Abstract
Severe acute respiratory syndrome-related coronavirus-2 (SARS-CoV-2) has resulted in a pandemic and continues to spread at an unprecedented rate around the world. Although a vaccine has recently been approved, there are currently few effective therapeutics to fight its associated disease in humans, COVID-19. SARS-CoV-2 and the related severe acute respiratory syndrome (SARS-CoV-1), and Middle East respiratory syndrome (MERS-CoV) result from zoonotic respiratory viruses that have bats as the primary host and an as yet unknown secondary host. While each of these viruses has different protein-based cell-surface receptors, each rely on the glycosaminoglycan, heparan sulfate as a co-receptor. In this study we compare, for the first time, differences and similarities in the structure of heparan sulfate in human and bat lungs. Furthermore, we show that the spike glycoprotein of COVID-19 binds 3.5 times stronger to human lung heparan sulfate than bat lung heparan sulfate.
Keywords: Heparan sulfate, Disaccharide composition, Molecular weight, SARS-CoV-2 virus, Spike protein RBD
1. Introduction
Bats are the only mammals capable of sustained flight and possess many notable characteristics such as echolocation, ultraviolet vision, and exceptionally long lifespans (Brunet-Rossinni & Austad, 2005; Suga & Shimozawa, 1974; Winter, López, & von Helversen, 2003). There are more than 1,200 species of bats grouped as Megachiroptera, which includes flying foxes and Microchiroptera, a more diverse group that include pollen, fruit, insect, hematogenous, and fish-eating bats. Many bats live in close proximity to humans and some species have become hosts for zoonotic viral pathogens (Olival et al., 2017).
Bats have received considerable attention in recent years as potential vectors of several deadly zoonotic viruses (Olival et al., 2017), most recently the SARS-CoV-2 virus (Fig. 1 ). It has long been known that bats serve as vectors of most lyssaviruses and the flavivirus Rio Bravo (de Oliveira Figueiredo et al., 2020; Price, 1978). Severe acute respiratory syndrome (SARS-CoV-1), Middle East respiratory syndrome (MERS-CoV), and most recently COVID-19 have intensified this interest given the public health significance. Apart from lyssaviruses, it is unclear to what degree these highly pathogenic viruses result in clinical disease in bats (Guito et al., 2021). This lack of significant pathology would allow for more interspecies spread, including transmission to humans. Several studies have demonstrated higher level of interferons present in bats as compared to other animals, including humans, as cytokines generated by host immune cells that enhance the initial immune response towards infection promoting rapid viral clearance or persistent infections (Brook et al., 2020; Pavlovich et al., 2018; Schountz, Baker, Butler, & Munster, 2017). Thus, in zoonotic infection bats may be the initial host serving as a viral reservoir.
Fig. 1.
The transmission of SARS-CoV-2 virus from bat to human directly or through some unknown intermediate hosts.
Coronaviruses (CoV) are well-known viruses that infect humans, domestic, and agriculture animals. Interspecies transmission has been documented for some coronaviruses including human coronavirus (HCoV-OC43), porcine epidemic diarrhea (PED), severe acute diarrhea syndrome (SADS), MERS-CoV, SARS-CoV-1, and recently SARS-CoV-2 (Banerjee, Kulcsar, Misra, Frieman, & Mossman, 2019). SARS-CoV-1, MERS-CoV, and SARS-CoV-2 are beta-corona viruses, which can cause severe respiratory disease and death in humans (Sahin et al., 2020).
Viruses infect by binding to receptors on the surface of host cells and penetrating or injecting their nucleic acid that is then replicated through transcription-translation by the host cell (Kumar, Sharma, Barua, Tripathi, & Rouse, 2018). Host cell receptors are typically membrane bound proteins but often require glycan-based co-receptors (Maginnis, 2018; Taylor, McGuckin, Wesselingh, & Rogers, 2018). Proteoglycans, comprised of a core protein with covalently attached glycosaminoglycan side chains, often serve as glycan co-receptors in viral infections (Kamhi, Joo, Dordick, & Linhardt, 2013). Heparan sulfate co-receptors are known to be involved in fusion activation of SARS-CoV and MERS-CoV and act together with their protein-based angiotensin-converting enzyme 2 (ACE2), and dipeptidyl peptidase 4 cell surface receptors, respectively (Lang et al., 2011; Li et al., 2005; Raj et al., 2013). SARS-CoV-2 was also recently confirmed to utilize ACE2 and host cell proteases during host cell entry (Hoffmann et al., 2020). Moreover, recent studies by our laboratory (Jin et al., 2020; Kim et al., 2020; Kwon et al., 2020; Tandon et al., 2020) and others (Clausen et al., 2020; Tree et al., 2020) confirmed that SARS-CoV-2 also relies on heparan sulfate as a co-receptor.
Since the beta-coronaviruses SARS-CoV-1, MERS-CoV and SARS-CoV-2 utilize heparan sulfate as their co-receptor for infecting respiratory epithelial cells and appear to have originated in bats, we comparatively examined the structure of lung-derived bat and human heparan sulfate. We hypothesized that bat lung heparan sulfate should act as a co-receptor since bats can serve as a reservoir for SARS-CoV-2 but that the interaction of spike glycoprotein should be weaker for of bat heparan sulfate than human heparan sulfate. While there have been some studies on human lung heparan sulfate (Uhl et al., 2020) and that from other species such as mouse (Warda et al., 2006), to our knowledge there have been no studies on bat-derived heparan sulfate. The current study describes the first isolation and characterization of bat lung heparan sulfate and compares its binding for SARS-CoV-2 spike protein, specifically assessing the receptor binding domain (RBD) and further utilizing a SARS-CoV-2 spike lentivirus pseudotype to human lung heparan sulfate.
2. Materials and methods
2.1. Materials
Eight bat lungs (4 male and 4 female) were harvested from rabies negative wild Big Brown bats (Eptesicus fuscus) submitted for routine rabies testing. Tissue was autoclaved to ensure any other potential pathogens that may be present in these bats would be rendered non-infectious. Previous studies have demonstrated that GAGs are stable under standard conditions used in autoclaving; indeed, heparin is routinely sterilized by autoclaving. Additionally, 6 human lung tissue samples derived from male cadavers were obtained from the University of Vermont (UVM) autopsy services under appropriate institutional guidelines. Porcine intestinal heparin (∼18 kDa) was from Celsus (Cincinnati, OH, USA). 4-deoxy-α-l-threo-hex-4-enopyranosyluronic acid (ΔUA)-N-acetyl-d-glucosamine (GlcNAc) (HS 0S); ΔUA-N-sulfo-d-glucosamine (GlcNS) (HS NS); ΔUA-GlcNAc6S (HS 6S); ΔUA2S-GlcNAc (HS 2S); ΔUA-GlcNS6S (HS NS6S); ΔUA2S-GlcNS (HS NS2S); ΔUA2S-GlcNAc6S (HS 2S6S); ΔUA2S-GlcNS6S (HS TriS); ΔUA-N-acetyl-d-galactosamine (GalNAc) (CS 0S); ΔUA2S-GalNAc (CS 2S); ΔUA-GalNAc4S (CS 4S); ΔUA-GalNAc6S (CS 6S); ΔUA2S-GalNAc4S (CS 2S4S); ΔUA2S-GalNAc6S (CS 2S6S); ΔUA-GalNAc4S6S (CS 4S6S); ΔUA2S-GalNAc4S6S (CS TriS); ΔUA-GlcNAc (hyaluronan, HA) were purchased from Iduron (Manchester, UK). 2-aminoacridone (AMAC) was purchased from Sigma-Aldrich (St. Louis, MO, USA). Actinase E was from Kaken Biochemicals (Tokyo, Japan).
2.2. SARS-CoV-2 Spike protein receptor binding domain (RBD)
A codon-optimized construct encoding amino acids 318–537 (RVQ…VNK) of the SARS-CoV-2 Spike protein (PDB 6VXX) was synthesized and cloned into the pTwist CMV BetaGlobin WPRE Neo vector (Twist Biosciences, San Francisco, CA). The synthesized gene also encoded a human Ig kappa leader sequence on the amino terminus of the protein and a GS linker and 6 × HIS tag on the carboxy terminus. The plasmid was produced in DH5α E. coli and transfected into Expi293 F cells (ThermoFisher) using 25,000 Da linear polyethylenimine (Polysciences, Inc.). Six days following transfection, supernatant was harvested and recombinant RBD was purified via passage over a HisTrap HP column (GE Healthcare). Protein concentration was measured by BCA (Thermo Scientific) and purification was verified by SDS-PAGE.
2.3. SARS-CoV-2 spike lentivirus pseudotype
HEK293 T cells (ATCC # CRL3216) were cultured in DMEM (Corning Inc) supplemented with 10 % fetal bovine serum (FBS, Fisher Scientific) at 37 °C with 5 % CO2. HEK293 T cells (2 × 106) were plated in a 100-mm tissue culture dish and transfected the next day when they were approximately 75 % confluent with a combination of the following plasmids: 9 μg of pLV-eGFP (a gift from Pantelis Tsoulfas, Addgene plasmid # 36083; http://n2t.net/addgene:36083; RRID:Addgene_36083), 9 μg of psPAX2 (a gift from Didier Trono, Addgene plasmid # 12260; http://n2t.net/addgene:12260; RRID:Addgene_12260), and 3 μg of pCAGGS-S (SARS-CoV-2) (Catalog No. NR-52310: BEI Resources) or VSV-G (a gift from Tannishtha Reya, Addgene plasmid # 14888; http://n2t.net/addgene:14888; RRID:Addgene_14888) as control. Polyethylenimine (PEI) reagent (Millipore Sigma, #408727) was used for transfection following manufacturer’s protocols. Next day, the cells were checked for transfection efficiency under a fluorescent microscope, indicated by GFP fluorescence. The supernatants from cell culture at 24 h were harvested and stored at 4 °C and 10 mL of complete media (DMEM containing 10 % FBS) was added to the plates. The supernatant from cell culture at 48 h was harvested and combined with the 24 h supernatant for each sample. The combined supernatants were spun in a tabletop centrifuge for 5 min at 2,000 g to pellet the residual cells and then passed through a 0.45 μm syringe filter. Aliquots were frozen at −80 °C. New HEK293 T cells plated in 12 well tissue culture dishes were infected with the harvested virus (supernatant) with a dilution range of 102 to 107. Virus (pLV-S) titers were calculated by counting the GFP positive cells in the dilution with 20–100 GFP positive cells.
2.4. General procedure for glycosaminoglycan (GAG) recovery and analysis
Lung tissues were washed in cold phosphate-buffered saline (PBS) at 4 °C, then rinsed by chloroform: methanol (2: 1, vol/vol). Fat was further removed by blending bat and human tissues with three solvent mixtures, chloroform: methanol (2: 1, 1: 1 and 1: 2, vol/vol), each centrifuged, removed liquid, left overnight at room temperature, and defatted tissue is dried in a fume hood. Defatted sample were suspended (5–10 %, wt/vol) in water and proteolyzed at 55 °C with 10 % (wt/wt) of a nonspecific protease actinase E (20 mg/mL) 24 h, boiled to deactivate actinase E and then freeze-dried. The resulting material was made up to 8 M urea and 2% (wt/vol) 3-((3-cholamidopropyl)-dimethyl-ammonio)-1-propanesulfonate (CHAPS) buffer at pH 8.3 and any insoluble residue was removed by centrifugation. Vivapure Maxi Q strong anion exchange (SAX) spin columns (Sartorius, Goettingen, Germany) were equilibrated with 8 M urea and 2% CHAPS buffer and sample was loaded onto the Maxi Q columns and washed with 8 M urea and 2 % CHAPS followed by 200 mM aqueous NaCl. The GAGs were released by washing the column with 16 % (wt/vol) NaCl. The eluent was desalted using a 3-kDa molecular weight cut-off (MWCO) spin column (MilliporeSigma, MA, USA), the GAG sample was freeze-dried and stored frozen. About one-fifth of the recovered GAG sample was used for disaccharide analysis independently and about four-fifths of the recovered GAGs were combined as bat or human groups for HS purification.
For disaccharide compositional analyses, the digestion was performed in a 3-kDa MWCO spin column; 200 μL of digestion buffer (50 mM ammonium acetate containing 2 mM calcium chloride, pH 7.4) was added to the filter unit. 10 mU of recombinant heparin lyase I, II, III (Linhardt, Turnbull, Wang, Loganathan, & Gallagher, 1990) and recombinant chondroitin lyase ABC (Zhang et al., 2009) were added to each sample and mixed well. The samples were all placed in a water bath at 37 °C overnight, after which disaccharides were collected by centrifugation. Lyophilized disaccharides were AMAC-labeled by adding 10 μL of 0.1 M AMAC in dimethyl sulfate/acetic acid (17: 3, vol/vol) incubated at room temperature for 10 min, followed by adding 10 μL of 1 M aqueous sodium cyanoborohydride and incubating for 1 h at 45 °C. The known concentration unsaturated disaccharide standards were similarly AMAC-labeled and used for each run as an external standard. After the AMAC-labeling reaction, the samples were centrifuged, and each supernatant was recovered and analyzed by a modification of a previously published method (Bhattacharyya et al., 2018). High performance liquid chromatography (HPLC) was performed on an Agilent 1200 LC system at 45 °C using an Agilent Poroshell 120 ECC18 (2.7 μm, 3.0 × 30 mm) column. Mobile phase A (MPA) was 50 mM ammonium acetate aqueous solution and the mobile phase B (MPB) was methanol. The mobile phase passed through the column at a flow rate of 300 μL/min. The gradient was 0–10 min, 5–45 % B; 10–10.2 min, 45–100 % B; 10.2–14 min, 100 % B; 14–22 min, 100–5 % B. Injection volume was 2 μL. A triple-quadrupole mass spectrometry system equipped with an electrospray ionization (ESI) source (Thermo Fisher Scientific) was used as a detector. The on-line mass spectral (MS) analysis was at the MRM mode. MS parameters were as follows: negative ionization mode with a spray voltage of 3000 V, a vaporizer temperature of 300 °C, and a capillary temperature of 270 °C. The data analysis was performed through Xcalibur software.
For HS recovery, the bat or human lung GAGs were added to 300 mU of recombinant chondroitin lyase ABC and also performed in a 3-kDa MWCO spin column. After incubating at 37 °C for overnight, the digested CS and HA disaccharides were removed by centrifugation, and the remaining chondroitin lyase ABC was removed by boiling and precipitation.
2.5. Interaction between SARS-CoV-2 spike protein RBD and recovered HS
The interaction behavior of recovered HS with SARS-CoV-2 Spike protein RBD was measured using surface plasmon resonance (SPR) on a BIAcore 3000 system (GE Healthcare, Uppsala, Sweden) based on a similar publication (Yan et al., 2020). Commercial heparin was used as a positive control. Briefly, biotinylated heparin and recovered HS were immobilized to the streptavidin (SA) chip respectively based on the manufacturer's protocol. Successful immobilization was confirmed by the observation of an over 400-resonance unit (RU) increase in the sensor chip. The SARS-CoV-2 Spike protein RBD was re-suspended in HBS-EP buffer (0.01 M HEPES, 0.15 M NaCl, 3 mM EDTA, 0.005 % surfactant P20 (vol/vol), pH 7.4). Different dilutions of the SARS-CoV-2 Spike protein RBD were injected at a flow rate of 30 μL/min. At the end of injection, the same buffer was flowed over the sensor surface to facilitate dissociation. After dissociation, the sensor surface was regenerated by injecting 30 μL of 2 M NaCl. The response was monitored as a function of time (sensorgram) at 25 °C.
2.6. Competition studies on spike lentivirus pseudotype binding to HS
Competitive SPR experiments (Zhang, Datta, Dordick, & Linhardt, 2020), using recovered HS, were performed to compare their binding ability to SARS-CoV-2 spike lentivirus pseudotype. SARS-CoV-2 spike lentivirus pseudotype (2 × 103 virus units/mL) was separately pre-incubated with a gradient of concentrations of bat or human lung HS in HBS-EP buffer and then injected over the heparin chip at a flow rate of 30 μL/min. After each injection and association, regeneration was performed by injecting 30 μL of 2 M NaCl. The “Inhibition (%)” is calculated by the ratio of highest RU in sensorgram with or without additional HS.
2.7. Polyacrylamide gel electrophoresis (PAGE) analysis of recovered HS
Bat lung HS and human lung HS were analyzed by PAGE. Briefly, 10 μg samples were loaded onto a 15 % resolving gel containing 14.08 % (wt/vol) acrylamide, 0.92 % (wt/vol) N, N′-methylenebis-acrylamide and 5 % (wt/vol) sucrose. All monomer solutions were prepared in resolving buffer (0.1 M boric acid, 0.1 M Tris, 0.01 M disodium EDTA). A 5 % stacking gel was also prepared in resolving buffer and contained 4.75 % (wt/vol) acrylamide and 0.25 % (wt/vol) N, N′-methylene-bis-acrylamide. A 10 cm × 7 mm diameter resolving gel column was cast from 4 mL of 15 % resolving gel monomer solution mixed with 4 μL of tetramethylethylenediamine (TEMED) and 12 μL of 10 % ammonium persulfate (APS). A stacking gel was cast from 1 mL 5% stacking gel monomer solution mixed with 1 μL of TEMED and 30 μL of 10 % APS. Phenol red dye prepared in 50 % (wt/vol) sucrose was added to the sample for visualization of the ion front during electrophoresis. Bovine heparin oligosaccharides ladder (10 μg), dp10 (1 μg) and dp18 (1 μg) with known molecular weight were used as standard. The gels were stained with Alcian blue, digitized, and their molecule weights were determined as previously described (Edens et al., 1992). The gel, visualized with Alcian blue staining was digitized with UN-SCAN-IT (Silk Scientific).
3. Results and discussion
3.1. Structure and function of GAGs
GAGs are linear and anionic polysaccharides composed of repeating disaccharide units. Despite their simple saccharidic backbone, most GAGs have complex modification on their repeating disaccharide units, such as the presence and variable position of sulfo groups (Esko & Selleck, 2002; Gandhi & Mancera, 2008; Song, Zhang, & Linhardt, 2020). Three main families of GAGs are heparin / HS, CS and HA, and their chemical structures are shown in Fig. 2 .
Fig. 2.
Disaccharide structure of heparin, HS, CS (DS (CS-type B) also contains some iduronic acid residues) and HA, where X = H or SO3, Y = Ac, SO3 or H.
Heparin and HS are probably the most complex of the GAGs. HS is found within the cell, on the cell surface, or secreted into the extracellular matrix (ECM). HS has been recognized as key players in cell signaling, growth, adhesion, migration and host-pathogen interactions (Capila & Linhardt, 2002; Linhardt & Toida, 2004). The structures of heparin and HS are based on l-iduronic acid (IdoA) or d-glucuronic acid (GlcA) linked to N-sulfoglucosamine (GlcNS) or N-acetylglucosamine (GlcNAc) (Fig. 2). Both heparin and HS have variable repeating units with different levels of sulfo modification and heparin is more highly sulfated than HS. The major sequence in heparin is a trisulfated disaccharide unit of the structure →4) 2-O-sulfo-α-l-IdoA (1→4) 6-O-sulfo-α-d-GlcNS (1→, while the major sequence in heparan sulfate is a non-sulfated disaccharide unit of the structure →4) α-d-GlcA (1→4) GlcNAc (1→).
CS has been shown involved in various biological processes including morphogenesis, central nervous system development and signal transduction. CS is particularly abundant in bone, tendons, blood vessels, nerve tissue and cartilage, although it is also found on the cell surfaces and in the ECM. The structure of CS is based on GlcA linked to GalNAc. Different types of CS (CS A-E) are based on the sulfo group modifications and epimerization of the C5-position of the GlcA residue to an IdoA residue in their disaccharide units. CSA has a →4) β-d-GlcA (1→3) 4-O-sulfo-β-d-GalNAc (1→ repeating unit. CSB, also known as dermatan sulfate (DS), has a →4) α-l-IdoA (1→3) 4-O-sulfo-β-d-GalNAc (1→ repeating unit. CSC has a →4) β-d-GlcA (1→3) 6-O-sulfo-β-d-GalNAc (1→ repeating unit. CSD has a →4) 2-O-sulfo-β-d-GlcA (1→3) 6-O-sulfo-β-d-GalNAc (1→ repeating unit. CSE has a →4) β-d-GlcA (1→3) 4,6-di-O-sulfo-β-d-GalNAc (1→ repeating unit.
HA is the simplest GAG without sulfo groups. It is primarily found in connective tissue and is a prominent component of the ECM. The structure of HA is based on a single repeating unit, →4) β-d-GlcA (1→3) β- GlcNAc (1→.
3.2. Disaccharide composition of recovered GAGs
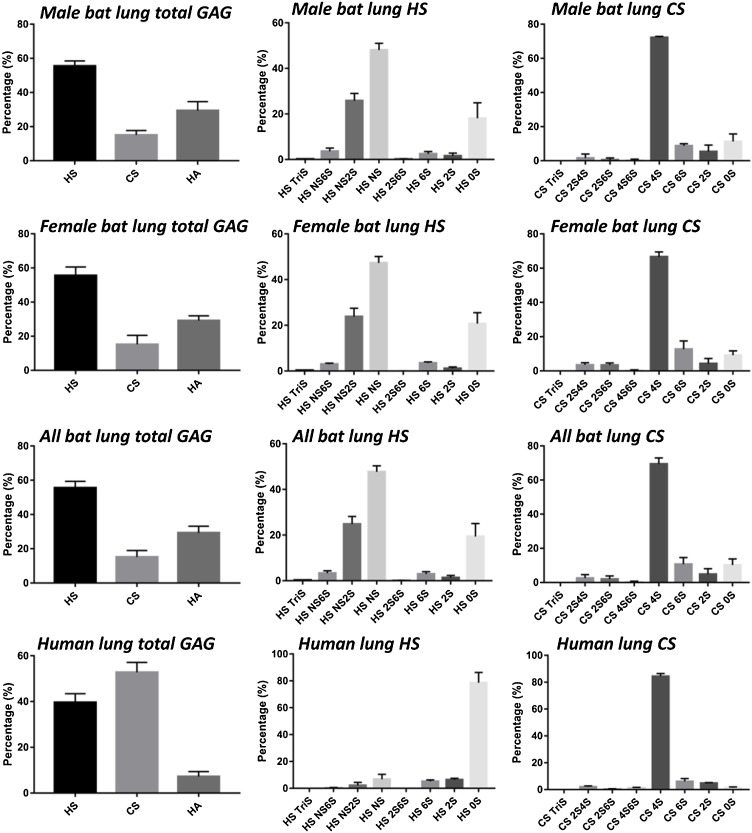
HS, CS and HA disaccharide repeating units can be quantitated by treating with polysaccharide lyases (Linhardt, 2001) followed by LC–MS analysis. The total GAG composition of bat lung (male, female, and combined male and female) and human lung (male) were determined and are shown in Fig. 3 . The result indicate that male bat lung and female bat lung contain very similar composition of total GAGs, corresponding to ∼56 % HS, ∼29 % HA and ∼15 % CS. However, human lung (male) exhibits a very different GAG composition than bat lung. Approximately half the GAG in human lung is CS, while HS corresponds to ∼40 % and only ∼7 % is the HA. Female human lungs were found to be similar to male lungs (data not shown). Furthermore, there was little difference in the bat lung GAG compositions between the male and female bats.
Fig. 3.
Composition of total GAGs, HS and CS, were derived from bat lungs (male, female and combined) and human lungs (male). All bat lung data were the average of those from male and female bats. Mean ± std. dev. is shown. N = 4 male bat lungs, 4 female bat lungs, and 6 male human lungs.
The HS and CS disaccharide composition of GAGs from bat and human lung are shown in Fig. 3 and Table 1 . Male and female bat lungs exhibited similar HS disaccharide composition which is not surprising as the HS disaccharide composition of male and female mouse lung HS are also similar (Warda et al., 2006). HS is primarily comprised of ∼48 % HS NS, ∼25 % HS NS 2S and ∼19 % HS 0S disaccharide units. In contrast, healthy human lung HS is mainly comprised of HS 0S, corresponding to ∼79 %. The HS disaccharide composition can be further assessed by calculating the percentage of disaccharides containing N-sulfo (NS) and N-acetyl (NAc) groups. The results demonstrate that both male and female bat lung HS had ∼76 % NS disaccharides with ∼24 % NAc disaccharides, however, human lung HS contains only ∼10 % NS disaccharides with ∼90 % NAc disaccharides. Finally, the total sulfation level (% of S/disaccharide) was very different when comparing bat lung HS and human lung HS. Bat lung HS had 1.1 sulfo groups/disaccharide repeating unit while less than a quarter of the disaccharides from human lung HS contained a sulfo group. The disaccharide composition of human lung HS is somewhat different than that reported by the detection method in the study of Clausen et al. (2020). These differences can be ascribed either to different analytical methods of biological variability associated with human tissues.
Table 1.
Composition of HS and CS from bat lung and human lung.
| HS TriS | HS NS6S | HS NS2S | HS NS | HS 2S6S | HS 6S | HS 2S | HS 0S | Total NS a | Total NAc b | Total sulfation c | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Male bat lung HS | 0.16 ± 0.10 % | 3.63 ± 1.41 % | 25.8 ± 3.19 % | 48.1 ± 2.94 % | 0.16 ± 0.16 % | 2.45 ± 1.06 % | 1.56 ± 1.24 % | 18.1 ± 6.82 % | 77.7 ± 6.47 % | 22.3th±5.11 % | 112 ± 15.1 % |
| Female bat lung HS | 0.34 ± 0.06 % | 3.02 ± 0.45 % | 23.8 ± 3.62 % | 47.3 ± 2.80 % | 0.03 ± 0.06 % | 3.53 ± 0.51 % | 1.15 ± 0.67 % | 20.8 ± 4.77 % | 74.5 ± 4.78 % | 25.5 ± 4.78 % | 107 ± 12.4 % |
| All bat lung HS | 0.25 ± 0.12 % | 3.32 ± 1.02 % | 24.8 ± 3.34 % | 47.7 ± 2.69 % | 0.09 ± 0.13 % | 2.99 ± 0.97 % | 1.35 ± 0.95 % | 19.4 ± 5.63 % | 76.1 ± 5.54 % | 23.9 ± 4.89 % | 109 ± 14.0 % |
| Human lung HS | 0.12 ± 0.21 % | 0.43 ± 0.55 % | 2.44 ± 2.37 % | 6.75 ± 3.82 % | 0.00 ± 0.00 % | 5.12 ± 0.97 % | 6.27 ± 1.11 % | 78.9 ± 7.74 % | 9.74 ± 6.81 % | 90.3 ± 6.81 % | 24.2 ± 12.4 % |
| CS TriS | CS 2S4S | CS 2S6S | CS 4S6S | CS 4S | CS 6S | CS 2S | CS 0S | / | / | Total sulfation d | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Male bat lung CS | 0.00 ± 0.00 % | 1.54 ± 2.37 % | 0.54 ± 1.08 % | 0.30 ± 0.59 % | 72.2 ± 0.68 % | 8.74 ± 1.22 % | 5.34 ± 3.81 % | 11.3 ± 4.41 % | / | / | 163 ± 13.9 % |
| Female bat lung CS | 0.00 ± 0.00 % | 3.53 ± 1.26 % | 3.44 ± 1.20 % | 0.21 ± 0.42 % | 66.7 ± 2.83 % | 12.7 ± 4.77 % | 4.28 ± 2.92 % | 9.15 ± 2.53 % | / | / | 164 ± 18.7 % |
| All bat lung CS | 0.00 ± 0.00 % | 2.54 ± 2.06 % | 1.99 ± 1.87 % | 0.25 ± 0.48 % | 69.4 ± 3.53 % | 10.7 ± 3.86 % | 4.81 ± 3.19 % | 10.2 ± 3.52 % | / | / | 164 ± 22.5 % |
| Human lung CS | 0.06 ± 0.04 % | 2.11 ± 0.58 % | 0.44 ± 0.08 % | 1.12 ± 0.52 % | 84.2 ± 1.76 % | 5.80 ± 1.82 % | 4.67 ± 0.12 % | 1.55 ± 0.49 % | / | / | 185 ± 7.41 % |
%S = 0×%0S + 1×%4S + 1×%6S + 1×%2S + 2×%2S4S + 2×%2S6S + 2×%4S6S + 3×%Tris. Values represent means ± standard deviations of 4 male bat lungs, 4 female bat lungs, and 6 male human lungs.
The total NS indicates all the HS disaccharides containing N-sulfo (NS) group.
The total NAc indicates all the HS disaccharides containing N-acetyl (NAc) group.
The total sulfation of HS (%S) is calculated by the following formula. %S = 0×%0S + 1×%NS + 1×%2S + 1×%6S + 2×%NS6S + 2×%NS2S + 2×%2S6S + 3×%Tris.
The total sulfation of CS (%S) is calculated by the following formula.
The composition of bat lung CS was next examined. Male and female bat lung CS showed similar disaccharide compositions and mainly (∼69 %) consisted of CS 4S disaccharide unit with the remainder consisting of CS 6S, CS 2S and CS 0S. Human lung CS was also composed mainly (∼84 %) of CS 4S with ∼6 % CS 6S and ∼5 % CS 2S disaccharide units. Thus, human lung and bat lung CS were similar. Additionally, the sulfation level of CS was greater than that of HS in both bat and human lungs. The average bat lung CS disaccharide had ∼1.6 sulfates and ∼1.9 sulfo groups/disaccharide were found in human lung CS. It should be noted that since our analysis relies on chondroitin lyase ABC we cannot distinguish CS type A from DS.
3.3. Recovered HS binding with SARS-CoV-2 spike protein RBD
Previous studies (Clausen et al., 2020; Jin et al., 2020; Kim et al., 2020; Kwon et al., 2020; Tandon et al., 2020; Tree et al., 2020) have demonstrated that HS and other sulfated polysaccharides effectively inhibit SARS-CoV-2 infection of cultured cells in vitro by binding the S-protein RBD. HS recovered from both male and female bats and male human lung tissues and from porcine intestinal heparin (positive control) were immobilized on a SA Biacore chip to compare binding affinity for SARS-CoV-2 spike protein RBD. The results of this direct binding study are shown in Fig. 4 and Table 2 . Heparin, as a positive control, exhibited a high binding affinity for SARS-CoV-2 spike protein RBD (KD = 105 nM). Bat lung HS and human lung HS both showed somewhat weaker binding affinities with SARS-CoV-2 spike protein RBD than did heparin with KD values of 703 nM and 194 nM, respectively. Human lung HS exhibited stronger binding affinity with SARS-CoV-2 spike protein RBD than did bat lung HS even though human lung HS had a lower sulfation level than bat lung HS. This is of interest as most GAG protein interactions results from ionic interactions between anionic sulfo groups and cationic amino acids (arginine and lysine) in the GAG-binding protein (Capila & Linhardt, 2002). Furthermore, the SARS-CoV-2 spike protein RBD contains Cardin-Weintraub motifs consistent with such ionic interactions (Cardin & Weintraub, 1989; Kim et al., 2020). The kinetic constants of these interactions presented in Table 2 clearly demonstrate a much weaker interaction with SARS-CoV-2 spike protein RBD for bat lung HS than for heparin and human lung HS.
Fig. 4.
SPR sensorgrams with fitted curve. Gradient concentrations of SARS-CoV-2 spike protein RBD interact with immobilized (A) heparin, (B) bat lung HS and (C) human lung HS.
Table 2.
Kinetic constants of interactions between SARS-CoV-2 spike protein RBD and immobilized heparin or heparan sulfate.
| ka (1/Ms)a | kd (1/s)a | KD (M) | |
|---|---|---|---|
| Heparin | 8.09 × 103 ± 151 | 8.49 × 104 ± 1.66 × 10−5 | 1.05 × 10−7 |
| Bat lung HS | 974 ± 43.1 | 6.85 × 104 ± 1.76 × 10−5 | 7.03 × 10−7 |
| Human lung HS | 5.71 × 103 ± 174 | 1.11 × 103 ± 2.20 × 10−5 | 1.94 × 10−7 |
The standard deviation was from the global fitting (n = 3).
3.4. Recovered HS binding with SARS-CoV-2 spike lentivirus pseudotype
An additional study of SARS-CoV-2 virus binding to bat lung HS and human lung HS was undertaken using a SARS-CoV-2 spike lentivirus pseudotype. An SPR competition experiment was used in this study and the results are shown in Fig. 5 . These studies indicate that SARS-CoV-2 spike lentivirus pseudotype could strongly bind to immobilized heparin resulting in an increase in response units (RU) in SPR sensorgram. Pre-incubation of SARS-CoV-2 spike lentivirus pseudotype with either bat lung HS or human lung HS reduced the RU in SPR sensorgram in a concentration-dependent fashion. Based on the inhibition curves the concentration value at which these two HS samples resulted in 50 % inhibition (IC50) was calculated. The IC50 for bat lung HS was 18.2 ± 2.3 μg/mL (± std. deviation, n = 3) and the IC50 for human lung HS was 10.6 ± 3.6 μg/mL at a SARS-CoV-2 spike lentivirus pseudotype concentration of 2 × 103 virus units/mL. These results clearly demonstrate that human lung HS has stronger binding ability with SARS-CoV-2 spike lentivirus pseudotype than does bat lung HS.
Fig. 5.
Competition of SARS-CoV-2 spike lentivirus pseudotype (2 × 103 virus units/mL) binding to immobilized heparin by gradient concentrations of (A) bat lung HS with (B) corresponding inhibition curve and (C) human lung HS with (D) corresponding inhibition curve (± std. deviations, n = 3). SARS-CoV-2 spike lentivirus pseudotype was pre-incubated with gradient concentrations of additional lung HS prior to interact with immobilized heparin. (E) Scheme of the SPR competition experiments.
3.5. Molecular weight analysis of recovered HS
The HS samples were next analyzed by PAGE using a 15 % polyacrylamide gel to characterize the size the bat lung HS and human lung HS chains (Fig. 6 , Table 3 ). The weight-average molecular weight (Mw) of bat lung HS was 6,969 Da which was much smaller than the 12,215 Da determined for the human lung HS. Indeed, most HS chains derived from animal tissues, such as 11,700 Da for porcine intestinal HS, have molecular weights above 10,000 Da.
Fig. 6.
A total of 15 % carbohydrate PAGE analysis of bat lung HS (lane 1) and human lung HS (lane 2). Bovine heparin oligosaccharide ladder, dp10 and dp 18 were used as standard.
Table 3.
Molecule information of bat lung HS and human lung HS.
| Mw | Mn | PDI | |
|---|---|---|---|
| Bat lung HS | 6,969 Da | 7,805 Da | 0.89 |
| Human lung HS | 12,215 Da | 9,674 Da | 1.26 |
In our previous studies on the interaction of SARS-CoV-2 spike protein RBD with heparin (Mw 18,000) and low molecular weight heparin (Mw 5,000), the low molecular weight heparin showed a 200-fold reduction in binding affinity by competition SPR and of inhibiting SARS-CoV-2 infection of Vero cells (Kim et al., 2020; Kwon et al., 2020). Moreover, a study on sulfated fucoidans (heparin analogs) showed a similar molecular weight dependence of the ability to inhibit viral infection (Kwon et al., 2020). The current study seems to suggest that chain size is more important than sulfation level for extracellular HS to bind with SARS-CoV-2 virus. We hypothesize that the short chain size bat lung HS might reduce the severity of lung infection by SARS-CoV-2 virus in bats simply by resulting in less virus binding and subsequent cell entry. In contrast, the longer chain size of human lung HS may promote lung infection in humans by SARS-CoV-2 virus. Additional studies will certainly be required to test this hypothesis. It is noteworthy that other viruses, which infect bats and humans, such as rabies (Kamhi et al., 2013), use heparan sulfate as co-receptor and chain size dependence should also be tested on such zoonotic pathogens. Moreover, future studies are planned assess chemokine binding to these heparan sulfate chains.
4. Conclusion
In the present study, we extracted and analyzed the total GAGs from bat lung and human lung tissues. The results indicate that male and female bat lungs are similar with respect to GAG composition and that bat lung total GAG is comprised of more HS than human lung total GAG. Further, the sulfation level of bat lung HS is much higher than that of human lung HS. However, SARS-CoV-2 spike protein RBD binds 3.5-times more strongly to human lung HS than bat lung HS. SPR competition studies also showed that human lung HS binds more strongly to a SARS-CoV-2 spike lentivirus pseudotype. Further analysis demonstrated that the molecular weight of human lung HS is 2-times of that of bat lung HS. Thus, we infer that molecular weight may be more important than sulfation level for lung HS binding to SARS-CoV-2 virus spike protein. In addition further studies will be required to understand the precise relationship between HS chain size (including minimum chain length), degree of sulfation, and structural motif (sulfate positions and uronic acid epimer and positions) required for binding to spike glycoprotein.
CRediT authorship contribution statement
Lufeng Yan: Methodology, Investigation, Writing - original draft. Yuefan Song: Methodology. Ke Xia: Methodology. Peng He: Methodology. Fuming Zhang: Conceptualization, Methodology, Investigation. Shiguo Chen: Writing - review & editing. Robert Pouliot: Methodology. Daniel J. Weiss: Investigation, Writing - review & editing. Ritesh Tandon: Methodology. John T. Bates: Investigation. Dallas R. Ederer: Investigation. Dipanwita Mitra: Investigation. Poonam Sharma: Investigation. April Davis: Investigation. Robert J. Linhardt: Conceptualization, Methodology, Writing - review & editing, Resources, Supervision, Project administration, Funding acquisition.
Declaration of Competing Interest
The authors report no declarations of interest.
Acknowledgements
This research was supported by grants from the National Institutes of Health Grant CA231074 (RJL) and DK111958 (RJL) and by WEISS18P0 (DJW), Cystic Fibrosis Foundation.
References
- Banerjee A., Kulcsar K., Misra V., Frieman M., Mossman K. Bats and coronaviruses. Viruses. 2019;11(1):41. doi: 10.3390/v11010041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya S., Feferman L., Han X., Ouyang Y., Zhang F., Linhardt R.J., et al. Decline in arylsulfatase B expression increases EGFR expression by inhibiting the protein-tyrosine phosphatase SHP2 and activating JNK in prostate cells. The Journal of Biological Chemistry. 2018;293(28):11076–11087. doi: 10.1074/jbc.RA117.001244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brook C.E., Boots M., Chandran K., Dobson A.P., Drosten C., Graham A.L., et al. Accelerated viral dynamics in bat cell lines, with implications for zoonotic emergence. eLife. 2020;9 doi: 10.7554/eLife.48401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunet-Rossinni A.K., Austad S.N. In: Handbook of the biology of aging. sixth edition. Masoro E.J., Austad S.N., editors. Academic Press; Burlington: 2005. Chapter 9 - Senescence in wild populations of mammals and birds; pp. 243–266. [Google Scholar]
- Capila I., Linhardt R.J. Heparin–Protein interactions. Angewandte Chemie International Edition. 2002;41(3):390–412. doi: 10.1002/1521-3773(20020201)41:3<390::aid-anie390>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- Cardin A.D., Weintraub H.J. Molecular modeling of protein-glycosaminoglycan interactions. Arteriosclerosis. 1989;9(1):21–32. doi: 10.1161/01.atv.9.1.21. [DOI] [PubMed] [Google Scholar]
- Clausen T.M., Sandoval D.R., Spliid C.B., Pihl J., Perrett H.R., Painter C.D., et al. SARS-CoV-2 infection depends on cellular heparan sulfate and ACE2. Cell. 2020;183(4):1043–1057. doi: 10.1016/j.cell.2020.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Oliveira Figueiredo P., Stoffella-Dutra A.G., Barbosa Costa G., Silva de Oliveira J., Dourado Amaral C., Duarte Santos J., et al. Re-emergence of yellow fever in Brazil during 2016-2019: Challenges, lessons learned, and perspectives. Viruses. 2020;12(11):1233. doi: 10.3390/v12111233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edens R.E., Al-Hakim A., Weiler J.M., Rethwisch D.G., Fareed J., Linhardt R.J. Gradient polyacrylamide gel electrophoresis for determination of molecular weights of heparin preparations and low-molecular-weight heparin derivatives. Journal of Pharmaceutical Sciences. 1992;81(8):823–827. doi: 10.1002/jps.2600810821. [DOI] [PubMed] [Google Scholar]
- Esko J.D., Selleck S.B. Order out of Chaos: Assembly of ligand binding sites in heparan sulfate. Annual Review of Biochemistry. 2002;71(1):435–471. doi: 10.1146/annurev.biochem.71.110601.135458. [DOI] [PubMed] [Google Scholar]
- Gandhi N.S., Mancera R.L. The structure of glycosaminoglycans and their interactions with proteins. Chemical Biology & Drug Design. 2008;72(6):455–482. doi: 10.1111/j.1747-0285.2008.00741.x. [DOI] [PubMed] [Google Scholar]
- Guito J.C., Prescott J.B., Arnold C.E., Amman B.R., Schuh A.J., Spengler J.R., et al. Asymptomatic infection of marburg virus reservoir bats is explained by a strategy of immunoprotective disease tolerance. Current Biology. 2021;31(2):257–270. doi: 10.1016/j.cub.2020.10.015. [DOI] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181(2):271–280. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin W., Zhang W., Mitra D., McCandless M.G., Sharma P., Tandon R., et al. The structure-activity relationship of the interactions of SARS-CoV-2 spike glycoproteins with glucuronomannan and sulfated galactofucan from Saccharina japonica. International Journal of Biological Macromolecules. 2020;163:1649–1658. doi: 10.1016/j.ijbiomac.2020.09.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamhi E., Joo E.J., Dordick J.S., Linhardt R.J. Glycosaminoglycans in infectious disease. Biological Reviews. 2013;88(4):928–943. doi: 10.1111/brv.12034. [DOI] [PubMed] [Google Scholar]
- Kim S.Y., Jin W., Sood A., Montgomery D.W., Grant O.C., Fuster M.M., et al. Characterization of heparin and severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2) spike glycoprotein binding interactions. Antiviral Research. 2020;181 doi: 10.1016/j.antiviral.2020.104873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar N., Sharma S., Barua S., Tripathi B.N., Rouse B.T. Virological and immunological outcomes of coinfections. Clinical Microbiology Reviews. 2018;31(4) doi: 10.1128/CMR.00111-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon P.S., Oh H., Kwon S.-J., Jin W., Zhang F., Fraser K., et al. Sulfated polysaccharides effectively inhibit SARS-CoV-2 in vitro. Cell Discovery. 2020;6(1):50. doi: 10.1038/s41421-020-00192-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang J., Yang N., Deng J., Liu K., Yang P., Zhang G., et al. Inhibition of SARS pseudovirus cell entry by lactoferrin binding to heparan sulfate proteoglycans. PloS One. 2011;6(8) doi: 10.1371/journal.pone.0023710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Zhang C., Sui J., Kuhn J.H., Moore M.J., Luo S., et al. Receptor and viral determinants of SARS-coronavirus adaptation to human ACE2. The EMBO Journal. 2005;24(8):1634–1643. doi: 10.1038/sj.emboj.7600640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linhardt R.J. Analysis of glycosaminoglycans with polysaccharide lyases. Current Protocols in Molecular Biology. 2001 doi: 10.1002/0471142727.mb1713bs48. Chapter 17, Unit17.13B. [DOI] [PubMed] [Google Scholar]
- Linhardt R.J., Toida T. Role of glycosaminoglycans in cellular communication. ChemInform. 2004;35(39) doi: 10.1021/ar030138x. [DOI] [PubMed] [Google Scholar]
- Linhardt R.J., Turnbull J.E., Wang H.M., Loganathan D., Gallagher J.T. Examination of the substrate specificity of heparin and heparan sulfate lyases. Biochemistry. 1990;29(10):2611–2617. doi: 10.1021/bi00462a026. [DOI] [PubMed] [Google Scholar]
- Maginnis M.S. Virus-receptor interactions: The key to cellular invasion. Journal of Molecular Biology. 2018;430(17):2590–2611. doi: 10.1016/j.jmb.2018.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olival K.J., Hosseini P.R., Zambrana-Torrelio C., Ross N., Bogich T.L., Daszak P. Host and viral traits predict zoonotic spillover from mammals. Nature. 2017;546(7660):646–650. doi: 10.1038/nature22975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlovich S.S., Lovett S.P., Koroleva G., Guito J.C., Arnold C.E., Nagle E.R., et al. The egyptian rousette genome reveals unexpected features of bat antiviral immunity. Cell. 2018;173(5):1098–1110. doi: 10.1016/j.cell.2018.03.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price J.L. Serological evidence of infection of Tacaribe virus and arboviruses in Trinidadian bats. The American Journal of Tropical Medicine and Hygiene. 1978;27(1 Pt 1):162–167. doi: 10.4269/ajtmh.1978.27.162. [DOI] [PubMed] [Google Scholar]
- Raj V.S., Mou H., Smits S.L., Dekkers D.H.W., Müller M.A., Dijkman R., et al. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature. 2013;495(7440):251–254. doi: 10.1038/nature12005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahin U., Muik A., Derhovanessian E., Vogler I., Kranz L.M., Vormehr M., et al. COVID-19 vaccine BNT162b1 elicits human antibody and TH1 T cell responses. Nature. 2020;586(7830):594–599. doi: 10.1038/s41586-020-2814-7. [DOI] [PubMed] [Google Scholar]
- Schountz T., Baker M.L., Butler J., Munster V. Immunological control of viral infections in bats and the emergence of viruses highly pathogenic to humans. Frontiers in Immunology. 2017;8:1098. doi: 10.3389/fimmu.2017.01098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y., Zhang F., Linhardt R.J. Analysis of the glycosaminoglycan chains of proteoglycans. Journal of Histochemistry and Cytochemistry. 2020 doi: 10.1369/0022155420937154. 0022155420937154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suga N., Shimozawa T. Site of neural attenuation of responses to self-vocalized sounds in echolocating bats. Science. 1974;183(4130):1211. doi: 10.1126/science.183.4130.1211. [DOI] [PubMed] [Google Scholar]
- Tandon R., Sharp J.S., Zhang F., Pomin V.H., Ashpole N.M., Mitra D., et al. Effective inhibition of SARS-CoV-2 entry by heparin and enoxaparin derivatives. Journal of Virology, JVI. 2020 doi: 10.1128/JVI.01987-20. 01987-01920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor S.L., McGuckin M.A., Wesselingh S., Rogers G.B. Infection’s sweet tooth: How glycans mediate infection and disease susceptibility. Trends in Microbiology. 2018;26(2):92–101. doi: 10.1016/j.tim.2017.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tree J.A., Turnbull J.E., Buttigieg K.R., Elmore M.J., Coombes N., Hogwood J., et al. Unfractionated heparin inhibits live wild-type SARS-CoV-2 cell infectivity at therapeutically relevant concentrations. British Journal of Pharmacology. 2020 doi: 10.1111/bph.15304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhl F.E., Zhang F., Pouliot R.A., Uriarte J.J., Rolandsson Enes S., Han X., et al. Functional role of glycosaminoglycans in decellularized lung extracellular matrix. Acta Biomaterialia. 2020;102:231–246. doi: 10.1016/j.actbio.2019.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warda M., Toida T., Zhang F., Sun P., Munoz E., Xie J., et al. Isolation and characterization of heparan sulfate from various murine tissues. Glycoconjugate Journal. 2006;23(7–8):555–563. doi: 10.1007/s10719-006-7668-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter Y., López J., von Helversen O. Ultraviolet vision in a bat. Nature. 2003;425(6958):612–614. doi: 10.1038/nature01971. [DOI] [PubMed] [Google Scholar]
- Yan L., Xia K., Yu Y., Miliakos A., Chaturvedi S., Zhang F., et al. Unique cell surface mannan of yeast pathogen Candida auris with selective binding to IgG. ACS Infectious Diseases. 2020;6(5):1018–1031. doi: 10.1021/acsinfecdis.9b00450. [DOI] [PubMed] [Google Scholar]
- Zhang F., Datta P., Dordick J.S., Linhardt R.J. Evaluating heparin products for heparin-induced thrombocytopenia using surface plasmon resonance. Journal of Pharmaceutical Sciences. 2020;109(2):975–980. doi: 10.1016/j.xphs.2019.10.040. [DOI] [PubMed] [Google Scholar]
- Zhang Z., Park Y., Kemp M.M., Zhao W., Im A.R., Shaya D., et al. Liquid chromatography-mass spectrometry to study chondroitin lyase action pattern. Analytical Biochemistry. 2009;385(1):57–64. doi: 10.1016/j.ab.2008.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]