Abstract
Renal epidermal growth factor receptor (EGFR) signaling is activated in models of diabetic nephropathy (DN), and inhibition of the EGFR signaling pathway protects against the development of DN. We have now determined that in cultured podocytes, high glucose led to increases in activation of EGFR signaling but decreases in autophagy activity as indicated by decreased beclin-1 and inhibition of LC3B autophagosome formation as well as increased rubicon (an autophagy inhibitor) and SQSTM1 (autophagy substrate). Either genetic (small interfering [si]EGFR) or pharmacologic (AG1478) inhibition of EGFR signaling attenuated the decreased autophagy activity. In addition, rubicon siRNA knockdown prevented high glucose–induced inhibition of autophagy in podocytes. We further examined whether selective EGFR deletion in podocytes affected the progression of DN in type 2 diabetes. Selective podocyte EGFR deletion had no effect on body weight or fasting blood sugars in either db/db mice or nos3−/−; db/db mice, a model of accelerated type 2 DN. However selective podocyte EGFR deletion led to relative podocyte preservation and marked reduction in albuminuria and glomerulosclerosis, renal proinflammatory cytokine/chemokine expression, and decreased profibrotic and fibrotic components in nos3−/−; db/db mice. Podocyte EGFR deletion led to decreased podocyte expression of rubicon, in association with increased podocyte autophagy activity. Therefore, activation of EGFR signaling in podocytes contributes to progression of DN at least in part by increasing rubicon expression, leading to subsequent autophagy inhibition and podocyte injury.
Introduction
Diabetes is the most common cause of chronic kidney disease and end-stage renal disease, accounting for approximately one-half of all cases (1,2). Although understanding of the underlying pathophysiologic mechanisms of diabetic nephropathy (DN) has advanced remarkably in the past decade, much remains to be learned about factors that promote or retard the development and progression of diabetic renal injury.
Podocytes are a primary target in the development of DN. Studies in patients with type 2 diabetes and type 2 diabetes mouse models have demonstrated that progressive glomerulopathy in diabetes is associated with compromise of the glomerular filtration barrier and progressive loss of podocytes (3,4). As terminally differentiated cells, podocytes are dependent on autophagy for maintenance of cellular integrity, and previous studies have identified decreased podocyte autophagy as an important factor in the pathogenesis of DN (5–11).
The epidermal growth factor receptor (EGFR, aka ErbB1/HER1) is a member of a family of receptor tyrosine kinase ErbB receptors (12,13). EGFR is widely expressed in the mammalian kidney, including the podocyte (14,15). EGFR activation leads to phosphorylation on specific tyrosine residues within its cytoplasmic tail that serve as docking sites for a variety of signaling molecules activating intracellular pathways, including the MAP kinase, JAK/STAT, src kinase, and PI3K pathways, that regulate cell proliferation, differentiation, and apoptosis (12,13,16).
In our previous studies, we found that administration of EGFR tyrosine kinase inhibitors can slow progression of DN in experimental animals (17), and in a model of type 1 diabetes induced by streptozotocin, podocyte-specific deletion of the epidermal growth factor receptor (EGFR) ameliorated glomerular injury (18). In the current studies, we have determined that activation of EGFR signaling in high glucose–treated podocytes led to increased expression of rubicon and subsequent inhibition of autophagy activity. In a model of accelerated severe type 2 DN, selective EGFR deletion in podocytes significantly ameliorated the progression of glomerulopathy and tubulointerstitial fibrosis, and this protection was associated with increased podocyte integrity and maintenance of autophagy through inhibition of rubicon expression.
Research Design and Methods
Animal Studies
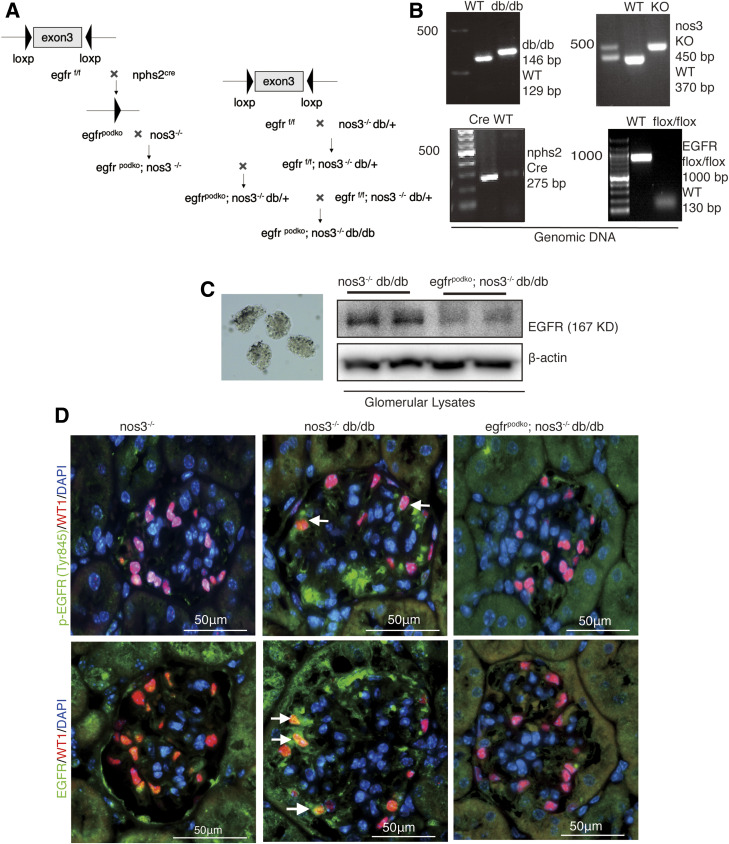
All animal experiments were performed in accordance with the guidelines of the Institutional Animal Care and Use Committee of Vanderbilt University. Egfrflox/flox, nphs2-Cre; egfrf/f mice, and nos3−/− db/db mice were generated as we previously described (15,18,19). For investigation of the role of podocyte EGFR in the development of DN, podocyte EGFR was deleted in classic type 2 diabetes (db/db mice) and in accelerated type 2 diabetes (BKS db/db with nos3 knockout [nos3−/−; db/db]. The strategy to generate db/db mice with selective podocyte EGFR deletion is as follows: 1) nphs2-Cre; egfrf/f mice were generated, 2) egfrf/f; db/+ mice were generated, 3) nphs2-Cre; egfrf/f mice were crossed with egfrf/f; db/+ mice to generate nphs2-Cre; egfrf/f; db/+ mice, and finally 4) nphs2-Cre; egfrf/f; db/+ mice were crossed with egfrf/f; db/+ to generate egfrf/f; db/db (designated as db/db) mice and nphs2-Cre; egfrf/f; db/db (designated as egfrpodKO db/db) mice, and they were sacrificed at 40 weeks of age. The strategy to generate nos3−/−; db/db mice with selective podocyte EGFR deletion is as follows (Fig. 3A and B): 1) nphs2-Cre; egfrf/f; nos3−/− mice were generated, 2) egfrf/f; nos3−/−; db/+ mice were generated, 3) nphs2-Cre; egfrf/f; nos3−/− mice were crossed with egfrf/f; nos3−/−; db/+ mice to generate nphs2-Cre; egfrf/f; nos3−/−; db/+ mice, and finally 4) nphs2-Cre; EGFRf/f; nos3−/−; db/+ mice were crossed with egfrf/f; nos3−/−; db/+ mice to generate egfrf/f; nos3−/−; db/db (designated as nos3−/− db/db) mice and nphs2-Cre; egfrf/f; nos3−/−; db/db (designated as egfrpodKO nos3−/− db/db) mice, and they were sacrificed at 20 weeks of age. All mice were on the BKS background. Both male and female mice were used for experiments. Genotyping was performed by PCR before initiation of the experiments and after the sacrifice of the animals.
Figure 3.
Confirmation of selective EGFR deletion in podocytes in nos3−/− db/db mice. A: schematic of generating podocyte-specific deletion of egfr using Nphs2-Cre mice and Cre-Lox technique. B: NOS3 knockout, db/db, EGFR deletion of exon 3, and Cre expression were verified by PCR using genomic DNA as the template. C: EGFR attenuation in podocytes was confirmed by immunoblotting of isolated glomerular lysates. D: With mice at 20 weeks of age, phosphorylated EGFR (Try845) and EGFR were colocalized with most of the WT1-positive (a marker of podocyte nuclei) cells (arrows) in egfrf/f; nos3−/−; db/db (nos3−/−; db/db) mice. In contrast, most of the WT1-positive cells were phosphorylated EGFR (Try845) and EGFR negative or had very weak EGFR staining (arrowheads) in nphs2-Cre; egfrf/f; nos3−/−; db/db (egfrpodKO; nos3−/−; db/db), confirming efficient and selective EGFR deletion in podocytes. bp, base pairs; WT, wild type.
Primers
Primers are as follows: for nos3 knockout mice, 5′-CTC CAA CTT AGT GCA GGT CT-3′ and 5′-ATG GTT GCC TTC ACA CGC TT-3′ for wild-type allele and 5′-CTC CAA CTT AGT GCA GGT CT-3′ and 5′-CTT CCT CGT GCT TTA CGG TA-3′ for knockout type allele; for Leprdb mice, 5′-AGA ACG GAC ACT CTT TGA AGT CTC-3′ and 5′-AAC CAT AGT TTA GGT TTG TTT C-3′ for wild-type allele and 5′-AGA ACG GAC ACT CTT TGA AGT CTC-3′ and 5′-CAA TTC AGT GTA AAC CAT AGT TTA GGT TTG TTT A-3′ for knockout type allele; for nphs2-Cre, 5′- GCATAACCAGTGAAACAGCATTGCTG-3′ and 5′-GGACATGTTCAGGGATCGCCAGGCG-3′; for egfrflox/flox, 5′-CTTTGGAGAACCTGCAGATC-3′ and 5′-CTGCTACTGGCTCAAGTTTC-3′ for verification of the EGFR gene floxed mice and 5′-ACACTAGCACTGACTGCTGG-3′ and 5′-CTGCTACTGGCTCAAGTTTC-3′ for verification of the EGFR gene wild-type mice.
Isolation of Glomeruli Using Dynabeads M-450
Mouse glomeruli were isolated as previously described (18). Briefly, mice were anesthetized by an injection of Nembutal sodium solution (50 mg/kg i.p.) and perfused with 4 × 107 Dynabeads diluted in 20 mL PBS through the heart. The kidneys were removed, minced, and digested in collagenase II (1 mg/mL, C6885; Sigma-Aldrich) and DNase I (50 units/mL, D5025; Sigma-Aldrich) at 37°C for 5 min with agitation. The tissue suspension was pressed through a 100-μm cell strainer. Glomeruli containing Dynabeads were gathered by a magnetic particle concentrator and washed with ice-cold PBS. The isolated glomeruli were lysed and then followed by immunoblotting analysis.
Cell Culture
Immortalized mouse podocyte cells were cultured as previously described (20). Briefly, cells were maintained at 33°C in RPMI medium containing 100 units/mL IFNγ and 10% FBS and induced to differentiate by shifting them to 37°C and culturing in RPMI medium containing 10% FBS without IFNγ for 10 days. The differentiated podocytes were made quiescent in RPMI medium containing 5.5 mmol/L glucose and 1% FBS for 16 h. Then the cells were treated with medium containing 25 mmol/L glucose (high glucose) with or without a selective EGFR tyrosine kinase inhibitor, AG1478, for additional 24 h. AG1478 (1 μmol/L) was added 30 min before high-glucose treatment.
siRNA Knockdown of EGFR, Rubicon, and Beclin-1
Mouse EGFR ON-TARGETplus SMARTpool siRNA (L-040411-00-0005) or negative control siRNA (Thermo Fisher Scientific, Lafayette, CO) and mouse rubicon siRNA (SC-146442; Santa Cruz Biotechnology), mouse Beclin-1 siRNA (SignalSilence Beclin-1 siRNA I, no. 6222; Cell Signaling Technology), or control siRNA (SC-37007; Santa Cruz Biotechnology) were transfected into differentiated mouse podocytes by the Lipofectamine method (Lipofectamine RNAiMAX Transfection Reagent; Invitrogen, Carlsbad, CA) as previously described (21). After transfection for 48 h, the cells were made quiescent in RPMI medium containing 5.5 mmol/L glucose and 1% FBS for 24 h followed by treatment with 25 mmol/L glucose for 24 h. Transient transfection efficiency was verified by immunoblotting and immunofluorescence staining as shown in Supplementary Figs. 1 and 2.
Measurements of Blood Glucose, HbA1c, Urinary Albumin Excretion, and Plasma Creatinine
Fasting blood glucose was evaluated with a B-glucose analyzer (HemoCue, Lake Forest, CA) on saphenous vein samples from conscious mice at 2:00 p.m. after fasting for 6 h initiated at 8:00 a.m. Glycated hemoglobin (HbA1c) levels were measured by a DCA Vantage HbA1c analyzer. Urinary albumin and creatinine excretion were determined with use of Albuwell M kits (Exocell, Philadelphia, PA). Albuminuria was expressed as urinary albumin concentration–to–creatinine concentration ratio (ACR) (µg/mg). Quantification of endogenous creatinine was performed on an ultra-performance liquid chromatography (UPLC)–tandem mass spectrometry system. Briefly, after centrifugation for the mixture of plasma, D3-creatinine internal standard, and UPLC solvent (20 mmol/L HCOONH4) at 20,000g, the supernatant was injected into UPLC (Waters) coupled with a TSQ Vantage Triple Quadrupole mass spectrometer (Thermo Fisher Scientific) running on positive electrospray ionization process mode. Creatinine was well resolved on a 2.1 × 50 mm 300-SCX column at 0.3 mL/min flow rate.
Antibodies
Rat anti-mouse F4/80 (MCA497R, a marker of macrophages; 1:50) and CD3 (MCA1477, a marker of T lymphocytes; 1:100) were purchased from AbD Serotec (now Bio-Rad Laboratories); goat anti-human connective tissue growth factor (CTGF) (cat. no. SC-14939, 1:500) was from Santa Cruz Biotechnology; rabbit anti–Wilms tumor protein (WT1) (a marker of podocytes) (ab89901, 1:50), goat anti-EGFR (E1282, 1:500), and mouse acta2 (anti–α-smooth muscle actin [α-SMA], a marker of myofibroblasts) (A5228, 1:1,000) were from Sigma-Aldrich; rabbit col1a1 (anti–collagen I) (600-401-103-01, 1:100) and col4a1 (collagen IV) (600-401-106-01, 1:100) were purchased from Rockland Immunochemicals; mouse anti–4-hydroxynonenal (4-HNE) (a marker of oxidative stress) (MAB3249, clone no. 198960; 1:1,000) was from R&D; LC3B (NB100-2220 and NB600-1384, 1:1,000) was from Novus Biologicals; beclin-1 (3495, 1:1,000), SQSTM1/p62 (23214S, 1:1,000), phosphorylated RPS6 (Ser240/244) (5364s, 1:1,000), RPS6 (2217s, 1:1,000), and α-tubulin (1:1,000) were from Cell Signaling Technology; and phosphorylated EGFR was purchased from Santa Cruz Biotechnology (SC-57542, 1:100). GAPDH (1:1,000) was from Life Technologies Corporation, and β-actin was from Sigma-Aldrich. The information regarding rubicon antibodies that we used is as follows: 1) rabbit monoclonal antibody, clone D9F7, immuned with a synthetic peptide corresponding to residues surrounding Leu210 of human rubicon protein, detects a 130-kDa band in both human and mouse samples (cat. no. 8465, 1:1,000; Cell Signaling Technology); 2) rabbit polyclonal antibody against human amino acids 922–972, detects a 109-kDa band in human samples (ab92388, 1:1,000; Abcam); 3) rabbit polyclonal antibody against mouse amino acids 750–850, detects a 106-kDa band in human and mouse samples (ab156052, 1:1,000; Abcam); and 4) rabbit polyclonal antibody against a 16–amino acid peptide near the amino terminus of human rubicon, detects a band ∼100 kDa in human and mouse and rat samples (PA5-38017, 1:1,000; Invitrogen). Of note, the rubicon antibodies used by Sun et al. (22) included cat. no. 8645 from Cell Signaling Technology, which detected a 130-kDa band.
Real-time PCR
Total kidney RNAs were isolated with use of Trizol reagent (Invitrogen). We used a SuperScript III First-Strand Synthesis System kit (Invitrogen) to synthesize cDNA from equal amounts of total RNA from each sample. Quantitative RT-PCR was performed with TaqMan real-time PCR (7900HT; Applied Biosystems). The Master Mix and all gene probes were also purchased from Applied Biosystems. The probes used in the experiments included mouse S18 (Mm02601778), collagen I (col1a1) (Mm00801666), collagen IV (col4a1) (Mm01210125), α-SMA (acta2) (Mm01546133), TGF-β (Mm00441726), IL-6 (Mm00446190), TNF-α (Mm99999068), IL-1α (Mm00439621), IL-1β (Mm00434228), IL-23 (Mm00518984), IRF5 (Mm00496477), NOS2 (Mm00440502), CCL3 (Mm00441258), arginase 1 (Arg1) (Mm00475991), IL4Rα (Mm01275139), mannose receptor (CD206, Mm01329362), FIZZ1 (RELMα, Mm00445109), and YM1 (Chi3l3) (Mm00657889).
Immunoblotting Analysis
The cultured mouse podocytes and kidney tissue were homogenized with lysis buffer (10 mmol/L Tris–HCl [pH 7.4], 50 mmol/L NaCl, 2 mmol/L EGTA, 2 mmol/L EDTA, 0.5% Nonidet P-40, 0.1% SDS, 100 μmol/L Na3VO4, 100 mmol/L NaF, 0.5% sodium deoxycholate, 10 mmol/L sodium pyrophosphate, 1 mmol/L phenylmethylsulfonyl fluoride, 10 μg/mL aprotinin, and 10 μg/mL leupeptin) and centrifuged at 15,000g for 20 min at 4°C. The cultured cells were lysed with the same lysis buffer. The BCA protein assay kit (Thermo Fisher Scientific, Waltham, MA) was used to measure the protein concentration of each sample. Western analysis was performed as previously described (17).
Immunohistochemical and Immunofluorescent Staining
Immunostaining was carried out as previously described (17). For mouse primary antibodies, the M.O.M. kit was used for reduction of endogenous mouse Ig staining. For tissue section fluorescent staining, antigen retrieval was performed by boiling of sections in citric acid buffer, pH 6.0 (100 mmol/L), for 3 × 5 min. After antigen retrieval, the sections were blocked for 1 h with PBS containing 10% normal donkey serum, and then primary antibody diluted in PBS containing 1% normal donkey serum, 1% BSA, 0.3% Triton X-100, and 0.001% sodium azide was added for 16 h. Cultured differentiated podocytes were fixed with 4% paraformaldehyde, blocked with 10% normal donkey serum for 1 h, and then incubated with primary antibodies overnight. After washing with PBS, fluorescent-labeled secondary antibody was added. Staining on the slides was imaged with a Nikon TE300 fluorescence microscope and a spot-cam digital camera (Diagnostic Instruments, Sterling Heights, MI), followed by quantification with ImageJ software (National Institutes of Health, Bethesda, MD). Fluorescence intensity were calculated in >30 fields per mice or 10 fields per cell slides and expressed as arbitrary units.
Histological Analysis
Periodic acid Schiff–stained slides were evaluated for glomerulosclerosis index without knowledge of the identity of the various groups. A semiquantitative index was used to evaluate the degree of glomerular sclerosis. Each glomerulus in a single section was graded from 0 to 4, where 0 represents no lesion and 1, 2, 3, and 4 represent sclerosis, involving ≤25, 25–50, 50–75, or ≥75% of the glomerular tuft area, respectively (17,23).
Statistics
Data are presented as means ± SD. Comparisons between groups were made with use of a two-tailed unpaired Student t test or one-way ANOVA with Bonferroni post hoc test. P < 0.05 was considered statistically significant.
Data and Resource Availability
The data generated during and/or analyzed during the current study are available from the corresponding author upon reasonable request.
Results
Podocyte EGFR Activation in Response to High Glucose Exposure Increased the Expression of an Autophagy Inhibitor, Rubicon
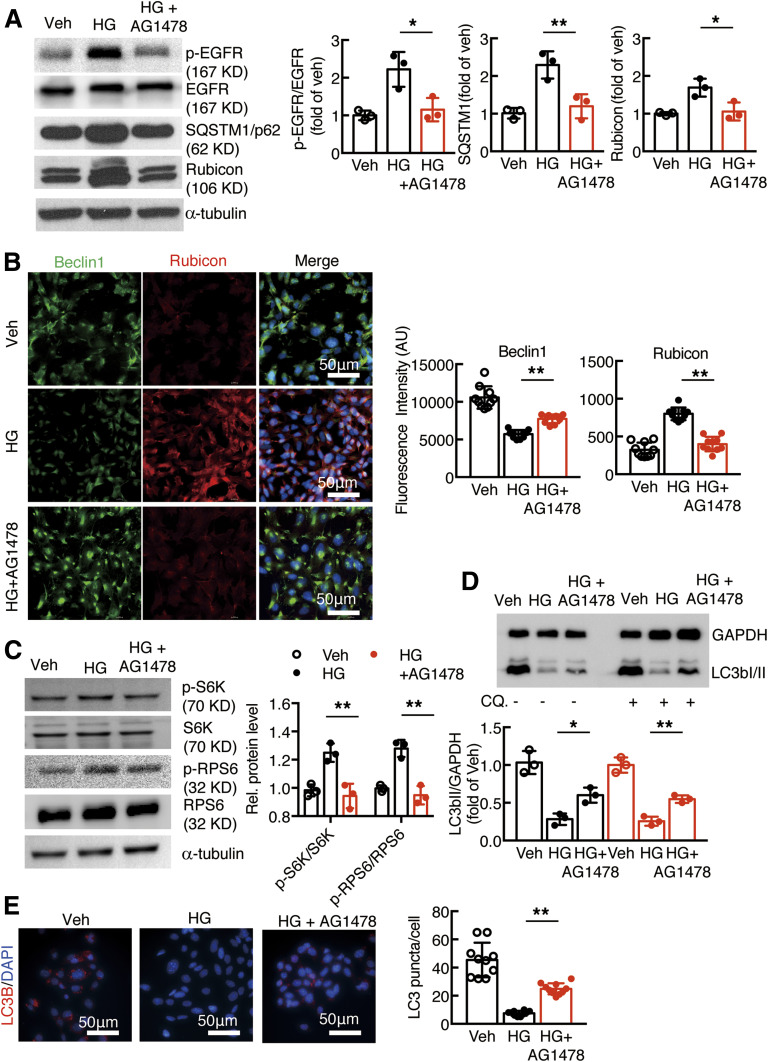
Autophagy is a protective mechanism for the terminally differentiated podocyte (24), and inhibition of EGFR signaling increases kidney autophagy activity (17). A mouse podocyte cell line was used to investigate the role of EGFR activation in regulation of podocyte autophagy activity. High-glucose treatment for 16 h led to increases in the expression levels of phosphorylated EGFR, SQSTM1 (p62, autophagy substrate), and rubicon (RUN domain and cysteine-rich domain–containing, beclin-1–interacting protein) but decreases in beclin-1 levels. All of these parameters were reversed by AG1478, an EGFR inhibitor (Fig. 1A and B). EGFR activation inhibits autophagy in part through activation of mTOR-p70 S6K and RPS6 signaling (17). High-glucose treatment for 16 h led to increased expression levels of phosphorylated p70S6K and phosphorylated RPS6, which were reversed by AG1478 (Fig. 1C).
Figure 1.
EGFR activation increased rubicon expression and inhibited autophagic flux in cultured podocytes. A: Exposure of podocytes to high glucose (HG) led to activation of EGFR signaling, and increases in SQSTM1 and rubicon expression were inhibited with AG1478, an EGFR tyrosine kinase inhibitor. B: High glucose–induced beclin-1 inhibition and rubicon stimulation were inhibited with AG1478 (C). AG1478 inhibited HG-induced phosphorylated (p-)S6K and phosphorylated S6 activation. D and E: AG1478 increased LC3B abundance (N = 10 fields per group) and LC3B autophagosomes in podocytes. For measurement of LC3B II turnover, the podocytes were examined without (−) or with (+) treatment with 50 μmol/L chloroquine (CQ.) for 16 h. *P < 0.05 and **P < 0.01 vs. high glucose group, N = 3 independent repeats, one-way ANOVA with Bonferroni post hoc test. AU, arbitrary units; Veh, vehicle.
Under normal conditions podocytes exhibit high levels of basal autophagy and autophagic flux. The LC3b abundance was significantly decreased in cultured podocytes in response to exposure to high glucose both without and with administration of 50 μmol/L chloroquine, a lysosomal inhibitor. Immunofluorescent staining confirmed that high glucose–induced beclin-1 inhibition and rubicon elevation were reversed by inhibition of EGFR tyrosine kinase activation with AG1478. AG1478 also reversed the high glucose–mediated decrease in autophagosome formation (Fig. 1D and E).
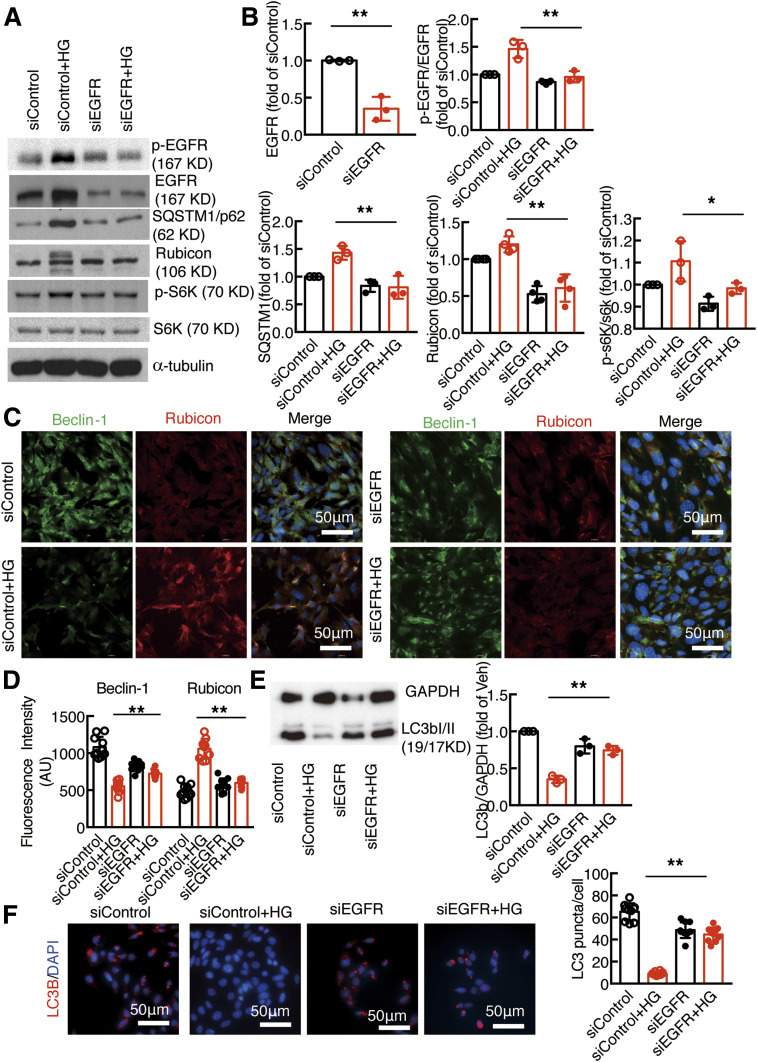
To avoid possible off-target effects of AG1478, we also used siRNA to knock down EGFR. As indicated in Fig. 2A, EGFR expression was effectively knocked down with EGFR siRNA compared with control siRNA in podocytes. Knocking down EGFR expression attenuated high glucose–induced increases in the expression levels of phosphorylated EGFR, SQSTM1, rubicon, and phosphorylated p70S6K (Fig. 2A and B). Immunofluorescent staining showed that high glucose–induced decreases in beclin-1 expression and increases in rubicon expression were attenuated by EGFR knockdown (Fig. 2C). Knocking down EGFR in podocytes increased autophagosome formation (Fig. 2D and E).
Figure 2.
Knockdown of EGFR attenuated increased rubicon expression and promoted autophagic flux in cultured podocytes. A and B: siRNA knockdown of podocyte EGFR led to less activation of EGFR signaling and decreased SQSTM1, rubicon, and phosphorylated (p-)S6K expression. C and D: High glucose–induced beclin-1 inhibition and rubicon stimulation in podocytes were abolished by siEGFR. E and F: siRNA knockdown of podocyte EGFR increased LC3B abundance after exposure to high glucose compared with control siRNA, which promoted autophagosome production. *P < 0.05 and **P < 0.01 vs. siControl group with high glucose (HG), N = 3 independent repeats, one-way ANOVA with Bonferroni post hoc test. AU, arbitrary units.
As rubicon exists in different isoforms/variants, we used different antibodies to validate rubicon expression (see research design and methods). As indicated in Supplementary Fig. 1, different rubicon antibodies detected the bands suggested by the manufacturers, which were markedly decreased by siRNA knockdown, confirming the specificity of the antibody and existence of different rubicon variants. Furthermore, the specificity of the beclin-1 antibody was confirmed by siRNA knockdown of beclin-1 in podocytes as shown in Supplementary Fig. 2.
EGFR Deletion in Podocytes In Vivo
For study of the potential role of podocyte EGFR in the development of type 2 DN, podocyte EGFR was selectively deleted in two mouse lines. The first mouse line was db/db mice with selective podocyte EGFR deletion: nphs2-Cre; EGFRf/f; db/db (egfrpodKO; db/db) mice and their corresponding controls, egfrf/f; db/db (db/db) mice. The second mouse line was an accelerated model of type 2 diabetes model with selective podocyte EGFR deletion: nphs2-Cre; egfrf/f; nos3−/−; db/db (egfrpodKO; nos3−/−; db/db) mice and their corresponding controls, egfrf/f; nos3−/−; db/db (nos3−/−; db/db) mice (Fig. 3A). The effectiveness of EGFR deletion in podocytes was confirmed by determination of genomic DNA (Fig. 3B), immunoblotting of glomerular lysates (Fig. 3C), and immunofluorescent staining of EGFR expression (Fig. 3D). Most WT1-positive (Wilms tumor protein 1, a podocyte nuclear marker) cells were also EGFR positive in 20-week-old nos3−/−; db/db mice. In contrast, most WT1-positive podocytes were devoid of EGFR staining or only had faint EGFR staining in 20-week-old egfrpodKO; nos3−/−; db/db mice.
Podocyte EGFR Deletion Had No Effect on Body Weight and Blood Glucose
Our previous studies indicated that either genetic or pharmacologic inhibition of EGFR decreased gain of body weight and blood glucose in nos3−/−; db/db mice (23). We investigated whether selective EGFR deletion in podocytes in db/db mice affected body weight gains as well as blood glucose and HbA1c. As indicated in Supplementary Fig. 3, selective EGFR deletion in podocytes had no effect on gain of body weight or blood glucose and HbA1c in 8- to 40-week-old db/db mice (n = 11 in db/db group and n = 13 in egfrpodKO; db/db group). Similarly, selective EGFR deletion in podocytes had no effect on gain of body weight or blood glucose and HbA1c in 8- to 20-week-old nos3−/−; db/db mice (8 weeks old: n = 19 in nos3−/−; db/db group and n = 25 in egfrpodKO; nos3−/−; db/db group, 20 weeks old: n = 16 in nos3−/−; db/db group and n = 22 in egfrpodKO; nos3−/−; db/db group) (Supplementary Fig. 4).
Podocyte EGFR Deletion Attenuated the Development of Diabetic Nephropathy
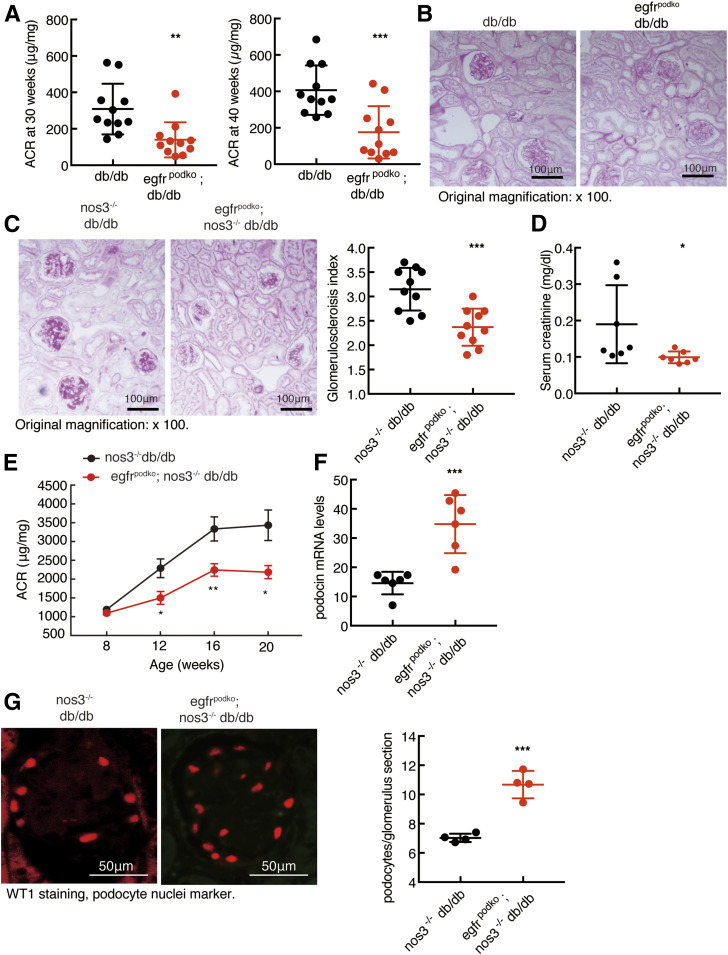
db/db mice developed moderate albuminuria at 30 and 40 weeks old, and podocyte EGFR deletion attenuated albuminuria at both time points (ACR: [means ± SD] 309 ± 42 vs. 140 ± 29 µg/mg at 30 weeks old, P < 0.01, and 407 ± 41 vs. 144 ± 44 µg/mg at 40 weeks old, P < 0.001; n = 11 for all groups] (Fig. 4A). Histology was examined at 40 weeks of age for db/db and egfrpodko; db/db mice. Histologically, there was minimal renal injury in db/db mice and egfrpodKO; db/db mice (Fig. 4B).
Figure 4.
Podocyte EGFR deficiency attenuated the development of DN in type 2 diabetes. A and B: egfrf/f; db/db (db/db) mice had moderate albuminuria at 30 and 40 weeks of age (A) as well as mesangial expansion at 40 weeks of age (B), and EGFR deletion in podocytes attenuated these parameters. **P < 0.01, ***P < 0.001 (n = 11 in each group). Original magnification: ×100. C: EGFR deletion in podocytes decreased glomerulosclerosis in nos3−/−; db/db mice as indicated by periodic acid Schiff staining (right panel shows quantitative glomerular sclerosis index). ***P < 0.001, n = 10 in each group. D: EGFR deletion in podocytes decreased serum creatinine. *P < 0.05, n = 7 in each group. E: The progressive increases in albuminuria seen in egfrf/f; nos3−/−; db/db (nos3−/−; db/db) mice were attenuated in nphs2-Cre; egfrf/f; nos3−/−; db/db (egfrpodKO; nos3−/−; db/db) mice. *P < 0.05, **P < 0.01 vs. the corresponding nos3−/−; db/db group (n = 16 in nos3−/−; db/db group and n = 12 in egfrpodKO; nos3−/−; db/db group). F: Podocyte EGFR deficiency markedly slowed the loss of podocytes in nos3−/−; db/db mice, as indicated by increased renal mRNA levels of nphs2, a specific marker of podocytes. ***P < 0.001, n = 6 in each group. G: Podocyte EGFR deficiency significantly slowed the loss of podocytes in nos3−/−; db/db mice, as indicated by fluorescent red WT1 staining, a marker of podocyte nuclei. Right panel shows podocyte number in each glomerulus section from different groups. ***P < 0.001, n = 4 in each group, two-tailed unpaired Student t test.
We have previously reported that albuminuria was apparent as early as 8 weeks old and progressively increased until 20 weeks old in nos3−/−; db/db mice (4,23,25). Histology was examined at 20 weeks of age for nos3−/−; db/db and egfrpodko; nos3−/−; db/db mice. The severe glomerulosclerosis seen in nos3−/−; db/db mice was markedly attenuated by podocyte EGFR deletion (glomerulosclerosis index: 2.37 ± 0.12 vs. 3.15 ± 0.14, P < 0.001; n = 10) (Fig. 4C). Furthermore, egfrpodKO; nos3−/−; db/db mice had lower serum creatinine compared with nos3−/−; db/db mice (Fig. 4D). Podocyte EGFR deletion markedly attenuated the progressive increases in albuminuria in nos3−/−; db/db mice (ACR: 3,434 ± 418 vs. 2,185 ± 175 µg/mg at 20 weeks old, P < 0.05; N = 13 in nos3−/−; db/db group and n = 11 in egfrpodKO; nos3−/−; db/db group) (Fig. 4E). Podocyte EGFR deletion led to increased renal mRNA levels of nphs2, a specific marker of podocytes (34.83 ± 4.06 vs. 14.60 ± 1.58 of nos3−/−; db/db mice, P < 0.001; n = 6) (Fig. 4F). In addition, the number of WT1-positive cells (marker of podocytes) was significantly increased, indicating preservation of podocyte number with selective podocyte EGFR deletion (10.7 ± 0.5 vs. 7.0 ± 0.2 podocytes/glomerulus section, P < 0.001; n = 4 in each group) (Fig. 4G). Therefore, in the following studies, we focused the effects of podocyte EGFR deletion on the development of DN in nos3−/−; db/db mice.
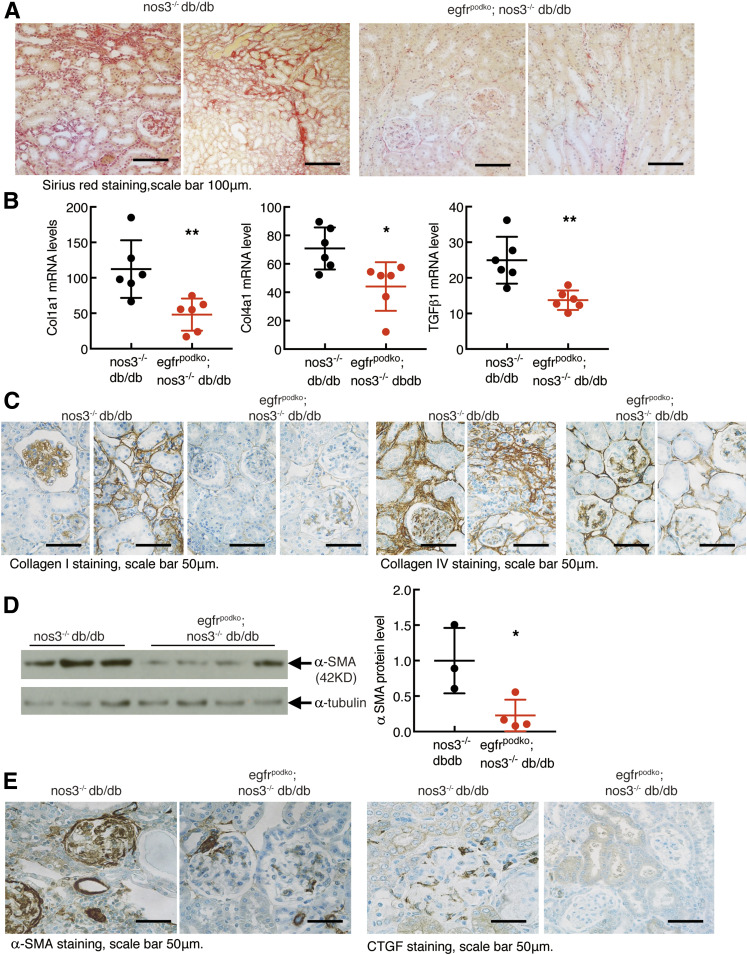
EgfrpodKO; nos3−/−; db/db mice exhibited less tubulointerstitial fibrosis, as indicated by decreased Sirius red staining (Fig. 5A). Total kidney mRNA levels for col1α1 (collagen I), col4α1 (collagen IV), and tgfb1 (TGF-β) were all significantly inhibited with selective podocyte deletion of EGFR (Fig. 5B). Immunohistochemical staining showed decreases in both glomerular and tubulointerstitial collagen I and collagen IV expression in egfrpodKO; nos3−/−; db/db mice (Fig. 5C). Renal protein expression of acta2 (α-SMA, a marker of myofibroblasts), and the profibrotic factor, CTGF, was also decreased in EgfrpodKO; nos3−/−; db/db mice (Fig. 5D and E).
Figure 5.
Podocyte EGFR deficiency decreased renal fibrosis in glomerulus and tubulointerstitium in nos3−/− db/db mice. Both egfrf/f; nos3−/−; db/db (nos3−/−; db/db) mice and nphs2-Cre; egfrf/f; nos3−/−; db/db (EgfrpodKO; nos3−/−; db/db) mice were sacrificed at 20 weeks old. A: EGFR deletion in podocytes led to markedly decreased renal fibrosis in glomerulus and tubulointerstitium, as indicated by Sirius red staining. Original magnification: ×200. B: EGFR deletion in podocytes led to decreased renal mRNA levels of profibrotic and fibrotic components, including Col1a1 (collagen I), Col4a1 (collagen IV), and Tgfb1 (TGF-β1). **P < 0.01, n = 6 in each group. C–E: EGFR deletion in podocytes also decreased renal protein expression of col1a1, col4a1, acta2, and CTGF in both glomerulus and tubulointerstitium. *P < 0.05, n = 3 in nos3−/−; db/db group and n = 4 in egfrpodKO; nos3−/−; db/db group. Two-tailed unpaired Student t test.
EgfrpodKO; nos3−/−; db/db Mice Had Decreased Renal Inflammation
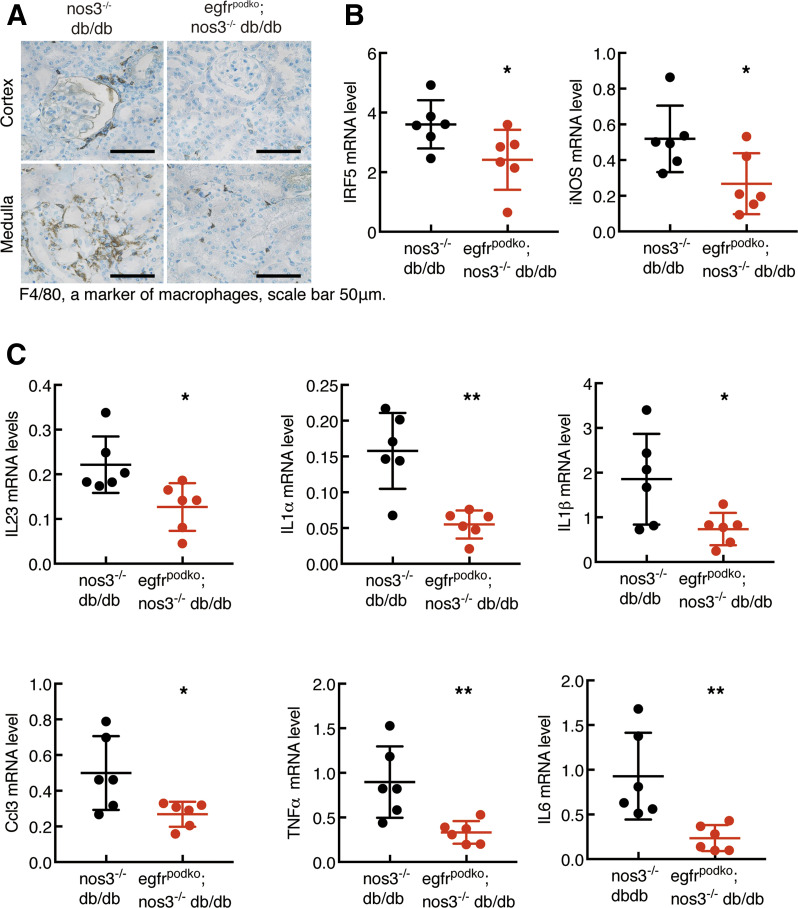
At 20 weeks of age, egfrpodKO; nos3−/−; db/db mice had less renal infiltration of F4/80+ macrophages (Fig. 6A) and CD3+ lymphocytes (Supplementary Fig. 5) and decreased renal oxidative stress, as indicated by 4-HNE staining (Supplementary Fig. 6). Renal mRNA levels of interferon regulatory factor 5 (IRF5) (a key regulator of macrophage M1 polarization) and NOS2 (a classic marker of M1 macrophages) were significantly reduced in egfrpodKO; nos3−/−; db/db mice (Fig. 6B). Renal mRNA levels of a panel of proinflammatory Th1/M1 cytokines/chemokines, including IL23α, IL1α, IL-1β, CCL3, TNF-α, and IL6, were all significantly decreased in egfrpodKO; nos3−/−; db/db mice (Fig. 6C). In contrast, mRNA levels of alternatively activated M2 macrophage markers, including CD206, IL4Rα, Arg1 (arginase 1), YM1, and FIZZ1, were not altered (Supplementary Fig. 7).
Figure 6.
Podocyte EGFR deficiency decreased renal macrophage infiltration and renal proinflammatory cytokines in nos3−/− db/db mice. Both egfrf/f; nos3−/−; db/db (nos3−/−; db/db) mice and nphs2-Cre; egfrf/f; nos3−/−; db/db (egfrpodKO; nos3−/−; db/db) mice were sacrificed at 20 weeks of age. A: EGFR deletion in podocytes led to decreased renal macrophage infiltration, as indicated by F4/80 staining, a marker of macrophages. Original magnification: ×400. B: EGFR deletion in podocytes decreased renal mRNA levels of IRF5, a transcription factor inducing macrophage M1 polarization, and NOS2, a marker of M1 macrophages. *P < 0.05, n = 6 in each group. C: EGFR deletion in podocytes led to decreased renal mRNA levels of proinflammatory cytokines/chemokines, including IL23, IL1α, IL1β, CCL3, TNFα, and IL6. *P < 0.05, **P < 0.01 (n = 6 in each group), two-tailed unpaired Student t test.
EgfrpodKO; nos3−/−; db/db Mice Had Increased Glomerulus Autophagy
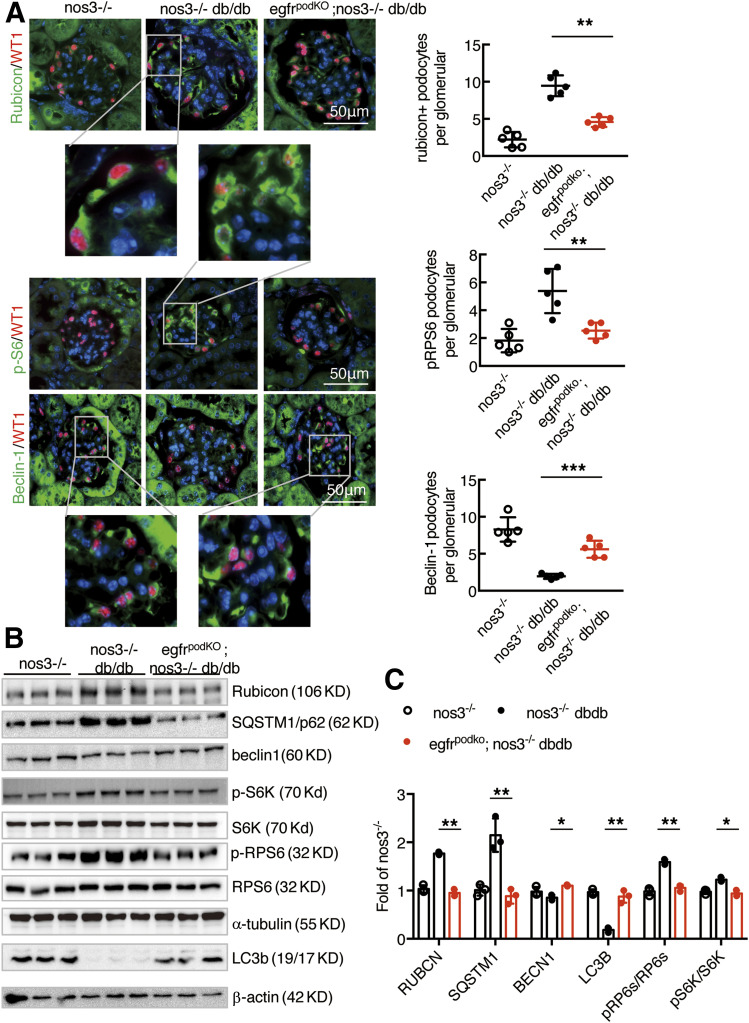
Genetic inhibition of autophagy in podocytes leads to glomerulopathy (26). Beclin-1 is essential for autophagy (27–29). Immunofluorescent staining showed low beclin-1 levels in glomeruli of nos3−/−; db/db mice but increased beclin-1 levels in glomeruli of egfrpodKO; nos3−/−; db/db mice. In contrast, immunofluorescent staining also indicated that LC3B puncta, a hallmark of autophagy, were increased in glomeruli of egfrpodKO; nos3−/−; db/db mice. In contrast, SQSTM1/p62, a substrate for autophagy, was decreased, indicating increased autophagic activity. Rubicon expression was higher, but beclin-1 expression was lower, in the same glomerulus from nos3−/−; db/db mice, while rubicon expression was lower but beclin-1 expression was higher in egfrpodKO; nos3−/−; db/db mice (Fig. 7A and Supplementary Fig. 8). Immunoblotting confirmed that glomerular beclin-1 was significantly increased in egfrpodKO; nos3−/−; db/db mice compared with nos3−/−; db/db mice, while rubicon and SQSTM1/p62 expressions were decreased (Fig. 7B and C).
Figure 7.
Podocyte EGFR deficiency led to increased autophagy in podocytes in nos3−/− db/db mice. Both nos3−/−; db/db (nos3−/−; db/db) mice and nos3−/−; db/db (EgfrpodKO; nos3−/−; db/db) mice were sacrificed at 20 weeks old. A: Representative kidney sections were stained and indicated that podocyte EGFR deletion decreased rubicon, SQSTM1, and phosphorylated (p)RPS6 (Ser240/244) protein expression and increased renal beclin-1. Quantification for rubicon, pRPS6/RPS6, and beclin-1 (colocalized with WT1, a podocyte nucleus marker) glomerulus represents results from 30 glomeruli each from five mice. **P < 0.01. B and C: Western blot of glomerular lysates obtained from nos3−/−; db/db (nos3−/−; db/db) mice and nos3−/−; db/db (egfrpodKO; nos3−/−; db/db) mice for rubicon, beclin-1, phosphorylated RPS6/RPS6 protein expression, LC3B I/II turnover and SQSTM1 abundance. *P < 0.05, **P < 0.01, n = 3 in nos3−/− group, n = 3 in nos3−/−; db/db group, and n = 3 in egfrpodKO; nos3−/−; db/db group; one-way ANOVA with Bonferroni post hoc test.
Selective EGFR deletion in podocytes led to decreases in renal mTORC activation, as indicated by decreased phosphorylation of the phosphorylated RPS6KB/p70S6k substrate RPS6 (ribosomal protein S6) levels in egfrpodKO; nos3−/−; db/db mice (Fig. 7A and B).
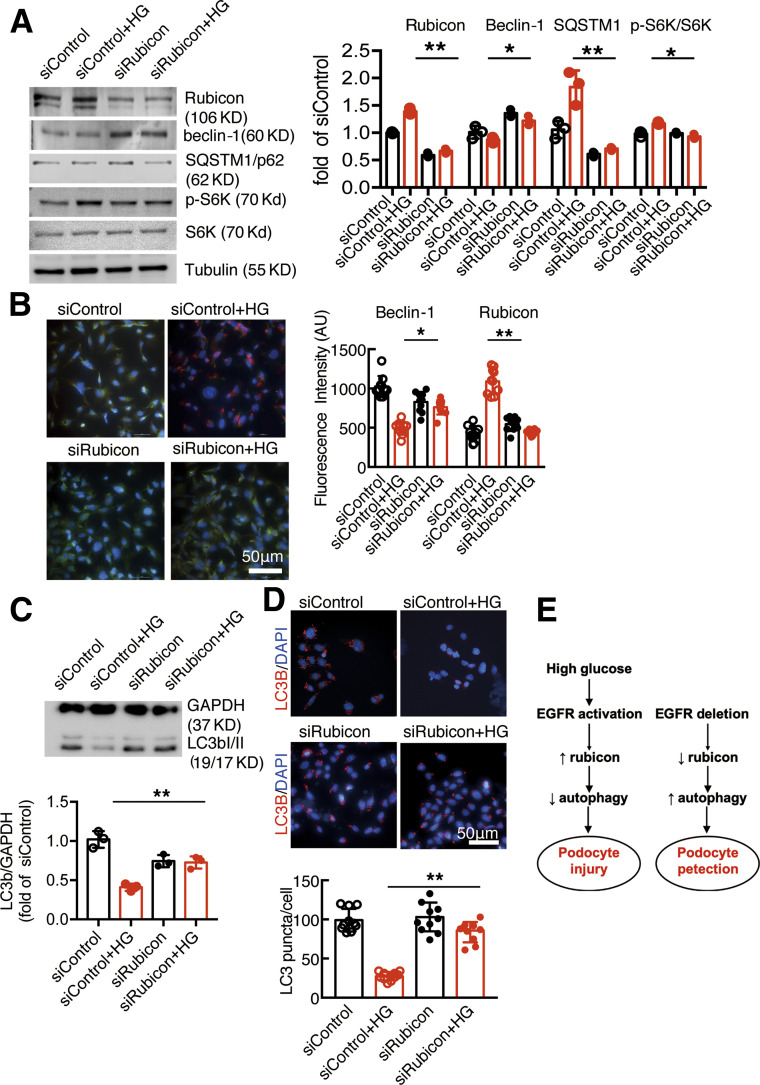
To investigate further the role of rubicon in EGFR-mediated inhibition of podocyte autophagy, we knocked down rubicon expression in cultured podocytes using siRNA and treated these cells with high glucose. As indicated in Fig. 8A, rubicon protein levels were markedly decreased in small interfering (si)Rubicon podocytes compared with siControl podocytes at baseline and in the presence of high glucose. Knocking down rubicon led to increased beclin-1 expression and autophagosome accumulation and decreased SQSTM1 expression in the presence of high glucose. Immunofluorescent staining also confirmed that high glucose–induced rubicon upregulation and beclin-1 downregulation were abolished with inhibition of rubicon expression in podocytes (Fig. 8B). Knocking down of rubicon also prevented high glucose–induced inhibition of autophagy influx (Fig. 8C and D).
Figure 8.
Rubicon knocking down attenuated EGFR-mediated inhibition of podocyte autophagy. A: Knocking down of rubicon increased beclin-1 but decreased SQSTM1 and phosphorylated (p-)s6K levels in podocyte in the presence of high glucose (HG). B: High glucose–induced beclin-1 inhibition and rubicon stimulation in podocytes were abolished by siRubicon. C and D: Knocking down of rubicon increased autophagy flux in podocytes in the presence of high glucose. *P < 0.05 and **P < 0.01 vs. siControl with high glucose. N = 3 independent repeats. E: Left: High glucose activates podocyte EGFR, leading to increased rubicon and subsequent inhibition of autophagy and podocyte injury. Right: EGFR inhibition leads to decreased rubicon and subsequent activation of autophagy and podocyte protection. One-way ANOVA with Bonferroni post hoc test. AU, arbitrary units.
Discussion
The present studies demonstrated that in a mouse model of type 2 DN, EGFR expression and activation in podocytes were mediators of accelerated kidney injury and that selective deletion of podocyte EGFR significantly ameliorated the progression. Deletion of podocyte EGFR preserved podocyte number and integrity, with decreased glomerulosclerosis, as well as other indicators of progressive kidney injury, including decreases in inflammatory cytokines, mediators of fibrosis, and tubulointerstitial collagen deposition. The amelioration of podocyte injury with selective EGFR deletion was associated with decreases in rubicon and SQSTM1/p62 expression in podocytes and subsequent increases in the activity of the autophagic pathway (Fig. 8E).
There is increasing evidence that autophagy is a protective mechanism for the terminally differentiated podocyte (24). Podocytes have high basal levels of autophagy (26), and we and others previously reported that inhibition of podocyte autophagy by targeting of autophagy-specific class III PI3K leads to progressive glomerulosclerosis (9,10). In mammals, ULK1 (Unc-51–like autophagy-activating kinase 1) regulates initiation of autophagy (30). Ulk1 is activated by AMPK phosphorylation of Ser317 and is inhibited by mTOR-dependent phosphorylation of Ser757 (31). mTOR activity increases in podocytes in diabetic mice and correlates with increased endoplasmic reticulum stress and progressive glomerulosclerosis (8). Renal mTOR activation in poorly controlled diabetes has been shown to result from AKT inhibition of TSC2, hyperglycemia-induced AMPK inhibition, and glycolysis-dependent Rheb activation of mTOR (32,33). In support of this, a recent study indicated that mTOR activity does not influence high basal levels of autophagy in podocytes in the absence of injurious stimuli. Instead, the AMPK/ULK1 pathway may play an important role in maintaining autophagy in podocytes (34). EGFR activation is a well-described mediator of mTOR activity through activation of the PI3K/AKT pathway (35). Our previous studies suggested that inhibition of EGFR tyrosine kinase activity with erlotinib slowed progression of DN in association with an increase in autophagy and a decrease in endoplasmic reticulum stress (17). In the current studies, we have demonstrated that podocyte-specific deletion of EGFR led to decreased phosphorylated RPS6, one of the important effectors of the mTORC1 pathway, in the kidneys of egfrpodKO; nos3−/−; db/db mice.
Selective podocyte EGFR depletion also increased expression of beclin-1 and LC3B and decreased SQSTM1, indicating increased autophagy. In the current study, we found that selective podocyte EGFR deletion decreased rubicon expression levels. Further studies in cultured podocytes indicated that exposure to high glucose led to activation of EGFR signaling in association with increased rubicon and SQSTM1 expression, which were inhibited by an EGFR tyrosine kinase inhibitor or by EGFR expression knockdown (Figs. 1 and 2).
Cross talk between EGFR and rubicon plays an important role in regulation of autophagy. EGFR signaling inhibits autophagy through multiple mechanisms. When the EGFR signaling pathway is activated, its several downstream molecules, including PI3K, AKT, and mTOR, are all activated to inhibit autophagy (36). Active EGFR can bind the autophagy protein beclin-1, leading to its multisite tyrosine phosphorylation and inactivation due to its increased binding to rubicon and decreased binding to the VPS34 lipid kinase (37). Interestingly, rubicon can also modulate EGFR signaling (22).
Tan et al. (38) reported an intriguing finding that with genetic or pharmacologic inhibition of EGFR tyrosine kinase activity, the receptor stimulated autophagy by releasing beclin-1 from inhibition by rubicon. Although this mechanism may partially underlie the effects seen in diabetic kidney models with EGFR kinase inhibitors (17), the current studies indicated that genetic deletion of EGFR expression could also stimulate podocyte autophagy, associated with increased expression of proautophagic factors and decreased expression of autophagy inhibitors.
In addition to promoting podocyte autophagy, selective decrease in podocyte EGFR expression also decreased inflammatory cell infiltration, inflammatory cytokine production in total kidney, and development of tubulointerstitial fibrosis even though there were no apparent differences in the extent of hyperglycemia between wild-type nos3−/−; db/db mice and egfrpodKO; nos3−/−; db/db mice. There is an ongoing argument about the relative role in DN of increased proteinuria versus direct toxic effects of hyperglycemia on the tubulointerstitium in the development of tubulointerstitial injury. The current studies suggest that either proteinuria per se or other factors either filtered or produced in the glomerulus may have a significant effect on development of tubulointerstitial fibrosis. Both circulating insulin levels and renal proinflammatory TNF-α expression are increased in type 2 diabetes. Interestingly, both TNF-α and insulin have been reported to transactivate EGFR in other cell types (39,40). The potential role of TNF-α and insulin in podocyte EGFR activation needs to be investigated in future studies.
In summary, selective EGFR deletion in podocytes decreased proteinuria and preserved podocyte integrity, increased autophagy and decreasing inflammation. These studies demonstrate that EGFR activation in podocytes in diabetes is a mediator of progressive diabetic kidney injury and indicate that inhibiting signaling downstream of receptor activation such as rubicon could provide a potential therapeutic option.
Article Information
Funding. These studies were supported by National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK), NIH, grants DK51265, DK95785, and DK62794 (to R.C.H. and M.-Z.Z.); NIDDK grant DK103067 and the Vanderbilt O’Brien Center (NIDDK grant P30DK114809) (to R.C.H. and M.-Z.Z.); U.S. Department of Veterans Affairs VA Merit Award 00507969 (R.C.H.); the Vanderbilt Center for Kidney Disease; and the National Natural Science Foundation of China (no. 8187030028 [to Y.P.]).
Duality of Interest. No potential conflicts of interest relevant to this article were reported.
Author Contributions. R.C.H. and M.-Z.Z. designed the study. Y.L., Y.P., S.C., K.S., Y.W., A.N., X.F., and S.W. performed the experiments. M.-Z.Z., Y.L., and Y.P. participated in figure preparation. R.C.H., M.-Z.Z., Y.L., and Y.P. wrote the manuscript. All authors read and approved the final version of manuscript. R.C.H. and M.-Z.Z. are the guarantors of this work and, as such, had full access to all the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis.
Footnotes
Y.L. and Y.P. made equal contributions.
This article contains supplementary material online at https://doi.org/10.2337/figshare.13252646.
References
- 1.Fineberg D, Jandeleit-Dahm KA, Cooper ME. Diabetic nephropathy: diagnosis and treatment. Nat Rev Endocrinol 2013;9:713–723 [DOI] [PubMed] [Google Scholar]
- 2.Wang X, Yao B, Wang Y, et al. Macrophage cyclooxygenase-2 protects against development of diabetic nephropathy. Diabetes 2017;66:494–504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wolf G, Chen S, Ziyadeh FN. From the periphery of the glomerular capillary wall toward the center of disease: podocyte injury comes of age in diabetic nephropathy. Diabetes 2005;54:1626–1634 [DOI] [PubMed] [Google Scholar]
- 4.Zhang MZ, Wang X, Yang H, et al. Lysophosphatidic acid receptor antagonism protects against diabetic nephropathy in a type 2 diabetic model. J Am Soc Nephrol 2017;28:3300–3311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yi M, Zhang L, Liu Y, et al. Autophagy is activated to protect against podocyte injury in adriamycin-induced nephropathy. Am J Physiol Renal Physiol 2017;313:F74–F84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Inoki K, Mori H, Wang J, et al. mTORC1 activation in podocytes is a critical step in the development of diabetic nephropathy in mice. J Clin Invest 2011;121:2181–2196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cina DP, Onay T, Paltoo A, et al. Inhibition of MTOR disrupts autophagic flux in podocytes. J Am Soc Nephrol 2012;23:412–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gödel M, Hartleben B, Herbach N, et al. Role of mTOR in podocyte function and diabetic nephropathy in humans and mice. J Clin Invest 2011;121:2197–2209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bechtel W, Helmstädter M, Balica J, et al. Vps34 deficiency reveals the importance of endocytosis for podocyte homeostasis. J Am Soc Nephrol 2013;24:727–743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen J, Chen MX, Fogo AB, Harris RC, Chen JK. mVps34 deletion in podocytes causes glomerulosclerosis by disrupting intracellular vesicle trafficking. J Am Soc Nephrol 2013;24:198–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fogo AB The targeted podocyte. J Clin Invest 2011;121:2142–2145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schlessinger J Ligand-induced, receptor-mediated dimerization and activation of EGF receptor. Cell 2002;110:669–672 [DOI] [PubMed] [Google Scholar]
- 13.Hynes NE, Lane HA. ERBB receptors and cancer: the complexity of targeted inhibitors. Nat Rev Cancer 2005;5:341–354 [DOI] [PubMed] [Google Scholar]
- 14.Zeng F, Singh AB, Harris RC. The role of the EGF family of ligands and receptors in renal development, physiology and pathophysiology. Exp Cell Res 2009;315:602–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen J, Chen JK, Nagai K, et al. EGFR signaling promotes TGFβ-dependent renal fibrosis. J Am Soc Nephrol 2012;23:215–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yarden RI, Wilson MA, Chrysogelos SA. Estrogen suppression of EGFR expression in breast cancer cells: a possible mechanism to modulate growth. J Cell Biochem Suppl 2001;(Suppl. 36):232–246 [DOI] [PubMed] [Google Scholar]
- 17.Zhang MZ, Wang Y, Paueksakon P, Harris RC. Epidermal growth factor receptor inhibition slows progression of diabetic nephropathy in association with a decrease in endoplasmic reticulum stress and an increase in autophagy. Diabetes 2014;63:2063–2072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen J, Chen JK, Harris RC. EGF receptor deletion in podocytes attenuates diabetic nephropathy. J Am Soc Nephrol 2015;26:1115–1125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao HJ, Wang S, Cheng H, et al. Endothelial nitric oxide synthase deficiency produces accelerated nephropathy in diabetic mice. J Am Soc Nephrol 2006;17:2664–2669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mundel P, Reiser J, Zúñiga Mejía Borja A, et al. Rearrangements of the cytoskeleton and cell contacts induce process formation during differentiation of conditionally immortalized mouse podocyte cell lines. Exp Cell Res 1997;236:248–258 [DOI] [PubMed] [Google Scholar]
- 21.Chen J, Harris RC. Interaction of the EGF receptor and the Hippo pathway in the diabetic kidney. J Am Soc Nephrol 2016;27:1689–1700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sun Q, Westphal W, Wong KN, Tan I, Zhong Q. Rubicon controls endosome maturation as a Rab7 effector. Proc Natl Acad Sci U S A 2010;107:19338–19343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li Z, Li Y, Overstreet JM, et al. Inhibition of epidermal growth factor receptor activation is associated with improved diabetic nephropathy and insulin resistance in type 2 diabetes. Diabetes 2018;67:1847–1857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fang L, Zhou Y, Cao H, et al. Autophagy attenuates diabetic glomerular damage through protection of hyperglycemia-induced podocyte injury. PLoS One 2013;8:e60546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang MZ, Wang S, Yang S, et al. Role of blood pressure and the renin-angiotensin system in development of diabetic nephropathy (DN) in eNOS-/- db/db mice. Am J Physiol Renal Physiol 2012;302:F433–F438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hartleben B, Gödel M, Meyer-Schwesinger C, et al. Autophagy influences glomerular disease susceptibility and maintains podocyte homeostasis in aging mice. J Clin Invest 2010;120:1084–1096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matsunaga K, Noda T, Yoshimori T. Binding rubicon to cross the rubicon. Autophagy 2009;5:876–877 [DOI] [PubMed] [Google Scholar]
- 28.Matsunaga K, Saitoh T, Tabata K, et al. Two Beclin 1-binding proteins, Atg14L and Rubicon, reciprocally regulate autophagy at different stages. Nat Cell Biol 2009;11:385–396 [DOI] [PubMed] [Google Scholar]
- 29.Zhong Y, Wang QJ, Li X, et al. Distinct regulation of autophagic activity by Atg14L and Rubicon associated with Beclin 1-phosphatidylinositol-3-kinase complex. Nat Cell Biol 2009;11:468–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Obermajer N, Wong JL, Edwards RP, Odunsi K, Moysich K, Kalinski P. PGE(2)-driven induction and maintenance of cancer-associated myeloid-derived suppressor cells. Immunol Invest 2012;41:635–657 [DOI] [PubMed] [Google Scholar]
- 31.Kim J, Kundu M, Viollet B, Guan KL. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol 2011;13:132–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brosius FC, Khoury CC, Buller CL, Chen S. Abnormalities in signaling pathways in diabetic nephropathy. Expert Rev Endocrinol Metab 2010;5:51–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Buller CL, Heilig CW, Brosius FC III. GLUT1 enhances mTOR activity independently of TSC2 and AMPK. Am J Physiol Renal Physiol 2011;301:F588–F596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bork T, Liang W, Yamahara K, et al. Podocytes maintain high basal levels of autophagy independent of mtor signaling. Autophagy 2020;16:1932–1948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chiu D, Ma K, Scott A, Duronio V. Acute activation of Erk1/Erk2 and protein kinase B/akt proceed by independent pathways in multiple cell types. FEBS J 2005;272:4372–4384 [DOI] [PubMed] [Google Scholar]
- 36.Botti J, Djavaheri-Mergny M, Pilatte Y, Codogno P. Autophagy signaling and the cogwheels of cancer. Autophagy 2006;2:67–73 [DOI] [PubMed] [Google Scholar]
- 37.Wei Y, Zou Z, Becker N, et al. EGFR-mediated Beclin 1 phosphorylation in autophagy suppression, tumor progression, and tumor chemoresistance. Cell 2013;154:1269–1284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tan X, Thapa N, Sun Y, Anderson RA. A kinase-independent role for EGF receptor in autophagy initiation. Cell 2015;160:145–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hobbs SS, Goettel JA, Liang D, et al. TNF transactivation of EGFR stimulates cytoprotective COX-2 expression in gastrointestinal epithelial cells. Am J Physiol Gastrointest Liver Physiol 2011;301:G220–G229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shin M, Yang EG, Song HK, Jeon H. Insulin activates EGFR by stimulating its interaction with IGF-1R in low-EGFR-expressing TNBC cells. BMB Rep 2015;48:342–347 [DOI] [PMC free article] [PubMed] [Google Scholar]