Abstract
Different groups have recently reported events of SARS-CoV-2 reinfection, where patients had a sequence of positive-negative-positive RT-PCR tests. However, such events could be explained by different scenarios such as intermittent viral shedding, bonafide re-infection or multiple infection with alternating predominance of different viruses. Analysis of minor variants is an important tool to distinguish between these scenarios. Using ARTIC network PCR amplification and next-generation sequencing, we obtained SARS-CoV-2 sequences from two timepoints (with a time span of 102 days) of a patient followed at the Brazilian National Cancer Institute. Within-host variant analysis evidenced three single nucleotide variants (SNVs) at the consensus viral sequence in the second timepoint that were already present in the first timepoint as minor variants. Another five SNVs found in the second timepoint were not detected in the first sample sequenced, suggesting an additional infection by a yet another new virus. Our observation shed light into the existence of different viral populations that are present in dynamic frequencies and fluctuate during the course of SARS-CoV-2 infection. The detection of these variants in distinct disease events of an individual highlights a complex interplay between viral reactivation from a pre-existing minority variant and reinfection by a different virus.
Keywords: SARS-CoV-2, Re-infection, Multiple infection, Reactivation, Intrahost variation
Growing evidence suggests that SARS-CoV-2 can re-infect individuals that recovered from an initial infection (Goldman et al., 2020; Gousseff et al., 2020; Gupta et al., 2020; Kam et al., 2020; Lafaie et al., 2020; Larson et al., 2020; To et al., 2020; Tomassini et al., 2020; Van Elslande et al., 2020). In a recent report, To and colleagues (To et al., 2020) described the first case of re-infection of a 33-yr old man that tested RT-PCR-positive for the virus after 142 days of the first documented episode of infection, with two negative RT-PCR tests in between both episodes. Re-infection was suggested by sequencing of different viruses from both timepoints (To et al., 2020).
A sequence of positive-negative-positive RT-PCR in consecutive samples of an individual can be explained by different scenarios (Lan et al., 2020). It can be due to intermittent viral shedding, with a unique viral entity causing both clinical episodes interspersed with an interval of undetectable viremia (Lu et al., 2020; Osman et al., 2020; Zheng et al., 2020). It can alternatively be resulted from a bonafide re-infection, where two distinct viral strains cause the two episodes. The third possibility is an initial infection by two or more viruses, in which one variant initially outgrows the other, is controlled by the host, and the second variant then outgrows the first due to waning immune responses or immune escape. While SARS-CoV-2 genome profiling of consensus sequences from two clinical episodes distinguishes between reactivation and re-infection, only the analysis of viral minor variants allows discrimination between the second and the third possibilities.
Waning immune responses over a few months after a first episode of SARS-COV-2 infection have been increasingly reported (Gaebler et al., 2021; Henss et al., 2021; Legros et al., 2021), and may explain sensitization of the host to a second infection event or reactivation of a previous infection. In an earlier report, even an IgM re-seroconversion after an IgG+/IgM- profile has been documented, suggesting a new immune priming event or at best an expansion of IgM+ memory B-cells (Bentivegna et al., 2020). Immunosuppressed patients, and in particular cancer patients with hematological disease are more prone to SARS-CoV-2 reinfection, virus reactivation or sustained viremia (repeated RT-PCR positive tests) due to their impaired immune responses against the virus (Liang et al., 2020; Luciani et al., 2020). Re-infection by a second viral strain different from the first may also be due to immune escape by differing epitopes, but this possibility has yet to be demonstrated. If formally proven, however, this could even implicate in changes in the efficacy of the currently developed vaccines against COVID-19.
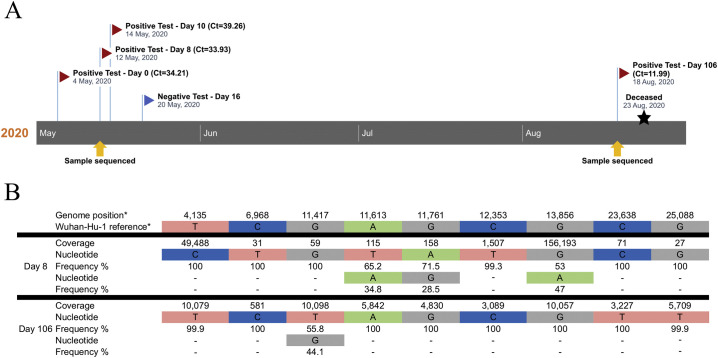
Herein we report the analysis of a 76-yr old female patient with chronic renal failure (with indwelling bladder catheter) and pyelonephritis followed-up at the Brazilian National Cancer Institute for a previous stage IIIb squamous cell carcinoma. The patient was hypertensive, with hypothyroidism and chronic obstructive pulmonary disease. The patient was admitted in late April 2020, and due to the pyelonephritis dialysis was started. On May 4th she presented cough and fever of 37.9 °C and a CT scan revealed small areas of ground glass and extensive consolidation area on the left base. Upon screening for SARS-CoV-2 infection, the patient had multiple RT-PCR-positive tests and severe COVID-19. She evolved with refractory bronchospasm and hypercapnia, as well as hypoxemia requiring endotracheal intubation. A negative RT-PCR test was obtained on May 20th (Fig. 1A). The patient recovered from the disease and was discharged in early August, but then returned on Aug 16th unconscious with severe sepsis (complicated ICU and pneumonia) and a new chest CT showing worsening of the consolidation image seen in May and scattered new areas of ground glass in both lungs. On the day of admission, the patient tested positive for SARS-CoV-2 again with low Ct value in the RT-PCR test (11.99), suggesting a high viral load (Fig. 1A). Due to her worsening condition, poor prognosis and inadequate family and social support, the assistant team deliberated non-invasive support. The patient died in the following day.
Fig. 1.
Timeline of naso- and oropharyngeal swab RT-PCR assays and results on samples collected across the COVID-19 clinical history of the patient studied. (A) Patient had consecutive weak positive results in the first clinical episode (Ct values of CDC protocol RT-PCR depicted in the Figure), and for one of them (day 8 after COVID-19 diagnosis) PCR and DNA sequencing was successful. Sequencing was also successful for the sample at day 106 post-diagnosis (second clinical episode). Sequenced samples are depicted with yellow arrows below the timeline. (B) Intrahost nucleotide frequency in the divergent positions identified between the samples collected at days 8 and 106 post-diagnosis. *, genomic coordinates based on the Wuhan-Hu-1 reference sequence.
We applied the ARTIC network (http://www.artic.network) PCR protocol to obtain SARS-CoV-2 near full-length genomes and sequenced the PCR-positive products in a MiSeq platform (2 × 231-bp; Illumina, Inc.). Resulting reads were assembled to the Wuhan-Hu-1 SARS-CoV-2 genome (Genbank acc# MN908947) and consensus extracted using Geneious R11 (Biomatters, Ltd., Auckland, New Zealand). Consensuses from the two timepoints were compared and divergent positions were manually inspected at read level to evaluate the nucleotide distribution and to detect minor variants. We also investigated minor variations using LoFreq v2.1.5 through the variants workflow available at Galaxy web platform (Afgan et al., 2016; Baker et al., 2020; Wilm et al., 2012). Only the variants detected in both methodologies with frequency higher than 1% were considered for further analysis. Polymorphism phase was evaluated at read level for positions with distances within the multiplex fragment length (400-bp).
Despite the high Ct value of the RT-PCR test (suggestive of low SARS-CoV-2 load), the patient's sample at day 8 post-COVID-19 diagnosis successfully provided viral genomic sequences spanning approximately 40% of the genome. On the other hand, the full-length genomic sequence of the virus present at day 106 was obtained, as viral load of that sample appeared to be much higher according to the Ct value of the RT-PCR test (11.99). We have compared the two viral sequences at the genomic fragments available for both timepoints. A total of nine single nucleotide variations (SNVs) were observed between the two sequences (Fig. 1B). Of those, four SNVs have nucleotide frequency fluctuations between the two timepoints, while five SNVs differed completely between the two major strains, with no polymorphisms observed in the samples. The analysis of the viral populations at those positions showed that three SNVs present as minor variants at the first timepoint (frequency between 34.8 and 53% at the viral population) became fixed the in major variant of the second timepoint (frequency of 100%). We were able to identify that two of them (positions 11,613 and 11,761 related to Wuhan-Hu-1 reference genome) are in the same phase, as they were simultaneously present in single sequencing reads. The third polymorphism (13,856), more than 2-Kb apart from the first two, was sequenced in a different DNA fragment and, consequently, its phase could be determined. One SNV present as major in the first strain (frequency of 100% at the viral population) became the minor variant in the second (frequency of 44.1%). The existence of minor polymorphisms observed at the first timepoint and their fixation in the second consensus strain suggest reactivation of pre-existing viral sequences in the second disease event. Considering a 30 kb genome and the proofreading activity of SARS-CoV-2 polymerase, the probability of a new infection by a new virus harboring the same polymorphisms by chance is infinitesimal. Conversely, the presence of a previous major polymorphism as a minor variant in the second timepoint could represent a vestige from the former virus. Alternative explanations for this observation include a recombination event from the major viral strains from the first and second timepoints or viral evolution over time, induced by selective pressures at this specific nucleotide (or encoded amino acid) position.
Due to technical limitations of the ARTIC network protocol, we are unable to determine the phase of all polymorphisms observed, and we cannot ensure that all minor and major variants belong to specific viral entities. For the same reason, given that the multiplex PCR is based on the amplification of overlapping 400-bp fragments with different efficacy across the viral genome and between different genomes, biases in the relative frequency of minor variations can occur, precluding the interpretation of having polymorphisms with similar frequency as being in phase (from the same viral entity).
Based on the results described above, we hypothesize that two different SARS-CoV-2 strains were circulating at the time of the first clinical episode, one as a major and the other as a minor variant. Upon patient's recovery, both variants were controlled but persisted at low levels. Three months later, the minor variant outgrew to high levels upon an immunosuppressive event in the patient. In this scenario, the minor viral population present in a dual infection in the first timepoint was present as major variant in the second timepoint. In addition, the five polymorphisms observed at the second timepoint that were absent in the first suggest the presence of a yet another new virus. A recombination event between these two viruses prior to the last timepoint could represent one possible explanation for the viral profile observed at that timepoint. However, we should recognize that our analysis is not able to detect recombination events. From these data, we conclude that multiple infection and variant selection over time occur in SARS-CoV-2 infections, and it cannot be distinguished from viral re-infection unless a thoroughly analysis of intrahost viral diversity is carried out.
Author contributions
JDS, LRG, BMA and MAS conceptualization. JDS, LRG, BMA, PSC and ACPS data curation. JDS, LRG, BMA and PSC formal analysis. CC, JA, JPBV, and MAS funding acquisition. JDS, LRG, BMA and MAS writing original draft. JDS, LRG, BMA, PSC, ACPS, CC, JA, JPBV and MAS writing, review and editing.
Funding
This work was supported by the Carlos Chagas Filho Rio de Janeiro State Science Foundation (FAPERJ) (Brazil); the Swiss Bridge Foundation (Switzerland); intramural grants of the National Institutes of Health (NIH, U.S.A.); and the Brazilian National Cancer Institute (INCA, Brazil).
Acknowledgments
We would like to thank the participants of INCA-COVID-19 Task Force, clinical staff and patients from the Brazilian National Cancer Institute (INCA) for providing conditions and samples that enabled the conduction of this study.
References
- Afgan E., Baker D., van den Beek M., Blankenberg D., Bouvier D., Čech M., Chilton J., Clements D., Coraor N., Eberhard C., Grüning B., Guerler A., Hillman-Jackson J., Von Kuster G., Rasche E., Soranzo N., Turaga N., Taylor J., Nekrutenko A., Goecks J. The galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2016 update. Nucleic Acids Res. 2016;44(W1):W3–W10. doi: 10.1093/nar/gkw343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker D., van den Beek M., Blankenberg D., Bouvier D., Chilton J., Coraor N., Coppens F., Eguinoa I., Gladman S., Grüning B., Keener N., Larivière D., Lonie A., Kosakovsky Pond S., Maier W., Nekrutenko A., Taylor J., Weaver S. No more business as usual: agile and effective responses to emerging pathogen threats require open data and open analytics. PLoS Pathog. 2020;16 doi: 10.1371/journal.ppat.1008643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentivegna E., Sentimentale A., Luciani M., Speranza M.L., Guerritore L., Martelletti P. New IgM seroconversion and positive RT-PCR test after exposure to the virus in recovered COVID-19 patient. J. Med. Virol. 2020 doi: 10.1002/jmv.26160. Jun 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaebler C., Wang Z., Lorenzi J.C.C., Muecksch F., Finkin S., Tokuyama M., Cho A., Jankovic M., Schaefer-Babajew D., Oliveira T.Y., Cipolla M., Viant C., Barnes C.O., Bram Y., Breton G., Hägglöf T., Mendoza P., Hurley A., Turroja M., Gordon K., Millard K.G., Ramos V., Schmidt F., Weisblum Y., Jha D., Tankelevich M., Martinez-Delgado G., Yee J., Patel R., Dizon J., Unson-O'Brien C., Shimeliovich I., Robbiani D.F., Zhao Z., Gazumyan A., Schwartz R.E., Hatziioannou T., Bjorkman P.J., Mehandru S., Bieniasz P.D., Caskey M., Nussenzweig M.C. Evolution of antibody immunity to SARS-CoV-2. Nature. 2021;(Jan 18) doi: 10.1038/s41586-021-03207-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman J.D., Wang K., Roltgen K., Nielsen S.C.A., Roach J.C., Naccache S.N., Yang F., Wirz O.F., Yost K.E., Lee J.Y., Chun K., Wrin T., Petropoulos C.J., Lee I., Fallen S., Manner P.M., Wallick J.A., Algren H.A., Murray K.M., Su Y., Hadlock J., Jeharajah J., Berrington W.R., Pappas G.P., Nyatsatsang S.T., Greninger A.L., Satpathy A.T., Pauk J.S., Boyd S.D., Heath J.R. Reinfection with SARS-CoV-2 and failure of Humoral immunity: a case report. medRxiv. 2020 doi: 10.1101/2020.09.22.20192443. Sep 25;2020.09.22.20192443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gousseff M., Penot P., Gallay L., Batisse D., Benech N., Bouiller K., Collarino R., Conrad A., Slama D., Joseph C., Lemaignen A., Lescure F.X., Levy B., Mahevas M., Pozzetto B., Vignier N., Wyplosz B., Salmon D., Goehringer F., Botelho-Nevers E. Clinical recurrences of COVID-19 symptoms after recovery: viral relapse, reinfection or inflammatory rebound? J Infec. 2020;81(5):816–846. doi: 10.1016/j.jinf.2020.06.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta V., Bhoyar R.C., Jain A., Srivastava S., Upadhayay R., Imran M., Jolly B., Divakar M.K., Sharma D., Sehgal P., Ranjan G., Gupta R., Scaria V., Sivasubbu S. Asymptomatic reinfection in two healthcare workers from India with genetically distinct SARS-CoV-2. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa1451. Sep 23; ciaa1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henss L., Scholz T., von Rhein C., Wieters I., Borgans F., Eberhardt F.J., Zacharowski K., Ciesek S., Rohde G., Vehreschild M., Stephan C., Wolf T., Hofmann-Winkler H., Scheiblauer H., Schnierle B.S. Analysis of Humoral immune responses in patients with severe acute respiratory syndrome coronavirus 2 infection. J. Infect. Dis. 2021;223(1):56–61. doi: 10.1093/infdis/jiaa680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kam Y.W., Ahmed M.Y., Amrun S.N., Lee B., Refaie T., Elgizouli K., Fong S.W., Renia L., Ng L.F. Systematic analysis of disease-specific immunological signatures in patients with febrile illness from Saudi Arabia. Clin. Transl. Immunol. 2020;9 doi: 10.1002/cti2.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafaie L., Célarier T., Goethals L., Pozzetto B., Grange S., Ojardias E., Annweiler C., Botelho-Nevers E. Recurrence or relapse of COVID-19 in older patients: a description of three cases. J. Amer. Geriatr. Soc. 2020;68(10):2179–2183. doi: 10.1111/jgs.16728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan L., Xu D., Ye G., Xia C., Wang S., Li Y., Xu H. Positive RT-PCR test results in patients recovered from COVID-19. JAMA. 2020;323:1502–1503. doi: 10.1001/jama.2020.2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson D., Brodniak S.L., Voegtly L.J., Cer R.Z., Glang L.A., Malagon F.J., Long K.A., Potocki R., Smith D.R., Lanteri C., Burgess T., Bishop-Lilly K.A. A case of early re-infection with SARS-CoV-2. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa1436. Sep 19; ciaa1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legros V., Denolly S., Vogrig M., Boson B., Siret E., Rigaill J., Pillet S., Grattard F., Gonzalo S., Verhoeven P., Allatif O., Berthelot P., Pélissier C., Thiery G., Botelho-Nevers E., Millet G., Morel J., Paul S., Walzer T., Cosset F.L., Bourlet T., Pozzetto B. A longitudinal study of SARS-CoV-2-infected patients reveals a high correlation between neutralizing antibodies and COVID-19 severity. Cell. Mol. Immunol. 2021;6:1–10. doi: 10.1038/s41423-020-00588-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang W., Guan W., Chen R., Wang W., Li J., Xu K., Li C., Ai Q., Lu W., Liang H., Li S., He J. Cancer patients in SARS-CoV-2 infection: a nationwide analysis in China. Lancet Oncol. 2020;21:335–337. doi: 10.1016/S1470-2045(20)30096-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J., Peng J., Xiong Q., Liu Z., Lin H., Tan X., Kang M., Yuan R., Zeng L., Zhou P., Liang C., Yi L., du Plessis L., Song T., Ma W., Sun J., Pybus O.G., Ke C. Clinical, immunological and virological characterization of COVID-19 patients that test re-positive for SARS-CoV-2 by RT-PCR. EBioMedicine. 2020;59:102960. doi: 10.1016/j.ebiom.2020.102960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luciani M., Bentivegna E., Spuntarelli V., Lamberti P.A., Cacioli G., Del Porto F., Sesti G., Martelletti P., De Biase L. Recurrent COVID-19 pneumonia in the course of chemotherapy: consequence of a weakened immune system? J. Med. Virol. 2020 doi: 10.1002/jmv.26701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osman A.A., Al Daajani M.M., Alsahafi A.J. Re-positive COVID-19 PCR test: could it be a reinfection? New Microbes New Infect. 2020;37:100748. doi: 10.1016/j.nmni.2020.100748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- To, K.K, Hung I.F., Ip J.D., Chu A.W., Chan W.M., Tam A.R., Fong C.H., Yuan S., Tsoi H.W., Ng A.C., Lee L.L., Wan P., Tso E., To, W.K, Tsang D., Chan K.H., Huang J.D., Kok K.H., Cheng V.C., Yuen K.Y. COVID-19 re-infection by a phylogenetically distinct SARS-coronavirus-2 strain confirmed by whole genome sequencing. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa1275. Aug 25; ciaa1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomassini S., Kotecha D., Bird P.W., Folwell A., Biju S., Tang J.W. Setting the criteria for SARS-CoV-2 reinfection - six possible cases. J. Infec. 2020;(20):30546. doi: 10.1016/j.jinf.2020.08.011. Aug 12; S0163-4453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Elslande J., Vermeersch P., Vandervoort K., Wawina-Bokalanga T., Vanmechelen B., Wollants E., Laenen L., André E., Van Ranst M., Lagrou K., Maes P. Symptomatic SARS-CoV-2 reinfection by a phylogenetically distinct strain. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa1330. Sep 5; ciaa1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilm A., Aw P.P., Bertrand D., Yeo G.H., Ong S.H., Wong C.H., Khor C.C., Petric R., Hibberd M.L., Nagarajan N. LoFreq: a sequence-quality aware, ultra-sensitive variant caller for uncovering cell-population heterogeneity from high-throughput sequencing datasets. Nucleic Acids Res. 2012;40:11189–11201. doi: 10.1093/nar/gks918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J., Zhou R., Chen F., Tang G., Wu K., Li F., Liu H., Lu J., Zhou J., Yang Z., Yuan Y., Lei C., Wu X. Incidence, clinical course and risk factor for recurrent PCR positivity in discharged COVID-19 patients in Guangzhou, China: a prospective cohort study. PLoS Neglect. Trop. Dis. 2020;14 doi: 10.1371/journal.pntd.0008648. [DOI] [PMC free article] [PubMed] [Google Scholar]