Abstract
World is familiar with the viral pathogen Severe Acute Respiratory Syndrome Coronavirus 2 (SARS CoV-2). The principle working enzymes of SARS CoV-2 have been identified as main proteases 3Cl pro which act as main regulators for SARS infection. The need for therapy is required immediately pertaining to the vulnerable conditions. Protein-ligand studies are imperative for understanding the functioning of biological interactions as they are crucial in providing a hypothetical origin for the design and unearthing of novel drug targets. Phytoconstituents from Glycyrrhiza glabra, earlier reported to be anticancerous in nature were used as repurposed drugs against SARS CoV-2 main protease 3Clpro. We analyzed the molecular interactions of protein-phytocompounds, by AutoDock Vina 4.2 tools. The best interactions of each algorithm were subjected to molecular dynamic (MD) simulations to have an insight of the molecular dynamic mechanisms involved. Selected phytoconstituents gave a good score for binding affinity with the main protease 6LU7 of SARS CoV-2 as compared to the antiviral drugs already being used in the disease therapy. DehydroglyasperinC(-8.7,-8.1,-6.7,-7.1)kcal/mol, Licochalcone D(-8.4,-8.2,-7.1,-7.9) kcal/mol, Liquiritin(-8.6,-9.0,-7.2,-7.8) kcal/mol have showed energy interactions with 3Clpro better than many FDA approved repurposed drugs; Remdesvir, Favipiravir, and Hydroxychloroquine. MD Simulation also corelates our findings for molecular docking studies.
Keywords: SARS CoV-2, 3Cl pro, Molecular docking, Phytoconstituents, Glycyrrhiza glabra, MD Simulations
Graphical abstract

1. Introduction
1.1. SARS CoV-2
The recently appeared SARS-CoV-2 has been seen to cause a major outburst of coronavirus disease particularly, acute respiratory distress syndrome (Liu et al., 2020). Because of its vast appearance in nearly all parts of the globe, WHO has classified it as a pandemic with a name of COVID-19 (Cucinotta and Vanelli, 2020). The COVID-19 pandemic has caused panic and significant economic damage across the world as the infection causing virus SARS-CoV-2 has resulted in 74,299,042 cases and 1,669,982 deaths globally as of Dec 12, 2020 (WHO, 2020). India became the second country with the highest number of COVID-19 cases after the United States and Brazil. With around 10 005 000 cases reported and 145 600 deaths in the country, India has become the hot spot for the survival of the virus (Kaushik et al., 2020) (Kannan et al., 2020). The disease has instigated an extensive general fear, intimidating universal health security. So far, no licensed FDA approved antiviral drugs or vaccines are available against COVID-19 even though numerous clinical trials are in progress to test likely therapies (Xu et al., 2020) (Ahmed et al., 2020). Although many drugs are being repurposed for probable therapeutics.
SARS CoV-2 characterizes the βCoronaviruses family. It is the seventh recognized human coronavirus from the same family after 229 E, NL63, OC43, HKU1, MERS-CoV and SARS-CoV 1 (Walls et al., 2020) (Shamsi et al., 2020). These Coronaviruses are single-stranded positive-sense RNA viruses and have 5′-cap and 3′-poly-A tail. The SARS-CoV-2 genome is 30 kb long and consists of six open reading frames (ORFs). The first ORF (ORF1a/b) is the largest and encodes 16 non-structural proteins (nsp 1-16). ORFs at the 3′end encodes four main structural proteins i.e. spike (S), membrane (M), envelope (E), and nucleocapsid (N) proteins respectively(Tai et al., 2020). Translation is divided into two phases; the early phase in which the viral replicase complex is assembled, and the late phase, where structural and nonstructural proteins are translated. The maturation of the RNA polymerase which is necessary for replication is accomplished through proteolytic processing by mainly three cysteine peptidases in which two are papain-like proteases (PLpro) and the third is the main protease, also known as the 3 Chymotrypsin-like protease (3Cl pro).
1.2. Phytoconstituents from Glycyrrhiza glabra as inhibitors for target viral proteins acting as antiviral compounds
Medicinal plants have been recognized and used throughout human history. All plants yield phytoconstituents as share of their usual metabolic activities which mediate their effect on the human body through courses similar to those of conventional drugs. Hence, herbal remedies are not in contrast with the functioning of conventional drugs. This authorizes herbal medications not only to have beneficial pharmacology, but also gives them the potential to have lesser side effects than conventional drugs (Hejazi et al., 2018).
Phytoconstituents present in rhizomes of Glycyrrhiza glabra are accountable for its authoritative antioxidant activity via free radical scavenging, hydrogen-donating and anti-lipid peroxidative capabilities. It is reported that Glycyrrhiza glabra extract also inhibits the growth of viruses, including herpes simplex, varicella zoster, influenza virus etc. It is found to have a significant antiviral activity. We tried in our studies to see molecular in silico docking interactions with SARS CoV 2 with various available compounds extracted from the ethyl acetate extract of Glycyrrhiza glabra (Hejazi et al., 2017).
The SARS-CoV-2 has approximately 80% of the genetic sequence similarity to SARS-CoV-1 having the same cell entry receptor, (ACE2). Moreover, pathogenesis of SARS-CoV-2 shares many clinical resemblances with that of SARS-CoV 1. Assuming the parallels between SARS-CoV 1 and SARS-CoV-2, we can take advantage from the clinical facts displayed by SARS-CoV 1 to handle SARS-CoV-2 produced disease (COVID-19). Research on SARS-CoV 1 has documented a variety of agents with the potential to treat SARS-CoV 1 infection. Glycyrrhizin compound from Glycyrrhiza glabra is a triterpene saponin with numerous biological functions and pharmacological effects and is one of the most promising candidates as it was found to be active against SARS-CoV 1 in vitro. Considering the experience and lessons of the fight against SARS, Glycyrrhiza glabra constituents might be promising candidates for treatment of COVID-19 and deserves further evaluation (Armanini et al., 2020) (Luo et al., 2020). We have taken compounds from the ethanol extract of Glycyrrhiza glabra. Glycyrrhizin compound from G glabra is a triterpene saponin with numerous biological functions and pharmacological effects and is one of the most promising candidates as it was found to be active against SARS-CoV 1 in vitro. But we have found in our studies that although glycyrrhizin is an effective compound against SARS CoV-1, it was not following certain rules for being a druggable compound. In this research, we intent to focus a possible positive effect on Covid-19 infections of DehydroglyasperinC, Licochalcone D and Liquiritin, the phytoconstituents of ethanol extract of Glycyrrhiza glabra L. All these compounds have a powerful anti-inflammatory effect, stabilizing the inflammatory storm caused in SARS CoV 2.
1.3. Docking interactions
As we all are aware of the current emergent situations of the contagion, computational molecular docking methods are a substitute to exhaustive lab based high-throughput screening of lead compounds as drug candidates. Identification of small chemical molecules that evidently target viral replication machinery have documented highest potential on the way to antiviral drug discovery (Malik, 2020). As the viral replication is the most important step for the survival of virus in the host cell therefore if any drug molecule that is structured to target as an inhibitor to the replication machinery could be a source of effective antiviral drug. So, keeping this in mind, researchers are trying to achieve their results by targeting specific proteases of virus which are responsible for the activation of replicase and polymerase enzymes. These proteases have been earlier recognized as cysteine like proteases or 3CL pro in SARS CoV 1 also. The structural elucidation of SARS CoV 2 has also shown the presence of 3CL pro in it. It processes the RNA polymerase by its extensive proteolytic cleavage into two polyproteins pp1ab and pp1a which further form many essential proteins of the replication as well as structural domains (Park, 2020). The cleavage sites existing on the polyproteins are highly conserved sequences having similarities with SARS CoV-1 and MERS CoV (Xu et al., 2020). This resemblance suggests the origin of SARS-CoV-2 from its previous counterparts which helped the scientific community for the identification of compounds with potential to inhibit its replication.
So as to take charge of the threat instigated by SARS-CoV-2, we propose vital druggable enzyme target, namely 3C-like proteinase crystal structures family mainly 6LU7(main protease in complex with an inhibitor), 6M03 (main protease in apo form), 6M2N (main protease in complex with an inhibitor), 1P9S (SARS CoV-1 main proteinase basis for design of anti-SARS Drugs) for interacting with the plant based probable drug like compounds from G. glabra. These compounds have been earlier found to be antioxidative, anti-inflammatory and anti-cancerous in nature. On the whole we can say that the present study utilises the systematic drug repurposing approach to recognize antiviral compounds from plants which can act as promising inhibitors against 3Cl pro and also identify their inhibitory process by extensive in silico approaches.
2. Methodology
2.1. Molecular docking
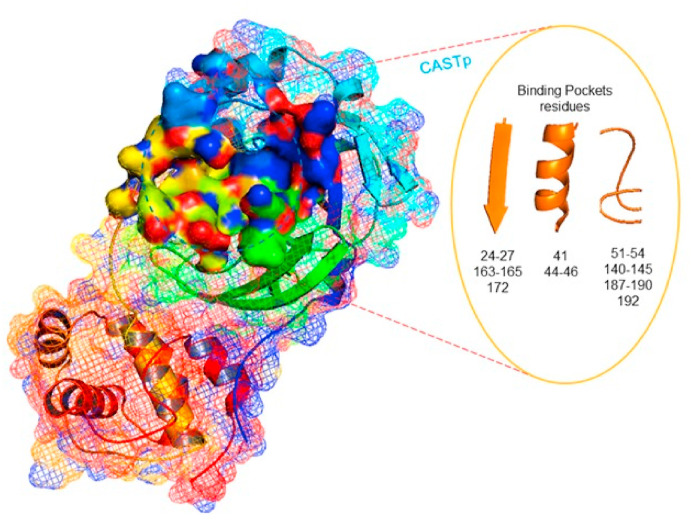
The very first step for molecular docking studies is to prepare ligand and receptor according to the virtual screening tools. Structure-based rational drug design is an important tool to discover probable inhibitors against an enzyme. Earlier in our studies, we have performed the hot sohxlet extraction of dry parts of G. glabra in various organic solvents. Several extracts were tested for their preliminary pharmacological activities and out of those relevant few were selected for their GC-MS analysis (Hejazi et al., 2017). Those extracts which were found to be antioxidative, anti-inflammatory and anticancerous in nature were further taken for our studies of drug repurposing for the SARS CoV-2. Various virtual screening tools are utilised for the purpose of analysing and seeing the interactions such as MGL Tools, Auto Dock Tools 4.2, Discovery studio and Molinspiration. All the tools were used for finding out the Gibbs Free Energy for binding and analysing drug candidates as high-affinity inhibitors against SARS-CoV-2 main protease 3Cl pro having crystal structures PDBIDs, 6LU7, 6M03, 6M2N. SARS CoV-1 main protease 1P9S is taken as comparison of the resulting interactions. Crystal structures were downloaded from Protein Data Bank and mechanism of inhibition was investigated and important amino acid residues were identified in the binding pocket. SARS-CoV-2 main protease consists of 306 amino acid residues. Crystal structure analysis suggests that Thr25, Thr26, Leu27, His41, Ser46, Met49, Tyr54, Phe140, Leu141, Asn142, Gly143, Cys145, His163, Met165, Glu166, Leu 167, Pro 168, Phe 185, Asp187, Gln189, Thr190, Ala 191, and Gln 192 are found in the active site pocket of SARS-CoV-2 main protease. Fig. 1 shows 6LU7 3CLpro main protease.
Fig. 1.
Three dimensional structure of 3CLpro main protease PDB ID 6LU7.
2.1.1. Preparation of ligand and receptor
-
(i)
Preparing a protein
In this step, we created a PDBQT file of our protein (or receptor) that contains hydrogen atoms, as well as partial charges. There are many different existing methods to estimate partial charges. We used the so-called “Gasteiger charges”. Gasteiger charges needs only data of the topology of a molecule and is thus prevalent for its simple and fast calculation. To protein structure file in PDB format, hydrogen atoms were added to prepare the receptor PDBQT file.
-
(ii)
Preparing a ligand
This step is quite similar to the preparation of the receptor. Here also, we created a PDBQT file from a ligand molecule, which is also in PDB format.
-
(iii)
Generating a grid parameter file and scoring functions
Now, 3D space that AutoDock considers for docking is defined, typically, a volume around the potential binding site of a receptor. We have used Grid parameters of centre_x = −12.855 centre_y = 16.974, centre_z = 0.66.782 and size x = 24, y = 20, z = 22 for 6LU7. For scoring functions Autodock makes use of the genetic algorithm known as Lamarkian genetic algorithm. It also calculates empirical binding free energy force field for predicting binding free energies and binding constants, for the interacting ligands. We have examined the existing crystal structures of SARS-CoV-2 3CL pro with the PDB IDs 6LU7,6M03, 6M2N. and SAS CoV-1 1P9S. All the protein structures were energetically minimized before using for docking. The entire internal energy (bonds energy, torsional angles, non-bonded atoms and electrostatic energy) of enzymes were calculated. Pymol Graphic Viewer was used for viewing the interactions between ligands and the receptor enzymes.
2.2. MD simulations
All the MD simulations were performed using GROMACS 5.1.2 Bio-Simulation package installed in Multi-core enabled Linux Ubuntu system. The force field GROMOS96 43a2 was used to determine the dynamic behaviour and different calculations of proteins and ligand which were used in this in silico studies. The gmx grep module was used to extract ligand files from the docked complexes.
PRODRG server was used to generate the topology and force-field parameters of ligands (Jalily Hasani and Barakat, 2016). The water model SPC216 was used to solvate the system (protein) and prepare the difference using different micro-molecules to see the inhibition or stability of macromolecules (proteins). As a control, MD simulation of 100 ns was carried in water at 298 K. All atoms of macromolecules (proteins) including micromolecules (ligands), were equilibrated in cubic box, having size of approximately 10.5 × 10.5 × 10.5 Å. Protein was immersed into water and well equilibrated and overlapping molecules were deleted. Each system was energy minimized with steepest descent, up to a tolerance of 1000 kJ mol−1nm−1 to remove all bad contacts. The overall charge on the system was neutralized with addition of cationic and anionic concentration of Na+Cl−. To run the simulation, based on the size and three-dimensional spatial orientation of protein, the dimension (x, y, and z) of simulation box was defined. All the systems were prepared in define box, and protein was placed in centre of box, padded around with water and co-solvents.
The steepest-descent algorithm along with conjugate gradient were used for the energy minimization process. The system was equilibrated with two ensemble processes, NVT and NPT. Before we start production run we have to prepare or establish the pre-define condition like pH and temperature. All these informations were encapsulated in NVT, NPT and MD parameter files. These physiochemical conditions play a vital role for protein structural and conformational changes. Once, the system is prepared the system is now ready to run. After the production run we have binary trajectory file for further analysis. The resulting trajectories were analyzed using gmx energy, gmx confirms, gmx rms, gmx gyrate, gmx rmsf, gmx hbond, make_ndx, gmx do_dssp and gmx sasa utilities of GROMACS. All graphical presentation of the 3D models were prepared using PyMol and Visual Molecular Dynamics (Lemkul et al., 2010).
MD simulation study has been executed to investigate the conformational stability of internal structure and biological functions (Pant et al., 2020). To get structural insights and to understand the binding mechanism of Dehydroglyaspirin, Isoliquiritine, Licochalcone and Liquiritine in active site of 6LU7 in water at 298 K, we monitored the time evolution plot of all atom RMSD, Rg, RMSF, SASA, h-bond and secondary structural content.
3. Results
3.1. Molecular docking
From our in silico studies, we were able to find out compounds from the plant which are able to interact with the SARS CoV 2 main protease 3Cl pro very efficiently, acting as an inhibitor the proteolytic functions of the protease. The selected compounds of the plant were also docked with the SARS CoV-1 3Cl pro as a comparision of the efficiency of the plant compounds. As a result we found the docking interactions with both the enzymes similar in effectiveness, opening new parameters for the drug discovery. The following plant compounds with their numbers, 1,2-o-isopropylidene-beta-l-idofuranurono-6,3-lactone(4), 2-Methyl-1(2H)-phthalazinone(29), Benzene propanoicacid_3,5-bis(1,1-dimethylethyl)-4-hydroxy(47), Capsaicin(51), Piperine(75), Isoliquiritin apioside(83), Dehydroglyasperin C(84), Licochalcone D(85), Glycyrrhizin(86) and Liquiritin (87) showed a significant energy of binding with the enzyme 3Cl pro of SARS CoV-2 and 3Cl pro of SARS CoV-1 represented in Supplementary Table 1. All compounds showed many hydrophobic interactions and hydrogen binding. Best results are given by the following compounds:
Isoliquiritin apioside (83) showed a binding energy of −7.8 kcal/mol having 7 hydrogen bonds with amino acid residues Thr26, Thr45, Ser46, Tyr54, Glu166, Asp187, 3 C_H bonds with residues Thr25, Gly143 and Gln189 and 2 hydrophobic interactions with His41 and Cys 145.
Dehydroglyasperin C (84) showed docking score of −7.1 kcal/mol and has 3 H bonds with residues Gly143, Cys145, Gln189, 1 C–H bond with His 164 and 3 hydrophobic interactions with Leu27, His41, Met165.
Licochalcone D (85) showed docking score of −7.9 kcal/mol and has 4 H bonds with residues Gly143, Ser144, His163, Thr190, 2 C–H bonds with Phe140, Glu166 and 4 hydrophobic interactions with His41, Met49, Cys145, Met165.
Liquiritin (87) showed docking score of −7.8 kcal/mol and has 6 H bonds with residues Thr45, Ser46, Phe140, Leu141, Ser144, Cys145 and 1 C–H bond with Thr25.
Although Glycyrrhizin (86) gave a strong binding but the compound was found to violate Lipinsky and Veber s Rule of five of ADMET predictors and drug likeness, and owing to its cytotoxicity, we will not recommend its use as a druggable compound.
All interactions are shown in Fig. 2, Fig. 3, Fig. 4, Fig. 5 respectively.
Fig. 2.
Comparison of various FDA approved drugs docking energy scores with plant compounds energy scores as docked in Auto dock.
Fig. 3.
Cartoon representation of 6LU7 binding with compounds a. Isoliquiritin apioside(83), b. Dehydroglyasperin C(84), c. Licochalcone D(85), d. Liquiritin (87).
Fig. 4.
Cartoon representation of 6LU7 showing hydrogen bond acceptors and hydrogen bond donors binding with compounds a. Isoliquiritin apioside(83), b. Dehydroglyasperin C(84), c. Licochalcone D(85), d. Liquiritin (87).
Fig. 5.
Cartoon representation of 6LU7 showing hydrophobic interactions binding with compounds a. Isoliquiritin apioside(83), b. Dehydroglyasperin C(84), c. Licochalcone D(85), d. Liquiritin (87).
Physiochemical predictors are represented in Table 1 . The best interacting residues with the PDB ID 6LU7are represented in Table 2 . We also docked the plant compounds with the 3Cl pro protease of SARs CoV-1 PDBID 19PS so as to compare our resultant interactions. All interactions are represented in Supplementary Table 1.
Table 1.
Physicochemical, pharmacokinetics and drug-likeness properties of compounds from G.glabra.
| Descriptions | (4) 1,2-o-isopropylidene-beta-l-idofuranurono-6,3-lactone |
(8) 1_2_benzenedicarboxylic acid_ bis(2methylpropyl) ester |
(47) Benzenepropanoic_acid, _3,5-bis(1,1-dimethylethyl)-4-hydroxy |
(51) Capsaicin |
(75) Piperine |
(83) Isoliquiritin_apioside |
(84) Dehydroglyasperin_C |
(85) Licochalcone D |
(86) Glycyrrhizin |
(87) Liquiritin |
|---|---|---|---|---|---|---|---|---|---|---|
| Physiochemical properties | ||||||||||
| M.W. | 216.19 | 278.34 | 278.3 | 305.4 | 285.3 | 550.5 | 354.40 | 354.40 | 822.9 | 418.3 |
| rotB | 0 | 8 | 5 | 10 | 4 | 9 | 4 | 6 | 7 | 4 |
| HBA | 6 | 4 | 3 | 3 | 3 | 13 | 5 | 5 | 16 | 9 |
| HBD | 1 | 0 | 2 | 2 | 0 | 8 | 3 | 3 | 8 | 5 |
| mRef. | 44.77 | 77.84 | 83.36 | 90.52 | 85.48 | 130.8 | 102.47 | 102.53 | 202.8 | 101.6 |
| TPSA | 74.22 | 52.60 | 57.53 | 58.56 | 38.77 | 215.8 | 79.15 | 86.99 | 267.0 | 145.9 |
| miLogP | 1.75 | 3.31 | 2.86 | 3.15 | 3.42 | 0.98 | 2.86 | 2.95 | 1.89 | 2.21 |
| Pharmacokinetics | ||||||||||
| GI abs. | High | High | High | High | High | Low | High | High | Low | Low |
| BBB | No | Yes | Yes | Yes | Yes | No | No | No | No | No |
| 2C9 | No | No | No | No | Yes | No | No | Yes | No | No |
| 2D6 | No | No | No | Yes | No | No | Yes | No | No | No |
| 3A4 | No | No | No | Yes | No | No | No | Yes | No | No |
| Log KP (cm/s) | −8.01 | −5.08 | −4.81 | −5.62 | −5.58 | −9.55 | −5.44 | −5.25 | −9.33 | −8.58 |
| Drug likeness | ||||||||||
| RO5 | Yes | Yes | Yes | Yes | Yes | No | Yes | Yes | No | Yes |
| Bscore | 0.55 | 0.55 | 0.56 | 0.55 | 0.55 | 0.17 | 0.55 | 0.55 | 0.11 | 0.55 |
TPSA = Total Polar Surface Area, HBA=Hydrogen bond acceptor, HBD=Hydrogen Bond Donor, mRef = Molar refractivity, MilogP = Molinspiration octanol-water partition coefficient, BBB=Blood brain Barrier, 2C9 = Cytochromes P450 isoform, 2D6 = Cytochromes P450 isoform, 3A4 = Cytochromes P450 isoform, Log KP = skin-water permeability coefficient, Bscore = Bioactivity Score.
Table 2.
PDB ID 6LU7 main protease interactions with selected plant compounds from G.glabra on the basis of their physiochemical properties and FDA approved drugs found in docking studies.
| Compounds | AutoDock score (kcal/mol) | H-bonds |
C–H bonds |
Hydrophobic Interaction |
|||
|---|---|---|---|---|---|---|---|
| No. | Amino acids | No. | Amino acids | No. | Amino acids | ||
| Plant compounds | |||||||
| 83 Isoliquiritin apioside |
−7.8 | 7 | Thr26 Thr45 Ser46 Tyr54 Glu166 Asp187 |
3 | Thr25 Gly143 Gln189 |
2 | His41 Cys145 |
| 84 DehydroglyasperinC |
−7.1 | 3 | Gly143 Cys145 Gln189 |
1 | His 164 | 3 | Leu27 His41 Met165 |
| 85 Licochalcone D |
−7.9 | 4 | Gly143 Ser144 His163 Thr190 |
2 | Phe140 Glu166 |
4 | His41 Met49 Cys145 Met165 |
| 87 Liquiritin |
−7.8 | 6 | Thr45 Ser46 Phe140 Leu141 Ser144 Cys145 |
1 | Thr25 | NA | NA |
| FDA approved Drugs | |||||||
| Remdesivir | −7.8 | 3 | Glu166 Arg188 Thr190 |
NA | NA | 3 | His41 Met49 Met165 |
| Favipiravir | −5.1 | 5 | Phe140 Gly143 Ser144 Cys145 Glu166 |
1 | His 172 | 2 | Leu141 Cys145 |
| Hydroxychloroquine | −6.5 | 3 | Ser144 Arg188 Gln189 |
3 | Asn142 His163 Glu166 |
2 | His41 Met165 |
3.2. MD simulation
3.2.1. Average potential energy of system
The average potential energy of free 6LU7, and their complexes Dehydroglyaspirin, Isoliquiritine apioside, Licochalcone and Liquiritine were monitored to ascertain the equilibration of the systems before the MD analysis. At a constant temperature (298 K) we have observed a constant fluctuation in each system which suggests a stable and accurate MD simulation (Table 3 ).
Table 3.
Average potential energy of system.
| Ligand Name | Pysiochemical condition | Average potential Energy (kJ/mol) |
|---|---|---|
| DehydroglyaspirinC | 298 K & pH = 7.0 | −1.34540e+06 |
| Isoliquiritine apioside | 298 K & pH = 7.0 | −1.80663e+06 |
| Licochalcone | 298 K & pH = 7.0 | −1.32104e+06 |
| Liquiritine | 298 K & pH = 7.0 | −1.35533e+06 |
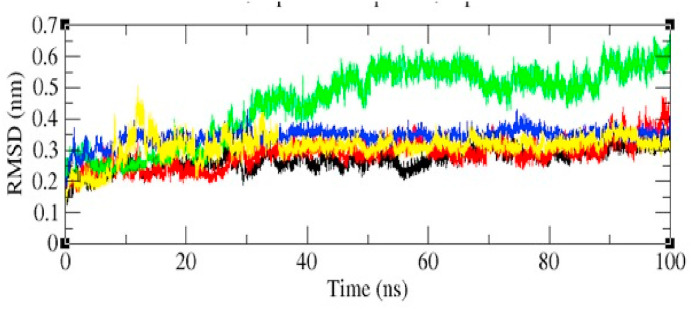
3.2.2. Root means square distance (RMSD)
The binding of a ligand of the protein can generally causes a significant conformational change in the structure. RMSD is calculated as a function of time, with respect to the initial conformation and is shown in Fig. 6 and Table 4 . The RMSD parameter was estimated to see whether the structure of a protein is stable and close to the experimental structure. The average RMSD value for 6LU7 was found to be 0.272 nm (seeFig. 7).
Fig. 6.
RMSD Vs Time plot.
Table 4.
Root means square distance (RMSD).
| S. no | Protein and ligands | Average values of RMSD (nm) |
|---|---|---|
| 1 | 6LU7 and DehydroglyaspirinC | 0.287 |
| 2 | 6LU7 and Isoliquiritine apioside | 0.341 |
| 3 | 6LU7 and Licochalcone | 0.453 |
| 4 | 6LU7 and Liquiritine | 0.311 |
Fig. 7.
Backbone residual fluctuations (RMSF) plots.
The RMSD plot clearly suggested that the protein and Dehydroglyaspirin complex is the most stable complex than the rest of the other complexes, because of the lesser deviations from its native conformation i.e., the wild type. However, in the case in the case of 6LU7-Licochalcone complex which is depicted in the green colour in the figure showed to have less fluctuation till 0–25 ns of MD trajectories, afterward the values of RMSD 6LU7-Licochalcone was increased.
To calculate the average fluctuation of all residues during the stimulation, the root mean square fluctuation (RMSF) of 6LU7 while ligand binding were plotted as a function of residue number (Fig. 2). RMSF plot showed that residual fluctuations are present at several regions. These residual fluctuations were found to be minimized upon binding of Dehydroglyaspirin or wild type protein and maximized with Licochalcone.
3.2.3. Radius of gyration
Radius of gyration (R g) was calculated to check the stability of the protein in a biological system. A stable protein is supposed to have the higher Rg value due to less tight packing. The average Rg values for free 6LU7, 6LU7-Dehydrogly Aspirin, 6LU7-Isoliquiritine apioside, 6LU7-Licochalcone, and 6LU7-liquiritine were found to be 2.11 nm, 2.12 nm, 2.08 nm, 2.09 nm and 2.11 nm, respectively. Rg plot suggested that the 6LU7 attained more tight packing in its native state and when bound to Dehydrogly-Aspirin than Licochalcone (Fig. 8 ) and rest of them in between of these two. However, such differences in the R g values are not significant.
Fig. 8.
A time evolution Rg.
3.2.4. Solvent accessible surface area (SASA)
Estimation of SASA provides information about the conformational changes in protein upon ligand binding. The average SASA values for 6LU7, 6LU7-DehydroglyaspirinC, 6LU7-Isoliquiritine apioside, 6LU7-Licochalcone and 6LU7-Liquiritine complexes were also monitored during 100 ns MD stimulations. The average SASA values for 6LU7, 6LU7-Dehydroglyaspirin C, 6LU7-Isoliquiritine apioside, 6LU7-Licochalcone and 6LU7-Liquiritine complexes were found to be 156.69 nm2, 156.182 nm2, 156.561 nm2, 191.161 nm2 and 157.646 nm2 respectively. It is shown in Fig. 9 .
Fig. 9.
Solvent accessible surface area Vs Time Plot.
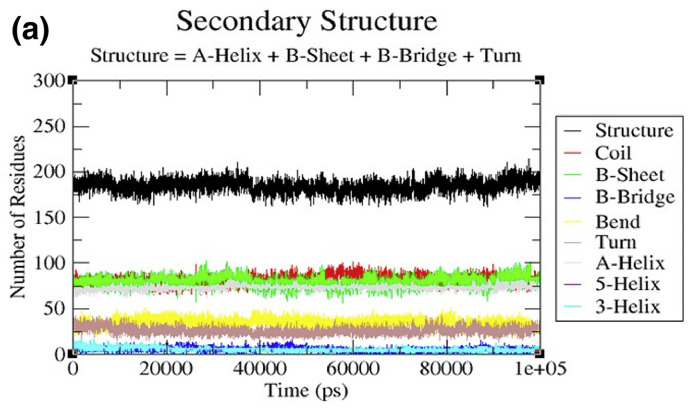
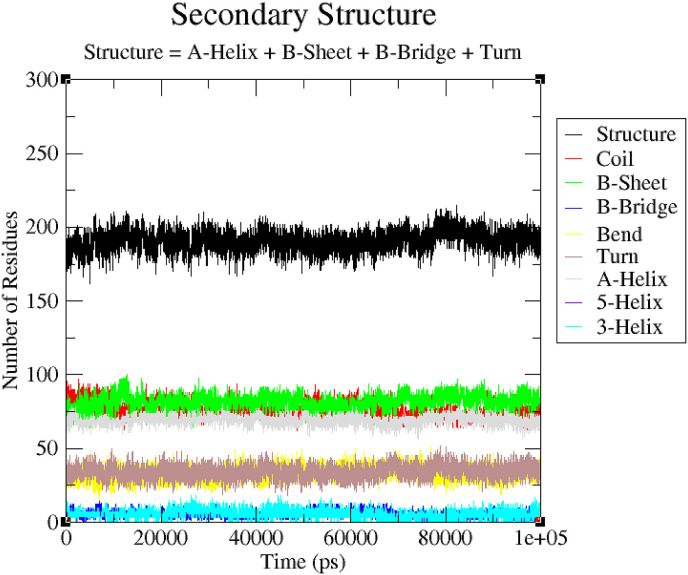
3.2.5. Secondary structure changes upon ligand binding
Secondary structure changes upon ligand binding were analyzed to compute the secondary structure content of the protein as a function of time. The secondary structure assignments in protein such as α-helix, beta-sheet and turn were broken into individual residues for each time step. The average numbers of residues involved in the formation of secondary structure were slightly decreased due to increase in percentage of coils, and slightly decrease in bends and α-helices (Table 5 ). The β-sheet was increased in cases of 6LU7-Dehydroglyaspirin, 6LU7-Liquiritine, 6LU7-Licochalcone respectively. There was decrease in percentage of β-sheet complex but no major change in secondary structure content of 6LU7 was seen during simulations. It is shown in Fig. 10a, Fig. 10b, Fig. 10c, Fig. 10d a 10b 10c and 10d respectively.
Table 5.
Percentage of protein secondary structure.
| Percentage of protein secondary structure (SS %) | ||||||||
|---|---|---|---|---|---|---|---|---|
| Complex | Structure* | Coil | β-sheet | β -bridge | Bend | Turn | α-helix | 310 -helix |
| 6LU7 | 60 | 26 | 26 | 2 | 11 | 8 | 24 | 2 |
| 6LU7-DehydroglyaspirinC | 60 | 27 | 27 | 1 | 11 | 9 | 23 | 2 |
| 6LU7-Lichochalcone | 60 | 26 | 27 | 2 | 12 | 8 | 23 | 2 |
| 6LU7-Liquiritin | 62 | 25 | 27 | 2 | 11 | 11 | 23 | 2 |
Fig. 10a.
Secondary structure plots. The secondary structure analysis indicating the structural elements present of 6LU7.
Fig. 10b.
Secondary structure plots. The secondary structure analysis indicating the structural elements present of 6LU7-Dehydroglyaspirin.
Fig. 10c.
Secondary structure plots. The secondary structure analysis indicating the structural elements present of 6LU7-Lichochalcone.
Fig. 10d.
Secondary structure plots. The secondary structure analysis indicating the structural elements present of 6LU7-Liquiritin.
3.2.6. Hydrogen bond interactions
Hydrogen bond interactions between a protein and ligand compounds provides a specificity and directionality of interaction that is a fundamental aspect of molecular recognition (Hubbard and Kamran Haider, 2001). .Hydrogen bonds between protein and ligands paired within 0.35 nm in 6LU7-Dehydroglyaspirin, 6LU7 Isoliquiritine, 6LU7-Licochalcon and 6LU7-Liquiritine at the given solvent condition were calculated during the simulations to validate the specificity and stability of docked complexes. During the analysis, it was found that the Dehydroglyaspirin and Licochalcone bind to binding pocket of 6LU7 with 2–5 hydrogen bonds, while Liquiritine binds to the active pocket of 6LU7 with 5–8 hydrogen bonds which is supporting our molecular docking analysis. Basically here, the stability of hydrogen bonds between 6LU7 and ligands was evaluated during the course of simulation time. Interaction analysis is shown in Fig. 11 .
Fig. 11.
The average number of hydrogen bonds as a function of time. (a) Red, (b) green, and (c) blue colours represent the number of hydrogen bonds for 6LU7- Dehydroglyaspirin, 6LU7-Licochalcone and 6LU7- Liquiritine complexes, respectively. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
4. Discussion
Undoubtedly, the danger from the usage of an accepted drug should be suggestively lesser than from the disease itself. Hence, the medication must be least cytotoxic to the body cells. This statistic significantly confuses the hunt of the drug molecule. It is quite likely that at the phase of huge clinical trials, it could be revealed that the drug is quite risky in terms of cytotoxicity to its own cells, a hazard that is unparalleled with the danger of the disease itself. However, remdesivir, favipiravir hydroxychloroquine and their combinations are at present the most hopeful drugs for use in COVID-19 treatment. Remdesivir (GS-5734) is an antivirus drug that was discovered to fight the Ebola virus disease epidemic in 2010. This nucleotide prodrug is also active against the, the SARS-CoV 1 and MERS-CoV) (Manning et al., 2020) (University, 2020) (Athari et al., 2020). Favipiravir is a pyrazine carboxamide molecule, which inhibits RNA polymerase of the viral machinary without affecting cellular synthesis (Du and Chen, 2020) (Kim et al., 2018) (Goldhill et al., 2018). Chloroquine and hydroxychloroquine antimalarial drugs also used in rheumatoid arthritis and systemic lupus arithromatosis treatments are recently been found helpful in treating SARS CoV 2. Both drugs are highly toxic, particularly, cardiotoxic (Pastick et al., 2020) (Saleh et al., 2020) (Şimşek Yavuz and Ünal, 2020).
Efforts are being made to find suitable, effective and harmless therapeutics. Keeping in mind the use of plant compounds to be the safest having least cytotoxicity than the chemically synthesized molecules, we have proposed the use of phytoconstituents from the plant G. glabra which is a traditional plant also having numerous health benefits.
For a drug to be absorbed it must be soluble and polar. Hydrophobic properties play a crucial role for a drug to penetrate several biological barriers and influence the anticipated place of action without having any cytotoxic effect to the own body cells. So comparison of few molecular descriptors is important, i.e., physicochemical properties and bioactivity scores (drug-likeness). This gives a quick approach for antiviral drug optimization and it forms the basis of the very well known Lipinski “rule of five” (RO5), which can be simply recognized by means of the open access tool Molinspiration (Walters, 2012) (Benet et al., 2016). The significance of RO5 (MW < 500, cLogP < 5, number of hydrogen-bond acceptors hba <10 and number of hydrogen-bond donors hbd < 5) is linked with the solubility and permeation of in vivo body barriers. So the descriptors are filters that assist in selection of drug-like compounds. Selected 10 plant compounds gave a good following of descriptors.
Similarly, few pharmacological targets do not have high affinity and selectivity ligands that comply with Lipinski s rule of five, so they have to be reliant beyond the RO5 chemical space with the much-expected consequences on the ADMET properties (absorption, distribution, metabolism, excretion and toxicity) of the ligands. Besides that, Veber's rule says that good bioavailability is more expected for compounds having ≤10 rotatable bonds (rotb), total hba + hbd < 12 and topological polar surface area (TPSA) ≤ 140 A◦(Kowalska et al., 2018) (Kar et al., 2020). Our compounds from the plant G glabra follow all the rules to be a probable drug like candidate.
Rotatable bonds describe the flexibility and compliance for effective interaction with the precise binding pocket of the receptor. TPSA descriptor defines the passive transport through membranes which can predict the transport of drugs to the blood-brain barrier. TPSA values less than 140 A◦ are associated with good cell membrane permeability(Votano, 2005) (Vihinen, 2020). Table 1 shows all the descriptors of our compounds and all plant compounds are found to follow all of them very precisely.
One of the primary tools in the hypothetical study of biological molecules is the technique of molecular dynamics simulations (MD). This computational technique calculates the time dependent conduct of a molecular system (Hospital et al., 2015). MD simulations have provided detailed statistics on the fluctuations and conformational changes of biological molecules as it is used to investigate their structure, dynamics and thermodynamics. Macromolecules, especially proteins, adopt multiple conformations that are linked to their biological functions which are best studied by molecular dynamic simulation tools. Dynamics plays a key role in studying the functionality of proteins or enzymes as these experience substantial conformational variations while carrying out their function (Naqvi et al., 2019).
The dynamic structural states of proteins were covered in magnitude between 10−11 and 10−6 m and time-scales from 10−12 s to 10−5 s. All MD simulations were processed with ligand bounded SARS-CoV-2 3CLpro for the timescale of 100 ns for understanding the dynamic behaviour in an aqueous environment and is performed in a similar procedure for both the protein and protein and substrate complex and the values of RMSD were plotted (Gharaghani et al., 2013). The SARS-CoV-2 3CLpro complexed with all the ligands showed a stable conformation throughout the simulations. This substrate complex is dynamically stable, we did not see any sudden surge or sliding from its position. This clearly says that, the substrate-bound complex is dynamically stable due to the tight amino acid contributions. The RMSD plot shows that all the compounds are stable with their constant RMSD values. Also the secondary structure complexes of 6LU7 and all the interacted phytocompounds were found to be in dynamically stable conformations except Isoliquiritin apioside which was not in a stable conformation. This might be due to its not binding in the active cleft of the enzyme. Rest of the compounds (DehydroglyasperinC, Lichochalcone and Liquiritin) were binding tightly with the enzyme complex thus can hinder its functionality in replication and protein synthesis required for the virus. Owing to this we can propose the use of these (DehydroglyasperinC, Lichochalcone and Liquiritin) compounds as potential drugs against the main protease. Further experiments in the lab are needed to prove these trials on cell lines and animals.
5. Conclusion
With the quickly increasing pandemic of COVID-19 triggered by the novel and challenging SARS-CoV-2 coronavirus, there is an imperative necessity for new therapies and prevention approaches that can support disease spread and decrease mortality rate. Inhibition of viral replication machinery and thereby inhibiting formation of multiple copies, establish reasonable therapeutic avenues. These days, molecular modelling and molecular docking are the elementary requirements of medicinal industries for drug discovery and progress which enable time-saving and are economic and effective making use of computer-aided drug discovery. In conclusion, the free available structure of the human SARS-CoV-2 3CL pro protein was downloaded and was recruited to study its structure and function along with its interaction with several phytoconstituents by means of computational methods so as to find a probable inhibitor to the enzyme 3Cl pro. Compounds Dehydroglyasperin_C (84), Licochalcone D (85), Liquiritin (87) have given significant binding efficiency by Auto Dock Vina tools with 3Cl pro PDB ID 6LU7 following all drug like characteristics. These compounds were also found to be dynamically stable as studied in our MD Simulation studies. We strongly recommend our phytoconstituents for the clinical trials against SARS CoV 2 and further investigations. These could provide a valid substitute for the various available antivirals with are heavily cytotoxic also. Here our approach was to repurpose the already available secondary metabolites of the plant G glabra to our use for the inhibition of replication machinery of the SARS CoV-2 with a lower level of cellular toxicity. However, our in silico approach has to be confirmed by the various in vitro and in vivo experiments.
CRediT authorship contribution statement
Iram Iqbal Hejazi: Original idea of the manuscript has been formulated by the author. Md Amjad Beg: Docking studies have been done by this author. Md Ali Imam: MD Simulations have been done by the author. Fareeda Athar: Helped in designing the idea. Asimul Islam: Helped in designing the idea.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Handling editor: Dr. Jose Luis Domingo
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.fct.2021.112057.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Ahmed S.F., Quadeer A.A., McKay M.R. Preliminary identification of potential vaccine targets for the COVID-19 coronavirus (SARS-CoV-2) based on SARS-CoV immunological studies. Viruses. 2020;12(3) doi: 10.3390/v12030254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armanini D., Fiore C., Bielenberg J., Sabbadin C., Bordin L. Coronavirus-19: possible therapeutic implications of spironolactone and dry extract of Glycyrrhiza glabra L. (Licorice) Front. Pharmacol. 2020;11 doi: 10.3389/fphar.2020.558418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Athari S.Z., Mohajeri D., Nourazar M.A., Doustar Y. vol. 9. 2020. Updates on coronavirus (COVID-19) and kidney; pp. 1–6. (Journal of Nephropathology). Issue 4. [DOI] [Google Scholar]
- Benet L.Z., Hosey C.M., Ursu O., Oprea T.I. vol. 101. 2016. BDDCS, the Rule of 5 and drugability; pp. 89–98. (Advanced Drug Delivery Reviews). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cucinotta D., Vanelli M. vol. 91. 2020. WHO declares COVID-19 a pandemic. (Acta Biomedica). Issue 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Y.X., Chen X.P. Clinical Pharmacology and Therapeutics. 2020. Favipiravir: pharmacokinetics and concerns about clinical trials for 2019-nCoV infection; pp. 242–247. [DOI] [PubMed] [Google Scholar]
- Gharaghani S., Khayamian T., Ebrahimi M. Molecular dynamics simulation study and molecular docking descriptors in structure-based QSAR on acetylcholinesterase (AChE) inhibitors. SAR QSAR Environ. Res. 2013;24(9) doi: 10.1080/1062936X.2013.792877. [DOI] [PubMed] [Google Scholar]
- Goldhill D.H., Te Velthuis A.J.W., Fletcher R.A., Langat P., Zambon M., Lackenby A., Barclay W.S. The mechanism of resistance to favipiravir in influenza. Proc. Natl. Acad. Sci. U. S. A. 2018;115(45):11613–11618. doi: 10.1073/pnas.1811345115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hejazi I.I., Khanam R., Mehdi S.H., Bhat A.R., Moshahid Alam Rizvi M., Islam A., Thakur S.C., Athar F. New insights into the antioxidant and apoptotic potential of Glycyrrhiza glabra L. during hydrogen peroxide mediated oxidative stress: an in vitro and in silico evaluation. Biomed. Pharmacother. 2017;94:265–279. doi: 10.1016/j.biopha.2017.06.108. [DOI] [PubMed] [Google Scholar]
- Hejazi I.I., Khanam R., Mehdi S.H., Bhat A.R., Rizvi M.M.A., Thakur S.C., Athar F. Antioxidative and anti-proliferative potential of Curculigo orchioides Gaertn in oxidative stress induced cytotoxicity: in vitro, ex vivo and in silico studies. Food Chem. Toxicol. 2018;115 doi: 10.1016/j.fct.2018.03.013. [DOI] [PubMed] [Google Scholar]
- Hospital A., Goñi J.R., Orozco M., Gelpí J.L. vol. 8. 2015. Molecular dynamics simulations: advances and applications. (Advances and Applications in Bioinformatics and Chemistry). Issue 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jalily Hasani H., Barakat K. Methods and Algorithms for Molecular Docking-Based Drug Design and Discovery. 2016. Methods and algorithms for molecular docking-based drug design and discovery. [Google Scholar]
- Kannan S., Shaik Syed Ali P., Sheeza A., Hemalatha K. COVID-19 (Novel Coronavirus 2019) - recent trends. Eur. Rev. Med. Pharmacol. Sci. 2020;24(4):2006–2011. doi: 10.26355/eurrev_202002_20378. [DOI] [PubMed] [Google Scholar]
- Kar P., Sharma N.R., Singh B., Sen A., Roy A. Natural compounds from Clerodendrum spp. as possible therapeutic candidates against SARS-CoV-2: an in silico investigation. J. Biomol. Struct. Dyn. 2020 doi: 10.1080/07391102.2020.1780947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaushik S., Kaushik S., Sharma Y., Kumar R., Yadav J.P. VirusDisease. 2020. The Indian perspective of COVID-19 outbreak. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.A., Seong R.K., Kumar M., Shin O.S. Favipiravir and ribavirin inhibit replication of Asian and African strains of zika virus in different cell models. Viruses. 2018;10(2):1–15. doi: 10.3390/v10020072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalska M., Fijalkowski K., Nowaczyk A. The biological activity assessment of potential drugs acting on cardiovascular system using Lipinski and Veber Rules. J. Educ. 2018;8(12):184–191. doi: 10.5281/zenodo.2066519. [DOI] [Google Scholar]
- Lemkul J.A., Allen W.J., Bevan D.R. Practical considerations for building GROMOS-compatible small-molecule topologies. J. Chem. Inf. Model. 2010;50(12) doi: 10.1021/ci100335w. [DOI] [PubMed] [Google Scholar]
- Liu Y.C., Kuo R.L., Shih S.R. COVID-19: the first documented coronavirus pandemic in history. Biomed. J. 2020 doi: 10.1016/j.bj.2020.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo P., Liu D., Li J. Pharmacological perspective: glycyrrhizin may be an efficacious therapeutic agent for COVID-19. Int. J. Antimicrob. Agents. 2020;55(6) doi: 10.1016/j.ijantimicag.2020.105995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik Y.A. Malaysian Journal of Pathology. 2020. Properties of coronavirus and SARS-CoV-2. [PubMed] [Google Scholar]
- Manning T.J., Thomas-Richardson J., Cowan M., Beard T. Drug Discovery Today. 2020. Vaporization, bioactive formulations and a marine natural product: different perspectives on antivirals; pp. 956–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naqvi A.A.T., Mohammad T., Hasan G.M., Hassan M.I. Advancements in docking and molecular dynamics simulations towards ligand-receptor interactions and structure-function relationships. Curr. Top. Med. Chem. 2019;18(20) doi: 10.2174/1568026618666181025114157. [DOI] [PubMed] [Google Scholar]
- Pant S., Singh M., Ravichandiran V., Murty U.S.N., Srivastava H.K. Peptide-like and small-molecule inhibitors against Covid-19. J. Biomol. Struct. Dyn. 2020 doi: 10.1080/07391102.2020.1757510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S.E. vol. 63. 2020. Epidemiology, virology, and clinical features of severe acute respiratory syndrome-coronavirus-2 (SARS-COV-2; Coronavirus Disease-19) pp. 119–124. (Korean Journal of Pediatrics). Issue 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastick K.A., Okafor E.C., Wang F., Lofgren S.M., Skipper C.P., Nicol M.R., Pullen M.F., Rajasingham R., McDonald E.G., Lee T.C., Schwartz I.S., Kelly L.E., Lother S.A., Mitjà O., Letang E., Abassi M., Boulware D.R. vol. 7. 2020. Review: hydroxychloroquine and chloroquine for treatment of SARS-CoV-2 (COVID-19) pp. 1–9. (Open Forum Infectious Diseases). Issue 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleh M., Gabriels J., Chang D., Soo Kim B., Mansoor A., Mahmood E., Makker P., Ismail H., Goldner B., Willner J., Beldner S., Mitra R., John R., Chinitz J., Skipitaris N., Mountantonakis S., Epstein L.M. Effect of chloroquine, hydroxychloroquine, and azithromycin on the corrected QT interval in patients with SARS-CoV-2 infection. Circulation. Arrhythmia and Electrophysiology. 2020;13(6):496–504. doi: 10.1161/CIRCEP.120.008662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamsi A., Mohammad T., Anwar S., AlAjmi M.F., Hussain A., Md Tabish R., Islam A., Md Imtaiyaz H. Glecaprevir and Maraviroc are high-affinity inhibitors of SARS-CoV-2 main protease: possible implication in COVID-19 therapy. Biosci. Rep. 2020;40(6) doi: 10.1042/BSR20201256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Şimşek Yavuz S., Ünal S. vol. 50. 2020. Antiviral treatment of covid-19; pp. 611–619. (Turkish Journal of Medical Sciences). Issue SI-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai W., He L., Zhang X., Pu J., Voronin D., Jiang S., Zhou Y., Du L. Characterization of the receptor-binding domain (RBD) of 2019 novel coronavirus: implication for development of RBD protein as a viral attachment inhibitor and vaccine. Cell. Mol. Immunol. 2020 doi: 10.1038/s41423-020-0400-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- University C.M. ClinicalTrials; 2020. A Trial of Remdesivir in Adults with Severe COVID-19. [Google Scholar]
- Vihinen M. Solubility of proteins. ADMET and DMPK. 2020. [DOI] [PMC free article] [PubMed]
- Votano J.R. vol. 8. 2005. Recent uses of topological indices in the development of in silico ADMET models. (Current Opinion in Drug Discovery and Development). Issue 1. [PubMed] [Google Scholar]
- Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181(2) doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters W.P. vol. 7. 2012. Going further than Lipinski's rule in drug design; pp. 99–107. (Expert Opinion on Drug Discovery). Issue 2. [DOI] [PubMed] [Google Scholar]
- WHO . WHO; 2020. Brazil: WHO Coronavirus Disease (COVID-19) Dashboard | WHO Coronavirus Disease (COVID-19) Dashboard.https://covid19.who.int/region/amro/country/br [Google Scholar]
- Xu J., Zhao S., Teng T., Abdalla A.E., Zhu W., Xie L., Wang Y., Guo X. Systematic comparison of two animal-to-human transmitted human coronaviruses: SARS-CoV-2 and SARS-CoV. Viruses. 2020;12(2) doi: 10.3390/v12020244. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.