST11 Klebsiella pneumoniae with whole-genome sequencing. Clin Microbiol Infect. 2015;21(11):1001–1007. doi: 10.1016/j.cmi.2015.07.001 [DOI] [PubMed] [Google Scholar]
- 27.Li H, Zhang J, Liu Y, et al. Molecular characteristics of carbapenemase-producing Enterobacteriaceae in China from 2008 to 2011: predominance of KPC-2 enzyme. Diagn Microbiol Infect Dis. 2014;78(1):63–65. doi: 10.1016/j.diagmicrobio.2013.10.002 [DOI] [PubMed] [Google Scholar]
- 28.Qi Y, Wei Z, Ji S, Du X, Shen P, Yu Y. ST11, the dominant clone of KPC-producing Klebsiella pneumoniae in China. J Antimicrob Chemother. 2011;66(2):307–312. doi: 10.1093/jac/dkq431 [DOI] [PubMed] [Google Scholar]
- 29.Zou C, Wei J, Shan B, Chen X, Wang D, Niu S. In vitro activity of ceftazidime-avibactam and aztreonam-avibactam against carbapenem-resistant Enterobacteriaceae isolates collected from three secondary hospitals in Southwest China Between 2018 and 2019. Infect Drug Resist. 2020;13:3563–3568. doi: 10.2147/IDR.S273989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wei J, Zou C, Wang D, Huang A, Niu S. Genetic diversity and in vitro activity of ceftazidime/avibactam and aztreonam/avibactam against imipenem-resistant Enterobacteriaceae isolates in Southwest China: a single-center study. J Glob Antimicrob Resist. 2020;22:448–451. doi: 10.1016/j.jgar.2020.04.023 [DOI] [PubMed] [Google Scholar]
- 31.Deleo FR, Chen L, Porcella SF, et al. Molecular dissection of the evolution of carbapenem-resistant multilocus sequence type 258 Klebsiella pneumoniae. Proc Natl Acad Sci U S A. 2014;111(13):4988–4993. doi: 10.1073/pnas.1321364111 [DOI] [PMC free article] [PubMed] [Google Scholar]