Abstract
The field of nanotechnology and personalised medicine is undergoing drastic changes in the approach and efficiency of experimentation. The COVID-19 pandemic has spiralled into mass stagnation of major laboratories around the globe and led to increased investment into remote systems for nanoparticle experiments. A significant number of laboratories now operate using automated systems; however, the extension to nanoparticle preparation and artificial intelligence–dependent databases holds great translational promise. The strive to combine automation with artificial intelligence (AI) grants the ability to optimise targeted therapeutic nanoparticles for unique cell types and patients. In this perspective, the current and future trends of automated approaches to nanomedicine synthesis are discussed and compared with traditional methods.
Graphical abstract
Keywords: Nanotechnology, Artificial intelligence, Automation, Personalised medicine, Microfluidics
Effect of the global pandemic on science
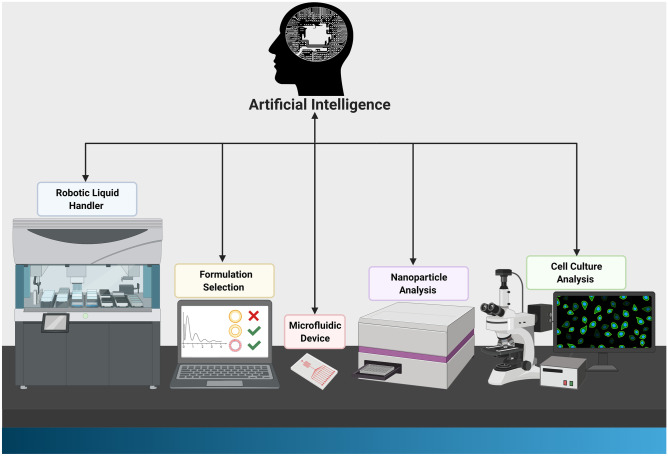
The detrimental effect of the global pandemic has revolutionised experimental methods and academic learning in all sciences. For the first time, hands-on experimental applications were brought to a standstill due to the inability to access laboratories. Furthermore, 1.3 billion school and university students in 186 countries cannot attend classrooms and are currently pursuing education online [1]. However, while this shift allowed students to learn from any point on the globe, the main complication arises when students are not able to enter laboratories and acquire experience in performing experiments. The recent demand for remote experimentation using robotic appliances can potentially establish a working model for students and scientists despite the global pandemic. Nanoparticle (NP) synthesis is typically carried out using manual manipulations and methods requiring more staff, resources and time. A fully functional robotic system will allow overcoming these hurdles by creating a ‘closed-loop’ for NP synthesis, immediate analysis and optimisation (Fig. 1). By integrating AI into software-operated lab machinery and establishing a remote access to a computer, anyone from any part of the world will be able to participate in experimental procedures and gain valuable research skills. With this, however, comes a separate responsibility of maintaining the integrity of the computational system, protecting the users’ Intellectual Property (IP) and reducing the threat of unauthorised network access. Mitigation of network security breaches require multiple network protection measures, such as strengthened user access control, up-to-date and standardised software, two-factor authentication, firewall access restrictions, enabled network level authentication and employee network security protocol training [2].
Fig. 1.
Schematic diagram of an AI-automated workstation. A functional AI-guided robotic nanoparticle synthesis and analysis system will start with genetic analysis of individual patients and personalised treatment selection. The nanoparticle will then be formulated and tested in cells and organs-on-chip using automated systems. The AI system will ultimately be able to determine the best formulation and treatment for individual patients for maximum response and optimal outcome
Nanotechnology in drug and siRNA delivery
Implementation of NPs dates back to the 1970s when nanoscale liposomes were loaded with medication and delivered to diseased cells [3–7]. The application of nanotechnology to numerous fields is becoming increasingly realised, especially in personalised medicine [8–10]. NPs grant the ability to overcome biological barriers and allow effective delivery of drugs and other compounds whilst promising to preferentially target drugs to specific biomarkers in individual patients [11–15]. However, the formulation of NPs comes with numerous challenges [16]. The properties of NPs are determined by several key characteristics, including size, surface morphology and charge, chemistry and drug release profile [17–19]. Subtle changes in the formulation process and composition can alter the properties of the nanomedicine leading to unwanted consequences and hurdles in reproducing experiments [18–21]. Hence, having better control of the stages involved in manufacturing NPs is crucial. Aside from the delivery of small-molecule drugs, NPs are increasingly being used in the delivery of biologics, including small interfering RNA (siRNA) and proteins [22–29]. NPs help to overcome the challenge of biological instability and poor cellular penetration of genetic payloads [30–32]. The applications of siRNA-nanoparticle complexes encompass numerous diseases like cancer and viral infections [33–37]. There are various siRNA and drug combinations complexed with different compositions of lipid NPs (LNPs) and other NPs [38–40]. Therefore, careful selection and formulation of particles is key in the complexation step. Establishing a system that screens for optimal nanoparticle preparation will pave the way for more efficient siRNA and drug delivery and its implementation as personalised medicine tool.
Microfluidics and automated synthesis of nanoparticles
Microfluidics, the study and manipulation of fluids at micro- and nanoscale levels, allows improved control of particle characteristics and ensuring that variation between synthesised batches is reduced [41]. Due to these benefits, it is becoming increasingly used within laboratories. Microfluidic chips are a type of microfluidic device that allows for tiny amounts of liquids containing particles to be processed and visualised [42]. The chips are a combination of micro-pumps, micro-valves, micro-mixers, micro-separators and micro-sized channels (diameters ranging from 1 to 1000µm) [42, 43]. The pumps move liquid within the channels in the chip at micro-level flow rates, allowing control over physical or chemical reactions. The liquids may contain either cells or nanoparticles. These microfluidic devices have opened an extensive range of research possibilities that were previously unachievable. For example, microfluidic chips were used to study the migration of lung cancer cells under different cancer invasion microenvironments [44].
Notably, research and industrial operations have established efficient ways in utilising microfluidics for large-scale NP production [45–51]. Currently, microfluidics is most commonly used for the synthesis of LNPs for RNA/DNA delivery [52, 53]. However, the production of LNPs, using microfluidics, is still being further refined [54, 55]. For example, investigation into utilising a microfluidic approach to produce droplet-stabilised giant unilamellar vesicles (dsGUVs) has shown promising results compared with conventional methods of synthetic cell synthesis [56]. In addition, shaping NPs’ configuration, using microfluidic techniques, is gaining traction and providing a deeper understanding of NP synthesis and physiochemical assembly properties of the molecular building blocks [39, 57]. For example, PRINT, a nanofabrication technique, granted the ability to generate particles with full control of the size, shape and surface chemistry, which affects their cellular internalisation and intracellular trafficking [58]. Achieving particle synthesis automation by combining a robotic liquid handler with microfluidic devices holds great promise for the future of laboratories and industries.
Liquid handling and robotics
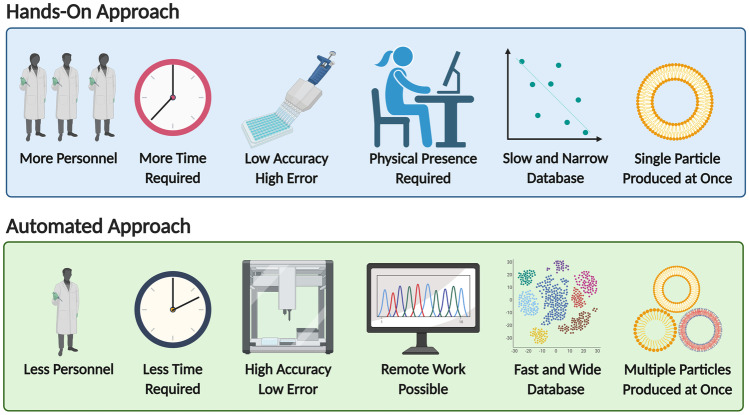
Automation has become a focal point for biomedical research. An essential part of any nanotechnology lab is the process of liquid handling. Historically, liquid handling has been a manual task whereby the user controlled liquid reagents through manual pipetting. When considering manual tasks in practical experimentation, it is often associated with human error and time and space limitation [59]. A monotonous task is an example of when a human error can occur, leading to a wide range of errors [60]. In many scenarios when unexpected results are received, the first question is whether there were sample and fluid handling errors [61]. A study performed by Schwarze et al. [62] showed that 15% of the cost related to genome sequencing is due to laboratory personnel errors. In efforts to reduce human errors, robotic liquid handlers (LiHas) are becoming increasingly utilised. Robotic LiHas can perform simple human functions such as aspiration and dispensing of liquids into different tubes and wells, whilst maintaining high precision and accuracy [63]. However, the range of robotic LiHa functions can be expanded into a multifunctional workstation for chemical and nanoparticle synthesis. Expansion of the functions can be achieved through the integration of robotic arms and various laboratory devices, such as centrifuges, microplate wasters and readers, heat sealers, heaters and shakers, bar code readers, storage devices and incubators [63]. Thus, robotic LiHas can be optimised for numerous techniques, including ELISA, PCR, genomic research, thin layer chromatography (TLC) spotting, solid-phase extraction (SPE), liquid-liquid extraction, nucleic acid preparation and cell-nanoparticle tests [64, 65]. Furthermore, there are dual robot arm liquid handlers that can emulate similar movements of a human, thus giving it a high number of degrees of freedom. In addition, acting as a 1:1 representation of a manual, human-operated, experiment, while in reality being an automated procedure [66]. During the current pandemic, robotic LiHas provide advantages and improved safety when it relates to testing of SARS-CoV-2 (COVID-19) [67]. Reducing the exposure of the laboratory technicians to COVID-19 samples is an excellent display of a programmable LiHa advantage. Such systems provide consistent performance, increased throughput and improved accuracy and precision, whilst reducing human error. The automatisation of NP synthesis using the LiHa possesses significant advantages, in comparison with the manual preparation methods. Robotic systems allow preparation of multiple formulations in a short period of time, characterising their activity on a biological specimen and optimising the nanoparticle formulation in an automated manner (Fig. 3).
Fig. 3.
Graphical comparison of the hands-on and automated approaches to experimentation
Automated microscopy
A crucial part of establishing an automated nanotechnology laboratory resides in the application of nanomedicines to cells, to test particle uptake and their effect on cell morphology and viability. In particular, automated confocal microscopes containing incubators allow live-cell imaging and characterisation of fluorescent components [68]. Despite the high accuracy and precision, manual confocal fluorescence microscopy is a time-consuming task when analysing hundreds of samples [69]. The need for automated microscopy has increased when the possibility of measuring a 384-well-plate in a single field-of-view in only several minutes became a reality [70]. In addition, automated microscopes are capable of producing high-quality 3D images with higher acquisition rate and much wider fields of view allowing encompassing the whole microplate instead of focusing only on individual wells [71]. This allows users to observe minuscule structures within the cell and on the cell’s surface [72, 73]. Incorporating a fluorescent microscope into a robotic workstation can improve the efficiency of acquired results and determination of nanoparticle trafficking within, and effect on, each cell [74]. Features such as wide field of view camera and laser autofocus improve quality and speed of image acquisition and enable imaging even in non-glass–bottom plates [73]. The advantage of being able to work with live cells can further improve experimentation in terms of multiple treatments and recording changes in cells over time. Furthermore, rapid automated analysis of collected data following image acquisition can be a potential improvement of current systems. Acquiring large datasets of microscopic images and incorporating machine learning into biotechnological analysis allows for the maintenance and improvement of existing databases and subsequently applying the knowledge to many disease models and personalised approach to medicine [75].
Artificial intelligence in research
Nanomedicine offers new ways for preventing and treating diseases. However, reaching the full potential of nanomedicine is still yet to be fully realised [76]. The use of automation is a step in the right direction of manufacturing nanoscale drugs but is only one piece of the puzzle. In order to improve and ensure that nanomedicine will achieve its desired result, computational analysis of large amounts of data must take place [77]. Therefore, the next step is integrating AI and machine learning into the evaluation and formulation of nanoscale drugs [78, 79]. While traditional computational methods require deep understanding of the physical, chemical and biological knowledge to construct relevant and accurate computational models [77], AI algorithms require only training datasets that can be produced by automated synthesis practices, or sourced from the literature [80, 81]. Providing large datasets of experimental results related to the subject of study allows the algorithm to produce accurate prediction models that then can be translated into improved nano-formulations.
There are multiple areas where machine learning can be integrated into nanomedicine applications [77, 82, 83]. For example, machine learning can be used to improve the understanding of how the structure of a nanoparticle affects its characteristics as well as its interaction with targeting tissues and cells. A study, using machine learning, of the adverse effects of nanoparticle properties conducted by Puzyn et al. predicted cytotoxicity of 17 different metal oxide NPs to Escherichia coli [82]. Alternatively, the AI subfield can help determine the correlation between drug dosage and therapeutic outcomes. A recent study obtained gene expression profiles from 82 breast cancer patients and trained a machine learning algorithm to predict complete pathologic responses with an accuracy of 92% [84]. It should also be noted that there are algorithm approaches that can be used when there is not enough data available to train the algorithm. However, it is imperative that the algorithm choice is appropriate for the experiment. An example of utilising a machine learning algorithm that does not require large sets of data is artificial neural network (ANN) [85]. ANN was used to predict optimal size and drug release in a recent study conducted by Baghaei, B. et al. about polylactic-co-glycolic acid (PLGA) NPs [85]. The machine learning algorithm reduced the prediction error from 28.0 to 2.93% and from 19.4 to 2.99%, for particle size and initial burst release of PLGA NPs, respectively [85]. The study shows that ANN integration into NP drug release experimentation provides an improved alternative to traditional computational measurements. However, to further advance the field, pathologists and AI experts need to work closely together to produce improved AI systems with accurate outcomes using automated robotic systems for particle synthesis. Integrating AI-driven data analysis and processing will allow faster and cheaper drug discovery, screening and application in both laboratories and industry [86].
The future of remote science and AI in nanotechnology automation
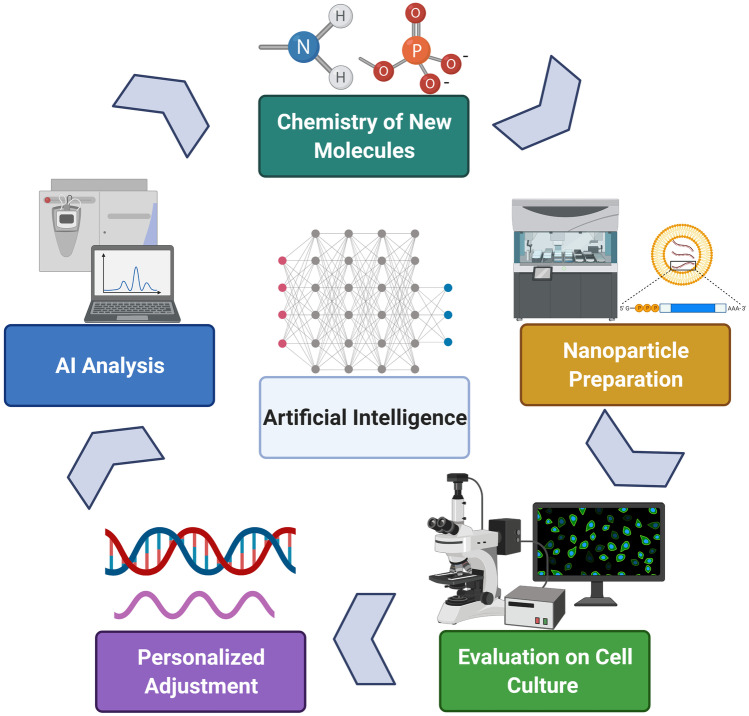
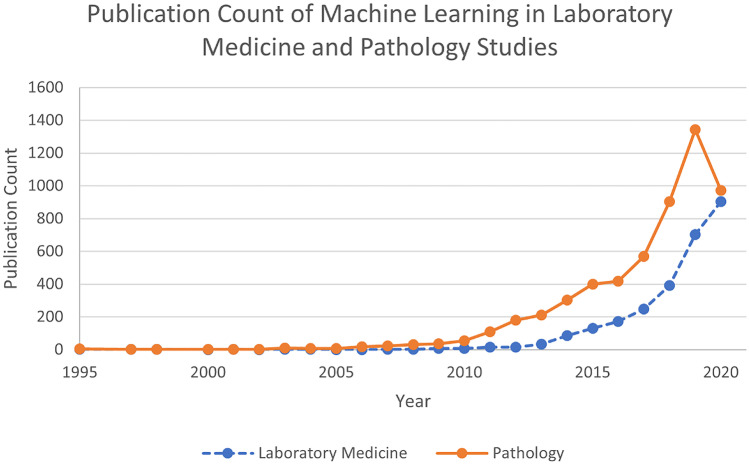
AI is integrating into clinical and laboratory research, as seen in Fig. 2. However, various barriers are affecting the development and adoption rate of machine learning in laboratories. One example is the limitation of sufficient datasets to inform machine learning results. To rectify, this will require changing the structure of chemical laboratories, opening them to students and experts from numerous fields. These new team members will work alongside chemists, biologists, bio-chemical engineers and biotechnology engineers (and others) to achieve optimal performance, relevance and results. Overall, the automated approach of nanoparticle preparation and utilisation holds great promise to create an effective method of conducting research and improving the field of personalised medicine, as seen on Fig. 3 that describes key differences between the approaches.
Fig. 2.
In the field of pathology, AI is already making major changes. Similar impact of AI can occur in the field of nanomedicine synthesis and prediction. Graphical representation of PubMed results based on two search queries—[“Machine Learning” AND “Laboratory Medicine” and “Machine Learning” AND “Pathology”]
Recent modifications of nanotechnology are rapidly evolving and adopting novel technological formats using machine learning and integrated analytical systems. Artificial intelligence demonstrates significant potential in “closing the loop” of nanoparticle synthesis, characterisation, refinement and testing or predicting activity in vitro and in vivo. The journey initiated from experimental laboratories is still lined with multiple challenges and obstacles along the way. However, recent studies and advancements on AI integration, automation and nanotechnology glimmer with great confidence, indicating that we are heading into the right direction.
Acknowledgements
Graphical abstract, Figs. 1 and 3 were created with BioRender.com
Author contribution
Egor Egorov and Calvin Pieters wrote the manuscript, reviewed literature and constructed the figures. Manuscript revision by Jeny Shklover and Hila Korach-Rechtman. Conceptualisation, supervision and funding acquisition by Avi Schroeder. All the authors commented on previous versions of the manuscript. All the authors read and approved the final manuscript.
Funding
This project has received funding from the European Union’s Horizon 2020 research and innovation programme under grant agreement No 680242-ERC-[Next-Generation Personalized Diagnostic Nanotechnologies for Predicting Response to Cancer Medicine]. Technion Integrated Cancer Center (TICC), the Russell Berrie Nanotechnology Institute, the Lorry I. Lokey Interdisciplinary Center for Life Sciences & Engineering, The Israel Ministry of Economy for a Kamin Grant (52752, 69230); the Israel Ministry of Science Technology and Space – Office of the Chief Scientist (3-11878); Israel Innovation Authority for Nofar Grant (67967), the Israel Science Foundation (1778/13, 1421/17); the Israel Ministry of Science & Technology (3-16963, 3-17418); the Israel Cancer Association (2015-0116); Leventhal 2020 COVID19 Research Fund (ATS #11947), the German-Israeli Foundation for Scientific Research and Development for a GIF Young grant (I-2328-1139.10/2012); the European Union FP-7 IRG Program for a Career Integration Grant (908049); the Phospholipid Research Center Grant (ASC-2018-062/1-1); the Louis family Cancer Research Fund, a Mallat Family Foundation Grant; The Unger Family Fund; a Carrie Rosenblatt Cancer Research Fund, A. Schroeder acknowledges Alon and Taub Fellowships. Calvin Pieters acknowledges the Sherman fellowship.
Declarations
Consent for publication
The authors give consent for the publication of the perspective.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Egor Egorov and Calvin Pieters are equally contributed to this publication
References
- 1.Unesco. 1.3 billion learners are still affected by school or university closures, as educational institutions start reopening around the world, says UNESCO. UNESCO. 2020.
- 2.Commission FT: securing remote access to your network. https://www.ftc.gov/tips-advice/business-center/small-businesses/cybersecurity/securing-remote-access-to-your-network (2019). Accessed 23 Jan 2021.
- 3.Hrkach J, Langer R. From micro to nano: evolution and impact of drug delivery in treating disease. Drug Deliv Transl Res. 2020;10(3):567–570. doi: 10.1007/s13346-020-00769-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ventola CL. Progress in nanomedicine: approved and investigational nanodrugs. Pharmacol Ther. 2017;42(12):742–755. [PMC free article] [PubMed] [Google Scholar]
- 5.Mg K, Krenn V, F H. History and possible uses of nanomedicine based on nanoparticles and nanotechnological progress. J Nanosci Nanotechnol. 2015;06. 10.4172/2157-7439.1000336.
- 6.Marty JJ, Oppenheim RC, Speiser P. Nanoparticles - a new colloidal drug delivery system. 1978. [PubMed]
- 7.New RRC, Chance ML, Thomas SC, Peters W. Antileishmanial activity of antimonials entrapped in liposomes. Nature. 1978;272(5648):55–56. doi: 10.1038/272055a0. [DOI] [PubMed] [Google Scholar]
- 8.Stark WJ. Nanoparticles in biological systems. Angew Chem Int Ed. 2011;50(6):1242–1258. doi: 10.1002/anie.200906684. [DOI] [PubMed] [Google Scholar]
- 9.Farokhzad OC, Jon S, Khademhosseini A. Tran T-NT, LaVan DA, Langer R. Nanoparticle-aptamer bioconjugates cancer research. 2004;64(21):7668. doi: 10.1158/0008-5472.CAN-04-2550. [DOI] [PubMed] [Google Scholar]
- 10.Anselmo AC, Mitragotri S. Nanoparticles in the clinic. Bioengineering & Translational Medicine. 2016;1(1):10–29. doi: 10.1002/btm2.10003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li S-D, Huang L. Pharmacokinetics and biodistribution of nanoparticles. Mol Pharm. 2008;5(4):496–504. doi: 10.1021/mp800049w. [DOI] [PubMed] [Google Scholar]
- 12.Prabhakar U, Maeda H, Jain RK, Sevick-Muraca EM, Zamboni W, Farokhzad OC, et al. Challenges and key considerations of the enhanced permeability and retention effect for nanomedicine drug delivery in oncology. Can Res. 2013;73(8):2412. doi: 10.1158/0008-5472.CAN-12-4561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Henry N, Clouet J, Le Visage C, Weiss P, Gautron E, Renard D, et al. Silica nanofibers as a new drug delivery system: a study of the protein–silica interactions. J Mater Chem B. 2017;5(16):2908–2920. doi: 10.1039/C7TB00332C. [DOI] [PubMed] [Google Scholar]
- 14.Zhao Z, Ukidve A, Krishnan V, Fehnel A, Pan DC, Gao Y, et al. Systemic tumour suppression via the preferential accumulation of erythrocyte-anchored chemokine-encapsulating nanoparticles in lung metastases. Nat Biomed Eng. 2020 doi: 10.1038/s41551-020-00644-2. [DOI] [PubMed] [Google Scholar]
- 15.Tadros AR, Romanyuk A, Miller IC, Santiago A, Noel RK, O'Farrell L, et al. STAR particles for enhanced topical drug and vaccine delivery. Nat Med. 2020;26(3):341–347. doi: 10.1038/s41591-020-0787-6. [DOI] [PubMed] [Google Scholar]
- 16.Suh J, Dawson M, Hanes J. Real-time multiple-particle tracking: applications to drug and gene delivery. Adv Drug Deliv Rev. 2005;57(1):63–78. doi: 10.1016/j.addr.2004.06.001. [DOI] [PubMed] [Google Scholar]
- 17.Timko BP, Arruebo M, Shankarappa SA, McAlvin JB, Okonkwo OS, Mizrahi B, et al. Near-infrared-actuated devices for remotely controlled drug delivery. Proc Natl Acad Sci USA. 2014;111(4):1349–1354. doi: 10.1073/pnas.1322651111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Santos HA, Riikonen J, Salonen J, Mäkilä E, Heikkilä T, Laaksonen T, et al. In vitro cytotoxicity of porous silicon microparticles: effect of the particle concentration, surface chemistry and size. Acta Biomater. 2010;6(7):2721–2731. doi: 10.1016/j.actbio.2009.12.043. [DOI] [PubMed] [Google Scholar]
- 19.Gref R, Minamitake Y, Peracchia MT, Trubetskoy V, Torchilin V, Langer R. Biodegradable long-circulating polymeric nanospheres. Science. 1994;263(5153):1600–1603. doi: 10.1126/science.8128245. [DOI] [PubMed] [Google Scholar]
- 20.Zylberberg C, Matosevic S. Pharmaceutical liposomal drug delivery: a review of new delivery systems and a look at the regulatory landscape. Drug Delivery. 2016;23(9):3319–3329. doi: 10.1080/10717544.2016.1177136. [DOI] [PubMed] [Google Scholar]
- 21.Leong HS, Butler KS, Brinker CJ, Azzawi M, Conlan S, Dufes C, et al. On the issue of transparency and reproducibility in nanomedicine. Nat Nanotechnol. 2019;14(7):629–635. doi: 10.1038/s41565-019-0496-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schulz A, Jaksch S, Schubel R, Wegener E, Di Z, Han Y, et al. Drug-induced morphology switch in drug delivery systems based on poly(2-oxazoline)s. ACS Nano. 2014;8(3):2686–2696. doi: 10.1021/nn406388t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yin H, Song C-Q, Dorkin JR, Zhu LJ, Li Y, Wu Q, et al. Therapeutic genome editing by combined viral and non-viral delivery of CRISPR system components in vivo. Nat Biotechnol. 2016;34(3):328–333. doi: 10.1038/nbt.3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Akinc A, Zumbuehl A, Goldberg M, Leshchiner ES, Busini V, Hossain N, et al. A combinatorial library of lipid-like materials for delivery of RNAi therapeutics. Nat Biotechnol. 2008;26(5):561–569. doi: 10.1038/nbt1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schiffelers RM, Ansari A, Xu J, Zhou Q, Tang Q, Storm G, et al. Cancer siRNA therapy by tumor selective delivery with ligand-targeted sterically stabilized nanoparticle. Nucleic Acids Res. 2004;32(19):e149-e. 10.1093/nar/gnh140. [DOI] [PMC free article] [PubMed]
- 26.Samaridou E, Walgrave H, Salta E, Alvarez DM, Castro-Lopez V, Loza M, et al. Nose-to-brain delivery of enveloped RNA - cell permeating peptide nanocomplexes for the treatment of neurodegenerative diseases. Biomaterials. 2020;230:119657. doi: 10.1016/j.biomaterials.2019.119657. [DOI] [PubMed] [Google Scholar]
- 27.Bohr A, Tsapis N, Foged C, Andreana I, Yang M, Fattal E. Treatment of acute lung inflammation by pulmonary delivery of anti-TNF-alpha siRNA with PAMAM dendrimers in a murine model. Eur J Pharm Biopharm. 2020;156:114–120. doi: 10.1016/j.ejpb.2020.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Conniot J, Scomparin A, Peres C, Yeini E, Pozzi S, Matos AI, et al. Immunization with mannosylated nanovaccines and inhibition of the immune-suppressing microenvironment sensitizes melanoma to immune checkpoint modulators. Nat Nanotechnol. 2019;14(9):891–901. doi: 10.1038/s41565-019-0512-0. [DOI] [PubMed] [Google Scholar]
- 29.Zinger A, Koren L, Adir O, Poley M, Alyan M, Yaari Z, et al. Collagenase nanoparticles enhance the penetration of drugs into pancreatic tumors. ACS Nano. 2019;13(10):11008–11021. doi: 10.1021/acsnano.9b02395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sahay G, Alakhova DY, Kabanov AV. Endocytosis of nanomedicines. J Control Release. 2010;145(3):182–195. doi: 10.1016/j.jconrel.2010.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kanasty R, Dorkin JR, Vegas A, Anderson D. Delivery materials for siRNA therapeutics. Nat Mater. 2013;12(11):967–977. doi: 10.1038/nmat3765. [DOI] [PubMed] [Google Scholar]
- 32.Tatiparti K, Sau S, Kashaw SK, Iyer AK. siRNA Delivery strategies: a comprehensive review of recent developments. Nanomaterials. 2017;7(4). 10.3390/nano7040077. [DOI] [PMC free article] [PubMed]
- 33.Shi J, Kantoff PW, Wooster R, Farokhzad OC. Cancer nanomedicine: progress, challenges and opportunities. Nat Rev Cancer. 2017;17(1):20–37. doi: 10.1038/nrc.2016.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nie C, Parshad B, Bhatia S, Cheng C, Stadtmüller M, Oehrl A, et al. Topology-matching design of an influenza-neutralizing spiky nanoparticle-based inhibitor with a dual mode of action. Angew Chem Int Ed. 2020;59(36):15532–15536. doi: 10.1002/anie.202004832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lares MR, Rossi JJ, Ouellet DL. RNAi and small interfering RNAs in human disease therapeutic applications. Trends Biotechnol. 2010;28(11):570–579. doi: 10.1016/j.tibtech.2010.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 37.Tanaka T, Decuzzi P, Cristofanilli M, Sakamoto JH, Tasciotti E, Robertson FM, et al. Nanotechnology for breast cancer therapy. Biomed Microdevice. 2009;11(1):49–63. doi: 10.1007/s10544-008-9209-0. [DOI] [PubMed] [Google Scholar]
- 38.Dahlman JE, Barnes C, Khan OF, Thiriot A, Jhunjunwala S, Shaw TE, et al. In vivo endothelial siRNA delivery using polymeric nanoparticles with low molecular weight. Nat Nanotechnol. 2014;9(8):648–655. doi: 10.1038/nnano.2014.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brandl M. Liposomes as drug carriers: a technological approach. Biotechnology Annual Review. Elsevier; 2001. p. 59–85. [DOI] [PubMed]
- 40.Vicent MJ, Duncan R. Polymer conjugates: nanosized medicines for treating cancer. Trends Biotechnol. 2006;24(1):39–47. doi: 10.1016/j.tibtech.2005.11.006. [DOI] [PubMed] [Google Scholar]
- 41.Hussain MT, Tiboni M, Perrie Y, Casettari L. Microfluidic production of protein loaded chimeric stealth liposomes. Int J Pharm. 2020;590:119955. doi: 10.1016/j.ijpharm.2020.119955. [DOI] [PubMed] [Google Scholar]
- 42.uFluidix: What is microfluidics. https://www.ufluidix.com/resources/definitions/. Accessed 20 Jan 2021.
- 43.Fallahi H, Zhang J, Phan HP, Nguyen NT. Flexible microfluidics: fundamentals, recent developments, and applications. Micromachines (Basel). 2019;10(12). 10.3390/mi10120830. [DOI] [PMC free article] [PubMed]
- 44.Anguiano M, Castilla C, Maška M, Ederra C, Peláez R, Morales X, et al. Characterization of three-dimensional cancer cell migration in mixed collagen-Matrigel scaffolds using microfluidics and image analysis. PLoS One. 2017;12(2):e0171417. doi: 10.1371/journal.pone.0171417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Roces CB, Lou G, Jain N, Abraham S, Thomas A, Halbert GW, et al. Manufacturing considerations for the development of lipid nanoparticles using microfluidics. Pharmaceutics. 2020;12(11). 10.3390/pharmaceutics12111095. [DOI] [PMC free article] [PubMed]
- 46.Hamano N, Bottger R, Lee SE, Yang Y, Kulkarni JA, Ip S, et al. Robust microfluidic technology and new lipid composition for fabrication of curcumin-loaded liposomes: effect on the anticancer activity and safety of cisplatin. Mol Pharm. 2019;16(9):3957–3967. doi: 10.1021/acs.molpharmaceut.9b00583. [DOI] [PubMed] [Google Scholar]
- 47.Hrkach J, Von Hoff D, Mukkaram Ali M, Andrianova E, Auer J, Campbell T, et al. Preclinical development and clinical translation of a PSMA-targeted docetaxel nanoparticle with a differentiated pharmacological profile. Sci Transl Med. 2012;4(128):128ra39. 10.1126/scitranslmed.3003651. [DOI] [PubMed]
- 48.Rudyak VY, Krasnolutskii SL. Dependence of the viscosity of nanofluids on nanoparticle size and material. Phys Lett A. 2014;378(26):1845–1849. doi: 10.1016/j.physleta.2014.04.060. [DOI] [Google Scholar]
- 49.Ahn J, Ko J, Lee S, Yu J, Kim Y, Jeon NL. Microfluidics in nanoparticle drug delivery; From synthesis to pre-clinical screening. Adv Drug Deliv Rev. 2018;128:29–53. doi: 10.1016/j.addr.2018.04.001. [DOI] [PubMed] [Google Scholar]
- 50.Rapp BE. Chapter 9 - Fluids. In: Rapp BE, editor. Microfluidics: modelling, mechanics and mathematics. Oxford: Elsevier; 2017. pp. 243–263. [Google Scholar]
- 51.Hung L-H, Lee A. Microfluidic devices for the synthesis of nanoparticles and biomaterials. Journal of Medical and Biological Engineering. 2006;27:1–6. [Google Scholar]
- 52.Sahay G, Querbes W, Alabi C, Eltoukhy A, Sarkar S, Zurenko C, et al. Efficiency of siRNA delivery by lipid nanoparticles is limited by endocytic recycling. Nat Biotechnol. 2013;31(7):653–658. doi: 10.1038/nbt.2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen D, Love KT, Chen Y, Eltoukhy AA, Kastrup C, Sahay G, et al. Rapid discovery of potent siRNA-containing lipid nanoparticles enabled by controlled microfluidic formulation. J Am Chem Soc. 2012;134(16):6948–6951. doi: 10.1021/ja301621z. [DOI] [PubMed] [Google Scholar]
- 54.Barenholz Y. Liposome application: problems and prospects. Curr Opin Colloid Interface Sci. 2001;6(1):66–77. doi: 10.1016/S1359-0294(00)00090-X. [DOI] [Google Scholar]
- 55.Chen D, Love KT, Chen Y, Eltoukhy AA, Kastrup C, Sahay G, et al. Rapid discovery of potent siRNA-containing lipid nanoparticles enabled by controlled microfluidic formulation. J Am Chem Soc. 2012 doi: 10.1021/ja301621z. [DOI] [PubMed] [Google Scholar]
- 56.Staufer O, Antona S, Zhang D, Csatári J, Schröter M, Janiesch J-W, et al. Microfluidic production and characterization of biofunctionalized giant unilamellar vesicles for targeted intracellular cargo delivery. Biomaterials. 2021;264:120203. doi: 10.1016/j.biomaterials.2020.120203. [DOI] [PubMed] [Google Scholar]
- 57.Hwang DK, Oakey J, Toner M, Arthur JA, Anseth KS, Lee S, et al. Stop-flow lithography for the production of shape-evolving degradable microgel particles. J Am Chem Soc. 2009;131(12):4499–4504. doi: 10.1021/ja809256d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gratton S, Ropp P, Pohlhaus P, Luft J, Madden V, Napier M, et al. The effect of particle design on cellular internalization pathways. Proc Natl Acad Sci USA. 2008;105:11613–11618. doi: 10.1073/pnas.0801763105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kohn LT, Corrigan JM, Donaldson MS. To err is human: building a safer health system. Washington (DC) 2000. [PubMed]
- 60.Io M. To Err is human: building a safer health system. Washington, DC: The National Academies Press; 2000. [PubMed] [Google Scholar]
- 61.Ewen KR, Bahlo M, Treloar SA, Levinson DF, Mowry B, Barlow JW, et al. Identification and analysis of error types in high-throughput genotyping. Am J Hum Genet. 2000;67(3):727–736. doi: 10.1086/303048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schwarze K, Buchanan J, Fermont JM, Dreau H, Tilley MW, Taylor JM, et al. The complete costs of genome sequencing: a microcosting study in cancer and rare diseases from a single center in the United Kingdom. Genet Med. 2020;22(1):85–94. doi: 10.1038/s41436-019-0618-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tecan: Fluent® Automation Workstation. https://lifesciences.tecan.com/fluent-laboratory-automation-workstation?p=tab--2. Accessed 25 Nov 2020.
- 64.LabCompare: Liquid Handling Equipment. https://www.labcompare.com/General-Laboratory-Equipment/312-Liquid-Handling-Equipment/. Accessed 25 Nov 2020 2020.
- 65.ThermoFisher: Spinnaker Microplate Robot. https://www.thermofisher.com/order/catalog/product/SPK0001#/SPK0001. Accessed 25 Nov 2020.
- 66.Fleischer H, Baumann D, Joshi S, Chu X, Roddelkopf T, Klos M, et al. Analytical measurements and efficient process generation using a dual-arm robot equipped with electronic pipettes. Energies. 2018;11(10):2567. doi: 10.3390/en11102567. [DOI] [Google Scholar]
- 67.Cresswell K, Ramalingam S, Sheikh A. Can robots improve testing capacity for SARS-CoV-2? J Med Internet Res. 2020;22(8):e20169-e. 10.2196/20169. [DOI] [PMC free article] [PubMed]
- 68.BioTek. Lionheart FX Automated microscope - overview.
- 69.Cutrona MB, Simpson JC. A high-throughput automated confocal microscopy platform for quantitative phenotyping of nanoparticle uptake and transport in spheroids. Small. 2019;15(37):1902033. doi: 10.1002/smll.201902033. [DOI] [PubMed] [Google Scholar]
- 70.Vision O. What is automated confocal microscopy for life sciences? Vision Online. 2019.
- 71.Elliott AD. Confocal microscopy: principles and modern practices. Curr Protoc Cytom. 2020;92(1):e68. doi: 10.1002/cpcy.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Swaim WD. Overview of Confocal Microscopy. In: Oliver C, Jamur MC, editors. Immunocytochemical methods and protocols. methods in molecular biology. Totowa, NJ: Humana Press; 2010. p. 187–201.
- 73.Griffin C: BioTek announces new wide field of view camera for Cytation™ 5. https://www.biotek.com/about/news/biotek-announces-new-wide-field-of-view-camera-for-cytation-5/ (2019). Accessed 25 Nov 2020.
- 74.Cavallo-Medved D, Rudy D, Blum G, Bogyo M, Caglic D, Sloane BF. Live-cell imaging demonstrates extracellular matrix degradation in association with active cathepsin B in caveolae of endothelial cells during tube formation. Exp Cell Res. 2009;315(7):1234–1246. doi: 10.1016/j.yexcr.2009.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Recursion: Recursion. https://www.recursionpharma.com/ (2020). Accessed 25 Nov 2020.
- 76.Venditto VJ, Szoka FC., Jr Cancer nanomedicines: so many papers and so few drugs! Adv Drug Deliv Rev. 2013;65(1):80–88. doi: 10.1016/j.addr.2012.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Adir O, Poley M, Chen G, Froim S, Krinsky N, Shklover J, et al. Integrating artificial intelligence and nanotechnology for precision cancer medicine. Adv Mater. 2020;32(13):1901989. doi: 10.1002/adma.201901989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mitchell TM. Machine Learning. 1997.
- 79.Rashidi HH, Tran NK, Betts EV, Howell LP, Green R. Artificial intelligence and machine learning in pathology: the present landscape of supervised methods. Academic pathology. 2019;6:2374289519873088-. 10.1177/2374289519873088. [DOI] [PMC free article] [PubMed]
- 80.Shamay Y, Shah J, Işık M, Mizrachi A, Leibold J, Tschaharganeh DF, et al. Quantitative self-assembly prediction yields targeted nanomedicines. Nat Mater. 2018;17(4):361–368. doi: 10.1038/s41563-017-0007-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Labouta HI, Asgarian N, Rinker K, Cramb DT. Meta-analysis of nanoparticle cytotoxicity via data-mining the literature. ACS Nano. 2019;13(2):1583–1594. doi: 10.1021/acsnano.8b07562. [DOI] [PubMed] [Google Scholar]
- 82.Puzyn T, Rasulev B, Gajewicz A, Hu X, Dasari TP, Michalkova A, et al. Using nano-QSAR to predict the cytotoxicity of metal oxide nanoparticles. Nat Nanotechnol. 2011;6(3):175–178. doi: 10.1038/nnano.2011.10. [DOI] [PubMed] [Google Scholar]
- 83.Regitnig P, Müller H, Holzinger A. Expectations of artificial intelligence for pathology. In: Holzinger A, Goebel R, Mengel M, Müller H, editors. Artificial Intelligence and Machine Learning for Digital Pathology: State-of-the-Art and Future Challenges. Cham: Springer International Publishing; 2020. pp. 1–15. [Google Scholar]
- 84.Hess KR, Anderson K, Symmans WF, Valero V, Ibrahim N, Mejia JA, et al. Pharmacogenomic predictor of sensitivity to preoperative chemotherapy with paclitaxel and fluorouracil, doxorubicin, and cyclophosphamide in breast cancer. J Clin Oncol. 2006;24(26):4236–4244. doi: 10.1200/jco.2006.05.6861. [DOI] [PubMed] [Google Scholar]
- 85.Baghaei B, Saeb MR, Jafari SH, Khonakdar HA, Rezaee B, Goodarzi V, et al. Modeling and closed-loop control of particle size and initial burst of PLGA biodegradable nanoparticles for targeted drug delivery. J Appl Polym Sci. 2017;134(33):45145. doi: 10.1002/app.45145. [DOI] [Google Scholar]
- 86.AI-Enabled Phenotypic Screens Reveal Clinically Relevant Drugs. 2017.