Abstract
Prime editing enables diverse genomic alterations to be written into target sites without requiring double-strand breaks or donor templates. The design of prime-editing guide RNAs (pegRNAs), which must be customized for each edit, can however be complex and time-consuming. Compared with single guide RNAs (sgRNAs), pegRNAs have an additional 3’ extension composed of a primer binding site and a reverse-transcription template. Here, we report a web tool, which we named pegFinder (http://pegfinder.sidichenlab.org), for the rapid design of pegRNAs from reference and edited DNA sequences. pegFinder can incorporate sgRNA on-target and off-target scoring predictions into its ranking system, and nominates secondary nicking sgRNAs for increasing editing efficiency. Cas9 variants with expanded targeting ranges are also supported. To facilitate downstream experimentation, pegFinder produces a comprehensive table of candidate pegRNAs, along with oligonucleotide sequences for cloning.
Clustered regularly interspaced short palindromic repeats (CRISPR)-based technologies have been widely adopted as powerful tools for targeted genomic manipulation1. Recently, a new CRISPR-based strategy for precision genome editing was developed that enables diverse genomic alterations to be directly written into target sites without requiring double-strand breaks (DSBs) or donor templates2. Termed prime editing, this approach involves two key components: 1) a catalytically impaired CRISPR-associated protein 9 (Cas9) nickase fused to a reverse transcriptase (PE2), and 2) a multifunctional prime editing guide RNA (pegRNA) that specifies the target site and further acts as a template for reverse transcription (RT). pegRNAs are similar to single guide RNAs (sgRNAs), but additionally have a customizable extension on the 3’ end. The 3’ extension is composed of a RT template that encodes the desired edit and a primer binding site (PBS) that anneals to the target genomic site to prime the RT reaction2. Secondary nicking sgRNAs can also be employed to potentially increase the efficiency of prime editing by nicking the opposite strand, thus favoring the edited strand during heteroduplex resolution (PE3)2. To enhance specificity and to reduce the probability of generating unwanted DSBs, the secondary nicking sgRNAs can designed such that they only become active after successful prime editing has occurred (PE3b).
These additional components considerably increase the complexity of pegRNA design compared to standard sgRNAs. In particular, the precise lengths of the RT template and PBS sequence have both been demonstrated to significantly effect prime editing efficiency2-4. Inspired by the variety of tools that have been developed for identifying candidate sgRNAs in a target DNA sequence5-11, we sought to create a user-friendly application for designing pegRNAs. We therefore developed pegFinder, a streamlined web tool that rapidly designs and ranks candidate pegRNAs for a user-specified genetic modification (Fig. 1a). The pegFinder web portal is freely available at http://pegfinder.sidichenlab.org (Supplementary Fig. 1).
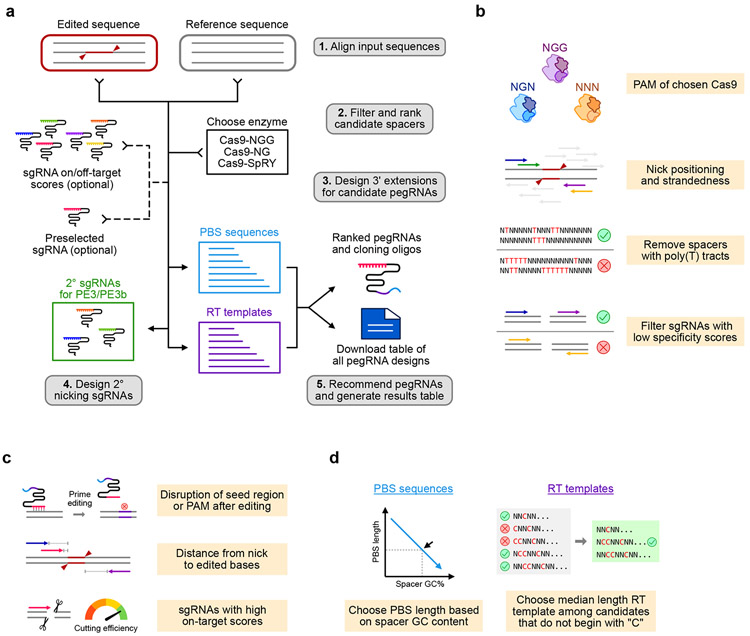
Fig. 1 ∣. pegFinder: A pegRNA designer for CRISPR prime editing.
a, Schematic of the pegFinder workflow for designing CRISPR prime editing pegRNAs. b, Criteria by which pegFinder identifies candidate pegRNA spacers that are suitable for the desired genomic edit. c, Overview of the factors considered by pegFinder when ranking and selecting candidate pegRNA spacers. d, Design principles by which pegFinder prioritizes PBS sequences and RT templates that compose the 3’ extension of pegRNAs.
Results and Discussion
pegFinder simply requires two inputs: 1) the wildtype/reference DNA sequence of the target site, and 2) the edited/desired DNA sequence (Supplementary Fig. 2 and Supplementary Table 1). For consideration of predicted on-target/off-target scores, the user can optionally include the results from the Broad sgRNA designer (https://portals.broadinstitute.org/gpp/public/analysis-tools/sgrna-design)6,7 or CRISPRscan (https://www.crisprscan.org/?page=sequence)8 using the wildtype DNA sequence as input (Fig. 1a). If desired, a preselected sgRNA spacer sequence can also be specified. After validating the inputs, pegFinder first identifies the differences between the wildtype and edited DNA sequences by performing a Needleman-Wunsch alignment with affine gap penalties12. Using the alignment, pegFinder chooses a single sgRNA spacer from the eligible candidates. pegFinder prioritizes spacers whose target sites would be disrupted after prime editing, further considering the distance between the nick site and the desired edits (Fig. 1b-c). If provided, pegFinder will factor in on-target/off-target scores when selecting candidate spacers.
pegFinder then identifies an appropriate RT template and PBS sequence to generate the desired edit by evaluating the positioning of the edited bases and the GC content of the sgRNA, respectively (Fig. 1d and Supplementary Fig. 2). Additionally, pegFinder identifies secondary sgRNAs that nick 40-150nt away (default setting) on the opposite strand from the primary sgRNA (PE3), as well as nicking sgRNAs that only become active after prime editing has occurred (PE3b). To facilitate rapid experimental implementation, pegFinder further generates oligonucleotide sequences that can be directly used to clone the pegRNAs into standard plasmid vectors (Supplementary Fig. 2).
Given that prime editing has only recently been developed, the rules governing pegRNA design are incompletely understood. Thus, optimization of pegRNAs for each experimental application may be necessary. Since the efficiency of prime editing has been shown to vary widely depending on the length and/or base composition of the RT template and PBS2-4, pegFinder reports RT templates and PBS sequences of varying lengths (Supplementary Fig. 2). pegFinder also generates a downloadable table containing a comprehensive catalogue of the pegRNA candidates for each of the top-ranked sgRNA spacers, along with the corresponding cloning oligo sequences (Supplementary Table 2). This table can be directly used to generate pegRNA libraries that exhaustively test the various combinations of sgRNA spacers, PBS sequences, and RT templates for a desired edit. In this manner, pegFinder can facilitate downstream optimization of prime editing experiments.
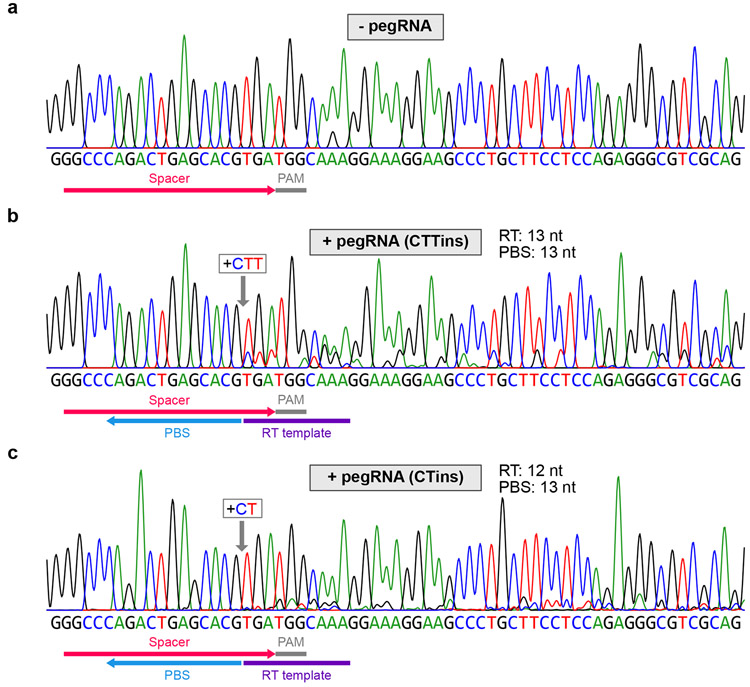
To validate the design algorithm, we first cross-referenced the pegRNA designs recommended by pegFinder with experimental data using prime editing in human cells2, murine cells3 and plants4, finding that pegFinder successfully identified functional pegRNAs in these systems (Supplementary Note). We then used pegFinder to design two different pegRNAs targeting the human HEK3 locus (also known as LINC01509). The first pegRNA was designed to insert “CTT” into the same genomic position as one of the pegRNAs described in the original prime editing study2, and thus served as a positive control benchmark. With only minimal user input, pegFinder designed a candidate CTTins pegRNA that was largely identical in sequence to the CTTins pegRNA described previously2, demonstrating the accuracy of the algorithm. The oligonucleotide sequences generated by pegFinder were then directly used for ligation cloning (Methods). In comparison to control cells co-transfected with PE2 and empty vector (Fig. 2a), cells co-transfected with PE2 and the pegFinder-designed CTTins pegRNA showed evidence of prime editing, as determined by analysis of minor peaks in the sequencing chromatograms (Fig. 2b). We similarly used pegFinder to design a pegRNA for inserting “CT” into the HEK3 locus; this particular edit (CTins) was not performed in the original study2. Using the constructs produced by pegFinder, we observed evidence of prime editing in cells transfected with the CTins pegRNA (Fig. 2c), experimentally demonstrating the functionality of pegFinder-designed pegRNAs.
Fig. 2 ∣. Experimental validation of pegRNAs designed by pegFinder.
a, Sequencing chromatogram of the HEK3 target site in cells transfected with vector control. b, Sequencing chromatogram of the HEK3 target site in cells transfected with a pegRNA designed to insert “CTT” into the +1 position, along with a secondary nicking sgRNA. c, Sequencing chromatogram of the HEK3 target site in cells transfected with a pegRNA designed to insert “CT” into the +1 position, along with a secondary nicking sgRNA. Representative data are shown, from three independent experiments.
Together, these data showcase the simplicity and utility of pegFinder for pegRNA design. pegFinder is a convenient tool for researchers in diverse fields to rapidly harness the versatility of prime editing.
Methods
Development of pegFinder algorithm and web server
The pegFinder core algorithm was developed in Perl. The web portal was implemented in the Mojolicious - Perl real-time web framework.
Inputs for pegFinder
pegFinder minimally requires two inputs. First, the wildtype DNA sequence of the region of interest is needed. Since candidate sgRNAs must be found within this wildtype sequence, we recommend >100nt flanks around the desired edit site. These sequences can be readily retrieved through genome browsers such as UCSC, IGV, or Ensembl BioMart. Second, the edited DNA sequence should be obtained by modifying the wildtype sequence to incorporate the desired alterations. Note that pegFinder expects the wildtype and edited sequences to share identical 5’ and 3’ ends, and will notify the user when this is not the case. As an example, consider a 200nt wildtype DNA sequence in which the user wishes to insert a 10nt sequence after the 100th nucleotide. The edited sequence should then be 100nt flank – 10nt insertion – 100nt flank, where the 5’ and 3’ 100nt flanks around the insert correspond exactly to the 200nt wildtype sequence. Thus, we recommend generating the edited DNA sequence by directly modifying the wildtype DNA input, as doing so ensures that the 5’ and 3’ flanks will remain identical.
By default, pegFinder bases its pegRNA designs on using wildtype Cas9 with an NGG protospacer adjacent motif (PAM). pegFinder also supports the use of Cas9-NG or Cas9-SpRY variants13, which expand the potential targeting range of prime editors, though such constructs have not yet been experimentally tested. When using Cas9-NGG as the CRISPR enzyme, pegFinder can further incorporate predicted on-target/off-target scores from sgRNA designer tools, with the caveat that these scoring algorithms were trained on gene knockout data, and thus may not be relevant for prime editing experiments. pegFinder can use the results from the Broad sgRNA designer tool6,7 (https://portals.broadinstitute.org/gpp/public/analysis-tools/sgrna-design) or CRISPRscan8 (https://www.crisprscan.org/?page=sequence), with the wildtype DNA sequence described above as the input query. For the Broad designer, the CRISPR enzyme should be selected as SpyoCas9 (NGG), and the appropriate target genome should be selected. Note also that all unpicked sequences should be reported. The tab-delimited results file that is produced (“sgRNA Picking Results”) can be saved and uploaded to pegFinder. For CRISPRscan, the correct target species should be chosen, the enzyme should be set as Cas9-NGG, and the option to find sgRNAs from both T7 and Sp6 promoters should be selected. The tab-delimited file that is produced can be saved and uploaded to pegFinder. As another alternative, if the user wishes to specify a preselected sgRNA, the 20nt spacer sequence can be entered directly into pegFinder. If a preselected sgRNA was specified, pegFinder will validate whether the chosen sgRNA is correctly positioned to produce the desired edits.
Alignment of wildtype and edited DNA sequences
The first step of pegFinder is to align the wildtype and edited DNA sequences using the Needleman-Wunsch algorithm, with an affine gap penalty function12. The positions of gaps and mismatched bases are noted, which are then used to select candidate sgRNAs.
Selection of primary nicking sgRNA spacers for pegRNAs
If no preselected sgRNA spacer was specified, pegFinder will identify candidate sgRNA spacers de novo. After determining the position range of the desired alterations, pegFinder searches for spacers that could potentially mediate the desired prime editing outcome. pegFinder designates sgRNA spacers on the “sense” strand as potential candidates if these conditions are met: 1) the sgRNA cut position is upstream of the 5’-most edited base, 2) the 3’-most edited base is within 150nt of the cut position (50nt for Cas9-NG and 20nt for Cas9-SpRY, given the vastly increased number of candidate spacers for these variants), 3) the spacer does not contain 5 or more consecutive thymidines that would terminate U6 transcription, and if applicable, 4) the sgRNA spacer has a total “off-target” Tier I Bin I + Bin II score ≤ 1 (as determined by the Broad sgRNA designer), or the number of “seed off-targets” is 0 (as determined by CRISPRscan). While a Tier I score of 1 on the Broad designer would normally indicate a perfect-match off-target site, in this context it would simply correspond to the genomic locus of the input sequence, and thus can be ignored. Similarly, for sgRNA spacers on the “antisense” strand, pegFinder designates a spacer as a candidate if: 1) the sgRNA cut position is downstream of the 3’-most edited base (in the sense orientation), 2) the 5’-most edited base (sense orientation) is within 150nt of the cut position (50 nt for Cas9-NG and 20nt for Cas9-SpRY), 3) no poly(T) tracts, and if applicable, 4) the sgRNA has a total “off-target” Tier I Bin I + Bin II score ≤ 1 (Broad sgRNA designer) or a “seed off-targets” count of 0 (CRISPRscan).
pegFinder then ranks the candidate sgRNA spacers based on their distance to the first edited base, prioritizing spacers whose target sites would be disrupted upon successful prime editing (i.e. mutations in the seed region and/or PAM). As disruption of the pegRNA target site would reduce the probability of repeated editing, it is suggested that such spacers would have higher prime editing efficiency. When incorporating on-target prediction scores from sgRNA designer tools, pegFinder additionally prioritizes sgRNA spacers with higher on-target efficacy. If there are candidate spacers with high on-target scores ≥ 0.5 (Broad designer) or ≥ 25 (CRISPRscan), pegFinder will choose the spacer with the shortest distance to the closest edit position, again prioritizing spacers that would no longer be functional upon successful prime editing. In the event of a tie, pegFinder chooses the sgRNA with the higher on-target score, if provided.
Selection of RT templates and PBS sequences
After selecting a primary sgRNA (or directly using the user-specified preselected sgRNA), pegFinder then extracts candidate RT templates and PBS sequences that can be incorporated into the 3’ extension of pegRNAs. For designing RT templates, pegFinder uses the edited/desired sequence and extracts the DNA between the primary nick site and the farthest edited base (the 3’-most base if using a sense strand sgRNA, or the 5’-most base if antisense), plus an additional 1nt. If the resultant sequence is < 10nt, pegFinder will report candidate RT templates ranging from 10-17nt in length. If the distance between the primary nick site and the farthest edited base is ≥ 10nt, pegFinder will report RT templates ranging from +1 to +7nt of the nick to edit distance. In all cases, pegFinder will flag RT templates that have a “C” as their 5’ most base (corresponding to a “G” as the final templated base), since it was previously demonstrated that such RT templates exhibit lower efficiency for prime editing, potentially due to base-pairing interactions with the sgRNA scaffold2. By default, pegFinder will then select a single RT template by choosing the template representing the median length among the candidates that do not begin with “C”, choosing the shorter template if there are an even number of candidates. If no RT templates exist that do not begin with “C”, pegFinder will select the template of median length among all candidates. Since the length of the RT template may require further optimization, all candidate RT templates are also reported by pegFinder.
To design PBS sequences, pegFinder extracts sequences 8-17nt in length from the reverse complement of the primary sgRNA sequence, moving backwards from the −1 position (the position before the cut site). Following the recommendations in the original study2, pegFinder selects a single PBS length based on the GC content of the spacer. Specifically, pegFinder uses the following formula: recommended PBS length = 24 – (GC% / 5), with a min-max of 8-17 nt. All PBS sequences 8-17nt are reported by pegFinder to facilitate experimental optimization.
Design of oligonucleotide sequences for cloning
After choosing a primary sgRNA spacer, secondary sgRNA spacer, RT template, and PBS sequence, pegFinder additionally generates oligonucleotide sequences that can be directly utilized for ligation cloning of the designed pegRNAs and/or sgRNAs. The oligos designed by pegFinder are intended for ligation cloning through an adaption of the lentiGuide-Puro protocol 14 (see section below). Of note, pegRNA/sgRNA sequences that do not begin with a “G” on the 5’ end will automatically have a “G” appended in the cloning oligonucleotide sequences to facilitate transcription from the standard U6 promoter.
pegFinder output table for experimental optimization
While pegFinder automatically recommends a single pegRNA design for the specified edit, pegFinder also produces a downloadable table containing a library of pegRNA designs for each of the top-ranked spacers. The user can further specify how many of the top candidate spacers will be included in the results table (default set to 3 spacers, each with a complete set of 3’ extension candidates). Cloning-ready oligonucleotide sequences are also provided for each of these designs, facilitating experimental optimization. If, for instance, subsequent experiments demonstrate that the sgRNA spacer originally chosen by pegFinder is inefficient at inducing prime editing, the user can readily refer to the pegFinder results table and test alternative sgRNA/pegRNA designs.
Selection of secondary nicking sgRNA spacers
Nicking the opposite strand can increase the efficiency of prime editing (PE3)2. To design “secondary” nicking sgRNAs to be used for PE3, pegFinder identifies sgRNA spacers de novo, based on the chosen CRISPR enzyme. pegFinder considers an sgRNA to be a candidate for secondary nicking if: 1) the sgRNA targets the strand opposite of the primary nicking sgRNA, 2) the secondary nick occurs 40-150nt away (default range, can be specified by user) from the primary nick, 3) no poly(T) tracts that would terminate U6 transcription, and if applicable, 4) the sgRNA has a sum “off-target” Tier I Bin I + Bin II score ≤ 1 (Broad designer) or the number of “seed off-targets” is 0 (CRISPRscan). pegFinder then selects the sgRNA spacer that nicks closest to ± 50nt from the primary nicking sgRNA. When incorporating on-target efficacy predictions, pegFinder chooses the sgRNA spacer with the highest on-target score among the candidate secondary nicking sgRNAs. All candidate secondary nicking sgRNA spacers are returned by pegFinder for ease of experimental optimization.
To increase the specificity of prime editing, Anzalone et al. described a variation of the PE3 system that uses secondary nicking sgRNAs which are only active after prime editing has occurred (PE3b). When possible, pegFinder also designs edit-specific PE3b secondary nicking sgRNAs.
pegRNA/sgRNA cloning protocol
Using the oligonucleotide sequences produced by pegFinder, each forward/reverse oligo pair was annealed in T4 Ligation Buffer (NEB), with T4 PNK to phosphorylate the oligos. The recipient pegRNA expression vector (pU6-pegRNA-GG-acceptor vector; Addgene #132777, a gift from David Liu) was digested by BsaI (NEB). After diluting the oligo duplexes 1:100, the primary nicking sgRNA, the invariant scaffold, and the 3’ extension were ligated together into the digested vector using Quick Ligase (NEB) to generate the complete pegRNA plasmid. Similarly, the secondary nicking sgRNA diluted duplex was ligated into a standard U6 sgRNA vector (such as the lentiGuide-Puro vector; Addgene #52963). Note that when using alternative plasmids for performing pegRNA/sgRNA cloning, the overhangs produced by pegFinder may need to be customized to match the sticky ends following plasmid digestion.
Experimental validation of pegRNAs
For the +1 CTT insertion at HEK3, the following pegRNA was used: 5’ GGCCCAGACTGAGCACGTGAGTTTTAGAGCTAGAAATAGCAAGTTAAAATAAGGCTAGTCCGTTATCAACTTGAAAAAGTGGCACCGAGTCGGTGCTCTGCCATCAAAGCGTGCTCAGTCTG 3’.
For the +1 CT insertion at HEK3, the following pegRNA was used: 5’ GGCCCAGACTGAGCACGTGAGTTTTAGAGCTAGAAATAGCAAGTTAAAATAAGGCTAGTCCGTTATCAACTTGAAAAAGTGGCACCGAGTCGGTGCTCTGCCATCAAGCGTGCTCAGTCTG 3’.
The oligo sequences provided by pegFinder were then cloned into the appropriate expression vectors, as described above. Experimental validation of the pegRNAs was performed as described previously, with minor modifications2. HEK293T cells (ATCC) were seeded on 24-well plates and transfected 16 hours later at 60-80% confluency with 2 ul Lipofectamine 2000 (ThermoFisher), 1.5 ug pCMV-PE2 plasmid (Addgene #132775), 500 ng pegRNA plasmid (cloned into Addgene #132777), and 200 ng secondary nicking sgRNA plasmid. 50 ng sfGFP-N1 (Addgene #54737) was also included to assess transfection efficiency. Cells were harvested 2 days post-transfection and genomic DNA (gDNA) was purified with the QIAamp DNA Blood Mini Kit (Qiagen). The genomic region surrounding the pegRNA target site was then amplified by PCR with the following primers, using 200 ng input gDNA: Forward, AGGGAAACGCCCATGCAATTAGTCT; Reverse, CTAGCCCCTGTCTAGGAAAAGCTGTC.
PCR was performed using Phusion Flash High-Fidelity polymerase (ThermoFisher) with the following settings: 98°C for 2 min, then 35 cycles of [98°C for 1 s, 60°C for 5 s, and 72°C for 5 s], followed by 72°C extension for 2 min. The resulting PCR amplicons were gel-purified (Qiagen) and processed for Sanger sequencing (Applied Biosystems 3730xL DNA Analyzer).
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
The main data supporting the results in this study are available within the paper and its Supplementary Information. For the pegRNAs that were experimentally tested in this study, all relevant information is provided as Supplementary Information. This information can be used to recreate the pegRNA designs described here, via the pegFinder web portal (http://pegfinder.sidichenlab.org).
Code availability
The custom code is available at GitHub (https://github.com/rdchow/pegfinder). The web portal is accessible at http://pegfinder.sidichenlab.org.
Supplementary Material
Acknowledgements
We thank S. Eisenbarth for support. RDC is supported by the Yale NIH MSTP training grant (T32GM136651) and an NIH NRSA fellowship from NCI (F30CA250249). JSC is supported by the Yale MSTP training grant from NIH (T32GM136651) and an NIH NSRA fellowship from NHLBI (F30HL149151). SC is supported by Yale SBI/Genetics Startup Fund, NIH/NCI/NIDA (DP2CA238295, 1R01CA231112, U54CA209992-8697, R33CA225498, RF1DA048811), DoD (W81XWH-20-1-0072 / BC190094), AACR (499395, 17-20-01-CHEN), Cancer Research Institute (CLIP), V Foundation, Ludwig Family Foundation, Sontag Foundation (DSA), Blavatnik Family Foundation and Chenevert Family Foundation.
Footnotes
Competing interests
The authors declare no competing interests. For full disclosure, S.C. is a co-founder, funding recipient and scientific advisor of EvolveImmune Therapeutics; the company has no relation to this study.
Supplementary information is available for this paper at https://doi.org/10.1038/s41551-020-00622-8.
Reprints and permissions information is available at www.nature.com/reprints.
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Pickar-Oliver A & Gersbach CA The next generation of CRISPR–Cas technologies and applications. Nat Rev Mol Cell Biol 20, 490–507 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anzalone AV et al. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature 576, 149–157 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu Y et al. Efficient generation of mouse models with the prime editing system. Cell Discovery 6, 1–4 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lin Q et al. Prime genome editing in rice and wheat. Nature Biotechnology 38, 582–585 (2020). [DOI] [PubMed] [Google Scholar]
- 5.Meier JA, Zhang F & Sanjana NE GUIDES: sgRNA design for loss-of-function screens. Nature Methods 14, 831–832 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Doench JG et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat. Biotechnol. 34, 184–191 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Listgarten J et al. Prediction of off-target activities for the end-to-end design of CRISPR guide RNAs. Nature Biomedical Engineering 2, 38 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moreno-Mateos MA et al. CRISPRscan: designing highly efficient sgRNAs for CRISPR-Cas9 targeting in vivo. Nat. Methods 12, 982–988 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haeussler M et al. Evaluation of off-target and on-target scoring algorithms and integration into the guide RNA selection tool CRISPOR. Genome Biology 17, 148 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Labun K et al. CHOPCHOP v3: expanding the CRISPR web toolbox beyond genome editing. Nucleic Acids Res 47, W171–W174 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Park J, Bae S & Kim J-S Cas-Designer: a web-based tool for choice of CRISPR-Cas9 target sites. Bioinformatics 31, 4014–4016 (2015). [DOI] [PubMed] [Google Scholar]
- 12.Needleman SB & Wunsch CD A general method applicable to the search for similarities in the amino acid sequence of two proteins. Journal of Molecular Biology 48, 443–453 (1970). [DOI] [PubMed] [Google Scholar]
- 13.Walton RT, Christie KA, Whittaker MN & Kleinstiver BP Unconstrained genome targeting with near-PAMless engineered CRISPR-Cas9 variants. Science 368, 290–296 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sanjana NE, Shalem O & Zhang F Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods 11, 783–784 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The main data supporting the results in this study are available within the paper and its Supplementary Information. For the pegRNAs that were experimentally tested in this study, all relevant information is provided as Supplementary Information. This information can be used to recreate the pegRNA designs described here, via the pegFinder web portal (http://pegfinder.sidichenlab.org).