Abstract
With the realization that mechanical forces mediate many biological processes and contribute to disease progression, researchers are focusing on developing new methods to understand the role of mechanotransduction in biological systems. Despite recent advances in stretching devices that analyze the effects of mechanical strain in vitro, there are still possibilities to develop new equipment. For example, many of these devices tend be expensive, whereas few have been designed to assess the effects of mechanical strain driven by the extracellular matrix (ECM) to epithelial cell monolayers and to cell-cell adhesion. In this chapter, we introduce a cost-efficient, user-friendly, 3D-printed stretching device that can be used to test the effects of mechanical strain on cultured epithelial cells. Evaluation of the device using speckle-tracking shows homogeneous strain distribution along the horizontal plane of membranes at 2.5% and 5% strains, supporting the reliability of the device. Since cell-cell junctions are mechanosensitive protein complexes, we hereby used this device to examine effects on cell-cell adhesion. For this, we used colon epithelial Caco2 cell monolayers that well-differentiate in culture and form mature adherens junctions. Subjecting Caco2 cells to 2.5% and 5% strain using our device resulted in significant reduction in the localization of the core adherens junction component E-cadherin at areas of cell-cell contact and its increased translocation to the cytoplasm, which in agreement with other methodologies showing that increased ECM-driven strain negatively affects cell-cell adhesion. In summary, we here present a new, cost-effective, homemade device that can be reliably used to examine effects of mechanical strain on epithelial cell monolayers and cell-cell adhesion, in vitro.
Keywords: Mechanical strain, Cell stretching, Cell-cell adhesion, Caco2 cells, 3D printing, E-cadherin, Adherens junctions
1. Introduction
Mechanical forces are crucial in numerous biological processes. Mechanical stimuli are converted into biochemical signals through signaling pathways that ultimately drive processes such as development and wound healing [1, 2]. Research in recent years has been focused on mechanical stress as a catalyst to disease, including tumor progression [3, 4]. For example, fibrosis, characterized by the excessive accumulation of extracellular matrix (ECM), is a major precursor to cancer [5, 6]. During fibrosis, ECM undergoes extensive remodeling, thereby introducing atypical physical conditions such as high stiffness and increased physical strain [3, 4], which alter biomechanical signaling and promote abnormal cell behavior [7-11]. Strain, defined as deformation due to stress [12, 13], is a major mode where cells experience mechanical stress via physical forces such as compaction and tension [14]. Therefore, the development of methodologies that allow the studying of mechanical regulation of biological processes has been an emerging field. A simple method to model cell deforming and its effects on cell signaling and behavior is by stretching cells, in vitro [3].
A number of commercially available or homemade stretching apparatuses that are either computer assisted or manually controlled have been developed. These devices can apply steady or cyclic stretch either uniaxially or biaxially over a predetermined duration [15-18]. Although computer-controlled devices can be accurate and precise compared to manually controlled devices, they can be costly. On the other hand, homemade apparatuses are most cost-efficient but can be hard to replicate, or they may produce inconsistent results, without published detailed protocols accompanying them. Depending on the frequency, load, and type of stretch, results of strain experiments can be vastly varying, which also calls for well-controlled and fully detailed published methodologies [19]. Disclosing all the characteristics of the stretch methodology when analyzing and comparing results is essential, especially when considering different studies or studies based on different protocols. Here, we introduce in full detail a homemade, manually controlled device that applies a steady uniaxial stretch over a set-time period. It is cost-efficient, easy to assemble, and user-friendly.
We particularly introduce this device to study cell-cell adhesion. Cell-cell and cell-extracellular matrix (ECM) interactions are mediated by mechanosensitive complexes that transmit physical signals to the inner cell environment and vice versa [19-23]. Thus, there is growing interest on cell-cell adhesion components, such as cadherin complexes, as readouts to assess the effects of physical forces in cellular homeostasis [24, 25]. Furthermore, cumulative evidence portrays E-cadherin-based adhesion complexes as signaling epicenters regulating epithelial cell behavior, further underscoring their significance as mechanoresponsive cellular components [26-30]. E-cadherin is the epithelial-specific cadherin and the most abundant cadherin in the human body. However, most models studying mechanical strain on cells have been focused on single-cell, non-epithelial, cell models [31-34], rather than in epithelial monolayers, where E-cadherin expression is predominant and critical. For the above reasons, we focused developing this protocol for the study of E-cadherin-based cell-cell junctions in Caco2 colon epithelial cells [35]. Caco2 cells are widely used as a model of the well-differentiated epithelium, and they abundantly express E-cadherin [36, 37]. Indeed, cyclic stretch has been previously noted to disturb cell-cell junctions and enzymatic activity of Caco2 cells [38]. In the present protocol, we developed a stretching device using 3D printing and standardized it using a speckle-tracking method. We then used it to analyze distribution of E-cadherin under increased strain. Results show significant and consistent changes in E-cadherin subcellular localization under different conditions of strain. We anticipate that our methodology will provide a useful tool for the study of the effects of mechanical strain in E-cadherin-based cell-cell junction complexes, as well as its effects on epithelial monolayer integrity, barrier function, cellular signaling, and cell behavior.
2. Materials
2.1. 3D Printing
Material: Fromlabs Clear resin, V4 (FLGPCL04).
Software: PreForm, Form 2 [39].
3D printing hardware: Formlabs, Form 2 Stereolithography (SLA) resin-based 3d printer.
Washing buffers: >90% Isopropyl alcohol (IPA), Form cure-Formlabs.
Flush cutters.
Dremel: 4300-9/64.
Taps and Dies: DWA1450.
2.2. Cell Culture
Caco2 cells: ATCC-HTB-37.
Silicone Membrane: Specialty Manufacturing, Inc. (SMImfg.com). Silicone Sheeting 0.010″ NRV G/G 40D 12″ × 12″. Lot number SMI8042512. Dimension of each strip is 2 cm × 11 cm. Membrane is cut according to the number of strips required.
1.5 μg/cm2 Laminin: Sigma, L2020 Laminin from Engelbreth-Holm-Swarm murine sarcoma basement membrane. Laminin is diluted in Hank’s Balanced Salt Solution (HBSS): Fisher Scientific 14170-120 as required (see Note 1).
Cell culture medium: MEM, Fisher Scientific MT10010CV-MEM W/GLN HI GLU PYR was supplemented with 10% FBS, Fisher Scientific A3160502, 1 mM sodium pyruvate, Invitrogen 11360070-100 mM and 1× non-essential amino-acid supplement, Invitrogen 11140050-100X.
PBS: Fisher Scientific 21040CV.
70% ethanol.
150 cm dishes: Corning 12556003.
Tweezers: Millipore XX62000006P.
Glass Staining Dishes for 50 Slides: DWK Life Sciences Wheaton, Fisher Scientific 08813C.
2.3. Immunofluorescence
Ice cold Methanol: Fisher Scientific A4524.
Antibodies: Primary antibody used in the study was E-cadherin 4A2C7, Life Technologies 180223. Secondary antibody used in the study was Alexa Fluor 488 anti-mouse, Life Technologies A21202, with a 1:500 dilution. Dilute both primary and secondary in antibody diluent: Dako S302281-2 at a 1:500 dilution.
Blocking solutions: Dako X090930-2.
Mounting solution: Polysciences, Aqua Poly/Mount-18606.
Coverslips: VWR48393-106-22x60.
Mounting slide: Denville Science M1011.
3. Methods
3.1. 3D Design and Printing of the Stretching Apparatus
Prototyping of the apparatus is carried out using PreForm: Form 2 software [39].
Stretching apparatus is printed using Fromlabs Clear resin-V4 (FLGPCL04) and with Formlabs: Form 2 Stereolithography (SLA) resin-based 3d printer.
Wash in >90% IPA for 15 min in Form Wash station.
Cure in Form cure for 15 min at 60 °C in Form Cure station.
Remove any support material with the use of flush cutters and a Dremel.
Clean the threads using taps and dies.
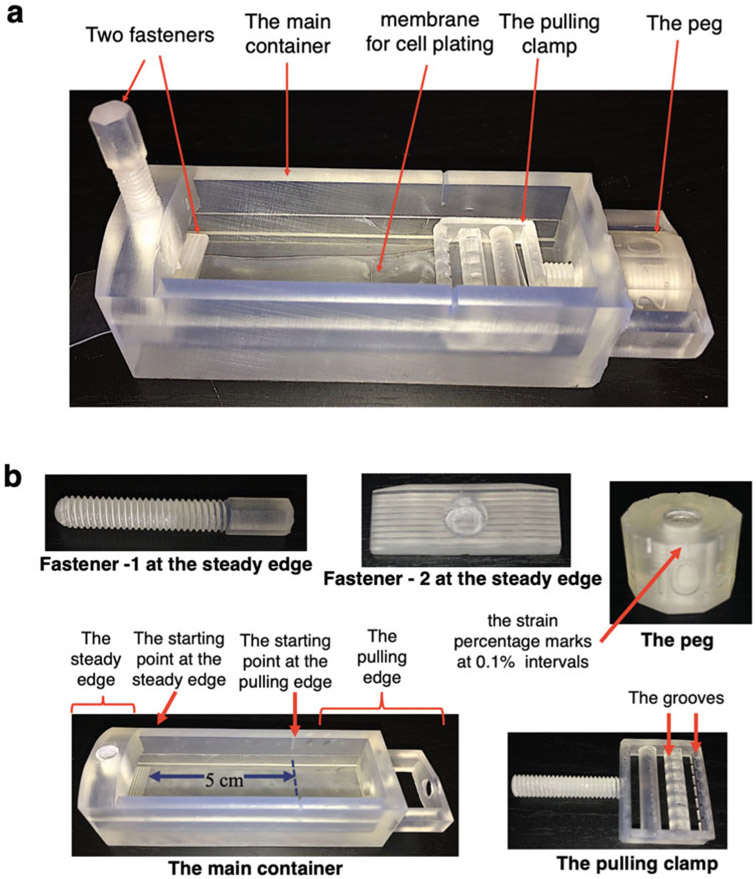
The stretching apparatus has five parts: two fasteners on the steady edge, one peg, one pulling clamp, and the main container. A starting point is marked in a side of the main container where the length from the steady edge to the starting point is 5 cm. This device is designed permitting the tensile strain equation shown below (Eq. 1), where ΔL is the change in length and L0 is the starting length [40]. Then, the ratio between ΔL and L0 is converted to a percentage to obtain the strain percentage (Eq. 2). Hence, at 0% strain the length of the membrane or L0 is 5 cm.
| (1) |
| (2) |
Two fasteners are designed to keep the membrane stable and locked at the steady edge. Thus, the membrane does not move at this end. The uniaxial strain is applied by pulling the clamp via rotating the peg. The peg is marked with numbers that represent 0.1% strain. A complete rotation of the peg is approximately equal to 1% strain. Thus, the change in length or ΔL is 5 mm. Another starting point is marked on top of the container at the pulling end. The pulling camp has multiple grooves to keep the membrane intact while pulling. At 0% strain, the pulling clamp is in line with the starting point marked at the side of the main container, and the 0% mark on the peg is in line with the starting point marked on top of the main container (Fig. 1a, b).
Fig. 1.
The stretching device. (a) The fully assembled device is comprised of five parts: (b) two fasteners, the peg, the pulling clamp, and the main container. The distance from the starting point at the steady edge to the starting point at the pulling edge (L0) is 5 cm. The peg is marked with 0.1% strain intervals. The grooves in the pulling clamp ensures the membrane is intact and stable during the strain
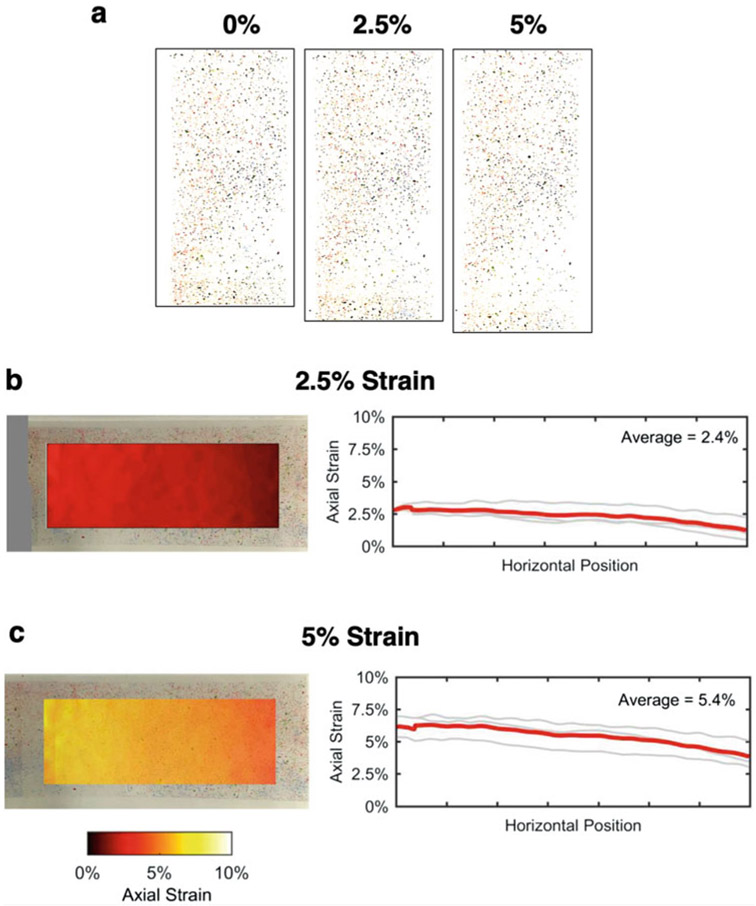
In order to validate that the stretching platform was applying a uniform stretch field to our cell cultures, we used standard two-dimensional digital image correlation (DIC) techniques. Briefly, culture membranes were marked by a scattered pattern of pencil shavings and imaged before and after 2.5% and 5% strain (Fig. 2a). Images were then fed into an open-source, MATLAB-based DIC algorithm (Ncorr.com) [41], which automatically registers each deformed image to the reference (undeformed) image, identifies particle patterns, measures displacements of particles between deformation steps, and then calculates the full deformation gradient tensor (F) and resulting Engineering strains (defined as the stretch ratio – 1, and corresponding to the macro-scale Tensile strain from Eq. 1 above). We repeated all strain calculations and averaged across three replicates (Fig. 2b, c).
Fig. 2.
Experimental platform stretching quantification. (a) Pencil shavings were scattered throughout the membranes and were put under 2.5% and 5% strain with 0% being the resting condition. Engineering strain (= stretch ratio – 1) along the axis of the stretching device (x/horizontal direction) was quantified by tracking fiduciary markers before and after 2.5% (b), and 5% (c) strains, measuring the resulting displacements, and calculating strains accordingly. Each strain condition was repeated three times, and, in all cases, the resulting strain fields showed little to no spatial variation across x/horizontal and y/vertical positions. As expected, transverse strains (y/vertical direction) and shear strains were very small relative to axial strains (data not shown)
3.2. Cell Culture
Wash cut membranes and tweezers with 70% Ethanol and leave in the cell culture hood under UV light on for 20 min until Ethanol is dried out.
Switching to bright light, carefully place the membranes inside a 150 mm cell culture dish using tweezers (see Notes 2 and 3).
Cover the membrane surfaces with diluted Laminin solution [42] in HBSS as calculated (see Note 1) and leave in the cell culture hood for 1 h (see Notes 4 and 5).
Remove the excess fluid and air dry the coated surfaces for 10 min.
Evenly plate 10.0 × 106 Caco2 cells using cell culture medium to homogeneously cover the whole 150 mm plate and leave the plate in the cell culture incubator at 37 °C with 5% CO2 until confluent (see Note 6).
3.3. Cell Stretching and Fixing Process
Autoclave the glass containers before the experiment begins.
Wash the disassembled stretching apparatuses with 70% ethanol and leave in the cell culture hood with UV light on for 20 min until Ethanol is dried out (for different strain conditions, different apparatuses are required).
Bring the confluent cell culture dish inside the cell culture hood.
Wash the cells with PBS once and add fresh medium.
Carefully place one membrane with cells at a time in each of pulling clamp of the apparatus using tweezers.
Place the pulling clamp with the membrane inside the main container and drag until the starting point is met by rotating the peg. It is important that the scale of the peg is at zero when the pulling clamp is at the starting point.
Lock the free end of the membrane to the steady end using the fasteners.
Turn the peg until the desired strain is met. For example, a 2.5 turn of the peg approximates to 2.5% strain, and the starting point is 0% strain.
Place the apparatuses with membranes inside the glass container (two apparatuses can fit in one container) and fill cell culture media until the membranes are fully covered (approximately 100 mL).
Place the closed glass container inside the cell culture cell hood at 37 °C with 5% CO2 for 2 h.
After 2 h, while the apparatuses are inside the glass container, remove media and wash the membranes with PBS once (see Note 7).
Add ice cold methanol into the glass container until the membranes are fully covered and place the glass container at −20 °C for 8 min.
Remove the methanol and wash membranes three times with PBS.
Carefully remove membranes from the apparatuses and place in a new 150 mm dish with PBS (see Note 8).
3.4. Immunofluorescence Staining
Block the membranes with blocking reagent for 1 h at room temperature and stain with primary antibodies diluted in antibody diluent overnight at 4 °C (for processing of multiple strips see Note 9).
Wash three times with PBS and stain with fluorescently labeled secondary antibodies diluted in antibody diluent for 1 h at room temperature.
Wash two times with PBS, co-stain with DAPI diluted in PBS, and wash once again with PBS.
Add drops of mounting solution on to the mounting slides and place the membranes with cells side up.
Add a drop of mounting solution on top of the membrane and place coverslip on top.
Seal the edges of coverslip with nail polish and dry overnight at room temperature in the dark.
3.5. Imaging and Analysis
Image the slides using confocal microscopy – for this study a Leica SP5 confocal microscope was used with 63× objective and an additional 1.5× zoom. Z-stacks are 0.5 μ in thickness.
Open the Z-stacks in ImageJ software [43] and create maximum projections of Z-stacks.
Adjust image settings to using Image → Brightness Contrast option. It should be noted that once the brightness and contrast parameters are set, they should be applied to all the images in an experiment.
Set the measurement parameters by Analyze → Set measurements → Click on Area and Mean gray value.
Select the “Free hand line” option and draw a line of a specific length (~10 μm in this experiment) along the membrane.
Select Analyze → Measure. This will provide the area and the mean color value.
Repeat steps 5 and 6 for at least additional three times on different areas of the Z-stack image to obtain a representative value. Repeat these steps as necessary to measure cytoplasmic and nuclear color values. It is important to keep the length of the line consistent between the Z-stacks.
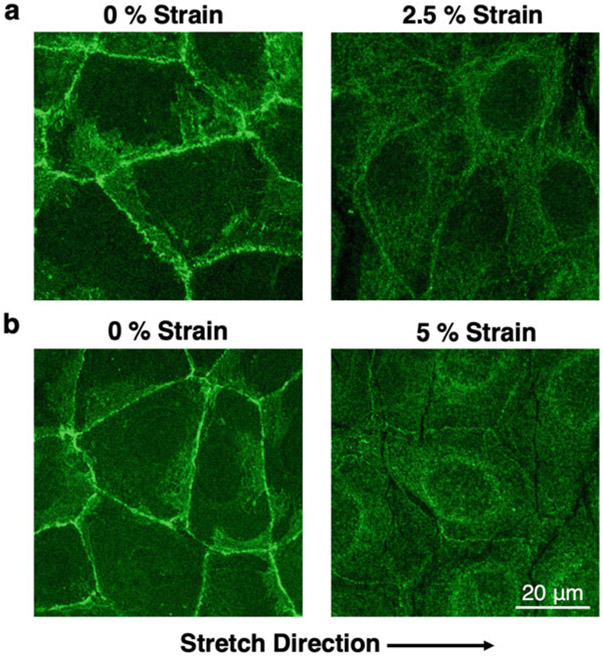
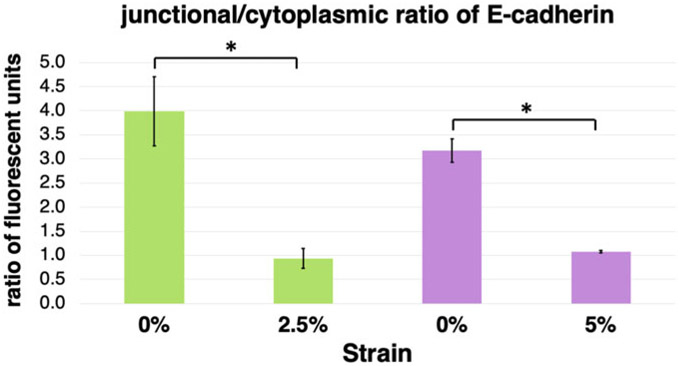
Table 1, Figs. 3 and 4 show results obtained through the procedure described above. There are significant changes in the E-cadherin localization at 2.5% and 5% strain as measured from the obtained immunofluorescence images (Fig. 3). At both strains E-cadherin junctional localization was decreased, while the cytoplasmic localization was increased (Table 1) resulting in overall decreased junctional/cytoplasmic ratio (Fig. 4).
Table 1.
E-cadherin junctional and cytoplasmic immunofluorescence intensity units of Caco2 cells under different conditions of strain (n = 3 representative fields; mean ± standard deviation)
| Strain (%) | Junctional | Cytoplasmic | Junctional/cytoplasmic ratio |
|---|---|---|---|
| 0 | 87.498 ± 12.634 | 21.935 ± 3.918 | 3.989 ± 0.715 |
| 2.5 | 31.350 ± 4.910 | 34.494 ± 7.632 | 0.934 ± 0.205 |
| 0 | 183.363 ± 5.634 | 57.979 ± 3.171 | 3.171 ± 0.245 |
| 5 | 70.472 ± 10.470 | 65.731 ± 11. 337 | 1.076 ± 0.029 |
Fig. 3.
E-cadherin localization is affected by strain. Caco2 cells were fixed at different strains and were stained for E-cadherin by immunofluorescence. Each strain was repeated three times with a 0% control. Representative images shown here, where E-cadherin primarily localizes at areas of cell-cell contact at the 0% resting condition. However, at 2.5% (a) and 5% (b) strains, localization of E-cadherin at areas of cell-cell contact is significantly reduced, while the cytoplasmic localization is increased. At 5% strain, a perinuclear E-cadherin localization was also observed, which was included in the cytoplasmic fraction for the calculations
Fig. 4.
The ratio of junctional vs cytoplasmic E-cadherin localization is decreased upon application of strain in Caco2 cells. Graphical representation of the E-cadherin junctional and cytoplasmic immunofluorescence quantification of Caco2 cells under different conditions of strain presented in Table 1. Upon application of strain, the ratio of junctional to cytoplasmic localization of E-cadherin is significantly reduced (n = 3 representative fields; mean ± standard deviation; *P < 0.05, student’s t-test)
4. Notes
Calculations for Laminin dilution can be performed as follows: area of one strip is 22 cm2 (2 cm × 11 cm). Thus, 33 μg (22 cm2 × 1.5 μg/l cm2) of Laminin is required to coat one strip. Considering the original concentration of Laminin vial is 1.5 mg/mL and 1 mL of HBSS is required to cover the area of one strip, the dilution is 22 μL of Laminin in 1 mL of HBSS. (33 μg/1.5 mg/mL).
Generally, up to three 2 cm × 11 cm membranes can be placed inside one 150 mm dish. If more membranes are placed, the area of the membranes that touches the bottom of dish will be decreased. This will negatively affect coating the membranes homogeneously.
Membranes should never dry and must be kept in medium or PBS at all times after fixation. Membranes should only be dried completely under the UV light, right before beginning of cell culture.
Sometimes, the diluted Laminin solution may leak out from the membranes into the cell culture dish bottom. This will negatively affect the membrane coating. In this situation, place the membranes in the bottom of the cell culture dish and cover the whole 150 mm plate with diluted Laminin for an even distribution. This will allow for a homogenous coating of the membranes.
We have identified Laminin as the substrate where the junctional localization of E-cadherin agrees with published work in well-differentiated Caco2 cells and tissues by us and others [29, 42, 44-46]. Nevertheless, other ECM proteins such as Collagens and Fibronectin can be used according to different cell types.
10 × 106 [47] is half the cells at confluency of a 150 mm dish according to Fisher Scientific protocols. If cells are not attaching and growing properly, the seeding density can be changed accordingly. It is also important to note that seeding density varies depending on the cell line.
Do not accidentally stretch the membranes while handling and placing inside and taking out of the apparatus.
Fixed membranes can be immersed in PBS, and stored at 4 °C until ready to be stained.
For multiple stains, membranes can be cut in 3–4 pieces accordingly, while they are still in PBS, before the blocking step.
Acknowledgments
This work was supported by: NIH P20 GM130457-01A1 (to AK); NIH P20 GM103444 (SC BioCRAFT pilot Award to AK); Concern Foundation’s Conquer Cancer Now Award (to AK); NIH P20 GM121342 (to WJR).
References
- 1.Miller CJ, Davidson LA (2013) The interplay between cell signalling and mechanics in developmental processes. Nat Rev Genet 14 (10):733–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barnes LA, Marshall CD, Leavitt T, Hu MS, Moore AL, Gonzalez JG, Longaker MT, Gurtner GC (2018) Mechanical forces in cutaneous wound healing: emerging therapies to minimize scar formation. Adv Wound Care (New Rochelle) 7(2):47–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Broders-Bondon F, Nguyen Ho-Bouldoires TH, Fernandez-Sanchez M-E, Farge E (2018) Mechanotransduction in tumor progression: the dark side of the force. J Cell Biol 217(5):1571–1587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kalli M, Stylianopoulos T (2018) Defining the role of solid stress and matrix stiffness in cancer cell proliferation and metastasis. Front Oncol (2234-943X (Print)) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim ER (2014) Colorectal cancer in inflammatory bowel disease: the risk, pathogenesis, prevention and diagnosis. World J Gastroenterol 20(29):9872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gordon IO, Agrawal N, Willis E, Goldblum JR, Lopez R, Allende D, Liu X, Patil DY, Yerian L, El-Khider F, Fiocchi C, Rieder F (2018) Fibrosis in ulcerative colitis is directly linked to severity and chronicity of mucosal inflammation. Aliment Pharmacol Ther 47 (7):922–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Basson MD, Li GD, Hong F, Han O, Sumpio BE (1996) Amplitude-dependent modulation of brush border enzymes and proliferation by cyclic strain in human intestinal Caco-2-monolayers. J Cell Physiol 168(2):476–488 [DOI] [PubMed] [Google Scholar]
- 8.Ali MY, Saif MTA (2014) Substrate stiffness mediated metastasis like phenotype of colon cancer cells is independent of cell to gel adhesion. Cell Mol Bioeng 7(4):532–543 [Google Scholar]
- 9.Fernández-Sánchez ME, Barbier S, Whitehead J, Béalle G, Michel A, Latorre-Ossa H, Rey C, Fouassier L, Claperon A, Brullé L, Girard E, Servant N, Rio-Frio T, Marie H, Lesieur S, Housset C, Gennisson J-L, Tanter M, Ménager C, Fre S, Robine S, Farge E (2015) Mechanical induction of the tumorigenic β-catenin pathway by tumour growth pressure. Nature 523(7558):92–95 [DOI] [PubMed] [Google Scholar]
- 10.Röper J-C, Mitrossilis D, Stirnemann G, Waharte F, Brito I, Fernandez-Sanchez M-E, Baaden M, Salamero J, Farge E (2018) The major β-catenin/E-cadherin junctional binding site is a primary molecular mechanotransductor of differentiation in vivo. Elife 7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ciasca G, Papi M, Minelli E, Palmieri V, De Spirito M (2016) Changes in cellular mechanical properties during onset or progression of colorectal cancer. World J Gastroenterol 22 (32):7203–7214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thompson WR, Scott A, Loghmani MT, Ward SR, Warden SJ (2016) Understanding mechanobiology: physical therapists as a force in mechanotherapy and musculoskeletal regenerative rehabilitation. Phys Ther 96(4):560–569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rawlinson SCF (2017) Mechanical responsiveness of distinct skeletal elements: possible exploitation of low weight-bearing bone. In: Rawlinson SCF (ed) Mechanobiology: exploitation for medical benefit. Wiley, Hoboken, pp 131–138 [Google Scholar]
- 14.Kalli M, Stylianopoulos T (2018) Defining the role of solid stress and matrix stiffness in cancer cell proliferation and metastasis. Front Oncol 8:55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Atcha H, Davis CT, Sullivan NR, Smith TD, Anis S, Dahbour WZ, Robinson ZR, Grosberg A, Liu WF (2018) A low-cost mechanical stretching device for uniaxial strain of cells: a platform for pedagogy in mechanobiology. J Biomech Eng 140(8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shao Y, Tan X, Novitski R, Muqaddam M, List P, Williamson L, Fu J, Liu AP (2013) Uniaxial cell stretching device for live-cell imaging of mechanosensitive cellular functions. Rev Sci Instrum 84(11):114304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Seriani S, Del Favero G, Mahaffey J, Marko D, Gallina P, Long CS, Mestroni L, Sbaizero O (2016) The cell-stretcher: a novel device for the mechanical stimulation of cell populations. Rev Sci Instrum 87(8):084301. [DOI] [PubMed] [Google Scholar]
- 18.USA, S. Cell Stretching Systems (2020) https://strexcell.com/cell-stretching-system/
- 19.Hoffman BD, Grashoff C, Schwartz MA (2011) Dynamic molecular processes mediate cellular mechanotransduction. Nature 475 (7356):316–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lecuit T, Yap AS (2015) E-cadherin junctions as active mechanical integrators in tissue dynamics. Nat Cell Biol 17(5):533–539 [DOI] [PubMed] [Google Scholar]
- 21.Lecuit T (2010) Alpha-catenin mechanosensing for adherens junctions. Nat Cell Biol 12 (6):522–524 [DOI] [PubMed] [Google Scholar]
- 22.Schwartz MA (2010) Integrins and extracellular matrix in mechanotransduction. Cold Spring Harb Perspect Biol 2(12):a005066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ross TD, Coon BG, Yun S, Baeyens N, Tanaka K, Ouyang M, Schwartz MA (2013) Integrins in mechanotransduction. Curr Opin Cell Biol 25(5):613–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Borghi N, Sorokina M, Shcherbakova OG, Weis WI, Pruitt BL, Nelson WJ, Dunn AR (2012) E-cadherin is under constitutive actomyosin-generated tension that is increased at cell-cell contacts upon externally applied stretch. Proc Natl Acad Sci U S A 109 (31):12568–12573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Benham-Pyle BW, Pruitt BL, Nelson WJ (2015) Cell adhesion. Mechanical strain induces E-cadherin-dependent Yap1 and beta-catenin activation to drive cell cycle entry. Science 348(6238):1024–1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Daulagala AC, Bridges MC, Kourtidis A (2019) E-cadherin beyond structure: a signaling hub in Colon homeostasis and disease. Int J Mol Sci 20(11) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kourtidis A, Lu R, Pence LJ, Anastasiadis PZ (2017) A central role for cadherin signaling in cancer. Exp Cell Res 358(1):78–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kourtidis A, Necela B, Lin WH, Lu R, Feathers RW, Asmann YW, Thompson EA, Anastasiadis PZ (2017) Cadherin complexes recruit mRNAs and RISC to regulate epithelial cell signaling. J Cell Biol 216(10):3073–3085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kourtidis A, Ngok SP, Pulimeno P, Feathers RW, Carpio LR, Baker TR, Carr JM, Yan IK, Borges S, Perez EA, Storz P, Copland JA, Patel T, Thompson EA, Citi S, Anastasiadis PZ (2015) Distinct E-cadherin-based complexes regulate cell behaviour through miRNA processing or Src and p120 catenin activity. Nat Cell Biol 17(9):1145–1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nair-Menon J, Daulagala AC, Connor DM, Rutledge L, Penix T, Bridges MC, Wellslager B, Spyropoulos DD, Timmers CD, Broome AM, Kourtidis A (2020) Predominant distribution of the RNAi machinery at apical Adherens junctions in colonic epithelia is disrupted in Cancer. Int J Mol Sci 21(7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen CH, Marymont JV, Huang MH, Geyer M, Luo ZP, Liu X (2007) Mechanical strain promotes fibroblast gene expression in presence of corticosteroid. Connect Tissue Res 48(2):65–69 [DOI] [PubMed] [Google Scholar]
- 32.Manuyakorn W, Smart DE, Noto A, Bucchieri F, Haitchi HM, Holgate ST, Howarth PH, Davies DE (2016) Mechanical strain causes adaptive change in bronchial fibroblasts enhancing Profibrotic and inflammatory responses. PLoS One 11(4):e0153926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Matsumoto T, Yung YC, Fischbach C, Kong HJ, Nakaoka R, Mooney DJ (2007) Mechanical strain regulates endothelial cell patterning in vitro. Tissue Eng 13(1):207–217 [DOI] [PubMed] [Google Scholar]
- 34.Zeiger AS, Liu FD, Durham JT, Jagielska A, Mahmoodian R, Van Vliet KJ, Herman IM (2016) Static mechanical strain induces capillary endothelial cell cycle re-entry and sprouting. Phys Biol 13(4):046006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lea T (2015) Caco-2 cell line. In: Verhoeckx K, Cotter P, López-Expósito I, Kleiveland C, Lea T, Mackie A, Requena T, Swiatecka D, Wichers H (eds) The impact of food bioactives on health. Springer, Cham, pp 103–111 [PubMed] [Google Scholar]
- 36.Grasset E, Bernabeu J, Pinto M (1985) Epithelial properties of human colonic carcinoma cell line Caco-2: effect of secretagogues. Am J Physiol 248(5 Pt 1):C410–C418 [DOI] [PubMed] [Google Scholar]
- 37.Hidalgo IJ, Raub TJ, Borchardt RT (1989) Characterization of the human colon carcinoma cell line (Caco-2) as a model system for intestinal epithelial permeability. Gastroenterology 96(3):736–749 [PubMed] [Google Scholar]
- 38.Samak G, Gangwar R, Crosby LM, Desai LP, Wilhelm K, Waters CM, Rao R (2014) Cyclic stretch disrupts apical junctional complexes in Caco-2 cell monolayers by a JNK-2-, c-Src-, and MLCK-dependent mechanism. Am J Physiol Gastrointest Liver Physiol 306(11):G947–G958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Formlabs (2020) https://formlabs.com/software/
- 40.Ling SJ, Sanny J, Moebs W (eds) University Physics.Open Stax Rice University, Houston [Google Scholar]
- 41.Blaber J, Adair B, Antoniou A (2015) Ncorr: open-source 2D digital image correlation Matlab software. Exp Mech 55:1105–1122 [Google Scholar]
- 42.Basson MD, Turowski G, Emenaker NJ (1996) Regulation of human (Caco-2) intestinal epithelial cell differentiation by extracellular matrix proteins. Exp Cell Res 225(2):301–305 [DOI] [PubMed] [Google Scholar]
- 43.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez JY, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A (2012) Fiji: an open-source platform for biological-image analysis. Nat Methods 9(7):676–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kourtidis A, Anastasiadis PZ (2016) PLE KHA7 defines an apical junctional complex with cytoskeletal associations and miRNA-mediated growth implications. Cell Cycle 15 (4):498–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jokhadar ŠZ, Šuštar V, Svetina S, Batista U (2009) Time lapse monitoring of CaCo-2 cell shapes and shape dependence of the distribution of integrin β1 and F-actin on their basal membrane. Cell Commun Adhes 16 (1–3):1–13 [DOI] [PubMed] [Google Scholar]
- 46.Zhang X, Cromwell JW, Kunjummen BD, Yee D, Garcia-Aguilar J (2003) The α2 and α3 integrins are required for morphologic differentiation of an intestinal epithelial cell line. Surgery 133(4):429–437 [DOI] [PubMed] [Google Scholar]
- 47.ThermoScientific (2020) Useful numbers for cell culture. https://www.thermofisher.com/us/en/home/references/gibco-cell-culture-basics/cell-culture-protocols/cell-culture-useful-numbers.html [Google Scholar]