Abstract
Purpose of review
We discuss new paradigms for understanding the immunopathology of MS through the recent development of high throughput genetic analysis, emergence of numerous candidate biomarkers and the broadening of the treatment arsenal.
Recent findings
The recent use of genome wide association studies provide new tools for a better understanding of Multiple Sclerosis etiology. GWASs have identified many genes implicated in immune regulation and the next step will be to elucidate how those genetic variations influence immune cells function to drive disease development and progression. Furthermore, patient care has seen the emergence of new biomarkers for monitoring disease progression and response to treatment. Finally, the introduction of numerous immunomodulatory treatments will likely improve clinical outcome of MS patients in the future.
Summary
breakthroughs in the field of Multiple Sclerosis have led to a better understanding of the physiopathology of the disease, follow up and treatment of the patients that develop relapsing remitting MS. The next challenge for MS will be to press forward to model and decipher MS progression, which will help both to develop therapeutics and generate knowledge about mechanisms of neurodegeneration.
Keywords: autoimmunity, Multiple Sclerosis, GWAS, Natalizumab, biomarkers
Introduction
MS is a multifocal demyelinating disease with progressive neurodegeneration caused by an autoimmune response to self-antigens. Clinical symptoms vary based on the site of neurologic lesions and often correlate with invasion of inflammatory cells across the blood-brain barrier with resulting demyelination and edema [1] (figure 1). Development of MS is the result of both genetic predisposition and environmental triggers. In this review we discuss new paradigms for understanding the immunopathology of MS through the recent development of high throughput genetic analysis, emergence of numerous candidate biomarkers and the broadening of the treatment arsenal.
Figure 1.
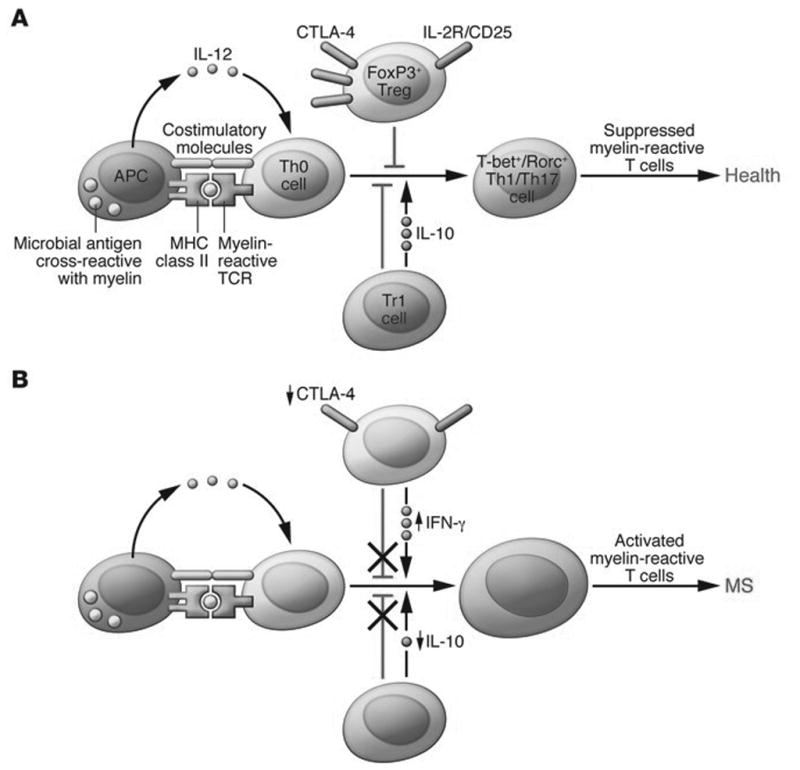
Defects in peripheral immune regulation lower the activation barrier for autoreactive T cells. (A) In normal homeostasis, APCs digest microbial antigens or self proteins and present them to naive T cells in the context of co-stimulatory molecules. An appropriate cytokine milieu can drive differentiation of these naive autoreactive T cells to a Th1 or Th17 cell phenotype; however, these potentially pathogenic T cells are not activated due to the actions of peripheral regulatory immune cell populations, such as FoxP3+ Tregs and Tr1 cells. Via the actions of co-inhibitory molecules and cytokines such as IL-10 and TGF-β, autoreactive T cells become anergic and autoimmune disease is prevented. Other mechanisms, such as thymic deletion and lack of co-stimulatory molecules on APCs, are also involved in controlling autoreactive T cells. (B) MS patients have defects in peripheral immune regulation, including higher expression of co-stimulatory molecules on APCs, lower CTLA-4 levels, and lower IL-10 production. Additionally, MS patients have an increased frequency of IFN-γ–secreting Tregs relative to healthy controls. Thus, the barrier for activation of autoreactive T cells is lowered for MS patients. Activated myelin-reactive T cells can then adhere to and extravasate across the choroid plexus and BBB, where they can initiate an inflammatory milieu that gives license to further waves of inflammation and eventual epitope spreading. Reproduced with permission from [1]
Genetics
1. The GWAS era
Genome-wide association studies (GWASs) using SNPs from the HapMap project allowed the use of an unbiased approach in scanning the whole genome and identifying SNPs associated with disease [2–5]. The MHC region was quickly identified as associated to MS susceptibility due to larger odd ratios [6]. Nine GWASs and a meta-analysis were performed and, in total, identified approximately 14 regions with genome-wide significance [7–14], including several previously identified associations [7, 15]. A GWAS with over 9,000 cases of MS replicated many of these previously suggested associations and identified an additional 29 novel susceptibility loci [16]. GWAS have now defined 194 genetic variants that are associated with MS, with that number likely to rise to over 400 (IMSGC, unpublished data). To date, approximately 1000 patients with progressive MS have been GWAS, and those subjects cannot be differentiated from patients with relapsing remitting MS. However, studies are planned to investigate larger cohorts of patients with PPMS.
2. Refinement of the GWASs discoveries
An inherent difficulty in GWAS studies is that linkage disequilibrium (LD) in the risk loci makes it difficult to pinpoint the most-likely causative variant. Currently, multiple fine-mapping efforts are underway to determine the most-likely causative SNP in many of these regions, but these have required novel tools to both fine-map and replicate these loci.
Having variants spread across the entire genome improves the breadth of the screen, but makes identifying individual variants within a region difficult. The Immunochip was designed to have dense clusters of variants around the SNPs called for by the original GWAS studies, thereby allowing close interrogation of all autoimmunity associated loci to date [17]. Fine-mapping the variants that were originally identified by GWAS identified 48 novel loci that had not reached the GWAS significance cutoff, but did so with the more targeted immunochip [18]. In addition to the Immunochip, a second targeted Illumina chip, the “MS chip” was developed to replicate recent GWAS hits. The MS chip also allows for identification of rare variants associated with MS that might have a high odds ratio and impact on disease.
As the Immunochip provided dense genotyping of MS associated Loci, this allows for an unbiased screen for most-likely causal SNPs for MS. The Wellcome trust pioneered a Bayesian analysis that was able to determine credible SNPs from three different diseases [19]. We have recently developed a new algorithm for fine mapping causal variants based on genetic evidence [20]. This model, called Probabilistic Identification of Causal SNPs (PICS), is a Bayesian algorithm modeled on dense genotyping data from the recent Immunochip study of MS [18]. Using the PICS algorithm, we were able to predict causal variants for GWAS data even when dense genotyping data was not available based on imputation to the 1000 genomes project [21].
3. From genetics to functional immunology
The next step upon us is characterizing phenotypic differences associated with variants in disease states. Several allelic variants in genes for cytokine receptors and co-stimulatory molecules have been associated with defects in Treg homeostasis. Our group and others have previously described that despite normal frequency of Tregs in MS patients compared to healthy controls, there is a loss of functional suppression by Tregs [22, 23]. Treg functional plasticity may also contribute to the autoimmune disease, as several studies have demonstrated that Tregs can produce inflammatory cytokines under certain conditions [24–27].
While many allelic variants associated with disease have been described, each variant alone carries a small increase in disease risk. As such, it is likely that multiple variants together represent a cumulative burden greater than the individual variants alone, falling within a limited number of signalling cascades primarily associated with immune responses and cytokine signalling [16]. In particular, variants within the NFκB signaling cascade as well as the STAT3/4/5 signalling cascades are highly represented in MS. It has been shown that CD4 cells from RRMS patients exhibit altered STAT3 signalling after IL6 stimulation [28]. In addition, we have demonstrated that naïve CD4 cells from MS patients exhibit constitutive activation of p65 NFκB [29]. This pathway is of special interest for MS as it is involved in both inflammation and neurodegeneration processes, thus possibly contributing to long-term disease progression [30].
4. Genetics, disease progression, and response to treatment
GWAS only identify variants associated with susceptibility to disease. The next question in MS genetics is determining what genetic variants are also associated with progression, severity, and response to treatment. A recent study compares genetic variants to CSF IgG levels and oligoclonal bands (OCBs) in 6950 MS patients from nine countries. Two regions, the MHC locus and the immunoglobulin heavy chain locus (IGHC), showed significant association to both IgG index and OCBs [31]. This confirmed a number of earlier, small cohort studies suggesting association of MHC and IGHC loci in CSF IgG and OCBs [32–38].
Biomarkers
1. Biomarkers of disease conversion
The McDonald criteria for MS diagnosis require two or more clinical episodes with two or more lesions on MRI appearing in separate loci over time [39–41]. Patients with clinically isolated syndrome (CIS) have a variable chance of conversion to relapsing remitting MS (RRMS) depending upon the MRI lesion load [42]. Some patients progress from RRMS to irreversible progressive disability called secondary progressive MS (SPMS). Approximately 10–15% of patients exhibit progressive disease after initial symptoms without relapses termed primary progressive MS (PPMS) [1]. It has been shown that early therapeutic intervention delays long-term disease progression [43, 44]. As such, determining those individuals presenting with optic neuritis or CIS with high risk of developing RRMS, SPMS, or PPMS would allow earlier treatment and improved outcomes. The presence of gadolinium enhancing lesions at CIS and OCBs in the CSF has been shown to be predictive biomarkers of subjects with high risk of progression to MS.
2. Biomarkers currently in clinical use
The presence of OCBs and high IgG levels in the CSF of MS patients was first shown in 1957 [45, 46]. Recently, renewed interest has been sparked in OCBs as the presence of OCBs in CSF is predictive of conversion from CIS to MS [47, 48]. As such, while it is no longer required for diagnosis of MS, testing for OCBs still represents a useful tool for ruling out other possible diagnoses and for prognostication of CIS conversion [49].
While it is clear that white matter lesions on MRI are indicative of progression from CIS to a clinically defined MS, the correlation between clinical disability (as measured by EDSS) and T2-weighted white matter lesion load varies broadly between different studies [50–53]. Presence of Gadolinium-enhancing lesions on magnetic resonance Imaging (MRI) in MS is indicative of active inflammation and lesion burden [54, 55]. Number and size of enhancing MRI lesions are predictive of both onset and severity of relapses [54–57], however there is a weak or absent correlation between Gadolinium-enhancing lesions and cognitive decline in RRMS [58]. More recently, studies have shown a positive correlation between overall grey matter atrophy and cognitive dysfunction suggesting that grey matter atrophy, rather than white matter lesion load, may be a useful biomarker for prediction of clinical severity [59].
Natalizumab (Tysabri) is a monoclonal antibody targeted to the α4 integrin. Blockade of α4β1 results in diminished T cell trafficking to the CNS and reduces relapse rate by 68% [60]. However, progressive multifocal leukoencephalopathy (PML) emerged as a rare adverse event from natalizumab treatment, generally occurring late (N 24 months) after initiating treatment [61]. PML is caused by reactivation of a latent JC virus in immunocompromised individuals. This leads to a debilitating encephalopathy that is often fatal. Previous infection with the JC virus can now be determined by seropositivity for JC viral antibodies prior to initiating and during natalizumab treatment. Only two cases of PML have been reported that were seronegative for JCV antibody [61, 62], making JCV antibodies an extremely useful clinical biomarker for assessing the risk of PML. However, while 50% of MS patients [62, 63] are JCV Ab seropositive, less than 1% will develop PML [64], obviating the need for more specific predictive markers of PML.
3. Potential biomarkers
CNS neurofilaments (Nfl) are released after axonal damage. Both the heavy chain and light chains are associated with axonal damage in MS and interestingly NF-M has been shown to be targeted in the CNS during EAE, the mouse model for MS [65, 66]. Recent studies have demonstrated that Nfl levels in CSF are increased in both RRMS and progressive MS compared to healthy control subjects [67]. In patients with RRMS, Nfl is increased at all disease stages, but fluctuates consistent with clinical course and the presence of active lesions by MRI [68]. Several studies suggest that Nfl levels could be a prognostic biomarker for an aggressive disease course and high risk for secondary progression [69–72] and correlate with treatment response to Fingolimod, natalizumab, and rituximab [43, 73, 74].
Other markers of neuronal and glial cell damage have been shown to be elevated in MS patients compared to healthy controls. Glial fibrillary acidic protein (GFAP), myelin basic protein (MBP), S100β (an astrocyte proliferation marker), tau, NCAM, NGF, CNTF and ferritin expression in the CSF have also been suggested as potential biomarkers [75–79]. However, they are non-specific and in the modern era with MRI, they are not useful as biomarkers.
CD163 is a monocyte/macrophage specific membrane marker. Upon activation, macrophages cleave CD163 from the surface and shed soluble CD163 (sCD163) that can be detected in the blood and CSF [80, 81]. Two studies demonstrated increased sCD163 levels in the blood [82] and the CSF of MS patients [83]. YKL-40 (Chitinase-3-like 1) is an activation marker for glia, macrophages, vascular smooth muscle cells, airway epithelia, and chondrocytes [84]. Elevated YKL-40 levels have been found in the serum of many inflammatory conditions [85–87]. In MS, YKL-40 levels in the CSF were found to be significantly higher in CIS patients that converted to MS as compared to patients that remained as CIS and has been correlated with shorter time to MS conversion and more rapid progression [72, 84, 88].
Osteopontin (OPN) is an early activation marker on T cells with a role in T cell costimulation and IFNγ expression [89]. OPN is highly expressed within MS lesions [89, 90] and is significantly higher in MS blood and CSF than healthy controls. Over the course of a five years follow up study, OPN correlated to disease severity and relapse rate [91–95]. High levels of OPN in the CSF also correlated to disease severity in PPMS [96]. However, there is disagreement on OPN levels as a prognostic biomarker of disease severity as two studies in MS have not found this association [97, 98].
Both CD4 and CD8 T cells are present in MS lesions and are believed to play a central role in disease development. Identification of a single inciting antigen triggering activation of myelin reactive T cells may not be possible, as the original targeted antigens may be unique to each patient and evolve throughout disease progression due to epitope spreading. Though, myelin-reactive T cells were observed repeatedly in MS patients and determining the inflammatory profiles of those cells may represent a biomarker for disease and disease progression. We and others originally described increased frequencies of myelin-reactive CD4 and CD8 cells in the peripheral blood from MS patients [99–101]. We have identified a phenotypic and transcriptional profile of myelin reactive T cells unique to MS patients compared to healthy donors [102, 103] suggesting the possibility that unique, MS-specific profiles of T cell libraries may identify individuals that will convert from initial diagnosis and CIS to MS.
Treatments
1. The first treatments
In 1993, FDA approved IFN-β1b to treat relapsing forms of MS [104, 105]. These agents reduce disease severity quite well for some patients, and might delay progression and improve survival [106–109]. Long-term follow-up of patients in a pivotal trial of intramuscular IFN-β1a revealed a poor prognosis for patients who exhibited active radiographic disease during the two years on drug [110]. This finding raised the hypothesis that patients with MS might exhibit a similarly mixed response to therapies as do patients with a range of other autoimmune disorders. Observational studies using different IFN-β preparations confirmed that this effect was shared among all members of the IFN-β drug class [111]. Yet, IFN-β therapy in SPMS patients fails to alter disease progression [112].
For glatiramer acetate, distinctions between good responders and poor responders were not as clearcut as with IFN-β [113]. Interferons are endogenous regulatory cytokines that increase or decrease transcriptional initiation for hundreds of genes in a cell-type-dependent fashion [114]. Therefore, in patients with MS who had good or poor responses to treatment, bioinformatic analysis of patterns in interferon-induced gene expression might predict clinical responses to IFN-β. These data could, in turn, yield insights into pathogenic mechanisms [115, 116].
2. Natalizumab and Fingolimod
Natalizumab has undergone a complex, and continuing, process of integration into clinical practice [117, 118]. Administered by monthly intravenous infusion, natalizumab exerts impressive inhibitory effects on the inflammatory aspects of MS, with >65% reduction in relapses during two years of treatment, and >90% suppression of new inflammatory MRI lesions [60, 119], with PML being the major side effect as discussed above [120].
Given the overall rarity of PML, this entirely unexpected complication was clearly caused by natalizumab, provoking immediate voluntary suspension of the drug’s distribution, though eventually no other PML cases were found to be incubating in the study population [121]. The mechanism by which natalizumab causes PML remains unknown [122]. Natalizumab-PML is not associated with generalized immunosuppression, and may be mechanism-driven [123]. The search for host factors that predispose to PML besides JCV infection continues.
The other second-era drug, fingolimod, is a prodrug that is converted in vivo to a sphingosine-1-phosphate (S1P) analogue. Fingolimod downregulates S1P receptor 1 on leukocytes and the endothelium, trapping naive and central memory T lymphocytes in lymph nodes. Treatment with fingolimod thereby suppresses MS disease activity, with 55–60% lower relapse rates and an impressive reduction of MRI-visible activity compared to placebo [124–127]. Fingolimod induces lymphopenia, unassociated with opportunistic infections beyond disseminated herpes zoster. For this reason, patients must have documented varicella zoster virus (VCV) immunity to be considered for fingolimod [128]. Unfortunately, both fingolimod and natalizumab fails to show efficacy in PPMS treatment [129]. These data suggests that while relapsing remitting MS represents primarily an inflammatory process of the adoptive immune system, secondary progressive MS may represent CNS activation of the innate immune system.
3. The future of MS therapy
This period is characterized by the introduction of medications including both small-molecules and biologics. Two oral immunomodulatory medications (teriflunomide and dimethyl-fumarate) have been approved [130–133]. After phase III trials, alemtuzumab—a leukocyte-depleting CD52 antibody —was approved [134–136]. Other agents, including laquinimod [137, 138] and daclizumab (an anti-CD25 antibody)[139–142], ocrelizumab and ofatumumab (both CD20 antibodies targeting B Lymphocytes), are undergoing advanced clinical testing. Recent data suggests the potential usefulness of Ocrelizumab as a first line drug in the treatment of both early RRMS and PPMS [143, 144].
Each agent tested so far in the third era has had a distinct mechanism of action, and yet has shown efficacy in double-blinded controlled trials. The variety of agents shown to reduce relapse frequency and MRI-monitored disease activity is consistent with recent chromatid mapping studies of allelic variants associated with MS. These studies indicated that multiple cell types— including Th17 cells, FOXP3 regulatory T cells, B cells and macrophages—are involved in MS disease pathogenesis [20].
Conclusions
In the past five years, potent insights have been made into the genetic basis of the disease, its environmental associations, its characterization by MRI, and its susceptibility to treatment by immunomodulation. This progress now poises the MS community for advancement to the prized objective of alleviating the burden exerted by progressive MS.
Key points.
High throughput analysis of the genetic burden in MS have allowed the identification of numerous genes and pathways involved in the regulation of immune functions.
Beyond MRI, OCBs and JC virus monitoring, new biomarkers are being characterized that will likely become useful in the clinics to follow disease progression and response to treatment.
Several immunomodulatory molecules with various immune targets have proven successful for MS treatment, broadening the therapeutic arsenal and helping a better understanding of MS physiopathology.
Acknowledgments
We would like to thank the Poste d’accueil from the French Institue of Health INSERM for supporting PPA’s research.
Financial support and sponsorship:
This research was supported by grants from the Poste d’accueil INSERM (France), the National Institute of Allergy and Infectious Disease (AI045757, AI046130, AI070352, and AI039671), the National Institute of Neurological Disorders and Stroke (NS067305 and F31NS086434), the National Institute of General Medical Sciences (GM093080), the National Multiple Sclerosis Society (CA1061-A-18), the Penates Foundation, and the Nancy Taylor Foundation for Chronic Disease.
Footnotes
Conflicts of interest:
DAH has been on Scientific Advisory Boards for the following companies over the past two years: Bristol-Myers Squibb, EMD Serono, Genzyme, Sanofi-Aventis, US Inc., MedImmune, Novartis Pharmaceuticals, Roche, and Teva Neuroscience. The remaining author has no conflicts of interest.
References
- 1.Nylander A, Hafler DA. Multiple sclerosis. Journal of Clinical Investigation. 2012;122(4):1180–8. doi: 10.1172/JCI58649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tishkoff SA, Verrelli BC. Role of evolutionary history on haplotype block structure in the human genome: implications for disease mapping. Current Opinion in Genetics & Development. 2003;13(6):569–75. doi: 10.1016/j.gde.2003.10.010. [DOI] [PubMed] [Google Scholar]
- 3.Frazer KA, Ballinger DG, Cox DR, et al. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449(7164):851–61. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hindorff LA, Sethupathy P, Junkins HA, et al. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proceedings of the National Academy of Sciences. 2009;106(23):9362–7. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Welter D, MacArthur J, Morales J, et al. The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Research. 2013;42(D1):D1001–D6. doi: 10.1093/nar/gkt1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.International Multiple Sclerosis Genetics C. Hafler DA, Compston A, et al. Risk alleles for multiple sclerosis identified by a genomewide study. The New England journal of medicine. 2007;357(9):851–62. doi: 10.1056/NEJMoa073493. [DOI] [PubMed] [Google Scholar]
- 7.Australia New Zealand Multiple Sclerosis Genetics C. Genome-wide association study identifies new multiple sclerosis susceptibility loci on chromosomes 12 and 20. Nat Genet. 2009;41(7):824–8. doi: 10.1038/ng.396. [DOI] [PubMed] [Google Scholar]
- 8.Burton PR, Clayton DG, Cardon LR, et al. Association scan of 14,500 nonsynonymous SNPs in four diseases identifies autoimmunity variants. Nature Genetics. 2007;39(11):1329–37. doi: 10.1038/ng.2007.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Comabella M, Craig DW, Camiña-Tato M, et al. Identification of a Novel Risk Locus for Multiple Sclerosis at 13q31.3 by a Pooled Genome-Wide Scan of 500,000 Single Nucleotide Polymorphisms. PLoS ONE. 2008;3(10):e3490. doi: 10.1371/journal.pone.0003490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baranzini SE, Wang J, Gibson RA, et al. Genome-wide association analysis of susceptibility and clinical phenotype in multiple sclerosis. Human Molecular Genetics. 2008;18(4):767–78. doi: 10.1093/hmg/ddn388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jakkula E, Leppä V, Sulonen A-M, et al. Genome-wide Association Study in a High-Risk Isolate for Multiple Sclerosis Reveals Associated Variants in STAT3 Gene. The American Journal of Human Genetics. 2010;86(2):285–91. doi: 10.1016/j.ajhg.2010.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sanna S, Pitzalis M, Zoledziewska M, et al. Variants within the immunoregulatory CBLB gene are associated with multiple sclerosis. Nature Genetics. 2010;42(6):495–7. doi: 10.1038/ng.584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aulchenko YS, Hoppenbrouwers IA, Ramagopalan SV, et al. Genetic variation in the KIF1B locus influences susceptibility to multiple sclerosis. Nature Genetics. 2008;40(12):1402–3. doi: 10.1038/ng.251. [DOI] [PubMed] [Google Scholar]
- 14.Nischwitz S, Cepok S, Kroner A, et al. Evidence for VAV2 and ZNF433 as susceptibility genes for multiple sclerosis. Journal of Neuroimmunology. 2010;227(1–2):162–6. doi: 10.1016/j.jneuroim.2010.06.003. [DOI] [PubMed] [Google Scholar]
- 15.De Jager PL, Jia X, Wang J, et al. Meta-analysis of genome scans and replication identify CD6, IRF8 and TNFRSF1A as new multiple sclerosis susceptibility loci. Nature Genetics. 2009;41(7):776–82. doi: 10.1038/ng.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.International Multiple Sclerosis Genetics C, Wellcome Trust Case Control C. Sawcer S, et al. Genetic risk and a primary role for cell-mediated immune mechanisms in multiple sclerosis. Nature. 2011;476(7359):214–9. doi: 10.1038/nature10251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cozen W, Li D, Best T, et al. A genome-wide meta-analysis of nodular sclerosing Hodgkin lymphoma identifies risk loci at 6p21.32. Blood. 2011;119(2):469–75. doi: 10.1182/blood-2011-03-343921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.International Multiple Sclerosis Genetics C. Beecham AH, Patsopoulos NA, et al. Analysis of immune-related loci identifies 48 new susceptibility variants for multiple sclerosis. Nat Genet. 2013;45(11):1353–60. doi: 10.1038/ng.2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hunt KA, Smyth DJ, Balschun T, et al. Rare and functional SIAE variants are not associated with autoimmune disease risk in up to 66,924 individuals of European ancestry. Nature Genetics. 2011;44(1):3–5. doi: 10.1038/ng.1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20**.Farh KK-H, Marson A, Zhu J, et al. Genetic and epigenetic fine mapping of causal autoimmune disease variants. Nature. 2014;518(7539):337–43. doi: 10.1038/nature13835. First study to determine causal variants from genomic association studies and link it to epigenetic modifications in multiple autoimmune an inflammatory diseases. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Genomes Project C. Abecasis GR, Auton A, et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491(7422):56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Viglietta V. Loss of Functional Suppression by CD4+CD25+ Regulatory T Cells in Patients with Multiple Sclerosis. Journal of Experimental Medicine. 2004;199(7):971–9. doi: 10.1084/jem.20031579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haas J, Hug A, Viehöver A, et al. Reduced suppressive effect of CD4+CD25high regulatory T cells on the T cell immune response against myelin oligodendrocyte glycoprotein in patients with multiple sclerosis. European Journal of Immunology. 2005;35(11):3343–52. doi: 10.1002/eji.200526065. [DOI] [PubMed] [Google Scholar]
- 24.Koenen HJPM, Smeets RL, Vink PM, et al. Human CD25highFoxp3pos regulatory T cells differentiate into IL-17-producing cells. Blood. 2008;112(6):2340–52. doi: 10.1182/blood-2008-01-133967. [DOI] [PubMed] [Google Scholar]
- 25.Beriou G, Costantino CM, Ashley CW, et al. IL-17-producing human peripheral regulatory T cells retain suppressive function. Blood. 2009;113(18):4240–9. doi: 10.1182/blood-2008-10-183251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ayyoub M, Deknuydt F, Raimbaud I, et al. Human memory FOXP3+ Tregs secrete IL-17 ex vivo and constitutively express the TH17 lineage-specific transcription factor ROR t. Proceedings of the National Academy of Sciences. 2009;106(21):8635–40. doi: 10.1073/pnas.0900621106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dominguez-Villar M, Baecher-Allan CM, Hafler DA. Identification of T helper type 1 like, Foxp3+ regulatory T cells in human autoimmune disease. Nature Medicine. 2011;17(6):673–5. doi: 10.1038/nm.2389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schneider A, Long SA, Cerosaletti K, et al. In Active Relapsing-Remitting Multiple Sclerosis, Effector T Cell Resistance to Adaptive Tregs Involves IL-6-Mediated Signaling. Science Translational Medicine. 2013;5(170):170ra15–ra15. doi: 10.1126/scitranslmed.3004970. [DOI] [PubMed] [Google Scholar]
- 29*.Housley WJ, Fernandez SD, Vera K, et al. Genetic variants associated with autoimmunity drive NF B signaling and responses to inflammatory stimuli. Science Translational Medicine. 2015;7(291):291ra93–ra93. doi: 10.1126/scitranslmed.aaa9223. This study describes the functional consequences of disease associated genetic variants on the NFκB signalling pathway. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sarnico I, Lanzillotta A, Benarese M, et al. NF-kappaB dimers in the regulation of neuronal survival. International review of neurobiology. 2009;85:351–62. doi: 10.1016/S0074-7742(09)85024-1. [DOI] [PubMed] [Google Scholar]
- 31**.Goris A, Pauwels I, Gustavsen MW, et al. Genetic variants are major determinants of CSF antibody levels in multiple sclerosis. Brain. 2015;138(3):632–43. doi: 10.1093/brain/awu405. This study is the first to demonstrate a link between disease associated genetic variants and a biological marker of disease activity. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Buck D, Albrecht E, Aslam M, et al. Genetic variants in the immunoglobulin heavy chain locus are associated with the IgG index in multiple sclerosis. Annals of Neurology. 2012;73(1):86–94. doi: 10.1002/ana.23749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Idiman E, Ozakbas S, Dogan Y, Kosehasanogullari G. The significance of oligoclonal bands in multiple sclerosis: Relevance of demographic and clinical features, and immunogenetic backgrounds. Journal of Neuroimmunology. 2009;212(1–2):121–4. doi: 10.1016/j.jneuroim.2009.04.014. [DOI] [PubMed] [Google Scholar]
- 34.Imrell K, Landtblom AM, Hillert J, Masterman T. Multiple sclerosis with and without CSF bands: Clinically indistinguishable but immunogenetically distinct. Neurology. 2006;67(6):1062–4. doi: 10.1212/01.wnl.0000237343.93389.35. [DOI] [PubMed] [Google Scholar]
- 35.Kikuchi S, Fukazawa T, Niino M, et al. HLA-related subpopulations of MS in Japanese with and without oligoclonal IgG bands. Neurology. 2003;60(4):647–51. doi: 10.1212/01.wnl.0000048202.09147.9e. [DOI] [PubMed] [Google Scholar]
- 36.Leone MA, Barizzone N, Esposito F, et al. Association of Genetic Markers with CSF Oligoclonal Bands in Multiple Sclerosis Patients. PLoS ONE. 2013;8(6):e64408. doi: 10.1371/journal.pone.0064408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Romero-Pinel L, Martínez-Yélamos S, Bau L, et al. Association of HLA-DRB1*15 allele and CSF oligoclonal bands in a Spanish multiple sclerosis cohort. European Journal of Neurology. 2011;18(10):1258–62. doi: 10.1111/j.1468-1331.2011.03379.x. [DOI] [PubMed] [Google Scholar]
- 38.Yoshimura S, Isobe N, Matsushita T, et al. Genetic and Infectious Profiles Influence Cerebrospinal Fluid IgG Abnormality in Japanese Multiple Sclerosis Patients. PLoS ONE. 2014;9(4):e95367. doi: 10.1371/journal.pone.0095367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McDonald WI, Compston A, Edan G, et al. Recommended diagnostic criteria for multiple sclerosis: Guidelines from the international panel on the diagnosis of multiple sclerosis. Annals of Neurology. 2001;50(1):121–7. doi: 10.1002/ana.1032. [DOI] [PubMed] [Google Scholar]
- 40.Polman CH, Reingold SC, Banwell B, et al. Diagnostic criteria for multiple sclerosis: 2010 Revisions to the McDonald criteria. Annals of Neurology. 2011;69(2):292–302. doi: 10.1002/ana.22366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Polman CH, Reingold SC, Edan G, et al. Diagnostic criteria for multiple sclerosis: 2005 revisions to the “McDonald Criteria”. Annals of Neurology. 2005;58(6):840–6. doi: 10.1002/ana.20703. [DOI] [PubMed] [Google Scholar]
- 42.Miller D, Barkhof F, Montalban X, et al. Clinically isolated syndromes suggestive of multiple sclerosis, part I: natural history, pathogenesis, diagnosis, and prognosis. The Lancet Neurology. 2005;4(5):281–8. doi: 10.1016/S1474-4422(05)70071-5. [DOI] [PubMed] [Google Scholar]
- 43.Gunnarsson M, Malmeström C, Axelsson M, et al. Axonal damage in relapsing multiple sclerosis is markedly reduced by natalizumab. Annals of Neurology. 2010;69(1):83–9. doi: 10.1002/ana.22247. [DOI] [PubMed] [Google Scholar]
- 44.Jones JL, Anderson JM, Phuah CL, et al. Improvement in disability after alemtuzumab treatment of multiple sclerosis is associated with neuroprotective autoimmunity. Brain. 2010;133(8):2232–47. doi: 10.1093/brain/awq176. [DOI] [PubMed] [Google Scholar]
- 45.Yahr MD, Goldensohn SS, Kabat EA. Further studies on the gamma globulin content of cerebrospinal fluid in multiple sclerosis and other neurological diseases. Annals of the New York Academy of Sciences. 1954;58(5):613–24. [Google Scholar]
- 46.Lowenthal A, Vansande M, Karcher D. The differential diagnosis of neurological diseases by fractionating electrophoretically the CSF gamma-globulins. J Neurochem. 1960;6:51–6. doi: 10.1111/j.1471-4159.1960.tb13448.x. [DOI] [PubMed] [Google Scholar]
- 47.Ignacio RJ, Liliana P, Edgardo C. Oligoclonal bands and MRI in clinically isolated syndromes: predicting conversion time to multiple sclerosis. Journal of Neurology. 2010;257(7):1188–91. doi: 10.1007/s00415-010-5490-y. [DOI] [PubMed] [Google Scholar]
- 48*.Kuhle J, Disanto G, Dobson R, et al. Conversion from clinically isolated syndrome to multiple sclerosis: A large multicentre study. Mult Scler. 2015;21(8):1013–24. doi: 10.1177/1352458514568827. This multicenter study demonstrates OCBs is predictive of CIS conversion to MS. [DOI] [PubMed] [Google Scholar]
- 49.Stangel M, Fredrikson S, Meinl E, et al. The utility of cerebrospinal fluid analysis in patients with multiple sclerosis. Nature Reviews Neurology. 2013;9(5):267–76. doi: 10.1038/nrneurol.2013.41. [DOI] [PubMed] [Google Scholar]
- 50.Barkhof F. MRI in multiple sclerosis: correlation with expanded disability status scale (EDSS) Multiple Sclerosis. 1999;5(4):283–6. doi: 10.1177/135245859900500415. [DOI] [PubMed] [Google Scholar]
- 51.Fisniku LK, Brex PA, Altmann DR, et al. Disability and T2 MRI lesions: a 20-year follow-up of patients with relapse onset of multiple sclerosis. Brain. 2008;131(3):808–17. doi: 10.1093/brain/awm329. [DOI] [PubMed] [Google Scholar]
- 52.Zivadinov R, Leist TP. Clinical Magnetic Resonance Imaging Correlations in Multiple Sclerosis. Journal of Neuroimaging. 2005;15:10S–21S. doi: 10.1177/1051228405283291. [DOI] [PubMed] [Google Scholar]
- 53.Li DKB, Held U, Petkau J, et al. MRI T2 lesion burden in multiple sclerosis: A plateauing relationship with clinical disability. Neurology. 2006;66(9):1384–9. doi: 10.1212/01.wnl.0000210506.00078.5c. [DOI] [PubMed] [Google Scholar]
- 54.Bruck W, Bitsch A, Kolenda H, et al. Inflammatory central nervous system demyelination: correlation of magnetic resonance imaging findings with lesion pathology. Ann Neurol. 1997;42(5):783–93. doi: 10.1002/ana.410420515. [DOI] [PubMed] [Google Scholar]
- 55.Katz D, Taubenberger JK, Cannella B, et al. Correlation between magnetic resonance imaging findings and lesion development in chronic, active multiple sclerosis. Annals of Neurology. 1993;34(5):661–9. doi: 10.1002/ana.410340507. [DOI] [PubMed] [Google Scholar]
- 56.Khoury SJ, Guttmann CRG, Orav EJ, et al. Longitudinal MRI in multiple sclerosis: Correlation between disability and lesion burden. Neurology. 1994;44(11):2120. doi: 10.1212/wnl.44.11.2120. [DOI] [PubMed] [Google Scholar]
- 57.Smith ME, Stone LA, Albert PS, et al. Clinical worsening in multiple sclerosis is associated with increased frequency and area of gadopentetate dimeglumine-enhancing magnetic resonance imaging lesions. Annals of Neurology. 1993;33(5):480–9. doi: 10.1002/ana.410330511. [DOI] [PubMed] [Google Scholar]
- 58.Rocca MA, Amato MP, De Stefano N, et al. Clinical and imaging assessment of cognitive dysfunction in multiple sclerosis. The Lancet Neurology. 2015;14(3):302–17. doi: 10.1016/S1474-4422(14)70250-9. [DOI] [PubMed] [Google Scholar]
- 59.Geurts JJG, Calabrese M, Fisher E, Rudick RA. Measurement and clinical effect of grey matter pathology in multiple sclerosis. The Lancet Neurology. 2012;11(12):1082–92. doi: 10.1016/S1474-4422(12)70230-2. [DOI] [PubMed] [Google Scholar]
- 60.Polman CH, O’Connor PW, Havrdova E, et al. A randomized, placebo-controlled trial of natalizumab for relapsing multiple sclerosis. The New England journal of medicine. 2006;354(9):899–910. doi: 10.1056/NEJMoa044397. [DOI] [PubMed] [Google Scholar]
- 61.Antoniol C, Stankoff B. Immunological Markers for PML Prediction in MS Patients Treated with Natalizumab. Front Immunol. 2015;5 doi: 10.3389/fimmu.2014.00668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Outteryck O, Zephir H, Salleron J, et al. JC-virus seroconversion in multiple sclerosis patients receiving natalizumab. Multiple Sclerosis Journal. 2013;20(7):822–9. doi: 10.1177/1352458513505353. [DOI] [PubMed] [Google Scholar]
- 63.Olsson T, Achiron A, Alfredsson L, et al. Anti-JC virus antibody prevalence in a multinational multiple sclerosis cohort. Multiple Sclerosis Journal. 2013;19(11):1533–8. doi: 10.1177/1352458513477925. [DOI] [PubMed] [Google Scholar]
- 64.Plavina T, Subramanyam M, Bloomgren G, et al. Anti-JC virus antibody levels in serum or plasma further define risk of natalizumab-associated progressive multifocal leukoencephalopathy. Annals of Neurology. 2014;76(6):802–12. doi: 10.1002/ana.24286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Krishnamoorthy G, Saxena A, Mars LT, et al. Myelin-specific T cells also recognize neuronal autoantigen in a transgenic mouse model of multiple sclerosis. Nat Med. 2009;15(6):626–32. doi: 10.1038/nm.1975. [DOI] [PubMed] [Google Scholar]
- 66.Ramadan A, Lucca LE, Carrié N, et al. In situ expansion of T cells that recognize distinct self-antigens sustains autoimmunity in the central nervous system. Brain. doi: 10.1093/brain/aww032. in press. [DOI] [PubMed] [Google Scholar]
- 67.Kuhle J, Malmeström C, Axelsson M, et al. Neurofilament light and heavy subunits compared as therapeutic biomarkers in multiple sclerosis. Acta Neurologica Scandinavica. 2013;128(6):e33–e6. doi: 10.1111/ane.12151. [DOI] [PubMed] [Google Scholar]
- 68*.Bielekova B, McDermott MP. Will CSF biomarkers guide future therapeutic decisions in multiple sclerosis? Neurology. 2015;84(16):1620–1. doi: 10.1212/WNL.0000000000001506. This reference together with 68–72 illustrate the rapid discovery of several CSF related biomarkers, which might be useful in monitoring response to treatment and disease progression. [DOI] [PubMed] [Google Scholar]
- 69.Disanto G, Adiutori R, Dobson R, et al. Serum neurofilament light chain levels are increased in patients with a clinically isolated syndrome. Journal of Neurology, Neurosurgery & Psychiatry. 2015 doi: 10.1136/jnnp-2014-309690. [DOI] [PubMed] [Google Scholar]
- 70.Salzer J, Svenningsson A, Sundstrom P. Neurofilament light as a prognostic marker in multiple sclerosis. Multiple Sclerosis Journal. 2010;16(3):287–92. doi: 10.1177/1352458509359725. [DOI] [PubMed] [Google Scholar]
- 71.Modvig S, Degn M, Roed H, et al. Cerebrospinal fluid levels of chitinase 3-like 1 and neurofilament light chain predict multiple sclerosis development and disability after optic neuritis. Multiple Sclerosis Journal. 2015;21(14):1761–70. doi: 10.1177/1352458515574148. [DOI] [PubMed] [Google Scholar]
- 72.Martinez MAM, Olsson B, Bau L, et al. Glial and neuronal markers in cerebrospinal fluid predict progression in multiple sclerosis. Multiple Sclerosis Journal. 2015;21(5):550–61. doi: 10.1177/1352458514549397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kuhle J, Disanto G, Lorscheider J, et al. Fingolimod and CSF neurofilament light chain levels in relapsing-remitting multiple sclerosis. Neurology. 2015;84(16):1639–43. doi: 10.1212/WNL.0000000000001491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Romme Christensen J, Ratzer R, Bornsen L, et al. Natalizumab in progressive MS: Results of an open-label, phase 2A, proof-of-concept trial. Neurology. 2014;82(17):1499–507. doi: 10.1212/WNL.0000000000000361. [DOI] [PubMed] [Google Scholar]
- 75.Petzold A. Markers for different glial cell responses in multiple sclerosis: clinical and pathological correlations. Brain. 2002;125(7):1462–73. doi: 10.1093/brain/awf165. [DOI] [PubMed] [Google Scholar]
- 76.Cohen SR, Herndon RM, McKhann GM. Radioimmunoassay of Myelin Basic Protein in Spinal Fluid. New England Journal of Medicine. 1976;295(26):1455–7. doi: 10.1056/NEJM197612232952604. [DOI] [PubMed] [Google Scholar]
- 77.Whitaker JN. Myelin encephalitogenic protein fragments in cerebrospinal fluid of persons with multiple sclerosis. Neurology. 1977;27(10):911. doi: 10.1212/wnl.27.10.911. [DOI] [PubMed] [Google Scholar]
- 78.Avsar T, Korkmaz D, Tutuncu M, et al. Protein biomarkers for multiple sclerosis: semi-quantitative analysis of cerebrospinal fluid candidate protein biomarkers in different forms of multiple sclerosis. Multiple Sclerosis Journal. 2012;18(8):1081–91. doi: 10.1177/1352458511433303. [DOI] [PubMed] [Google Scholar]
- 79.Massaro AR, Tonali P. Cerebrospinal fluid markers in multiple sclerosis: an overview. Multiple Sclerosis. 1998;4(1):1–4. doi: 10.1177/135245859800400101. [DOI] [PubMed] [Google Scholar]
- 80.Davis BH, Zarev PV. Human monocyte CD163 expression inversely correlates with soluble CD163 plasma levels. Cytometry. 2004;63B(1):16–22. doi: 10.1002/cyto.b.20031. [DOI] [PubMed] [Google Scholar]
- 81.Stilund M, Reuschlein A-K, Christensen T, et al. Soluble CD163 as a Marker of Macrophage Activity in Newly Diagnosed Patients with Multiple Sclerosis. PLoS ONE. 2014;9(6):e98588. doi: 10.1371/journal.pone.0098588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Fabriek BO, Møller HJ, Vloet RPM, et al. Proteolytic shedding of the macrophage scavenger receptor CD163 in multiple sclerosis. Journal of Neuroimmunology. 2007;187(1–2):179–86. doi: 10.1016/j.jneuroim.2007.04.016. [DOI] [PubMed] [Google Scholar]
- 83.Stilund M, Gjelstrup MC, Petersen T, et al. Biomarkers of Inflammation and Axonal Degeneration/Damage in Patients with Newly Diagnosed Multiple Sclerosis: Contributions of the Soluble CD163 CSF/Serum Ratio to a Biomarker Panel. PLOS ONE. 2015;10(4):e0119681. doi: 10.1371/journal.pone.0119681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Canto E, Tintore M, Villar LM, et al. Chitinase 3-like 1: prognostic biomarker in clinically isolated syndromes. Brain. 2015;138(4):918–31. doi: 10.1093/brain/awv017. [DOI] [PubMed] [Google Scholar]
- 85.Peltomaa R, Paimela L, Harvey S, et al. Increased level of YKL-40 in sera from patients with early rheumatoid arthritis: a new marker for disease activity. Rheumatology International. 2001;20(5):192–6. doi: 10.1007/s002960100115. [DOI] [PubMed] [Google Scholar]
- 86.Vos K. Raised human cartilage glycoprotein-39 plasma levels in patients with rheumatoid arthritis and other inflammatory conditions. Annals of the Rheumatic Diseases. 2000;59(7):544–8. doi: 10.1136/ard.59.7.544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.IV, SJJ, APP, PM Serum YKL-40, a Potential New Marker of Disease Activity in Patients with Inflammatory Bowel Disease. Scand J Gastroenterol. 2003;38(6):599–605. doi: 10.1080/00365520310000537. [DOI] [PubMed] [Google Scholar]
- 88.Comabella M, Fernandez M, Martin R, et al. Cerebrospinal fluid chitinase 3-like 1 levels are associated with conversion to multiple sclerosis. Brain. 2010;133(4):1082–93. doi: 10.1093/brain/awq035. [DOI] [PubMed] [Google Scholar]
- 89.Chabas D. The Influence of the Proinflammatory Cytokine, Osteopontin, on Autoimmune Demyelinating Disease. Science. 2001;294(5547):1731–5. doi: 10.1126/science.1062960. [DOI] [PubMed] [Google Scholar]
- 90.Sinclair C, Mirakhur M, Kirk J, et al. Up-regulation of osteopontin and alphaBeta-crystallin in the normal-appearing white matter of multiple sclerosis: an immunohistochemical study utilizing tissue microarrays. Neuropathol Appl Neurobiol. 2005;31(3):292–303. doi: 10.1111/j.1365-2990.2004.00638.x. [DOI] [PubMed] [Google Scholar]
- 91.Comabella M, Pericot I, Goertsches R, et al. Plasma osteopontin levels in multiple sclerosis. Journal of Neuroimmunology. 2005;158(1–2):231–9. doi: 10.1016/j.jneuroim.2004.09.004. [DOI] [PubMed] [Google Scholar]
- 92.Szalardy L, Zadori D, Simu M, et al. Evaluating biomarkers of neuronal degeneration and neuroinflammation in CSF of patients with multiple sclerosis osteopontin as a potential marker of clinical severity. Journal of the Neurological Sciences. 2013;331(1–2):38–42. doi: 10.1016/j.jns.2013.04.024. [DOI] [PubMed] [Google Scholar]
- 93.Vogt MHJ, Floris S, Killestein J, et al. Osteopontin levels and increased disease activity in relapsing remitting multiple sclerosis patients. Journal of Neuroimmunology. 2004;155(1–2):155–60. doi: 10.1016/j.jneuroim.2004.06.007. [DOI] [PubMed] [Google Scholar]
- 94.Vogt MHJ, Lopatinskaya L, Smits M, et al. Elevated osteopontin levels in active relapsing-remitting multiple sclerosis. Annals of Neurology. 2003;53(6):819–22. doi: 10.1002/ana.10606. [DOI] [PubMed] [Google Scholar]
- 95.Wen S-R, Liu G-J, Feng R-N, et al. Increased levels of IL-23 and osteopontin in serum and cerebrospinal fluid of multiple sclerosis patients. Journal of Neuroimmunology. 2012;244(1–2):94–6. doi: 10.1016/j.jneuroim.2011.12.004. [DOI] [PubMed] [Google Scholar]
- 96.Bornsen L, Khademi M, Olsson T, et al. Osteopontin concentrations are increased in cerebrospinal fluid during attacks of multiple sclerosis. Multiple Sclerosis Journal. 2010;17(1):32–42. doi: 10.1177/1352458510382247. [DOI] [PubMed] [Google Scholar]
- 97.Kivisakk P, Healy BC, Francois K, et al. Evaluation of circulating osteopontin levels in an unselected cohort of patients with multiple sclerosis: relevance for biomarker development. Multiple Sclerosis Journal. 2013;20(4):438–44. doi: 10.1177/1352458513503052. [DOI] [PubMed] [Google Scholar]
- 98.Runia TF, van Meurs M, Nasserinejad K, Hintzen RQ. No evidence for an association of osteopontin plasma levels with disease activity in multiple sclerosis. Multiple Sclerosis Journal. 2014;20(12):1670–1. doi: 10.1177/1352458514528765. [DOI] [PubMed] [Google Scholar]
- 99.Martin R. A myelin basic protein peptide is recognized by cytotoxic T cells in the context of four HLA-DR types associated with multiple sclerosis. Journal of Experimental Medicine. 1991;173(1):19–24. doi: 10.1084/jem.173.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ota K, Matsui M, Milford EL, et al. T-cell recognition of an immuno-dominant myelin basic protein epitope in multiple sclerosis. Nature. 1990;346(6280):183–7. doi: 10.1038/346183a0. [DOI] [PubMed] [Google Scholar]
- 101.Pette M, Fujita K, Kitze B, et al. Myelin basic protein-specific T lymphocyte lines from MS patients and healthy individuals. Neurology. 1990;40(11):1770. doi: 10.1212/wnl.40.11.1770. [DOI] [PubMed] [Google Scholar]
- 102.Raddassi K, Kent SC, Yang J, et al. Increased Frequencies of Myelin Oligodendrocyte Glycoprotein/MHC Class II-Binding CD4 Cells in Patients with Multiple Sclerosis. The Journal of Immunology. 2011;187(2):1039–46. doi: 10.4049/jimmunol.1001543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103**.Cao Y, Goods BA, Raddassi K, et al. Functional inflammatory profiles distinguish myelin-reactive T cells from patients with multiple sclerosis. Science Translational Medicine. 2015;7(287):287ra74–ra74. doi: 10.1126/scitranslmed.aaa8038. This study demonstrates for the first time that myelin-specific autoreactive CD4 T cells harbor a specific inflammatory profile in MS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Hauser SL, Dawson DM, Lehrich JR, et al. Intensive Immunosuppression in Progressive Multiple Sclerosis. New England Journal of Medicine. 1983;308(4):173–80. doi: 10.1056/NEJM198301273080401. [DOI] [PubMed] [Google Scholar]
- 105.Paty DW, Li DKB. Interferon beta-1b is effective in relapsing-remitting multiple sclerosis: II. MRI analysis results of a multicenter, randomized, double-blind, placebo-controlled trial. Neurology. 1993;43(4):662. doi: 10.1212/wnl.43.4.662. [DOI] [PubMed] [Google Scholar]
- 106.Trojano M, Pellegrini F, Fuiani A, et al. New natural history of interferon-β-treated relapsing multiple sclerosis. Annals of Neurology. 2007;61(4):300–6. doi: 10.1002/ana.21102. [DOI] [PubMed] [Google Scholar]
- 107.Trojano M, Pellegrini F, Paolicelli D, et al. Real-life impact of early interferonβ therapy in relapsing multiple sclerosis. Annals of Neurology. 2009;66(4):513–20. doi: 10.1002/ana.21757. [DOI] [PubMed] [Google Scholar]
- 108.Scalfari A, Knappertz V, Cutter G, et al. Mortality in patients with multiple sclerosis. Neurology. 2013;81(2):184–92. doi: 10.1212/WNL.0b013e31829a3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Goodin DS, Reder AT, Ebers GC, et al. Survival in MS: A randomized cohort study 21 years after the start of the pivotal IFN -1b trial. Neurology. 2012;78(17):1315–22. doi: 10.1212/WNL.0b013e3182535cf6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Bermel RA, You X, Foulds P, et al. Predictors of long-term outcome in multiple sclerosis patients treated with interferon beta. Annals of Neurology. 2013;73(1):95–103. doi: 10.1002/ana.23758. [DOI] [PubMed] [Google Scholar]
- 111.Prosperini L, Gallo V, Petsas N, et al. One-year MRI scan predicts clinical response to interferon beta in multiple sclerosis. European Journal of Neurology. 2009;16(11):1202–9. doi: 10.1111/j.1468-1331.2009.02708.x. [DOI] [PubMed] [Google Scholar]
- 112.Kuhle J, Hardmeier M, Disanto G, et al. A 10-year follow-up of the European multicenter trial of interferon beta-1b in secondary-progressive multiple sclerosis. Mult Scler. 2015 doi: 10.1177/1352458515594440. [DOI] [PubMed] [Google Scholar]
- 113.McGraw CA, Lublin FD. Interferon beta and glatiramer acetate therapy. Neurotherapeutics. 2013;10(1):2–18. doi: 10.1007/s13311-012-0163-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Borden EC, Sen GC, Uze G, et al. Interferons at age 50: past, current and future impact on biomedicine. Nature Reviews Drug Discovery. 2007;6(12):975–90. doi: 10.1038/nrd2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Comabella M, Lunemann JD, Rio J, et al. A type I interferon signature in monocytes is associated with poor response to interferon- in multiple sclerosis. Brain. 2009;132(12):3353–65. doi: 10.1093/brain/awp228. [DOI] [PubMed] [Google Scholar]
- 116.Ottoboni L, Keenan BT, Tamayo P, et al. An RNA Profile Identifies Two Subsets of Multiple Sclerosis Patients Differing in Disease Activity. Science Translational Medicine. 2012;4(153):153ra31–ra31. doi: 10.1126/scitranslmed.3004186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Brown B. Natalizumab in the treatment of multiple sclerosis. Therapeutics and Clinical Risk Management. 2009:585. doi: 10.2147/tcrm.s5218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Ransohoff RM. Natalizumab for Multiple Sclerosis. New England Journal of Medicine. 2007;356(25):2622–9. doi: 10.1056/NEJMct071462. [DOI] [PubMed] [Google Scholar]
- 119.Rudick RA, Stuart WH, Calabresi PA, et al. Natalizumab plus interferon beta-1a for relapsing multiple sclerosis. The New England journal of medicine. 2006;354(9):911–23. doi: 10.1056/NEJMoa044396. [DOI] [PubMed] [Google Scholar]
- 120.Langer-Gould A, Atlas SW, Green AJ, et al. Progressive Multifocal Leukoencephalopathy in a Patient Treated with Natalizumab. New England Journal of Medicine. 2005;353(4):375–81. doi: 10.1056/NEJMoa051847. [DOI] [PubMed] [Google Scholar]
- 121.Yousry TA, Major EO, Ryschkewitsch C, et al. Evaluation of patients treated with natalizumab for progressive multifocal leukoencephalopathy. The New England journal of medicine. 2006;354(9):924–33. doi: 10.1056/NEJMoa054693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Ransohoff RM. PML risk and natalizumab: more questions than answers. The Lancet Neurology. 2010;9(3):231–3. doi: 10.1016/S1474-4422(10)70025-9. [DOI] [PubMed] [Google Scholar]
- 123.Bloomgren G, Richman S, Hotermans C, et al. Risk of Natalizumab-Associated Progressive Multifocal Leukoencephalopathy. New England Journal of Medicine. 2012;366(20):1870–80. doi: 10.1056/NEJMoa1107829. [DOI] [PubMed] [Google Scholar]
- 124.Cohen JA, Barkhof F, Comi G, et al. Oral fingolimod or intramuscular interferon for relapsing multiple sclerosis. The New England journal of medicine. 2010;362(5):402–15. doi: 10.1056/NEJMoa0907839. [DOI] [PubMed] [Google Scholar]
- 125.Kappos L, Radue EW, O’Connor P, et al. A placebo-controlled trial of oral fingolimod in relapsing multiple sclerosis. The New England journal of medicine. 2010;362(5):387–401. doi: 10.1056/NEJMoa0909494. [DOI] [PubMed] [Google Scholar]
- 126.Pelletier D, Hafler DA. Fingolimod for Multiple Sclerosis. New England Journal of Medicine. 2012;366(4):339–47. doi: 10.1056/NEJMct1101691. [DOI] [PubMed] [Google Scholar]
- 127**.He A, Spelman T, Jokubaitis V, et al. Comparison of switch to fingolimod or interferon beta/glatiramer acetate in active multiple sclerosis. JAMA Neurol. 2015;72(4):405–13. doi: 10.1001/jamaneurol.2014.4147. Large retrospective study documents efficacy of Fingolimod treatment in RRMS. [DOI] [PubMed] [Google Scholar]
- 128*.Arvin AM, Wolinsky JS, Kappos L, et al. Varicella-Zoster Virus Infections in Patients Treated With Fingolimod. JAMA Neurology. 2015;72(1):31. doi: 10.1001/jamaneurol.2014.3065. This study details VZV infection as the major side effect of Fingolimod therapy. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Lublin F, Miller DH, Freedman MS, et al. Oral fingolimod in primary progressive multiple sclerosis (INFORMS): a phase 3, randomised, double-blind, placebo-controlled trial. Lancet. 2016 doi: 10.1016/S0140-6736(15)01314-8. [DOI] [PubMed] [Google Scholar]
- 130.Gold R, Kappos L, Arnold DL, et al. Placebo-controlled phase 3 study of oral BG-12 for relapsing multiple sclerosis. The New England journal of medicine. 2012;367(12):1098–107. doi: 10.1056/NEJMoa1114287. [DOI] [PubMed] [Google Scholar]
- 131.Fox RJ, Miller DH, Phillips JT, et al. Placebo-Controlled Phase 3 Study of Oral BG-12 or Glatiramer in Multiple Sclerosis. New England Journal of Medicine. 2012;367(12):1087–97. doi: 10.1056/NEJMoa1206328. [DOI] [PubMed] [Google Scholar]
- 132.Kappos L, Gold R, Miller DH, et al. Effect of BG-12 on contrast-enhanced lesions in patients with relapsing- remitting multiple sclerosis: subgroup analyses from the phase 2b study. Multiple Sclerosis Journal. 2011;18(3):314–21. doi: 10.1177/1352458511421054. [DOI] [PubMed] [Google Scholar]
- 133.Kappos L, Gold R, Miller DH, et al. Efficacy and safety of oral fumarate in patients with relapsing-remitting multiple sclerosis: a multicentre, randomised, double-blind, placebo-controlled phase IIb study. The Lancet. 2008;372(9648):1463–72. doi: 10.1016/S0140-6736(08)61619-0. [DOI] [PubMed] [Google Scholar]
- 134.Cohen JA, Coles AJ, Arnold DL, et al. Alemtuzumab versus interferon beta 1a as first-line treatment for patients with relapsing-remitting multiple sclerosis: a randomised controlled phase 3 trial. The Lancet. 2012;380(9856):1819–28. doi: 10.1016/S0140-6736(12)61769-3. [DOI] [PubMed] [Google Scholar]
- 135.Coles AJ, Twyman CL, Arnold DL, et al. Alemtuzumab for patients with relapsing multiple sclerosis after disease-modifying therapy: a randomised controlled phase 3 trial. The Lancet. 2012;380(9856):1829–39. doi: 10.1016/S0140-6736(12)61768-1. [DOI] [PubMed] [Google Scholar]
- 136.Investigators CT, Coles AJ, Compston DA, et al. Alemtuzumab vs. interferon beta-1a in early multiple sclerosis. The New England journal of medicine. 2008;359(17):1786–801. doi: 10.1056/NEJMoa0802670. [DOI] [PubMed] [Google Scholar]
- 137.Comi G, Jeffery D, Kappos L, et al. Placebo-Controlled Trial of Oral Laquinimod for Multiple Sclerosis. New England Journal of Medicine. 2012;366(11):1000–9. doi: 10.1056/NEJMoa1104318. [DOI] [PubMed] [Google Scholar]
- 138.Comi G, Pulizzi A, Rovaris M, et al. Effect of laquinimod on MRI-monitored disease activity in patients with relapsing-remitting multiple sclerosis: a multicentre, randomised, double-blind, placebo-controlled phase IIb study. The Lancet. 2008;371(9630):2085–92. doi: 10.1016/S0140-6736(08)60918-6. [DOI] [PubMed] [Google Scholar]
- 139.Gold R, Giovannoni G, Selmaj K, et al. Daclizumab high-yield process in relapsing-remitting multiple sclerosis (SELECT): a randomised, double-blind, placebo-controlled trial. The Lancet. 2013;381(9884):2167–75. doi: 10.1016/S0140-6736(12)62190-4. [DOI] [PubMed] [Google Scholar]
- 140.Wynn D, Kaufman M, Montalban X, et al. Daclizumab in active relapsing multiple sclerosis (CHOICE study): a phase 2, randomised, double-blind, placebo-controlled, add-on trial with interferon beta. The Lancet Neurology. 2010;9(4):381–90. doi: 10.1016/S1474-4422(10)70033-8. [DOI] [PubMed] [Google Scholar]
- 141.Bielekova B, Richert N, Howard T, et al. Humanized anti-CD25 (daclizumab) inhibits disease activity in multiple sclerosis patients failing to respond to interferon beta. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(23):8705–8. doi: 10.1073/pnas.0402653101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142*.Kappos L, Wiendl H, Selmaj K, et al. Daclizumab HYP versus Interferon Beta-1a in Relapsing Multiple Sclerosis. The New England journal of medicine. 2015;373(15):1418–28. doi: 10.1056/NEJMoa1501481. Phase 3 trial study demonstrating decreased relapse frequency following Daclizumab treatment in RRMS patients. [DOI] [PubMed] [Google Scholar]
- 143.Media release from Roche on Ocrelizumab phase III trials. 2015 Available from: http://www.roche.com/media/store/releases/med-cor-2015-10-08.htm.
- 144**.Montalban X, Hemmer B, Rammohan K, et al. Efficacy and safety of ocrelizumab in primary progressive multiple sclerosis - results of the placebo-controlled, double-blind, Phase III ORATORIO study. ECTRIMS Online Library. 2015;(Oct 10) First trials of anti-CD20 ocrelizumab therapy show promising outcome for the treatment of both RRMS and PPMS. [Google Scholar]


